Abstract
RAGE (Receptor for Advanced Glycation End-Products) has emerged as a major receptor that mediates vascular inflammation. Signaling through RAGE by damage-associated molecular pattern molecules often leads to uncontrolled inflammation that exacerbates the impact of the underlying disease. Oligomerization of RAGE is believed to play an essential role in signal transduction, but the molecular mechanism of oligomerization remains elusive. Here we report that RAGE activation of Erk1/2 phosphorylation on endothelial cells in response to a number of ligands depends on a mechanism that involves heparan sulfate-induced hexamerization of the RAGE extracellular domain. Structural studies of the extracellular V-C1 domain-dodecasaccharide complex by X-ray diffraction and small-angle X-ray scattering revealed that the hexamer consists of a trimer of dimers, with a stoichiometry of 2:1 RAGE:dodecasaccharide. Mutagenesis studies mapped the heparan sulfate binding site and the interfacial surface between the monomers, and demonstrated that electrostatic interactions with heparan sulfate and inter-monomer hydrophobic interactions work in concert to stabilize the dimer. The importance of oligomerization was demonstrated by inhibition of signaling with a new epitope-defined monoclonal antibody that specifically targets oligomerization. These findings indicate that RAGE-heparan sulfate oligomeric complexes are essential for signaling and that interfering with RAGE oligomerization might be of therapeutic value.
INTRODUCTION
The Receptor for Advanced Glycation End-Products (RAGE) is a promiscuous pattern recognition receptor that interacts with a variety of ligands. RAGE was first discovered as the receptor for advanced glycation end-products (AGE) generated by the Amadori rearrangement of non-enzymatically glycated proteins 1. Additional ligands include many damage-associated molecular pattern proteins such as S100 proteins and High Mobility Group protein B1 (HMGB1) that are released from the nucleus or cytoplasm in response to inflammation or cellular necrosis 2, 3. Activation of RAGE by its ligands often leads to prolonged inflammation that exacerbate tissue damage in many disease settings, including diabetes, atherosclerosis, multiple sclerosis, ischemic-reperfusion injury and sepsis 4–8. Therefore, there is great interest in developing agents to antagonize RAGE signaling as a way to suppress uncontrolled inflammation.
RAGE is a multi-domain protein consisting of three extracellular immunoglobulin domains (V-C1-C2, also known as sRAGE), a single transmembrane domain, and a short cytoplasmic tail 9. The two most N-terminal immunoglobulin domains, V and C1, are responsible for most if not all ligand binding 10–13. The short cytoplasmic tail lacks intrinsic kinase activity, but serves an essential role in signal transduction by recruiting adaptor proteins, such as Diaphanous-1 and Toll-Interleukin 1 Receptor Domain containing Adaptor Protein (TIRAP), or the extracellular signal-regulated kinase Erk1 14–16. RAGE oligomerization has been observed in situ at the cell surface using fluorescence resonance energy transfer and chemical crosslinking17, 18. Immunoprecipitation and in vitro binding assays further support that RAGE can self-associate 10, 12, 18, 19. Moreover, disrupting oligomerization of endogenous RAGE with sRAGE results in greatly reduced mitogen-activated protein kinase phosphorylation and nuclear factor NF-κB activation, suggesting that targeting RAGE oligomerization might have therapeutic potential 18. Key questions that remain unanswered concern the mechanism of RAGE oligomerization and the organization of the oligomers.
Here we report that heparan sulfate is an integral part of the functional RAGE signaling complex and functions by promoting RAGE oligomerization. We show that dodecasaccharides derived from heparin, a highly sulfated form of heparan sulfate, support formation of a stable RAGE hexamer, with a stoichiometry of 2:1 RAGE:oligosaccharide. We further present the crystal structure of the RAGE hexamer in complex with dodecasaccharides, which is arranged as a trimer of dimers. The solution structure of RAGE hexamer was also determined by small-angle X-ray scattering (SAXS) and shows a high degree of agreement with the crystal structure. Finally, these structural insights led to the successful identification of an epitope-defined monoclonal antibody that inhibits RAGE signaling by hindering RAGE oligomerization.
RESULTS and DISCUSSION
Endothelial heparan sulfate is essential for RAGE signaling
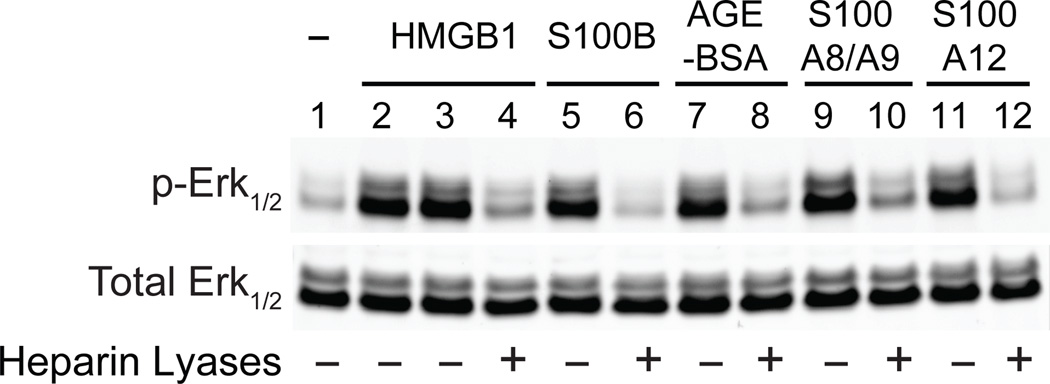
RAGE is a transmembrane receptor that binds many damage-associated molecular pattern proteins. Previous studies showed that RAGE on the cell surface exists as an oligomer of unknown stoichiometry17, 18, but a stable RAGE oligomer has not been reported to form in vitro, which greatly hinders biophysical characterization of RAGE oligomerization. Recently, we showed that RAGE exists in a complex with heparan sulfate 20, a ubiquitous polysaccharide found covalently linked to membrane and matrix proteoglycans. To gain better insight into the role that heparan sulfate plays in receptor architecture and function, we first tested a variety of ligands that activate RAGE. Some of these ligands bind to heparan sulfate (HMGB1, S100A8/A9 and S100A12), whereas others do not (S100b, AGE-BSA) 21–23. Nevertheless, RAGE signaling by all five ligands depends on heparan sulfate, as measured by the loss of Erk1/2 phosphorylation after removal of cell surface heparan sulfate (Fig. 1). Erk1/2 phosphorylation was chosen as the readout of RAGE signaling because it is associated with RAGE activation in endothelial cells 7, 20. Thus, the requirement for heparan sulfate for signaling appears to be an intrinsic property of RAGE.
Figure 1. Endothelial heparan sulfate is required for RAGE signaling.
Erk1/2 phosphorylation of primary human endothelial cells was measured by immunoblotting before (lane 1) and after stimulation by RAGE ligands (lanes 2–12). In lane 2, endothelial cells were stimulated for 15 min with a mutated form of HMGB1 that does not bind heparan sulfate. Cells were stimulated with wild-type HMGB1 (lanes 3 and 4), S100B (lanes 5 and 6), AGE-BSA (lanes 7 and 8), S100A8/A9 (lanes 9 and 10) or S100A12 (lanes 11 and 12). Selected samples were treated with heparin lyases I, II and III for 15 min prior to adding ligand as indicated.
