Abstract
3-[5-{2-(2,3-Dihydroxyprop-1-yl)-o-carboran-1-yl}pentan-1-yl]thymidine (N5-2OH) is a first generation 3-carboranyl thymidine analog (3CTA) that has been intensively studied as a boron-10 (10B) delivery agent for neutron capture therapy (NCT). N5-2OH is an excellent substrate of thymidine kinase 1 and its favorable biodistribution profile in rodents led to successful preclinical NCT of rats bearing intracerebral RG2 glioma. The present study explored cellular influx and efflux mechanisms of N5-2OH, as well as its intracellular anabolism beyond the monophosphate level. N5-2OH entered cultured human CCRF-CEM cells via passive diffusion, whereas the multidrug resistance-associated protein 4 appeared to be a major mediator of N5-2OH monophosphate efflux. N5-2OH was effectively monophosphorylated in cultured murine L929 [thymidine kinase 1 (TK1+)] cells whereas formation of N5-2OH monophosphate was markedly lower in L929 (TK1–) cell variants. Further metabolism to the di- and triphosphate forms was not observed in any of the cell lines. Regardless of monophosphorylation, parental N5-2OH was the major intracellular component in both TK1+ and TK1– cells. Phosphate transfer experiments with enzyme preparations showed that N5-2OH monophosphate, as well as the monophosphate of a second 3-carboranyl thymidine analog [3-[5-(o-carboran-1-yl)pentan-1-yl]thymidine (N5)], were not substrates of thymidine monophosphate kinase. Surprisingly, N5-diphosphate was phosphorylated by nucleoside diphosphate kinase although N5-triphosphate apparently was not a substrate of DNA polymerase. Our results provide valuable information on the cellular metabolism and pharmacokinetic profile of 3-carboranyl thymidine analogs.
Introduction
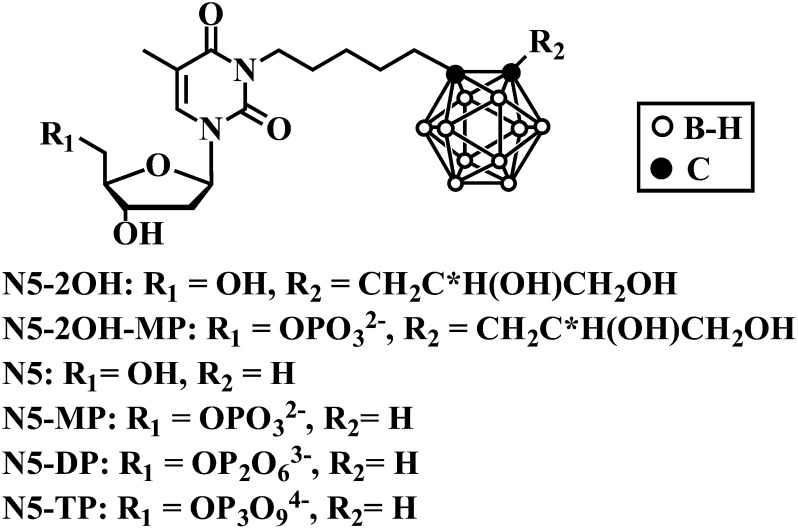
A class of nucleoside bioconjugates designated as 3-carboranyl thymidine analogs (3CTAs) has been intensively studied in recent years as potential boron-delivery agents for neutron capture therapy (NCT). Presumably, these agents accumulate selectively in malignant cells by thymidine kinase 1 (TK1)-mediated trapping through 5′-monophosphorylation (Khalil et al., 2013). TK1 is a deoxynucleoside kinase that is primarily active during the S-phase of the cell cycle (Arnér and Eriksson, 1995). The first generation 3CTA 3-[5-{2-(2,3-dihydroxyprop-1-yl)-o-carboran-1-yl}pentan-1-yl]thymidine (N5-2OH; Fig. 1) is an excellent substrate of TK1 and exhibits high uptake and retention in TK1-expressing tumor cells in vivo (Al-Madhoun et al., 2004; Barth et al., 2004, 2008). The favorable in vivo profile of N5-2OH led to successful preclinical NCT of rats bearing intracerebral RG2 glioma (Barth et al., 2008). However, almost nothing is known about cellular influx and efflux mechanisms of N5-2OH or its intracellular anabolism beyond the monophosphate level. The purpose of the present study was to gain information on these aspects of cellular 3CTA metabolism and pharmacokinetics because they are important in assessing the basic clinical potential of these agents.
Fig. 1.
Structures of N5 and N5-2OH and their phosphates.
Many purine and pyrimidine nucleosides enter cells by mediated transport processes (Plunkett and Saunders, 1991). Understanding transport mechanisms for nucleosides is important for evaluating clinical results because they have been implicated in resistance to nucleoside analog prodrugs, such as gemcitabine and capecitabine (Damaraju et al., 2003). There are four members of the equilibrative nucleoside transporter family (hENT1/2/3/4) and three of the concentrative nucleoside transporter family (hCNT1/2/3) (Cass et al., 1998). In addition, organic anion transporter (OAT) proteins mediate the exchange and transport of organic anions and show broad substrate selectivity including some cyclic nucleotides and antiviral nucleosides (Koepsell and Endou, 2004). Upon entering cells, nucleosides are converted to triphosphates by a succession of cellular kinases before incorporation into nucleic acids by DNA polymerase. Monophosphorylation is carried out by nucleoside kinases (NKs), diphosphorylation by nucleoside monophosphate kinases (NMPKs), and triphosphorylation by nucleoside diphosphate kinase (NDPK) (Arnér and Eriksson, 1995; Parks and Agarwal, 1973; Kreimeyer et al., 2001; Eriksson et al., 2002; Pasti et al., 2003). These phosphorylation steps are also critical in the activation of all biomedically relevant nucleoside analog prodrugs (Galmarini et al., 2001; Deville-Bonne et al., 2010). Three members of the family of human multidrug resistance-associated proteins (MRP4/5/8) are able to transport nucleoside monophosphate analogs out of cells against concentration gradients. The presence of MRP4/5 is associated with resistance to several nucleoside analogs used in anticancer and antiviral therapies, including zidovudine (AZT), adefovir, ganciclovir, abacavir, and gemcitabine (Borst et al., 2004; Pastor-Anglada et al., 2005; Ritter et al., 2005).
