Abstract
Cells sense and interpret chemical gradients, and respond by localized responses that lead to directed migration. An open microfluidic device (OMD) was developed to provide quantitative information on both the gradient and morphological changes that occurred as cells crawled through various microfabricated channels. This device overcame problems that many current devices have been plagued with, such as complicated cell loading, media evaporation and channel blockage by air bubbles. We used a micropipette to set up stable gradients formed by passive diffusion and thus avoided confounding cellular responses produced by shear forces. Two versions of the OMD are reported here: one device that has channels with widths of 6, 8, 10 and 12 μm, while the other has two large 100 μm channels to minimize cellular interaction with lateral walls. These experiments compared the migration rates and qualitative behavior of Dictyostelium discoideum cells responding to measurable cAMP and folic acid gradients in small and large channels. We report on the influence that polarity has on a cell’s ability to migrate when confined in a channel. Polarized cells that migrated to cAMP were significantly faster than the unpolarized cells that crawled toward folic acid. Unpolarized cells in wide channels often strayed off course, yet migrated faster than unpolarized cells in confined channels. Cells in channels farthest from the micropipette migrated through the channels at rates similar to cells in channels with higher concentrations, suggesting that cell speed was independent of mean concentration. Lastly, it was found that the polarized cells could easily change migration direction even when only the leading edge of the cell was exposed to a lateral gradient.
Introduction
Chemotaxis is a fascinating cellular behavior characterized by the movement of cells up or down a chemical concentration gradient. Directed cell migration plays a critical role in the cell movements that take place during embryo development and are necessary for the day-to-day function of the immune system.1,2 Chemotaxis also regulates many pathological conditions, including those that involve allergic inflammation and tumor metastasis.3–9 The social amoeba Dictyostelium discoideum has played an important role in revealing the mechanisms that control directional sensing and migration. These amoeboid cells are dependent on this process throughout their life cycle, exhibiting chemotaxis toward folic acid during the vegetative state and responding to cyclic adenosine monophosphate (cAMP) during their developmental cycle. Growing D. discoideum can feed on folic acid secreted by Gram-negative bacteria such as Escherichia coli and Klebsiella aerogenes. When these amoeboid-shaped cells are starved, they launch a developmental program during which cells aggregate and form a multicellular fruiting body. In the early stages of this process, starved cells begin to secrete and respond to cAMP, become elongated, and over the course of several hours polarize with a distinct front and rear. Vegetative Dictyostelium cells, on the other hand, remain very unpolarized, even while migrating up a folic acid concentration gradient. While the physiological conditions of these two cellular states are fundamentally different, the signaling mechanisms regulating their directed movement are thought to be very similar.
Chemotaxing cells can measure and respond to small changes in a chemical gradient, detecting a difference of ~5% receptor occupancy over the length of a cell.10 Studies from a number of research groups have characterized the localization of signaling molecules of Dictyostelium cells responding to cAMP gradients. The initial components of the cAMP circuit, such as serpentine receptors and heterotrimeric G-proteins, remain uniformly distributed along the membrane.11–13 Receptor occupancy and the heterotrimeric G-protein activation mimic the gradient of chemoattractant.10,12 The signaling molecules that regulate phosphoinositide turnover display early signs of a localized response during cAMP gradient sensing.14,15 The small G-protein Ras is critical at this stage and is enriched at the leading edge and promotes the localized activation of key chemotactic effectors, such as PI 3-kinase (PI3K).16,17 PI3K, in turn, is recruited to the anterior plasma membrane of the cell and interacts with its substrate, PI(4,5)P2 (PIP2) and gives rise to PI(3,4,5)P3 (PIP3).18,19 The tumor suppressor PTEN undergoes reciprocal movements and is redistributed to the lateral and posterior membrane of the cell.20,21 Moreover, actin polymerization and pseudopod extension preferentially occur in areas where the membrane has high PI3K activity. Positive feedback between the cytoskeleton and the regulators of PIP2 and PIP3 production at the rear and front of the cell, respectively, amplifies these morphological responses, even in the absence of a cAMP gradient.22,23 Interestingly, these signaling molecules also are highly regulated during cell division,24 phagocytosis25–27 and macropinocytosis.27,28 The regulation of the phosphoinositides appears to be a key signaling component for numerous cell processes that regulate changes in the cytoskeleton. These critical and diverse morphological changes are regulated by a system, which contains a great deal of redundancy, with no one component being required for the circuit to function adequately.29–31
Recent work has shed new light on the target of rapamycin complex 2 (TORC2), another part of the signaling pathway that is important for Dictyostelium chemotaxis.32,33 The TORC2 signaling complex is essential for activating mammalian protein kinase B homologs, PKBR1 and PKBA. PKBA contains a Pleckstrin homology (PH) binding domain that triggers the association of the protein with the plasma membrane when PIP3 levels rise. PKBR1 lacks the PH domain and instead has a myristoylation modification and is constitutively localized to the plasma membrane. Recently, the PKB substrates were identified and their phosphorylation states in response to cAMP were analyzed.33 PKBR1 activation was PIP3-independent and was activated through small G proteins. Recent results suggest that the PKB substrates are phosphorylated by folic acid stimulation in a manner similar to those seen with cAMP addition.34
While there is substantial scientific literature describing the mechanisms that regulate cAMP-mediated chemotaxis, the actual data describing the cellular movements that take place during folic acid-mediated cell migration are quite limited.30,35–37 Studies have documented the responses of cells to uniform increments of folate, but little is known about the morphological changes that take place when cells are migrating in a gradient of folic acid.24,38–43 Folic acid appears to bind to a putative seven-transmembrane receptor since cells lacking the heterotrimeric Ga4 subunit are unable to respond to folate.44 To aid in the investigation of the cellular properties of cells migrating in response to a gradient of folic acid, a protocol was developed for creating cells that show strong responses to the chemoattractant folic acid (K. Srinivasan, et al., manuscript in preparation). This method will also allow the direct comparison of chemotactic responses between vegetative cells and developed cells to their cognate chemoattractants. We developed a novel microfluidic device to aid in these experiments. This open microfluidic device (OMD) allowed the direct, simultaneous observation of both cell types in a highly reproducible and quantitative manner.
The standard method for visualizing chemotaxing eukaryotic cells, including D. discoideum, has been to perform a micropipette assay in an open chamber on an inverted microscope.15 William Pfeffer first used a pipette assay to study the chemotaxis of bacteria and flagellates in the late 1880s.45 Typically, today’s devices have a one-micron diameter opening at the tip of a micropipette, which is used to set up a diffusion gradient across a microscope coverslip. Cells then crawl on top of the glass and make their way toward the micropipette. Other methods used for chemotaxis assays make use of devices such as the Boyden chamber (or more recently the Transwell Assay),46 Zigmond chamber,47 or the Dunn chamber.48 While all three devices are capable of making linear gradients, these gradients change over time and cannot easily be controlled. In addition, only the Zigmond and Dunn chambers allow the viewing of cells during their migration.