Heparin-derived oligosaccharide induces a stable RAGE oligomer
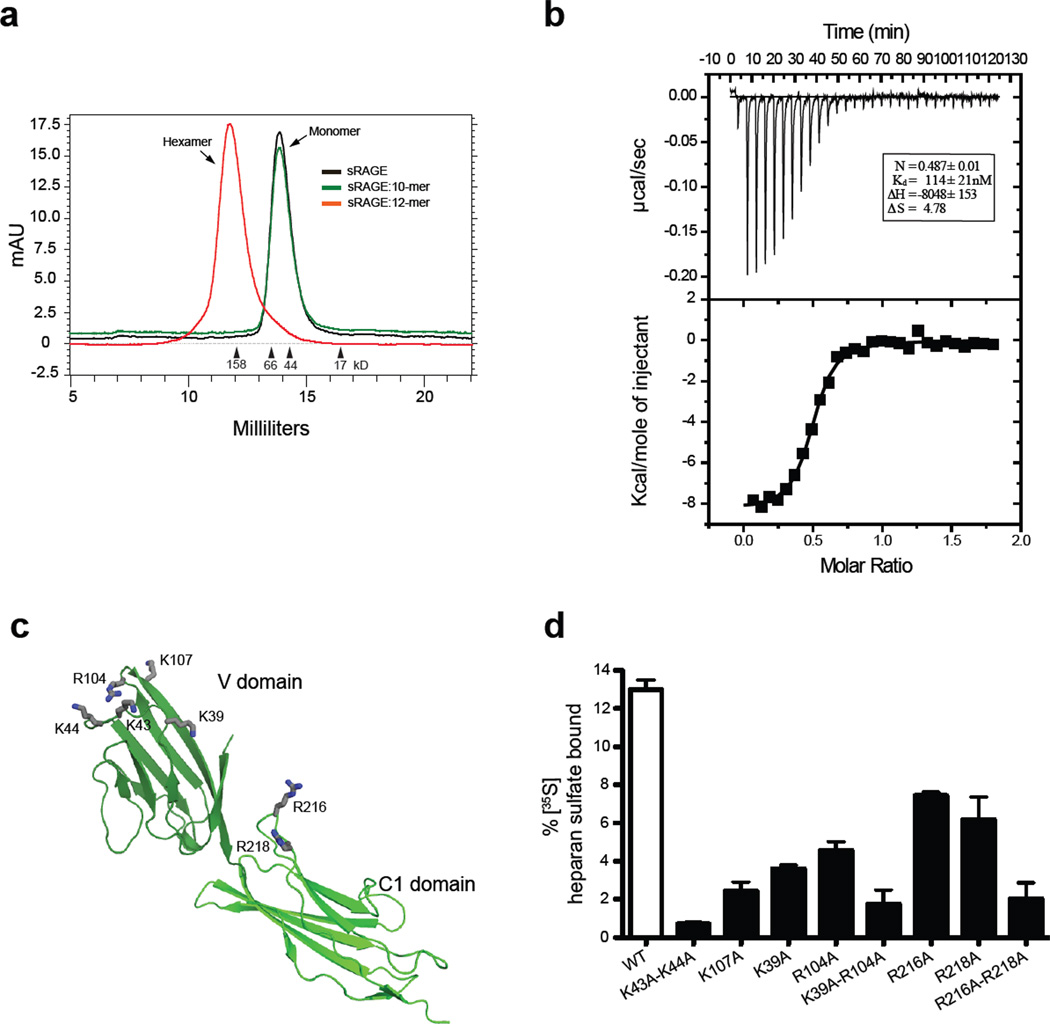
To test if heparan sulfate promotes RAGE oligomerization, we mixed recombinant sRAGE (V-C1-C2 domains) with heparin-derived oligosaccharides of defined size and examined RAGE oligomerization by size exclusion chromatography (SEC). Heparin is related in structure to heparan sulfate and thus mimics the behavior of heparan sulfate in most systems. By itself, sRAGE eluted from the column as a monomer, with an apparent molecular mass larger than its predicted mass (~41 kDa vs. 32 kDa), presumably due to its elongated shape 9, 10. However, when we mixed sRAGE with heparin-derived dodecasaccharide (dp12), sRAGE eluted as a single peak of ~200 kDa (Fig. 2a). The formation of this oligomer has a strict dependence on the length of the oligosaccharide, as exemplified by a total lack of oligomerization by heparin decasaccharide (dp10) (Fig. 2a). We also found that V-C1 domains were sufficient to support the formation of a stable oligomer (Supplemental Fig. 1a).
Figure 2. Heparin-derived dodecasaccharides induce a stable oligomer of RAGE.
(a) Mixtures of sRAGE and heparin-derived decasaccharides or dodecasaccharides were resolved by SEC. (b) Binding of dodecasaccharides was monitored by isothermal calorimetry by titrating sRAGE with dodecasaccharide. (c) Cartoon representation of RAGE V-C1 domain (PDB-id: 3CJJ). Amino residues demonstrated to interact with heparan sulfate are shown in stick representation (gray). (d) Binding of wild-type and various RAGE mutants to 35S-labeled endothelial heparan sulfate as determined by filter binding assay (n = 3, error bars represent the standard deviation). The extent of binding was quantified by dividing the counts retained by the membrane by the total input counts.
We next examined the stoichiometry of the sRAGE/oligosaccharide complex by Isothermal titration calorimetry (ITC). Fitting of the thermogram for titration of the dodecasaccharide into sRAGE yielded a dissociation constant (Kd) of 114 ± 18 nM and a stoichiometry of 0.49 ± 0.01 (Fig. 2b), suggesting the stoichiometry of the oligomeric complex is two RAGE molecules to one oligosaccharide. Using multiple-angle light scattering (MALS) coupled with SEC, we determined the complex has a molecular mass of 196 ± 5 kDa, very close to the predicted molecular mass of an sRAGE hexamer plus 3 oligosaccharides (202 kDa, Supplemental Fig. 1b).
Location of the heparan sulfate-binding site of RAGE
To identify the amino acid residues that are responsible for binding of RAGE to heparan sulfate, a systematic site-directed mutagenesis study of the 22 conserved lysine and arginine residues in V-C1 was performed (Supplemental Fig. 2). Alanine substitution mutants showed greatly reduced binding to heparin-Sepharose, displaying a reduction of 50 –110 mM in salt concentration required for elution (Supplemental Table 1). Seven critical residues were identified in this way, five located in the V domain (K39, K43, K44, R104 and K107), and two located in the C1 domain (R216 and R218, Fig. 2c).
We also found that these mutants showed greatly reduced binding to endothelial heparan sulfate in solution, and the effect was especially dramatic with double mutants (Fig. 2d). However, these mutants bound the ligand HMGB1 with a Kd of 8–13 nM (10 nM for wild-type RAGE) and to S100b with a Kd of 63–90 nM (83 nM for wild-type RAGE) (Supplemental Table 2). The preservation of HMGB1 and S100b binding affinity suggested that these ligands and heparan sulfate interact with separate surfaces of RAGE. We also found that all heparin sulfate-binding mutants displayed substantially reduced hexamer formation (70–100% reduction compared to wild-type RAGE) when mixed with oligosaccharide (Supplemental Fig. 3).
