Abstract
DNA repair competency is one determinant of sensitivity to certain chemotherapy drugs, such as cisplatin. Cancer cells with intact DNA repair can avoid the accumulation of genome damage during growth and also can repair platinum-induced DNA damage. We sought genomic signatures indicative of defective DNA repair in cell lines and tumors, and correlated these signatures to platinum sensitivity. The number of sub-chromosomal regions with allelic imbalance extending to the telomere (NtAI) predicted cisplatin sensitivity in-vitro, and pathologic response to preoperative cisplatin treatment in patients with triple-negative breast cancer (TNBC). In serous ovarian cancer treated with platinum-based chemotherapy, higher NtAI forecast better initial response. We found an inverse relationship between BRCA1 expression and NtAI in sporadic TNBC and serous ovarian cancers without BRCA1 or BRCA2 mutation. Thus, accumulation of tAI is a marker of platinum sensitivity and suggests impaired DNA repair.
Keywords: DNA damage, allelic imbalance, chemotherapy sensitivity, triple negative breast cancer
Introduction
Cell lines carrying BRCA1 or BRCA2 mutations are more sensitive to killing by the platinum salts cisplatin and carboplatin than wild-type cells (1, 2). Breast and ovarian cancers in patients carrying BRCA1 or BRCA2 mutations are likewise sensitive to platinum-based chemotherapy (3, 4). The majority of breast cancers arising in women with a germline BRCA1 mutation lack expression of estrogen and progesterone receptors or amplification of the HER2-neu gene (“triple-negative”). BRCA1-related breast cancers share a number of phenotypic characteristics with sporadic triple-negative breast cancer (TNBC) (5-7). Both tumor types share a common pattern of genomic abnormalities and have high global levels of chromosomal aberrations including allelic imbalance (AI), the unequal contribution of maternal and paternal DNA sequences with or without changes in overall DNA copy number (8-10). Since they have in common genomic aberrations suggesting a shared lesion in genomic integrity control, it is reasonable to posit sporadic TNBC that has accumulated high levels of AI might share the sensitivity to platinum-based chemotherapy that characterizes BRCA1-associated cancer.
These observations prompted a clinical trial, Cisplatin-1, in which 28 patients with operable TNBC were treated preoperatively with cisplatin monotherapy. Preoperative treatment in Cisplatin-1 resulted in greater than 90% tumor reduction in 10 of 28 (36%) patients, including pathologic complete response (pCR) in 6 women, 2 of whom had BRCA1-associated cancers (11). A second trial, Cisplatin-2, accrued 51 patients with TNBC who received the same preoperative cisplatin regimen as Cisplatin-1, but in combination with the angiogenesis inhibitor bevacizumab (12). Response rate in Cisplatin-2 were similar to Cisplatin-1. In the second trial, a greater than 90% tumor reduction was observed in 17 of 44 women (39%) completing treatment. In Cisplatin-2, 8 patients carried a germline BRCA1 or BRCA2 mutation, of which 4 patients achieved a pCR or near pCR to the cisplatin-bevacizumab regimen. In both trials, all patients had research sequencing to determine their germline BRCA1 and BRCA2 status. We compared the number of various chromosomal abnormalities including AI present in tumor biopsies obtained before therapy to pathologically determined tumor response to cisplatin, alone or in combination with bevacizumab, assessed by examination of the post-treatment surgical specimen.
Chromosomal abnormalities such as regions of allelic imbalance, other than those resulting from whole chromosome gain or loss, might result from improper repair of DNA double-strand breaks during tumor development. If so, then a genome-wide count of abnormal chromosomal regions in tumors may indicate the degree of DNA repair incompetence, independent of knowledge of any specific causative DNA repair defect. We hypothesized that the number of chromosomal regions of AI in tumors would predict sensitivity to drugs that induce DNA crosslinks such as cisplatin.
We first sought associations between various measures of sub-chromosomal abnormalities and sensitivity to cisplatin in breast cancer cell lines and found the most accurate predictor to be AI extending to the telomeric end of the chromosome (NtAI). Finally, we tested if NtAI was associated with treatment response in patient tumor samples in the Cisplatin-1 and Cisplatin-2 TNBC trials and in The Cancer Genome Atlas (TCGA) public data set of serous ovarian cancer, a cancer routinely treated with platinum-based therapy. In an effort to understand more about the processes leading to telomeric allelic imbalances, we mapped the location of their breakpoints and observed a striking association of these breakpoints with regions of the genome that are difficult to replicate, common copy number variants (CNVs). Further, a subset of high NtAI tumors display low BRCA1 mRNA levels. These observations begin to suggest models of how tAI may occur.
Results
Cisplatin sensitivity correlates with burden of telomeric allelic imbalance in breast cancer cell lines
We obtained single nucleotide polymorphism (SNP) genotype array data from the Wellcome Trust Sanger Institute for a set of established BRCA1 wild-type breast cancer cell lines for which we had determined cisplatin sensitivity (13) (Fig. 1A). Allele copy number was determined from the SNP array data and AI detected using ASCAT analysis (10) (Fig. S1). We tested for association between the IC50 values for cisplatin and each of three summary measures of chromosomal alteration: the number of chromosome regions with AI (NAI, Fig. S2A), the number of regions with copy number gains (NGain, Fig. S2B), and the number of regions with copy number loss (NLoss, Fig. S2C). None of these measures were correlated with cisplatin sensitivity in the cell lines.
