Abstract
Mycobacterium tuberculosis (Mtb) causes tuberculosis, one of the leading causes of fatal infectious diseases worldwide. Cell-cell recognition between the pathogen Mtb and its host are mediated in part by glycosylated proteins. So far, glycoproteins in Mtb are understudied and for only very few glycoproteins glycosylation sites have been described, e.g., alanine and proline rich secreted protein apa, superoxide dismutase SODC, lipoprotein lpqH and MPB83/MPT83. In this study, glycosylated proteins in Mtb culture filtrate were investigated using liquid chromatography-mass spectrometry approaches and bioinformatic analyses. To validate the presence of glycoproteins, several strategies were pursued including collision induced dissociation, high energy collision dissociation and electron transfer dissociation techniques, and bioinformatics analyses involving a neutral loss search for glycosylated moieties. After extensive data curation, we report glycosylation sites for thirteen Mtb glycoproteins using a combination of mass spectrometry techniques on a dataset collected from culture filtrate proteins. This is the first glycoproteomics study identifying glycosylation sites on mycobacterial culture filtrate proteins (CFP) on a global scale.
Keywords: Mycobacterium tuberculosis, O-linked glycoproteins, mannose, LC-MS/MS, HCD
1. Introduction
Worldwide, tuberculosis has one of the highest mortalities of any infectious diseases and thus continues to be a major public health threat[1]. The causative agent, Mycobacterium tuberculosis (Mtb), has a complex relationship with its host that is mediated in part by secreted, glycosylated proteins. For instance, it has been proposed that mannose receptors on host cells might directly interact with mannosylated Mtb proteins to enter the macrophages for survival [2]. While it is now no longer questioned that bacteria including mycobacteria produce glycoproteins [3], our current knowledge about the glycoproteins of Mtb is very limited. In fact, since the first indication that Mtb is reactive towards ConA lectins in 1989 [4], glycosylation sites for only four Mtb glycoproteins have so far been described. Mass spectrometric analysis of purified alanine and proline rich secreted protein APA, digested with subtilisin, identified Thr49, Thr57, Thr66 and Thr316 as mannosylated sites [5, 6]. It was also found that changes in the mannosylation pattern led to a reduced stimulatory T-lymphocyte response, pointing to the biological importance of the sugar moiety [7]. A combination of chymotrypsin and trypsin digestion was necessary to determine the glycosylation sites in overexpressed superoxide dismutase SodC [8] by mass spectrometry, indicating a N-terminal clustering of Ser and Thr mannosylation sites at Thr45, Thr46, Ser48, Thr51, Ser53, and Ser56. Both APA and SodC had sites containing multiple mannose residues such as mannobiose, mannotriose etc., SodC with up to 9 mannose residues [5, 6, 8]. For the characterization of the lipoprotein LpqH site-directed mutagenesis of several Thr residues (Thr34, Thr35, Thr36, Thr40, and Thr41) was used in combination with loss of ConA binding [9] . The M. bovis cell surface lipoprotein MPB83 was found to be mannosylated by mass spectrometry [10]. Detailed MS analysis showed a combination of mono-, di- and tri-mannosylations at Thr48 and Thr49. The protein sequence of the MPT83 is identical to the bovine MPB83, suggesting that the same Thr residues are modified in Mtb.
When using a recombinant expression system together with ConA binding to determine sequence patterns in O-glycosylation sites, Herrmann et al. identified eight glycoproteins including apa, SodC, LpqH, MPT83, and lipoproteins LppN, LppQ, phosphate-binding protein PstS1, and amino acid ABC transporter/probable glutamine-binding lipoprotein GlnH [11]. Expression and ConA binding analysis of the predicted glycosylation sites indicated that O-glycosylation sites are rich in Pro, Gly and Ala [11]. Recent proteomics approaches have identified additional glycoprotein candidates, but so far, little progress has been made in characterizing glycosylation sites in new glycoproteins [2, 12–14] .
To identify potentially glycosylated proteins in Mtb secreted proteome, we investigated the culture filtrate proteins of Mtb using liquid chromatography-mass spectrometry approaches and bioinformatic analyses. Overall, we report glycosylation sites for thirteen Mtb glycoproteins using a combination of mass spectrometry techniques on a dataset of 900,000 spectra collected from CFP. To validate the presence of glycoproteins, several strategies were pursued including collision induced dissociation (CID), high energy collision dissociation (HCD) and electron transfer dissociation (ETD) techniques, and bioinformatics analyses involving a neutral loss search for glycosylated moieties. Taken together, this is the first glycoproteomics study identifying glycosylation sites on mycobacterial CFP proteins on a global scale. The number of verified Mtb glycoproteins has been tripled in this study.
2. Material and methods
Chemicals and reagents
Acetonitrile and water, (Chromasolv LC-MS quality), formic acid (99%), methyl α-D-mannopyranoside, 1-ethyl-3 (3-dimethylaminopropyl) carbodiimide, glycine, dithiothreitol (DTT), iodoacetamide, Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and urea were supplied by Sigma-Aldrich, St. Louis, MO. Calcium chloride, manganese chloride, and sodium chloride were from Mallinckrodt, Hazelwood, MO, TRIS-HCl was from MP Biomedicals, Santa Ana, CA. Lysyl endopeptidase (Lys-C) Wako USA Richmond, Va. Trypsin (modified sequencing grade) was from Promega, Madison, WI. All other chemicals and reagents were of the highest purity available. CFP (05.CS.93.1.12.5.CFP) was obtained from the Biodefense and Emerging Infections Research Resources Repository (Manassas, VA).
Proteolytic digestion
Protein samples were proteolytically digested in solution as follows: lyophilized protein samples (10–20 µg) were resolubilized in 40 µL of freshly prepared 8 M urea, 100 mM TRIS•HCl, pH 8.5 and reduced by incubation with a final concentration of 3 mM TCEP in LC-MS water for 20 min at RT. Reduced cysteines were alkylated by addition of 10 mM iodoacetamide and incubated for 15 min at RT in the dark. Proteolysis was initiated with 0.1 µg Lysyl endopeptidase (Lys-C) and allowed to proceed for 3 hours at room temperature in the dark. The sample was diluted to a final concentration of 2 M urea by the addition of 100 mM TRIS•HCl, pH 8.5 and adjusted to 1 mM CaCl2. Next, trypsin (0.5 µg, sequencing grade) was added to the mixture and incubated overnight (ca. 18 h) at room temperature in the dark. The digestion was quenched by the addition of formic acid to a final concentration of 5%. The digested peptides were desalted and concentrated into one fraction with a C8 peptide macrotrap (Bruker-Michrom Bioresources, Auburn CA) on an Alliance 2795 (Waters, Milford MA). The collected material was lyophilized and resuspended in 0.2% formic acid (Sigma, St. Louis MO).