With the advent of microfluidics, numerous groups are making use of devices that contain microchannels for generating chemical gradients. These channels are on the order of the physical scale of biological cells and have several applications for cell culture as well as single cell analysis.49–51 Such devices can provide cells with three-dimensional environments that may be more typical of what they experience in nature.52–55 Microfluidic cell culture environments have been touted as being uniquely suited for achieving a level of quantification and gradient control that is required for correlating observed cell responses with specific gradient characteristics. While their small size gives them certain advantages, active mixing gradient generators require constant fluid flow and can often fail to form reproducible gradients in circumstances where air bubbles or other obstructions intrude. Fluid flow subjects cells to confounding or damaging shear and drag forces, and can cause the gradient upstream to differ from the gradient downstream.56,57 It is possible to avoid shear forces by creating microfluidic devices that rely on passive diffusion to establish a gradient without flow, and these devices can readily include structural elements designed to restrict the motion of cells.58 Indeed, several groups have investigated a number of cell types using microfabricated channels that responded to passive gradients.59,60 However, closed microfluidic devices often require a complicated cell loading process or are difficult to keep in working condition and free of air bubbles. It would therefore be valuable to have a microfluidic device that allows easy cell loading and generates a stable passive gradient such as that described above with the micropipette, particularly for cells that migrate as rapidly as D. discoideum.
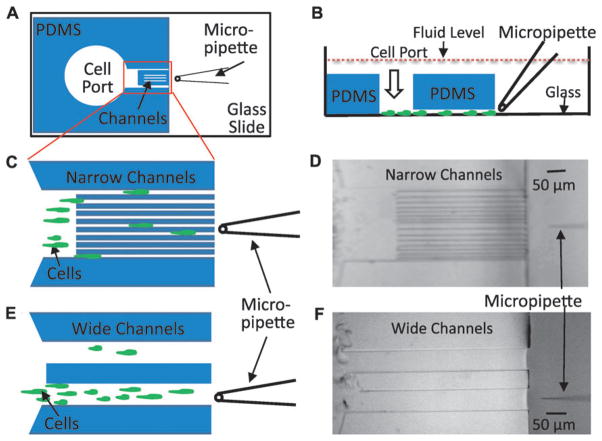
This manuscript describes the OMD, which combines a simple micropipette with the complex spatial configurations that can be achieved with a microfabricated polydimethylsiloxane (PDMS) structure. Cells were loaded into an open port on the inside of the PDMS device, which had a set of parallel channels that connected this port to the outside of the device (Fig. 1). The micropipette could be manipulated on the open outside region, and the length of the channels was short enough that gradient equilibration occurred quickly. We tested the OMD on D. discoideum cells to determine their ability to chemotax to cAMP and/or folic acid gradients. We wished to compare the relative speeds of cells chemotaxing to either folic acid or cAMP and used this device to study the contributions of cell polarity to the ability of cells to migrate directionally.
Fig. 1.
The open microfluidic device (OMD) for studying gradient sensing and cell migration. (A) Schematic diagram of an open microfluidic device viewed from the bottom. The PDMS block containing cell port and channels is plasma bonded to the glass coverslip of a one-well chamber. A micropipette filled with chemoattractant is placed in front of the narrow channel region. (B) A side view of the device in 1A depicting cells loaded through the cell port and crawling on the glass surface toward the micropipette. Note that entire device is submerged in buffer solution so that hydrostatic pressure is essentially equivalent on the inside and outside of the device. (C) Magnified cartoon of the narrow channel region with cells crawling between the different-sized channels toward the micropipette placed at the center of the device. In the actual device, there are 16 channels with each size represented 4 times. The channel widths are 6 μm, 6 μm, 8 μm, 8 μm, 10 μm, 10 μm, 12 μm, 12 μm, 6 μm, 6 μm, 8 μm, 8 μm, 10 μm, 10 μm, 12 μm and 12 μm. (D) A bright field image of the device with the narrow channel region described in 1C was obtained using a 10× objective. (E) Schematic of two wide channel regions with a cartoon that depicts cells crawling toward the micropipette. The width of each wide channel is 100 μm. The micropipette is placed at the center of the bottom wide channel. (F) Bright field image acquired using a 10× objective of the device with the wide channels.
The experimental setups described here were remarkably simple and highly reproducible. Cell loading was trivial (cells were pipetted in and sank to the bottom of an open chamber) and chemoattractant gradients were highly controllable by adjusting the pumping pressure of the micopipette. For each experiment, cells were lured from the “open” part of the device into the channels by a passive chemoattractant gradient. Quantitative studies performed within the same device analyzed the effects of different-sized channels on cell migration rates. A careful analysis of the chemoattractant gradients with fluorescent chemoattractants and dyes demonstrated the usefulness of having low ceiling heights in the channel region. The small optical z plane in which the chemoattractant resided and the greatly reduced out-of-focus fluorescence afforded by having a PDMS superstructure rather than open fluid permitted more reliable measurements of the gradients than would be possible with a simple micropipette system in an open dish. The effects of microfluidic channel widths on migration rates of unpolarized cells and of developed cells were observed in the presence of folic acid and cAMP, respectively. We speculated that the narrow channels might suppress lateral pseudopod extension and increase the probability that anterior pseudopods would extend toward the gradient of folic acid in the unpolarized cells. This, in turn, would increase the chemotactic speed relative to unpolarized cells in wide channels. We also reasoned that the polarized cells within the channels would migrate toward the source of attractant faster than the unpolarized cells in narrow and wide channels. The results suggest that the narrow channels do partially suppress pseudopod extension in unpolarized cells but do not increase the migration speed of the cell relative to cells in wide channels. Polarized cells migrate significantly faster than unpolarized cells toward their respective chemoattractants. These elongated cells are also highly sensitive to chemoattractants at their anterior end. Our experimental results suggest that the anterior of the cell, alone, has the capacity to sense the gradient and direct the cell up the concentration gradient.
Results
Device design and layout
Open-faced channels are microfabricated in PDMS and then plasma bonded to a glass coverslip to form the closed channels (Fig. 1A and B). The gradient is formed by a micropipette attached to a micromanipulator and pump. The glass surface forms the bottom of the device where the cells are attached and viewed. The PDMS structure defines the ceiling and the walls of the device. Two devices were created (see Fig. 1C–F). One scheme had two very wide channels (100 μm) while the other device was created with a tandem repeat of four different widths (6, 8, 10 and 12 μm, respectively). The 100 μm width simulated a region where cells could migrate without the confinement of the narrow channels. The ceiling height of both devices was approximately 15 μm. The height of the PDMS (2 mm) was less than that of the fluid depth in the chamber. This design feature was key for the prevention of convective flow due to differences in hydrostatic pressure between the inside and outside of the device. Cells, loaded by gentle micropipetting into the cell port, chemotaxed through the channels to the other side of the device and ultimately to the chemoattractant-filled micropipette.
