Abstract
Variations and similarities in our individual genomes are part of our history, our heritage, and our identity. Some human genomic variants are associated with common traits such as hair and eye color, while others are associated with susceptibility to disease or response to drug treatment. Identifying the human variations producing clinically relevant phenotypic changes is critical for providing accurate and personalized diagnosis, prognosis, and treatment for diseases. Furthermore, a better understanding of the molecular underpinning of disease can lead to development of new drug targets for precision medicine. Several resources have been designed for collecting and storing human genomic variations in highly structured, easily accessible databases. Unfortunately, a vast amount of information about these genetic variants and their functional and phenotypic associations is currently buried in the literature, only accessible by manual curation or sophisticated text mining technology to extract the relevant information. In addition, the low cost of sequencing technologies coupled with increasing computational power has enabled the development of numerous computational methodologies to predict the pathogenicity of human variants. This review provides a detailed comparison of current human variant resources, including HGMD, OMIM, ClinVar, and UniProt/Swiss-Prot, followed by an overview of the computational methods and techniques used to leverage the available data to predict novel deleterious variants. We expect these resources and tools to become the foundation for understanding the molecular details of genomic variants leading to disease, which in turn will enable the promise of precision medicine.
Keywords: Human Disease Variants, Function Prediction, Databases for Human Variants
1. Introduction
The decreasing cost of next-generation sequencing technologies has enabled the study of human genetic variation on a large scale. Rapid advancement of these techniques foreshadows the use of whole-genome or whole-exome sequencing towards the goal of personalized medicine or, more broadly, precision medicine, targeting not just individuals but also sub-populations that differ in their susceptibility to disease or drug treatment[1]. This promise has already come to fruition in anecdotal cases such as leveraging individual genomes for the treatment or prevention of a variety of diseases ranging from rare disorders[2, 3] to common diseases such as cancer[4]. Despite these advancements, systematic implementation of such diagnoses remains elusive even though the Human Genome Project has been complete for over a decade [5]. A major impediment for utilizing personal genomics for medical applications is navigating through “Big Data” to extract the relevant information[6, 7]. This technical need will only increase as the cost of sequencing technologies declines. For example, in the analysis of more than a thousand individual genomes from the 1000 Genomes Project[8], a recent study has estimated that each individual has 10,000–11,000 variants that result in an amino acid change, 10,000–12,000 variants that result in a change to a synonymous codon, and 410–460 insertions and deletions when compared to the reference genome, with an estimated 25–50% of rare non-synonymous variants being deleterious[9]. These variations underscore the vast genetic differences among individuals and present a challenge for studying the molecular underpinnings of disease and drug susceptibility. Thus, to enable the study of genotypic and phenotypic relationships, particularly for complex diseases, it is imperative that the wealth of knowledge on the molecular causes of disease be made accessible to the scientific and medical community. To this end, several organizations and research groups have created resources and techniques to gather and distribute relevant data and developed computational methods and algorithms to predict the phenotypic relevance of variants. In this review, we focus on resources and methods related to human variants and diseases including disease-causing variants, disease-associated variants, and variants computationally predicted to be deleterious, all of which we will refer to as “disease variants”. However, two areas of research in complex diseases are outside the scope of this review due to the extensive coverage elsewhere, namely Genome Wide Association Studies (GWAS) and resources and approaches focusing on cancer. GWAS focuses on associating haplotypes in large sequencing studies with disease susceptibility. This approach has had several complications due to incomplete penetrance and linkage disequilibrium, as discussed in other reviews[10–12]. Cancer-specific resources including COSMIC[13] and TCGA[14] have empowered research centered specifically on acquired somatic variants in cancer and how to detect those that play a significant role in tumor development, proliferation, or response to treatment [15–17].
This review is divided into three sections. We first introduce current repositories for human disease variant information and their standards for annotation and provide a detailed comparison of four of the most comprehensive of these resources, including HGMD[18], OMIM[19], ClinVar[20], and UniProt/Swiss-Prot[21]. Despite the vast amount of data manually accumulated in these resources, much of the information about variants remains buried in the literature. Thus, the second part of this review provides an overview of techniques used for text mining of disease variant information from the scientific literature. Finally, due to the vast space of possible variants and the fact that rare disease variants are less likely to have been previously reported, we expect a large number of disease-variant relationships to be absent in current databases and/or to not even be known or published. Thus, the third part of this review focuses on the computational methods and techniques to predict new, deleterious variants. These efforts will become the foundation for understanding disease at a molecular level, facilitating the promise of precision medicine.
2. Variant Databases and Resources
In this section, we provide an overview of the current repositories for disease variants and a detailed comparison of four of the most comprehensive databases. We further discuss the implications of this comparison in regards to the diversity of focus and purpose of each resource.
2.1 Overview of Variant Databases
The advent of next-generation sequencing technologies has produced an abundance of genetic data, elucidating the vast genetic differences among individuals. However, there still remains much to be understood about how these variations impact human health. Thus, it is imperative that these data be organized and readily available for use in scientific research. A variety of disparate databases have been created to provide genetic variant information for genomic regions ranging in size from individual genes to entire genomes. The largest of these resources is the NCBI’s dbSNP database[22], which contains over 50 million unique genetic variations as of build 137. The dbSNP entries are derived from researcher-submitted data and are augmented by the 1000 Genomes Project[8], a study that aims to capture over 95% of variants in genomic regions accessible by current high-throughput sequencing technologies. However, the phenotypic effects of the majority of the variants in the dbSNP database remain unknown, as only 25,318 (less than 3%) of exonic missense variants from dbSNP are known to be involved in disease. The current explosion in the amount of available genetic data is expected to persist as sequencing projects such as the Personal Genome Project[23] continue to genotype new individuals. However, the biomedical goal of utilizing these data will be limited by our understanding of molecular genetics and our approaches for analyzing such data.
In contrast to resources that aim to capture all types of human genetic differences, several resources focus specifically on genetic variants with a known phenotypic impact. Vital to the success of such resources is a team of manual curators tasked with gleaning the relevant information from published literature or ensuring the quality of user-submitted data. For example, the PharmGKB[24] resource tracks known pharmacodynamic/pharmacokinetic pathways, drug-gene relationships, and 654 human variants linked to drug response. Other databases focus on variants known to influence disease. Among these resources, locus-specific databases (LSDBs) aim to cover all known variations for individual genes or loci[25]. These resources have recently shifted towards more standardized formats due to the development of specific quality control measures and standardized nomenclature[26–28] as well as software geared towards supporting manual curators such as LOVD[29] and UMD[30]. Similar databases were built with a focus on collecting all variants pertaining to a specific disease (e.g. caBIG[31], PDmutDB[32], AutDB[33], and IDbases[34] for cancer, Parkinson’s disease, autism, and immunodeficiency, respectively). In addition, MITOMAP[35], MitoDat[36], and mtDB[37] provide variant annotation for mitochondrial genes. However, this federation of LSDBs, disease-specific databases, and mitochondrial databases are not connected and cover only specific genomic regions, making them unsuitable in their current form for the analysis of human genetic variation at the genomic level. Nevertheless, these databases represent a valuable asset to the community and, with sufficient coordination, could one day rival any similar effort by a single organization to capture such information. On the other hand, several variant databases aim to provide comprehensive resources for disease variants. One such resource, Online Mendelian Inheritance in Man (OMIM) (http://www.omim.org)[19], a compendium of disease-related literature for each gene and disease, consists of 16,069 disease variants manually curated from scientific publications. In addition, the NCBI has recently released a new resource, the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/)[20], containing a collection of clinically relevant variants from OMIM, GeneReviews[38], LSDBs, and contributing testing laboratories. The ClinVar database currently contains 67,555 reports, of which, 13,465 are classified as clinically significant. Additionally, the UniProt/Swiss-Prot knowledgebase (http://swissvar.expasy.org/)[21], part of the UniProt database[39], contains 66,723 variants. Of these, 23,229 have been classified as disease variants, 37,446 as polymorphic variants, and 6,365 as unclassified. Lastly, the Human Gene Mutation Database (HGMD) (http://www.hgmd.org/)[18] is another manually curated resource and is divided into a freely available, or “Public” version, and a proprietary, or “Professional” version, respectively consisting of 80,173 and 109,521 disease variants with one or more references in the literature (as of April 2013). This includes exonic missense variants, variants from promoter regions, splice site variants, insertions, deletions, and complex rearrangements. An overview of the total number of records for each variant type contained in each variant resource can be found in Table 1.
Table 1. Total disease variants of each type for each resource.