Heparan sulfate stabilizes a hydrophobic dimeric interface of RAGE
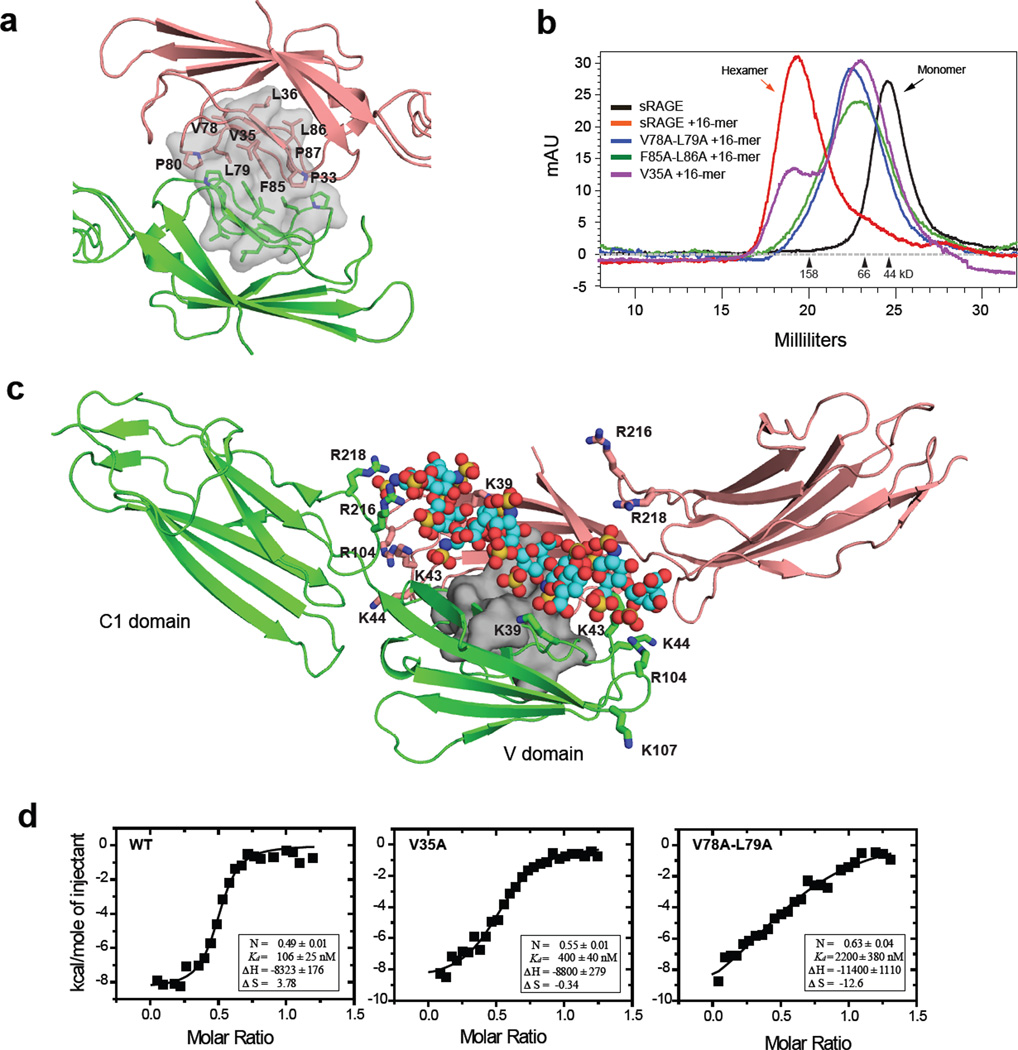
Examination of the crystal structures of the monomeric RAGE V-C1 domain (PDB-id: 3CJJ and 3O3U) revealed a hydrophobic patch (P33, V35, L36, V78, L79, P80, F85, L86, and P87) that is involved in crystal packing between two molecules (Fig. 3a). To test if these residues mediate oligomerization, we prepared three mutants in the center of the hydrophobic patch (V35A, V78A-L79A, F85A-L86A). Remarkably, all three mutants showed severe defects in hexamerization. The single mutation, V35A, supported partial hexamer formation when mixed with oligosaccharide, but the majority of the protein eluted as a broad peak that overlaps with the elution position of the monomer, suggesting that the oligomer was destabilized (Fig. 3b). The effect was more pronounced with the two double mutants, V78A-L79A and F85A-L86A, which eluted in SEC exclusively as a broad peak (Fig. 3b). MALS analysis of the V78A-L79A mutant suggested that the broad peak contained mostly species with molecular weights in the range of 32 to 70 kD, consistent with the idea of a monomer/dimer equilibrium (Supplemental Fig. 1b).
Figure 3. A hydrophobic dimeric interface is stabilized by heparin.
(a) Cartoon diagram of the V-C1 dimer (salmon and green) showing the proposed dimeric interface (gray surface) deduced from the crystal structure of hRAGE V-C1 (pdb-id: 3CJJ). The relevant hydrophobic residues are shown in stick representation. (b) Mutants of sRAGE were incubated with oligosaccharide at 1:1 molar ratio and the mixtures were resolved by SEC. (c) The proposed heparan sulfate binding cleft of the dimer. Monomers are shown in green or salmon. Amino acid residues that bind to heparan sulfate are shown in stick representation on the corresponding monomer. A space-filling model of the dodecasaccharide is modeled into the binding cleft (carbon in cyan, oxygen in red, nitrogen in blue and sulfate in yellow). The hydrophobic interface between the dimers is represented as a grey molecular surface. (d) Binding of dodecasaccharide to wild-type, V35A or V78A-L79A RAGE V-C1 domain was assessed by Isothermal calorimetry.
All three oligomerization mutants showed wild-type level of binding to both HMGB1 and S100b (Supplemental Table 2), and eluted from heparin-Sepharose at a similar salt concentration as wild-type RAGE (Supplemental Table 1). These findings suggested that the mutants probably maintain wild-type like folding and that the defects in hexamerization most likely arose due to destabilized hydrophobic interactions between subunits. Based on the cumulative results, we propose that the basic building block of RAGE oligomers is a dimmer stabilized by the identified hydrophobic patch.
Examination of the dimer structure (extracted from 3CJJ) showed that two separate heparin binding sites from each subunit converge into a large positively charged cleft that would accommodate a negatively charged heparan sulfate oligosaccharide (Fig. 3c). Therefore, it seemed likely that heparan sulfate might function to stabilize the RAGE dimer. Thus, we proposed that heparan sulfate binding and the protein-protein hydrophobic interaction are energetically coupled and that both contribute to the formation of a stable dimer. If this model were correct, one would expect that alanine mutants of the hydrophobic patch, which decrease dimer formation, would decrease the binding affinity of RAGE to heparan sulfate. We tested this idea by measuring binding of V35A and V78A-L79A mutants to oligosaccharide by ITC. As predicted by the model, V35A, which has a moderate effect on oligomerization, showed 4-fold lower affinity for dodecasaccharide compared to wild-type RAGE (400 ± 40 vs. 106 ± 25 nM, Fig. 3d). In comparison, V78A-L79A displayed a 20-fold lower affinity for dodecasaccharide compared to wild-type (2200 ± 380 vs. 106 ± 25 nM, Fig. 3d). The stoichiometry of binding of RAGE to dodecasaccharide also deviated from the value of 0.5 observed for wild-type RAGE, giving values of 0.55 and 0.64 for V35A and V78A-L79A, respectively. These findings support the idea that dimer formation creates the optimal surface for binding to heparan sulfate.