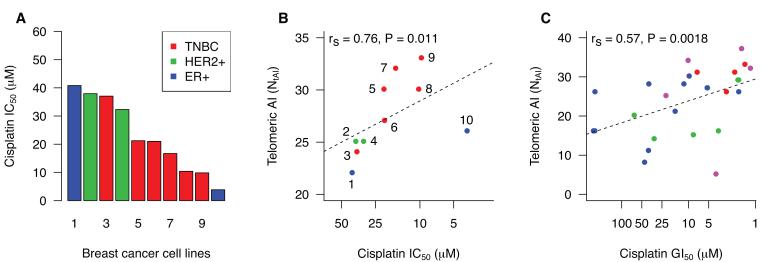
Figure 1. Chromosomal aberrations and cisplatin sensitivity in vitro.
The relationship between NtAI and cisplatin sensitivity was analyzed in breast cancer cell lines. A and B: 10 cell lines were included in this study, 1: CAMA-1, 2: HCC1954, 3:MDA-MB-231, 4: MDA-MB-361, 5: HCC1187, 6:BT-549, 7: HCC1143, 8: MDA-MB-468, 9: BT-20, 10: T47D. A. IC50 values for each of the 10 cell lines. A proliferation assay was used to assess viability after 48 hours of cisplatin exposure, and IC50 was determined from the dose response curves. B. Relationship between NtAI and cisplatin sensitivity. Breast cancer subtype is indicated as follows: ER− HER2−, red; HER2+, green, ER+ HER2−, blue. C. Relationship between NtAI and cisplatin sensitivity as determined by GI50 in breast cancer cell lines from Heiser et al. (18). Reported transcriptional subtype is indicated as follows: basal, red; claudin-low, pink; ERBB2Amp, green; luminal, blue. See supplemental methods for cell line identifiers.
Known defects in DNA double strand break repair, including loss of BRCA1, cause the spontaneous formation of triradial and quadriradial chromosome structures, which are cytologic indications of aberrant chromosome recombination (14-16). The resolution of these chromosome rearrangements at mitosis can result in large regions of AI and/or copy number changes extending from the crossover to the telomere (15, 17). More generally, several error-prone repair processes potentially employed by cells with defective DNA repair cause chromosome cross-over or copy choice events that result in allelic loss or copy number change extending from the site of DNA damage to the telomere. We therefore looked for an association between cisplatin sensitivity and the number of contiguous regions of AI, copy gain, or copy loss that either extended to a telomere and did not cross the centromere (telomeric regions) or did not extend to a telomere (interstitial regions) (Fig. S1, Fig. 1B and Fig. S3). The number of regions of telomeric AI was the only summary genomic measure that was significantly associated with cisplatin sensitivity in the breast cancer cell lines (rs = 0.76 P = 0.011, Fig. 1B); the correlation between NtAI and cisplatin sensitivity was stronger when the analysis was restricted to the triple negative breast cancer lines (Fig. 1B, red circles; rs = 0.82 P = 0.0499). A similar relationship was observed between NtAI and cisplatin sensitivity as measured by GI50 in a recently published study of breast cancer cell lines (rs = 0.57 P = 0.0018, Fig. 1C) (18). Of all the drugs tested in this study, NtAI was most highly correlated to cisplatin sensitivity.
Tumors sensitive to cisplatin-based chemotherapy have higher levels of telomeric allelic imbalance
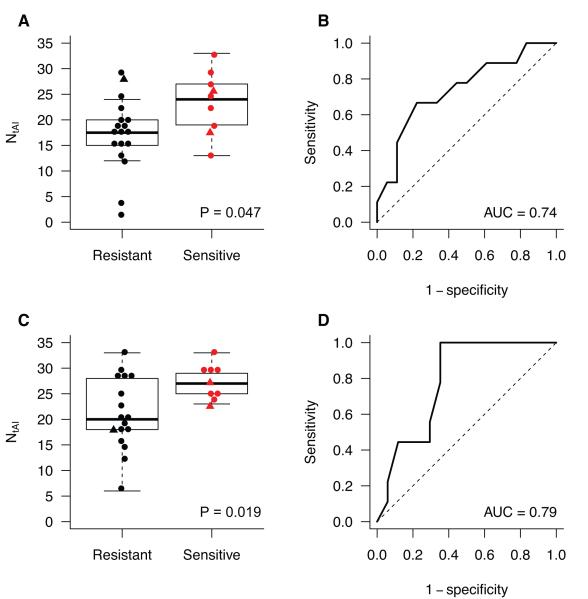
We then investigated whether the association between NtAI in clinical tumor samples and cisplatin sensitivity was present in the Cisplatin-1 trial. Sensitivity was measured by pathologic response determined after pre-operative treatment (11). Molecular inversion probe SNP genotype data from pretreatment tumor samples (n = 27) were evaluated by ASCAT analysis to determine NtAI. We compared tumors with a reduction of at least 90% in the content of malignant cells (cisplatin sensitive) to tumors with limited or no response to cisplatin (cisplatin resistant, defined by tumor reduction of less than 90%). Cisplatin sensitive tumors had significantly higher NtAI (median 24 versus 17.5, P = 0.047, Fig. 2A). We tested the ability of NtAI to predict cisplatin response by calculating the area under the receiver operating characteristic (ROC) curve (AUC). ROC analysis showed that higher NtAI was associated with cisplatin sensitivity (AUC = 0.74, CI 0.50-0.90, Fig. 2B).
Figure 2. NtAI and cisplatin response in breast cancer.
In two clinical trials, TNBC patients were given preoperative cisplatin (Cisplatin-1, Fig. 2 A-B) or cisplatin and bevacizumab (Cisplatin-2, Fig. 2 C-D). Cisplatin resistant tumors are indicated in black, cisplatin sensitive tumors are indicated in red. Tumors with germline mutations in BRCA1/2 are indicated with triangles. A and C. Box plots showing NtAI distribution in cisplatin resistant and sensitive tumors. B and D. Receiver operating characteristic curves showing the ability of NtAI to predict for sensitivity to cisplatin.
