Abstract
Chronic lymphocytic leukemia (CLL) patients with deletion of chromosome 17p, where the tumor suppressor p53 gene is located, often develop more aggressive disease with poor clinical outcomes. To investigate the underlying mechanisms responsible for the highly malignant phenotype of 17p- CLL and to facilitate the in vivo evaluation of potential drugs against CLL with p53 deletion, we have created a mouse model with TCL1-Tg:p53−/− genotype. The TCL1-Tg:p53−/− mice develop B-cell leukemia at very early age and follow an aggressive path of disease development that resembles human CLL with 17p deletion, with an early appearance of CD5+/IgM+ B cells in the peritoneal cavity, spleen and bone marrow. These TCL1-Tg:p53−/− leukemia cells exhibit higher survival capacity and are more resistant to drug treatment than the leukemia cells from the TCL1-Tg:p53wt (TCL1-Tg) mice. Analysis of microRNA expression reveals that the p53 deletion resulted in a significant decrease of miR-15a and miR-16-1, leading to a substantial elevated expression of Mcl-1. Primary leukemia cells from CLL patients with 17p deletion also show a decrease in miR-15a/miR-16-1 and an increase in Mcl-1 expression. Our study has created a novel CLL mouse model, and suggests that the p53/miR15a/16-Mcl-1 axis may contribute to the aggressive phenotype and drug resistance in CLL cells with loss of p53.
Keywords: Leukemia, p53, TCL1, mouse model, Drug resistance
Introduction
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia in the Western countries is characterized by aberrant accumulation of neoplastic CD5+ B cells (1). Despite major progress in our understanding of CLL biology and the pathological processes and the development of new drugs in the recent years, CLL remains an incurable disease (2). Cytogenetic alterations such chromosomal aberrations are rather common in CLL and have been observed in the majority (approximately 80%) of CLL patients (3). Some of the cytogenetic changes are associated with poor prognosis and aggressive disease progression. Chromosome 17p deletion (17p-) is the most recognized cytogenic alteration in CLL associated with resistance to chemotherapy and poor clinical outcomes (4). Since the tumor suppressor p53 gene is located in human chromosome 17p (5), it is suspected that the loss of p53 function in CLL cells with 17p- may be responsible for the poor prognosis of this subgroup of CLL patients (6–7). Interestingly, recent study suggests a very high concordance (over 70%) in 17p deletion and mutations in the remaining p53 allele (8). Furthermore, p53 dysfunction may also arise via alternative mechanisms such as functional inactivation, which may explain certain CLL with poor prognosis but without apparent structural changes in p53 gene such as 17p-deletion or mutations (9). Thus, it is clear that the loss of p53 function has profound effect on the CLL disease progress and treatment resistance. However, the underlying mechanisms remain to be elucidated.
Animal models are important tools to investigate disease processes and the associated pathological mechanisms in vivo. Currently there are several CLL mouse models, which include TCL1 transgenic mice (10), APRIL transgenic mice (11), Bcl-2 transgenic mice (12), the miR-155 mouse model (13), the NZB mouse model with miR-16 alteration (14), and the miR-29 transgenic mice (15). The TCL1-Tg mouse model, which was created by the insertion of the human TCL1 gene under the control of the immunoglobulin heavy chain variable region promoter and immunoglobulin heavy chain enhancer, represents a commonly used and well-characterized mouse model that develops leukemia resembling human CLL (10). Previous studies have shown that over-expression of B-cell lymphoma-2 (Bcl-2) family members in many cases of CLL and this is correlated with resistance to therapy and a poor prognosis (16). In particular, the myeloid cell leukemia-1 (Mcl-1), one of the Bcl-2 family proteins, has been demonstrated as an important anti-apoptotic protein in CLL both in vitro and in vivo (17). It has been shown that Mcl-1 promotes CLL cell survival by inhibiting the intrinsic Bak/Bax-mediated apoptotic pathway (18). Loss of p53 function in cancer cells has also been associated with decrease in apoptotic response and drug resistance (19), and mice with p53−/− genotype are highly susceptible to the development of a variety of tumors (20). However, currently it is unclear if there is a link between the loss of p53 and over-expression of Mcl-1 in CLL cells.
In the present study, we generated a mouse colony with TCL1 transgenic and p53-deletion (TCL1-Tg:p53−/−) genotype by crossing the TCL1-Tg mice with p53−/− mice. The TCL1-Tg:p53−/− mice develop leukemia that resembles human aggressive CLL disease around 3–4 months. The leukemia cells from TCL1-Tg:p53−/− mice exhibited higher proliferation, higher survival capacity, and more resistant to drug treatment with fludarabine (F-ara-A) than the leukemia cells from the TCL1 transgenic mice. We further demonstrated that the loss of p53 led to a significant increase of Mcl-1 expression, likely through the expression of miR15a and miR-16-1. The association between the loss of p53, the decrease in miR15a and miR-16-1 expression, and the increase in Mcl-1 was further confirmed in primary leukemia cells from CLL patients with chromosome 17p deletion. This study provides in vivo evidence to support that p53→miR15a/16-1→Mcl-1 axis may contribute to the pathogenesis of aggressive CLL.
Methods
Reagents
9-β-D-arabinofuranosyl-2-fluoro-adenine (F-ara-A, the nucleoside form of fludarabine), oxaliplatin, chlorambucil, propidium iodide (PI), and PCR primers were purchased from Sigma-Aldrich (St. Louis, MO). Ficoll-lite Lympho H was from Atlanta Biological (Lawrenceville, GA). CD19 microbeads were purchased from MACS Miltenyi Biotech Inc. (Auburn, CA). ACK lysis buffer and Annexin V-FITC were from BD Biosciences (San Jose, CA). TUNEL staining kit was obtained from Roche Applied Science (Indianapolis, IN). Antibodies against Bcl-XL, Bcl-2 and β-actin were purchased from Cell Signaling Technology Inc. (Danvers, MA). Anti-Mcl1 antibody was from Santa Cruz Biotechnology (Santa Cruz, CA).
Isolation of CLL cells and cytotoxicity assays
Primary leukemia cells (white blood cells) were isolated from the peripheral blood samples of CLL patients diagnosed according to the NCI criteria (21). Proper informed consents under a research protocol approved by the Institutional Review Board (IRB) of MD Anderson Cancer Center were obtained from all patients before the collection of blood samples. Specimens from CLL patients with or without 17p deletion were all used for comparison. CLL cells were isolated from blood samples by density gradient centrifugation as described previously (22), and incubated in RPMI 1640 medium supplemented with 10% FBS and Penicillin (100 U/ml) + Streptomycin (100 ug/ml) overnight before testing drug sensitivity by incubation with F-ara-A or oxaliplatin for 48h. Peritoneal cavity (PC) cells and splenocytes were isolated and treated with ACK cell lysis buffer for 2 minutes on ice to remove red blood cells. After lysis, RPMI medium with 10% FBS was added to the cells to stop the lysis. Afterwards, the cells were washed once by PBS and filtered through cell strainer with 40 μM nylon mesh (Fisher Scientific, Pittsburgh, PA) for single cell preparation and cultured in the same medium as primary leukemia cells. B cells were purified from white blood cells by using CD19 microbeads, and incubated in RPMI 1640 medium supplemented with 10% FBS and Penicillin (100 U/ml) + Streptomycin (100 ug/ml). At the same day, those B cells were treated with F-ara-A or oxaliplatin for 48h. Cell viability and cellular sensitivity to drug treatment in vitro were determined by flow cytometry after double staining of 1×106 cells with annexinV-FITC and PI as previously described (23).