Lectin coupled magnetic beads
Concanavalin A (ConA; EY Laboratories, San Mateo, CA) was dissolved in coupling buffer (10 mM potassium phosphate, 150 mM sodium chloride, pH 5.5). Magnetic beads (Bioclone San Diego, CA) with both a carboxy-terminated and amine-terminated functionality was used. ConA was bound to the magnetic beads with the addition of 1-ethyl-3 (3-dimethylaminopropyl) carbodiimide and reacted at room temperature on a rotisserie for 24 hours at a pH of 4.5 – 6.0. The beads were washed three times with wash/storage buffer (10 mM TRIS-HCl pH 7.5, 500 mM NaCl, 0.1% BSA (w/v), 1 mM EDTA, 0.1% NaN3) and blocked with 1 M Glycine, pH 8 for 2 hours and washed again three times.
Lectin Affinity Purification
Lyophilized peptides were resuspended in lectin loading buffer (20 mM TRIS-HCl pH 7.4, 150 mM NaCl, 1 mM MnCl2, 1 mM CaCl2) and rotated for 2 hours at room temperature, washed 3 times, and eluted with the 200 mM methyl α-D-mannopyranoside.
LC-MS
LC-MS analysis was performed on a nanoLC coupled to an Orbitrap, Orbitrap Elite or LQTFT (Thermo Fisher Scientific, Waltham, MA). The LC gradient was 2–30% buffer B (80% ACN, 0.2% formic acid) on a 75 µm ID silica capillary column packed in house with 15 cm of C18AQ, 3 µm, 120 Å (Dr. Maisch, Ammerbuch-Entringen, Germany) in 160 min analysis. The mass spectrometer was operated in different data dependent modes with the scan event 1 performed in the Orbitrap with an Automated Gain Control (AGC) injection target of 1e6 and a maximum injection time of 200 milliseconds (ms) for the full scan. The following dependent scan setups were used: 1) 20 dependent scans in the IT; rapid scan mode; IT AGC target was 5,000 ions with a maximum injection time of 50 ms; 2) data dependent NL MS3; 3 dependent scans on NL of hexose using CID with 50K resolution in the FT; FT AGC target was 5e5 ions; 3) 20 dependent scans using HCD with 15K resolution in the Orbitrap; FT AGC target was 1e6 ions with a maximum injection time of 200 ms; 4) 10 dependent scans using ETD; FT AGC target was 1e6 ions with a maximum injection time of 200 ms; ETD reagent AGC target was 2e5 ions with a maximum injection time of 200 ms and an activation time of 100 ms with supplemental activation enabled.
Ions from m/z = 400 to 1600 were surveyed. Charge state screening and rejection were enabled so that charge states of the precursor ion ≥ +2 were accepted. Injection waveforms were enabled. Dynamic exclusion was also enabled for the maximum list size of 500 for a duration of 90 seconds. At least three technical replicates were run for each sample.
Data analysis
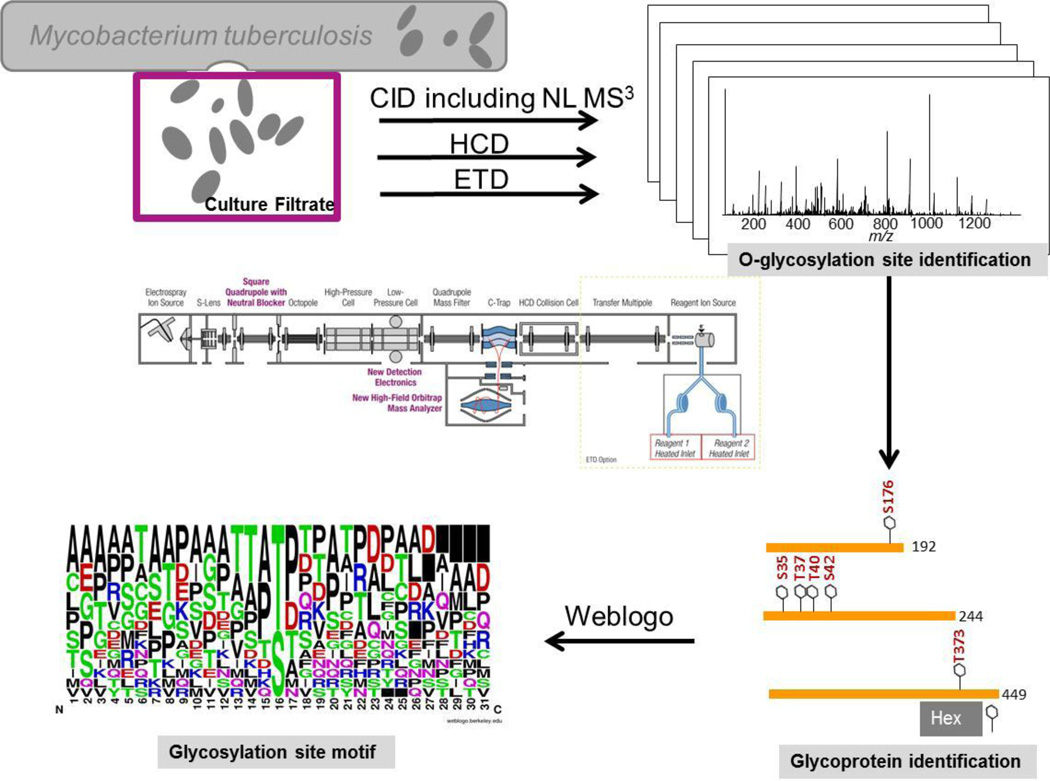
Raw data were analyzed with MaxQuant (version 1.3.0.5)[15–17] against a Mycobacterium tuberculosis database (downloaded from Uniprot on Nov28th, 2012 containing 3977 Mtb sequences) and a contaminant database (247 entries). A decoy database was constructed by MaxQuant on-the-fly to determine the false discovery rate (FDR). Trypsin ([KR][^P]) was specified as the proteolytic enzyme with up to two missed cleavages. Carboxyamidomethyl modification of cysteine (57.0215 Da) was specified as a fixed modification. Variable modifications included mono (162.0528 Da), di (324.1056 Da), and tri (486.1584 Da) hexose additions on serine and threonine (neutral losses of the hexoses were also specified), oxidation of methionine (15.9949 Da), and protein N-terminus acetylations (42.0106 Da). Peptides were searched with a tolerance of 6 ppm after MS1 recalibration in MaxQuant and an MS/MS tolerance of 0.5 Da (ion trap) and 30 ppm (FT) and a peptide and protein FDR of 1%. Weblogo was used to create a weblogo [18]. The unique sequences of all peptides containing Oglycosylation sites were submitted to WebLogo and the resulting sequence atlas was shown.