Crystal structure of mRAGE in presence of oligosaccharide
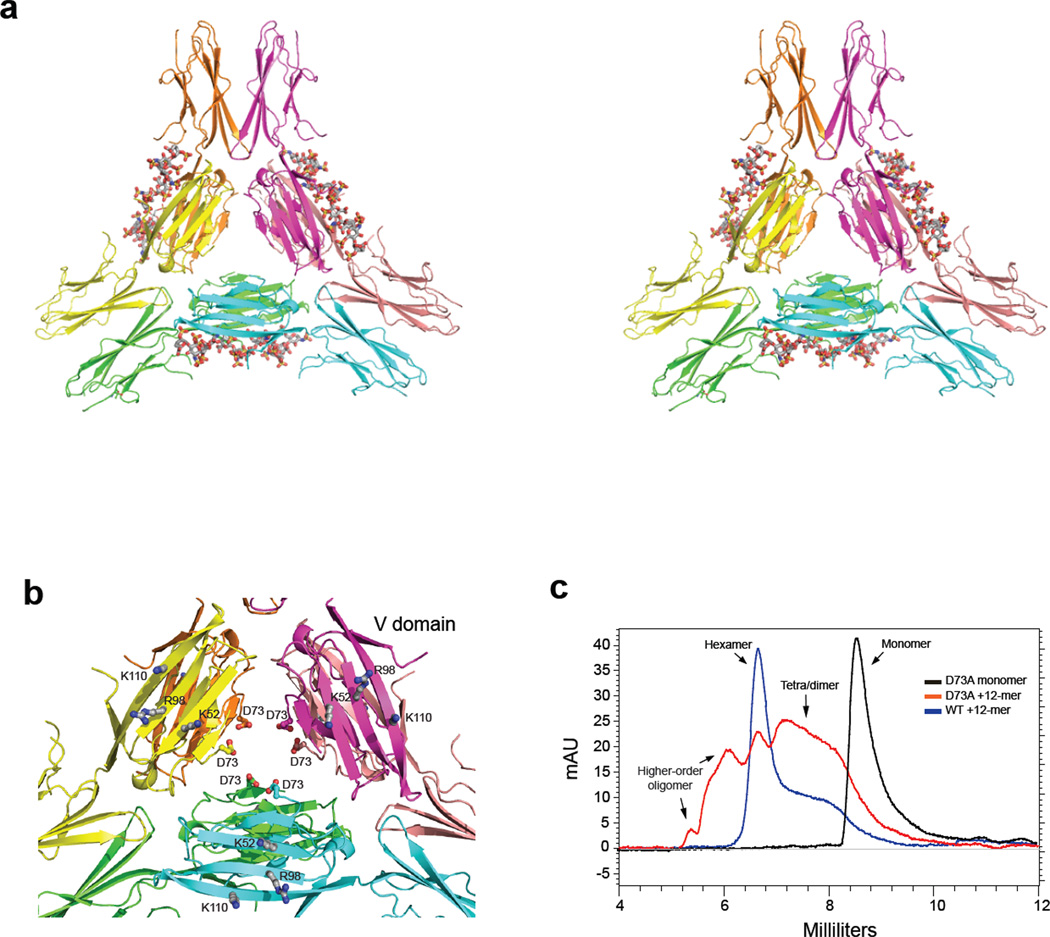
To obtain deeper insight into the oligomerization of RAGE, mouse RAGE V-C1 (mV-C1) was crystallized in complex with dodecasaccharide. The structure was determined at a resolution of 3.5 Å (Fig. 4a and Supplemental Fig. 4) in space group P4132 with one molecule of mV-C1 in the asymmetric unit (Supplemental Table 3). Lattice packing revealed dimeric interactions between symmetry related V domains similar to that seen in the lattice packing of human RAGE V-C1 (hV-C1, PDB-id 3CJJ). The same dimeric interactions were also present in recently deposited structures of hV-C1 in the presence of DNA (PDB-id 3S58 and 3S59). The overall fold of mV-C1 closely resembles that of hV-C1 with an RMSD of 0.98 Å over 176 residues. One difference exists: residues P213–R228 of the terminal strand of the C1 domain form a strand swap with a neighboring hexamer in the mV-C1 crystal lattice. In our structure, three dimers come together to form a hexamer with the V domains situated in the center forming a hub and the C1 domains extending outward like six spokes resembling a wheel. This arrangement differs from crystal packing in previously reported V-C1 structures obtained in the absence of heparin.
Figure 4. Crystal structure of the RAGE hexamer.
(a) Stereo view of the cartoon representation of the crystal structure of the hexameric mRAGE V-C1 domain. The three dimmers are colored in green and cyan, yellow and orange, and salmon and magenta, respectively. Dodecasaccharides (in sticks) were modeled into the heparan sulfate binding clefts based on the observed sugar density in the crystal structure. The C-terminal strands, involved in the strand-swap, from neighboring molecules in the crystal are displayed to give a canonical view of the C1 domains. (b) The central part of the hexamer consisting of the V-domains is shown. Residues involved in ligand binding (K52, R98 and K110) are shown as ball and stick model and are located at the surface of the hexamer. The aspartic acids at position 73 of each monomer are oriented inward at the core of the hexamer. (c) D73A mutant or wild-type sRAGE were incubated with oligosaccharide at 2:1 molar ratio and resolved by SEC.
Although the electron density maps reveal no clear density for a bound dodecasaccharide, there are regions of discontinuous difference density within the proposed heparan sulfate binding cleft (Supplemental Fig. 5a). The absence of clearly defined density suggests the dodecasaccharide is bound heterogeneously to V-C1. Interestingly, superposition of the DNA bound hV-C1 structure (PDB-id: 3S59) with the crystallographic dimer of mV-C1 reveals many residues that make contact with the DNA (K39, K43, K107, R216, and R218) are very near the regions of positive difference density in the mV-C1 structure supporting their role in dodecasaccharide binding (Supplemental Fig. 5b). In the DNA bound crystal structure there is a gap of ~20 Å and four base pairs across the dimerization interface where there are no hydrogen bond interactions with the DNA. The lack of interactions in this region could contribute to the disorder of the less rigid polysaccharide to mV-C1.
The crystal structure of the hexameric complex is consistent with previous RAGE ligand binding studies in that the proposed S100b, AGE and lysophosphatidic acid binding surface, which includes K52, R98 and K110, is exposed to solvent (Fig. 4b) 10, 13, 24. The architecture of the hexamer indicates that the three dimers are tethered together by interacting with each other within the core of the hexamer. Unfortunately, the majority of a key loop (65SPQGGGPWDS74) that is centrally located at the interface is disordered. The conformations of this loop are different among three crystal structures of isolated RAGE V-C1 (3CJJ, 3S59 and 3O3U), which suggests that the loop is flexible 25. The loop is presumably involved in inter-dimer interactions. A conserved D73 residue (D72 in mouse) is located at the end of the disordered loop (Fig. 4b). The alanine mutant of this residue was detrimental to formation of the hexamer (Fig. 4c), consistent with the possible role of D73 as a hexamer stabilizer.
The crystal structure shows clearly that the dimer is the basic building unit of the RAGE hexamer. The dimer possesses four discrete functional surfaces with almost no overlap. The flat ligand binding surfaces are located at the ends of the V domain. The hydrophobic dimerization interface is embedded centrally in the core of the dimer. The heparan sulfate-binding residues line a positive cleft between dimer-related subunits. The residues involved in inter-dimer interactions lie on the opposite side of the heparan sulfate-binding surface (Fig. 3a and 3c). In the absence of heparin, RAGE does not dimerize in solution, suggesting that the hydrophobic interactions at the dimer interface must be too weak to sustain a dimer without an additional stabilizing factor. The small hydrophobic dimer interface is only 300 Å2, much less than the 1200 – 2000 Å2 interface commonly found at protein–protein interfaces 26. Our mutational data indicates that hydrophobic and electrostatic interactions work synergistically to stabilize the dimer interface.
