Abstract
This review outlines the use of expressed protein ligation (EPL) to study protein structure, function and stability. EPL is a chemoselective ligation method that allows the selective ligation of unprotected polypeptides from synthetic and recombinant origin for the production of semi-synthetic protein samples of well-defined and homogeneous chemical composition. This method has been extensively used for the site-specific introduction of biophysical probes, unnatural amino acids, and increasingly complex post-translational modifications. Since it was introduced 10 years ago, EPL applications have grown increasingly more sophisticated in order to address even more complex biological questions. In this review, we highlight how this powerful technology combined with standard biochemical analysis techniques has been used to improve our ability to understand protein structure and function.
Keywords: Chemical ligation, Protein α-thioesters, Intein, Protein splicing, Circular polypeptides, Post-translational modifications, Isotopic labeling
Introduction
The field of protein engineering has always played a key role in biological science, biomedical research, and biotechnology [1]. The development of recombinant DNA and heterologous expression techniques has allowed for the routine preparation of many proteins. These techniques, however, are limited to the introduction of the 20 naturally genetically encoded amino acids. In many cases, the introduction of chemical modifications, such as post-translational modifications (PTMs), un-natural amino acids or biophysical probes that are impossible to prepare by standard ribosomal synthesis, is desirable. During the last two decades, and thanks to the efforts of numerous chemists, an array of different techniques has been developed for the site-specific modification of proteins. These range, for example, from classical bioconjugation techniques [2] to more sophisticated approaches such as the use of nonsense suppression mutagenesis.
The use of chemical ligation techniques has also emerged during the last decade as another powerful approach for the chemical engineering of proteins (see [1] for a recent and extensive review). Chemical ligation utilizes efficient reactions between unprotected peptides to form a stable peptide bond in a chemoselective way between the α-carboxyl group of one of the peptide fragments and the α-amino group of the other peptide fragment.
A defining point, however, was established when native chemical ligation (NCL) was independently introduced by Kent and Tam in 1994 [3, 4]. In this reaction, two fully unprotected peptides, one containing a C-terminal α-thioester group and the other an N-terminal Cys, react chemoselectively under neutral conditions with the formation of a native peptide bond at the ligation site. This type of thioester-based chemistry was first pioneered by Wieland in the 1950s for the synthesis of small Cys-containing peptides [5, 6]. Since its introduction in 1994, NCL has been widely used for the chemical synthesis of a multitude of natural and chemically modified medium-sized proteins (see [7] for a recent review).
The major strength of NCL is that it combines the ability of chemical (peptide) synthesis to access any desired modification with the flexibility of recombinant DNA technology to produce any size of protein, thus permitting the semisynthesis of even large proteins. The NCL of a synthetic peptide thioester with a recombinant N-terminal Cys-containing protein was reported by Verdine and co-workers [8].
The NCL of recombinant polypeptide α-thioesters and synthetic N-terminal Cys-containing polypeptides was first reported in 1998 independently by the Muir and Xu groups, and was named expressed protein ligation (EPL) [9] and intein-mediated protein ligation (IPL) [10], respectively.
Since its introduction in 1998, EPL has been applied to the engineering of many classes of protein from both eukaryotic and prokaryotic organisms (see [11–13] for recent reviews). These include kinases, phosphatases, transcription factors, polymerases, ion channels, cytoplasmic and membrane signaling proteins as well as antibodies. A variety of chemical modifications have been introduced into these proteins allowing the answering of questions that would be difficult to respond to by other means.
Most recently, new types of chemical ligation involving protein splicing are also emerging for the chemical engineering of proteins (see [14, 15] for recent reviews). Intein-mediated protein trans-splicing is based on the use of split inteins that mediate the linking of N- and C-terminal exteins by a native peptide bond in trans with concomitant removal of the intein complex. This naturally occurring post-translational modification is a self-processing event that only requires the polypeptide fragments to be linked to be fused to a split intein, thus providing a extremely powerful technique for chemical modification of proteins in vitro and also in vivo [14]. The use of protease-catalyzed protein splicing has also been introduced recently for the chemical engineering of proteins [14].
The goal of this review is to provide a recent overview of EPL and its applications, with a particular emphasis in those involved in the study of protein structure function and stability. These include introduction of complex protein modifications (such as lipidation or glycosylation), protein immobilization, and biosynthesis of topologically altered proteins, among others.
Native chemical ligation
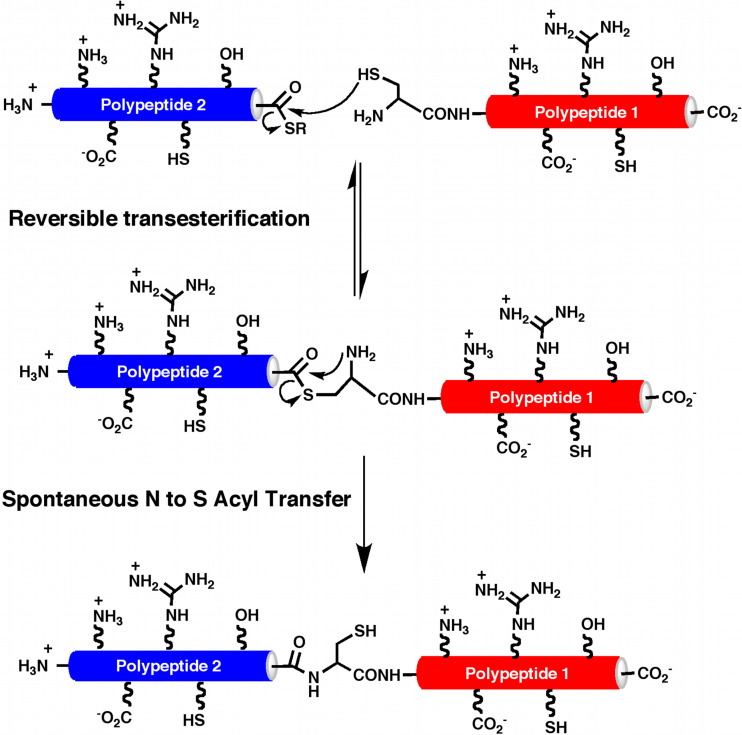
Native chemical lLigation (NCL) is an exquisitely specific ligation reaction that has been extensively used for the total synthesis, semi-synthesis, and engineering of different proteins [16–19]. In this reaction, two fully unprotected polypeptides, one containing a C-terminal α-thioester group and the other an N-terminal Cys residue, react chemoselectively under neutral aqueous conditions with the formation of a native peptide bond (Fig. 1). The initial step in this ligation involves the formation of a thioester-linked intermediate, which is generated by a trans-thioesterification reaction involving the α-thioester moiety of one fragment and the N-terminal Cys thiol group of the other fragment. This intermediate then spontaneously rearranges to produce a peptide bond at the ligation site.
Fig. 1.
Principle of native chemical ligation (NCL)
The presence of other sulfhydryl groups from Cys residues within the peptide fragments does not affect the reaction since the trans-thioesterification step is reversible, and only the N-terminal Cys residue contains an α-amino group that reacts irreversibly with the thioester moiety to give the corresponding peptide bond. The speed of the NCL strongly depends on the nature of the amino acid present at the C-terminal of the thioester, with Gly the one that makes the reaction faster, while β-branched amino acids make the reaction proceed more slowly and with lower yields [20]. The nature of the thioester also plays an important role on the efficiency of the reaction, and usually aryl-thioesters are preferred with respect to alkyl thioesters [1].
Synthesis of peptide α-thioesters
Several solid-phase methods are available for the chemical synthesis of peptide α-thioesters. The most general uses tert-butyloxycarbonyl (Boc)-based solid-phase peptide synthesis (SPPS) [3, 4, 20–23]. This approach uses acid-base deprotections to which thioester linkers are stable. However, the final cleavage step typically involves the use of the highly toxic and corrosive anhydrous HF, which is not well suited for the synthesis of phospho- [9, 24] and glyco-peptides [25–27].
The commonly used 9-fluorenylmethoxycarbonyl (Fmoc)–based methodology uses repeated base treatments, which renders this strategy incompatible with thioester linkers. However, this approach allows the incorporation of acid-sensitive groups, such as phosphates, carbohydrates, and prenylated moieties [11]. Consequently, several technologies have been developed to allow the synthesis of peptide α-thioesters by Fmoc-based SPPS [13, 28]. Although none of the techniques is as robust as Boc-based SPPS, the use of safety-catch linkers (or “masked thioesters”) is quite promising [25, 29–32].
Synthesis of N-terminal Cys-containing peptides
N-Cys peptides can be chemically synthesized by routine SPPS using either Boc- or Fmoc-based SPPS [7, 28]. It should be noted, however, that the use of side-chain protecting groups that generate formaldehyde, such as the benzyloxymethyl (Bom) or tert-butyloxymethyl (Bum) groups, may give rise to alkylation of the N-terminal Cys residue to produce an NCL unreactive thiazolidine [33]. The thiazolidine group can be removed, however, by treatment with methoxamine to yield the free N-terminal Cys residue [34].
Expressed protein ligation
Despite the simplicity and robustness of NCL, which has resulted in its widespread application [18], this extremely powerful technology is limited at present to the synthesis of small to moderately sized proteins, mainly because of the current restriction of SPPS to peptides of ≈60 amino acids in length and the difficulties associated with performing multiple NCL ligation steps.
A way to overcome this size limitation is by combining the NCL approach with recombinant protein production. This can be accomplished via two different approaches. First, a synthetic peptide thioester can be reacted by NCL with a recombinant protein with an N-terminal Cys residue [8], which allows the introduction of chemically modified peptides at the N-terminus of recombinant proteins. The other possibility involves the ligation of a synthetic N-terminal Cys peptide with a recombinant protein α-thioester protein. The latter approach was first reported in 1998 [9, 10, 35, 36] and was termed expressed protein ligation (EPL) or, less frequently, intein-mediated protein ligation.