In the Cisplatin-2 trial, cisplatin sensitive tumors (n=9) had significantly higher NtAI than resistant tumors (n=17, median 27 versus 20, P = 0.019, Fig. 2C). NtAI was also associated with response to cisplatin and bevacizumab by ROC analysis (AUC = 0.79, CI 0.55-0.93, Fig. 2D). The association between NtAI and cisplatin sensitivity remained significant when cases with BRCA1 or BRCA2 mutation were excluded and only BRCA normal cases were analyzed (P = 0.030 and P = 0.023 in Cisplatin-1 and Cisplatin-2, respectively). Therefore, in two separate pre-operative trials in breast cancer, in which treatment sensitivity was assessed by a quantitative measure of pathologic response, NtAI reliably forecast the response to cisplatin-based treatment.
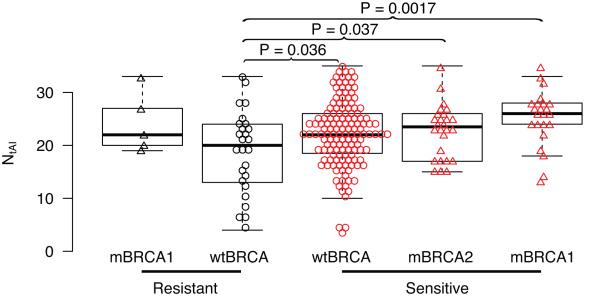
To test if the NtAI metric indicates platinum sensitivity in cancers other than breast, we determined the association between NtAI and initial treatment response in The Cancer Genome Atlas (TCGA) cohort of serous ovarian cancer patients that had received adjuvant platinum and taxane chemotherapy (19). Among the ovarian cancers without mutation in BRCA1 or BRCA2 (wtBRCA), the platinum sensitive tumors had significantly higher NtAI than platinum-resistant cancers (median 22 versus 20, P = 0.036, Fig. 3), and were predictive of treatment response by ROC analysis (AUC = 0.63, CI 0.50-0.76, Fig. S4). The ovarian cancers with somatic or germline mutation in BRCA1 or BRCA2 that were sensitive to platinum therapy had even higher NtAI (median = 26, P = 0.0017 and median 23.5, P = 0.037 versus resistant wtBRCA, respectively, Fig. 3). All of the BRCA2 mutated cancers were platinum sensitive; however, 5 BRCA1 mutated tumors were resistant to platinum therapy yet appeared to have relatively high levels of NtAI. Thus high NtAI is characteristic of serous ovarian cancer with known mutation in either BRCA1 or BRCA2; high NtAI is also found in a subset of sporadic cancers without BRCA mutations where it is predictive of platinum sensitivity.
Figure 3. NtAI and cisplatin response in serous ovarian cancer.
Box plots showing NtAI distribution in platinum sensitive and resistant tumors in cancers without BRCA1 or BRCA2 mutations (wtBRCA) and for cancers with germline or somatic mutation in BRCA1 (mBRCA1) or in BRCA2 (mBRCA2). Red indicate sensitive samples, triangles indicate samples with germline or somatic mutations in BRCA1 or BRCA2. Significant differences between resistant wtBRCA and sensitive groups are indicated. In addition, significant differences were found between sensitive wtBRCA and sensitive mBRCA2 (P = 0.047), and sensitive wtBRCA and sensitive mBRCA1 (P = 0.014).
Locations of NtAI-associated chromosomal breaks are not random
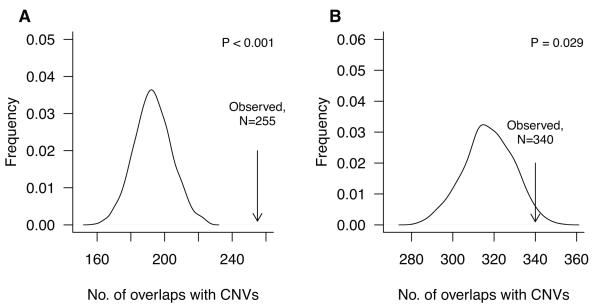
To understand the processes leading to tAI better, we mapped the location of the chromosome breakpoints defining the boundary of the tAI regions. We observed many breakpoints were located in very close proximity to each other (Fig. S5), suggesting a non-random distribution of DNA breaks causing telomeric allelic imbalance. Recurrent chromosomal translocation breakpoints may be associated with regions of repeated DNA sequence that may cause stalled replication forks, an increased frequency of DNA breaks, and subsequent rearrangement by non-allelic homologous recombination or other similar mechanisms (20, 21). Copy number variants (CNVs) are highly homologous DNA sequences for which germline copy number varies between healthy individuals (22, 23). CNVs have been proposed to facilitate the generation of chromosomal alterations, similar to fragile sites (21, 24, 25). We compared the number of observed breaks within 25 kB of a CNV to the frequency expected by chance alone, based on permuted data. In the Cisplatin-1 cohort, of 517 NtAI breakpoints, 255 (49%) were associated with overlapping CNVs. Similarly, in the cisplatin-2 cohort, out of 599 NtAI breakpoints, 340 (57%) were associated with CNVs. In both trials, the observed number of NtAI breaks associated with CNVs was significantly higher than expected by chance (Fig. 4, A-B). Thus many of the breakpoints leading to telomeric AI in TNBC occur near CNVs suggesting stalled replication forks, replication stress, or other CNV-associated mechanisms may be involved in the genesis of telomeric AI.
Figure 4. Enrichment of common CNVs in tAI chromosomal breakpoints from TNBC.
Association of tAI breakpoints with common CNV loci based on computational simulations that compared the expected number of breakpoints containing CNVs with the observed number in total cases in Cisplatin-1 (A) and Cisplatin-2 (B).