Overview of the total number of disease variants of each type (i.e., exonic missense variants, splice site, insertions, deletions, stop-gain, stop-loss, and variants from promoter regions) contained in the ClinVar[20], HGMD[18], OMIM[19], and UniProt/Swiss-Prot[21] resources. Variants were considered only after filtering each database based on several criteria and redundant variants were removed if they were not annotated for a unique gene, wild type, mutated type, and position.
| Name | ClinVar | OMIM | UniProt/Swiss-Prot | HGMD Public | HGMD Professional |
|---|---|---|---|---|---|
| Exonic Missense Variants | 10,420 | 10,967 | 22,912 | 44,427 | 60,058 |
| Splice Site Variants | 332 | 896 | N/A | 9,455 | 12,665 |
| Insertions or Deletions | 264 | 1,666 | 317 | 13,396 | 19,089 |
| Stop-gain Variants | 2,344 | 2,521 | N/A | 11,110 | 14,972 |
| Stop-loss Variants | 32 | 19 | N/A | 32 | 74 |
| Variants from Promoter Regions or UTRs | 73 | N/A | N/A | 1,753 | 2,663 |
| Total | 13,465 | 16,069 | 23,229 | 80,173 | 109,521 |
In addition to the aforementioned resources, there exist several other databases that can be utilized in the study of molecular causes of disease. Various resources have been created with the intent of annotating variants with information regarding their functional or phenotypic impact (PMD[40], PhenCode[41], HumDiv/HumVar[42], and NewHumVar/HumVarProf[43]) or resulting structural stability (ProTherm[44] and ProNit[45]). These are derivative databases, constructed from one or more variant databases with the intent of annotating specific variants with functional or phenotypic relevance. These datasets and those tracked by VariBench[46], a suite of variant datasets that can be used as a benchmark, contain both positive and negative cases and can thus be used to train computational methods. Additionally, it has been shown that model organisms can be informative for the study of human diseases or drug responses by studying not only the function of orthologous genes[47], but also individual variants in conserved regions[48, 49]. Thus, species-specific resources such as SGD (Saccharomyces Genome Database)[50] for yeast or MGD (Mouse Genome Database)[51] for mice as well as the OMIA (Online Mendelian Inheritance in Animals)[52], which aims to encompass the phenotypically relevant variations in all model organisms, are expected to play a significant role in the study of human disease in the coming years. The variant databases discussed in this review are all of particular interest in the study of human genetic variation and, hence, several tools have been created to visualize or annotate these variants with relevant information including the UCSC genome browser[53], the NCBI’s annotation tracks[54], MutDB[55], Domain Mapping of Disease Mutations (DMDM)[56], SNPedia[57], SNP@Domain[58], StSNP[59], PicSNP[60], PupaSuite[61], FancyGene[62], AltAnalyze[63], and HOPE[64].
2.2 Standards for the Documentation of Variants
In the interest of enabling large-scale analyses of the available data, it is imperative that standards are maintained for storing and accessing variant information. To this end, members of the Human Genome Variation Society (HGVS) have formulated guidelines and recommendations on several topics related to the documentation of genetic variants, especially for the nomenclature of gene variations and guidelines for variation databases[65]. Importantly, when describing genes and proteins, the standards recommend that only HGNC gene symbols[66] be used when referencing genes and that the DNA reference sequence should preferably be a Locus Reference Genomic sequence (LRG)[28]. When no LRG is available, a RefSeq[67] record should be used, listing both accession and version number (e.g., NM_007294.3). The intent of this standardization is to reduce the effort needed to compare variants, as mapping between protein resources such as RefSeq, UniProt/Swiss-Prot, Ensembl[68], and PDB[69] can be difficult. Mapping variants between these disparate resources requires either using BLAST[70] or a similar sequence alignment algorithm to align the residues of equivalent sequences from other databases. Alternatively, there exist various identifier mapping resources (e.g., UniProt’s internal identifier mapping[71] or BioMart[72]). However, these may still require the residues to be aligned as the reference sequences for each resource could differ.
Nearly all of the databases discussed in this review are accessible via web interfaces maintained by the respective parties. However, it is not feasible to conduct large-scale analyses using this format as it becomes a bottleneck when acquiring data. To enable high-throughput distribution, each resource allows the full database to be downloaded, but these formats vary. For example, the “Public” version of HGMD is only accessible using the web interface, while the “Professional” version is distributed in relational database format. OMIM and UniProt/Swiss-Prot both maintain their own flat-file formats, which can be accessed at http://omim.org/downloads and http://www.uniprot.org/docs/humsavar, respectively. Alternatively, both the OMIM and UniProt/Swiss-Prot databases are accessible via Perl or Python APIs.
With the advent of high-throughput sequencing studies, several new file formats for standardized variant storage have been proposed. Recently, the SAM /BAM file format[73] has been used as a standard for storing next-generation read alignments. This format, though useful in storing raw sequence data, contains an abundance of redundant information. Hence, two competing variant formats, VCF[74] and GVF[75], address this issue by condensing the information into relevant data for each variant such as chromosomal location, reference and alternate alleles, and patients or samples containing each allele. For example, one of the several distribution methods provided by the ClinVar database is a VCF file that can be obtained from NCBI’s FTP site. The advantage of this format is that it enables the use of numerous tools designed to annotate, visualize, or manipulate these files such as VCFtools[74], Annovar[76], PLATO[77], PLINK[78], SnpEff[79], and AnnTools[80]. Alternatively, the ClinVar database (or specific subsets) is also available via NCBI’s E-Utilities service or by using the custom queries from NCBI’s website[81].
Several ontologies have been created with the purpose of providing standardized and systematic methods for describing variants or for facilitating their extraction from the literature. For instance, VariO[82] allows the annotation of effects, consequences, and mechanisms of DNA, RNA, and Protein variations along with additional functional, structural, or epigenetic properties. In addition, the Disease Ontology[83], while intended for the broader study of disease, annotates terms with OMIM identifiers, and can thus be utilized for the analysis of associated variants. Finally, the Mutation Impact Extraction Ontology (MIEO)[84] is an open-source tool which assists in documenting how variants and their associated properties and phenotypic effects are described in natural language, which can be used to facilitate the automated extraction of variants from the literature.
2.3 Methods of Variant Database Comparison
Knowing the content of the manually curated disease variant databases and how they overlap is key for utilizing this information in research. To compare the existing resources for the storage and visualization of human variants, the dbSNP database was acquired on March 20th, 2013, HGMD (release 2013.1) and ClinVar were downloaded on April 11th, 2013, and OMIM and UniProt/Swiss-Prot were obtained on April 9th, 2013. Once downloaded, the ClinVar VCF file was mapped to associated genes and proteins using the annotate_variation tool from ANNOVAR[76]. Records from ClinVar were only included if they were annotated as having a clinical significance of 4 or greater. A total of 261 variant records from OMIM and 173 from ClinVar were removed because the disease description contained the words “polymorphism,” “somatic,” or “unknown significance,” as these entries may not be disease related. The OMIM database was further filtered, removing 543 records due to nonstandard variant annotations. Namely, nonstandard variant entries were removed if the recorded variant itself (as opposed to the disease description or gene name) contained the words “haplotype,” “variant,” “expansion,” “multiple,” “fusion,” “mutation,” “repeat,” “methyl,” “breakpoint,” “conversion,” “complex,” “residues,” “silenced,” “tract,” or contained symbols that are not used in standard HGVS format. OMIM also contains several phenotypes that are not related to disease and 44 of these entries were removed from the analysis (this list of phenotypes was provided by Susan Bell, Jackson Labs, personal communication). In regard to the HGMD database, this analysis only considers variants from the “mutation,” “prom,” “splice,” “deletion,” “insertion,” “indel,” “grosins,” “grosdel,” and “complex” tables from the MySQL database. When comparing these resources, duplicate exonic missense variants were removed by considering only those annotated for a unique gene and variant in HGVS format.
2.4 Results of Variant Database Comparison
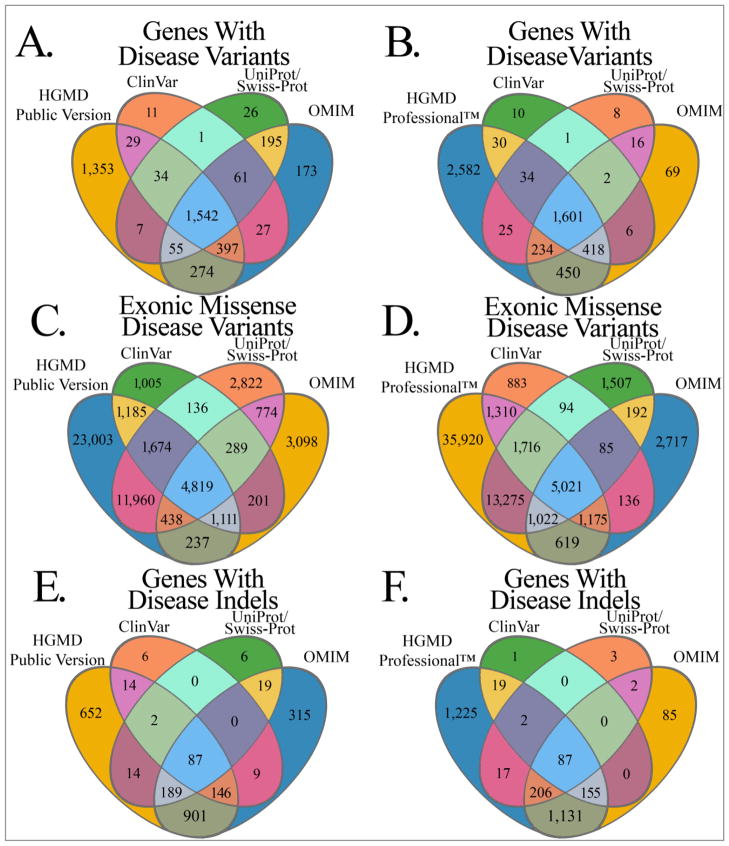
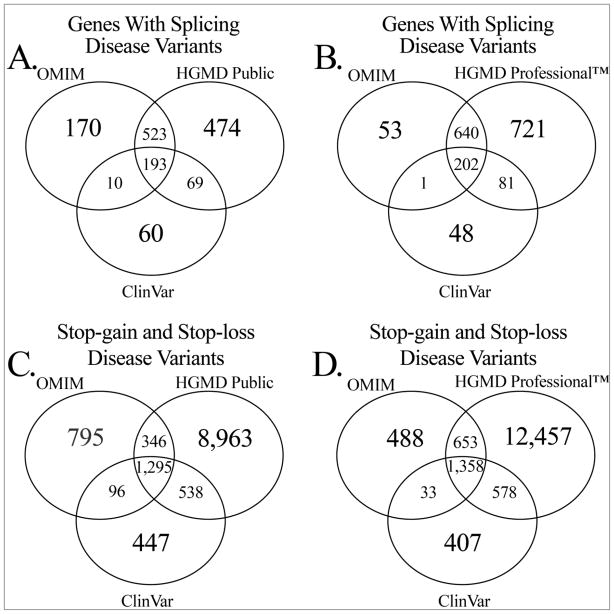
The results of comparing entire genes, exonic missense variants, and insertions/deletions across all current databases of human disease variants are depicted in Figure 1. However, the UniProt/Swiss-Prot knowledgebase does not track variants that affect splicing, stop-gain, or stop-loss variants, so a separate comparison is shown in Figure 2 for these variant types. Since HGMD Professional™ is available by subscription only, we distinguished between the public and professional versions and compared only publicly available resources in Figures 1A, 1C, 1E, 2A, and 2C. The results presented for HGMD were generated using the “mutation” MySQL table for Figures 1C and 1D, the “deletion,” “insertion,” “indel,” “grosins,” “grosdel,” and “complex” MySQL tables for Figures 1E and 1F, and the “splice” MySQL table for Figures 2A and 2B.