In the present study, we have investigated intracellular uptake and retention of N5-2OH and its metabolites in cultured human CCRF-CEM (TK1+) and murine L929 (TK1+) cells and their TK1-deficient variants. The impact of various cellular influx and/or efflux transporters [e.g., hENT1/2, hCNT1/2/3, organic cation transporters (OCTs), OATs, MRP4/5, P-glycoprotein (P-gp)] on cellular pharmacokinetics of N5-2OH and its metabolites was also explored. To study possible further metabolism of N5-2OH-monophosphate (MP), experiments with thymidine monophosphate kinase (TMPK), uridine monophosphate–cytidine monophosphate kinase (UMP-CMPK), and NDPK were performed. The capacity of Klenow DNA polymerase I to incorporate the triphosphate form of 3-[5-(o-carboran-1-yl)pentan-1-yl]thymidine (N5; Fig. 1), a 3CTA that is structurally related to N5-2OH (Khalil et al., 2013), into oligonucleotide templates during processive DNA synthesis was also explored.
Materials and Methods
The synthesis of N5/N5-2OH and their phosphates (Fig. 1) are described in Supplemental Data. Nitrobenzylmercaptopurine ribonucleoside (NBMPR), dilazep, unlabeled nucleosides, and other chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Yeast nitrogen base was from Difco Laboratories Inc. (Detroit, MI). [3H]Uridine ([3H]Urd) (35.5 Ci/mmol) and [3H]N5-2OH (59.1 and 117.5 Ci/mmol) were from Moravek Biochemicals (Brea, CA). [γ−32P]ATP was from PerkinElmer Life and Analytical Sciences (Boston, MA). Tissue culture (96-well) plates, cell culture media, horse serum, and fetal bovine serum (FBS) were from Gibco (Life Technologies, Grand Island, NY). Ecolite was from ICN Pharmaceuticals (Montreal, QC, Canada). The Cell Titer 96 Aqueous One Solution Cell Proliferation Assay Kit was from Promega (Madison, WI). Trypsin-EDTA and penicillin/streptomycin mixture (PeSt) were purchased from SVA (Uppsala, Sweden). Human recombinant TK1 and TMPK were prepared as described previously (Lunato et al., 1999; Carnrot et al., 2008); human recombinant cytosolic UMP-CMPK and human erythrocyte NDPK were purchased from OriGene Technologies Inc. (Rockville, MD) and Sigma-Aldrich, respectively. Light paraffin oil and silicone 550 oil were purchased from Sigma-Aldrich.
Cell Cultures and Preparation of Cell Extracts.
Murine fibroblast cell lines, TK1 positive ATCC-CCL-34 (L929 TK1+) and TK1 negative ATCC-CCL-1.3 (L929 TK1–), and the human T-lymphoblast CCRF-CEM (ATCC-CCL-119) cell line (CEM TK1+) were purchased from the American Type Culture Collection (ATCC, Manassas, VA). A subline of CCRF-CEM, deficient in thymidine kinase activity (CEM TK1–), was originally provided by B. Ullman (Oregon State University, Corvallis, OR). Cell cultures were maintained according to ATCC recommendations and grown at 37°C in a humidified incubator in the presence of 5% CO2. L929 TK+ cells were grown in Eagle’s minimal essential medium supplemented with 10% horse serum, 2 mM l-glutamine and 1% PeSt. L929 TK– cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, 4 mM l-glutamine, 1% PeSt; CCRF-CEM TK1+ and TK1– cells were grown as suspension cultures in RPMI 1640 with 10% FBS, 10 mM Hepes, 1 mM sodium pyruvate, and 1% PeSt. For uptake experiments CCRF-CEM and L929 cells were seeded at cell densities of 5 × l05 cells/ml in growth media for 24 hours to ensure actively proliferating cells with population densities not exceeding 2 × 106 cells/ml.
Metabolic Studies.
Prior to metabolic assays, L929 cells were washed once in ice-cold phosphate-buffered saline (PBS) and resuspended in growth medium containing 10 μM 3H-labeled nucleoside solution (1 part 3H-labeled nucleoside and 9 parts cold nucleoside) of N5-2OH or thymidine (dThd). Following 2-hour incubation at 37°C in a humidified incubator in the presence of 5% CO2, medium was removed from L929 cells, and cells were washed with ice-cold PBS twice, trypsinized with 0.5% trypsin-EDTA for 5 minutes, and collected by centrifugation at 600g for 5 minutes at 4°C. After centrifugation, cell pellets were resuspended with 1 ml of PBS, cell numbers were determined, and cells were recentrifuged. Cell pellets were resuspended in 200 μl of 70% MeOH/30% 20 mM EDTA on ice, frozen in liquid nitrogen, and immediately sonicated (3 × 1 minute) on ice. Supernatants were removed after centrifugation at 16,000g for 10 minutes at 4°C and frozen in dry ice, and MeOH/EDTA was evaporated by lyophilization. Samples were dissolved in the high-performance liquid chromatography (HPLC) mobile phase, and 200-μl aliquots were used for reversed-phase HPLC on a HPLC C18 column (250 × 4.6 mm; 5-μm particles) to analyze intracellular levels of [3H]N5-2OH and [3H]N5-2OH-monophosphate (MP). The mobile phase was A = 0.1% trifluoroacetic acid in water, B = 0.1% trifluoroacetic acid in acetonitrile at a flow rate of 1 ml/min. The gradient program used was as follows: 0–27 minutes, 35–70% B; 27–28 minutes, 70–35% B; 28–40 minutes, 35% B.
Intracellular [3H]dThd, [3H]dThd-MP, and [3H]dThd-DP levels in cell extracts were analyzed by DEAE-Sepharose chromatography. Cells were trypsinized with 0.5% trypsin-EDTA and centrifuged at 600g for 5 minutes. Cell pellets were resuspended in prechilled lysis buffer containing 10 mM Tris base, 50 mM NaCl, 1 mM MgCl2, 15 mM NaF, 10 mM dithiothreitol (DTT), 0.5% Nonidet P-40, and 10 mM EDTA and incubated on ice for 30 minutes. The cell extract was centrifuged twice at 16,000g for 10 minutes at 4°C. The supernatant was diluted with 0.5 M NH4HCO3 to a final concentration of 0.5 mM and applied to DEAE-Sepharose. [3H]dThd-MP eluted at a concentration of 50–75 mM NH4HCO3, and [3H]dThd-DP eluted at a concentration of 100–150 mM NH4HCO3. Combined eluates containing either [3H]dThd-MP or [3H]dThd-DP were concentrated by freeze drying and redissolved in water.