SAXS analysis of RAGE hexamer in solution
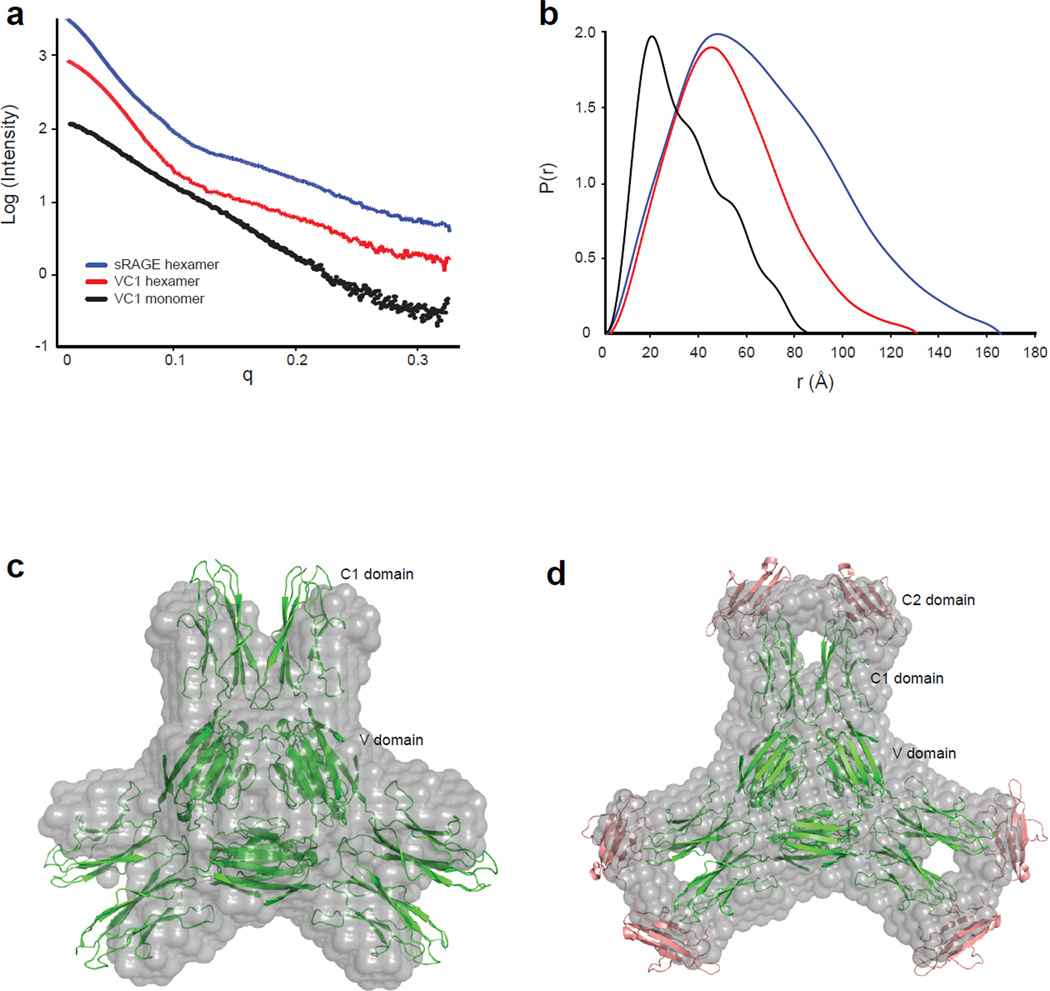
The technique of small angle x-ray scattering was used to assess whether the structure observed in the crystalline state is retained in solution. The scattering profile, I(q), of V-C1 and the V-C1/oligosaccharide complex and the sRAGE/oligosaccharide complex reveals well-folded architecture, since plots of I(q) vs. q reveal the fine structure expected for relatively fixed spatial arrangements of domains (Fig. 5a) 27, 28. The distance distribution function, P(r), and Guinier plot was determined for all three data sets, from which real space RG values were extracted (Fig. 5b and Supplemental Table 4). MW estimations of the complexes based on scattering curves are also consistent with hexamer (Supplemental Table 4).
Figure 5. Solution structure of RAGE oligomer.
(a) Raw SAXS scattering curves of the monomeric V-C1 domain, the hexameric V-C1 domain/dodecasaccharide complex and the hexameric sRAGE/dodecasaccharide complex. (b) P(r) pair function analysis of the scattering curves. Profiles are colored as in (a). (c) The GASBOR generated ab initio model of hexameric V-C1 is shown in gray and is superimposed on the crystal structure of V-C1 hexamer shown in green. (d) An ab initio model of hexameric sRAGE superimposed with the crystal structure of V-C1 hexamer (green) and the structure of C2 domain (PDB: 2ENS, salmon) is shown.
The theoretical scattering curve back-calculated from the crystal structure of hexameric V-C1 domain displays good agreement with the experimental scattering curve with a value of 2.5 for the Chi fitting parameter (Supplemental Fig. 6). The experimental and simulated curves exhibit excellent overlap up until q = 0.1, which suggests the two structures have similar overall shape but that the average conformation in solution is different from the crystal structure. This observation is consistent with isolation of a specific low-energy conformation in the crystal from the ensemble of structures that are populated in solution.
We further generated low-resolution models of the complexes using the ab initio envelope reconstruction program GASBOR. The average GASBOR model of multiple runs based on P3 symmetry is presented in Fig. 5c (V-C1 complex) and 5d (sRAGE complex). The overall shape of V-C1 hexamer model bears striking similarity to our crystal structure, most notably the discrete representation of the six spokes that correspond to the C1 domains. Alignment of the GASBOR envelope of V-C1 hexamer to the crystal structure using SUPCOM showed a high degree of agreement (ρf = 1.02, Fig. 5c). The GASBOR envelope of the sRAGE hexamer closely resembles that of the V-C1 hexamer, except that additional density is visible at the tip of the spokes. This additional volume nicely accommodates the C2 domains as shown in Fig. 5d.
The combined results of the SAXS and crystallographic studies reveal that RAGE exists as a hexamer in the presence of heparan sulfate. This configuration resembles a wheel-like structure with the V domains forming the central hub, the C1 domains forming the spokes of the wheel, and the C2 domains forming a rim at the periphery. Although we lack conclusive evidence for oligosaccharide binding in the crystal, the lack of crystal formation in the absence of dodecasaccharide (under identical crystallization conditions) and positive difference-electron density in solvent channels of the crystal suggests the presence of heparin.
Our structural characterization of RAGE oligomerization provides insight relevant to how RAGE transduces signals across the cell membrane. Our data indicate that the RAGE hexamer lies parallel to the cell surface, with one or more heparan sulfate chains arrayed around the perimeter. In this orientation, the ligand-binding residues on the V domain (K52, R98 and K110) are fully exposed and would be available for ligand binding. It was reported that S100b interact with this basic surface by electrostatic interactions possibly through a negatively charged surface 10. In addition, S100b forms oligomers and an octamer structure of S100b has been reported 12. Interestingly, the diameter of the octameric S100b matches quite well to the center hub formed by the V domains (Supplemental Fig. 7), suggesting that S100b could interact with RAGE in a multivalent manner. This orientation would also allow the C2 domains, which are connected to V-C1 through a flexible loop, and the transmembrane domains to orient perpendicular to the hexamer. We propose that such arrangement poises RAGE in a functionally ready state for signaling.
RAGE is an example of a growing number of receptors that depend on heparan sulfate for activity, which includes fibroblast growth factor (FGF) and vascular endothelial growth factor (VEGF) receptors. Although RAGE bears many structural similarities to these receptors, it is unique in that the formation of RAGE hexamer depends only on heparan sulfate and exists in the absence of ligand, whereas the functional forms of FGF and VEGF receptors depend on ligand-induced dimerization and association with heparan sulfate 29–32. These differences highlight the versatility of the heparan sulfate in regulating receptor activation. Notably, our study stresses that heparan sulfate can function entirely at the receptor level, without having to directly interact with its cognate ligands. Historically most heparan sulfate-binding receptors were only discovered after their heparan sulfate-binding ligands were known. We now need to reexamine the possibility that many more receptors exist that depend on direct interaction with heparan sulfate for activity.