Figure 1. Coverage comparison of genes, exonic missense variants, and indels between variant resources.
Comparison of OMIM, UniProt/Swiss-Prot, ClinVar, HGMD Public, and HGMD Professional™ for the genes (1A and 1B), the exonic missense variants (1C and 1D), and the insertions/deletions (1E and 1F) annotated with disease variants in each database. The area of each section of the venn diagrams is not proportional to size and the colors are for aesthetic purposes only.
Figure 2. Indel and splice site variant comparison between variant resources.
Comparison of the OMIM, ClinVar, HGMD Public, and HGMD Professional™ resources for genes annotated with disease variants that effect splicing sites (2A and 2B) and genes annotated with stop-loss or stop-gain disease variants (2C and 2D).
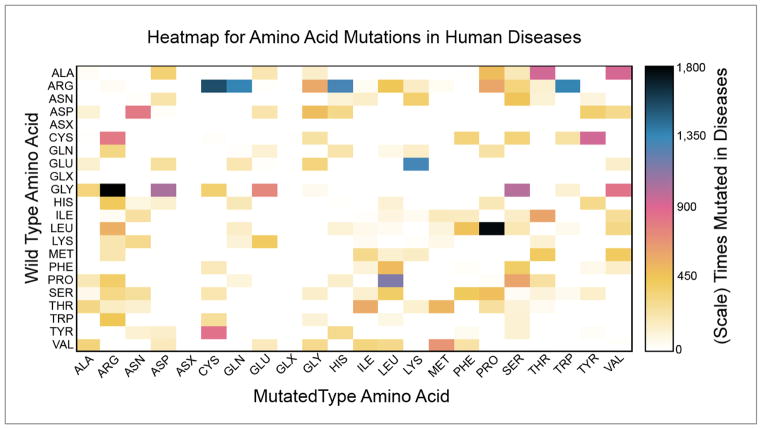
The compilation of these disparate databases provided an opportunity to analyze all known amino acid changes that result in disease, which is illustrated in Figure 3. This information could be informative for understanding the underlying molecular causes of disease when combined with other properties of amino acids such as frequency of occurrence and amino acid versatility. The three most common amino acid variants that result in a human disease recorded in these databases are Leucine to Proline, Glycine to Arginine, and Arginine to Cysteine. On one hand, it is not surprising that Leucine and Glycine are involved as wild type residues, as these are two of the most frequently occurring amino acids in the UniProt protein database[85]. On the other hand, the frequency of occurrence of Arginine is average but the Cysteine mutated type is not frequently found in proteins while being the most functionally versatile protein residue[86].
Figure 3. Heatmap of amino acid variants in human diseases.
Depiction of the observed frequency of wild type to mutated type transitions implicated in human diseases. The missense variants analyzed were a non-redundant collection compiled using the OMIM, HGMD, UniProt/Swiss-Prot, and ClinVar resources.
2.5 Discussion of Variant Database Comparison
The comparison of variant databases is a complex task due to differences in inclusion criteria, quality filters, amount and quality of annotation, and discrepancies in the reference sequences used. Despite the many difficulties, enumerated below, a quantitative comparison of these resources provides an overview of their relative size, overlap, and uniqueness. The results of this analysis show that HGMD Professional™ is currently the largest resource for human disease variants. However, because of the differences in scope, context, and additional information provided, each of the reviewed resources is unique and useful. For example, UniProt/Swiss-Prot is accompanied by a host of contextual information for each variant, allowing the user to analyze relevant information such as genes, proteins, post-translational modifications, taxonomic lineage, and Gene Ontology[87] terms. Additionally, the ClinVar resource facilitates the collection of variant data via a user submission system, which is a promising framework for collaborating with LSDBs and research laboratories. Likewise, OMIM is accompanied by a large, narrative-style explanation of each variant, disease, and gene, which is useful for medical practitioners.
Each resource maintains different criteria for the types of variants that will be included in the database. Thus, while we expect some overlap between the different databases, we also expect them to contain several unique entries as the databases ultimately have different purposes. For example, HGMD states that it includes “the first example of all mutations causing or associated with human inherited disease, plus disease-associated/functional polymorphisms reported in the literature”[88]. In contrast, for the OMIM database: “For most genes, only selected mutations are included. Criteria for inclusion include the first mutation to be discovered, high population frequency, distinctive phenotype, historic significance, unusual mechanism of mutation, unusual pathogenetic mechanism, and distinctive inheritance (e.g., dominant with some mutations, recessive with other mutations in the same gene). Most of the allelic variants represent disease-causing mutations. A few polymorphisms are included, many of which show a positive correlation with particular common disorders”[89]. Thus, since the OMIM database intends to capture disease at the level of individual genes, and includes only selected variants for each gene, it may, in many cases, purposefully exclude variants included in other databases. Similarly, HGMD does not include neutral polymorphisms, somatic variants, variants of unknown significance, and variants implicated in GWAS that may only be in linkage disequilibrium with actual disease-causing variants. In our analysis, 848 of these types of variants were removed from the OMIM database for the purpose of comparing against HGMD and others. In addition, ClinVar entries were considered to be disease variants only if they were annotated as having known clinical significance (i.e., those with a “CLNSIG” value greater than or equal to 4), which resulted in 51,847 records being removed. An additional 173 records were removed from ClinVar because the description contained the words “polymorphism,” “somatic,” or “unknown significance”. Similarly, UniProt/Swiss-Prot contains 37,446 variants classified as polymorphisms and 6,365 with unknown significance that were removed for this analysis.
Furthermore, the following factors also influence the overlap between the compared databases, which, in some cases, is expected to be underestimated in our analysis. (1) Variants may have been excluded from a given database due to quality control filters, which vary between resources. (2) For genes located in the mitochondrial genome, HGMD maintains external links to MITOMAP but does not contain variant data that could be included in our analysis. (3) Some variants tracked by the ClinVar database were not available in the VCF file used in our analysis because their exact chromosomal locations have yet to be determined. The remaining set of variants contained in ClinVar (but not in the VCF file) should comprise the full set of OMIM variants, plus entries from several LSDBs (from personal communication, Donna Maglott, National Center for Biotechnology Information, National Institutes of Health). (4) A disease variant contained in more than one database could appear to be different when one of them uses an alternate gene name or an outdated identifier or accession. For example, in OMIM, the gene TGIF1 is referred to by an alternate gene name (TGIF), resulting in several variants that could not be reconciled between databases. (5) Identical records between resources could also be missed due to differences in the reference genome or the recorded gene product (e.g. alternate isoforms, mature protein numbering, etc.) or when additional curation efforts to resolve the variants to sequence are made only in some of the databases. For instance, the KIF21A, IMPDH1, and CTSA genes contain several variants that are identical in OMIM, UniProt/Swiss-Prot, and ClinVar but are annotated with an apparently different protein position in HGMD. In all three examples, this inconsistency is due to alternate sequences being used as a reference by the different databases. (6) Variants could be annotated incorrectly in one or more of the databases (e.g., reversed wild type and mutated type amino acid changes, errors in the HGVS format, etc). (7) Criteria for variant classification (i.e., missense, splicing, indels, and complex rearrangements) vary between resources potentially resulting in variants being classified differently among resources.