Low BRCA1 mRNA is associated with high NtAI and sensitivity to cisplatin
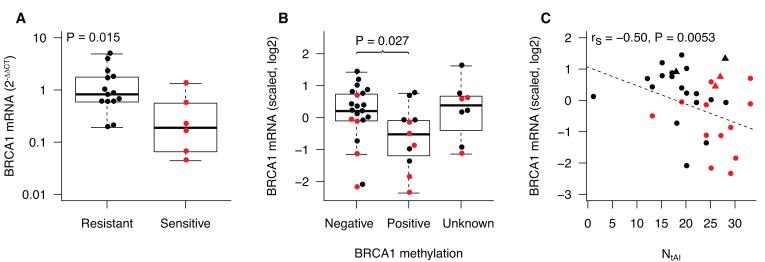
In our previous report of the Cisplatin-1 trial, we found an association between low BRCA1 transcript levels and better response to cisplatin (11). In the more recent Cisplatin-2 trial, BRCA1 transcript levels measured by qPCR are also associated with cisplatin response (P = 0.015, Fig. 5A). In a combined analysis of data from both trials, lower BRCA1 transcript levels are associated with methylation of the BRCA1 promoter (P = 0.027, Fig. 5B), though BRCA1 promoter methylation itself is not significantly associated with cisplatin response (P = 0.25, Fishers exact test). BRCA1 mRNA levels are inversely associated with NtAI in the two cisplatin trials (rs = −0.50, P = 0.0053, Fig. 5C). This finding suggests that dysfunction of a BRCA1-dependent process or other abnormality causing low BRCA1 mRNA may be responsible for the high level of telomeric allelic imbalance and also cisplatin sensitivity in many of these TNBCs.
Figure 5. Association between BRCA1 transcript level and cisplatin sensitivity, BRCA1 promoter methylation, and NtAI.
Red indicates tumors sensitive to cisplatin. Tumors with a germline mutation in BRCA1 or BRCA2 are excluded in A. and B., but included in C., represented as triangles. In B. and C., BRCA1 transcript levels measured by qPCR were scaled and combined by centering the values, and dividing by the standard deviation within each trial. A. BRCA1 transcripts in resistant and sensitive tumors in the Cisplatin-2 cohort. B. BRCA1 expression in tumors by methylation status of the BRCA1 promoter region in the combined Cisplatin-1 and Cisplatin-2 cohorts. C. Relationship of BRCA1 transcript level and NtAI in the combined Cisplatin-1 and Cisplatin-2 cohorts.
ROC analysis of the combined TNBC trials suggests that BRCA1 expression level or NtAI may give a similar predictive accuracy for cisplatin sensitivity (Fig. S6A). When high NtAI and low BRCA1 expression are combined in a predictive model, the positive predictive value and specificity of prediction improved considerably but the sensitivity was decreased relative to NtAI alone (Fig. S6B), suggesting that low BRCA1 expression does not account for all cisplatin sensitive tumors.
In the TNBC trials, we noted a few cisplatin sensitive tumors with high levels of NtAI but high BRCA1 mRNA, suggesting that alternative mechanisms may drive the generation of tAI in some tumors. Analysis of TCGA data of ER−/HER2− breast cancer and wtBRCA serous ovarian cancer demonstrate an inverse correlation between NtAI and BRCA1 expression. Yet in both cohorts there was a considerable subset of tumors with high NtAI and high BRCA1 expression (Fig S7A, B). Unlike NtAI, BRCA1 expression was not apparently different between sensitive and resistant wtBRCA serous ovarian cancers (Fig S7C). These findings suggest a model whereby high NtAI may represent a readout of DNA repair deficiency resulting from either low BRCA1 expression or from other known or unknown mechanisms (Fig. 6).
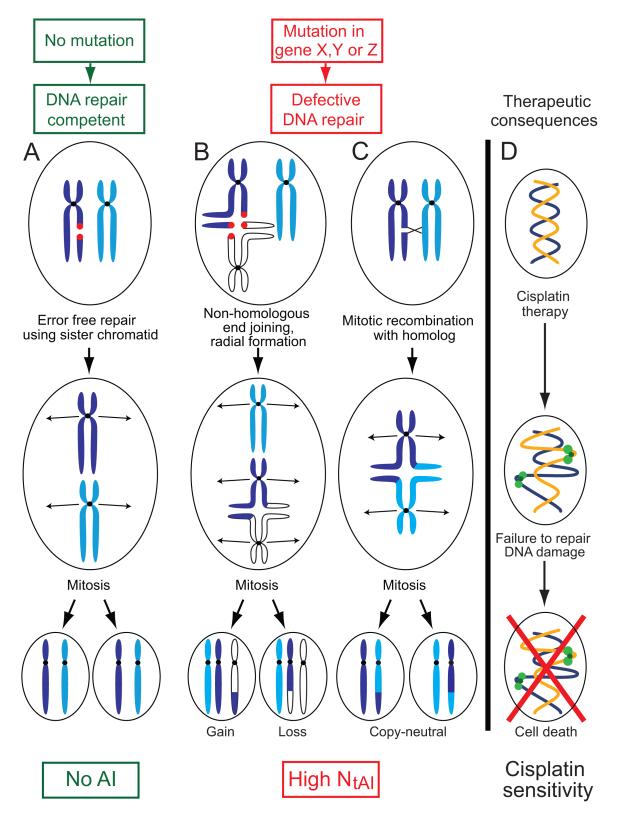
Figure 6. A model relating DNA repair to accumulation of telomeric AI and response to platinum agents.