Recombinant polypeptide α-thioesters
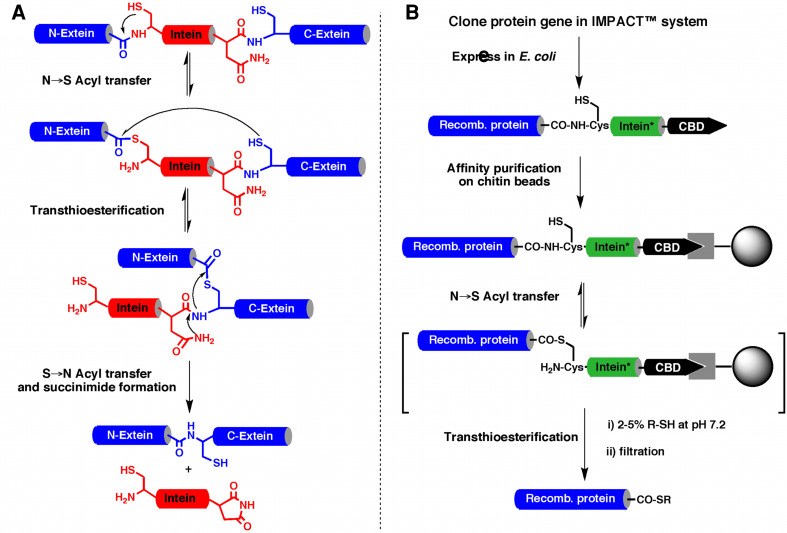
Recombinant protein α-thioesters can be obtained by using engineered inteins [9, 17, 35, 36]. Inteins are self-processing domains which mediate the naturally occurring process called protein splicing [37] (Fig. 2). Protein splicing is a cellular processing event that occurs post-translationally at the polypeptide level. In this multi-step process, an internal polypeptide fragment, called intein, is self-excised from a precursor protein and in the process ligates the flanking protein sequences (N- and C-exteins) to give a different protein. The current understanding of the mechanism is summarized in Fig. 2a and involves the formation of thioester/ester intermediates [37]. The first step in the splicing process involves an N → S or N → O acyl shift in which the N-extein is transferred to the thiol/alcohol group of the first residue of the intein. After the initial N → (S/O) acyl shift, a trans-esterification step occurs in which the N-extein is transferred to the side-chain of a second conserved Cys, Ser, or Thr residue, this time located at the junction between the intein and the C-extein. The amide bond at this junction is then broken as a result of succinimide formation involving a conserved Asn residue within the intein. In the final step of the process, a peptide bond is formed between the N-extein and C-extein following an (S/O) → N acyl shift (similar to the last step of NCL; see Fig. 1a). Mutation of the conserved Asn residue within the intein to Ala blocks the splicing process in midstream thus resulting in the formation of an α-thioester linkage between N-extein and the intein [37] (Fig. 2b). This thioester bond can be cleaved using an appropriate thiol through a trans-thioesterification step to give the corresponding recombinant polypeptide α-thioester.
Fig. 2.
Biosynthesis of recombinant polypeptide α-thioesters. a Scheme representing the proposed canonical mechanism for protein splicing mediated by a Cys-intein. b Expression and purification of recombinant polypeptide thioesters using a modified intein fusion protein. In the modified intein (represented by an asterisk) the last Asn residue of the intein has been mutated to Ala to prevent C-terminal cleavage and splicing. This mutation allows trapping a thioester intermediate that can be cleaved with a thiol to provide the corresponding thioester funtion
Several modified inteins are commonly used for this purpose and many are commercially available as E. coli expression vectors [38, 39]. One of the most generally useful inteins is the Mycobacterium xenopy DNA gyrase (Mxe GyrA) intein. This intein has shown several important features for this purpose: (1) it is relatively small (198 amino acids) and can be expressed very efficiently in E. coli; (2) it does not have special sequence preferences for the last residues of the N-extein fragment; (3) the thiolysis reaction can be performed in the presence of detergents [40, 41], and small amounts of denaturing agents [41] and organic solvents [40]; and (4) the GyrA intein can be efficiently refolded thus allowing the recovery of intein-fusion proteins from E. coli incusion bodies [40].
Recombinant proteins with N-terminal Cys residues
The introduction of N-terminal Cys residues into expressed proteins can be readily accomplished by cleaving (by proteolysis or auto-proteolysis) the appropriate fusion proteins. The simplest way to generate a recombinant polypeptide containing an N-terminal Cys residue is to introduce a Cys downstream to the initiating Met residue. Once the translation step is completed, the endogenous methionyl aminopeptidases (MAP) removes the Met residue, thereby generating in vivo an N-terminal Cys residue [42–46]. Other approaches involve the use of exogenous proteases. Verdine and co-workers added a Factor Xa recognition sequence immediately in front of the N-terminal Cys residue of the protein of interest [8]. After purification, the fusion protein was treated with the protease Factor Xa, which generated the corresponding N-terminal Cys protein. Tolbert and Wong have also shown that the cysteine protease from tobacco etch virus (TEV) can be used for the same purpose [47]. This protease is highly specific and can be overexpressed in E. coli. Other proteases that cleave at the C-terminal side of their recognition site, like enterokinase and ubiquitin C-terminal hydrolase, could also be used for the generation of N-terminal Cys residues. Proteins with N-terminal Cys can also be obtained by the convenient modification of expression vectors with the putative thrombin cleavage site LVPRG to LVPRC [48]. More recently, Hauser and Ryan [49] have used the N-terminal pelB leader sequence to direct newly synthesized fusion proteins to the E. coli periplasmic space where the corresponding endogenous leader peptidases [50, 51] can generate the desired N-terminal cysteine-containing protein fragment.
Finally, protein splicing can also be engineered to produce recombinant N-terminal Cys-containing polypeptides. Several inteins have already been mutated in such a way that cleavage at the C-terminal splice junction (i.e., between the intein and the C-extein; see Fig. 2b) can be accomplished in a pH- and temperature-dependent fashion [10, 52, 53].
Selected applications of expressed protein ligation
The initial scope of EPL was the site-specific introduction of chemical modifications at the C-terminus of recombinant proteins. Since then, this technology has been used for a variety of novel purposes, including the incorporation of novel chemical groups to evaluate novel bioconjugation techniques [54–57], protein immobilization on solid-supports [58, 59], polypeptide backbone cyclization [44, 60–62], incorporation of non-natural amino acids [40, 63] and optical probes [64, 65], isotopic-editing [41, 66–69], and semi-synthesis of prenylated proteins [70–72], among others. In this section, we will highlight just a few representative examples that serve to illustrate the power of this technology to allow the detailed analysis of protein function and structure.
Site-specific immobilization of polypeptides onto solid supports
Many experimental approaches in biology and biophysics as well as applications in diagnosis and drug discovery require proteins to be immobilized on solid substrates. Immobilized proteins are instrumental in identifying protein–protein, protein–DNA, and protein–small molecule interactions; they can also be used for a variety of diagnostic and profiling purposes (see [73] for a recent review).
Although enormous progress has been made in immobilizing DNA onto different types of solid supports, the immobilization of proteins has been a particularly challenging task, mainly due to the heterogeneous chemical nature of proteins and the marginal stability of the native, active tertiary structure over the denatured and inactive random coil structure.
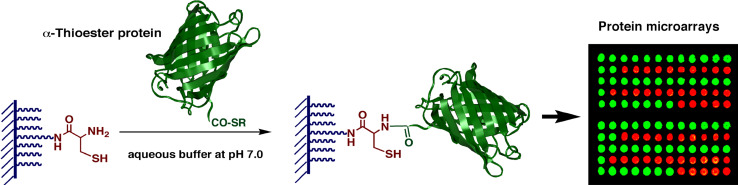
In 2003, we reported the use of EPL for the covalent and site-specific attachment of recombinant protein onto glass surfaces [58]. In this work, C-terminal α-thioesters of two fluorescent proteins (EGFP and DsRed) and the c-Crk SH3 domain were immobilized onto an N-terminal Cys-coated glass slide (Fig. 3). The reaction was highly selective allowing the covalent immobilization of folded and biologically active proteins through their C-termini.
Fig. 3.
Selective attachment of EGFP (green) and DsRed (red) α-thioesters onto a Cys-containing glass slide though EPL as an example of protein microarray production. Epifluorescence image of the glass slide after protein thioester spotting and incubation overnight. The slide was extensively washed with phosphate buffer solution to removed non-specifically bound proteins before imaging
Yao and co-workers have also used NCL and EPL for the selective immobilization of N-terminal Cys-containing polypeptides [74] and proteins [59] onto α-thioester-coated glass slides. In this case, the polypeptide–proteins were site-specifically immobilized through their N-termini, which may be convenient in cases where the C-terminal immobilization, described earlier, affects the activity of the protein.
EPL has also been used for the site-specific introduction of reactive groups such as alkynes [56, 75] and azides [55, 57], that can be used later for the site-specific immobilization of proteins onto modified solid supports using Staudinger and/or Cu(I)-catalized Huisgen 1,3-dipolar cycloaddition reactions (see [1, 76] for recent reviews).
Yao and co-workers have also used EPL for the site-specific biotinylation of proteins in order to be selectively immobilized onto streptavidin/avidin-coated solid supports [77, 78]. This reaction was performed by either in vivo or in vitro cleavage of the corresponding modified-intein fusion protein with N-Cys biotinylated peptides. More recently, Beck-Sickinger and co-workers have used a similar approach for the immobilization of the biotinylated aldoreductase AKR1A [79]. Investigation of the kinetic parameters of the immobilized enzyme showed they were comparable to those of the wild-type enzyme in solution and 60- to 300-fold greater than those of the randomly immobilized enzymes. Furthermore, the enzyme was surprisingly stable. No loss of activity was observed for over a week, and even after 50 days more than 35% of activity was maintained.