3. Variant Extraction from the Literature
Resources that focus on the relationship between variants and disease require manual curation of the associated literature before the information can be included in their databases. The process of translating published literature into a usable format is very costly in terms of time and money, and, with over 22 million citations stored in NCBI’s PubMed as of 2013, it has become increasingly difficult for manually curated databases to keep up with the literature. Automated text mining of such variants and their associations can solve the problem of retrieving associations from the literature and effectively keeping up with the current exponential growth. Here, text mining can be used to extract genes, variants, and their associations to diseases, drugs, and other processes. The initial task is to extract genes and their variants from text. Early work in gene and variant extraction showed high precision for variant extraction but low recall. Rebholz-Schuhmann et al. developed Mutation Extraction from Medline Abstracts (MEMA)[90] to extract genes and variants from text. MEMA matches variants using specific regular expressions and matches genes using HUGO[91] gene symbols and alternate names. While precision was high for extracted variant-gene pairs (93.4%), recall was only 35.2%. Around the same time, Horn et al. introduced MuteXt[92] to extract genes and variant pairs from abstracts and full text (when available) and specifically applied their system to G protein-coupled receptors (GPCRs) and nuclear hormone receptors (NRs) superfamilies. Variants were extracted using regular expressions and a gene dictionary was constructed using names from UniProt/Swiss-Prot for the GPCR and NR superfamilies. MuteXt achieved a precision of 88.4% and a recall of 82.9% for these two superfamilies. However, gene name recognition is a difficult text-mining task. For example, some genes are much easier to find in text due to more standardized naming schemes. So, while the application of MuteXt to specific superfamilies was shown to be successful, such results may not be accurate for variants in other genes and thus will require additional manual curation to eliminate false positives[93]. Additional tools have been created for the task of extracting gene and variant mentions in text (MutationFinder (variant only)[94], Mutation GraB[95], Mutator[96], Osiris[97], Mutation Miner[98], CoagMDB[99]). However, additional methods are still needed to find phenotypic associations for variants in text.
To address the task of disease variant extraction, Erdogmus et al. created Mutation Gene eXtractor (MuGeX)[100]. MuGeX uses regular expressions to find variant mentions and then disambiguates variants (e.g., protein or gene variants) using supervised machine learning. Extracted variant-gene pairs from an abstract corpus related to Alzheimer disease were compared to the Alzheimer variants for APP, PSEN1, and PSEN2 genes in the Alzheimer Disease & Frontotemporal Dementia Mutation Database (AD&FTDMDB) (http://www.molgen.vib-ua.be/ADMutations)[101]. MuGeX was able to correctly extract 97.4% of known variants in AD&FTDMDB that were present in the Alzheimer abstract corpus. While this result is promising, the comparison with AD&FTDMDB only included variants from three genes and did not evaluate the correctness of disease association of all extracted variants from the Alzheimer Disease corpus. Also, with only variants from three genes, there is little variation within the comparison set, which could lead to fewer opportunities for variant extraction errors and/or gene recognition errors. To further address this task, Doughty et al. developed the Extractor of MUtations (EMU)[102] to extract disease variants from text. EMU uses regular expressions to find variants in text and an extensive dictionary of gene symbols to find gene mentions. For variant-gene and variant-gene-disease mentions, EMU achieved a precision of 74% and 61% for variant-gene and variant-gene-disease extraction for breast cancer, respectively. Similar results were found for prostate cancer. These analyses highlight the difficulty in associating variants to disease based only on co-occurrence in text. For example, an abstract may mention two diseases and one variant, and thus co-occurrence of disease and variant will yield one correct disease-variant match and one incorrect disease-variant match. These types of association have the potential for generating a significant number of false positives, as shown by EMU’s precision in extracting variant-gene-disease mentions, which will require further manual curation to obtain accurate disease variant data. Alternatively, natural language processing may be one solution to link variants to disease based on descriptions at the sentence level and thus reduce this source of false positives.
Another important text mining task associated with disease variants is that of extracting variants that influence drug response. To address this issue, Garten et al. developed Pharmspresso (using Textpresso[103]) to extract genes, variants, and drugs (and the relationships between them) from full text articles[104]. Pharmspresso was evaluated based on gene-only, variant-only, and drug-only extractions. Combining entity extractions (for example, drug and gene) decreased performance. Since EMU was created with disease associations in mind, Rance et al. extended EMU to find drug-associated variants in text[105]. The authors extracted variants using EMU and used MetaMap[106] and RxNorm[107] to identify drug mentions. When applied to drugs, EMU was found to be complimentary to current relation-centric approaches for pharmacogenomics information. However, this approach is still limited by drug-variant co-occurrences in text in order to establish a relationship between the two entities, as discussed above.
Additional methodologies have been developed to extract other types of human variant associations from text. Hakenberg et al. created SNPshot[108] to extract genes, variants, drugs, diseases, adverse drug reactions, allele frequencies, population frequencies, and the relations between them from text. SNPshot had a 90–92% precision for gene, drug, and disease mentions and a 76–84% precision for extracted relations. Specifically, gene-variant relations had a 58.8% precision and 73.4% recall. While SNPshot has a good recall for this task, it will still generate a significant number of false positives that will require further manual curation. EnzyMiner[109] extracts protein variants (using MuGeX) and their impact on targeted enzymes. Similar to disease variant extraction, EnzyMiner has difficulty associating variants to the correct enzyme when multiple enzymes are present in the same abstract and thus may benefit from natural language processing. Laurila et al. created a method to extract variants (using MutationFinder) and impact relations from abstracts[110]. Finally, Naderi et al. extended this work and developed the Open Mutation Miner[111] to extract variants and impact relations using an ontology model for variant impact relations.
While text mining has already been proven to be a useful biocuration tool for genetic variants in HGMD[112] and for post-translational modification (PTM) annotation in the UniProt/Swiss-Prot[113] knowledgebase, these techniques still face challenges with named entity recognition, variant grounding (linking variants to the correct gene in text), correctly matching variant to phenotype associations, and incorrectly annotated variants (e.g., spelling errors or different reference genomes/amino acid numbering systems) [93]. In addition, in many cases, only the abstract is used for text mining purpose and the relevant information is missed when it is contained in supplementary information or tables and figures within the original article. While the search for variants has yielded high precision and relatively high recall, future work is still needed for the extraction of more variant types such as insertion and deletion variants, intronic variants, and frameshift variants; more accurate gene and protein name recognition; and more precise associations to different phenotypes including disease.
4. Tools for Predicting Deleterious Variants
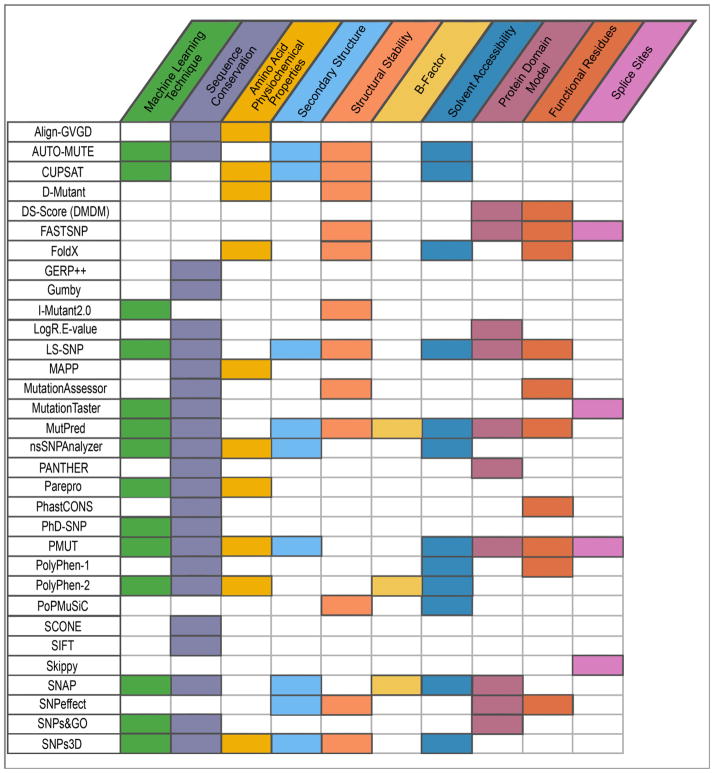
The recent explosion in the availability of genetic data due to technological advances has opened the door for large-scale molecular analyses of human disease. However, the body of known disease variants is incomplete and experimental approaches to determine the molecular underpinnings of disease can be time consuming. Thus, several computational methods have been proposed for determining which variants are likely to have functional impact and whether they contribute to changes in phenotype. A variety of these tools exist, using scoring metrics or machine learning techniques and leveraging existing variant data for their predictions. Of those approaches based on machine learning methods (Table 2), CUPSAT[114], I-Mutant2.0[115], LS-SNP[116], Parepro[117], PhD-SNP[43], SNPs&GO[118–120], and SNPs3D[121] use support vector machines (SVMs). MutPred[122] and nsSNPAnalyzer[123] use Random Forests; PMUT[124] and SNAP[125] use feed forward neural networks. PolyPhen-2[42] and MutationTaster[126] employ a Naïve Bayes approach. Additionally, AUTO-MUTE[127] has the option to use either SVM or Random Forest algorithms. The remaining methods, Align-GVGD[128], D-Mutant[129], DS-Score[130], FASTSNP[131], FoldX[132], GERP++[133], Gumby[134], LogR.E-value[135], MAPP[136], MutationAssessor[137], PANTHER[138], PhastCONS[139], PolyPhen-1[140], PopMuSiC[141], SCONE[142], SIFT[143], Skippy[144], and SNPeffect[145] are rule-based, meaning they use various measures to assess variants and employ standard criteria for determining if the variant will be deleterious. It is important to emphasize that, depending on the type of variant dataset used for training, the results of the predictions will vary from predicting disease (e.g., OMIM, HGMD, HumVar), structural disruption (e.g., ProTherm and ProNit), or functional disruption (PMD). Furthermore, to aggregate the predictions of this vast array of tools, a number of meta-analysis suites exist to annotate variants with the results of the aforementioned prediction methods (e.g., F-SNP[146], pfSNP[147], SNP Functional Portal[148], SNPit[149], and VISTA[150]) as well as tools that base their predictions on the combined results of selected sets of prediction tools (e.g., CONDEL[151], Meta-SNP[152], PolyDoms[153], PON-P[154], and Pro-Maya[155]).