Mouse genotyping and analysis cell surface antigens
The generation of Eu-TCL1 mice and their maintenance were described previously (10). The Eu-TCL1-Tg homozygous mice (B6C3 strain) were mated with p53−/− mice (C57BL/6) to generate TCL1-Tg:p53+/− mice, which were further mated to generate mice with TCL1-Tg:p53−/− genotype. The first generated TCL1-Tg:p53−/− mice were further crossed to generate more TCL1-Tg:p53−/− mice for studies. All mice were housed in the conventional barrier animal facility at the University of Texas MD Anderson Cancer Center and the animal study was carried out under a research protocol approved by the Institutional Animal Care and Use Committee (IACUC). For mouse genotyping, small segments of mouse tail tips were collected from littermates at the age of 3–4 weeks, and were digested in 200 μL tail lysis buffer (Viagen Biotech) with 5 μL proteinase K at 56 °C in a water bath for overnight, followed by a 5-minute incubation at 95°C and then cooled on ice. After removal of tissue debris by centrifugation, 2 μL supernatant was used in a PCR reaction for genotyping as described previously (10), and p53 genotyping protocol was provided by Chad Smith (Transgenic Core Facility of M.D. Anderson Cancer Center). Blood samples were collected from the mouse tails for white blood cell counting and analysis performed by the pathology service in the animal facility at MD Anderson Cancer Center. To analyze cell surface CD5 and IgM, single-cell suspensions were prepared from the mouse spleen, bone marrow, and the peritoneal cavity washout. The cells were stained for surface expression of CD5 and IgM using allophycocyanin (APC)-labeled anti-CD5 and FITC-labeled anti-IgM antibodies (Ebioscience).
Analysis of cell proliferation and apoptosis
Mouse spleen sections were fixed in neutral buffered 10% formalin solution for preparation of tissue slides. Cell proliferation was estimated by Ki67 immunostaining using Ki67 specific antibody and a horseradish peroxidast (HRP)-conjugated secondary antibody to reveal the diaminobenzidine (DAB) staining (Ki67 staining service ordered from the histology lab in M.D. Anderson Cancer Center, Houston). Terminal deoxynucleotidyl transferase deoxyuridine-triphosphatase nick-end labeling (TUNEL) assays were performed with an In Situ Cell Death Detection kit (Roche) according to manufacturer’s instruction and visualized under fluorescent microscopy. Annexin-V/propidium iodide (PI) double-staining and flow cytometry analysis were used to monitor cell death.
Analysis of microRNA expression
Total RNA was isolated from spleen cells from TCL1-Tg and TCL1-Tg:p53−/− mice or from human CLL cells with or without 17p deletion, using a microRNA isolation kit (Ambion). For analysis of microRNA expression profiles, 5 μg total RNA isolated from 3 mice or 3 CLL patient samples per group was analyzed using a commercial microRNA array (LC Sciences, Houston, TX). For real-time PCR analysis, total RNA from 1×107 splenocytes isolated from TCL1-Tg or TCL1-Tg:p53−/− mice (4 mice each genotype), purified using a RNeasy Mini kit (Qiagen), and quantified by Ultrospec 3300 pro UV/visible spectrophotometer. First-strand cDNA was synthesized from 0.5 μg total RNA using a commercial kit (RevertAid First Strand cDNA Synthesis Kit-Fermentas) according to the manufacturer’ instructions. Real-time PCR was performed using the 7900 GT sequence detection system (ABI PRISM). Each PCR was performed in a 25-μL volume on a 96-well optical plate for 2 minutes at 50 °C, followed by 10 minutes at 95 °C, then followed by 40 cycles of 95 °C for 30 seconds, 60 °C for 30 seconds and 72 °C for 1minute and a final 10 minutes at 72 °C. To independently validate the individual microRNA expression pattern from the microRNA array result, total RNA was converted to cDNA synthesis using Taqman MicroRNA Reverse Transcription kit and Taqman RT primers (Applied Biosystems). MicroRNA-specific real-time PCRs were performed using Taqman Universal PCR Master Mix and Taqman small assays according to the manufacturer’s recommended protocol. The relative expression of specific microRNAs was calculated by the delta (deltaCt) method.
Immunoblotting
Primary CLL cells were isolated from patient samples using Fico reagent as described above. Mouse splenocytes were purified by ACK lysis buffer. Cell number was determined by a Coulter Z2 particle count and size analyzer (Beckman Coulter, Inc., Fullerton, CA). The mouse splenocytes/PC cells or primary CLL cells/B cells with same amount were lysed in protein lysis buffer containing a cocktail of protease inhibitors. The nuclei and cell debris were removed by centrifugation at 4°C (13,000 rpm for 5 min), and the supernatants were collected as protein lysates. The protein lysates were then heated at 95°C for 5–15 min and separated by SDS-PAGE followed by Western blot analyses with antibodies specific for Bcl-XL, Mcl-1, Bcl-2, EZH2 and b-Myb.
Statistical analysis
Student t tests were used for testing the statistical difference between two groups of samples. Mouse survival curves by Kaplan-Meier plots were generated by Graphpad Prism software (GraphPad, San Diego, CA), and the statistical significance was analyzed by the log-rank (Mantel-Cox) test. A p value of less than 0.05 was considered statistically significant.
Results
TCL1-Tg:p53−/− mice develop aggressive CLL with early disease onset and short lifespan
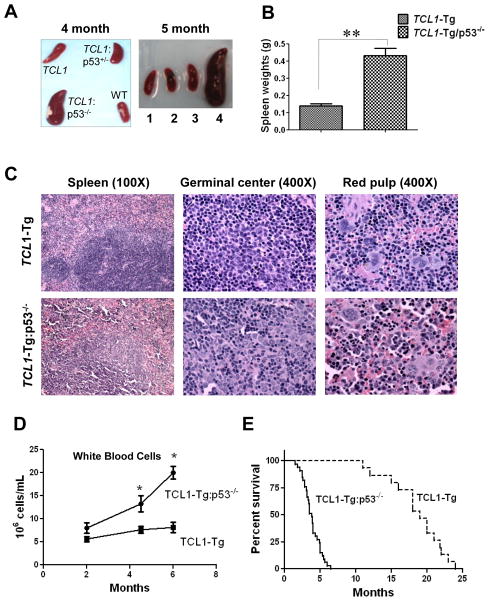
P53 is one of the most frequently mutated genes in cancers. In human B-CLL, loss of p53 function has been associated with accelerated disease progression, poor prognosis, and resistance to antitumor agents. Currently there is no CLL mouse model with loss of p53 for investigating the pathological process of this aggressive CLL. To create such a animal model, we used the well-characterized TCL1 transgenic CLL mice to cross breed with p53−/− mice to generate progenies harboring TCL1-Tg:p53−/− genotype. Figure 1A shows the weights of spleens from 5 TCL1-Tg and 5 TCL1-Tg:p53−/− mice. 17 mice in total 20 TCL1-Tg:p53−/− mice developed CLL disease with early disease onset at the age of approximately 3 month, with severe splenomegaly by 4–5 month (Fig 1B). In contrast, the spleens of 20 TCL1-Tg mice with wt p53 or p53+/− mice appeared relatively normal in size (Fig 1B).
Figure 1. Generation and characterization of TCL1-Tg:p53−/− mice.