Backbone cyclized polypeptides and proteins
A significant number of natural products with wide range of pharmacological activities derive from cyclic polypeptides. In fact, peptide cyclization is widely used in medicinal chemistry to improve the biochemical and biophysical properties of peptide-based drug candidates [80, 81]. Cyclization rigidifies the polypeptide backbone structure, thereby minimizing the entropic cost of receptor binding and also improving the stability of the topologically constrained polypeptide. Among the different approaches used to cyclize polypeptides, backbone, or head-to-tail, cyclization remains one of the most extensively used to introduce structural constraints into biologically active peptides.
Despite the fact that the chemical synthesis of cyclic peptides has been well explored and a number of different approaches involving solid-phase or liquid-phase exist [22, 82–85], the biosynthesis of cyclic polypeptides offers many advantages over purely synthetic methods. Using the tools of molecular biology, large combinatorial libraries of cyclic peptides may be generated and screened in vivo.
A typical chemical synthesis may generate ≈104 different molecules. It is not uncommon for a recombinant library to contain as many as ≈109 members. The molecular diversity generated by this approach is analogous to phage-display technology. Moreover, this approach takes advantage of the enhanced pharmacological properties of backbone-cyclized peptides as opposed to linear peptides or disulfide-stabilized polypeptides. Also, the approach differs from phage-display in that the backbone-cyclized polypeptides are not fused to or displayed by any viral particle or protein, but remain on the inside of the living cell where they can be further screened for biological activity. The complex cellular cytoplasm provides the appropriate environment to address the physiological relevance of potential leads.
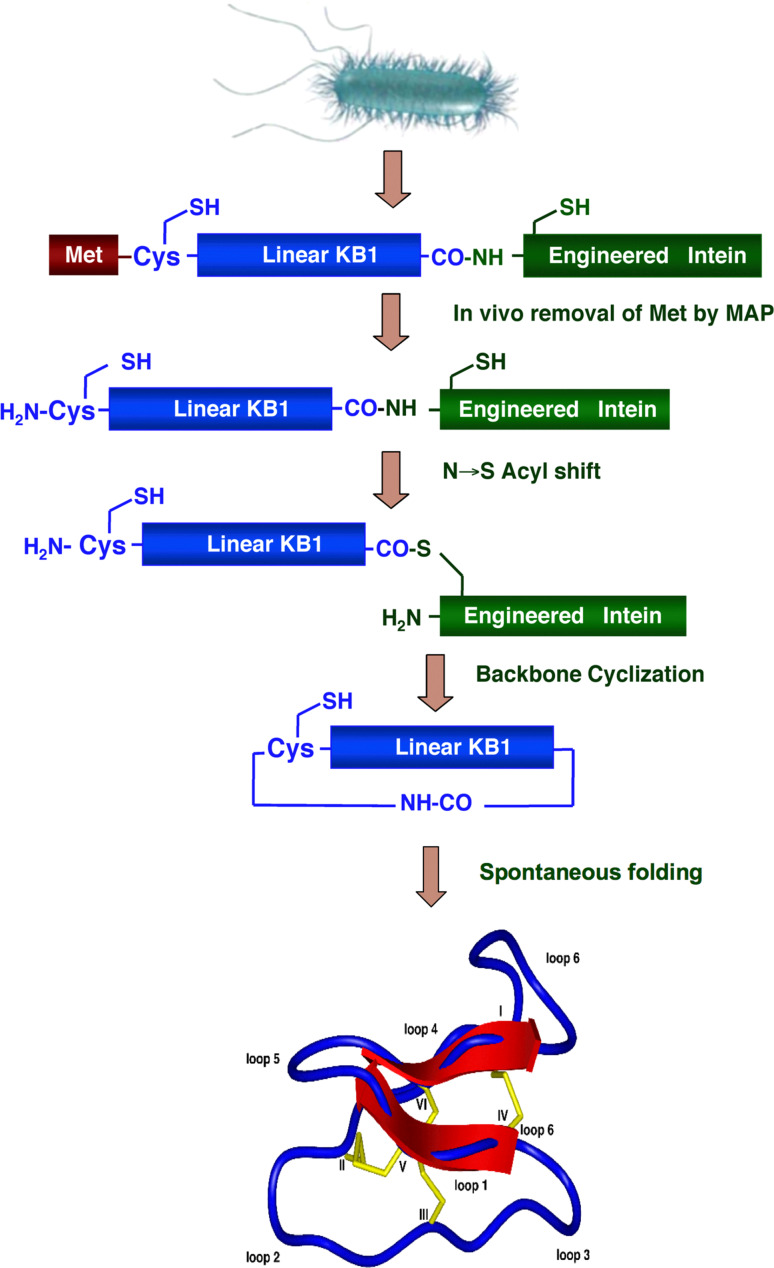
An attractive alternative approach to the biosynthesis of circular polypeptides is the use of an intramolecular version of EPL reaction. The approach employed for the biosynthesis of backbone-cyclized polypeptides using EPL is depicted in Fig. 4. The target polypeptide to be cyclized is fused at the N-terminus with a peptide leading sequence immediately followed by a Cys residue, and at the C-terminus with an engineered intein. The N-terminal leading sequence can be cleaved in vitro or in vivo by a proteolytic or self-proteolytic event thereby generating the required N-terminal Cys residue. This Cys residue then reacts in an intramolecular fashion with the α-thioester generated by the engineered intein at the C-terminus, thus providing a recombinantly generated backbone-cyclized polypeptide. This approach has been used for the in vitro and in vivo biosynthesis of different backbone-cyclized polypeptides [45, 60–62, 86].
Fig. 4.
Biosynthetic approach for in vivo production of cyclotides inside live E. coli cells using intramolecular EPL. Backbone cyclization of the linear cyclotide precursor is mediated by a modified protein splicing unit or intein. The cyclized product then folds spontaneously in the bacterial cytoplasm
The demonstration of this biosynthetic cyclization strategy was first reported in vitro and in vivo by Camarero and Muir using the N-terminal SH3 domain of the c-Crk protein as model protein [60, 87]. Iwai and Pluckthum [44] also reported the biosynthesis of a cyclized version of the β-lactamase protein using a similar approach.
More recently, we have applied the same approach for the biosynthesis of cyclotides inside living bacterial cells (Fig. 4) [62]. Cyclotides are small globular microproteins with a unique head-to-tail cyclized backbone, which is stabilized by three disulfide bonds [88] (Fig. 4). The number and positions of cysteine residues are conserved throughout the family, forming the cyclic cystine-knot motif (CCK) [88] that acts as a highly stable and versatile scaffold on which hyper-variable loops are arranged. This CCK framework gives the cyclotides exceptional resistance to thermal and chemical denaturation and enzymatic degradation. Moreover, several cyclotides have been found able to cross eukaryotic cell membranes [89]. All these unique properties make them ideal candidates for the development of peptide-based drugs [90]. Our group has recently developed and successfully used a bio-mimetic approach for the biosynthesis of several folded cyclotides inside cells by making use of intramolecular NCL in combination with modified protein splicing units [61, 62] (Fig. 4). This important finding makes possible the generation of large libraries of cyclotides (≈109) for high throughput cell-based screening and selection of specific sequences able to recognize particular biomolecular targets [61, 62].
Isotope-edited spectroscopy
The development of EPL has helped to overcome some of the size limitations associated with the structural analysis of proteins by nuclear magnetic resonance (NMR). Although observable, NMR signals from large proteins exhibit extreme spectral overlap, which cannot be resolved even in 3D- or 4D-NMR spectra [91]. A way to decrease such spectral complexity is to use samples where only selected amino acids are labeled with NMR active nuclei, thus editing out signals from the rest of the molecule. EPL allows the site-specific introduction of specific NMR-active isotopes within a protein, thus facilitating the assignment of resonances from the labeled residues [92]. In one recent example, Muir and co-workers were able to label the N-extein residue located at the intein junction of the GyrA intein with 13C and 15N using EPL [66]. This allowed the 1JNC coupling constant of the amide bond at the N-extein–intein junction to be measured on an active intein. The data indicated this peptide bond was highly polarized, thus supporting the idea that the first step in protein splicing is facilitated, in part, by destabilizing the scissile amide bond. Baransky and co-workers used EPL to introduce 13C-labeled amino acids at the carboxyl terminus of the α subunit of heterotrimeric G protein [67]. Analysis of the 13C resonances revealed the Gα carboxyl terminus is highly mobile in its GDP-bound state but adopts an ordered conformation upon activation of the subunit. The authors suggest that this conformation change may facilitate the release of the Gα subunit from the G-protein-coupled receptor.
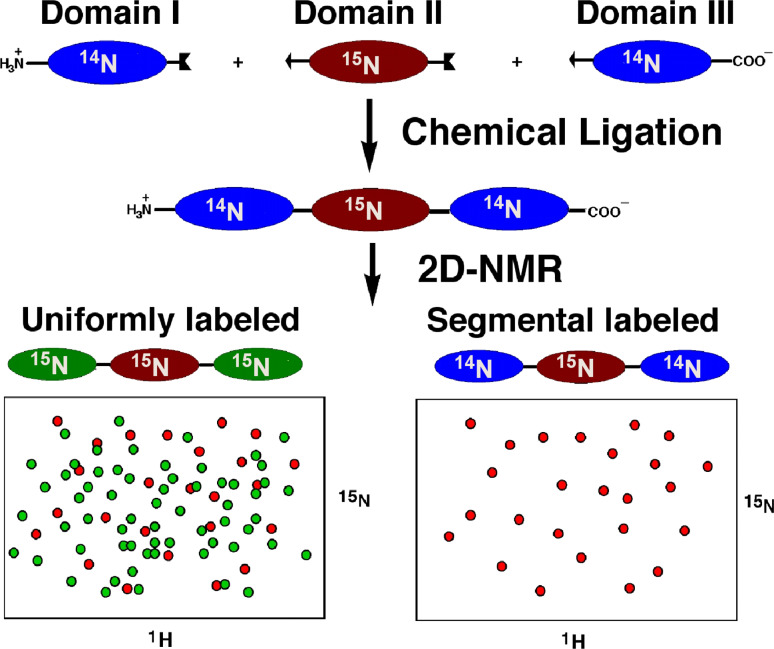
EPL also allows the generation of segmental isotopically labeled proteins for structural studies using NMR. Hence, by using EPL, is possible to ligate a uniformly labeled protein fragment to the rest of the unlabeled protein. This approach dramatically decreases the spectral complexity of the sample allowing analysis of the signals from the labeled fragment in the context of the whole protein (Fig. 5).