Targeting RAGE oligomerization with mAbs
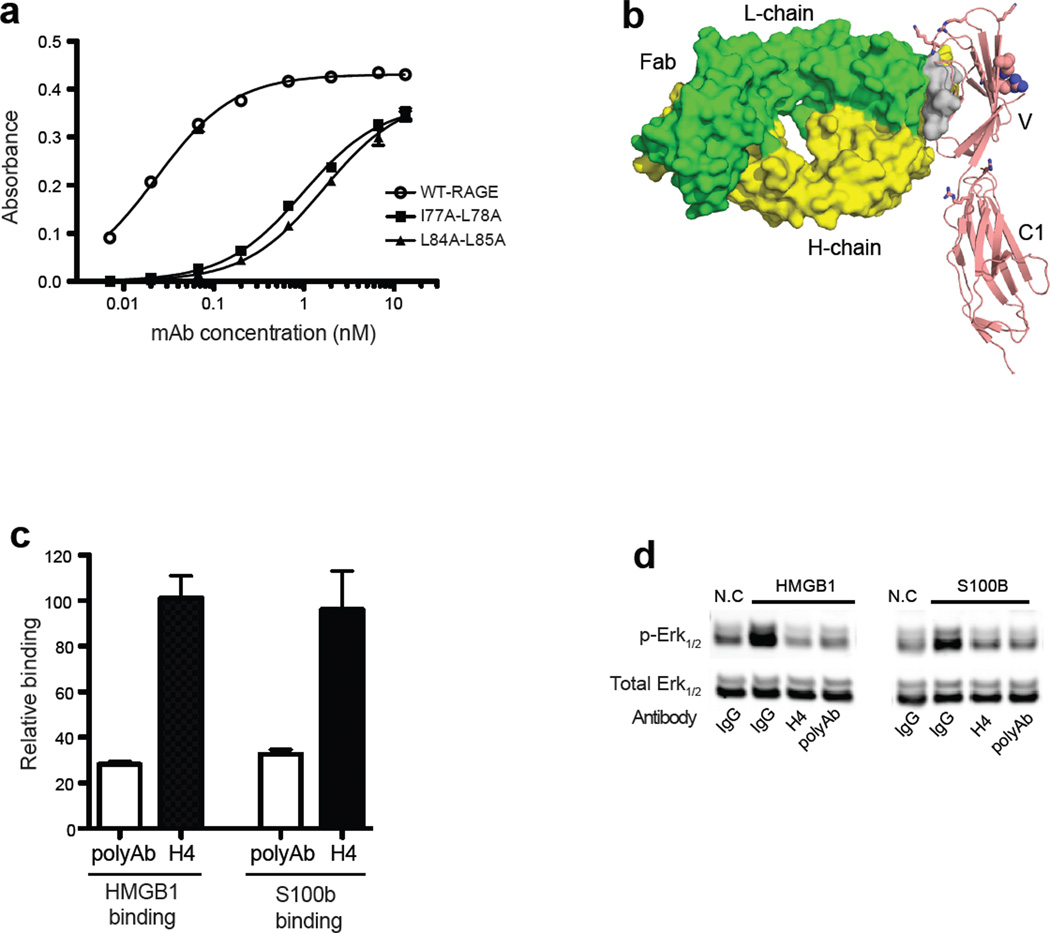
Our characterization of the receptor complex suggested that disrupting RAGE oligomerization might block RAGE signaling. In fact, this mode of inhibition might be a more universal way to block RAGE signaling because it should affect signaling by most if not all RAGE ligands. To test this hypothesis, we generated a collection of rabbit mAbs against mouse RAGE V-C1 domain. Ninety mAbs were identified and mapped against the various mutants of RAGE, which led to the identification of one mAb (H4) that specifically targets the hydrophobic interface. Binding of H4 to RAGE depends on amino acid residues I77, L78, L84 and L85 (mouse homologues of human V78, L79, F85 and L86). Double mutants involving these residues showed 50- to 70-fold reduction in affinity of the mAb:RAGE interaction (Fig. 6a), which suggests that H4 most likely directly targets the hydrophobic dimerization interface (a schematic view of the postulated H4:RAGE interaction is shown in Fig. 6b). mAb H4 has an extremely high affinity for mouse RAGE with a Kd of 23 pM, and it cross-reacts with human RAGE with a Kd of 0.9 nM. As predicted, binding of mAb H4 to RAGE did not interfere with ligand binding (consistent with the location of the ligand binding site on the opposite side of the V domain; Fig. 6b), whereas a rabbit polyclonal antibody substantially inhibited both HMGB1 and S100b binding (Fig. 6c). These properties allowed us to directly test the effect of blocking RAGE oligomerization on RAGE signaling. When primary human endothelial cells were stimulated with either HMGB1 or S100b in the presence of mAb H4 at 5 µg/ml, ERK1/2 phosphorylation was completely abolished (Fig. 6d). The inhibition of signaling by this oligomerization-specific mAb suggests that RAGE oligomerization is an important part of signaling. Pharmaceutical inhibition of RAGE oligomerization might therefore be a feasible approach for inhibition of RAGE signaling in pathological conditions.
Figure 6. Targeting the oligomerization interface with a mAb blocks RAGE signaling.
(a) Binding of anti-RAGE rabbit mAb H4 to immobilized wild-type mRAGE V-C1 domain, I77A-L78A (mouse equivalent of human V78A-L79A), and L84A-L85A (mouse equivalent of human F85AL86A) mutants were determined by ELISA. (b) Postulated binding model of H4 to RAGE. Fab fragment (PDB-id: 3GRW) of IgG is shown as surface representation with light chain in green and heavy chain in yellow. V-C1 is shown as cartoon in salmon. Also shown are the hydrophobic dimerization interface (gray surface), heparan sulfate binding residues (sticks) and ligand binding residues (spheres). (c) Binding of biotinylated mRAGE V-C1 domain to immobilized HMGB1 or S100b was measured in the absence or presence of anti-RAGE rabbit polyclonal Ab (polyAb) or H4 (both at 5 µg/ml). The percentage of inhibition was determined by comparing the absorbance value to standard binding curves of mRAGE V-C1 to HMGB1 or S100b. n = 3, the error bar represents SD. (d) Immunoblot analysis of Erk1/2 and phosphoErk1/2 in human endothelial cells after stimulation with HMGB1 or S100b. Cells were pre-incubated with non-specific rabbit IgG, H4, or anti-RAGE polyAb at 5 µg/ml for 30 min prior to stimulation.
Most receptors require oligomerization for activity, suggesting that drug-like compounds or mAbs that target subunit interfaces could be a viable strategy for therapeutic intervention. Indeed, several tumor suppressing mAbs (Cetuximab and Matuzumab) abrogate EGF receptor signaling by preventing receptor dimerization 33, 34. Similarly, a mAb that inhibits FGFR3 dimerization has proven effective in inhibiting FGFR3 signaling in several tumor models 35. Our development of an inhibitory mAb targeted to the dimer interface represents another example of effective targeting of receptor oligomerization. Theoretically, mAbs that specifically interfere with heparan sulfate binding would achieve the same effect by inhibiting formation of RAGE hexamers. Our success with this strategy might serve as a paradigm for developing mAbs to target other heparan sulfate-binding proteins that depend on heparan sulfate for oligomerization. Moreover, our experience with the dimer interface-specific mAb strongly suggests that the oligomerization interface could be explored as a target for computer-based virtual screening and drug design.
METHODS
Protein expression
The production of recombinant sRAGE and site-directed mutagenesis is described in Supplemental Methods.
Immunoblotting
Human dermal microvascular endothelial cells (HDMVEC, Lonza) were serum starved in DMEM for 5 hr prior to treatment with HMGB1 (50 ng/ml) 20, S100A8/A9 or S100A12 (5 µg/ml, gifts from Geetha Srikrishna), bovine S100b or AGE-BSA (5 µg/ml, EMD Biosciences). Selected samples were pretreated with heparin lyases as described 20. Cells were lysed and resolved by SDS-PAGE and blotted with antibodies to Erk1/2 and phospho-Erk1/2 (Cell Signaling). Bands were visualized on an Odyssey Infrared imaging system (Li-Cor).