Table 2. Machine learning methods and training sets for variant prediction.
Summary of variant prediction methods based on machine learning techniques. Each of these methods utilizes various conservation metrics, training sets, and machine learning techniques for the final prediction.
| Name | URL | Sequence Conservation Metric | Machine Learning Method | Variants used for Training |
|---|---|---|---|---|
| AUTO-MUTE [127] | http://proteins.gmu.edu/automute/ | None | SVM and Random Forest | ProTherm |
| CUPSAT [114] | http://cupsat.tu-bs.de/ | None | SVM (Multiple Regression) | ProTherm |
| I-Mutant2.0 [115] | http://gpcr2.biocomp.unibo.it/~emidio/I-Mutant2.0/I-Mutant2.0_Details.html | None | SVM | ProTherm |
| LS-SNP [116] | http://modbase.compbio.ucsf.edu/LS-SNP/ | SIFT | SVM | dbSNP |
| MutationTaster [126] | http://www.mutationtaster.org/ | Positional conservation in comparison to homologous sequences | Naive Bayes | OMIM, HGMD, common variants from dbSNP |
| MutPred [122] | http://mutpred.mutdb.org/ | Internal, position-specific conservation scorescore | Random Forest | UniProt/Swiss-Prot, HGMD, somatic cancer variantsvariants |
| nsSNPAnalyzer [123] | http://snpanalyzer.uthsc.edu/ | Substitution frequencies in multiple sequence alignments | Random Forest | UniProt/Swiss-Prot |
| Parepro [117] | http://www.mobioinfor.cn/parepro/ | Substitution frequencies of surrounding positions | SVM | HumVar, HumVarProf, and NewHumVar |
| PhD-SNP [43] | http://snps.biofold.org/phd-snp/phd-snp.html | Substitution frequencies in multiple sequence alignments | SVM | HumVar |
| PMUT [124] | http://mmb2.pcb.ub.es:8080/PMut/ | Substitution frequencies from PSI-BLAST | Feed Forward Neural Network | UniProt/Swiss-Prot |
| PolyPhen-2 [42] | http://genetics.bwh.harvard.edu/pph2/ | PSIC | Naive Bayes | HumDiv and HumVar |
| SNAP [125] | https://rostlab.org/services/snap/ | PSIC | Feed Forward Neural Network | PMD |
| SNPs&GO [118–120] | http://snps-and-go.biocomp.unibo.it/snps-and-go/ | subPSEC | SVM | UniProt/Swiss-Prot |
| SNPs3D [121] | http://www.snps3d.org/ | Substitution frequencies and Shannon entropy | SVM | HGMD, substitutions in homologous sequences |
Although the variant prediction tools differ in the particular model used and, thus, how the variant is ultimately interpreted, the types of information utilized for prediction are often common between methods (Figure 4). Importantly, a prominent feature used in most of these methods is a metric to quantify sequence conservation, namely SIFT[143], PSIC[156], subPSEC[157], FIS[137], or another method devised to assess amino acid substitution frequencies. SIFT (Sorting Intolerant From Tolerant) is a widely-used sequence conservation metric that makes use of Dirichlet priors calculated from multiple sequence alignments. The SIFT method is used as a machine learning attribute in LS-SNP, MutPred, and nsSNPAnalyzer, but is commonly used as an independent tool for the assessment of positional conservation. PSIC (position-specific independent counts), created for use in PolyPhen, is a similar method that formally addresses the problem of interdependence between sequences in an alignment, accounting for sequences in the alignment with high similarity. Another metric, subPSEC (substitution position-specific evolutionary conservation), used in PANTHER and SNPs&GO, emphasizes the divergence of specific functions within protein families by taking advantage of the PANTHER database, a collection of protein families and functionally related subfamilies. Additionally, FIS (functional impact score) is a metric used by MutationAssessor based on the evolutionary conservation of the mutated residue in protein families and, separately, in each of its subfamilies. Apart from the abovementioned sequence conservation metrics, a number of prediction methods utilize their own sequence conservation measurements derived from multiple sequence alignments, amino acid substitution frequencies, or amino acid similarity. While most of these prediction methods exploit one or more of these sequence conservation metrics in determining if a variant will be deleterious, the additional attributes used in the analysis differ among them. This includes amino acid physiochemical properties (e.g., hydrophobicity, charge, size), secondary structure (e.g., helices, sheets, coils), predicted structural stability, B-factor[158], solvent accessibility, protein domain models, functional residues (e.g., binding sites, active sites, hinge regions), and whether the variant affects splice sites, i.e., it occurs between the boundaries of exons and introns or it is otherwise predicted to disrupt the normal splicing of the open reading frame (ORF). It should be noted that having more features is not always an indication of a better prediction method, as some machine learning techniques can be improved by selecting only the features that are the most predictive. Several authors have reported formal comparisons of machine learning methods[159–163]. However, it is important to know that some machine learning methods may “overfit” for the set of variants on which it was trained, and their performance will likely be worse when new variants are introduced. Ideally, when comparing these methods, the test set of variants used should have no overlap with the variants used in training. Finding such a dataset can be problematic due to the overlap of variant databases, as previously discussed, and to the fact that each method is trained using a different set of variants.
Figure 4. Comparison of features utilized by techniques for predicting deleterious variants.
Depiction of the types of information used by different methods for predicting deleterious variants.
5. Final remarks
More than a decade after the completion of the human genome project[164, 165], the promise of a revolution in medicine has yet to be fulfilled[166]. While the ability to sequence individual genomes has only changed clinical practices for a few anecdotal cases[2, 4, 167], the insight we have gained into human genomics has proven to be an invaluable resource for biomedical research. For instance, the ENCODE project[168] has already identified biochemical functions for 80% of the human genome[169] including regions of transcription, transcription factor association, chromatin structure, and histone modification. In addition, the reduced cost of sequencing technology is enabling projects that can capture the genomes of thousands of patients. These large case-control studies are essential for boosting the statistical power needed to detect the genetic variants responsible for rare diseases and can provide the necessary knowledge for use in the clinical setting. The variant databases and resources for text mining the literature are key for translating these data into valuable information available to clinicians that will ultimately impact how patients are diagnosed and treated. For example, it has been shown that each individual could harbor 40–110 disease variants from HGMD[170], which is information that could be vital for diagnosis in the clinical setting The computational approaches reviewed in this article are based on functional properties of the variant independent of population prevalence and can thus predict whether new variations found in an individual are likely to be pathogenic. This is crucial for the analysis of rare variants, for which population studies have not yet been successful[171, 172]. However, methods for predicting deleterious variants rely on the completeness and organization of the known disease variant data and are limited by our current knowledge of disease mechanisms. Furthermore, even when predictions are correct, no recommendation for treatment can be readily suggested from the result of these variant analyses. Importantly, most of the prediction methods only predict whether a variant is deleterious, but do not identify the causative molecular disruption that results in disease, such as the disruption of PTMs[173–175] and catalytic residues[176]. Thus, translating the analysis of human variants performed in-silico into the clinical setting is still one of the main challenges towards the goal of precision medicine. Several ways of addressing this challenge exist, such as predicting the phenotype[177] or response to drug treatment that will result from the variant. However, current techniques are constrained by the limited knowledge regarding genotypic and phenotypic relationships that can be used as inference. In the coming years, we anticipate the incorporation of model organisms into the analysis of variants, as they can be more easily studied through directed mutagenesis. In a recent study, Peterson et al.[49] compared human disease variants against variants known to be phenotypically altering in yeast. The authors concluded that phenotypically altering variants from model organisms even as evolutionarily distant as yeast display a significant overlap with human disease variants. Moreover, methods for prioritizing disease genes, reviewed elsewhere, could also be incorporated into the analysis of human variants. For instance, using network analyses[178–180], ontological enrichment[181–183], affected cellular compartment[184, 185], gene function identification using text mining[186, 187], or inference to the function of orthologous sequences[47, 188, 189] have been shown to provide insight into the types of phenotypes for which the deleterious variant is important. The progression of disease variant analysis techniques is essential for utilizing genomic data for precision medicine. The rapid advancement of this field foretells the advent of genetic information as a household commodity and understanding such information could be the framework for answering some of the most fundamental questions about our ancestries, our behavior, and our health.
Highlights.
Evaluation of resources for the analysis of human genetic variants
When compared, HGMD, OMIM, Swiss-Prot and ClinVar all appear to contain unique human variants
Several automated methods exist for extracting variants from the literature
Computational methods for predicting novel deleterious variants are compared
This field must advance in order to enable precision medicine
Acknowledgments
We would like to acknowledge the organizations and research groups discussed in this review as well as those that we, unfortunately, might have missed, for the valuable resources and methods that have been made available to the community. Special thanks to David Cooper and Peter Stenson from HGMD, Donna Maglott from the NCBI, Joanna Amberger and Ada Hamosh from OMIM, and Ioannis Xenarios, Livia Famiglietti, Jerven Bollenman, and Sylvain Poux from UniProt/Swiss-Prot for support in accessing their resources and thoughtful feedback. We are also grateful to Susan Bell for providing a list of OMIM phenotypes that are not related to disease, Danilo Gonzalez and Daniel Almonacid for their help analyzing amino acid frequency in disease variants, and Emidio Capriotti, Sean Mooney, and Predrag Radivojac for valuable suggestions and feedback on the manuscript.