A. In DNA repair-competent cells, DNA breaks are repaired using error-free homologous recombination employing the identical sister chromatid as a template, resulting in no AI. B. and C. Compromised DNA repair favors the use of error-prone repair pathways, resulting in chromosome rearrangements and aberrant radial chromosome formation. After mitotic division, daughter cells will have imbalance in the parental contribution of telomeric segments of chromosomes (telomeric AI). B. Non-homologous end joining is one error-prone mechanism that joins a broken chromatid of one chromosome (dark blue) to the chromatid of another, usually nonhomologous, chromosome (white). Mitotic segregation results in cells with telomeric AI due to mono-allelic change in DNA copy number of the affected telomeric region. C. Mitotic recombination may result in rearrangements between homologous chromosomes (dark blue and light blue). Mitotic segregation results in cells with AI due to copy neutral loss of heterozygosity (LOH). D. The same compromise in DNA repair that causes telomeric AI may also result in the inability of the tumor cell to repair drug-induced DNA damage, leading to tumor sensitivity to drugs such as platinum salts.
Discussion
Our study utilized two preoperative clinical trials in women with triple negative breast cancer treated with cisplatin, in which pathologic response at the time of surgery provided an experimental endpoint. Sporadic triple negative breast cancers are heterogeneous in their responses to platinum salts, chemotherapeutic agents that depend in part on DNA repair defects for their cytotoxic activity (26, 27). Lesions in DNA repair caused by BRCA1 or BRCA2 dysfunction lead to platinum sensitivity; we reasoned that the types of chromosomal aberrations arising in the context of BRCA dysfunction might also be associated with platinum sensitivity in wtBRCA cancers. Based on results in cell lines, we chose to enumerate one such chromosomal abnormality, telomeric allelic imbalance in pre-treatment tumor genomes and to relate this to pathologic response after cisplatin therapy. NtAI was associated with response to platinum treatment in our TNBC cisplatin trials and in platinum treated serous ovarian cancer and suggests the burden of this genomic abnormality exposes an underlying deficiency of DNA repair in the platinum-sensitive subset of these cancers. Allelic imbalance propagated from a given chromosomal location to the telomere suggests the operation of error-prone processes giving rise to abnormal crossover or template switching events, rather than error-free DNA repair.
We found the breakpoints of tAI regions are non-random and enriched for CNVs. This pattern also suggests defective DNA repair. CNVs are associated with other repeat sequences such as Alu repeats, are concentrated in pericentromeric and subtelomeric regions, and are associated also with common fragile sites (28, 29). These repeat elements are thought to result in replication “slow zones” prone to replication stalling and formation of DNA double strand breaks (30, 31). Furthermore, downregulation of Rad51 or inhibition of BRCA1 increases the fragility at such sites when cells are under replication stress (32, 33). The observed association of low BRCA1 expression levels in many tumors with high NtAI suggests deficient homologous recombination, impaired S or G2/M checkpoint function, or a combination of these factors underlies the generation of this type of genomic abnormality.
Cisplatin forms inter-strand crosslinks on DNA that lead to stalled replication forks and DNA double stand breaks that must be repaired if the cell is to survive. It is likely these breaks are repaired using similar mechanisms to those employed at stalled replication forks and DNA breaks generated at sites of CNVs. Therefore, high pre-treatment NtAI identifies tumors unable to accurately repair breaks and restart stalled replication forks at sites of CNV. These same tumors are also unable to contend with stalled forks at sites of cisplatin crosslinks.
While allelic imbalance at sites of CNV may reflect inefficient error-free repair, other explanations should be considered. Both triple negative cohorts showed a significant relationship between NtAI and pathologic response to cisplatin chemotherapy. Nevertheless, there were patients in both trials whose tumors showed poor response to cisplatin therapy despite having high NtAI. Similarly, a few of the BRCA1-mutated ovarian cancers had high NtAI yet were resistant to platinum therapy. Since NtAI is a summation of ongoing and past DNA lesions, resistance mechanisms acquired after generation of tAI would confound the relationship between NtAI and response. In carriers of BRCA1 or BRCA2 mutations, some tumors that become resistant to platinum agents carry a reversion mutation that partially or completely restores BRCA1 or BRCA2 function and restores homologous recombination (26, 27, 34). Reversion has also been seen in a cell line with a BRCA2 mutation selected for PARP inhibitor resistance (27). Reversion mutations and in cis compensating mutations were observed in Fanconi anemia patients, resulting in improvement in their bone marrow function (35). Inactivation of TP53BP1 restores the balance between homologous recombination and non-homologous end joining in BRCA1-mutated cells and renders them resistant to PARP inhibitors (36, 37). Finally, drug transporters may prevent accumulation of platinum agents in tumor cells (38). Therefore, reversion of or compensation for a preexisting DNA repair defect may generate a tumor with high NtAI but resistance to platinum treatment; other platinum resistance mechanisms unrelated to DNA repair would have the same effect.
Our analysis begins to suggest an outline of the molecular taxonomy of TNBC and ovarian cancer with respect to DNA repair and drug sensitivity. Most platinum resistant breast or ovarian cancers are tumors with repair proficiency and low NtAI. Two subsets of wtBRCA tumors possess high NtAI and are sensitive to platinum-containing drugs. In one of these subsets, repair deficiency may be the consequence of low BRCA1 expression and in the other subset, repair may be crippled by mechanisms that do not depend upon BRCA1 expression. These observations will no doubt be further refined; inclusion of reversion mutations, compensations by other events in DNA repair pathways, other mechanisms of drug resistance, and other as yet unappreciated factors may help to enhance our prediction of drug sensitivity in the future.
In conclusion, a summary measure of telomeric chromosome aberrations in the tumor genome, NtAI, predicts sensitivity to platinum treatment. Our findings implicate NtAI as a marker of impaired DNA double-strand break repair. Assays to determine NtAI are feasible using formalin fixed paraffin embedded tumor material and recent algorithms such as ASCAT permit accurate determination of copy number and allelic imbalance in a majority of samples despite low tumor cell content. NtAI may prove useful in predicting response to a variety of therapeutic strategies exploiting defective DNA repair.