Analytical size exclusion chromatography (SEC)
Purified sRAGE monomer (100–200 µg) and size-defined heparin-derived oligosaccharides (Iduron) were incubated in 20 mM Tris, 200 mM NaCl, pH 7.7 at room temperature overnight. The complexes were resolved on a Superdex 200 SEC column (size 10/300 mm or 10/420 mm, GE Healthcare) or a SRT SEC-300 HPLC column (7.8/300 mm, Sepax). Disaccharide analysis provided by Iduron states that the tri-sulfated disaccharide (I2S6) represents about 75% of the disaccharides in the dodecasaccharide. The rest of the disaccharide species are mostly di-sulfated (I0S6 or I2S0). The method for SEC-MALS is described in Supplemental Methods.
Isothermal Titration Calorimetry (ITC)
ITC was performed on a MicroCal VP-ITC at 25 °C. Titrations were performed by injecting 10 µl (for a total of 28 injections) of 60 or 80 µM dodecasaccharide into 10 µM protein in the sample cells. Both solutions were dialyzed into 20 mM Tris, 150 mM NaCl at pH 7.7 prior to the experiment. Data were analyzed using Origin software (5.0) assuming single-site binding.
Heparin-Sepharose Chromatography
Purified RAGE mutants (200 µg) were applied to a 1-ml HiTrap heparin-Sepharose column (GE Healthcare) and eluted with a salt gradient from 150mM to 1M NaCl at pH 7.2 (HEPES buffer). The conductivity measurements at the peak of the elution were converted to the concentration of NaCl.
Nitrocellulous filter binding assay
Recombinant wild-type sRAGE or mutants (500 ng) were incubated with 10,000 counts of 35S-labeled microvascular endothelial lung heparin sulfate in 100 µl of PBS for one hour at 22 °C. The mixtures were applied to a pre-washed nitrocellulose membrane in a Bio-Rad 96 well blot apparatus and washed with 100 µl of PBS twice. Free [35S]heparan sulfate passed through the membrane and only the portions that bound to protein were retained. The membrane was eluted with 0.5 ml of HEPES buffer containing 1 M NaCl. Released [35S]heparan sulfate was measured by liquid scintillation counting. The extent of binding was quantified by dividing the counts retained by the membrane by the total input counts.
X-ray crystallography
Mouse RAGE V-C1/dodecaccharide complex was concentrated to 10.25 mg/ml in 20 mM Tris, 100 mM NaCl, at pH 7.7. Crystals were obtained by vapor diffusion using the hanging drop method by mixing 2.0 μL of the complex with 2.0 μL of 0.1 M Tris pH 8.5, 200 mM MgCl2, and 22% PEG 400. For data collection crystals were flash frozen in liquid nitrogen and placed into a stream of liquid nitrogen gas cooled to −180 °C. Diffraction data were collected to 3.5 Å on the Southeast Regional Collaborative Access Team 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. The structure was determined by molecular replacement in Phenix with the hRAGEV1C1 domain (pdb id code= 3CJJ) as the starting model 36. Refinement and model building were carried out using Phenix and COOT respectively 36, 37.
SAXS data acquisition
Scattering data were collected at the beamline 12.3.1 at the Lawrence Berkeley National Laboratory. Monodisperse sRAGE/dodecasaccharide and V-C1/dodecasaccharide complexes purified by SEC were concentrated to 1, 3, and 6 mg/ml in a buffer containing 20 mM Tris, 200 mM NaCl, 1% glycerol at pH 7.7. The scattering profile of each sample was collected with 3 exposures (0.5, 1 and 6 sec). SAXs data analysis is described in Supplemental Methods.
Rabbit monoclonal antibody (mAb)
Rabbits were immunized with mouse RAGE V-C1 domain. After the desired titer was obtained, the anti-serum was purified by immobilized RAGE to obtain the polyclonal anti-RAGE IgG. Splenocytes derived from the rabbits were fused with rabbit plasmacytoma cells 240E-W2 at Epitomics Inc. We screened 4,000 clones and obtained 90 positive clones by antigen binding ELISA (Supplemental Methods). Positive clones were subjected to epitope mapping by direct ELISA using immobilized heparan sulfate-binding mutants and dimerization mutants of RAGE. Mutants that showed reduced binding affinity to a given mAb compared to the wild-type RAGE defined residues that are part of the epitope. Rabbit mAb H4, which blocks the oligomerization of RAGE, was purified by Protein A chromatography from hybridoma supernatant.
Accession Codes
Coordinates for the RAGE V-C1/dodecaccharide complex structure have been deposited with the Protein Data Bank under the accession code 4IM8.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by grants 13BGIA14150008 from the American Heart Association (to D.X), P01 HL57345 and P01 HL107150 (to J.D.E), R56 AI091771 (to W.J.C) from the National Institutes of Health, the Division of Intramural Research of the National Institute of Environmental Health Sciences, National Institutes of Health (1 ZIA ES102645-03, to L.C.P), and by the Ludwig Institute for Cancer Research (to K.D.C). We would like to thank G. Srikrishna and H. Freeze for providing S100A8/A9 and S100A12 proteins, and SIBYLS beamline scientist M. Hammel and G. Hura for helpful discussions. SAXS data collection at the SIBYLS beamline of Advanced Light Source, Lawrence Berkeley National Laboratory, are supported in part by the DOE program Integrated Diffraction Analysis Technologies (IDAT) under Contract Number DE-AC02-05CH11231 with the U.S. Department of Energy.
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
AUTHOR CONTRIBUTIONS
D.X., L.P., W.J.C. and J.D.E. designed the research, D.X., L.P., J.Y and D.S performed the work and statistical analyses, D.X., L.P., J.Y., D.S., W.J.C., K.C., J.K and J.D.E analyzed and interpreted data, and D.X., W.J.C., L.P and J.D.E wrote the manuscript.
The authors declare no conflict of interest.