Funding
The National Institutes of Health (NIH) 1K22CA143148 to MGK (PI) and R01LM009722 to MGK (co-investigator) and the American Cancer Society, ACS-IRG grant to MGK (PI) supported this work. ED is funded by the National Library of Medicine training grant T15 LM007033.
Footnotes
Authors’ Contributions
All authors contributed to conceiving, writing, and editing the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Thomas A Peterson, Email: tpeters1@umbc.edu.
Emily Doughty, Email: edoughty@stanford.edu.
Maricel G Kann, Email: mkann@umbc.edu.
References
- 1.Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Engl J Med. 2012;366(6):489–91. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 2.Yong E. We Gained Hope. The Story of Lilly Grossman’s Genome. 2013 [cited 2013 May 2nd]; Available from: http://phenomena.nationalgeographic.com/2013/03/11/we-gained-hope-the-story-of-lilly-grossmans-genome/
- 3.A one-in-a million disease; a lifesaving bone marrow transplant - given twice. 2012 [cited 2013 May 23rd]; Available from: http://www.jsonline.com/features/health/a-gift-of-life--given-twice-6f4a2f3-140316093.html.
- 4.A genetic test solves a hereditary mystery and saves a life 2012. 2012 Dec 07; [Available from: http://www.theglobeandmail.com/news/national/time-to-lead/a-genetic-test-solves-a-hereditary-mystery-and-saves-a-life/article6131121/
- 5.Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300(5617):286–90. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 6.Schadt EE, et al. Cloud and heterogeneous computing solutions exist today for the emerging big data problems in biology. Nat Rev Genet. 2011;12(3):224. doi: 10.1038/nrg2857-c2. [DOI] [PubMed] [Google Scholar]
- 7.Butte AJ, Shah NH. Computationally translating molecular discoveries into tools for medicine: translational bioinformatics articles now featured in JAMIA. J Am Med Inform Assoc. 2011;18(4):352–3. doi: 10.1136/amiajnl-2011-000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 11.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2011;13(2):135–45. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Visscher PM, et al. Five years of GWAS discovery. Am J Hum Genet. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas. 2013 [cited 2013 July 2nd]; Available from: http://cancergenome.nih.gov/
- 15.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339(6127):1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331(6024):1553–8. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 17.McLeod HL. Cancer pharmacogenomics: early promise, but concerted effort needed. Science. 2013;339(6127):1563–6. doi: 10.1126/science.1234139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stenson PD, et al. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics. 2012;Chapter 1(Unit 1):13. doi: 10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- 19.Amberger J, Bocchini C, Hamosh A. A new face and new challenges for Online Mendelian Inheritance in Man (OMIM(R)) Hum Mutat. 2011;32(5):564–7. doi: 10.1002/humu.21466. [DOI] [PubMed] [Google Scholar]
- 20.About ClinVar. [cited 2013 February 3rd]; Available from: http://www.ncbi.nlm.nih.gov/clinvar/
- 21.Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31(1):365–70. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Personal Genomes Project. 2013 [cited 2013 May 5th]; Available from: http://www.personalgenomes.org/
- 24.Whirl-Carrillo M, et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther. 2012;92(4):414–7. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Claustres M, et al. Time for a unified system of mutation description and reporting: a review of locus-specific mutation databases. Genome Res. 2002;12(5):680–8. doi: 10.1101/gr.217702. [DOI] [PubMed] [Google Scholar]
- 26.Scriver CR, Nowacki PM, Lehvaslaiho H. Guidelines and recommendations for content, structure, and deployment of mutation databases. Hum Mutat. 1999;13(5):344–50. doi: 10.1002/(SICI)1098-1004(1999)13:5<344::AID-HUMU2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Scriver CR, Nowacki PM, Lehvaslaiho H. Guidelines and recommendations for content, structure, and deployment of mutation databases: II. Journey in progress. Hum Mutat. 2000;15(1):13–5. doi: 10.1002/(SICI)1098-1004(200001)15:1<13::AID-HUMU5>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 28.Dalgleish R, et al. Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2010;2(4):24. doi: 10.1186/gm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fokkema IF, et al. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32(5):557–63. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 30.Beroud C, et al. UMD (Universal Mutation Database): 2005 update. Hum Mutat. 2005;26(3):184–91. doi: 10.1002/humu.20210. [DOI] [PubMed] [Google Scholar]
- 31.The Cancer Biomedical Informatics Grid (caBIG): infrastructure and applications for a worldwide research community. Stud Health Technol Inform. 2007;129(Pt 1):330–4. [PubMed] [Google Scholar]
- 32.Nuytemans K, et al. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: a mutation update. Hum Mutat. 2010;31(7):763–80. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37(Database issue):D832–6. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piirila H, Valiaho J, Vihinen M. Immunodeficiency mutation databases (IDbases) Hum Mutat. 2006;27(12):1200–8. doi: 10.1002/humu.20405. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Pesini E, et al. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35(Database issue):D823–8. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemkin PF, et al. A World Wide Web (WWW) server database engine for an organelle database, MitoDat. Electrophoresis. 1996;17(3):566–72. doi: 10.1002/elps.1150170327. [DOI] [PubMed] [Google Scholar]
- 37.Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34(Database issue):D749–51. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.GeneTests Medical Genetics Information Resource (database online). 1993–2013. 2013 Apr 19; Available from: http://www.genetests.org.
- 39.Apweiler RMM, O’Donovan C, Magrane M, Alam-Faruque Y, Antunes R, Barrell D, Bely B, Bingley M, Binns D, Bower L, Browne P, Chan WM, Dimmer E, Eberhardt R, Fedotov A, Foulger R, Garavelli J, Huntley R, Jacobsen J, Kleen M, Laiho K, Leinonen R, Legge D, Lin Q, Liu W, Luo J, Orchard S, Patient S, Poggioli D, Pruess M, Corbett M, di Martino G, Donnelly M, van Rensburg P, Bairoch A, Bougueleret L, Xenarios I, Altairac S, Auchincloss A, Argoud-Puy G, Axelsen K, Baratin D, Blatter MC, Boeckmann B, Bolleman J, Bollondi L, Boutet E, Quintaje SB, Breuza L, Bridge A, deCastro E, Ciapina L, Coral D, Coudert E, Cusin I, Delbard G, Doche M, Dornevil D, Roggli PD, Duvaud S, Estreicher A, Famiglietti L, Feuermann M, Gehant S, Farriol-Mathis N, Ferro S, Gasteiger E, Gateau A, Gerritsen V, Gos A, Gruaz-Gumowski N, Hinz U, Hulo C, Hulo N, James J, Jimenez S, Jungo F, Kappler T, Keller G, Lachaize C, Lane-Guermonprez L, Langendijk-Genevaux P, Lara V, Lemercier P, Lieberherr D, de Oliveira Lima T, Mangold V, Martin X, Masson P, Moinat M, Morgat A, Mottaz A, Paesano S, Pedruzzi I, Pilbout S, Pillet V, Poux S, Pozzato M, Redaschi N, Rivoire C, Roechert B, Schneider M, Sigrist C, Sonesson K, Staehli S, Stanley E, Stutz A, Sundaram S, Tognolli M, Verbregue L, Veuthey AL, Yip L, Zuletta L, Wu C, Arighi C, Arminski L, Barker W, Chen C, Chen Y, Hu ZZ, Huang H, Mazumder R, McGarvey P, Natale DA, Nchoutmboube J, Petrova N, Subramanian N, Suzek BE, Ugochukwu U, Vasudevan S, Vinayaka CR, Yeh LS, Zhang J. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38(Database issue):D142–8. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawabata T, Ota M, Nishikawa K. The Protein Mutant Database. Nucleic Acids Res. 1999;27(1):355–7. doi: 10.1093/nar/27.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giardine B, et al. PhenCode: connecting ENCODE data with mutations and phenotype. Hum Mutat. 2007 doi: 10.1002/humu.20484. [DOI] [PubMed] [Google Scholar]
- 42.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22(22):2729–34. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- 44.Gromiha MM, et al. ProTherm, version 2.0: thermodynamic database for proteins and mutants. Nucleic Acids Res. 2000;28(1):283–5. doi: 10.1093/nar/28.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prabakaran P, et al. Thermodynamic database for protein-nucleic acid interactions (ProNIT) Bioinformatics. 2001;17(11):1027–34. doi: 10.1093/bioinformatics/17.11.1027. [DOI] [PubMed] [Google Scholar]
- 46.Sasidharan Nair P, Vihinen M. VariBench: a benchmark database for variations. Hum Mutat. 2013;34(1):42–9. doi: 10.1002/humu.22204. [DOI] [PubMed] [Google Scholar]
- 47.McGary KL, et al. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc Natl Acad Sci U S A. 2010;107(14):6544–9. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420(6915):520–62. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 49.Peterson TA, Park D, Kann MG. A protein domain-centric approach for the comparative analysis of human and yeast phenotypically relevant mutations. BMC Genomics. 2013;14(Suppl 3):S5. doi: 10.1186/1471-2164-14-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40(Database issue):D700–5. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bult CJ, et al. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38(Database issue):D586–92. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Online Mendelian Inheritance in Animals, OMIA. Faculty of Veterinary Science, University of Sydney; Available from: http://omia.angis.org.au/ [Google Scholar]
- 53.Meyer LR, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41(Database issue):D64–9. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.NCBI’s 1000 Genomes Annotation Tracks. Available from: http://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/www.ncbi.nlm.nih.gov/variation/
- 55.Mooney SD, Altman RB. MutDB: annotating human variation with functionally relevant data. Bioinformatics. 2003;19(14):1858–60. doi: 10.1093/bioinformatics/btg241. [DOI] [PubMed] [Google Scholar]