Values represent picomoles of [3H]N5-2OH and [3H]dThd metabolites per 106 cells and are from a representative experiment, which was repeated three times with similar results (Supplemental Figs. 1 and 2; Table 2).
TABLE 2.
Retention of intracellular [3H]N5-2OH and [3H]dThd metabolites in L929-TK1+ and L929-TK1– cells
The intracellular [3H]N5-2OH and [3H]N5-2OH-MP levels in cell extracts from L929 TK1+ and L929 TK1–were analyzed by HPLC. Intracellular [3H]dThd, [3H]dThd-MP, and [3H]dThd-DP levels in cell extracts from TK1+ and TK1– cells were analyzed by DEAE-Sepharose chromatography, as described in Materials and Methods. The values represent pmol of [3H]N5-2OH and [3H]dThd metabolites per 106 cells and are from a representative experiment, which was repeated three times with similar results.
| Cells | L929 TK1+ |
L929 TK1– |
||||
|---|---|---|---|---|---|---|
| N | MP | DP | N | MP | DP | |
| N5-2OH | 4.6 ± 0.5 | 1.5 ± 0.12 | ND | 6.3 ± 0.7 | 0.3 ± 0.02 | ND |
| dThd | 1.5 ± 0.16 | 5.8 ± 0.56 | 0.5 ± 0.03 | 1.4 ± 0.09 | 0.039 ± 0.004 | ND |
N, N5-2OH and dThd; MP, N5-2OH-MP and dTMP; DP, dTDP; ND, not detected.
Nucleoside Transport in Saccharomyces cerevisiae.
Yeast were separately transformed with plasmids (pYPhENT1, pYPhENT2, pYPhCNT1, pYPhCNT2, or pYPhCNT3) encoding hNTs (hENT1, hENT2, hCNT1, hCNT2, or hCNT3, respectively), as described elsewhere (Vickers et al., 2002; Zhang et al., 2003). Thus, yeast genetically manipulated in this way expresses each of the five human nucleoside transporters. Uptake of 1 μM [3H]Urd into yeast was measured, as previously described (Vickers et al., 2002; Zhang et al., 2003, 2005), using the semiautomated cell harvester (Micro96 HARVESTER; Skatron Instruments, Lier, Norway). Yeast strains were maintained in complete minimal medium (CMM) containing 0.67% yeast nitrogen base amino acids (as required to maintain auxotrophic selection), and 2% glucose. Yeast cells were grown in CMM 2% glucose to an absorbance at 600 nm (A600 = 0.5–1.0), washed twice, and resuspended to A600 = 4.0. Yeast were incubated at ambient temperature with graded concentrations (0–1 mM) of N5-2OH in the presence of 1 μM [3H]Urd in transport buffer (pH 7.4) containing 20 mM Tris, 3 mM K2HPO4, 1 mM MgCl2, 1.4 mM CaCl2, and 5 mM glucose with 144 mM NaCl. Yeast were also incubated with 1 μM [3H]Urd in the absence of test compound as positive controls. Values from cultures without test compound were used to calculate the “% control.” Urd self-inhibition was used as an internal control to define maximum inhibition of mediated transport. Transport reactions were initiated by rapid mixing of 50-μl yeast suspensions with or without graded concentrations of N5-2OH with 50 μl of 2 × [3H]nucleoside in 96-well microtiter plates. Yeast cells were collected on filter mats using a Micro96 Cell Harvester and rapidly washed with deionized water. Individual filter circles, corresponding to wells of the microtiter plates, were removed from filter mats with forceps and transferred to vials for quantification of radioactivity by scintillation counting.
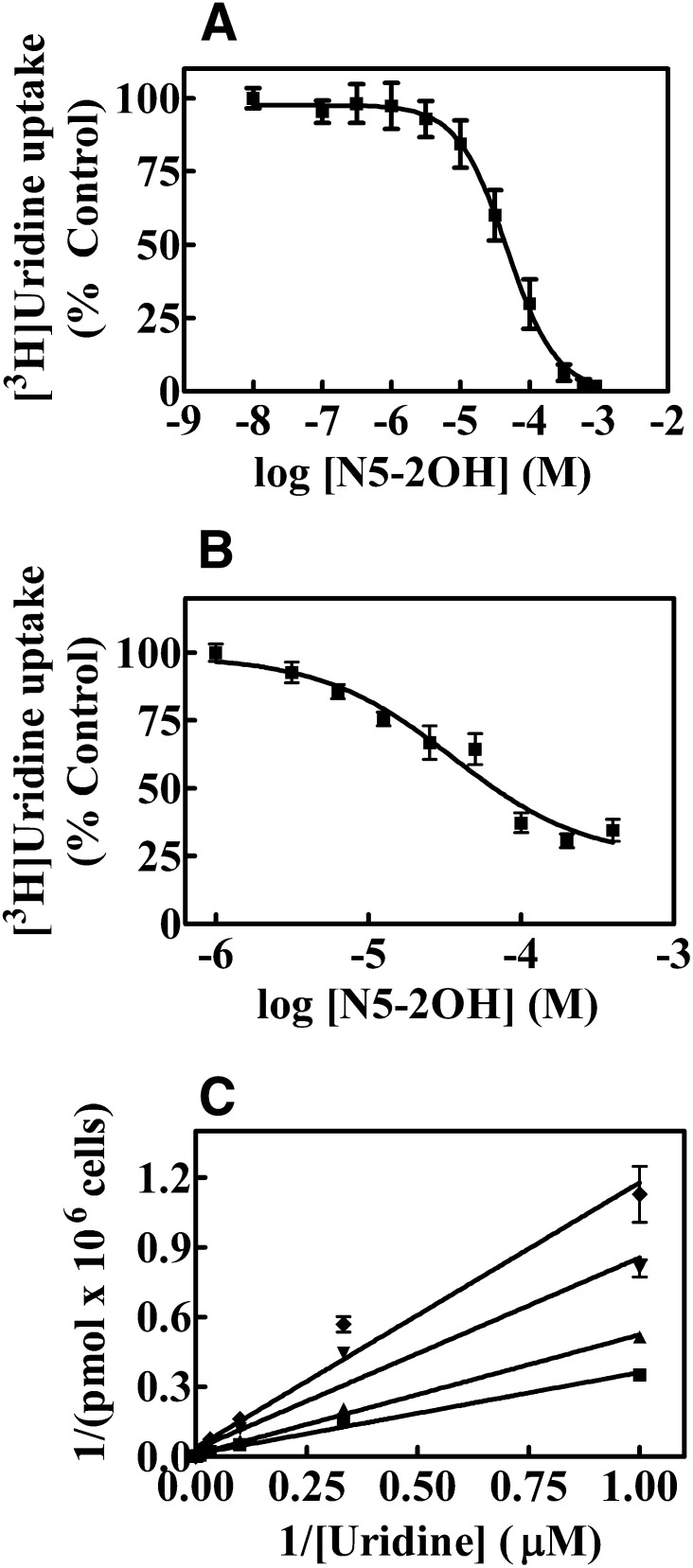
Data were subjected to nonlinear regression analysis using GraphPad Prism software (version 4.03; GraphPad Software Inc., San Diego, CA) to obtain the concentration of test compound that inhibited growth of treated cells by 50% relative to that of untreated cells (IC50 values). IC50 values were determined from concentration-effect curves. Each experiment was conducted with nine concentrations and six replicates per concentration and was repeated three times to obtain accurate IC50 values (Fig. 2A; Table 1).