Fig. 5.
Segmental isotopic labeling of a multidomain protein using EPL. This technique allows study the nuclear resonance signals from one domain (in this case the internal domain) in the context of the full length protein. The spectrum of the uniformly labeled sample, on the other hand, shows extensive signal overlap, therefore making difficult signal assignment and analysis of large proteins
Allain and co-workers have used an optimized on-column EPL protocol in combination with transverse relaxation-optimized NMR spectroscopy [68] to elucidate the structure of the two C-terminal RNA recognition motifs (RRM3 and RRM4) of the polypyrimidine tract binding protein (PTB). Although preliminary studies showed that these two domains do not interact in the free state, they interact extensively when bound to RNA. EPL allowed the production of PTB constructs where either RRM3 or RRM4 were labeled with 13C and 15N, thus allowing the rapid elucidation of the structure of the construct containing both domains. The final structure revealed a large interdomain interface, resulting in a very unusual positioning of the two RRM domains relative to one another. Based on these results, the authors suggest that this unusual structure induces the formation of RNA loops, which could repress splicing by sequestering either a short alternative-exon or a branch point within these RNA loops. The same authors have also used a similar EPL approach to study the intramolecular interactions in two other RRM-containing proteins, heterogeneous nuclear ribonucleoprotein L (hnRNP L) and Npl3 [69]. The results indicated that RRMs of hnRNP L interact, whereas those of Npl3 were independent.
A similar on column procedure has also been reported recently for the preparation of segmentally labeled constructs of the lipoprotein Apoliprotein E3 (apoE) [93] in order to elucidate its structure. This protein, involved in the catabolism of lipids, contains a 22-kDa N-terminal domain and a 10-kDa C-terminal domain linked by a protease-sensitive hinge region. A potential domain–domain interaction has been hypothesized between the two domains of apoE that seems to regulate its biological functions. Although the structure of the ApoE N-terminal domain in the lipid free state is known, there is no structure available to date for the apoE C-terminal domain and full-length apoE. Since apoE is a 299-residue helical protein, its NMR spectrum is significantly overlapped. To reduce NMR spectral complexity for a complete spectral assignment, the authors produced several segmental labeled apoEs, in which one domain was 13C/15N labeled whereas the other domain was deuterated.
Access to isotopically labeled proteins has also extended the scope of vibrational spectroscopy for the structural analysis of proteins. An example of this has recently been illustrated by Tatulian and co-workers who used EPL for the generation of a segmental 13C-labeled version of phospholipase A2 (PLA2) in combination with polarized attenuated total reflection Fourier transform infrared (ATR-FTIR) spectroscopy to assess the mode of interaction of this protein with membranes [94]. The use of a segmental labeled PLA2, where only two of the three α-helices of this protein were 13C-labeled, allowed them to assign the amide I signals from the labeled α-helices. This information was used to generate structural constraints that established the orientation of the membrane-bound PLA2. This approach is in principle quite general and it is likely to become a fundamental tool for determination and analysis of the structure of membrane proteins, which will undoubtedly provide valuable information on the molecular mechanisms of this important class of proteins.
More recently, EPL has also been used for the incorporation of the positron-emitting isotope 18F into leptin [95]. A two-step, site-specific ligation approach was developed for this purpose, in which an aminooxy-reactive group was incorporated at the C-terminus of leptin using EPL. This modified protein was subsequently derivatized with 18F-fluorobenzaldehyde using an aniline-accelerated radiochemical oximation reaction. The modified hormone was shown to be biologically active in vitro and in vivo, and it was applied to positronic emission tomography (PET) imaging in mice lacking a functional leptin gene.
Introduction of post-translational modifications for structural and biological studies
EPL has also made possible the detailed structural study of proteins containing specific post-translational modifications (PTMs). Homogeneous protein samples containing chemically well-defined PTMs are extremely difficult to generate by using standard protein expression methods. PTMs are extremely important in regulating protein function. One of the most common PTM found in eukaryotic cells is phosphorylation of Ser, Thr, and Tyr residues. It has been estimated that as many as a third of all mammalian proteins are phosphorylated. In fact, one of the first applications of EPL was to produce a semi-synthetic version of the kinase Csk where one of the C-terminal Tyr residues was site-specificially phoshorylated [9]. This seminal work served to illustrate the enormous potential of expressed protein ligation as a simple and powerful new method in protein engineering to introduce sequences of unnatural amino acids, post-translational modifications, and biophysical probes into proteins of any size.
More recently, EPL has been used to prepare phosphorylated versions of the TFG-β signaling proteins, Smad2 and Smad3 [24, 96, 97]. These proteins are mammalian transcription factors that upon phosphorylation are able to oligomerize and be translocated to the nucleus where they control transcription. The preparation of semi-synthetic Smad proteins specifically phosphorylated at specific Ser and Thr residues has allowed several high-resolution crystal structures to be determined [98–101]. These structural data have led to a detailed understanding of the interactions underlying the Smad homo- and hetero-oligomerization that involves the activation of this family of transcription factors.
Another type of PTM found in eukaryotic proteins is prenylation. This interesting PTM is found in Rab GTPases. These proteins are members of the class of monomeric GTPases and play a critical role in membrane trafficking. Rabs are anchored to the membrane by PTM with lipids at the C-terminus [71]. Lipid modifications, and in particular prenylation, are extremely challenging due to the problems associated with low solubility and stability of the lipidated protein. The development of effective ligation conditions and synthetic approaches for the production of prenylated proteins is a remarkable achievement (see [70] for an extensive review).
Waldmann and co-workers have extensively used EPL to prepare mono- and bi-prenylated versions of the small Rab/Ypt protein family member Ypt1 [102, 103]. The co-crystal structures of these proteins with Rab GDP dissociation inhibitor (RabGDI) were determined providing structural insight on how RabGDI is able to extract lipidated-Rabs from membranes to facilitate their translocation to acceptor membranes [104–106].
Another important field where EPL has made important contributions is in the study of the biological effects of PTMs on histones [11]. Histones have flexible N-terminal tails that are heavily post-translationally modified. These modifications regulate both the structure and function of chromatin in transcription, replication, repair, and condensation. Both the position and nature of the modifications control the biological effects, and numerous enzymes have been identified that are able to introduce or remove such modifications [107].
Several groups have recently used protein ligation techniques for preparing homogeneously modified full-length histones. Peterson and co-workers reported the preparation of a histone H3 variant with phosphorylated Ser at position 10. This homogeneously modified histone was then incorporated into nucleosomes arrays [108] and a series of enzymatic reactions involving the histone acetyltransferase Gcn5. Kinetic analysis revealed that Gcn5 had increased activity on the modified nucleosomes, while the Gcn5-containing SAGA complex was not stimulated by H3 phosphorylation in the context of nucleosomal arrays. By using a similar approach, a histone H4 variant with an ε-acetylated Lys residue at position 16 was also prepared by the same group [109]. The incorporation of this modified histone into nucleosomal arrays inhibited the formation of compact 30-nanometer-like fibers and impeded the ability of chromatin to form cross-fiber interactions. Furthermore, this acetylated histone also inhibited the ability of the adenosine triphosphate-utilizing chromatin assembly and remodeling enzyme ACF to mobilize a mononucleosome, indicating that this single histone modification modulates both higher order chromatin structure and functional interactions between a non-histone protein and the chromatin fiber.
McCafferty and co-workers also used chemical ligation to prepare several site-specifically-modified histones, including an acetylated and methylated H3 and an acetylated H4 [110]. In this work, the semisynthetic approach to generate the modified histones was extended by adding a desulfurization step after the ligation, thereby converting the Cys residue into Ala, allowing a traceless ligation. The modified histones were fully functional, as evidenced by their self-assembly into a higher order H3/H4 heterotetramer, their deposition into regular nucleosome arrays, and utilization as substrates for histone modifying enzymes.
Muir and co-workers have also developed a traceless semisynthetic approach for the preparation of ubiquitylated and sumoylated histone-derived peptides [111]. This approach was recently extended to the preparation of a modified full-length H2B histone [112]. Ubiquitylated H2B was incorporated into nucleosomes, and it was demonstrated that this modification stimulates intranucleosomal methylation of H3 Lys79 by the methyltransferase of hDot1L. According to the authors, this effect is mediated through the catalytic domain of hDot1L, most likely through allosteric mechanisms. This result demonstrates the direct biochemical evidence of crosstalk between two modifications on separate histone proteins within a nucleosome.
Another important PTM found in eukaryotic proteins is glycosylation. It is estimated that more than half of proteins present in nature are glycosylated. These carbohydrate chains play various essential roles, such as protein folding, cell adhesion, cell differentiation, and tumor metastasis. In contrast to nucleotide and amino acid sequences, the structure of sugar chains is not determined genetically, but solely by the activity of enzymes such as glycosyltransferases. As a result, the carbohydrate structure on the same amino acid sequence is highly variable, which is known as glycoform. Recent advances in the chemical synthesis of glycopeptides has allowed for the first time a reliable way to obtain homogeneous glycopeptides or glycoproteins (see [113] for a recent review).