Gradient measurements
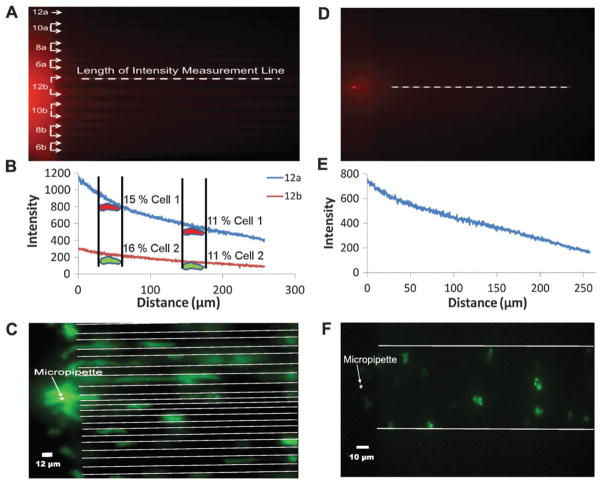
A number of studies have measured the relationship between the chemoattractant gradient and the responses of cells to these stimuli.61–64 These “input/output” experiments have demonstrated that cells can sense very shallow gradients and have the ability to amplify internal responses in order to move directionally. The OMD provided a more precisely measured gradient profile when compared to those measured using a micropipette alone since the gradients that the cell encountered in the OMD were measured directly without contributions from out-of-focus fluorescence. In an open dish, the observed fluorescence represents a projection into the optical plane of out-of-focus light from a radially-decaying spherical chemoattractant distribution. Hence in the OMD the gradients were approximately linear rather than 1/r2.65 A micropipette supplying Cy3-cAMP resulted in a stable gradient in both the narrow and wide channel regions within the microfluidic device. A pumping pressure of 50 hecto Pascals (hPa) was applied to the micropipette in these experiments. Using this pressure, a gradient was clearly visible (Fig. 2A and D). These gradients were measured in the various channels. Concentration profiles had similar slopes with different mean concentrations (Fig. 2B and D). As would be expected from the radial distribution of label around the micropipette tip, the highest fluorescence intensity was measured in the 12 μm channel nearest to the micropipette and gradually decreased for those channels farther away (see Fig. S1‡). This made the OMD useful in that it provided unique experimental conditions that varied in each channel. To demonstrate the differences in gradient profiles that cells might encounter at various points along two channels, we measured the relative changes in pixel intensities (Fig. 2B) that 30 μm long cells might encounter in a typical gradient like that depicted in Fig. 2A. The change from front to rear of both cells early on was similar (about 11%, cells on right) and later (15% for cell 1 and 16% for cell 2, both on left). The mean concentration eliciting a response, however, was dramatically different (several fold) in the two channels. Yet in our experiments, we detected little difference in speed between cells in channels near and far from the micropipette, suggesting that cells respond to the relative concentration gradient and not the absolute concentration (see Fig. S2 and Movie S1‡).
Fig. 2.
Gradients and cells in the OMD. (A) A Cy3-cAMP image of the narrow channel configuration with pumping pressure of 50 hPa. The micropipette was placed in the center of the channel region. The number corresponds to the size of the channel, in microns. For instance, channels labeled 12b are the two middle channels and are both 12 μm wide. The dotted white line drawn across the top 12b channel is representative of where pixel values were obtained for the two channels displayed in 2B. (B) Pixel intensity values as a function of distance for channels 12a and the top 12b are shown in 2A. The intensities were measured along the length of the representative dotted white line as shown in the center of 12b in 2A. Average intensities for the other channels are shown in Fig. S1.‡ We measured the relative change in chemoattractant a cell might see if it were at two different points in two different channels. The hypothetical change from front to rear of both cells early on was similar (about 11%, cells on right) and later (15% for cell 1 and 16% for cell 2, both on left). The mean concentration eliciting a response, however, was dramatically different (several fold) in the two channels, suggesting that the mean concentration has little if any effect on migration speeds. (C) Unpolarized and polarized cells crawling toward a micropipette containing both cAMP and FA in the narrow channel configuration. (D) Cy3-cAMP image of the wide (100 μm) channel with pumping pressure of 50 hPa. The micropipette was placed in the center of the wide channel. The white line drawn along the center depicts where the intensities were obtained. (E) Pixel intensities as a function of distance for the 100 μm wide channel region shown in 2D. The intensities were measured along the length of the white line. Five lines were drawn along the wide channel and the average intensities were used for the plot. (F) Unpolarized cells crawling toward a micropipette containing FA in the 100 μm wide channel.
Migration speeds of polarized cells versus unpolarized cells within channels
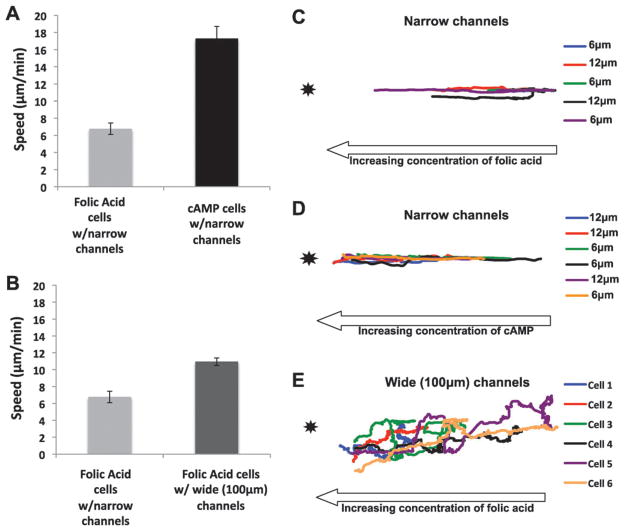
Chemoattractant delivery using a micropipette pressure of 50 hPa created a concentration gradient that can cause cells to migrate rapidly in both narrow and wide channels (Fig. 2C and F, and see Movie S2 and S3‡). This delivery setting, in conjunction with the OMD channel geometry, created controlled experimental conditions where cells could be “raced” against each other. For these assays, wild-type cells expressing PH-GFP, a biosensor for PI(3,4,5)P3, and LimE, a marker for actin polymerization, were starved and developed for 7 h and raced against wild-type, unpolarized cells expressing PH-GFP alone.66,67 The developed cells were polarized and responded to cAMP, while the unpolarized cells were grown in the presence of bacteria and responded to the chemoattractant folic acid. Fluorescent cells were mixed and loaded into the cell chamber within the OMD (Fig. 1A). A single micropipette was loaded with a 1: 1 ratio of 10 μM cAMP and 10 μM folic acid and was lowered on one side of the channels as depicted in Fig. 1C. Individual cells were tracked and their migration speed determined within the various channels (Fig. 3A). The polarized cells migrated to cAMP at 17 μm min−1 (±1 μm min−1) while the vegetative cells moved toward folic acid at 7 μm min−1 (±0.6 μm min−1). Thus, the polarized cells migrated significantly faster than the vegetative cells in identical gradients of their respective chemoattractants.
Fig. 3.