- 56.Peterson TA, et al. DMDM: domain mapping of disease mutations. Bioinformatics. 2010;26(19):2458–9. doi: 10.1093/bioinformatics/btq447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cariaso M, Lennon G. SNPedia: a wiki supporting personal genome annotation, interpretation and analysis. Nucleic Acids Res. 2012;40(Database issue):D1308–12. doi: 10.1093/nar/gkr798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han A, et al. SNP@Domain: a web resource of single nucleotide polymorphisms (SNPs) within protein domain structures and sequences. Nucleic Acids Res. 2006;34(Web Server issue):W642–4. doi: 10.1093/nar/gkl323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uzun A, et al. Structure SNP (StSNP): a web server for mapping and modeling nsSNPs on protein structures with linkage to metabolic pathways. Nucleic Acids Res. 2007;35(Web Server issue):W384–92. doi: 10.1093/nar/gkm232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang H, Fujita T. PicSNP: a browsable catalog of nonsynonymous single nucleotide polymorphisms in the human genome. Biochem Biophys Res Commun. 2001;287(1):288–91. doi: 10.1006/bbrc.2001.5576. [DOI] [PubMed] [Google Scholar]
- 61.Conde L, et al. PupaSuite: finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res. 2006;34(Web Server issue):W621–5. doi: 10.1093/nar/gkl071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rambaldi D, Ciccarelli FD. FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics. 2009;25(17):2281–2. doi: 10.1093/bioinformatics/btp381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emig D, et al. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38(Web Server issue):W755–62. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Venselaar H, et al. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15(1):7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 66.Wain HM, et al. Guidelines for human gene nomenclature. Genomics. 2002;79(4):464–70. doi: 10.1006/geno.2002.6748. [DOI] [PubMed] [Google Scholar]
- 67.Pruitt KD, et al. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40(Database issue):D130–5. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flicek P, et al. Ensembl 2012. Nucleic Acids Res. 2012;40(Database issue):D84–90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berman HM, et al. The future of the protein data bank. Biopolymers. 2013;99(3):218–22. doi: 10.1002/bip.22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Index of /pub/databases/uniprot/current_release/knowledgebase/idmapping/ [cited 2013 May 5th]; Uniprot’s Internal Identifier Mapping]. Available from: ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/idmapping/
- 72.Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database (Oxford) 2011;2011:bar049. doi: 10.1093/database/bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reese MG, et al. A standard variation file format for human genome sequences. Genome Biol. 2010;11(8):R88. doi: 10.1186/gb-2010-11-8-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grady BJ, et al. Finding unique filter sets in plato: a precursor to efficient interaction analysis in gwas data. Pac Symp Biocomput. 2010:315–26. [PMC free article] [PubMed] [Google Scholar]
- 78.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Makarov V, et al. AnnTools: a comprehensive and versatile annotation toolkit for genomic variants. Bioinformatics. 2012;28(5):724–5. doi: 10.1093/bioinformatics/bts032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Entrez Help [Internet] Bethesda (MD): National Center for Biotechnology Information (US); 2006. Jan 20, 2005-. Entrez Help. [Updated 2011 Dec 19]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK3837/ [Google Scholar]
- 82.Vihinen M. VariO. 2013 [cited 2013 April 27th]; Available from: http://variationontology.org/index.shtml.
- 83.Schriml LM, et al. Disease Ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012;40(Database issue):D940–6. doi: 10.1093/nar/gkr972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klein A, et al. Benchmarking infrastructure for mutation text mining. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.UniProtKB/Swiss-Prot protein knowledgebase release 2013_05 statistics. 2013 [cited 2013 May 20th]; Available from: http://web.expasy.org/docs/relnotes/relstat.html.
- 86.Holliday GL, et al. The chemistry of protein catalysis. J Mol Biol. 2007;372(5):1261–77. doi: 10.1016/j.jmb.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 87.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Human Gene Mutation Database Background. [cited 2013 March 12th]; Available from: http://www.hgmd.cf.ac.uk/docs/new_back.html.
- 89.OMIM Frequently Asked Questions (FAQs) [cited 2013 March 12th]; Available from: http://www.omim.org/help/faq-1.4.
- 90.Rebholz-Schuhmann D, et al. Automatic extraction of mutations from Medline and cross-validation with OMIM. Nucleic Acids Res. 2004;32(1):135–42. doi: 10.1093/nar/gkh162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gray KA, et al. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013;41(Database issue):D545–52. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Horn F, Lau AL, Cohen FE. Automated extraction of mutation data from the literature: application of MuteXt to G protein-coupled receptors and nuclear hormone receptors. Bioinformatics. 2004;20(4):557–68. doi: 10.1093/bioinformatics/btg449. [DOI] [PubMed] [Google Scholar]
- 93.Stenson PD, Cooper DN. Prospects for the automated extraction of mutation data from the scientific literature. Hum Genomics. 2010;5(1):1–4. doi: 10.1186/1479-7364-5-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caporaso JG, et al. MutationFinder: a high-performance system for extracting point mutation mentions from text. Bioinformatics. 2007;23(14):1862–5. doi: 10.1093/bioinformatics/btm235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee LC, Horn F, Cohen FE. Automatic extraction of protein point mutations using a graph bigram association. PLoS Comput Biol. 2007;3(2):e16. doi: 10.1371/journal.pcbi.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kuipers R, et al. Novel tools for extraction and validation of disease-related mutations applied to Fabry disease. Hum Mutat. 2010;31(9):1026–32. doi: 10.1002/humu.21317. [DOI] [PubMed] [Google Scholar]
- 97.Bonis J, Furlong LI, Sanz F. OSIRIS: a tool for retrieving literature about sequence variants. Bioinformatics. 2006;22(20):2567–9. doi: 10.1093/bioinformatics/btl421. [DOI] [PubMed] [Google Scholar]
- 98.Baker CJ, et al. Mutation Mining--A Prospector’s Tale. Information Systems Frontiers. 2006;8(1):47–57. [Google Scholar]
- 99.Saunders RE, Perkins SJ. CoagMDB: a database analysis of missense mutations within four conserved domains in five vitamin K-dependent coagulation serine proteases using a text-mining tool. Hum Mutat. 2008;29(3):333–44. doi: 10.1002/humu.20629. [DOI] [PubMed] [Google Scholar]
- 100.Erdogmus M, Sezerman OU. Application of automatic mutation-gene pair extraction to diseases. J Bioinform Comput Biol. 2007;5(6):1261–75. doi: 10.1142/s021972000700317x. [DOI] [PubMed] [Google Scholar]
- 101.Cruts M, Theuns J, Van Broeckhoven C. Locus-specific mutation databases for neurodegenerative brain diseases. Hum Mutat. 2012;33(9):1340–4. doi: 10.1002/humu.22117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Doughty E, et al. Toward an automatic method for extracting cancer- and other disease-related point mutations from the biomedical literature. Bioinformatics. 2011;27(3):408–15. doi: 10.1093/bioinformatics/btq667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2(11):e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garten Y, Altman RB. Pharmspresso: a text mining tool for extraction of pharmacogenomic concepts and relationships from full text. BMC Bioinformatics. 2009;10(Suppl 2):S6. doi: 10.1186/1471-2105-10-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rance B, et al. A mutation-centric approach to identifying pharmacogenomic relations in text. J Biomed Inform. 2012;45(5):835–41. doi: 10.1016/j.jbi.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nelson SJ, et al. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18(4):441–8. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hakenberg J, et al. A SNPshot of PubMed to associate genetic variants with drugs, diseases, and adverse reactions. J Biomed Inform. 2012;45(5):842–50. doi: 10.1016/j.jbi.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 109.Yeniterzi S, Sezerman U. EnzyMiner: automatic identification of protein level mutations and their impact on target enzymes from PubMed abstracts. BMC Bioinformatics. 2009;10(Suppl 8):S2. doi: 10.1186/1471-2105-10-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Laurila JB, et al. Algorithms and semantic infrastructure for mutation impact extraction and grounding. BMC Genomics. 2010;11(Suppl 4):S24. doi: 10.1186/1471-2164-11-S4-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Naderi N, Witte R. Automated extraction and semantic analysis of mutation impacts from the biomedical literature. BMC Genomics. 2012;13(Suppl 4):S10. doi: 10.1186/1471-2164-13-S4-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Stenson PD, et al. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1(1):13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Veuthey AL, et al. Application of text-mining for updating protein post-translational modification annotation in UniProtKB. BMC Bioinformatics. 2013;14(1):104. doi: 10.1186/1471-2105-14-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Parthiban V, et al. Structural analysis and prediction of protein mutant stability using distance and torsion potentials: role of secondary structure and solvent accessibility. Proteins. 2007;66(1):41–52. doi: 10.1002/prot.21115. [DOI] [PubMed] [Google Scholar]
- 115.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33(Web Server issue):W306–10. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karchin R, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21(12):2814–20. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- 117.Tian J, et al. Predicting the phenotypic effects of non-synonymous single nucleotide polymorphisms based on support vector machines. BMC Bioinformatics. 2007;8:450. doi: 10.1186/1471-2105-8-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Calabrese R, et al. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30(8):1237–44. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- 119.Capriotti E, et al. WS-SNPs&GO: a web server for predicting the deleterious effect of human protein variants using functional annotation. BMC Genomics. 2013;14(Suppl 3):S6. doi: 10.1186/1471-2164-14-S3-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Capriotti E, Altman RB. Improving the prediction of disease-related variants using protein three-dimensional structure. BMC Bioinformatics. 2011;12(Suppl 4):S3. doi: 10.1186/1471-2105-12-S4-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yue P, Moult J. Identification and analysis of deleterious human SNPs. J Mol Biol. 2006;356(5):1263–74. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 122.Li B, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25(21):2744–50. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bao L, Zhou M, Cui Y. nsSNPAnalyzer: identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res. 2005;33(Web Server issue):W480–2. doi: 10.1093/nar/gki372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ferrer-Costa C, et al. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21(14):3176–8. doi: 10.1093/bioinformatics/bti486. [DOI] [PubMed] [Google Scholar]