EPL and NCL have been extensively used for the preparation of homogenous glycoprotein samples. Berotzzi and co-workers prepared a variant of diptericin, an 82-residue antibacterial glycoprotein produced by insects in response to immunological challenge [25]. Native diptericin exists as a mixture of O-linked glycoforms; one of the simplest of them possesses single GalNAc residues at the two glycosylation sites, Thr10 and Thr54. This glycoform was prepared by NCL of two synthetic glycopeptides, each of which was generated Fmoc-based SPPS. The resulting glycoprotein was biologically active in bacterial growth inhibition assays. The same group also succeeded in the synthesis of Lymphotactin, a 93-residue chemokine containing eight sites of O-linked glycosylation, using a similar approach, except the thioester fragment was synthesized by Boc-based SPPS [114]. NCL was also used by Imperiali and co-workers for the preparation of an N-linked chitobiose glycoprotein analogue of Im7, an 87-residue protein [115]. Im7 is a member of a family of four homologous E colicin immunity proteins, the function of which is to inhibit the endonuclease domain of their specific bacterial colicin toxin. Im7, while not naturally glycosylated, was used as model system for the study of the effects of glycosylation on protein folding and stability. The reported results indicated that the folding mechanism of the glyosylated Im7 variant was not significantly altered over the unglycosylated analogue.
The use of EPL was first reported by Tolbert and Wong for the ligation of a 392-residue intein-generated α-thioester and N-Cys dipeptide functionalized with a single N-acetylgluocasmine residue [26]. EPL was also used for the semi-synthesis of GlyCAM-1, a mucin-like glycoprotein composed of 132 residues which functions as a ligand for l-selectin [116].
Incorporation of non-natural amino acids
An interesting example of using EPL for the incorporation of non natural amino acids onto proteins to explore their structure and function was reported by Muir and co-workers for the semi-synthesis of a potassium channel analog with a d-amino acid located at the selectivity filter [40, 63]. Potassium channels are integral membranes, which permit the rapid and selective conduction of potassium ions across biological membranes. The recent elucidation of the crystal structures of the bacterial potassium channel KcsA has led to unprecedented insights into the basis of ion selectivity versus sodium and other cations. The selectivity filter of KcsA includes Gly77, which upon mutation to Ala results in functional loss. Gly77 exists in a left-handed helical conformation. However, its precise contribution to potassium channel function had been unclear. It was considered that a d-amino acid at this position would maintain the left-handed helical conformation and that the Gly is effectively serving as a d-amino acid surrogate. To test this hypothesis, a KcsA analog with Gly77 replaced by D-Ala was prepared using EPL [40]. The crystal structure of this analog revealed that, in the presence of high [K+], the D-Ala replacement had no effect on the structure of the filter, which was expected. Interestingly, the D-Ala containing selectivity filter remained in the conductive conformation even at low [K+] and was able to conduct Na+ in the absence of K+ ions [63]. However, the channel was still completely selective for K+ in the presence Na+. The same authors also explored the replacement of a native peptide bond (Tyr78–Gly79) within the selectivity filter with an ester bond [117]. This subtle replacement is nearly isosteric but is expected to result in a reduction of the electronegativity of the carbonyl group by ≈50%. The crystal structure of this modified channel was studied under different conditions. However, the isosteric ester substitution did not have a significant effect on the backbone structure of the selectivity filter, but as anticipated, significantly reduced ion density at that particular location with the selectivity filter. These studies demonstrate how nature can use Gly in lieu of a d-amino acid in a protein to achieve a desired structure–function relationship. The semi-synthesis of the different analogs of this integral membrane protein and their assembly into a functional tetrameric form represents a technical milestone in the protein chemistry arena.
Incorporation of optical probes
Perhaps of all the potential chemical modifications of a protein, fluorescent labeling has been one the most widely used in biological research. Nature provides two natural fluorescent amino acids, Tyr and Trp, although Trp is the more sensitive and by far the most used to monitor protein folding transitions and ligand binding events. However, large proteins usually contain more than one Trp residue, which reduces the spectral resolution of the analysis. Protein ligation can expand the repertoire of fluorescent probes that can be introduced into proteins. There have been a large number of fluorescent-labeled proteins prepared by EPL [19, 118]. Most of these proteins have been used to study ligand binding events, either by change in the fluorescence of a single probe or by using fluorescence energy transfer (FRET) between donor and acceptor probes introduced site-specifically in the protein of interest.
An example of the first approach was reported by Muir and co-workers [65], who combined expressed protein ligation (EPL) and in vivo amino acid replacement of tryptophans with 7-azatryptophan (7AW, a tryptophan analogue) to produce a 7AW-labeled SH3 domain from the c-Crk-I adaptor protein. This was accomplished using a Trp auxotroph E. coli strain. The 7AW is isosteric with tryptophan, but its fluorescence excitation and emission properties are red-shifted. Chemical ligation of the 7AW-labeled SH3 domain to the c-Crk-I Src homology 2 (SH2) domain, via EPL, generated the multidomain protein, c-Crk-I, with a domain-specific label. Use of this non-invasive optical probe allowed the equilibrium stability and ligand-binding properties of the SH3 domain to be unambiguously studied in the context of the full-length protein.
Lorsch and co-workers have also used EPL for the site-specific labeling of elF1A and elF1 with fluorescein and rhodamine [119, 120]. These two translation initiation factors are required for the formation of the 43S mRNA-ribosomal subunit complex. Using a combination of fluorescence anisotropy and FRET measurements, it was revealed that elF1 and elF1A are close to each other when initially binding to the ribosome but elF1 dissociates after start codon recognition [120].
Cole and co-workers have also used EPL in combination with FRET spectroscopy to study serotonin N-acetyltransferase [121]. Serotonin N-acetyltransferase [arylalkylamine N-acetyltransferase (AANAT)] is a key circadian rhythm enzyme that drives the nocturnal production of melatonin in the pineal gland. EPL was used for the generation of fluorescent versions AANAT and the protein 14-3-3ζ, in order to develop a rapid fluorescence-based assay to study the AANNAT–14-3-3ζ interaction [121]. EPL was also used to generate doubly fluorescently labeled AANAT that could be used to assess the stability of this protein in a live cell using a real-time assay by fluorescence resonance energy transfer measured by microscopic imaging [121].
More recently, Zheng and co-workers [122] have used EPL to site-specifically label the histone acetyltransferases (HATs) PCAF and p300 with FRET quenchers. These labeled proteins were used to develop a novel assay for the identification and characterization of HAT inhibitors using both FRET and fluorescence polarization. HATs are an important class of epigenetic enzymes involved in chromatin restructuring and transcriptional regulation. This strategy should be useful in the search of new anticancer drugs that target the substrate interfaces of the HATs, as well as to find values in mechanistic study of HATs.
Conclusions and outlook
The exciting field of protein engineering has been dramatically enhanced since the capability of producing recombinant α-thioesters by modified inteins was combined with NCL. This new set of technologies has been applied to many problems in biochemistry and biophysics. The fundamental strength of this technology is that it allows the preparation of homogeneous samples of proteins with site-specific chemical modifications on a scale sufficient to be studied by standard analytical methods. In principle, any chemical modification can be introduced into proteins using EPL, as long as the final product is stable. Careful optimization of the conditions employed during the ligation reactions allows the preparation of extremely challenging molecules such as membrane proteins (potassium channel KcsA) or lipidated proteins (prenylated Rabs), for example. In this review, we have tried to show a representative sample of several applications of EPL to study protein structure and function; however, the number of applications is starting to grow exponentially. For example, EPL shows special promise in the area of nanotechnology [123], where this mild ligation approach could be used for the site-specific immobilization of proteins onto nanoparticles [124] and solid supports [58]. Another field where EPL would prove increasingly useful is the development of complex protein-based therapeutics. For example, EPL have been used for the in vivo and in vitro synthesis of several cyclotides [61, 62]. Cyclotides are extremely stable micro-proteins that show special promise for the development of a new type of protein-based therapeutics [125]. EPL will also continue to be used for the incorporation of increasingly complex PTMs, for example with polysaccharides such as GPI-anchors [126] or proteins with multiple modifications.
In summary, in spite of continuing challenges, the remarkable developments in protein semi-synthesis over the past decade assure a bright future ahead for the role of EPL in the protein engineering challenges of this century.