Analysis of cell migration in the OMD. (A) Migration speeds of unpolarized cells within narrow channels are significantly slower than polarized cells within narrow channels toward the chemoattractant source. The migration speed (μm min−1) was measured from a mixture of unpolarized and polarized cells, as described in the materials and methods, toward a micropipette loaded with 10 μM cAMP and folic acid. Ten cells from three different days were observed. The error bars represent the standard error of the mean where n = 30. (B) Unpolarized cells in the presence of narrow channels migrate slower than folic acid cells in the 100 μm wide channels toward folic acid. The migration speed (μm min−1) was measured for the vegetative cells in a device containing narrow channels and a device containing 100 μm wide channels toward 10 μM folic acid. Thirty cells were analyzed, taking ten cells each from three different days. The error bars represent the standard error of the mean where n = 30. (C) Unpolarized cells within the narrow channels migrate in a relatively straight line toward the attractant source. The unpolarized cells were tracked from the videos analyzed in Fig. 3A. Cells from the two innermost 6 and 12 μm channels in the device were analyzed. Five cells were analyzed from three different days. (D) Polarized cells within the narrow channels migrate in a straight line toward the attractant source. The polarized cells were tracked from the same videos analyzed in Fig. 3A. Cells from the two innermost 6 and 12 μm channels in the device were tracked. Six cells were analyzed from three different days. (E) Unpolarized cells in the wide channels migrate in a biased random walk toward the attractant source. The unpolarized cells were tracked from the same videos analyzed in Fig. 3B. Six cells were analyzed from two different days.
Migration speeds of unpolarized cells within and in the absence of channels
Unpolarized cells extend random pseudopods quite often even when migrating up a concentration gradient of folic acid. To determine whether confinement in the narrow channels would affect migration speeds, unpolarized cells were assayed as described above toward folic acid in the presence of narrow channels (Fig. 1C) and in wide channels (Fig. 1E). Cells in the wide channels moved at an average speed of 11 μm min−1 (±0.4 μm min−1) while those with narrow channels migrated at 7 μm min−1 (±0.6 μm min−1) (Fig. 3B). Surprisingly, these data indicate that the confinement of the narrow channels did not increase the migration speed of the unpolarized cells.
We also investigated whether the size of a narrow channel (6 μm versus 12 μm channels) would have an effect on the cell migration speed of unpolarized cells. The migration speeds of the unpolarized cell in 6 μm and 12 μm channels were measured (Fig. S2A‡). Unpolarized cells in the 6 μm and 12 μm channels had an average speed of 8.82 μm min−1 (±0.74 μm min−1) and 8.18 μm min−1 (±0.94 μm min−1), respectively. A time lapse video of polarized cells was also recorded. Cells in each of the channels exhibited similar relative speeds during an 18 min experiment (Fig. S2B–D and Movie S1‡). These data suggested that the size of the channel did not improve or impede relative migration speeds of the cells observed. They also provided further evidence that cells respond to the relative change in concentration of chemoattractant since the outer channels would have several fold lower mean concentrations (see Fig. 2B).
The migration path of cells under three different conditions was determined by cell tracking. Polarized cells moving toward cAMP and unpolarized cells migrating toward folic acid were tracked when confined within the middle 12 μm and 6 μm channels (Fig. 3C and D). These two channels were chosen since the mean concentration and slope most closely resembled those of the other device, which had wide channels. The polarized cells migrated in a relatively straight line (Fig. 3D). The unpolarized cells in narrow channels exhibited a migration path with a greater number of lateral movements and more path reversals when compared to the polarized cells migrating to cAMP (Fig. 3C). Lastly, the unpolarized cells in the absence of channels meandered toward the pipette rather than taking a direct route, as described above for both the polarized cells and the vegetative cells within the channels (Fig. 3E). Cells in the wide channels clearly strayed off course, yet they migrated faster than cells confined to narrow channels.
Spatial gradient sensing occurs primarily at the leading edge
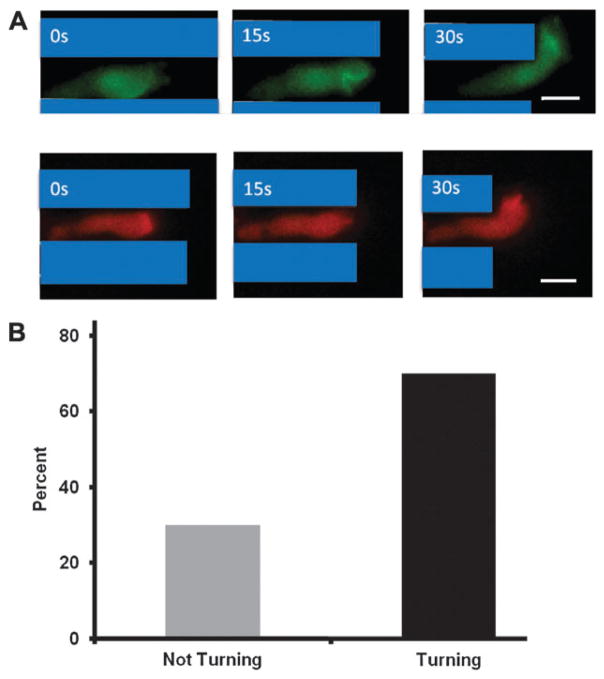
Polarized cells moved rapidly toward the micropipette while in the channel and also once they exited the channel on the side facing the micropipette. In fact, an interesting observation was made that demonstrated that cells could sense the change in the cAMP profile before fully exiting the channel (Fig. 4A). We determined the percentage of cells that turned toward the chemoattractant source while less than 50% of the cell was outside the channel. The thirty cells observed were divided into two classes: (1) cells that turned directly toward the attractant loaded micropipette with less than 50% of their cell bodies outside of the channels, and (2) cells that turned away from, or did not turn directly toward the micropipette with less than 50% of their cell bodies outside of the channels. A stunning 70% of cells meeting the first criteria turned directly toward the cAMP-loaded micropipette (Fig. 4B). Additionally, PH-GFP and LimE-RFP localized to the fraction of the leading edge that was facing the cAMP-loaded micropipette (Fig. 4A). These results demonstrated that the leading edge of the cell could sense a change in the concentration gradient across the midline of the cell without the help of the rear of the cell.
Fig. 4.
The turning behavior of cells as they exit the narrow channels. (A) The leading edge of a cell has the capacity to sense a cAMP gradient across the midline and turn toward the attractant source. Depicted is a representative cell that exhibited turning behavior toward the cAMP-loaded micropipette. This cell was expressing PH-GFP (green) and LimE-RFP (red). The snapshots in A, B and C were three consecutive frames 15 s apart. Scale bar is 10 μm. (B) 70% of polarized cells turned their leading edge toward the micropipette as the cells exited the channels. Thirty cells (10 cells from 3 different days) were analyzed. For a positive turning result to be scored, the cells were not in contact with other cells and had at least 50% or less of the cell body outside of the channel.
Materials and methods
Media and buffers
HL-5 and SM Agar were purchased from Formedium. For HL-5 liquid media; 22 g of Formedium HL-5 powder, 10 g of dextrose were added to 1 L of distilled H20 and then autoclaved. For SM media; 41.7 g of Formedium SM agar powder was added to 1 L of distilled H2O and autoclaved. Development buffer (DB) contained 5 mM Na2HPO4, 5 mM KH2PO4, 1 mM CaCl2, and 2 mM MgCl2.
Strains
Dictyostelium discoideum wild-type strain AX2 expressing GFP-tagged PH-CRAC (PH-GFP) was used for folic acid chemotaxing cells.15 RFP-tagged LimE (LimE-RFP)66 and PH-GFP expressing AX2 cells were used for cAMP chemotaxis. Klebsiella aerogenes were used as a food source for D. discoideum on SM agar plates.