Fig. 2.
Effects of N5-2OH on [3H]Urd uptake in yeast (A) and CEM cells (B). Yeast or CEM cells were incubated with 1 μM [3H]Urd for 10 minutes or 30 seconds, respectively, in the absence or presence of increasing N5-2OH concentrations (0–1000 μM). Shown are experiments performed with six and three replicates per concentration, respectively, for yeast and CEM cells, and data are expressed as mean ± S.D. Error bars are not shown where S.D. values are smaller than the size of the symbol. (C) Effect of increasing concentrations of N5-2OH on the uptake of [3H]Urd in CEM cells is shown. The effects of 0 (■), 5 (▲), 25 (▼) or 50 (♦) μM N5-2OH on Urd uptake rates were assessed and values with mean ± S.D. are shown in each panel. Each experiment was repeated three times.
TABLE 1.
Effects of N5-2OH on [3H]Urd uptake in S. cerevisiae producing recombinant hENT1, hENT2, hCNT1, hCNT2, or hCNT3
Shown are IC50 values that represent N5-2OH concentrations at which uptake of 1 μM [3H]Urd was inhibited by 50% over a fixed time period as determined by computer-generated concentration-effect curves.
| Transporters | IC50 ± S.E. |
|---|---|
| μM | |
| hENT1 | 44 ± 5 |
| hENT2 | 300 ± 47 |
| hCNT1 | 580 ± 50 |
| hCNT2 | 360 ± 90 |
| hCNT3 | 700 ± 30 |
Nucleoside Transport Assays in CEM Cells.
CCRF-CEM wild-type cells are known to only express hENT1 (Belt et al., 1993). Inhibition of 1 μM [3H]Urd uptake (30 seconds) was measured at ambient temperature in CEM cells in transport buffer (pH 7.4) containing 20 mM Tris, 3 mM K2HPO4, 1 mM MgCl2, 1.4 mM CaCl2, and 5 mM glucose with 144 mM NaCl in absence or presence of graded concentrations (0–0.5 mM) of N5-2OH. At the end of the uptake intervals, permeant-containing solutions were removed by aspiration, cells were spun down and quickly rinsed twice with transport buffer, and solubilized with 5% Triton X-100. Radioactivity in solubilized extracts was measured by liquid scintillation counting. Uptake values were expressed as picomoles per 106 cells and converted to %-control activity, and graphs were generated using GraphPad Prism. Each experiment was conducted three times with triplicate measurements for each condition (Figs. 2, B and C).
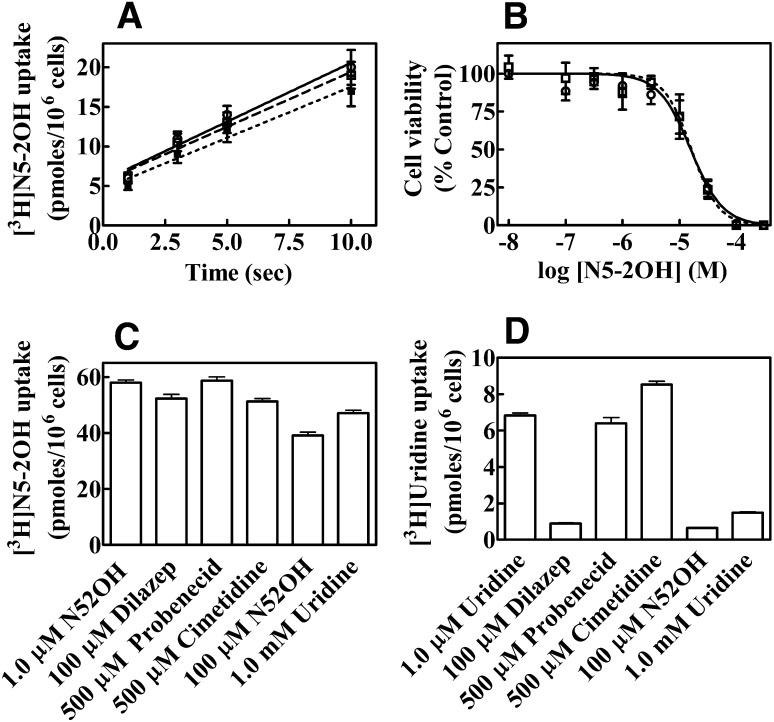
Inhibition of [3H]N5-2OH and [3H]Urd uptake was determined by exposing CEM cells to 1 μM [3H]N5-2OH or 1 μM [3H]Urd in absence or presence of either 100 μM dilazep (ENT inhibitor), 500 μM probenecid (OAT inhibitor), 500 μM cimetidine (OCT inhibitor), 100 μM N5-2OH, or 1 mM Urd for 1 minute, after which uptake was terminated by spinning cells though oil. Cells were washed by centrifugation and cell-associated radioactivity was determined by scintillation counting. Each experiment was conducted with six replicates per condition and was repeated three times with similar results (Fig. 3, C and D).
Fig. 3.