3. Results
The aim of this study was to identify glycosylation sites in Mtb CFP using nanoLC coupled tandem mass spectrometry. Our rationale was that mass spectrometers have significantly advanced since the first investigations in the nineties that glycosylation site identification directly from CFP seemed feasible. Since mannosylations have previously been described in Mtb, [2, 5–8, 10, 13, 14] we focused on simple hexose modifications such as mannosylations in this analysis. For the comprehensive analysis of hexosylated glycoproteins in CFP, we pursued the strategy outlined in Figure 1. CFP samples were analyzed by CID, NL MS3, HCD and ETD. ETD spectra were generated from CFP and ConA enriched CFP samples. Collected spectra were analyzed by MaxQuant with a peptide and protein FDR ≤ 1%.
Figure 1.
Workflow diagram of the strategy to identify O-linked glycopeptide and glycoproteins in Mtb.
While the MaxQuant analysis is generally very powerful, we found it was essential that additional manual curation is performed to assure correct site identification. This stringent curation resulted in 34 glycosylation site identifications of 13 glycoproteins. Table 1 summarizes the highest confidence glycosylation site identifications along with the technique that provided the highest score and lowest posterior error probability. Please note that in many cases multiple spectra support the identification and site localization.
Table 1.
High confidence glycoprotein and glycosylation site identifications using different dissociation techniques. Best matching spectra are available in Supplementary Data 1.
| Protein ID (Gene name) Protein name | |||||||
|---|---|---|---|---|---|---|---|
| CID/ ETD/ HCD |
Modified sequence identified by MaxQuant |
Locali zation prob. |
Position in proteins |
Comment | # Hex | PEP |
# MS/MS* |
| O06213 (MT2222, Rv2164c) Probable conserved proline rich membrane protein | |||||||
| HCD | _VT(he)PGPDDPAPPAR_ | 1 | 252 | 1 | 2.7E-03 | 1 | |
| O06354 (MT3595, Rv3491) Putative uncharacterized protein# | |||||||
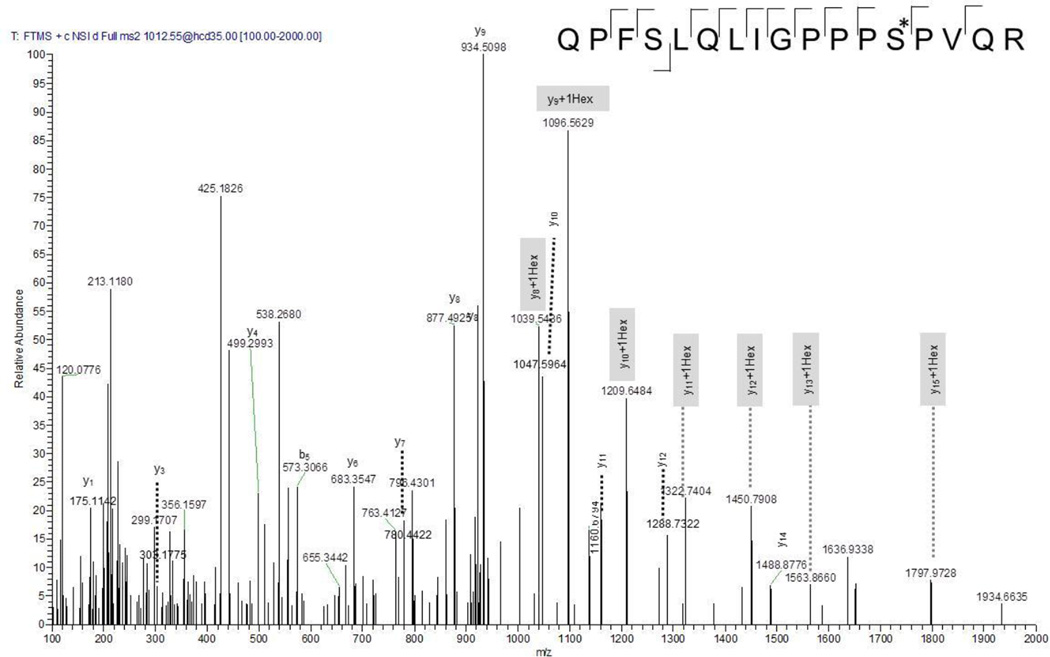
| HCD, ETD, CID | _QPFSLQLIGPPPS(he)PVQR_ | 1 | 176 | 1 | 3.7E-94 | 42 | |
| O07419 (Rv0175) Probable conserved MCE associated membrane protein | |||||||
| CID | _DCVAATQAPDAGAMS(he)ASMQK_ | 0.49 | 114 | 114 or | 4.2E-11 | 3 | |
| _DCVAATQAPDAGAMSAS(he)MQK_ | 0.84 | 116 | 116 | 1 | 4.2E-11 | ||
| O07745 (Rv1887) Putative uncharacterized protein | |||||||
| CID | _AVAPAVPPPPT(he)VT(he)PPVPAR_ | 1 | 308 | 1;2 | 2.0E-03 | 7 | |
| _AVAPAVPPPPT(he)VT(he)PPVPAR_ | 1 | 310 | 2 | 2.0E-03 | |||
| O53444 (MT1128, Rv1096) Possible glycosyl hydrolase# | |||||||
| ETD, CID | _DIPASEIPPLPNT(he)S(he)S(he)PK_ | 1 | 265 | 3 | 8.1E-03 | 2 | |
| _DIPASEIPPLPNT(he)S(he)S(he)PK_ | 1 | 266 | 3 | 8.1E-03 | |||
| _DIPASEIPPLPNT(he)S(he)S(he)PK_ | 1 | 267 | 3 | 8.1E-03 | |||
| P0C5C1 (blaA, Rv2068c) Beta-lactamase# | |||||||