Folic acid preparation
Folic acid powder was purchased from Fisher. Folic acid was prepared by taking 5.5 mg of folic acid and diluting it in 12.5 mL of deionized water and 13.5 mL of 2 N NaOH to make a 1.25 mM stock solution of folic acid. For folic acid chemotaxis, the 1.25 mM stock solution was diluted 1: 125 in deionized water to make a 10 μM final concentration.
cAMP Preparation
cAMP was purchased from Sigma. A 10 mM stock solution of cAMP was prepared in ddH2O. For cAMP development the 10 mM stock solution was diluted in 30 mL of development buffer (DB) to make a 2.5 μM cAMP solution. For cAMP chemotaxis, the 10 mM stock solution was diluted 1: 1000 in 1 mL of DB to make a 10 μM final concentration.
Vegetative cells
500 μL of 2 × 106 cells ml−1 PH-GFP expressing AX2 cells were mixed with ~1 × 109 cells mL−1 K. aerogenes bacteria from an SM agar plate and the mixture was plated on an SM agar plate. The mixture of cells was allowed to incubate at room temperature overnight. For experiments, the mixture of cells from the SM plate was washed several times in development buffer (DB) to wash off the bacteria. After washing, the D. discoideum cells were resuspended in 1 mL of DB.
Developing cells with cAMP
5 mL of 2 × 107 cells mL−1 of PH-GFP/LimE-RFP expressing AX2 cells were pulsed with 100 μL of 2.5 μM cAMP every 6 min for 7 h with constant agitation. An aliquot of cells was removed and 5 mM EDTA was added to the cells. The cells were then washed twice with DB to remove the EDTA.
Mixed folic acid (FA) and cAMP cells
75 μL of FA cells and 25 μL of cAMP cells were mixed together in the presence of 3 μM caffeine. Six μL of the cell mixture was added to the device.
Device design
The OMD was designed using the AutoCAD software package (Autodesk, San Rafael, CA). One design contained an array of parallel channels with widths of 6, 8, 10 and 12 μm, respectively. These channels are 50 μm away from the cell port. Another design contained only wide 100 μm channels in order to minimize lateral cell movement constraints. The large channel is directly coupled to the cell loading port.
Device fabrication processes
The device design was transferred to a chrome mask using a commercial printing process (Advance Reproductions, North Andover, MA). The chrome mask was then used to create a three-dimensional mold using photolithography techniques. In short, a silicon wafer was coated with the negative photoresist polymer SU-8 2010 (Microchem Corp., Newton, MA) which cross-links when exposed to a UV light source. The thickness of the SU-8 coating was controlled by spinning the wafer at the speed and duration recommended by the resist manufacture in order to create the desired height profile of 15 μm. The chrome mask was then placed on top of the coated wafer and exposed to a UV light source (Exfo Novacure 2100 UV). This caused the exposed regions on the resist to crosslink, which, in turn, were the only features that remained on the silicon wafer after a solvent solution of SU-8 developer removed all of the unexposed pre-polymer. The final silicon wafer mold thus consisted of a pattern of 15-micron-tall raised structures composed of solidified SU-8. This served as the mold to be used in the microfabrication process. PDMS (Sylgard 184, Dow Corning, Midland, MI) was poured onto the SU-8 and silicon mold and degassed to get rid of air bubbles, and was cured in an oven at a temperature of 65 °C overnight.68 The cured flexible PDMS device was peeled from the silicon wafer, and then the cell loading port was punched out using a hollow punch and the front of the device was cut using a sharp cutter to expose the channels that face the micropipette. A one-well chamber with a glass bottom (Lab-Tek, Rochester, NY), along with the PDMS slab containing the channels and the cell port, was placed in a plasma cleaner (Harrick Plasma © Plasma Cleaner, Ithaca, NY) for approximately 30–40 s. After removal from the plasma chamber, the side of the PDMS slab with the channel patterns was pressed onto the bottom of the one-well chamber where it formed a permanent seal. Immediately after the bonding process, the device was wetted using developmental buffer (DB) in order to preserve the hydrophilic nature of oxidized PDMS.
Gradient characterization
The gradient characterization was done using 1.0 μM of Cy3-cAMP, which was loaded into an Eppendorff femtotip and released in front of the channel structures. Fluorescent images were taken of the region. The intensity profile of the Cy3 fluorescence was measured using Image J’s Plot profile feature (U. S. National Institutes of Health, Bethesda, MD) and the intensity values were plotted as a function of distance using Microsoft Excel (Redmond, WA).
Gradient in the channels
To measure the gradient in the narrow channels, 1.0 μM Cy3-cAMP was pumped at 50 hPa in front of the narrow channels. The intensity was calculated using SlideBook software after subtracting the background intensity. The graph shows designations of “a” and “b” after the numbers. “a” stands for above and “b” stands for below the micropipette. For example, 6 “a” represents the average intensity values of the two 6 μm channels that are at the top of the image. The intensities were measured along the same distance for each channel (see Fig. 2B and Fig. S1‡). The change in Cy3-cAMP across cells in Fig. 2B was calculated as: Percent difference = Absolute value of (X1 − X2/(X1 + X2)/2) × 100.
Gradient stability over time
While typical chemotaxis assays generally only last 20–30 min, we did provide data of cells chemotaxing for several hours. The time limitation in these experiments is typically dependent on the clogging of the micropipette or the channels by the cells. To measure the stability of the gradient, 10 μM fluorescein (FITC) was pumped at 50 hPa in front of the wide channel region. FITC has a mass similar to cAMP (332.306 g mol−1 versus 329.06 g mol−1, respectively). Time lapse images were captured every 5 min for over two hours. The gradient was measured by drawing a line and measuring the pixel intensities in the center of the channel for all the time points using software provided by SlideBook. These data are provided as Fig. S3.‡
Chemotaxis assay using the open microfluidic device
The cells were added to the cell port using a P-20 micropipette (Fig. 1A). As the cells settled to the bottom of the device, a micromanipulator (InjectMan NI2, Eppendorf) was used to lower a micropipette (Femtotip, Eppendorf) filled with chemoattractant at the front of the device. The chemoattractant gradient was elicited from the micropipette using a pump (Eppendorf FemtoJet) at a pressure of 50 hPa. The diffusion coefficients of cAMP and folic acid are very similar to each other (2.7 ± 0.2 × 10−6 cm2 s−1 and 1.94 × 10−6 cm2 s−1, respectively).69,70 The micropipette was in position for at least 10 min before image capture was initiated. Cells were observed using a 40× PlanNeofluar 1.3 NA wide-field oil immersion objective.
Imaging
All imaging was conducted on a Marianas™ Workstation equipped with a Cool Snap CCD camera and an Extended QE, high-speed cooled CCD camera (Cascade II 512). We used an inverted, wide-field epifluorescence microscope (Zeiss Axio Observer Z1, Thornwood, NY). Images were taken using SlideBook software (Intelligent Imaging Innovations, Inc., Denver, CO). Time lapse images were taken every 15 s for approximately 2 h.