A Spleen weights shown for age- and sex-matched TCL1-Tg and TCL1-Tg:p53−/− mice at 4 months (n=5 per indicated genotype). **, p<0.01 between groups.
B Spleen size shown for age- and sex-matched WT, TCL1-Tg, TCL1-Tg:p53+/− and TCL1-Tg:p53−/− mice at 4 months and 5 months old of age (1. WT; 2. TCL1-Tg; 3. TCL1-Tg:p53+/−; 4. TCL1-Tg:p53−/−). Almost 100% TCL1-Tg:p53−/− mice older than 4 months had larger spleen compared to that of TCL1-Tg mice (n=20 per indicated strain).
C Hematoxylin-eosin (H&E) staining of the spleen from TCL1-Tg and TCL1-Tg:p53−/− mice at 4- momth of age. Mouse spleen tissues were fixed in 10% formalin buffered solution and embedded in paraffin. Tissue section from a representative 4-month TCL1-Tg mouse showed normal architecture (top panels), while the spleen from a representative TCL1-Tg:p53−/− mouse showed ill-defined lymphoid follicles (bottom panels) (n=6 per indicated strain).
D White blood cells (WBC) in the indicated mouse strains at different ages (n=5 per indicated age). *, p<0.05 between groups.
E Survival curve (Kaplan-Meier) of TCL1-Tg (n=20) and TCL1-Tg:p53−/− mice (n=33). Median survival time for the TCL1-Tg:p53−/− mice was 3.8 months compared to 19 months for TCL1-Tg mice (p<0.01).
Histological examination of the spleen sections of 6 4-month old TCL1-Tg mice showed normal tissue architecture, whereas the histological sections of the spleen from 6 4-month TCL1-Tg:p53−/− mice showed that the lymphoid follicles were ill-defined (Fig 1C). The germinal centers of the TCL1-Tg:p53−/− spleen exhibited histological features reminiscent of the proliferation centers characteristic of CLL/small lymphocytic lymphoma because of the presence of large lymphocytes with abundant eosinophilic cytoplasm. The red pulps in between the lymphoid follicles contained lymphoid cells with more abundant cytoplasm, granulocytes, and megakaryocytes compared with red pulps seen in TCL1-Tg mouse spleen (Fig 1C). Furthermore, blood smear revealed significantly expansion of the white blood cells (WBC) in TCL1-Tg:p53−/− mice compared to TCL1-Tg mice (Fig 1D). Most of TCL1-Tg:p53−/− mice died at the age of 3–5 months, while the most of the TCL1-Tg mice survive more than 12 months (Fig 1E). This was consistent with the clinical observations that CLL patients with 17p deletion have significantly shorter overall survival compared to the CLL patients without 17p deletion (24).
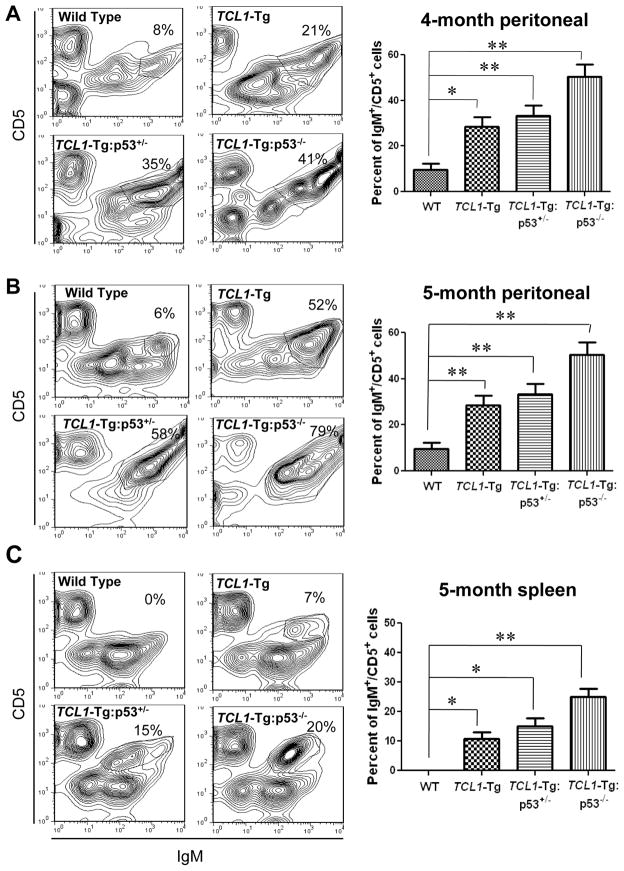
Since TCL1-Tg mice show features of human CLL with an increase in CD5+/IgM+ B cells in the spleen, PC and bone marrow (10), we compared CD5+/IgM+ cells in the spleen, PC and bone marrow of the wild-type mice, p53−/− mice, TCL1-Tg mice, TCL1-Tg:p53+/− mice, and TCL1-Tg:p53−/− mice. Flow cytometry analysis showed that at the age of 4 months, the wild-type mice showed 8% CD5+/IgM+ cells in the PC, the TCL1-Tg mice had 21% CD5+/IgM+ cells, and the TCL1-Tg:p53+/− and TCL1-Tg:p53−/− mice had 35% and 41% CD5+/IgM+ cells in PC respectively (Fig 2A). By 5 months, the CD5+/IgM+ cells in PC of TCL1-Tg:p53+/− and TCL1-Tg:p53−/− mice increase to 58% and 79%, respectively (Fig 2B). In the spleens of a 5-month old mice, the CD5+/IgM+ cells were undetectable in the wild-type mice, 7% in TCL1-Tg mice, 15% in TCL1-Tg:p53+/− mice, and 20% in TCL1-Tg:p53−/− mice (Fig 2C). These data seemed consistent with the early onset of CLL in TCL1-Tg:p53−/− mice (Fig 1). At 3–5 months, no CD5+/IgM+ cells were detected in bone marrow of those mice. Minimum CLL population was observed in bone marrow of TCL1-Tg mice at 7-month old of age, however around 20% CD5+/IgM+ cells were observed in bone marrow of 7-month old TCL1-Tg:p53+/− mice (Fig S2). In the spleens of 4-month old p53−/− mice without TCL1-Tg, the CD5+/IgM+ cells were undetectable, and only about 10% CD5+/IgM+ cells were observed in the PC of those mice (Fig S1).
Figure 2. TCL1-Tg:p53−/− mice had early onset of leukemia and increased CD5+/IgM+ B cells compared to TCL1-Tg mice.
A Flow cytometry analysis of PC cells from sex-matched different mouse strains collected at 4-month old of age (n=4 per indicated genotype), and quantitative bar graph shown on the right panel. *, p<0.05, **, p<0.01 between groups.
B Flow cytometry analysis of PC cells from sex-matched different mouse strains collected at 5-month old of age (n=4 per indicated genotype), and quantitative bar graph shown on the right panel. **, p<0.01 between groups.
C Flow cytometry analysis of splenocytes from sex-matched different mouse strains collected at 5-month old of age (n=4 per indicated genotype), and quantitative bar graph shown on the right panel. *, p<0.05, **, p<0.01 between groups.