Materials and Methods
Cell lines and drug sensitivity assays
Breast cancer cell lines were originally obtained from American Type Culture Collection and were most recently authenticated by Promega PowerPlex 1.2 short tandem repeat profiling at the DF/HCC microarray core laboratory in September 2011. Drug sensitivity measurements in breast cancer cell lines BT20, BT549, HCC1187, HCC1143, MDAMB-231, MDA-MB-468, HCC38, MDA-MB-453 (triple negative), CAMA-1, MCF7, T47D (ER positive), BT474, HCC1954 and MDA-MB-361 (HER2 positive) were originally generated for a separate study in which it was reported as “data not shown” in a recently published manuscript (13). Briefly, cells were exposed to a series of concentrations of various chemotherapeutic agents for 48 hours. Viable cell number was quantified using CellTiter 96 AQueous One Solution Cell Proliferation Assay according to the manufacturer’s instructions (Promega). Drug sensitivity was quantified as the dose of drug resulting in a 50% reduction of growth (IC50). We found MCF7 to be highly resistant to all of the chemotherapeutic agents tested, consistent with its reported caspase-3 deficiency and resistance to drug induced apoptosis (39). In our analyses with measures of genomic aberration, MCF7 was the only clear outlier and for these reasons, was excluded from our analyses.
Breast cancer cohorts and assessment of therapeutic response
For this study, subjects were included for analysis of response to cisplatin if they progressed on therapy or if they received at least 3 of 4 cycles of the planned cisplatin therapy, had received no other non-protocol therapy before surgery, and if an adequate amount of tumor was available from the pre-treatment biopsy. Therapeutic response was measured using the semiquantitative Miller-Payne grading system, which estimates the percent reduction in invasive tumor volume and cellularity based on pathological assessment of surgical samples after therapy (40). Cisplatin-1 consists of 28 mainly sporadic TNBC patients treated with preoperative cisplatin monotherapy, of whom 4 progressed on therapy and 24 completed 4 cycles of cisplatin therapy (11). Cisplatin-2 consists of 51 TNBC patients treated with preoperative cisplatin and bevacizumab, of which one patient progressed on therapy and 44 patients completed 4 cycles of cisplatin therapy prior to surgery (12). Two patients included in this study were taken to surgery after completing 3 cycles of cisplatin therapy due to the development of toxicity; in both cases there was no appreciable pathologic response in the excised tumor after 3 cycles of cisplatin.
Preparation of breast cancer samples
For both trials, core biopsies of tumor were obtained before initiation of treatment. Adequate tumor for analysis was present for 27 of 28 subjects in Cisplatin-1 and 37 of 51 subjects in Cisplatin-2. H&E stained tissue sections of pre-treatment core needle biopsies were examined microscopically; for all biopsies for which enrichment was deemed feasible, sections were manually microdissected using an 18-gauge needle. DNA was extracted by proteinase K and RNase A digestions, phenol/chloroform extraction, and ethanol precipitation. Paired normal DNA from patients was obtained from peripheral blood lymphocytes for all cases in Cisplatin-1 and from 10 cases in Cisplatin-2.
TCGA Ovarian and breast cancer cohorts
Public SNP array data, expression data, and clinical annotation data was obtained for the TCGA ovarian (19) and breast cancer cohorts from the TCGA web site (41). BRCA1 and BRCA2 mutation status for the ovarian cancers was obtained from cBIO data portal (42). In the ovarian cohort, we identified 218 samples with SNP data that passed ASCAT (see below), BRCA mutation status, and interpretable clinical annotations for treatment and outcomes indicating initial treatment with adjuvant platinum-based chemotherapy, predominantly the combination of carboplatin and docetaxel. We classified “treatment sensitive” as those annotated as partial or complete response to initial treatment and no progression or recurrence within 6 months of initial treatment (n = 187); “treatment resistant” were those annotated as stable or progressive disease on initial therapy or disease recurrence or progression within 6 months (n = 31). In the breast cohort, we identified 78 samples with matched gene expression and SNP data that passed ASCAT, which were classified as ER−/HER2− based on clustering of the ESR1 and ERBB2 gene (see supplementary methods).
Genotyping and copy number analysis
DNA was sent to Affymetrix, Inc. (Santa Clara, CA) for determination of genotypes using the molecular inversion probe based genotyping system, OncoScan FFPE Express (43). The commercial assay, which determines genotype of 330,000 SNPs was used for analysis of the Cisplatin-2 trial. An early version of the OncoScan assay which genotypes 42,000 SNPs was used for the Cisplatin-1 trial. Allele signal intensity and genotypes from the OncoScan genotyping assay were processed and provided to us by Affymetrix. The OncoScan SNP genotype data for the cisplatin therapy trials is submitted to the NCBI GEO database under accession GSE28330. Public SNP array raw data for the breast cancer cell lines were obtained from the Sanger Institute Catalogue Of Somatic Mutations In Cancer web site (44, 45), public SNP array data from an independent breast cancer cell line study, Heiser et al. (18), and public SNP array data from the TCGA ovarian (19) and breast cancer cohorts were preprocessed by the AROMAv2 and CalMaTe algorithms (46) and, when a paired normal samples was available, TumorBoost (47). Processed genotype data from OncoScan genotyping and public SNP array data was analyzed for allele-specific copy numbers and tumor cell content by the algorithm “Allele-specific copy number analysis of tumors”, ASCAT (10). ASCAT is designed to correct for normal cell contamination and tumor cell ploidy, but occasionally fails to fit a model to a given sample. In this study, ASCAT failed to process 3 of 14 cell lines from Sanger, 15 of 42 cell lines from Heiser et al., and 5 of 37 samples from the Cisplatin-2 trial. Allelic imbalance was defined as any time the copy number of the two alleles were not equal, and at least one allele was present (Fig S1). To ensure that all trial cases were comparable, we eliminated cases estimated by ASCAT to have less than 36% tumor cell content, the highest level of normal cell admixture in the Cisplatin-1 trial, which was the trial with an overall greater tumor purity. Thus we included all 27 samples with SNP array data from the Cisplatin-1 trial, 26 out of 32 samples with SNP array data that passed ASCAT from the Cisplatin-2 trial.