Cell speed and tracking
Cells were tracked frame-by-frame using the manual-tracking feature of Image J. 30 cells each were tracked for folic acid and cAMP cells in channels, and 16 folic acid cells were tracked in the wide channel. For cell turning, 30 cAMP chemotaxing cells were tracked in ImageJ.
Discussion
The OMD apparatus was created for studying the migration of cells in response to a passive chemical gradient. Two- and three-dimensional patterns within this device can be designed to study various properties of migrating cells to further our understanding of gradient sensing and the mechanical properties that regulate cell motility up or down a concentration gradient. While other groups have shown the migration of cells through channels in response to a passive gradient,59,60 our device produces stable gradients by using a micropipette. In addition, the cell loading and gradient generation in the OMD are completely independent of one another. Cells can be readily introduced into this system through an open well. The micropipette is a high-impedance controlled source of chemoattractant whose instantaneous position and flow rate can be readily controlled by the experimenter, whereas the devices that utilize reservoirs and multiple channels are generally subject to convective flows driven by small differences in hydrostatic pressure between the various reservoirs. The device by Butler et al.60 avoids the convection problem by using closed channels, but this eliminates the possibility of changing the local gradient, for example, at the opening to the migration channel, and limits the ability to load cells into the channel from the low side of the gradient. While these other devices are not capable of the rapid changes in gradients or concentrations that we can achieve, their geometries may be better suited for the study of cells that move or grow slowly. The OMD is ideal for rapidly moving cells, and is not subject to the hydrodynamic forces associated with flow gradient mixers.71,72 The OMD generated gradients of varying profiles and mean concentrations and quantified the responses of Dictyostelium amoebae toward folic acid or cAMP. For our biological assays, a mixed population of cells, as well as a combination of the chemoattractants cAMP and folic acid, was used to provide a direct comparison of migration rates while cells were under identical experimental conditions.
The micropipette was positioned at the very center of the device and produced a gradient in all of the channels. The concentration of chemoattractant entering the channels nearest the micropipette had a mean concentration that was higher than those farther away. While the absolute amount of chemoattractant differed between the channels, the relative change in concentration that a cell would experience varied very little, and the cells migrated with similar speeds. The experimental design allowed us to directly test the effects of cell confinement on migration rates, since the channels were alternated and similarly sized channels had both low and high mean concentrations. By varying the micropipette pumping pressure, different profiles were generated and their effects on chemotaxis were observed. The OMD was used to measure the migration speed of unpolarized vegetative cells and of polarized developed cells. We hypothesized that the narrow channels within the OMD would suppress lateral pseudopod extension in the unpolarized cells and as a result cause the cells to migrate toward the source of attractant faster than unpolarized cells in the wide channels. This was not observed. The migration speed of the unpolarized cells in the narrow channels was significantly less than that seen for both the unpolarized cells in the wide channels and the polarized cells responding to cAMP (Fig. 3A and B). We speculate that this result occurred because pseudopods from unpolarized cells were extinguished if they encountered a wall in the channel. Since the cells appeared to bias their pseudopods in the direction of the concentration gradient, only those pseudopods that randomly formed directly up the gradient, and in the center of the channel, lead to motility. This resulted in an overall decrease in motility to folic acid. This explanation was further supported when the cell migration tracks were measured for each of the cell conditions. The vegetative and cAMP-developed cells within the channels migrated in a relatively straight line (Fig. 3C and D). However, the vegetative cells in the absence of channels migrated in a much more random fashion (Fig. 3E).
The gradient sensing system regulates the spatiotemporal localization of signaling molecules at the front and rear of the cells.14 With the OMD, the micropipette formed a gradient via radial diffusion. As a consequence, the cells were exposed to a gradient vector that was parallel to the channels within the channels and a gradient vector that was perpendicular to the channels when the cells exit the channels. Analysis of polarized cells exiting the channels demonstrated that the leading edge of the cell was capable, by itself, of sensing a gradient (Fig. 4A and B). In addition, the sensing mechanism localized PI3K activity, leading to higher PIP3 levels and ultimately actin polymerization as shown by cells expressing PH-GFP and LimE-RFP, respectively. This demonstrated that the front of the cell could respond to the gradient independent of ligand binding at the rear of the cell. This is a unique finding since most reports on gradient sensing focus on the differences in attractant concentration and signaling responses between the front and the back of the cell.14,35,64,73 Previous studies in Dictyostelium have also shown that cells are more sensitive at the front than the rear22,67 and that immobile cells can respond to multiple stimuli simultaneously.64
Cells in a gradient will display stable, localized responses, but when given a global stimulus will show a uniform response and adapt.15 Numerous models, many containing feedback mechanisms, have been proposed to help explain these results and the ability of cells to amplify their response when in a chemoattractant gradient.22,74,75 One scheme, the Local Excitation and Global Inhibition (LEGI) model, provides a potential mechanism for these spatial temporal sensing responses.22,64,67 The LEGI model has two main components, a fast excitation process, and a slower global inhibitor. In a uniform stimulus of chemoattractant the fast local excitation processes increase proportionally to receptor occupancy. The inhibitory processes increase slowly as a function of the average ligand binding and eventually turn off the response. In a steady state gradient, the excitatory processes along the length of the cell are higher in the front of the cell than the back, while the inhibitory processes are proportional to the mean receptor occupancy and exceed excitatory signals in the rear.14,64,76 This will allow a persistent directional response toward a spatial gradient.
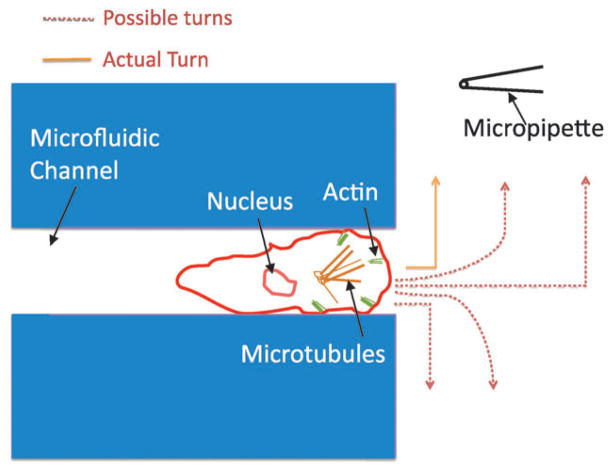
The results reported here demonstrate that cells can abruptly turn toward the chemoattractant source as they are exiting the channels. This suggests that the LEGI mechanism is capable of providing localized responses by measuring the gradient difference across the leading edge of the cell. This is consistent with previous data which showed that a cell can elicit amplified responses at opposing ends.64 In this report, however, it is important to note that the rear of the cell does not see a change in the gradient concentration across the short axis of the cell. This is quite different from previously reported micropipette assays where cells turn and chase the change in gradient concentration. The turning behavior reported here is depicted in Fig. 5, where the cartoon also displays several paths that the cell could have taken. The majority of cells observed in these experiments made an approximate 90° turn toward the micropipette upon exiting the channel. These results suggest that the leading edge of a cell, in addition to being more responsive to incremental changes in chemoattractant when exposed to a uniform stimulus, is also very sensitive to changes in the gradient concentration.
Fig. 5.