Loss of p53 in CLL cells promotes proliferation and cell survival
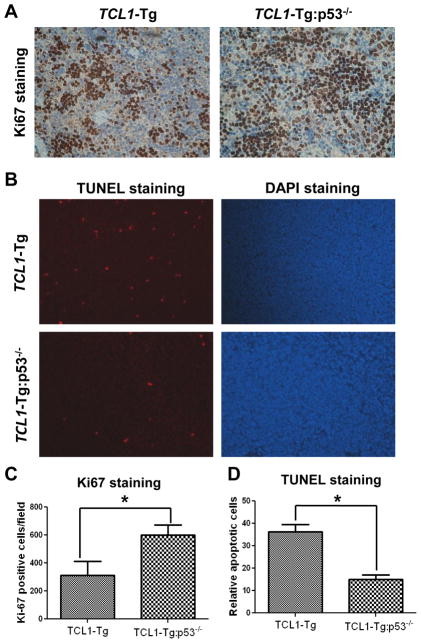
Recent study suggested that CLL cells in TCL1-Tg mice may undergo accelerated cell proliferation accompanied by elevated cell apoptosis, thereby displayed a low accumulation of CLL cells and slow disease progression (25). Since p53 plays a pivotal role in regulation of cell proliferation and apoptosis in response to various stimuli, we examined if a loss of p53 might affect CLL cells proliferation in TCL1-Tg mice. Immunostaining of spleen tissue slides with the proliferation marker Ki-67 revealed that Ki67-positive cells were significantly higher in the spleens of TCL1-Tg:p53−/− mice compared to that of TCL1-Tg mice (Fig 3A, 3C), suggesting an increase in proliferation of the splenocytes in TCL1-Tg:p53−/− mice.
Figure 3. p53 deficiency increased cell proliferation and elevated cell survival of leukemia cells from TCL1-Tg:p53−/− mice.
A Mouse splenic sections were stained with the proliferation marker Ki-67 for age and sex-matched TCL1-Tg and TCL1-Tg:p53−/− mice.
B Left panel, apoptosis in splenic sections shown by staining with TUNEL-TMR-Red. Right panel, DAPI staining indicates nuclear staining of total cells.
C Statistic analysis of the results for A showed average of Ki67-positive cells/field from splenic sections of indicated groups (n=3 per group). *, p<0.05 between groups.
D Statistic analysis of the results for B showed average of TUENL-positive cells/field from splenic sections of indicated groups (n=3 per group). *, p<0.05 between groups.
We then used TUNEL assay to compare in vivo apoptosis of splenocytes in TCL1-Tg and TCL1-Tg:p53−/− mice. TUNEL staining of the spleen tissue sections showed that TCL1-Tg:p53−/− mice had significantly less apoptotic cells in the spleen compared to that in the spleen of TCL1-Tg mice (Fig 3B, 3D). Consistently, annexin-V/PI double-staining of the splenocytes revealed that the isolated splenocytes from TCL1-Tg:p53−/− mice were less apoptotic when cultured in vitro for 24–72 h compared to the splenocytes isolated from TCL1-Tg mice cultured under identical conditions (data not shown). Taken together, these data suggest that the loss of p53 in TCL1-Tg mice seems to promote cell proliferation and decrease apoptosis. This might account for the much higher accumulation of CLL cells and rapid disease progression in TCL1-Tg:p53−/− mice.
Leukemia cells from TCL1-Tg:p53−/− mice or from CLL patients with 17p deletion are resistant to chemotherapeutic drugs
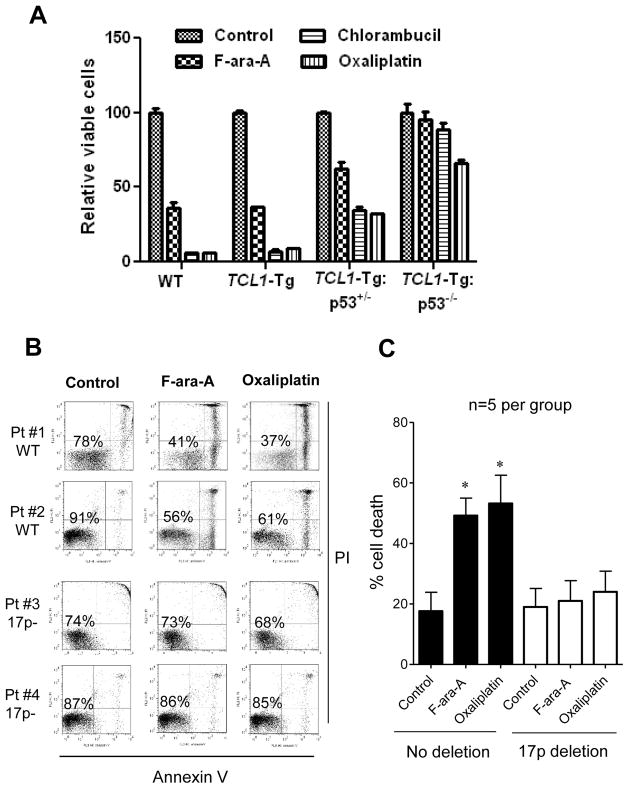
The observations that the leukemia cells isolated from TCL1-Tg:p53−/− mice exhibited less spontaneous apoptosis (Fig 3B, 3D) prompted us to speculate that the leukemia cells with loss of p53 might be less sensitive to apoptotic induction and thus might be more resistant to chemotherapeutic agents. To test this possibility, we isolated splenocytes from the wild-type control mice and from TCL1-Tg, TCL1-Tg:p53+/− and TCL1-Tg:p53−/− mice, and then treated the cells with several standard anti-CLL chemotherapeutic agents in culture (26). As shown in Figure 4A, the splenocytes from the control and TCL1-Tg mice exhibited similar sensitivity to F-ara-A (active form of fludarabine, 10 μM), chlorambucil (10 μM), and oxaliplatin (10 μM). The loss of one p53 allele (TCL1-Tg:p53+/−) caused a moderate decrease in drug sensitivity, whereas the loss of both p53 alleles (TCL1-Tg:p53−/−) led to a significant resistance to all three chemotherapeutic agents (Fig 4A).
Figure 4. p53 deletion makes leukemia cells more resistant to standard CLL drug treatment.
A Splenic cells from 4-month old indicated mouse strains were cultured in RPMI1640 medium and treated with 10uM F-ara-A, Chlorambucil and Oxaliplatin for 48h and Annexin V-PI analysis by FACS was performed to detect apoptosis on those splenocytes (n=5 per indicated strain).
B White blood cells were isolated from CLL patients’ blood and cultured in RPMI1640 medium containing 10% FBS and 1% PS. CLL patient samples with 17p WT (Pt #1 & Pt #2) or with >70% 17p deletion (Pt #3 & Pt #4) were treated with 10uM F-ara-A and 10uM Oxaliplatin for 48h, respectively. Cell apoptosis was analyzed by Annexin V-PI staining.
C The bar graph shows the mean ± SD of 5 patient samples, *, p<0.05 compared to the control.
To further confirm the correlation between the loss of p53 and drug resistance in primary CLL cells from patients, we compared the drug sensitivity of primary leukemia cells from CLL patients with or without 17p deletion. Flow cytometry analysis showed that CLL cells isolated from patients without 17p deletion were sensitive to F-ara-A (10 μM) and oxaliplatin (10 μM), which caused a loss of 30–50% cell viability during the 48-h drug incubation (Fig 4B). For instance, in patient #1 the control cell showed 78% viability, and drug treatment led to a substantial decrease of viable cells (37–41%). Similar results were observed in CLL cells from patient #2. In contrast, CLL cells with 17p deletion were highly resistant to F-ara-A and oxaliplatin (Fig 4B, patients #3 & #4). For instance, in CLL sample #4, the control sample without drug treatment showed 87% viable cells. After treatment with F-ara-A or oxaliplatin, the cell viability remained at 85–86%. Similar drug resistance was observed in patient sample #3. The Figure S3 showed similar drug resistance of 17p− B cells isolated from primary CLL cells. The results from experiments with leukemia cells from mice and from CLL patients consistently suggest that the loss of p53 lead to the drug resistance to standard chemotherapeutic drugs.