Effects of transport inhibitors on uptake of [3H]N5-2OH and [3H]Urd in CEM cells and toxicity of N5-2OH in CEM cells in absence or presence of NBMPR. (A) CEM cells incubated with [3H]N5-2OH in the absence (○, solid line) or presence of 0.1 μM NBMPR (□, dashed line), or 1 μM NBMPR (●, dotted line). Transport was measured over 10 seconds. (B) The effect of NBMPR on the toxicity of N5-2OH to CEM cells that were exposed to graded concentrations (0–300 μM) of N5-2OH in the absence (○, solid line) or presence of 1 μM NBMPR (□, dashed line) for 72 hours. Cell viability was measured by the MTS assay as described in Materials and Methods. Cytotoxicity was determined by expressing values as % control of untreated cells. The results (mean ± S.D.) shown are average of three experiments each conducted with six replicates per point and repeated three times. (C and D) Uptake of 1 μM [3H]N5-2OH (C) or 1 μM [3H]Urd (D) into CEM cells. Uptake was measured for 1 minute in the absence or presence of 100 μM dilazep (ENT inhibitor), 500 μM probenecid (OAT inhibitor), 500 μM cimetidine (OCT inhibitor), 100 μM N5-2OH, or 1 mM Urd and cell-associated radioactivity was quantitated. Values (pmol/106 cells, mean ± S.D.) plotted are from a representative experiment, which was repeated three times with similar results.
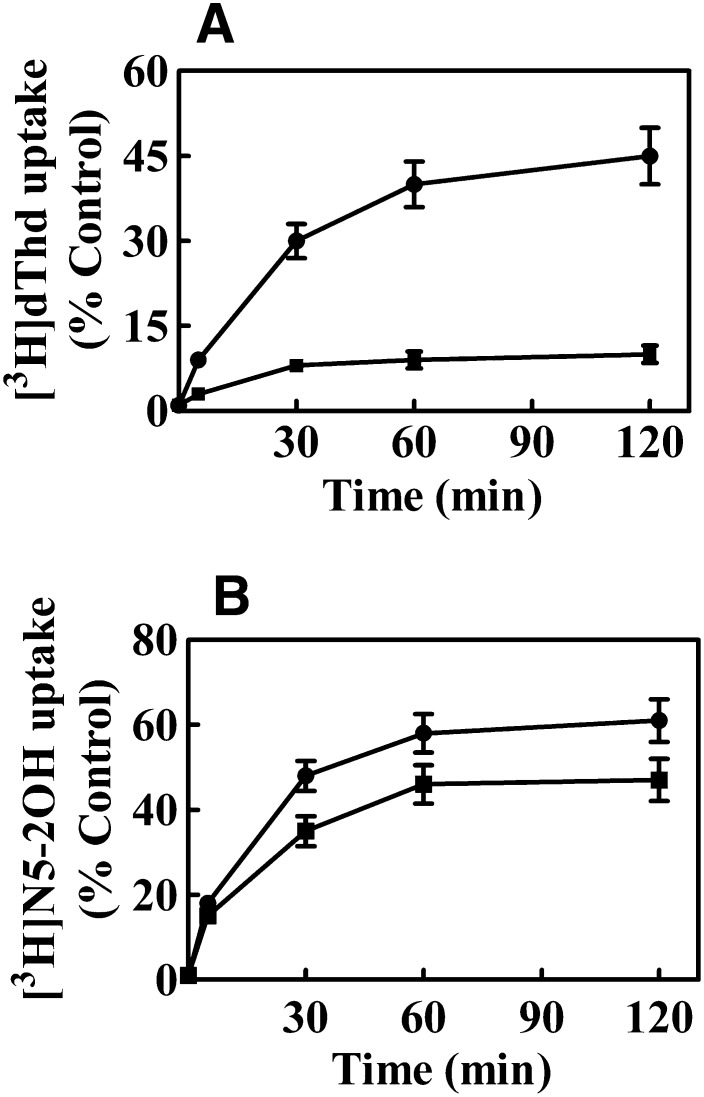
Nucleoside uptake was measured at ambient temperature in actively proliferating cells (2 × 106 cells/ml) using the oil-stop transport method (Harley et al., 1982). CEM TK1+ and CEM TK1– cells were grown as described above. Cells were harvested by centrifugation (600g, 5 minutes), washed twice with PBS, and then resuspended in 100 μl of transport buffer (5 × 106 cells). The time courses for initial rates of [3H]N5-2OH uptake in CEM TK1+ cells were determined using a rapid sampling procedure (10 seconds) that was initiated by addition of 100 μl of cells to 100 μl of [3H]N5-2OH solution to a final concentration of 1 μM and terminated by the addition of 100 μl of 1 mM N5-2OH solution followed by centrifugation (16,000g, 30 seconds) through transport oil (Fig. 3A). Cellular uptake of [3H]N5-2OH and [3H]dThd in CEM TK1+ and CEM TK1– cells were determined at 30, 60, 90, and 120 minutes using the procedure described above. Final nucleoside concentrations were 1 μM and termination was accomplished by addition of 1 mM N5-2OH or dThd solutions followed by centrifugation through transport oil (Fig. 4). Cell pellets were solubilized in 5% Triton X-100 and cell-associated radioactivity was determined by liquid scintillation counting.
Fig. 4.
Cellular uptake of [3H]dThd (A) and [3H]N5-2OH (B) into CEM-TK1+ (●) and CEM TK1– (■) cells during incubation with nucleosides for up to 2-hour time-courses of cellular uptake of [3H]N5-2OH and [3H]dThd were determined by addition of 100 μl of cells to 100 μl of labeled nucleoside solution to a final concentration of 1 μM and further processing as described in Materials and Methods. % Control is the mean counts per minute of cell-associated radioactivity compared with total counts per minute used for initiation of nucleoside transport (100%).
The role of hENT1-mediated uptake of N5-2OH into CEM TK1+ was studied in the presence of a range of concentrations of the hENT1 specific inhibitor NBMPR (0.1 and 1.0 μM). Cells were preincubated with inhibitors for 30 minutes, washed twice with PBS, and incubated with 1 μM [3H]N5-2OH for 1 hour (Fig. 3A).
Uptake values were expressed as picomoles per 106 cells (Fig. 3A) or % control (Fig. 4), which is the mean counts per minute of cell-associated radioactivity compared with total counts per minute used for initiation of nucleoside transport (100%). Graphs were generated using GraphPad Prism. All experiments were performed in triplicates.
Cytotoxicity Assays.
The Cell Titer 96 proliferation assay kit was used to quantify drug-induced cytotoxicity. Unless otherwise noted, CEM cells seeded in 96-well plates were exposed to graded concentrations (0–100 μM) of N5-2OH in the absence or presence of 1 μM NBMPR for 72 hours after which they were treated with 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) reagent for determination of cytotoxicity. IC50 values were calculated from nonlinear regression analyses of values plotted as percentages of control values against the logarithm of drug concentrations (Fig. 3B).