Turning strategy of cells exiting the channel. Cells that exited the channels had a number of possible directions to follow as they made their way toward the micropipette. Cells could have continued in a straight line, turned in the correct direction, or migrated 180° in the opposite direction. They could have also made more shallow turns in the correct or incorrect direction. They mostly migrated directly toward the micropipette, even while their trailing edge was still not exposed to the gradient that was perpendicular to the cell. Since the cells did not make a shallow turn, as might be expected if you averaged the proportion of the cell’s outer surface that is normally exposed to the chemoattractant, the rear of the cell contributed little or no spatial information to the turning profile of the cell.
In addition to similar questions outlined in this report using the model system Dictyostelium, the open microfluidic device could be used for addressing many biological questions related to directional sensing and cell migration in a wide range of prokaryotic and eukaryotic organisms. The OMD is very easy to use, with setup times on the order of a few minutes. Cells were loaded into an open chamber and allowed to settle. During this time the micropipette was loaded and positioned. Since there were no valves, flow or tubing within the PDMS device, there was very little that could go wrong, and there were no leaks, air bubbles or evaporation issues. The device allowed cells to chemotax toward a passive gradient without encountering shear forces. Future chemotaxis experiments can custom design the PDMS channels and alter the shape and size of the channels to observe the migratory properties of cells when confined by a number of different surfaces. These devices, combined with imaging techniques such as total internal reflection fluorescence microscopy (TIRFM), will help elucidate the basic principles by which cells orient and move directionally.
Supplementary Material
Insight, innovation, integration.
Gradient sensing and cell migration are processes critical for cell movements during embryonic development, the immune response and many disease states. Experimental platforms are needed to gain insight into the mechanisms that control directed cell migration. A quantitative technique is described here that exposes microfluidic channels to a passive gradient formed from a micropipette. This open microfluidic device was used to investigate the migration paths of cells with different polarized morphologies. This simple setup enabled the production of stable, measurable chemoattractant gradients within channels of varying sizes. Qualitative and quantitative measurements were made of both the chemical gradient and migration paths taken by Dictyostelium cells and provided fundamental knowledge about the ability of cells to sense passive gradients in confined channels.
Acknowledgments
This research has been supported in part by the Vanderbilt Institute for Integrative Biosystems Research and Education. Thanks to Allison Price for editing the manuscript. Thanks to Dictybase for providing cell lines and A. Muller-Taubenberger for providing LimE-RFP construct. Funding for GW and CJ provided by NIHGM080370.
Footnotes
Published as part of a themed issue on Mechanisms of Directed Cell Migration: Guest Editors David Beebe and Anna Huttenlocher.
Electronic supplementary information (ESI) available: Fig. S1–S3, Movie S1–S3. See DOI: 10.1039/c0ib00110d
References
- 1.Thelen M. Nat Immunol. 2001;2:129–134. doi: 10.1038/84224. [DOI] [PubMed] [Google Scholar]
- 2.Surmi BK, Hasty AH. Vasc Pharmacol. 2010;52:27–36. doi: 10.1016/j.vph.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lazennec G, Richmond A. Trends Mol Med. 2010;16:133–144. doi: 10.1016/j.molmed.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Paepe B, Creus KK, De Bleecker JL. Curr Opin Rheumatol. 2009;21:610–616. doi: 10.1097/BOR.0b013e3283317b31. [DOI] [PubMed] [Google Scholar]
- 5.Hansson GK. J Thromb Haemostasis. 2009;7(Suppl 1):328–331. doi: 10.1111/j.1538-7836.2009.03416.x. [DOI] [PubMed] [Google Scholar]
- 6.Johnson Z, Power CA, Weiss C, Rintelen F, Ji H, Ruckle T, Camps M, Wells TN, Schwarz MK, Proudfoot AE, Rommel C. Biochem Soc Trans. 2004;32:366–377. doi: 10.1042/bst0320366. [DOI] [PubMed] [Google Scholar]
- 7.Wu X, Lee VC, Chevalier E, Hwang ST. Curr Pharm Des. 2009;15:742–757. doi: 10.2174/138161209787582165. [DOI] [PubMed] [Google Scholar]
- 8.Dorsam RT, Gutkind JS. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 9.Condeelis J, Singer RH, Segall JE. Annu Rev Cell Dev Biol. 2005;21:695–718. doi: 10.1146/annurev.cellbio.21.122303.120306. [DOI] [PubMed] [Google Scholar]
- 10.Ueda M, Sako Y, Tanaka T, Devreotes P, Yanagida T. Science. 2001;294:864–867. doi: 10.1126/science.1063951. [DOI] [PubMed] [Google Scholar]
- 11.Jin T, Zhang N, Long Y, Parent CA, Devreotes PN. Science. 2000;287:1034–1036. doi: 10.1126/science.287.5455.1034. [DOI] [PubMed] [Google Scholar]
- 12.Janetopoulos C, Jin T, Devreotes P. Science. 2001;291:2408–2411. doi: 10.1126/science.1055835. [DOI] [PubMed] [Google Scholar]
- 13.Xiao Z, Zhang N, Murphy DB, Devreotes PN. J Cell Biol. 1997;139:365–374. doi: 10.1083/jcb.139.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janetopoulos C, Firtel RA. FEBS Lett. 2008;582:2075–2085. doi: 10.1016/j.febslet.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Charest PG, Firtel RA. Curr Biol. 2008;18:1587–1593. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kortholt A, van Haastert PJ. Cell Signalling. 2008;20:1415–1422. doi: 10.1016/j.cellsig.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 18.Merlot S, Firtel RA. J Cell Sci. 2003;116:3471–3478. doi: 10.1242/jcs.00703. [DOI] [PubMed] [Google Scholar]
- 19.Huang YE, Iijima M, Parent CA, Funamoto S, Firtel RA, Devreotes P. Mol Biol Cell. 2003;14:1913–1922. doi: 10.1091/mbc.E02-10-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iijima M, Devreotes P. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 21.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 22.Devreotes P, Janetopoulos C. J Biol Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, Meili R, Devreotes PN, Firtel RA. J Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janetopoulos C, Borleis J, Vazquez F, Iijima M, Devreotes P. Dev Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez-Paris J, Grove B, Buczynski G, Cardelli J. Mol Biol Cell. 1996;7:3448–3448. doi: 10.1091/mbc.7.10.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rupper A, Rodriguez-Paris J, Grove B, Cardelli J. Mol Biol Cell. 1998;9:2685. [Google Scholar]
- 27.Cardelli J. Traffic. 2001;2:311–320. doi: 10.1034/j.1600-0854.2001.002005311.x. [DOI] [PubMed] [Google Scholar]