| ETD | _PAST(he)T(he)LPAGADLADR_ | 0.93 | 36 | 2 | 1.5E-02 | 1 | |
| _PAST(he)T(he)LPAGADLADR_ | 0.99 | 37 | 2 | 1.5E-02 | |||
| P0C5C4 (Rv2744c) 35kDa protein | |||||||
| ETD | _GEALPAGGT(he)TATPR_ | 0.50 | 248 | 248, 249 or 251 |
1.7E-02 | 1 | |
| _GEALPAGGTT(he)ATPR_ | 0.50 | 249 | 1.7E-02 | ||||
| P71652 (MT2867.1, Rv2799) Probable membrane protein# | |||||||
| HCD, ETD, CID | _S(he)PIVAT(he)TDPSPFDPCRDIPFDVIQR_ | 1 | 68 | at least one Hex on 68, 73, 74, 77, but no more than 2 present at once |
1;2 | 2.5E-24 | 73 |
| _S(he)PIVAT(he)TDPSPFDPCRDIPFDVIQR_ | 0.87 | 73 | 2 | 3.2E-15 | |||
| HCD, ETD, CID | _SPIVAT(he)T(he)DPSPFDPCR_ | 0.94 | 74 | 2 | 1.3E-15 | ||
| _SPIVATT(he)DPS(he)PFDPCR_ | 0.99 | 77 | 1;2 | 2.5E-24 | |||
| P96243 (Rv3835) Uncharacterized membrane protein | |||||||
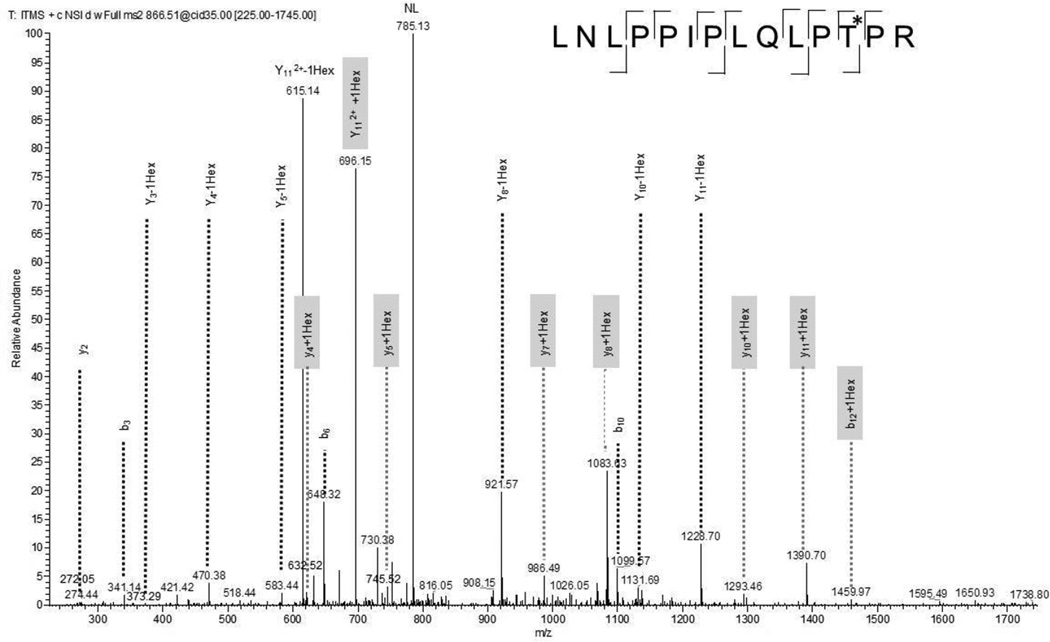
| CID | _LNLPPIPLQLPT(he)PR_ | 1 | 373 | 1 | 1.0E-12 | 3 | |
| Q11049 (lprA, Rv1270c) Putative lipoprotein# | |||||||
| CID, ETD | _AS(he)DT(he)AAT(he)AS(he)NGDAAM(ox)LLK_ | 1 | 35 | 1, 2 or 4 Hex between 35, 37, 40 and 42 |
4 | 1.5E-08 | 20 |
| _AS(he)DT(he)AAT(he)AS(he)NGDAAM(ox)LLK_ | 1 | 37 | 2;4 | 1.0E-41 | |||
| _AS(he)DT(he)AAT(he)AS(he)NGDAAM(ox)LLK_ | 1 | 40 | 1;2;4 | 1.0E-41 | |||
| _AS(he)DT(he)AAT(he)AS(he)NGDAAM(ox)LLK_ | 1 | 42 | 1;2;4 | 5.7E-09 | |||
| Q50675 (lppO, Rv2290) putative lipoprotein | |||||||
| HCD, ETD, CID | _T(he)ATPSESGTQTTR_ | 1 | 73 | 1 or 2 Hex between 73, 75, 77, 79, 81, 83, 84. |
1 | 7.0E-03 | 6 |
| _TAT(he)PSESGTQTTR_ | 0.79 | 75 | 1;2 | 3.6E-14 | |||
| _TATPS(he)ESGTQTTR_ | 0.18 | 77 | 8.8E-08 | ||||
| _TATPS(he)ESGTQTTR_ | 0.18 | 79 | 8.8E-08 | ||||
| _TATPS(he)ESGTQTTR_ | 0.18 | 81 | 8.8E-08 | ||||
| _TAT(he)PSESGTQT(he)TR_ | 0.65 | 83 | 2 | 8.8E-08 | |||
| _TATPS(he)ESGTQTTR_ | 0.18 | 84 | 8.8E-08 | ||||
| Q50906 (apa, Rv1860) Alanine and proline-rich secreted protein | |||||||
| HCD, ETD, CID | ALAESIRPLVAPPPAPAPAPAEPAPAPAPAGEV APT(he)PT(he)T(he)PTPQR_ |
0.93 | 313 | 1, 2 or 3 Hex between 313, 315, 316 and 318 |
1;2;3 | 2.4E-12 | 71 |
| ALAESIR… EVAPT(he)PT(he)T(he)PTPQR_ | 0.88 | 315 | 2;3 | 3.8E-12 | |||
| ALAESIR… EVAPT(he)PT(he)T(he)PTPQR_ | 0.88 | 316& | 2;3 | 3.3E-07 | |||
| ALAESIR… EVAPTPT(he)T(he)PT(he)PQR_ | 0.95 | 318 | 1;2;3 | 5.3E-24 | |||
| P71750 (Rv2394) Gamma-Glutamyltransferase# | |||||||
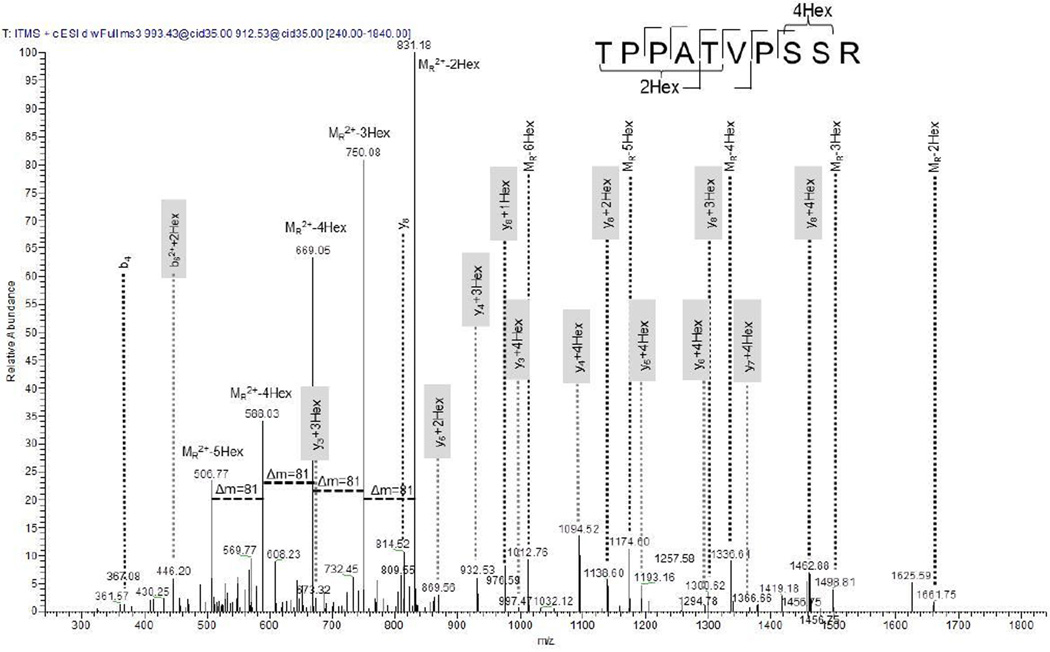
| NL MS3 | _TPPAT(dh)VPS(dh)S(dh)R_ | 0.88 | 51 | 6 Hex are potentially distributed between positions 47, 51, 54, and 55 |
6 | 5.0E-2 | 1 |
| _TPPAT(dh)VPS(dh)S(dh)R_ | 0.99 | 54 | 6 | 5.0E-2 | |||
| _TPPAT(dh)VPS(dh)S(dh)R_ | 0.99 | 55 | 6 | 5.0E-2 | |||
Legend: CID = collision induced dissociation, HCD = high energy collision dissociation, ETD = electron transfer dissociation, NL MS3 Neutral loss MS3, a CID technique, Localization prob. = Localization probability, # Hex = number of hexose(s) observed, PEP = posterior error probability