Loss of p53 in CLL cells promotes Mcl-1 expression associated with down-regulation of miR-15a and miR-16-1
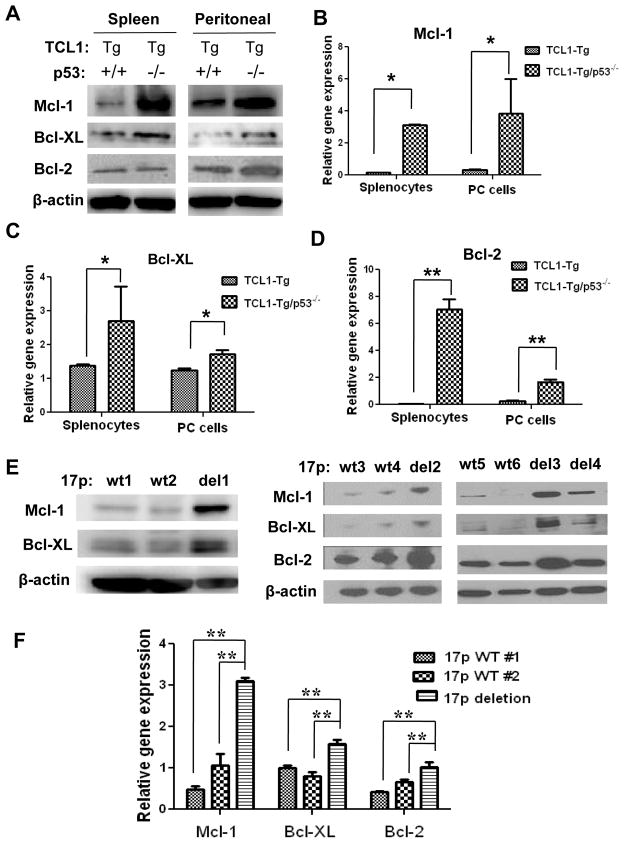
To investigate the mechanisms that contribute to the drug resistant phenotype in CLL cells lacking p53, we first compared the expression of the anti-apoptotic Bcl-2 family members including Bcl-2, Mcl-1, and Bcl-XL in CLL cells isolated from the spleens and PCs of the TCL1-Tg and TCL1-Tg:p53−/− mice. Western blot analysis showed that the expression of these anti-apoptotic molecules increased to various degrees in the p53-null cells (Fig 5A), with the elevation of Mcl-1 protein being the most prominent event, which was detected in leukemia cells isolated from spleen and PC of TCL1-Tg:p53−/− mice. Bcl-XL protein levels were also increased in both the splenocytes and PC cells from TCL1-Tg:p53−/− mice compared to TCL1-Tg mice. Interestingly, the increased Bcl-2 was observed in the PC cells but not in splenocytes (Fig 5A). Real-time RT-PCR analysis showed a significantly increase in mRNA expression of Mcl-1, Bcl-XL, and Bcl-2 in the splenocytes and PC cells from TCL1-Tg:p53−/− mice compared to those from TCL1-Tg mice (Figs 5B–5D).
Figure 5. Up-regulation of survival gene expression in leukemia cells from TCL1-Tg:p53−/− mice or from CLL patient samples with 17p deletion.
A Mcl-1, Bcl-XL and Bcl-2 protein expression shown by Western blot analysis (cells isolated from 4-month old mice; n=4 per group).
B Mcl-1 mRNA levels in mouse splenocytes and PC cells shown by RT-PCR. *, p<0.05 between groups (n=4 per indicated strain).
C Bcl-XL mRNA levels in mouse splenocytes and PC cells shown by RT-PCR. *, p<0.05 between groups (n=4 per indicated strain).
D Bcl-2 mRNA levels in mouse splenocytes and PC cells shown by RT-PCR. **, p<0.01 between groups (n=4 per indicated strain).
E Mcl-1, Bcl-XL and Bcl-2 protein levels in CLL cells from human CLL patients with or without 17p deletion (4 >70%17p deletion samples: del1-4; 6 17p WT samples: WT1-6).
F Mcl-1, Bcl-XL and Bcl-2 mRNA levels in CLL cells from patients with or without 17p deletion shown by RT-PCR. **, p<0.01 between groups.
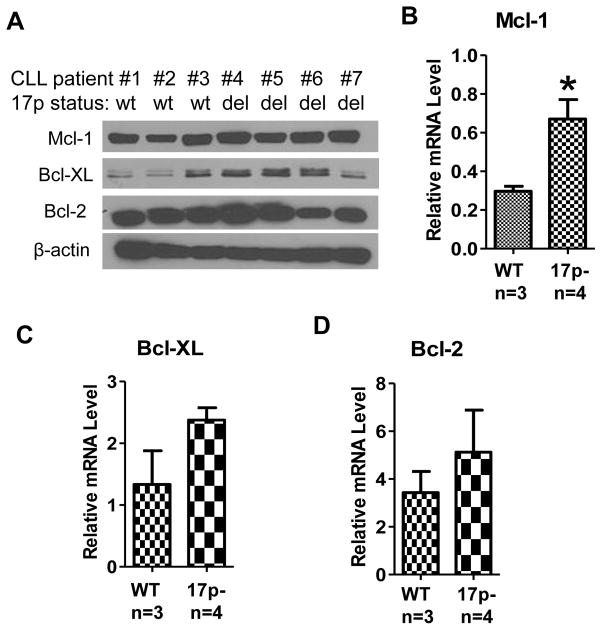
Importantly, the increase in Mcl-1, Bcl-XL, and Bcl-2 protein expression was also observed in primary CLL cells isolated from patients with 17p deletion (Fig 5E). Such increase was consistently observed in multiple patient samples. Real-time RT-PCR analysis showed that the mRNA expression of these three molecules was also increased in primary CLL cells with 17p deletion (Fig 5F). Consistently, both protein and mRNA levels of Mcl-1 and Bcl-XL were increased in 17p− B cells isolated from patient white blood cells (Figs 6A-C). These data suggest that the increased expression of Bcl-2 family/members occurred mainly at transcriptional level. The increase in Bcl-2 expression was observed only in some of the 17p− patient samples (Fig 6A, 6D). We analyzed the expression of Mcl-1, Bcl-XL, and Bcl-2 mRNA and proteins in purified CD19+ CLL cells from a total of 12 CLL patient samples (6 with 17p deletion and 6 without deletion), and consistently observed the up-regulation of Mcl-1 in CLL cells with 17p-deletion, whereas the expression of Bcl-XL and Bcl-2 was heterogeneous among the patient samples (Supplemental Fig S4).
Figure 6. Up-regulation of survival gene expression in CD19 positive B cells from CLL patient samples with 17p deletion.
A Mcl-1, Bcl-XL and Bcl-2 protein levels in CD19 positive B cells from CLL patients with or without 17p deletion (3 17p WT samples and 4 17p− samples).
B Mcl-1 mRNA levels in in CD19 positive B cells from CLL patients with or without 17p deletion (3 17p WT samples and 4 17p− samples).