References
- 1.Hackenberger CP, Schwarzer D. Chemoselective ligation and modification strategies for peptides and proteins. Angew Chem Int Ed Engl. 2008;47:10030–10074. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]
- 2.Camarero JA. New developments for the site-specific attachment of proteins to surfaces. Biophys Rev Lett. 2006;1:1–28. [Google Scholar]
- 3.Dawson PE, Muir TW, Clark-Lewis I, Kent SBH. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 4.Tam JP, Lu YA, Liu CF, Shao J. Peptide synthesis using unprotected peptides through orthogonal coupling methods. Proc Natl Acad Sci USA. 1995;92:12485–12489. doi: 10.1073/pnas.92.26.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wieland T, Bokelmann E, Bauer L, Lang HU, Lau H. Polypeptide synthesis. VIII. Formation of sulfur containing peptides by intramolecular migration of aminoacyl groups. Liebigs Ann Chem. 1953;583:129–149. [Google Scholar]
- 6.Wieland T (1988) Sulfur in biomimetic peptide synthesis. In: Kleinkauf Jaeniche VD (eds) The roots of modern biochemistry. Walter de Gruyter, Berlin, pp 213–221
- 7.Kent SB. Total chemical synthesis of proteins. Chem Soc Rev. 2009;38:338–351. doi: 10.1039/b700141j. [DOI] [PubMed] [Google Scholar]
- 8.Erlanson DA, Chytil M, Verdine GL. The leucine zipper domain controls the orientation of AP-1 in the NFAT•AP-1•DNA complex. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 9.Muir TW, Sondhi D, Cole PA. Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans TC, Benner J, Xu M-Q. The in Vitro ligation of bacterially expressed proteins using an intein from Metanobacterium thermoautotrophicum . J Biol Chem. 1999;274:3923–3926. doi: 10.1074/jbc.274.7.3923. [DOI] [PubMed] [Google Scholar]
- 11.Flavell RR, Muir TW. Expressed protein ligation (EPL) in the study of signal transduction, ion conduction, and chromatin biology. Acc Chem Res. 2009;42:107–116. doi: 10.1021/ar800129c. [DOI] [PubMed] [Google Scholar]
- 12.Pellois JP, Muir TW. Semisynthetic proteins in mechanistic studies: using chemistry to go where nature can’t. Curr Opin Chem Biol. 2006;10:487–491. doi: 10.1016/j.cbpa.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nat Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 14.Tsukiji S, Nagamune T. Sortase-mediated ligation: a gift from Gram-positive bacteria to protein engineering. Chembiochem. 2009;10:787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 15.Saleh L, Perler FB. Protein splicing in cis and in trans. Chem Rec. 2006;6:183–193. doi: 10.1002/tcr.20082. [DOI] [PubMed] [Google Scholar]
- 16.Evans TC, Jr, Xu MQ. Intein-mediated protein ligation: harnessing nature’s escape artists. Biopolymers. 1999;51:333–342. doi: 10.1002/(SICI)1097-0282(1999)51:5<333::AID-BIP3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 17.Camarero JA, Muir TW (1999) Native chemical ligation of polypeptides. Curr Protoc Protein Sci 15:18.4.1–18.4.21 [DOI] [PubMed]
- 18.Dawson PE, Kent SB. Synthesis of native proteins by chemical ligation. Annu Rev Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 19.Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 20.Hackeng TM, Griffin JH, Dawson PE. Protein synthesis by native chemical ligation: expanded scope by using straightforward methodology. Proc Natl Acad Sci USA. 1999;96:10068–10073. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hojo H, Aimoto S. Polypeptide Synthesis Using the S-Alkyl Thioester of a Partially Protected Peptide Segment. Synthesis of the DNA-Binding Domain of c-Myb Protein (142–193)-NH2. Bull Chem Soc Jpn. 1991;64:111–117. [Google Scholar]
- 22.Camarero JA, Cotton GJ, Adeva A, Muir TW. Chemical ligation of unprotected peptides directly form a solid support. J Pept Res. 1998;51:303–316. doi: 10.1111/j.1399-3011.1998.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 23.Camarero JA, Adeva A, Muir TW. 3-Thiopropionic acid as a highly versatile multidetachable thioester resin linker. Int J Pept Res Ther. 2000;7:17–21. [Google Scholar]
- 24.Huse M, Holford MN, Kuriyan J, Muir TW. Semisynthesis of hyperphosphorylated type I TGF beta receptor: addressing the mechanism of kinase activation. J Am Chem Soc. 2000;122:8337–8338. [Google Scholar]
- 25.Shin Y, Winans KA, Backes BJ, Kent SBH, Ellman JA, Bertozzi CR. Fmoc-based synthesis of Peptide-aThioesters: application to the total chemical synthesis of a glycoprotein by native chemical ligation. J Am Chem Soc. 1999;121:11684–11689. [Google Scholar]
- 26.Tolbert TJ, Wong C-H. Intein-mediated synthesis of proteins containing carbohydrates and other molecular probes. J Am Chem Soc. 2000;122:5421–5428. [Google Scholar]
- 27.Miller JS, Dudkin VY, Lyon GJ, Muir TW, Danishefsky SJ. Toward fully synthetic N-linked glycoproteins. Angew Chem Int Ed. 2003;42:431–434. doi: 10.1002/anie.200390131. [DOI] [PubMed] [Google Scholar]
- 28.Camarero JA, Mitchell AR. Synthesis of proteins by native chemical ligation using Fmoc-based chemistry. Protein Pept Lett. 2005;12:723–728. doi: 10.2174/0929866054864166. [DOI] [PubMed] [Google Scholar]
- 29.Ingenito R, Bianchi E, Fattori D, Pessi A. Solid phase synthesis of peptide C-terminal thioesters by Fmoc/t-Bu chemistry Source. J Am Chem Soc. 1999;121:11369–11374. [Google Scholar]
- 30.Camarero JA, Hackel BJ, de Yoreo JJ, Mitchell AR. Fmoc-based synthesis of peptide alpha-thioesters using an aryl hydrazine support. J Org Chem. 2004;69:4145–4151. doi: 10.1021/jo040140h. [DOI] [PubMed] [Google Scholar]
- 31.Blanco-Canosa JB, Dawson PE. An efficient Fmoc-SPPS approach for the generation of thioester peptide precursors for use in native chemical ligation. Angew Chem Int Ed Engl. 2008;47:6851–6855. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami T, Aimoto S. Peptide ligation via the in situ transformation of an amide into a thioester at a cysteine residue. Adv Exp Med Biol. 2009;611:117–118. doi: 10.1007/978-0-387-73657-0_52. [DOI] [PubMed] [Google Scholar]
- 33.Gesquièe J-C, Diesis E, Tartar A. Conversion of N-terminal cysteine to thiazolidine carboxylic acid during hydrogen fluoride deparotection of peptides containing N-bom protected histidine. J Chem Soc Chem Commun. 1990;20:1402–1403. [Google Scholar]
- 34.Pentelute BL, Gates ZP, Dashnau JL, Vanderkooi JM, Kent SB. Mirror image forms of snow flea antifreeze protein prepared by total chemical synthesis have identical antifreeze activities. J Am Chem Soc. 2008;130:9702–9707. doi: 10.1021/ja801352j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Severinov K, Muir TW. Expressed protein ligation, a novel method for studying protein–protein interactions in transcription. J Biol Chem. 1998;273:16205–16209. doi: 10.1074/jbc.273.26.16205. [DOI] [PubMed] [Google Scholar]
- 36.Evans TC, Benner J, Xu M-Q. Semisynthesis of cytotoxic proteins using a modified protein splicing element. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu M-Q, Perler FB. The mechanism of protein splicing and its modulation by mutation. EMBO J. 1996;15:5146–5153. [PMC free article] [PubMed] [Google Scholar]
- 38.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen CA, Pelletier JJ, Paulus H, Xu MQ. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 39.Chong S, Montello GE, Zhang A, Cantor EJ, Liao W, Xu M-Q, Benner J. Utilizing the C-terminal cleavage activity of a protein splicing element to purify recombinant proteins in a single chromatographic step. Nucleic Acid Res. 1998;26:5109–5115. doi: 10.1093/nar/26.22.5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valiyaveetil FI, MacKinnon R, Muir TW. Semisynthesis and folding of the potassium channel KcsA. J Am Chem Soc. 2002;124:9113–9120. doi: 10.1021/ja0266722. [DOI] [PubMed] [Google Scholar]
- 41.Camarero JA, Shektman A, Campbell E, Chlenov M, Gruber TM, Bryant DA, Darst SA, Cowburn D, Muir TW. Autoregulation of a bacterial sigma factor explored using segmental isotopic labeling and NMR. Proc Natl Acad Sci USA. 2002;99:8536–8541. doi: 10.1073/pnas.132033899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirel PH, Schmitter MJ, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dwyer MA, Lu W, Dwyer JJ, Kossiakoff AA. Biosynthetic phage display: a novel protein engineering tool combining chemical and genetic diversity. Chem Biol. 2000;7:263–274. doi: 10.1016/s1074-5521(00)00102-2. [DOI] [PubMed] [Google Scholar]
- 44.Iwai H, Pluckthun A. Circular b-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 1999;459:166–172. doi: 10.1016/s0014-5793(99)01220-x. [DOI] [PubMed] [Google Scholar]
- 45.Camarero JA, Fushman D, Cowburn D, Muir TW. Peptide chemical ligation inside living cells: in vivo generation of a circular protein domain. Bioorg Med Chem. 2001;9:2479–2484. doi: 10.1016/s0968-0896(01)00217-6. [DOI] [PubMed] [Google Scholar]
- 46.Cotton GJ, Ayers B, Xu R, Muir TW. Insertion of a synthetic peptide into a recombinant protein framework; a protein biosensor. J Am Chem Soc. 1999;121:1100–1101. [Google Scholar]