A minimum number of consecutive probes showing an aberration was required in order to call regions of AI and CNA with confidence. To ensure similar aberration detection across the three platforms that were used, the minimum number of probes required to define a region of aberration was set to be proportional to the overall SNP density of the platform. The probe densities of the platforms were 42,000/genome OncoScan (prototype), 330,000/genome OncoScan FFPE Express, and 900,000/genome SNP6.0 for an approximate ratio of 1:8:20. Minimum probe requirements of 25 probes for 42k OncoScan prototype, 200 probes for 330k OncoScan FFPE Express, and 500 probes for SNP6.0 platform were chosen based on optimizing for correlation of aberration measurement in a subset of samples with replicate data generated on both versions of the OncoScan platform (See also Supplementary Methods).
Telomeric AI and telomeric CNA are defined as regions that extend to one of the sub-telomeres but do not cross the centromere. Copy number of telomeric AI regions was defined as the mean copy number of the probes mapping to the region. Copy loss was defined as a mean of less than 1.5 copies and copy gain was defined as a mean of greater than 2.5 copies. Association between NtAI and response to cisplatin was measured by the AUC of the ROC curve for binary response. Correlation was determined by Spearman’s rank correlation coefficient. Statistical significance was assessed by Wilcoxon’s rank sum test. All P values are two-sided.
Enrichment of copy number variants at site of DNA breakpoints
The genomic location of common copy number variants (CNVs) was acquired from the Database of Genomic Variants (48). Mapping for HG17 and HG18 was acquired in order to match the SNP probe mapping of the 42K prototype and 330K commercial OncoScan platforms, respectively. CNVs were considered associated with a breakpoint if they overlapped within a 25 kb window on either side of the breakpoint. To test for enrichment, we performed 1000 permutations for each cohort, where we randomly shuffled the location of the DNA breakpoints based on the location of the SNP probes, and determined how many were associated with CNVs.
BRCA1 transcript quantitation and promoter methylation analysis
BRCA1 exon 16/17 and RPLP0 (control) quantitative polymerase chain reaction assay was performed as previously described (11) using amplified tumor cDNA generated using Ovation RNA Amplification System V2 kit (NuGen Technologies, Inc., San Carlos, CA). BRCA1 promoter methylation assay was performed as previously described (11). In order to combine the qPCR transcript data from the Cisplatin-1 and Cisplatin-2 trials, the data was scaled within each cohort by centering it and dividing by the standard deviation.
BRCA1 mRNA expression in public TCGA cohorts
Public normalized and summarized Agilent based gene expression data was acquired from the TCGA for all breast cancer samples (level 3). Raw Affymetrix CEL files were obtained for ovarian cancer samples (level 1). Expression data for all TCGA ovarian cancer samples were normalized and summarized using RMA, and the probe set “204531_s_at” was identified as the optimum probe set for measuring BRCA1 expression using the R package “JetSet” (49).
Supplementary Material
SIGNIFICANCE.
Mutations in BRCA genes cause defects in DNA repair that predict sensitivity to DNA damaging agents including platinum; however, some patients without BRCA mutations also benefit from these agents. NtAI, a genomic measure of unfaithfully repaired DNA, may identify cancer patients likely to benefit from treatments targeting defective DNA repair.
Acknowledgements
We would like to acknowledge Affymetrix scientists Malek Faham, Yuker Wang, and Victoria Carlson who performed the tumor genotype analysis of Cisplatin-1 using an early version of OncoScan FFPE Express under a research agreement with Affymetrix, Inc.
Financial Support: This work is partly supported by the Dana-Farber/Harvard SPORE in Breast Cancer, grant number CA089393, by an Avon supplement to 5 P30 CA06516 Dana-Farber/Harvard Cancer Center Support Grant, and by the V Foundation for Cancer Research. JEG, NT, ZCW, JDI, ZS and ALR are supported by the Breast Cancer Research Foundation. NJB, AE, QL and ZS are supported by The Danish Council for Independent Research Medical Sciences (FSS). DPS gratefully acknowledges support of the Cogan Family Foundation.
Footnotes
Conflicts of Interest: ALR, ZS, ZCW, DPS, ACE, NJB are inventors on a pending patent application based on this work.