C Bcl-XL mRNA levels in in CD19 positive B cells from CLL patients with or without 17p deletion (3 17p WT samples and 4 17p− samples).
D Bcl-2 mRNA levels in in CD19 positive B cells from CLL patients with or without 17p deletion (3 17p WT samples and 4 17p− samples).
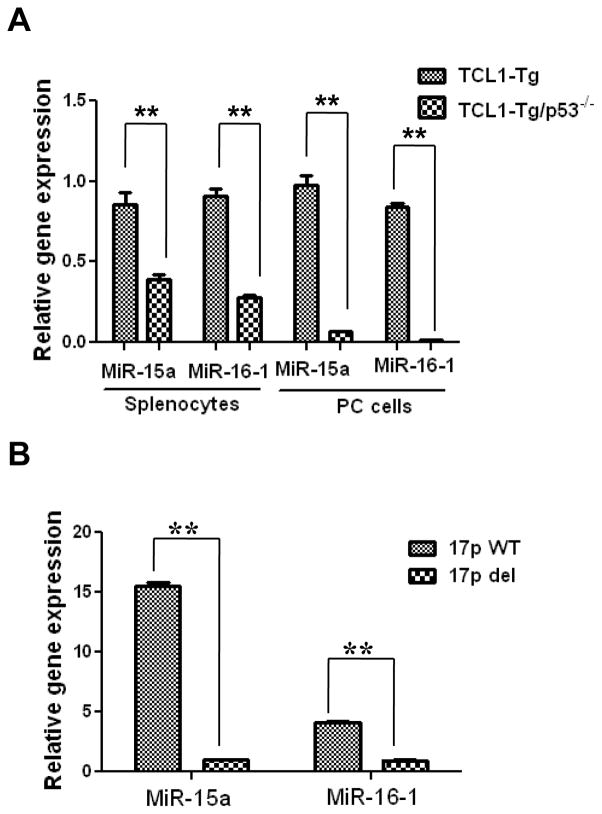
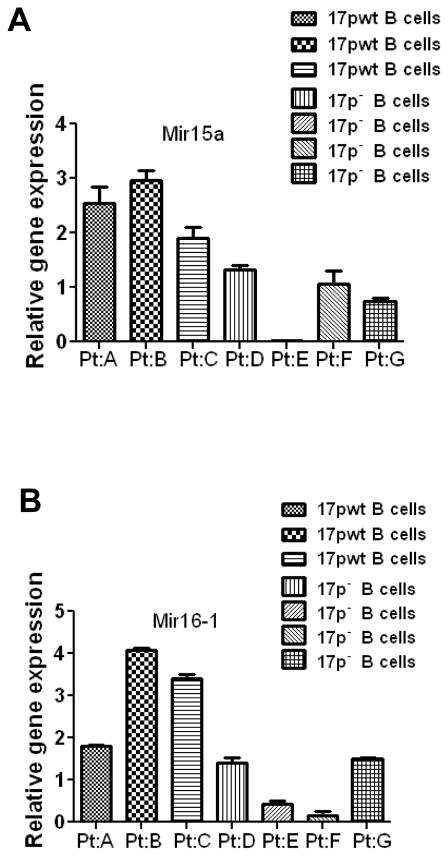
To further investigate the possible mechanism by which loss of p53 led to increased expression of multiple Bcl-2 family members, we speculated that since the expression of Bcl-2 multiple family members is known to be regulated by certain microRNAs (miR15a, miR-16) (27), it is possible that the loss of p53 might cause a decrease expression of miR15a/16-1 or other microRNAs, leading to over-expression of Mcl-1, Bcl-XL, and Bcl-2. To test this possibility, we first isolated total RNA from the splenocytes of TCL1-Tg and TCL1-Tg:p53−/− mice, and explored the expression profiles of microRNAs. Among the >300 microRNAs detected by the microarray analysis, miR-15a and miR-16*1 consistently showed a marked decrease in the splenocytes of TCL1-Tg:p53−/− mice compared to that of TCL1-Tg mice. The decrease of miR-15a and miR-16-1 in cells from TCL1-Tg:p53−/− mice was further confirmed by real-time RT-PCR using the leukemia cells isolated from the spleens and PCs of TCL1-Tg and TCL1-Tg:p53−/− mice. As shown in Fig 7A, there was a significant decrease in the expression of miR-15a and miR-16-1 in both splenocytes and PC cells from mice without p53. Importantly, the decrease in miR-15a/16 expression was further confirmed in primary CLL cells isolated from 5 patients with 17p deletion (Fig 7B). Consistently, similar results were observed in B cells isolated from human WBC (Fig 8). These data together suggest that suppression of miR15a/miR16-1 expression may be an important mechanism by which the loss of p53 enhances the expression of Mcl-1, Bcl-2 and Bcl-XL, leading to increased cell viability and drug resistance.
Figure 7. Down-regulation of tumor suppressor miR-15a/16-1 in leukemia cells from TCL1-Tg:p53−/− mice or from patient samples with 17p deletion.
A MiR-15a/16-1 expression in mouse splenocytes and PC cells shown by RT-PCR. **, p<0.01 between groups (cells isolated from 4-month old mice; n=4 per group).
B MiR-15a/16-1 expression in CLL cells from human patients with or without 17p deletion shown by RT-PCR. **, p<0.01 between groups (n=5 per group).
Figure 8. Down-regulation of tumor suppressor miR-15a/16-1 in CD19 positive B cell from CLL patient samples with 17p deletion.
A MiR-15a expression in CD19 positive B cells purified from patients with or without 17p deletion shown by RT-PCR (3 17p WT samples and 4 17p− samples).
B MiR-16-1 expression in CD19 positive B cells purified from patients with or without 17p deletion shown by RT-PCR (3 17p WT samples and 4 17p− samples).
Discussion
Recent progress in investigation of CLL biology and the development of new therapies such as F-ara-A-based regimens has led to significant improvements of therapeutic outcomes. However, many CLL patients, particularly those with loss of p53 function due to chromosome 17p deletion and p53 mutations, are refractory to the current therapeutic regimens with poor prognosis (24). The p53 gene (TP53) is among the most commonly mutated genes in human cancers (28–31). Loss of p53 function can be due to deletion or mutations of the gene that encodes for p53, epigenetic silencing, and functional inactivation. In CLL, chromosome 17p deletion and p53 mutations are well-documented mechanisms that lead to loss of p53 function associated with poor prognosis (24, 32). The exact mechanisms by which loss of p53 may lead to aggressive disease progression and poor clinical outcomes in CLL remain illusive. Based on the important role of p53 in cell cycle control and regulation of apoptosis (33), it is generally speculated that a loss of p53 function may result in impairment of cycle-cycle checkpoints and compromised apoptotic response, leading to disease progression and drug resistance. However, there has no conclusive evidence in vivo to support this notion, perhaps in part due to the lack of proper CLL animal model with loss of p53.