previously identified as a putative glycoprotein candidate by [14].
# MS/MS is number of high confidence MS/MS spectra.
3.1. Identified Glycoproteins
As shown in Table 1, glycosylation sites were identified for 13 glycoproteins. We could confirm the T316 glycosylation site of alanine and proline-rich secreted protein apa and additionally found one, two or three hexoses between Thr313, Thr315, Thr316 and T318 as glycosylation sites.
In addition, we identified glycosylation sites for the following proteins that have been identified in a recent screen using ConA [14]: the putative uncharacterized protein MT3595 (Rv3491; O06354), possible glycosyl hydrolase MT1128 (Rv1096; O53444), beta-lactamase blaA (Rv2068; P0C5C1), probable membrane protein MT2867.1 (Rv2799; P71652), putative lipoprotein LprA (Rv1270c; Q11049), and gamma-glutamyltransferase (Rv2391; P71750). On top of that, we identified glycosylation sites of six more glycoproteins listed in Table 1 in the culture filtrate. Interestingly, three of the glycoproteins are predicted lipoproteins (betalactamase, LprA, LppO).
With the exception of apa, most proteins identified as glycoproteins in this study have not been characterized. However, some functions can be annotated. For instance, the enzyme betalactamase mediates resistance to penicillin and other beta-lactam antibiotics by cleaving their beta-lactam ring [19]. The possible glycosyl hydrolase is also known as putative polysaccharide deacetylase. Because of its ability to deacetylate N-acetylglucosamine (GlcNAc) residues of the host, it may be viewed as a potential virulence factor [20]. Furthermore, deacetylases have been suggested to promote the evasion of the immune responses of the host through their ability to deacetylate GlcNAc residues [20].
3.2. CID spectra, including NL MS3
A common feature of the CID spectra is the occurrence of a neutral loss of the hexose (162.0528) from the precursor. In cases where only one potential glycosylation site is present in the peptide, the glycosylation site can still be assigned. As shown in Figure 2, assignment of the glycosylation site is also possible when the hexose fragmentation competes with the backbone fragmentation and two ion series exist for the glycosylated and deglycosylated peptide as is the case for the hexosylated LNLPDIPLQIPTPR peptide (aa 362–375) of the uncharacterized membrane protein Rv3835. The doubly charged precursor ion m/z = 866.0099 results in a mass of 1730.0053 Da, one hexose (162.0528 Da) higher than the molecular weight of the peptide, indicating one hexose being attached. Fragment ions that support the assignment of the hexosylated Thr373 are y4+Hex, y5+Hex, y7+Hex, y8+Hex, y10+Hex, y11+Hex and b12+Hex. The ion corresponding to y6+Hex was also seen at low abundance, but was not assigned since it has almost identical mass to the unmodified b5 ion (535.2722 vs. 535.3239).