- 125.Bromberg Y, Rost B. SNAP: predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35(11):3823–35. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schwarz JM, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–6. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 127.Mathe E, et al. Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res. 2006;34(5):1317–25. doi: 10.1093/nar/gkj518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tavtigian SV, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43(4):295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou H, Zhou Y. Distance-scaled, finite ideal-gas reference state improves structure-derived potentials of mean force for structure selection and stability prediction. Protein Sci. 2002;11(11):2714–26. doi: 10.1110/ps.0217002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Peterson TA, et al. Incorporating molecular and functional context into the analysis and prioritization of human variants associated with cancer. J Am Med Inform Assoc. 2012;19(2):275–83. doi: 10.1136/amiajnl-2011-000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yuan HY, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34(Web Server issue):W635–41. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schymkowitz J, et al. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33(Web Server issue):W382–8. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6(12):e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Prabhakar S, et al. Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 2006;16(7):855–63. doi: 10.1101/gr.4717506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clifford RJ, et al. Large-scale analysis of non-synonymous coding region single nucleotide polymorphisms. Bioinformatics. 2004;20(7):1006–14. doi: 10.1093/bioinformatics/bth029. [DOI] [PubMed] [Google Scholar]
- 136.Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15(7):978–86. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gonnelli G, Rooman M, Dehouck Y. Structure-based mutant stability predictions on proteins of unknown structure. J Biotechnol. 2012;161(3):287–93. doi: 10.1016/j.jbiotec.2012.06.020. [DOI] [PubMed] [Google Scholar]
- 142.Asthana S, et al. Analysis of sequence conservation at nucleotide resolution. PLoS Comput Biol. 2007;3(12):e254. doi: 10.1371/journal.pcbi.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–4. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Woolfe A, Mullikin JC, Elnitski L. Genomic features defining exonic variants that modulate splicing. Genome Biol. 2010;11(2):R20. doi: 10.1186/gb-2010-11-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.De Baets G, et al. SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucleic Acids Res. 2012;40(Database issue):D935–9. doi: 10.1093/nar/gkr996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36(Database issue):D820–4. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang J, et al. pfSNP: An integrated potentially functional SNP resource that facilitates hypotheses generation through knowledge syntheses. Hum Mutat. 2011;32(1):19–24. doi: 10.1002/humu.21331. [DOI] [PubMed] [Google Scholar]
- 148.Wang P, et al. SNP Function Portal: a web database for exploring the function implication of SNP alleles. Bioinformatics. 2006;22(14):e523–9. doi: 10.1093/bioinformatics/btl241. [DOI] [PubMed] [Google Scholar]
- 149.Shen TH, Carlson CS, Tarczy-Hornoch P. SNPit: a federated data integration system for the purpose of functional SNP annotation. Comput Methods Programs Biomed. 2009;95(2):181–9. doi: 10.1016/j.cmpb.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lukashin I, et al. VISTA Region Viewer (RViewer)--a computational system for prioritizing genomic intervals for biomedical studies. Bioinformatics. 2011;27(18):2595–7. doi: 10.1093/bioinformatics/btr440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88(4):440–9. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Capriotti E, Altman RB, Bromberg Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics. 2013;14(Suppl 3):S2. doi: 10.1186/1471-2164-14-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jegga AG, et al. PolyDoms: a whole genome database for the identification of non-synonymous coding SNPs with the potential to impact disease. Nucleic Acids Res. 2007;35(Database issue):D700–6. doi: 10.1093/nar/gkl826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Olatubosun A, et al. PON-P: integrated predictor for pathogenicity of missense variants. Hum Mutat. 2012;33(8):1166–74. doi: 10.1002/humu.22102. [DOI] [PubMed] [Google Scholar]
- 155.Wainreb G, et al. Protein stability: a single recorded mutation aids in predicting the effects of other mutations in the same amino acid site. Bioinformatics. 2011;27(23):3286–92. doi: 10.1093/bioinformatics/btr576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gong S, Blundell TL. Structural and functional restraints on the occurrence of single amino acid variations in human proteins. PLoS One. 2010;5(2):e9186. doi: 10.1371/journal.pone.0009186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stehr H, et al. The structural impact of cancer-associated missense mutations in oncogenes and tumor suppressors. Mol Cancer. 2011;10:54. doi: 10.1186/1476-4598-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Radivojac P, et al. Protein flexibility and intrinsic disorder. Protein Sci. 2004;13(1):71–80. doi: 10.1110/ps.03128904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vihinen M. How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genomics. 2012;13(Suppl 4):S2. doi: 10.1186/1471-2164-13-S4-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Potapov V, Cohen M, Schreiber G. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Eng Des Sel. 2009;22(9):553–60. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- 161.Vihinen M. Guidelines for reporting and using prediction tools for genetic variation analysis. Hum Mutat. 2013;34(2):275–82. doi: 10.1002/humu.22253. [DOI] [PubMed] [Google Scholar]
- 162.Yang Y, et al. Structure-based prediction of the effects of a missense variant on protein stability. Amino Acids. 2013;44(3):847–55. doi: 10.1007/s00726-012-1407-7. [DOI] [PubMed] [Google Scholar]
- 163.Thusberg J, Olatubosun A, Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32(4):358–68. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 164.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 165.Venter JC, et al. The sequence of the human genome. Science. 2001;291(5507):1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 166.Hall SS. Revolution postponed. Scientific American. 2010;303(4):60–67. doi: 10.1038/scientificamerican1010-60. [DOI] [PubMed] [Google Scholar]
- 167.Ashley EA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375(9725):1525–35. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306(5696):636–40. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 169.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xue Y, et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet. 2012;91(6):1022–32. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337(6090):100–4. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Radivojac P, et al. Gain and loss of phosphorylation sites in human cancer. Bioinformatics. 2008;24(16):i241–7. doi: 10.1093/bioinformatics/btn267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li S, et al. Loss of post-translational modification sites in disease. Pac Symp Biocomput. 2010:337–47. doi: 10.1142/9789814295291_0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mort M, et al. In silico functional profiling of human disease-associated and polymorphic amino acid substitutions. Hum Mutat. 2010;31(3):335–46. doi: 10.1002/humu.21192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Xin F, et al. Structure-based kernels for the prediction of catalytic residues and their involvement in human inherited disease. Bioinformatics. 2010;26(16):1975–82. doi: 10.1093/bioinformatics/btq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lehne B, Schlitt T. Breaking free from the chains of pathway annotation: de novo pathway discovery for the analysis of disease processes. Pharmacogenomics. 2012;13(16):1967–78. doi: 10.2217/pgs.12.170. [DOI] [PubMed] [Google Scholar]
- 178.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jesmin J, et al. Gene regulatory network reveals oxidative stress as the underlying molecular mechanism of type 2 diabetes and hypertension. BMC Med Genomics. 2010;3:45. doi: 10.1186/1755-8794-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: current approaches and outstanding challenges. PLoS Comput Biol. 2012;8(2):e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shah NH, Cole T, Musen MA. Chapter 9: Analyses using disease ontologies. PLoS Comput Biol. 2012;8(12):e1002827. doi: 10.1371/journal.pcbi.1002827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Beissbarth T. Interpreting experimental results using gene ontologies. Methods Enzymol. 2006;411:340–52. doi: 10.1016/S0076-6879(06)11018-6. [DOI] [PubMed] [Google Scholar]
- 184.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39(10):1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jiang BB, Wang JG, Wang Y. Gene prioritization for type 2 diabetes in tissue-specific protein interaction networks. Syst Biol. 2009;10801131:319–28. [Google Scholar]
- 186.Hirschman L, et al. Overview of BioCreAtIvE: critical assessment of information extraction for biology. BMC Bioinformatics. 2005;6(Suppl 1):S1. doi: 10.1186/1471-2105-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Altman RB, et al. Text mining for biology--the way forward: opinions from leading scientists. Genome Biol. 2008;9(Suppl 2):S7. doi: 10.1186/gb-2008-9-s2-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Staubert C, et al. Evolutionary aspects in evaluating mutations in the melanocortin 4 receptor. Endocrinology. 2007;148(10):4642–8. doi: 10.1210/en.2007-0138. [DOI] [PubMed] [Google Scholar]
- 189.Washington NL, et al. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7(11):e1000247. doi: 10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]