Efflux Studies.
CCRF-CEM wild-type are known to express efflux pump proteins approximately in the following order: MRP4 > > MRP5 > P-gp (Peng et al., 2008). For efflux studies CEM-TK1+ cells (1 × 106 per ml) were seeded in T75 flasks in growth media consisting of RPMI-1640 (without phenol red) with 10% FBS, 10 mM HEPES, 1 mM sodium pyruvate, and 1% PeSt and grown overnight. Cells were incubated for 1 hour with 1 μM [3H]N5-2OH, washed twice in ice-cold PBS, and then incubated in growth medium without [3H]N5-2OH for 150 minutes. To determine the amount of [3H]N5-2OH-MP in cells at the beginning of efflux, a portion of the cell-containing mixture was taken immediately, cells were pelleted by centrifugation for 1 minutes at 12,000g, resulting cell pellets were solubilized by 5% Triton X-100, and radioactivity was determined by scintillation counting. The total counts per minute in 1-ml portions of cell-free medium was determined by scintillation counting at different time points. Levels of [3H]N5-2OH and [3H]N5-2OH-MP in the growth media of CEM-TK1+ cells were analyzed by HPLC at the indicated timed points.
For efflux inhibition experiments, CEM-TK1+ cells were incubated with 1 μM nucleoside mixture, composed of 0.1 μM radiolabeled [3H]N5-2OH and 0.9 μM unlabeled N5-2OH, for 2 hours at 37°C in growth media as described above with either dipyridamole, indomethacin, or verapamil at a concentration of 100 μM. Cells were washed twice with ice-cold PBS, followed by addition of prewarmed media, and incubated for 2 hours at 37°C. [3H]N5-2OH-MP concentrations in the media from cells were determined by HPLC. Values shown are % counts per minute of [3H]N5-2OH-MP in media with inhibitors relative to 100% of control values without inhibitors (Fig. 6).
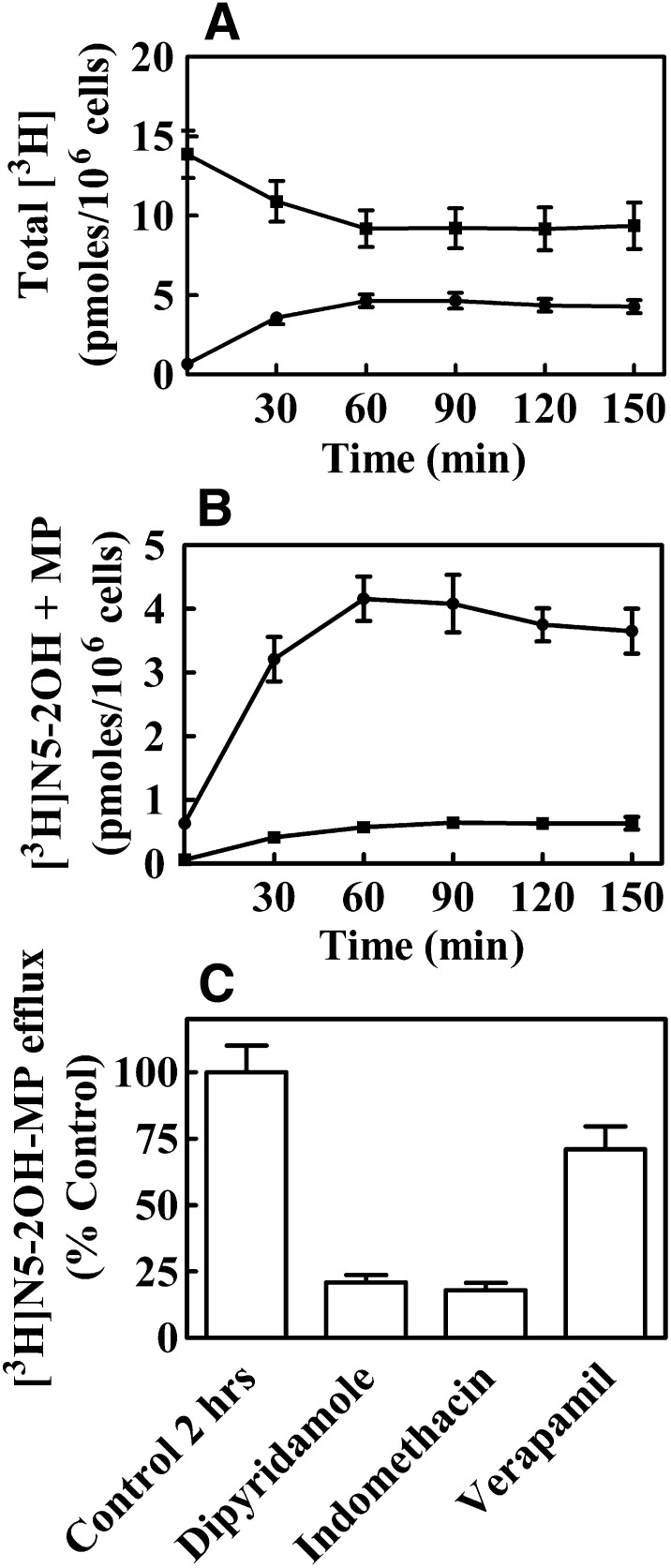
Fig. 6.
Time-dependent efflux of total [3H]N5-2OH metabolites (A) and [3H]N5-2OH monophosphate to the growth media (B) of CEM TK1+ cells. (C) Effects of transport inhibitors on the efflux of N5-2OH-MP from CEM TK1+ cells. CEM TK1+ cells were incubated 1 hour with 1 μM [3H]N5-2OH, washed with PBS, and then incubated in growth media without [3H]N5-2OH for 150 minutes. The total intracellular counts per minute (A, ■) and total counts per minute in free media (A, ●) were determined by scintillation counting at different time points after removal of the labeled nucleoside. N5-2OH (B, ●) and N5-2OH-MP (B, ■) in free media were analyzed by HPLC as described in Materials and Methods. Results shown are the average of three experiments. For the efflux inhibition experiments, cells were incubated with 1 μM labeled nucleoside solution [3H]N5-2OH for 2 hours at 37°C in growth media in the presence of 100 μM dipyridamole, indomethacin, or verapamil. [3H]N5-2OH-MP in media from cells was determined by HPLC as described in Materials and Methods. Values show the % of counts per minute of [3H]N5-2OH-MP in media with inhibitors to 100% of the control experiment without inhibitors in media.