Figure 2.
CID spectrum of tryptic peptide LNLPPIPLQLPT*PR (aa 362–375) of the uncharacterized membrane protein Rv3835 indicates monohexosylation at Thr373. Fragment ions that contain the hexose residue are highlighted in gray.
Neutral loss (NL)-dependent MS3 experiments have been successfully used in phosphorylation analysis [21]. As seen in the CID spectra, neutral losses can also occur in glycosylated peptides. Therefore, we conducted NL-triggered MS3 experiments for the characterization of glycosylation site identification. Shown in Figure 3 is the spectrum for the tryptic peptide TPATVPSSR (aa 47–56) of gamma-glutamyltransferase Rv2394. MS3 was performed on the NL of doubly charged m/z 993.43, indicating a total of 6 hexoses attached to the peptide. A series of singly and doubly charged neutral losses dominates the spectrum. The y3 ion was identified with three and four hexoses. This indicates that at least four hexoses are present at Ser54 and/or Ser55. It cannot be stated with absolute certainty whether one of the Ser is modified with four sugars or both are modified with one and three or two and two hexoses (or whether in fact a combination of these sugar modifications are present in this peptide). The b62++2Hex ion supports the presence of two hexoses at either Thr47 or Thr51. The presence of the unmodified b4 ion supports the interpretation that Thr51 is more likely modified with two hexoses, but the modification of Thr47 cannot be excluded at this point, because we cannot exclude that the sugar might have been fragmented before the backbone.
Figure 3.
NL MS3 spectrum of tryptic peptide TPPATVPSSR (aa 47–56) of the gamma-glutamyltransferase indicates a total of six hexoses. The spectrum is dominated by doubly and singly charged NL ions. Due to the complexity of the fragmentation pattern and possible combinations, a definite glycosite assignment cannot be made. For instance, the y3 ion was observed with four Hex indicating four Hex at SSR. Since the y2 ion was not observed, the possibility remains that either one or both Ser are modified. Fragment ions that contain the hexose residue are highlighted in gray.
3.3. HCD spectra
In contrast to CID spectra, HCD spectra provide high accuracy for the precursor and the fragment ions, which aids disambiguation in peptide identification and modification site localization when the sugar has not been lost. Figure 4 shows the MS/MS spectrum of the singly hexose modified QPFSLQLIGPPPS*PVQR peptide (aa164–180) of the putative uncharacterized protein MT3595 (O06354). As can be seen in Figure 4, the y-ion series y8+Hex, y9+Hex, y10+Hex, y11+Hex, y12+Hex, y13+Hex unambiguously support the localization of the hexose at Ser176. A similar ion series can also be seen for the unmodified peptide, indicating that the dissociation of the sugar competes with backbone fragmentation.
Figure 4.
HCD spectrum of tryptic peptide QPFSLQLIGPPPS*PVQR (aa 68–83) of the probable membrane protein Rv2799, previously identified as a glycoprotein candidate [14]. The monohexose could clearly be located at Ser79. Fragment ions that contain the hexose residue are highlighted in gray.
Previous investigations of glycosylation sites on GlcNAc-modified peptides revealed typical oxonium ions at m/z = 204.086 (and subsequent species exhibiting additional water losses) in the HCD spectra [22],[23]. The analog oxonium ion of a hexose such as mannose has an m/z = 163.0606. We therefore screened all HCD spectra but did not detect any oxonium ions at m/z = 163.0606. Similar observations were made by Darula et al. [24]. A possible explanation for this difference is that the oxonium ions formed from a hexose lack a nitrogen in contrast to their analog GlcNAc oxonium ions, and thus are less stable.
While powerful, HCD is not always able to provide a final glycosylation assignment. An illustrative example where a final conclusion can currently not be drawn is the peptide SPIVATTDPSPFDPC(57)R (aa 68–83) of the probable membrane protein Rv2799. The doubly charged precursor ion has an m/z = 1042.97, clearly indicating that two hexoses are added to the carbamidomethylated peptide. The b- and y-ion series also support the identification of the peptide with high accuracy. However, difficulties arise when trying to unambiguously assign the glycosylation site(s). Possible assignments are either a single hexose on two sites or two hexoses at one site. This leads to many possible theoretical fragments that are identical for all considered possibilities. In addition, at present, we cannot exclude that a dihexose would result in one neutral loss and the second hexose may remain on the peptide chain. There is also a possibility that multiple isomers are present resulting in overlaying fragmentation patterns. Several best matching scenarios are shown in Table 1. One HCD spectrum supports the localization of the glycosylation at Ser68 and Thr73, another HCD spectrum supports the localization of the glycosylation at Thr74 and Thr77. The most plausible explanation is that multiple glycosylation patterns exist.
3.4. ETD spectra
As a non-ergodic dissociation technique, electron transfer dissociation is generally regarded as the best choice for PTM localization [25]. We therefore also explored this fragmentation technique for the characterization of glycosylations in CFP. Of note, all ETD spectra that were assigned as best identification were from ConA enriched samples, indicating that an enrichment may further improve glycosylation site identification.