- 47.Tolbert TJ, Wong C-H. New methods for proteomic research: preparation of proteins with N-terminal cysteines for labeling and conjugation. Angew Chem Int Ed Engl. 2002;41:2171–2174. doi: 10.1002/1521-3773(20020617)41:12<2171::aid-anie2171>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 48.Liu D, Xu R, Dutta K, Cowburn D. N-terminal cysteinyl proteins can be prepared using thrombin cleavage. FEBS Lett. 2008;582:1163–1167. doi: 10.1016/j.febslet.2008.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hauser PS, Ryan RO. Expressed protein ligation using an N-terminal cysteine containing fragment generated in vivo from a pelB fusion protein. Protein Expr Purif. 2007;54:227–233. doi: 10.1016/j.pep.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dalbey RE, Lively MO, Bron S, van Dijl JM. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paetzel M, Dalbey RE, Strynadka NC. Crystal structure of a bacterial signal peptidase apoenzyme: implications for signal peptide binding and the Ser–Lys dyad mechanism. J Biol Chem. 2002;277:9512–9519. doi: 10.1074/jbc.M110983200. [DOI] [PubMed] [Google Scholar]
- 52.Southworth MW, Amaya K, Evans TC, Xu MQ, Perler FB. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques. 1999;27:110–114. doi: 10.2144/99271st04. [DOI] [PubMed] [Google Scholar]
- 53.Mathys S, Evans TC, Chute IC, Wu H, Chong S, Benner J, Liu XQ, Xu MQ. Characterization of a self-splicing mini-intein and its conversion into autocatalytic N- and C-terminal cleavage elements: facile production of protein building blocks for protein ligation. Gene. 1999;231:1–13. doi: 10.1016/s0378-1119(99)00103-1. [DOI] [PubMed] [Google Scholar]
- 54.Watzke A, Gutierrez-Rodriguez M, Kohn M, Wacker R, Schroeder H, Breinbauer R, Kuhlmann J, Alexandrov K, Niemeyer CM, Goody RS, Waldmann H. A generic building block for C- and N-terminal protein-labeling and protein-immobilization. Bioorg Med Chem. 2006;14:6288–6306. doi: 10.1016/j.bmc.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 55.Watzke A, Kohn M, Gutierrez-Rodriguez M, Wacker R, Schroder H, Breinbauer R, Kuhlmann J, Alexandrov K, Niemeyer CM, Goody RS, Waldmann H. Site-selective protein immobilization by staudinger ligation. Angew Chem Int Ed Engl. 2006;45:1408–1412. doi: 10.1002/anie.200502057. [DOI] [PubMed] [Google Scholar]
- 56.Lin PC, Ueng SH, Tseng MC, Ko JL, Huang KT, Yu SC, Adak AK, Chen YJ, Lin CC. Site-specific protein modification through Cu(I)-catalyzed 1, 2, 3-triazole formation and its implementation in protein microarray fabrication. Angew Chem Int Ed Engl. 2006;45:4286–4290. doi: 10.1002/anie.200600756. [DOI] [PubMed] [Google Scholar]
- 57.Kalia J, Raines RT. Reactivity of intein thioesters: appending a functional group to a protein. Chembiochem. 2006;7:1375–1383. doi: 10.1002/cbic.200600150. [DOI] [PubMed] [Google Scholar]
- 58.Camarero JA, Kwon Y, Coleman MA. Chemoselective attachment of biologically active proteins to surfaces by expressed protein ligation and its application for “protein chip” fabrication. J Am Chem Soc. 2004;126:14730–14731. doi: 10.1021/ja0456611. [DOI] [PubMed] [Google Scholar]
- 59.Girish A, Sun H, Yeo DS, Chen GY, Chua TK, Yao SQ. Site-specific immobilization of proteins in a microarray using intein-mediated protein splicing. Bioorg Med Chem Lett. 2005;15:2447–2451. doi: 10.1016/j.bmcl.2005.03.079. [DOI] [PubMed] [Google Scholar]
- 60.Camarero JA, Muir TW. Biosynthesis of a head-to-tail cyclized protein with improved biological activity. J Am Chem Soc. 1999;121:5597–5598. [Google Scholar]
- 61.Kimura RH, Tran AT, Camarero JA. Biosynthesis of the cyclotide Kalata B1 by using protein splicing. Angew Chem Int Ed Engl. 2006;45:973–976. doi: 10.1002/anie.200503882. [DOI] [PubMed] [Google Scholar]
- 62.Camarero JA, Kimura RH, Woo YH, Shekhtman A, Cantor J. Biosynthesis of a fully functional cyclotide inside living bacterial cells. Chembiochem. 2007;8:1363–1366. doi: 10.1002/cbic.200700183. [DOI] [PubMed] [Google Scholar]
- 63.Valiyaveetil FI, Leonetti M, Muir TW, Mackinnon R. Ion selectivity in a semisynthetic K+ channel locked in the conductive conformation. Science. 2006;314:1004–1007. doi: 10.1126/science.1133415. [DOI] [PubMed] [Google Scholar]
- 64.Scheibner KA, Zhang Z, Cole PA. Merging fluorescence resonance energy transfer and expressed protein ligation to analyze protein–protein interactions. Anal Biochem. 2003;317:226–232. doi: 10.1016/s0003-2697(03)00087-3. [DOI] [PubMed] [Google Scholar]
- 65.Muralidharan V, Cho J, Trester-Zedlitz M, Kowalik L, Chait BT, Raleigh DP, Muir TW. Domain-specific incorporation of noninvasive optical probes into recombinant proteins. J Am Chem Soc. 2004;126:14004–14012. doi: 10.1021/ja0466199. [DOI] [PubMed] [Google Scholar]
- 66.Romanelli A, Shekhtman A, Cowburn D, Muir TW. Semisynthesis of a segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at the N-extein–intein junction. Proc Natl Acad Sci USA. 2004;101:6397–6402. doi: 10.1073/pnas.0306616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anderson LL, Marshall GR, Crocker E, Smith SO, Baranski TJ. Motion of carboxyl terminus of Galpha is restricted upon G protein activation. A solution NMR study using semisynthetic Galpha subunits. J Biol Chem. 2005;280:31019–31026. doi: 10.1074/jbc.M503690200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vitali F, Henning A, Oberstrass FC, Hargous Y, Auweter SD, Erat M, Allain FH. Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labeling. EMBO J. 2006;25:150–162. doi: 10.1038/sj.emboj.7600911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skrisovska L, Allain FH. Improved segmental isotope labeling methods for the NMR study of multidomain or large proteins: application to the RRMs of Npl3p and hnRNP L. J Mol Biol. 2008;375:151–164. doi: 10.1016/j.jmb.2007.09.030. [DOI] [PubMed] [Google Scholar]
- 70.Goody RS, Durek T, Waldmann H, Brunsveld L, Alexandrov K. Application of protein semisynthesis for the construction of functionalized posttranslationally modified rab GTPases. Methods Enzymol. 2005;403:29–42. doi: 10.1016/S0076-6879(05)03004-1. [DOI] [PubMed] [Google Scholar]
- 71.Brunsveld L, Kuhlmann J, Alexandrov K, Wittinghofer A, Goody RS, Waldmann H. Lipidated ras and rab peptides and proteins-synthesis, structure, and function. Angew Chem Int Ed Engl. 2006;45:6622–6646. doi: 10.1002/anie.200600855. [DOI] [PubMed] [Google Scholar]
- 72.Reuther G, Tan KT, Vogel A, Nowak C, Arnold K, Kuhlmann J, Waldmann H, Huster D. The lipidated membrane anchor of full length N-Ras protein shows an extensive dynamics as revealed by solid-state NMR spectroscopy. J Am Chem Soc. 2006;128:13840–13846. doi: 10.1021/ja063635s. [DOI] [PubMed] [Google Scholar]
- 73.Camarero JA. Recent developments in the site-specific immobilization of proteins onto solid supports. Biopolymers. 2008;90:450–458. doi: 10.1002/bip.20803. [DOI] [PubMed] [Google Scholar]
- 74.Lesaicherre ML, Uttamchandani M, Chen GY, Yao SQ. Developing site-specific immobilization strategies of peptides in a microarray. Bioorg Med Chem Lett. 2002;12:2079–2083. doi: 10.1016/s0960-894x(02)00379-7. [DOI] [PubMed] [Google Scholar]
- 75.Govindaraju T, Jonkheijm P, Gogolin L, Schroeder H, Becker CF, Niemeyer CM, Waldmann H. Surface immobilization of biomolecules by click sulfonamide reaction. Chem Commun (Camb) 2008;32:3723–3725. doi: 10.1039/b806764c. [DOI] [PubMed] [Google Scholar]
- 76.Camarero JA, Kwon Y. Traceless and site-specific attachment of proteins onto solid supports. Int J Pept Res Ther. 2008;14:351–357. [Google Scholar]
- 77.Lesaicherre ML, Lue RY, Chen GY, Zhu Q, Yao SQ. Intein-mediated biotinylation of proteins and its application in a protein microarray. J Am Chem Soc. 2002;124:8768–8769. doi: 10.1021/ja0265963. [DOI] [PubMed] [Google Scholar]
- 78.Lue RY, Chen GY, Hu Y, Zhu Q, Yao SQ. Versatile protein biotinylation strategies for potential high-throughput proteomics. J Am Chem Soc. 2004;126:1055–1062. doi: 10.1021/ja037914g. [DOI] [PubMed] [Google Scholar]
- 79.Holland-Nell K, Beck-Sickinger AG. Specifically immobilised Aldo/Keto reductase AKR1A1 shows a dramatic increase in activity relative to the randomly immobilised enzyme. Chembiochem. 2007;8:1071–1076. doi: 10.1002/cbic.200700056. [DOI] [PubMed] [Google Scholar]
- 80.Hruby VJ, Al-Obeidi F. Emerging approaches in the molecular design of receptor-selective peptide ligands: conformational, topographical and dynamic considerations. J Biochem. 1990;268:249–262. doi: 10.1042/bj2680249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rizo J, Gierasch LM. Constrained peptides: models of bioactive peptides and protein substructures. Annu Rev Biochem. 1992;61:387–418. doi: 10.1146/annurev.bi.61.070192.002131. [DOI] [PubMed] [Google Scholar]
- 82.Camarero JA, Muir TW. Chemoselective backbone cyclization of unprotected peptides. J Chem Soc Chem Comm. 1997;27:1369–1370. [Google Scholar]
- 83.Zhang L, Tam JP. Synthesis and application of unprotected cyclic peptides as building blocks for peptide dendrimers. J Am Chem Soc. 1997;119:2363–2370. [Google Scholar]