REFERENCE
- 1.Brownlee M, Cerami A, Vlassara H. Advanced glycosylation end products in tissue and the biochemical basis of diabetic complications. The New England journal of medicine. 1988;318:1315–1321. doi: 10.1056/NEJM198805193182007. [DOI] [PubMed] [Google Scholar]
- 2.Basta G. Receptor for advanced glycation endproducts and atherosclerosis: From basic mechanisms to clinical implications. Atherosclerosis. 2008;196:9–21. doi: 10.1016/j.atherosclerosis.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 3.Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010;28:367–388. doi: 10.1146/annurev.immunol.021908.132603. [DOI] [PubMed] [Google Scholar]
- 4.Liliensiek B, Weigand MA, Bierhaus A, Nicklas W, Kasper M, Hofer S, Plachky J, Grone HJ, Kurschus FC, Schmidt AM, Yan SD, Martin E, Schleicher E, Stern DM, Hammerling GG, Nawroth PP, Arnold B. Receptor for advanced glycation end products (RAGE) regulates sepsis but not the adaptive immune response. J Clin Invest. 2004;113:1641–1650. doi: 10.1172/JCI18704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muhammad S, Barakat W, Stoyanov S, Murikinati S, Yang H, Tracey KJ, Bendszus M, Rossetti G, Nawroth PP, Bierhaus A, Schwaninger M. The HMGB1 receptor RAGE mediates ischemic brain damage. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:12023–12031. doi: 10.1523/JNEUROSCI.2435-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park L, Raman KG, Lee KJ, Lu Y, Ferran LJ, Jr, Chow WS, Stern D, Schmidt AM. Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation endproducts. Nat Med. 1998;4:1025–1031. doi: 10.1038/2012. [DOI] [PubMed] [Google Scholar]
- 7.Yan SF, Ramasamy R, Schmidt AM. The RAGE axis: a fundamental mechanism signaling danger to the vulnerable vasculature. Circ Res. 2010;106:842–853. doi: 10.1161/CIRCRESAHA.109.212217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan SS, Wu ZY, Zhang HP, Furtado G, Chen X, Yan SF, Schmidt AM, Brown C, Stern A, LaFaille J, Chess L, Stern DM, Jiang H. Suppression of experimental autoimmune encephalomyelitis by selective blockade of encephalitogenic T-cell infiltration of the central nervous system. Nat Med. 2003;9:287–293. doi: 10.1038/nm831. [DOI] [PubMed] [Google Scholar]
- 9.Dattilo BM, Fritz G, Leclerc E, Kooi CW, Heizmann CW, Chazin WJ. The extracellular region of the receptor for advanced glycation end products is composed of two independent structural units. Biochemistry. 2007;46:6957–6970. doi: 10.1021/bi7003735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch M, Chitayat S, Dattilo BM, Schiefner A, Diez J, Chazin WJ, Fritz G. Structural basis for ligand recognition and activation of RAGE. Structure. 2010;18:1342–1352. doi: 10.1016/j.str.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leclerc E, Fritz G, Weibel M, Heizmann CW, Galichet A. S100B and S100A6 differentially modulate cell survival by interacting with distinct RAGE (receptor for advanced glycation end products) immunoglobulin domains. J Biol Chem. 2007;282:31317–31331. doi: 10.1074/jbc.M703951200. [DOI] [PubMed] [Google Scholar]
- 12.Ostendorp T, Leclerc E, Galichet A, Koch M, Demling N, Weigle B, Heizmann CW, Kroneck PM, Fritz G. Structural and functional insights into RAGE activation by multimeric S100B. EMBO J. 2007;26:3868–3878. doi: 10.1038/sj.emboj.7601805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue J, Rai V, Singer D, Chabierski S, Xie J, Reverdatto S, Burz DS, Schmidt AM, Hoffmann R, Shekhtman A. Advanced glycation end product recognition by the receptor for AGEs. Structure. 2011;19:722–732. doi: 10.1016/j.str.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakaguchi M, Murata H, Yamamoto K, Ono T, Sakaguchi Y, Motoyama A, Hibino T, Kataoka K, Huh NH. TIRAP, an adaptor protein for TLR2/4, transduces a signal from RAGE phosphorylated upon ligand binding. PLoS One. 2011;6:e23132. doi: 10.1371/journal.pone.0023132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hudson BI, Kalea AZ, Del Mar Arriero M, Harja E, Boulanger E, D'Agati V, Schmidt AM. Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42. J Biol Chem. 2008;283:34457–34468. doi: 10.1074/jbc.M801465200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishihara K, Tsutsumi K, Kawane S, Nakajima M, Kasaoka T. The receptor for advanced glycation end-products (RAGE) directly binds to ERK by a D-domain-like docking site. FEBS Lett. 2003;550:107–113. doi: 10.1016/s0014-5793(03)00846-9. [DOI] [PubMed] [Google Scholar]
- 17.Xie J, Reverdatto S, Frolov A, Hoffmann R, Burz DS, Shekhtman A. Structural basis for pattern recognition by the receptor for advanced glycation end products (RAGE) J Biol Chem. 2008;283:27255–27269. doi: 10.1074/jbc.M801622200. [DOI] [PubMed] [Google Scholar]
- 18.Zong H, Madden A, Ward M, Mooney MH, Elliott CT, Stitt AW. Homodimerization is essential for the receptor for advanced glycation end products (RAGE)-mediated signal transduction. J Biol Chem. 2010;285:23137–23146. doi: 10.1074/jbc.M110.133827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarkany Z, Ikonen TP, Ferreira-da-Silva F, Saraiva MJ, Svergun D, Damas AM. Solution structure of the soluble receptor for advanced glycation end products (sRAGE) J Biol Chem. 2011;286:37525–37534. doi: 10.1074/jbc.M111.223438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu D, Young J, Song D, Esko JD. Heparan Sulfate Is Essential for High Mobility Group Protein 1 (HMGB1) Signaling by the Receptor for Advanced Glycation End Products (RAGE) J Biol Chem. 2011;286:41736–41744. doi: 10.1074/jbc.M111.299685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goyette J, Yan WX, Yamen E, Chung YM, Lim SY, Hsu K, Rahimi F, Di Girolamo N, Song C, Jessup W, Kockx M, Bobryshev YV, Ben Freedman S, Geczy CL. Pleiotropic Roles of S100A12 in Coronary Atherosclerotic Plaque Formation and Rupture. J Immunol. 2009;183:593–603. doi: 10.4049/jimmunol.0900373. [DOI] [PubMed] [Google Scholar]
- 22.Robinson MJ, Tessier P, Poulsom R, Hogg N. The S100 family heterodimer, MRP-8/14, binds with high affinity to heparin and heparan sulfate glycosaminoglycans on endothelial cells. J Biol Chem. 2002;277:3658–3665. doi: 10.1074/jbc.M102950200. [DOI] [PubMed] [Google Scholar]
- 23.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, Neurath MF, Slattery T, Beach D, McClary J, Nagashima M, Morser J, Stern D, Schmidt AM. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 24.Rai V, Toure F, Chitayat S, Pei R, Song F, Li Q, Zhang J, Rosario R, Ramasamy R, Chazin WJ, Schmidt AM. Lysophosphatidic acid targets vascular and oncogenic pathways via RAGE signaling. The Journal of experimental medicine. 2012;209:2339–2350. doi: 10.1084/jem.20120873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park H, Adsit FG, Boyington JC. The 1.5 A crystal structure of human receptor for advanced glycation endproducts (RAGE) ectodomains reveals unique features determining ligand binding. J Biol Chem. 2010;285:40762–40770. doi: 10.1074/jbc.M110.169276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Q Rev Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 27.Bernado P. Effect of interdomain dynamics on the structure determination of modular proteins by small-angle scattering. European biophysics journal : EBJ. 2010;39:769–780. doi: 10.1007/s00249-009-0549-3. [DOI] [PubMed] [Google Scholar]
- 28.Heller WT. Influence of multiple well defined conformations on small-angle scattering of proteins in solution. Acta crystallographica. Section D, Biological crystallography. 2005;61:33–44. doi: 10.1107/S0907444904025855. [DOI] [PubMed] [Google Scholar]
- 29.Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Schlessinger J, Plotnikov AN, Ibrahimi OA, Eliseenkova AV, Yeh BK, Yayon A, Linhardt RJ, Mohammadi M. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol Cell. 2000;6:743–750. doi: 10.1016/s1097-2765(00)00073-3. [DOI] [PubMed] [Google Scholar]
- 31.Brozzo MS, Bjelic S, Kisko K, Schleier T, Leppanen VM, Alitalo K, Winkler FK, Ballmer-Hofer K. Thermodynamic and structural description of allosterically regulated VEGFR-2 dimerization. Blood. 2012;119:1781–1788. doi: 10.1182/blood-2011-11-390922. [DOI] [PubMed] [Google Scholar]
- 32.Xu D, Fuster MM, Lawrence R, Esko JD. Heparan sulfate regulates VEGF165- and VEGF121-mediated vascular hyperpermeability. J Biol Chem. 2011;286:737–745. doi: 10.1074/jbc.M110.177006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leahy DJ. A molecular view of anti-ErbB monoclonal antibody therapy. Cancer cell. 2008;13:291–293. doi: 10.1016/j.ccr.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 34.Schmiedel J, Blaukat A, Li S, Knochel T, Ferguson KM. Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization. Cancer cell. 2008;13:365–373. doi: 10.1016/j.ccr.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qing J, Du X, Chen Y, Chan P, Li H, Wu P, Marsters S, Stawicki S, Tien J, Totpal K, Ross S, Stinson S, Dornan D, French D, Wang QR, Stephan JP, Wu Y, Wiesmann C, Ashkenazi A. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J Clin Invest. 2009;119:1216–1229. doi: 10.1172/JCI38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.