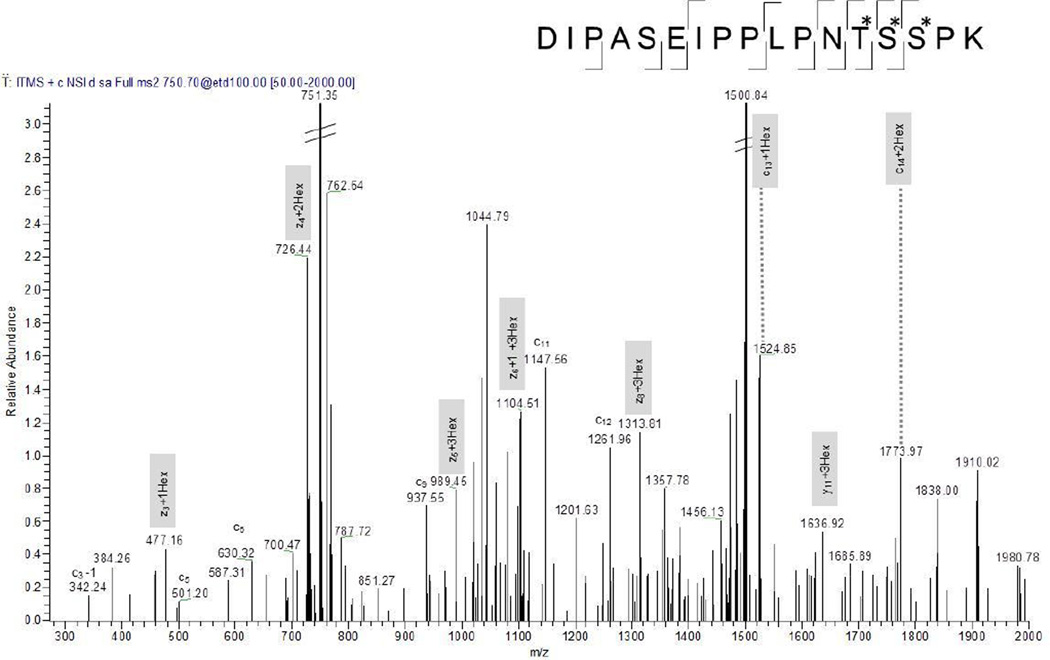
Figure 5 displays the ETD spectrum of the triply charged m/z 750.3648 of DIPASEIPPLPNT*S*S*PK (aa 254–270) of possible glycosyl hydrolase (O53444). Z3 ion with 1 Hex modification, z4 with 2 Hex modifications and z5 with 3 Hex modifications indicates that Thr 266, Ser267 and S268 are modified with one hexose each. This is further supported by c13 ion with 1 Hex and c14 ion with 2 Hex modifications. ETD thus allowed the characterization of three modified amino acids in one spectrum.
Figure 5.
ETD spectrum of tryptic peptide DIPASEIPPLPNT*S*S*PK (aa 254–270) of the possible glycosyl hydrolase. Three monohexoses could clearly be located at Thr266, Ser267, Ser268. Fragment ions that contain the hexose residues are highlighted in gray.
There are also examples in Table 1, where a definite assignment cannot be made using ETD. For instance, the GEALPAGGTTATPR peptide of the 35kDa protein Rv2744c was modified with a single Hex modification at Thr248, Thr249 or Thr251. An explanation for this ambiguity is the possibility that isomeric forms coexisted.
3.5. Glycosylation site motif
After extensive analysis of the mass spectra, we determined the amino acid composition of the glycosylation motifs of the high confidence identifications by submitting the unique sequences of all peptides containing a glycosylation site to WebLogo [18]. Figure 6 shows the resulting sequence logo, where the height of each amino acid indicates its relative frequency at this position. Roughly 60% showed the glycosylation site at the Thr residue and 40% at the Ser. The sequence logo also reveals a higher probability for the glycosylation sites to be at the C-terminus as indicated by the black boxes. In agreement with previous reports that investigated Oglycosylation sites [11, 24], we also found a relatively high propensity for Pro and Ala present. Whether this is a result of an enzymatic preference or of the higher tendency of Pro containing peptides to fragment and thus enable glycopeptide identification remains to be further studied.
Figure 6.
The O-glycosylation motif generated from the high confidence identifications indicates a higher likelihood of hydrophobic Pro and Ala, interspersed with some hydrophilic Thr around the O-glycosylation site. In addition, the glycosylation motifs seem to cluster at the C-terminus as indicated by the black boxes on the right.
4. Discussion
Using a global glycoproteomics strategy, we have been able to identify novel glycopeptides in novel mycobacterial glycoproteins. In all cases presented in this study, the confidence that the modified peptide is a glycopeptide is high as evidenced by the score and the assignment of the respective ion series. In many cases, glycosylation site identification was possible, but in some cases, the definite assignment of the glycosylation site(s) remains ambiguous at this point, particularly when multiple hexoses are attached to one peptide. Possible explanations for this ambiguity lie in the fragmenting nature of the glycosylation sites and the likely co-occurrence of isomeric glycopeptides. When the hexose modification fragments more easily than the backbone peptide, positional information was sometimes lost. This ambiguity was observed with all fragmentation techniques used, including ETD.
In addition, in many cases, clusters of glycosylations have been observed, where more than one hexose was present. Apparently, this complicates data interpretation, particularly when chimeric spectra were observed. Isobaric glycopeptides with positional isomers (e.g. hexose on S1 or S2) are unlikely to be separated by reversed phase chromatography, thus isobaric glycopeptides are expected to co-elute. When subjected to dissociation, fragments from the positional isomers are expected to be detected. In some cases only a minor fragment ion was observed for an isomeric glycopeptide, in other cases, a fragment was shared by three or more isomeric candidates. In these cases, the assignment remained ambiguous. It should be noted that this clustering of multiple hexoses has previously been observed, and could not be resolved even when recombinantly expressed proteins were investigated [5–8, 10]. A fact that complicates matters more is the possibility that hexoses can bind to each other in several positional isomeric forms e.g. 1–2, 1–3 or 1–6 binding) as has been observed by Michell et al. [10]. This may very well add additional complexity to the sugar modifications on Mtb that would be interesting to study. Presumably enriched proteins are currently still needed for such a comprehensive analysis. While the identification of novel mycobacterial glycoproteins has great scientific interest due to their likely biological implication in pathogen-host recognition events, progress in the field has been slow and definitive glycosylation sites have only been known for four mycobacterial glycoproteins. This study triples the number of known glycoproteins, many of which are clearly understudied and only putative names and functions are assigned. It can be expected that the characterization of these proteins as glycoproteins will spark further studies in this area.
5. Conclusions
In this study the CID, HCD and ETD fragmentation techniques were used to identify and characterize O-glycosylation in Mtb with high confidence. We have demonstrated that all techniques are capable of identifying O-glycosylation sites of so far uncharacterized mycobacterial glycoproteins. This analysis was focused on simple sugar modifications such as mannosylations. Sequence logo amino acid distribution analysis showed an enrichment of hydrophobic Pro and Ala around the glycosylation sites. This study more than tripled the characterization of glycosylation sites in known mycobacterial proteins.