- 28.Rupper A, Lee K, Knecht D, Cardelli J. Mol Biol Cell. 2001;12:2813–2824. doi: 10.1091/mbc.12.9.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoeller O, Kay RR. Curr Biol. 2007;17:813–817. doi: 10.1016/j.cub.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Veltman DM, Keizer-Gunnik I, Van Haastert PJ. J Cell Biol. 2008;180:747–753. doi: 10.1083/jcb.200709180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Insall R, Andrew N. Curr Opin Microbiol. 2007;10:578–581. doi: 10.1016/j.mib.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Lee S, Comer FI, Sasaki A, McLeod IX, Duong Y, Okumura K, Yates JR, Parent CA, Firtel RA. Mol Biol Cell. 2005;16:4572–4583. doi: 10.1091/mbc.E05-04-0342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamimura Y, Xiong Y, Iglesias PA, Hoeller O, Bolourani P, Devreotes PN. Curr Biol. 2008;18:1034–1043. doi: 10.1016/j.cub.2008.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao XH, Buggey J, Kimmel AR. J Cell Sci. 2010;123:983–992. doi: 10.1242/jcs.064022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korohoda W, Madeja Z, Sroka J. Cell Motil Cytoskeleton. 2002;53:1–25. doi: 10.1002/cm.10052. [DOI] [PubMed] [Google Scholar]
- 36.Laevsky G, Knecht DA. Biotechniques. 2001;31:1140–1142. 1144, 1146–1149. doi: 10.2144/01315rr03. [DOI] [PubMed] [Google Scholar]
- 37.Sameshima M, Imai Y, Hashimoto Y. Cell Motil Cytoskeleton. 1988;9:111–116. doi: 10.1002/cm.970090203. [DOI] [PubMed] [Google Scholar]
- 38.Kuwayama H, Ishida S, Van Haastert PJ. J Cell Biol. 1993;123:1453–1462. doi: 10.1083/jcb.123.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malchow D, Bohme R, Rahmsdorf HJ. Eur J Biochem. 2005;117:213–218. doi: 10.1111/j.1432-1033.1981.tb06324.x. [DOI] [PubMed] [Google Scholar]
- 40.de Wit RJ, Bulgakov R, Bominaar TA, Rinke de Wit TF. Biochim Biophys Acta, Mol Cell Res. 1987;930:1–9. doi: 10.1016/0167-4889(87)90149-2. [DOI] [PubMed] [Google Scholar]
- 41.Segall JE. J Muscle Res Cell Motil. 1988;9:481–490. doi: 10.1007/BF01738753. [DOI] [PubMed] [Google Scholar]
- 42.Rifkin JL, Goldberg RR. Cell Motil Cytoskeleton. 2006;63:1–5. doi: 10.1002/cm.20096. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y, Liu J, Segall JE. J Cell Sci. 1998;111(Pt 3):373–383. doi: 10.1242/jcs.111.3.373. [DOI] [PubMed] [Google Scholar]
- 44.Hadwiger JA, Lee S, Firtel RA. Proc Natl Acad Sci U S A. 1994;91:10566–10570. doi: 10.1073/pnas.91.22.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bunning E. Ahead of His Time: Wilhelm Pfeffer. Carlton University Press; Ottawa, Ontario, Canada: 1989. [Google Scholar]
- 46.Boyden S. J Exp Med. 1962;115:453–466. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zigmond SH. J Cell Biol. 1977;75:606–616. doi: 10.1083/jcb.75.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zicha D, Dunn G, Jones G. Methods Mol Biol. 1997;75:449–457. doi: 10.1385/0-89603-441-0:449. [DOI] [PubMed] [Google Scholar]
- 49.Young EWK, Beebe DJ. Chem Soc Rev. 2010;39:1036–1048. doi: 10.1039/b909900j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andersson H, van den Berg A. Sens Actuators, B. 2003;92:315–325. [Google Scholar]
- 51.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 52.Hegerfeldt Y, Tusch M, Brocker EB, Friedl P. Cancer Res. 2002;62:2125–2130. [PubMed] [Google Scholar]
- 53.Knight B, Laukaitis C, Akhtar N, Hotchin NA, Edlund M, Horwitz AR. Curr Biol. 2000;10:576–585. doi: 10.1016/s0960-9822(00)00486-3. [DOI] [PubMed] [Google Scholar]
- 54.Cukierman E, Pankov R, Stevens DR, Yamada KM. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 55.Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P. J Cell Biol. 2003;160:267–277. doi: 10.1083/jcb.200209006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker GM, Sai J, Richmond A, Stremier M, Chung CY, Wikswo JP. Lab Chip. 2005;5:611–618. doi: 10.1039/b417245k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keenan TM, Folch A. Lab Chip. 2008;8:34–57. doi: 10.1039/b711887b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skandarajah A, Janetopoulos CJ, Wikswo J, Samson P. 2010 in preparation. [Google Scholar]
- 59.Taylor AMB-J, Woo Rhee M, Cribbs S, Cotman DH, Li Jeong CWN. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Butler KLA, Agrawal V, Bilodeau N, Toner M, Tompkins M, Fagan RG, Irimia SD. PLoS One. 2010;5:e11921. doi: 10.1371/journal.pone.0011921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song Lea. Eur J Cell Biol. 2006;85:981–989. doi: 10.1016/j.ejcb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 62.Schneider IC, Haugh JM. J Cell Biol. 2005;171:883–892. doi: 10.1083/jcb.200509028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lauffenburger DA, Tranquillo RT, Zigmond SH. Methods Enzymol. 1988;162:85–101. doi: 10.1016/0076-6879(88)62067-2. [DOI] [PubMed] [Google Scholar]
- 64.Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Proc Natl Acad Sci U S A. 2004;101:8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gruver JS, Wikswo JP, Chung CY. Biophys J. 2008;95:4057–4067. doi: 10.1529/biophysj.108.130179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clarke M, Muller-Taubenberger A, Anderson KI, Engel U, Gerisch G. Mol Biol Cell. 2006;17:4866–4875. doi: 10.1091/mbc.E06-04-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parent CA, Devreotes PN. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 68.Camelliti P, Gallagher JO, Kohl P, McCulloch AD. Nat Protoc. 2006;1:1379–1391. doi: 10.1038/nprot.2006.203. [DOI] [PubMed] [Google Scholar]
- 69.Chen CN, Koutalos TY. Biophys J. 1999;76:2861–2867. doi: 10.1016/S0006-3495(99)77440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalimuthu PJ, SA Biosens Bioelectron. 2009;24:3575–3580. doi: 10.1016/j.bios.2009.05.017. [DOI] [PubMed] [Google Scholar]
- 71.Lin FS, Rhee W, Wang SW, Mittal S, Jeon S, NL Lab Chip. 2004;4:164–167. doi: 10.1039/b313600k. [DOI] [PubMed] [Google Scholar]
- 72.Walker GM, Sai J, Richmond A, Stremier M, Chung CY, Wikswo JP. Lab Chip. 2005;5:611–618. doi: 10.1039/b417245k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kimmel AR, Parent CA, Gough NR. Science’s STKE. 2004;2004:tr3. doi: 10.1126/stke.2342004tr3. [DOI] [PubMed] [Google Scholar]
- 74.Levine H, Kessler DA, Rappel WJ. Proc Natl Acad Sci U S A. 2006;103:9761–9766. doi: 10.1073/pnas.0601302103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fuller D, Chen W, Adler M, Groisman A, Levine H, Rappel WJ, Loomis WF. Proc Natl Acad Sci U S A. 2010;107:9656–9659. doi: 10.1073/pnas.0911178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma L, Janetopoulos C, Yang L, Devreotes PN, Iglesias PA. Biophys J. 2004;87:3764–3774. doi: 10.1529/biophysj.104.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.