Phosphate Transfer Assay.
The adenosine 5′-triphosphate transfer assay was performed with 0.05 μM [γ−32P]ATP (10 μCi/μl), 100 μM ATP, 50 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 100 mM KCl, 0.5 mg/ml bovine serum albumin (BSA), 10 mM DTT, and different concentrations of nucleoside analog. Reactions were initiated by adding either TK1, UMP-CMPK, TMPK, or NDPK followed by incubation at 37°C and termination by short boiling after different time points. Four microliters of reaction solutions were applied to polyethylenimine (PEI)-cellulose thin-layer chromatography (TLC) plates. Depending on the enzyme reaction, chromatography was performed with different buffers. For identification of nucleoside diphosphates, chromatography was performed with 0.2 M NaH2PO4 buffer. Triphosphate products of thymidine triphosphate (dTTP) and N5-triphosphate (TP) from the NDPK assay were separated with 0.4 M NaH2PO4 and 30% isopropanol. Phosphates were detected by autoradiography following 1-hour exposures of TLC plates to a phosphor imaging plate (BAS cassette 2040; Fujifilm, Tokyo, Japan) using the phosphorimaging system, Fuji BAS 2500/LAS 1000 (Fujifilm), in combination with an Image Reader V 1.7E (Supplemental Fig. 4; Fig. 5, A and B).
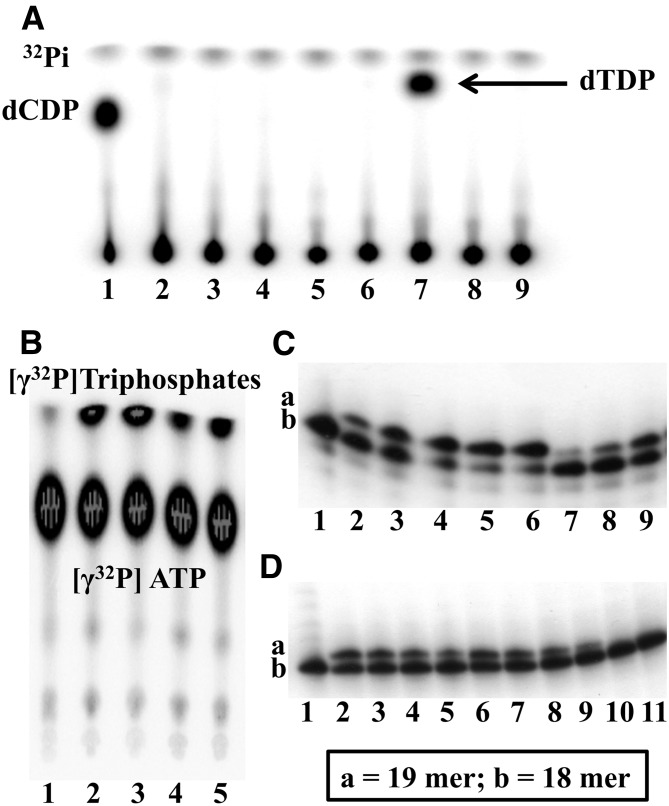
Fig. 5.
(A) Autoradiograms of a TLC analysis of the products from a [γ−32P]ATP phosphate transfer assays with UMP-CMPK (1–4) or TMPK (6–9) using dCMP (1, 6), dTMP (2, 7), N5-MP (3, 8), and N5-2OH-MP (4, 9) as substrates. Lane 5, [γ−32P]ATP only. (B) Autoradiogram of a TLC analysis of the diphosphate products from a [γ−32P]ATP phosphate transfer assay with NDPK using dTDP (2, 3) and N5-DP (4, 5) as substrates. The assays were performed for 30 (2, 4) and 60 (3, 5) minutes. [γ−32P]ATP (1) served as a control. (C and D) Autoradiograms of a gel analysis of the reaction products of dTTP, AZT-TP, and N5-TP with Klenow DNA polymerase I using a running start primer-template DNA. (C 1–6) Incorporation of dTTP into the running start primer at six different concentrations of dTTP (0, 1, 10, 25, 50, and 100 μM). (C 7–9) Incorporation of the dTTP product from a coupled synthesis using dThd and TK1, TMPK, and NDPK, as described in Materials and Methods. (Dilutions: C7 = 1/16, C8 = 1/8, C9 = 1/4.) The 16-mer primer template was incubated with T4 polynucleotide kinase and [γ−32P]ATP for 10 minutes at 37°C leading to formation of a 32Pi-labeled 18-mer nucleotide product (C1 and D1). Additions of dTTP (C 1–6) or dTTP (D 2–5), AZT-TP (D 6–9), and N5-TP (D 10, 11) from coupled reactions and Klenow DNA polymerase to the labeled 18-mer only leads to the formation of 32Pi-labeled 19-mer nucleotide products for dTTP and AZT-TP (Dilutions: D2/D6 = 0, D3/D7 = 1/2, D4/D8 = 1/4, D5/D9 = 1/8, D10 = 0, D = 1/2). Reactions were terminated by addition of 2 volumes of 90% formamide, 0.25 M EDTA. Products were analyzed on 15% polyacrylamide-7 M urea gels and detected by phosphorimaging (1 hour).
Coupled Synthesis of dTTP and AZT Triphosphate.
A coupled synthesis of dTTP and AZT triphosphate (AZT-TP) was performed using 200 μM of dThd or AZT with 0.05 μM [γ−32P]ATP (10 μCi/μl) and 100 μM ATP in phosphate transfer assay buffer [50 mM Tris-HCl (pH 7.6), 5 mM MgCl2, 100 mM KCl, 0.5 mg/ml bovine serum albumin, 10 mM DTT]. The reaction was initiated by adding a mixture of recombinant human TK1 (100 ng), TMPK (100 ng), UMP-CMPK (100 ng), and NDPK (100 ng) followed by incubation at 37°C and termination by boiling after different time points. For separation of nucleoside monophosphates and diphosphates in the coupled reactions, chromatography was performed for 8–12 hours using isobutyric acid: NH4OH/H2O (66:1:33) (v/v) as the mobile phase. Products of the kinase reactions were detected by autoradiography as described above (Supplemental Fig. 5; Fig. 5, C and D).
DNA Polymerase Running Start Assays.