- 84.Camarero JA, Pavel J, Muir TW. Chemical synthesis of a circular protein domain: evidence for folding-assisted cyclization. Angew Chem Int Ed. 1998;37:347–349. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 85.Shao Y, Lu WY, Kent SBH. A novel method to synthesize cyclic peptides. Tetrahedron Lett. 1998;39:3911–3914. [Google Scholar]
- 86.Evans TC, Benner J, Xu M-Q. The cyclization and polymerization of bacterially expressed proteins using modified sef-splicing inteins. J Biol Chem. 1999;274:18359–18363. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 87.Camarero JA, Fushman D, Sato S, Giriat I, Cowburn D, Raleigh DP, Muir TW. Rescuing a destabilized protein fold through backbone cyclization. J Mol Biol. 2001;308:1045–1062. doi: 10.1006/jmbi.2001.4631. [DOI] [PubMed] [Google Scholar]
- 88.Craik DJ, Daly NL, Bond T, Waine C. Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 89.Greenwood KP, Daly NL, Brown DL, Stow JL, Craik DJ. The cyclic cystine knot miniprotein MCoTI-II is internalized into cells by macropinocytosis. Int J Biochem Cell Biol. 2007;39:2252–2264. doi: 10.1016/j.biocel.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 90.Craik DJ, Simonsen S, Daly NL. The cyclotides: novel macrocyclic peptides as scaffolds in drug design. Curr Opin Drug Discov Devel. 2002;5:251–260. [PubMed] [Google Scholar]
- 91.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci USA. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu R, Ayers B, Cowburn D, Muir TW. Chemical ligation of folded recombinant proteins; segmental isotopic labeling of domains for NMR studies. Proc Natl Acad Sci USA. 1999;96:388–393. doi: 10.1073/pnas.96.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao W, Zhang Y, Cui C, Li Q, Wang J. An efficient on-column expressed protein ligation strategy: application to segmental triple labeling of human apolipoprotein E3. Protein Sci. 2008;17:736–747. doi: 10.1110/ps.073383708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tatulian SA, Qin S, Pande AH, He X. Positioning membrane proteins by novel protein engineering and biophysical approaches. J Mol Biol. 2005;351:939–947. doi: 10.1016/j.jmb.2005.06.080. [DOI] [PubMed] [Google Scholar]
- 95.Flavell RR, Kothari P, Bar-Dagan M, Synan M, Vallabhajosula S, Friedman JM, Muir TW, Ceccarini G. Site-specific (18)F-labeling of the protein hormone leptin using a general two-step ligation procedure. J Am Chem Soc. 2008;130:9106–9112. doi: 10.1021/ja801666z. [DOI] [PubMed] [Google Scholar]
- 96.Flavell RR, Huse M, Goger M, Trester-Zedlitz M, Kuriyan J, Muir TW. Efficient semisynthesis of a tetraphosphorylated analogue of the Type I TGFbeta receptor. Org Lett. 2002;4:165–168. doi: 10.1021/ol016859i. [DOI] [PubMed] [Google Scholar]
- 97.Ottesen JJ, Huse M, Sekedat MD, Muir TW. Semisynthesis of phosphovariants of Smad2 reveals a substrate preference of the activated T beta RI kinase. Biochemistry. 2004;43:5698–5706. doi: 10.1021/bi0498407. [DOI] [PubMed] [Google Scholar]
- 98.Wu JW, Hu M, Chai J, Seoane J, Huse M, Li C, Rigotti DJ, Kyin S, Muir TW, Fairman R, Massague J, Shi Y. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol Cell. 2001;8:1277–1289. doi: 10.1016/s1097-2765(01)00421-x. [DOI] [PubMed] [Google Scholar]
- 99.Huse M, Muir TW, Xu L, Chen YG, Kuriyan J, Massague J. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 100.Qin BY, Lam SS, Correia JJ, Lin K. Smad3 allostery links TGF-beta receptor kinase activation to transcriptional control. Genes Dev. 2002;16:1950–1963. doi: 10.1101/gad.1002002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chacko BM, Qin BY, Tiwari A, Shi G, Lam S, Hayward LJ, De Caestecker M, Lin K. Structural basis of heteromeric smad protein assembly in TGF-beta signaling. Mol Cell. 2004;15:813–823. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 102.Durek T, Alexandrov K, Goody RS, Hildebrand A, Heinemann I, Waldmann H. Synthesis of fluorescently labeled mono- and diprenylated Rab7 GTPase. J Am Chem Soc. 2004;126:16368–16378. doi: 10.1021/ja046164n. [DOI] [PubMed] [Google Scholar]
- 103.Brunsveld L, Watzke A, Durek T, Alexandrov K, Goody RS, Waldmann H. Synthesis of functionalized rab GTPases by a combination of solution- or solid-phase lipopeptide synthesis with expressed protein ligation. Chemistry. 2005;11:2756–2772. doi: 10.1002/chem.200401041. [DOI] [PubMed] [Google Scholar]
- 104.Rak A, Pylypenko O, Durek T, Watzke A, Kushnir S, Brunsveld L, Waldmann H, Goody RS, Alexandrov K. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302:646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 105.Pylypenko O, Rak A, Durek T, Kushnir S, Dursina BE, Thomae NH, Constantinescu AT, Brunsveld L, Watzke A, Waldmann H, Goody RS, Alexandrov K. Structure of doubly prenylated Ypt1:GDI complex and the mechanism of GDI-mediated Rab recycling. EMBO J. 2006;25:13–23. doi: 10.1038/sj.emboj.7600921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guo Z, Wu YW, Das D, Delon C, Cramer J, Yu S, Thuns S, Lupilova N, Waldmann H, Brunsveld L, Goody RS, Alexandrov K, Blankenfeldt W. Structures of RabGGTase-substrate/product complexes provide insights into the evolution of protein prenylation. EMBO J. 2008;27:2444–2456. doi: 10.1038/emboj.2008.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 108.Shogren-Knaak MA, Fry CJ, Peterson CL. A native peptide ligation strategy for deciphering nucleosomal histone modifications. J Biol Chem. 2003;278:15744–15748. doi: 10.1074/jbc.M301445200. [DOI] [PubMed] [Google Scholar]
- 109.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 110.He S, Bauman D, Davis JS, Loyola A, Nishioka K, Gronlund JL, Reinberg D, Meng F, Kelleher N, McCafferty DG. Facile synthesis of site-specifically acetylated and methylated histone proteins: reagents for evaluation of the histone code hypothesis. Proc Natl Acad Sci USA. 2003;100:12033–12038. doi: 10.1073/pnas.2035256100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chatterjee C, McGinty RK, Pellois JP, Muir TW. Auxiliary-mediated site-specific peptide ubiquitylation. Angew Chem Int Ed Engl. 2007;46:2814–2818. doi: 10.1002/anie.200605155. [DOI] [PubMed] [Google Scholar]
- 112.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hojo H, Nakahara Y. Recent progress in the field of glycopeptide synthesis. Biopolymers. 2007;88:308–324. doi: 10.1002/bip.20699. [DOI] [PubMed] [Google Scholar]
- 114.Marcaurelle LA, Mizoue LS, Wilken J, Oldham L, Kent SB, Handel TM, Bertozzi CR. Chemical synthesis of lymphotactin: a glycosylated chemokine with a C-terminal mucin-like domain. Chemistry. 2001;7:1129–1132. doi: 10.1002/1521-3765(20010302)7:5<1129::aid-chem1129>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 115.Hackenberger CP, Friel CT, Radford SE, Imperiali B. Semisynthesis of a glycosylated Im7 analogue for protein folding studies. J Am Chem Soc. 2005;127:12882–12889. doi: 10.1021/ja051855k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Macmillan D, Bertozzi CR. Modular assembly of glycoproteins: towards the synthesis of GlyCAM-1 by using expressed protein ligation. Angew Chem Int Ed Engl. 2004;43:1355–1359. doi: 10.1002/anie.200352673. [DOI] [PubMed] [Google Scholar]
- 117.Valiyaveetil FI, Sekedat M, MacKinnon R, Muir TW. Structural and functional consequences of an amide-to-ester substitution in the selectivity filter of a potassium channel. J Am Chem Soc. 2006;128:11591–11599. doi: 10.1021/ja0631955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Schwarzer D, Cole PA. Protein semisynthesis and expressed protein ligation: chasing a protein’s tail. Curr Opin Chem Biol. 2005;9:561–569. doi: 10.1016/j.cbpa.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 119.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 120.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 121.Szewczuk LM, Tarrant MK, Sample V, Drury WJ, 3rd, Zhang J, Cole PA. Analysis of serotonin N-acetyltransferase regulation in vitro and in live cells using protein semisynthesis. Biochemistry. 2008;47:10407–10419. doi: 10.1021/bi801189d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xie N, Elangwe EN, Asher S, Zheng YG. A dual-mode fluorescence strategy for screening HAT modulators. Bioconjug Chem. 2009;20:360–366. doi: 10.1021/bc800467a. [DOI] [PubMed] [Google Scholar]
- 123.Woo Y-H, Camarero JA. Interfacing ‘Hard’ and ‘Soft’ matter with exquisite chemical control. Curr Nanosci. 2006;2:93–103. [Google Scholar]
- 124.Becker CF, Marsac Y, Hazarika P, Moser J, Goody RS, Niemeyer CM. Functional immobilization of the small GTPase Rab6A on DNA-Gold nanoparticles by using a site-specifically attached poly(ethylene glycol) linker and thiol place-exchange reaction. Chembiochem. 2007;8:32–36. doi: 10.1002/cbic.200600422. [DOI] [PubMed] [Google Scholar]
- 125.Craik DJ, Cemazar M, Daly NL. The cyclotides and related macrocyclic peptides as scaffolds in drug design. Curr Opin Drug Discov Dev. 2006;9:251–260. [PubMed] [Google Scholar]
- 126.Paulick MG, Wise AR, Forstner MB, Groves JT, Bertozzi CR. Synthetic analogues of glycosylphosphatidylinositol-anchored proteins and their behavior in supported lipid bilayers. J Am Chem Soc. 2007;129:11543–11550. doi: 10.1021/ja073271j. [DOI] [PubMed] [Google Scholar]