We envision that these methods could be used for the identification and characterization of glycosylation sites in other mycobacterial subcellular fractions and be extended to other organisms.
Supplementary Material
Highlights.
-
►
Glycosylsation sites in Mtb were identified using CID, HCD and ETD
-
►
CID spectra were able to assign glycosylated peptide due to the competition between backbone and sugar fragmentation
-
►
Thirteen glycoproteins have been identified with high confidence
-
►
Glycosylation sites often cluster and seem to be rich in Pro and Ala
Acknowledgements
The Proteome Exploration Laboratory is funded by the Gordon and Betty Moore Foundation through Grant GBMF775, the Beckman Institute and +the NIH award SRR029594A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Organization WH. Global Tuberculosis Report. 2012 ISBN 978 92 4 156450 2.
- 2.Torrelles JB, Schlesinger LS. Diversity in Mycobacterium tuberculosis mannosylated cel lwall determinants impacts adaptation to the host. Tuberculosis (Edinb) 2010;90:84–93. doi: 10.1016/j.tube.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham RLJ, Hess S. Mass Spectrometry in the Elucidation of the Glycoproteome of Bacterial Pathogens. Curr Proteomics. 2010;7:57–81. [Google Scholar]
- 4.Espitia C, Mancilla R. Identification, isolation and partial characterization of Mycobacterium tuberculosis glycoprotein antigens. Clin Exp Immunol. 1989;77:378–383. [PMC free article] [PubMed] [Google Scholar]
- 5.Dobos KM, Khoo KH, Swiderek KM, Brennan PJ, Belisle JT. Definition of the full extent of glycosylation of the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Journal of Bacteriology. 1996;178:2498–2506. doi: 10.1128/jb.178.9.2498-2506.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobos KM, Swiderek K, Khoo KH, Brennan PJ, Belisle JT. Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infection and Immunity. 1995;63:2846–2853. doi: 10.1128/iai.63.8.2846-2853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horn C, Namane A, Pescher P, Riviere M, Romain F, Puzo G, et al. Decreased capacity of recombinant 45/47-kDa molecules (Apa) of Mycobacterium tuberculosis to stimulate T lymphocyte responses related to changes in their mannosylation pattern. The Journal of Biological Chemistry. 1999;274:32023–32030. doi: 10.1074/jbc.274.45.32023. [DOI] [PubMed] [Google Scholar]
- 8.Sartain MJ, Belisle JT. N-Terminal clustering of the O-glycosylation sites in the Mycobacterium tuberculosis lipoprotein SodC. Glycobiology. 2009;19:38–51. doi: 10.1093/glycob/cwn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann JL, O'Gaora P, Gallagher A, Thole JE, Young DB. Bacterial glycoproteins: a link between glycosylation and proteolytic cleavage of a 19 kDa antigen from Mycobacterium tuberculosis. EMBO J. 1996;15:3547–3554. [PMC free article] [PubMed] [Google Scholar]
- 10.Michell SL, Whelan AO, Wheeler PR, Panico M, Easton RL, Etienne AT, et al. The MPB83 antigen from Mycobacterium bovis contains O-linked mannose and (1-->3)-mannobiose moieties. The Journal of Biological Chemistry. 2003;278:16423–16432. doi: 10.1074/jbc.M207959200. [DOI] [PubMed] [Google Scholar]
- 11.Herrmann JL, Delahay R, Gallagher A, Robertson B, Young D. Analysis of post-translational modification of mycobacterial proteins using a cassette expression system. FEBS Lett. 2000;473:358–362. doi: 10.1016/s0014-5793(00)01553-2. [DOI] [PubMed] [Google Scholar]
- 12.Bell C, Smith GT, Sweredoski MJ, Hess S. Characterization of the Mycobacterium tuberculosis proteome by liquid chromatography mass spectrometry-based proteomics techniques: a comprehensive resource for tuberculosis research. Journal of Proteome Research. 2012;11:119–130. doi: 10.1021/pr2007939. [DOI] [PubMed] [Google Scholar]
- 13.Espitia C, Servin-Gonzalez L, Mancilla R. New insights into protein O-mannosylation in actinomycetes. Molecular BioSystems. 2010;6:775–781. doi: 10.1039/b916394h. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Zamorano M, Mendoza-Hernandez G, Xolalpa W, Parada C, Vallecillo AJ, Bigi F, et al. Mycobacterium tuberculosis glycoproteomics based on ConA-lectin affinity capture of mannosylated proteins. Journal of Proteome Research. 2009;8:721–733. doi: 10.1021/pr800756a. [DOI] [PubMed] [Google Scholar]
- 15.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 16.Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, Olsen JV, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nature Protocols. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 17.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. Journal of Proteome Research. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 18.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. Nature. 1940;146:837. [PubMed] [Google Scholar]
- 20.Milani CJ, Aziz RK, Locke JB, Dahesh S, Nizet V, Buchanan JT. The novel polysaccharide deacetylase homologue Pdi contributes to virulence of the aquatic pathogen Streptococcus iniae. Microbiology. 2010;156:543–554. doi: 10.1099/mic.0.028365-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villen J, Beausoleil SA, Gygi SP. Evaluation of the utility of neutral-loss-dependent MS3 strategies in large-scale phosphorylation analysis. Proteomics. 2008;8:4444–4452. doi: 10.1002/pmic.200800283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahne H, Gholami AM, Kuster B. Discovery of O-GlcNAc-modified Proteins in Published Large-scale Proteome Data. Molecular & Cellular Proteomics. 2012;11:843–850. doi: 10.1074/mcp.M112.019463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao P, Viner R, Teo CF, Boons GJ, Horn D, Wells L. Combining high-energy C-trap dissociation and electron transfer dissociation for protein O-GlcNAc modification site assignment. Journal of Proteome Research. 2011;10:4088–4104. doi: 10.1021/pr2002726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darula Z, Sherman J, Medzihradszky KF. How to dig deeper? Improved enrichment methods for mucin core-1 type glycopeptides. Molecular & Cellular Proteomics: MCP. 2012;11 doi: 10.1074/mcp.O111.016774. O111 016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, et al. The utility of ETD mass spectrometry in proteomic analysis. Biochimica et Biophysica Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.