In the present study, we generated a mouse colony with TCL1-Tg:p53−/− genotype, and demonstrated that these mice developed leukemia that resembles aggressive human CLL. The TCL1-Tg:p53−/− mice exhibited signs of CLL disease around 3 months, with early appearance of CD5+/IgM+ cells in the PC and spleen. The p53−/− mice without TCL1-Tg didn’t show much accumulation of CD5+/IgM+ cells in the spleen and PC. Most mice showed highly abnormal accumulation of WBC in the blood and developed severe splenomegaly at 3–4 month, and died before 6 months. This is in contrast with the TCL1-Tg mice, which develop CLL approximately at the age of 1 year, and the disease progresses slowly (10). In the TCL1-Tg:p53−/− mice, we observed a significant increase in lymphoid cell proliferation in the spleen and a decrease in apoptosis. This may explain why these mice had severe accumulation of leukemia cells and enlargement of the spleen at early age. Since p53 plays a key role in enhancing the expression of apoptotic molecules such as Bax and PUMA (34), loss of p53 function would significantly compromise this apoptotic pathway, leading to resistance to chemotherapeutic agents. In fact, we observed that the leukemia cells isolated from TCL1-Tg:p53−/− mice or from CLL patients with 17p deletion were highly resistance to standard anti-CLL drugs F-ara-A and oxaliplatin. Furthermore, CD19 positive B cells purified from CLL patients with 17p deletion were highly resistance to F-ara-A and oxaliplatin.
An important observation in this study was the significant up-regulation of Mcl-1 expression in the leukemia cells lacking p53. This was seen both in the leukemia cells from the TCL1-Tg:p53−/− mice and in primary CLL cells from patients with 17p deletion (Fig 5). Mcl-1 is a key anti-apoptotic protein in the Bcl-2 family, and this molecule is known to be particularly important for the survival of CLL cells (17). Thus, upregulation of Mcl-1 may play a major role in apoptosis resistance in CLL cells lacking p53, and the moderate increase in Bcl-XL and Bcl-2 expression may also contribute to the increased viability of leukemia cells in TCL1-Tg:p53−/− mice. Since the increase in Mcl-1 expression was observed at mRNA and protein levels, it is likely that loss of p53 may promote Mcl-1 expression mainly at the transcriptional level. This is consistent with the observation that p53 transcriptionally represses Mcl-1 (35–37).
Interestingly, the microRNA expression levels of miR15a and miR-16-1 were significantly decreased in CLL cells with loss of p53. This was observed both in the TCL1-Tg:p53−/− mouse model and in primary CLL cells isolated from patients with 17p deletion. Because Mcl-1 appears to be a target of miR-15a and miR-16-1 (38), the decrease in these microRNAs would release their suppression on Mcl-1 expression and thus lead to elevated MCL-1 protein level. Thus, it is possible that p53 might regulate Mcl-1 expression through modulating miR15a/16. It has been shown previously that microRNAs may be involved in CLL pathogenesis and prognosis due to their function as oncogenes or tumor suppressors (39). For example, miR-15a/miR-16-1 is located in chromosome 13q14.3, a region that is frequently mutated or deleted in CLL patients and may affect CLL cell survival and drug resistance (27). Intriguingly, p53 may regulate the expression of multiple microRNAs, many of which are closely involved in cell cycle regulation, proliferation and apoptosis (40). Our initial study using microRNA array to examine the effect of loss of p53 on microRNA expression in the TCL1-Tg:p53−/− mice had identified miR-15a and miR-16-1 being significantly down-regulated, which was further validated by real-time RT-PCR. Clearly, miR-15a/16-1 expression is significantly lower in splenocytes and PC cells from TCL1-Tg:p53−/− mice than that of TCL1-Tg mice. These observations together suggest that p53→miR-15a/16-1→Mcl-1 axis may be an important pathway in regulating CLL cell apoptosis and drug resistance. The important role of the p53→miR-15a/16-1→Mcl-1 axis in the development of aggressive CLL merits further investigation in clinical setting.
In summary, our study has generated TCL1-Tg:p53−/− mice, which develop aggressive CLL with an early disease onset, likely due to increase proliferation of the leukemia cells and decrease in apoptosis. The CLL cells lacking p53 exhibited low sensitivity to standard anti-CLL drugs. Our study also provided in vivo evidence that loss of p53 led to upregulation of Mcl-1 expression, likely though the down regulation of miR-15a and miR-16-1 and thus released their suppression on Mcl-1 expression, although the loss of p53 suppression on Mcl-1 transcription may also lead to increased MCL-1 protein. The TCL1-Tg:p53−/− mouse colony may serve as a valuable mouse model to further investigate the pathogenesis of aggressive CLL due to loss of p53 function. In addition, since these mice develop leukemia at early age and die within 6 months, this animal model may be useful in testing new drugs for their in vivo therapeutic activity against aggressive CLL with loss of p53 function. It should be noted, however, that the hallmark of human CLL with 17p deletion/p53 mutation is mainly manifested as drug resistance and poor treatment response, not so much as rapid disease progression, although aggressive disease course may be seen in some patients. This caveat should be noted when interpreting results from study using the TCL1-Tg:p53−/− mouse model.
Supplementary Material
Acknowledgments
The authors thank B.A. Hayes and R LaPushin for their assistance in handling CLL samples. This work was supported in part by grants CA085563, CA100428, and CA016672 from the National Institutes of Health, grant RP110322 from the Cancer Prevention and Research Institute of Texas (CPRIT), and a grant for the CLL Global Research Foundation.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Authorship
Contributions: J.L. designed and performed research, analyzed data and drafted the manuscript; G.C., L. F., H.P., F.W., W.Z., M.A.O., W.L. performed research and analyzed data; H.A. perform histology/pathology analysis; C.M.C contributed TCL1 transgenic mice; M.J.K. identified clinical specimens, designed research and interpreted data; P.H. directed the study design, data analysis/interpretation, and drafted the manuscript.
Supplemental information is available on the Leukemia website.