References
- 1.Samouelian V, Maugard CM, Jolicoeur M, Bertrand R, Arcand SL, Tonin PN, et al. Chemosensitivity and radiosensitivity profiles of four new human epithelial ovarian cancer cell lines exhibiting genetic alterations in BRCA2, TGFbeta-RII, KRAS2, TP53 and/or CDNK2A. Cancer Chemother Pharmacol. 2004;54:497–504. doi: 10.1007/s00280-004-0843-9. [DOI] [PubMed] [Google Scholar]
- 2.Tassone P, Tagliaferri P, Perricelli A, Blotta S, Quaresima B, Martelli ML, et al. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br J Cancer. 2003;88:1285–1291. doi: 10.1038/sj.bjc.6600859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrski T, Huzarski T, Dent R, Gronwald J, Zuziak D, Cybulski C, et al. Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat. 2009;115:359–363. doi: 10.1007/s10549-008-0128-9. [DOI] [PubMed] [Google Scholar]
- 4.Cass I, Baldwin RL, Varkey T, Moslehi R, Narod SA, Karlan BY. Improved survival in women with BRCA-associated ovarian carcinoma. Cancer. 2003;97:2187–2195. doi: 10.1002/cncr.11310. [DOI] [PubMed] [Google Scholar]
- 5.Turner NC, Reis-Filho JS, Russell AM, Springall RJ, Ryder K, Steele D, et al. BRCA1 dysfunction in sporadic basal-like breast cancer. Oncogene. 2007;26:2126–2132. doi: 10.1038/sj.onc.1210014. [DOI] [PubMed] [Google Scholar]
- 6.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lakhani SR, Reis-Filho JS, Fulford L, Penault-Llorca F, van der Vijver M, Parry S, et al. Prediction of BRCA1 status in patients with breast cancer using estrogen receptor and basal phenotype. Clin Cancer Res. 2005;11:5175–5180. doi: 10.1158/1078-0432.CCR-04-2424. [DOI] [PubMed] [Google Scholar]
- 8.Wang ZC, Lin M, Wei LJ, Li C, Miron A, Lodeiro G, et al. Loss of heterozygosity and its correlation with expression profiles in subclasses of invasive breast cancers. Cancer Research. 2004;64:64–71. doi: 10.1158/0008-5472.can-03-2570. [DOI] [PubMed] [Google Scholar]
- 9.Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silver DP, Richardson AL, Eklund AC, Wang ZC, Szallasi Z, Li Q, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol. 2010;28:1145–1153. doi: 10.1200/JCO.2009.22.4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ryan PD, Richardson AL, Tung NM, Isakoff S, Golshan M, Sgroi DC, et al. Neoadjuvant cisplatin and bevacizumab in triple negative breast cancer (TNBC): Safety and Efficacy. J Clin Oncol. 2009;27:551. [Google Scholar]
- 13.Li Y, Zou L, Li Q, Haibe-Kains B, Tian R, Li Y, et al. Amplification of LAPTM4B and YWHAZ contributes to chemotherapy resistance and recurrence of breast cancer. Nat Med. 2010;16:214–218. doi: 10.1038/nm.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silver DP, Dimitrov SD, Feunteun J, Gelman R, Drapkin R, Lu SD, et al. Further evidence for BRCA1 communication with the inactive X chromosome. Cell. 2007;128:991–1002. doi: 10.1016/j.cell.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 15.Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, et al. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Weaver Z, Linke SP, Li C, Gotay J, Wang XW, et al. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon 11 isoform-deficient cells. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 17.Vrieling H. Mitotic maneuvers in the light. Nat Genet. 2001;28:101–102. doi: 10.1038/88794. [DOI] [PubMed] [Google Scholar]
- 18.Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35:97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- 21.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 23.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 24.Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, Withers M, et al. Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet. 2003;72:1101–1116. doi: 10.1086/374385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 28.McVean G. What drives recombination hotspots to repeat DNA in humans? Philos Trans R Soc Lond B Biol Sci. 2010;365:1213–1218. doi: 10.1098/rstb.2009.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puliti A, Rizzato C, Conti V, Bedini A, Gimelli G, Barale R, et al. Low-copy repeats on chromosome 22q11.2 show replication timing switches, DNA flexibility peaks and stress inducible asynchrony, sharing instability features with fragile sites. Mutat Res. 2010;686:74–83. doi: 10.1016/j.mrfmmm.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 30.Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cha RS, Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 32.Arlt MF, Xu B, Durkin SG, Casper AM, Kastan MB, Glover TW. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol Cell Biol. 2004;24:6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz M, Zlotorynski E, Goldberg M, Ozeri E, Rahat A, le Sage C, et al. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005;19:2715–2726. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–2586. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalb R, Neveling K, Nanda I, Schindler D, Hoehn H. Fanconi anemia: causes and consequences of genetic instability. Genome Dyn. 2006;1:218–242. doi: 10.1159/000092510. [DOI] [PubMed] [Google Scholar]
- 36.Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burger H, Loos WJ, Eechoute K, Verweij J, Mathijssen RH, Wiemer EA. Drug transporters of platinum-based anticancer agents and their clinical significance. Drug Resist Updat. 2011 doi: 10.1016/j.drup.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Yang XH, Sladek TL, Liu X, Butler BR, Froelich CJ, Thor AD. Reconstitution of caspase 3 sensitizes MCF-7 breast cancer cells to doxorubicin and etoposide-induced apoptosis. Cancer Res. 2001;61:348–354. [PubMed] [Google Scholar]
- 40.Ogston KN, Miller ID, Payne S, Hutcheon AW, Sarkar TK, Smith I, et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. Breast. 2003;12:320–327. doi: 10.1016/s0960-9776(03)00106-1. [DOI] [PubMed] [Google Scholar]
- 41. The Cancer Genome Atlas website: http://tcga-data.nci.nih.gov/tcga/
- 42. cBioportal website: http://bit.ly/wpwRXd.
- 43.Wang Y, Moorhead M, Karlin-Neumann G, Wang NJ, Ireland J, Lin S, et al. Analysis of molecular inversion probe performance for allele copy number determination. Genome Biol. 2007;8:R246. doi: 10.1186/gb-2007-8-11-r246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. http://www.sanger.ac.uk/cosmic.
- 45.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bengtsson H, Wirapati P, Speed TP. A single-array preprocessing method for estimating full-resolution raw copy numbers from all Affymetrix genotyping arrays including GenomeWideSNP 5 & 6. Bioinformatics. 2009;25:2149–2156. doi: 10.1093/bioinformatics/btp371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bengtsson H, Neuvial P, Speed TP. TumorBoost: normalization of allele-specific tumor copy numbers from a single pair of tumor-normal genotyping microarrays. BMC Bioinformatics. 2010;11:245. doi: 10.1186/1471-2105-11-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. http://projects.tcag.ca/variation/
- 49.Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12:474. doi: 10.1186/1471-2105-12-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.