References
- 1.Caligaris-Cappio F, Gobbi M, Bofill M, Janossy G. Infrequent normal B lymphocytes express features of B-chronic lymphocytic leukemia. J Exp Med. 1982 Feb 1;155(2):623–628. doi: 10.1084/jem.155.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landis SH, Murray T, Bolden S, Wingo PA. Cancer statistics, 1998. CA Cancer J Clin. 1998 Jan-Feb;48(1):6–29. doi: 10.3322/canjclin.48.1.6. [DOI] [PubMed] [Google Scholar]
- 3.Zenz T, Mertens D, Dohner H, Stilgenbauer S. Importance of genetics in chronic lymphocytic leukemia. Blood Rev. 2011 May;25(3):131–137. doi: 10.1016/j.blre.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000 Dec 28;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 5.Leonard CJ, Canman CE, Kastan MB. The role of p53 in cell-cycle control and apoptosis: implications for cancer. Important Adv Oncol. 1995:33–42. [PubMed] [Google Scholar]
- 6.Zenz T, Krober A, Scherer K, Habe S, Buhler A, Benner A, et al. Monoallelic TP53 inactivation is associated with poor prognosis in chronic lymphocytic leukemia: results from a detailed genetic characterization with long-term follow-up. Blood. 2008 Oct 15;112(8):3322–3329. doi: 10.1182/blood-2008-04-154070. [DOI] [PubMed] [Google Scholar]
- 7.Zenz T, Frohling S, Mertens D, Dohner H, Stilgenbauer S. Moving from prognostic to predictive factors in chronic lymphocytic leukaemia (CLL) Best Pract Res Clin Haematol. 2010 Mar;23(1):71–84. doi: 10.1016/j.beha.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011 Jun 1;29(16):2223–2229. doi: 10.1200/JCO.2010.32.0838. [DOI] [PubMed] [Google Scholar]
- 9.de Viron E, Michaux L, Put N, Bontemps F, van den Neste E. Present status and perspectives in functional analysis of p53 in chronic lymphocytic leukemia. Leuk Lymphoma. 2012 Mar 1; doi: 10.3109/10428194.2012.660630. [DOI] [PubMed] [Google Scholar]
- 10.Bichi R, Shinton SA, Martin ES, Koval A, Calin GA, Cesari R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci U S A. 2002 May 14;99(10):6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Planelles L, Carvalho-Pinto CE, Hardenberg G, Smaniotto S, Savino W, Gomez-Caro R, et al. APRIL promotes B-1 cell-associated neoplasm. Cancer Cell. 2004 Oct;6(4):399–408. doi: 10.1016/j.ccr.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 12.Zapata JM, Krajewska M, Morse HC, 3rd, Choi Y, Reed JC. TNF receptor-associated factor (TRAF) domain and Bcl-2 cooperate to induce small B cell lymphoma/chronic lymphocytic leukemia in transgenic mice. Proc Natl Acad Sci U S A. 2004 Nov 23;101(47):16600–16605. doi: 10.1073/pnas.0407541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006 May 2;103(18):7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, et al. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007 Jun 15;109(12):5079–5086. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santanam U, Zanesi N, Efanov A, Costinean S, Palamarchuk A, Hagan JP, et al. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci U S A. 2010 Jul 6;107(27):12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson LE, Plunkett W, McConnell K, Keating MJ, McDonnell TJ. Bcl-2 expression in chronic lymphocytic leukemia and its correlation with the induction of apoptosis and clinical outcome. Leukemia. 1996 Mar;10(3):456–459. [PubMed] [Google Scholar]
- 17.Pepper C, Lin TT, Pratt G, Hewamana S, Brennan P, Hiller L, et al. Mcl-1 expression has in vitro and in vivo significance in chronic lymphocytic leukemia and is associated with other poor prognostic markers. Blood. 2008 Nov 1;112(9):3807–3817. doi: 10.1182/blood-2008-05-157131. [DOI] [PubMed] [Google Scholar]
- 18.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, et al. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005 Jun 1;19(11):1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zenz T, Mohr J, Edelmann J, Sarno A, Hoth P, Heuberger M, et al. Treatment resistance in chronic lymphocytic leukemia: the role of the p53 pathway. Leuk Lymphoma. 2009 Mar;50(3):510–513. doi: 10.1080/10428190902763533. [DOI] [PubMed] [Google Scholar]
- 20.Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CA, Jr, Butel JS, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992 Mar 19;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 21.Cheson BD, Bennett JM, Rai KR, Grever MR, Kay NE, Schiffer CA, et al. Guidelines for clinical protocols for chronic lymphocytic leukemia: recommendations of the National Cancer Institute-sponsored working group. Am J Hematol. 1988 Nov;29(3):152–163. doi: 10.1002/ajh.2830290307. [DOI] [PubMed] [Google Scholar]
- 22.Trachootham D, Zhang H, Zhang W, Feng L, Du M, Zhou Y, et al. Effective elimination of fludarabine-resistant CLL cells by PEITC through a redox-mediated mechanism. Blood. 2008 Sep 1;112(5):1912–1922. doi: 10.1182/blood-2008-04-149815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pelicano H, Feng L, Zhou Y, Carew JS, Hileman EO, Plunkett W, et al. Inhibition of mitochondrial respiration: a novel strategy to enhance drug-induced apoptosis in human leukemia cells by a reactive oxygen species-mediated mechanism. J Biol Chem. 2003 Sep 26;278(39):37832–37839. doi: 10.1074/jbc.M301546200. [DOI] [PubMed] [Google Scholar]
- 24.Dohner H, Fischer K, Bentz M, Hansen K, Benner A, Cabot G, et al. p53 gene deletion predicts for poor survival and non-response to therapy with purine analogs in chronic B-cell leukemias. Blood. 1995 Mar 15;85(6):1580–1589. [PubMed] [Google Scholar]
- 25.Enzler T, Kater AP, Zhang W, Widhopf GF, 2nd, Chuang HY, Lee J, et al. Chronic lymphocytic leukemia of Emu-TCL1 transgenic mice undergoes rapid cell turnover that can be offset by extrinsic CD257 to accelerate disease progression. Blood. 2009 Nov 12;114(20):4469–4476. doi: 10.1182/blood-2009-06-230169. [DOI] [PubMed] [Google Scholar]
- 26.Huang P, Sandoval A, Van Den Neste E, Keating MJ, Plunkett W. Inhibition of RNA transcription: a biochemical mechanism of action against chronic lymphocytic leukemia cells by fludarabine. Leukemia. 2000 Aug;14(8):1405–1413. doi: 10.1038/sj.leu.2401845. [DOI] [PubMed] [Google Scholar]
- 27.Aqeilan RI, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010 Feb;17(2):215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 28.Wang LL. Biology of osteogenic sarcoma. Cancer J. 2005 Jul-Aug;11(4):294–305. doi: 10.1097/00130404-200507000-00005. [DOI] [PubMed] [Google Scholar]
- 29.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991 Jul 5;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 30.Bennett WP, Hollstein MC, Hsu IC, Sidransky D, Lane DP, Vogelstein B, et al. Mutational spectra and immunohistochemical analyses of p53 in human cancers. Chest. 1992 Mar;101(3 Suppl):19S–20S. doi: 10.1378/chest.101.3_supplement.19s. [DOI] [PubMed] [Google Scholar]
- 31.Levine AJ, Momand J, Finlay CA. The p53 tumour suppressor gene. Nature. 1991 Jun 6;351(6326):453–456. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 32.Cordone I, Masi S, Mauro FR, Soddu S, Morsilli O, Valentini T, et al. p53 expression in B-cell chronic lymphocytic leukemia: a marker of disease progression and poor prognosis. Blood. 1998 Jun 1;91(11):4342–4349. [PubMed] [Google Scholar]
- 33.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009 Oct;9(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chipuk JE, Green DR. Dissecting p53-dependent apoptosis. Cell Death Differ. 2006 Jun;13(6):994–1002. doi: 10.1038/sj.cdd.4401908. [DOI] [PubMed] [Google Scholar]
- 35.Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011 Jul;12(7):439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- 36.Ploner C, Kofler R, Villunger A. Noxa: at the tip of the balance between life and death. Oncogene. 2008 Dec;27( Suppl 1):S84–92. doi: 10.1038/onc.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pietrzak M, Puzianowska-Kuznicka M. p53-dependent repression of the human MCL-1 gene encoding an anti-apoptotic member of the BCL-2 family: the role of Sp1 and of basic transcription factor binding sites in the MCL-1 promoter. Biol Chem. 2008 Apr;389(4):383–393. doi: 10.1515/BC.2008.039. [DOI] [PubMed] [Google Scholar]
- 38.Sampath D, Liu C, Vasan K, Sulda M, Puduvalli VK, Wierda WG, et al. Histone deacetylases mediate the silencing of miR-15a, miR-16, and miR-29b in chronic lymphocytic leukemia. Blood. 2012 Feb 2;119(5):1162–1172. doi: 10.1182/blood-2011-05-351510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calin GA, Croce CM. Chronic lymphocytic leukemia: interplay between noncoding RNAs and protein-coding genes. Blood. 2009 Nov 26;114(23):4761–4770. doi: 10.1182/blood-2009-07-192740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merkel O, Asslaber D, Pinon JD, Egle A, Greil R. Interdependent regulation of p53 and miR-34a in chronic lymphocytic leukemia. Cell Cycle. 2010 Jul 15;9(14):2764–2768. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.