To prepare primer templates for running start reactions, the 16-mer primer 5′-CGC CCA CGC GGC AGA G-3′ (Invitrogen, Carlsbad, CA) was 5′-end labeled with T4 polynucleotide kinase (as described in the manufacturer’s procedure) and [γ–32P]ATP, desalted on a PD SpinTrap G-25 column (GE Healthcare, Pataskala, OH) in TEN buffer (10 mM Tris-Cl, pH 8.0, 1 mM EDTA, 100 mM NaCl), and annealed in a 1:2 ratio to the respective 36-mer templates 3′-GCG GGT GCG CCT GCT CTT ACC TCT TCT CTC TTC TCT-5′ (Invitrogen, Carlsbad, CA) in TEN buffer by heating to 75°C for 5 minutes followed by slow cooling to room temperature. Incorporation of triphosphates into the primer template was carried out in a volume of 20 μl for 45 minutes at 37°C with the 5-IU Klenow fragment of Escherichia coli DNA polymerase I (New England BioLabs Inc., Ipswich, MA), 0.25 pmol of labeled primer-template, and different concentrations of deoxynucleoside triphosphates.
Incorporation of triphosphates from coupled reactions into primer templates was carried out in final volumes of 20 μl by adding 5 μl of the original reaction product or 5 μl of diluted reaction product. Four dilution ratios (1/2, 1/4, 1/8, 1/16) were generated by adding appropriate quantities of reaction buffer to the original reaction product (Fig. 5, C and D).
Results
Effects of N5-2OH on [3H]Urd Uptake in Yeast and CEM Cells.
The inhibitory activity of N5-2OH on transporter-mediated uptake of 1 μM [3H]Urd was determined using hENT1/2 and hCNT1/2/3-producing yeast, as described in Materials and Methods. Effects of N5-2OH on transporter-mediated Urd uptake were assessed in concentration-dependent inhibition of [3H]Urd transport experiments to determine IC50 values for each of the five recombinant transporters. Representative concentration-effect curves for inhibition of hENT1-mediated Urd transport by N5-2OH are shown in Fig. 2A. It was evident that N5-2OH produced dose-dependent inhibition of [3H]Urd transport at μM concentrations. IC50 values obtained from similar experiments with yeast producing each of the five transporters are summarized in Table 1. Results indicate that only hENT1 interacted significantly with N5-2OH.
Since N5-2OH inhibited hENT1-mediated [3H]Urd transport in yeast radiotracer experiments, its inhibition of hENT1-mediated transport of [3H]Urd was also evaluated in human CEM cells, which possess hENT1 as the major nucleoside transporter (Belt et al., 1993). Figure 2B shows effects of graded N5-2OH concentrations on 1 μM [3H]Urd transport in CEM cells. These concentration-effect studies yielded IC50 values of 36 ± 5 μM for N5-2OH (Fig. 2B) for inhibition of hENT1-mediated Urd transport. Figure 2C shows effects of fixed concentrations of N5-2OH on Urd transport rates. Data were analyzed using Lineweaver-Burk plot, which shows competitive inhibition of Urd transport by N5-2OH. Further analysis by Dixon plot gave a Ki value of 22 ± 1.6 μM.
Effects of Inhibitors on Uptake of 1 μM [3H]N5-2OH or 1 μM [3H]Urd and of NBMPR on Cytotoxicity of N5-2OH in CEM Cells.
Initial rates of uptake of 0.1 μM [3H]N5-2OH measured for up to 10 seconds with CEM cells are shown in Fig. 3A. Uptake appeared to be linear and the presence of either 0.1 μM or 1 μM NBMPR had no effect on initial rates, i.e., 1.33 ± 0.26 mol/s per 106 cells (without NBMPR) compared with 1.21 ± 0.28 mol/s per 106 cells with NBMPR (Fig. 3A). Results of cytotoxicity studies with NBMPR were consistent with the lack of effect on initial rates of uptake in that exposures to graded concentrations (0–100 μM) of N5-2OH in the absence or presence of NBMPR (1 μM) for 72 hours yielded similar results (Fig. 3B).
Uptake of 1 μM [3H]N5-2OH or 1 μM [3H]Urd into CEM cells was measured in the absence or presence of either 100 μM dilazep (ENT inhibitor), 500 μM probenecid (OAT inhibitor), 500 μM cimetidine (OCT inhibitor), 100 μM N5-2OH, or 1 mM Urd. Results obtained with N5-2OH (Fig. 3C) demonstrated minimal inhibition of uptake by dilazep, probenecid, cimetidine, or excess Urd. In contrast, Urd uptake (Fig. 3D) in CEM cells was inhibited by both dilazep and excess Urd as expected in the case of mediated uptake. Excess N5-2OH inhibited [3H]uridine uptake, indicating interaction with hENT1, whereas excess N5-2OH had a very small to negligible effect on uptake of [3H]N5-2OH uptake.
Uptake and Anabolism of N5-2OH in Cultured TK+ and TK– Cells.
Earlier studies with N5 and N5-2OH have shown a 5- to 10-fold difference in uptake and retention of these 3CTAs in cultured L929 TK+ and TK1– cells (Barth et al., 2004). By use of the same cell lines as well as CEM TK1+ and TK1– cells, but different experimental conditions and compound detection methods, we characterized intracellular [3H]N5-2OH metabolites. In accordance with results obtained with the TMPK enzyme assay (see below), only [3H]N5-2OH and its monophosphate were detected within cell extracts (Supplemental Figs. 1 and 2). The uptake mechanism of N5-2OH by CEM TK1+ and TK1– cells, both at long and short periods of incubation, was investigated. The time course of cellular uptake and metabolism of [3H]N5-2OH was followed for up to 120 minutes, as described in Materials and Methods. Uptake and retention of dThd and N5-2OH (and their metabolites) reached steady-state levels after 60 minutes of incubation in both CEM TK1+ and CEM TK1– cells (Fig. 4). Levels of [3H]dThd in TK1– cells were more than 10-fold lower than that in TK1+ cells (Fig. 4A), whereas the difference in [3H]N5-2OH levels between cell lines was less pronounced (Fig. 4B). The intracellular level of [3H]dThd found in TK1– cells after 60 minutes was 10%, whereas that in TK1+ cells was 45%. In contrast, intracellular levels of [3H]N5-2OH were approximately 40 and 55%, respectively.
In a chase experiment, a 2-hour labeling period with [3H]N5-2OH was followed by 24 hours incubation in compound-free medium, resulting in a retention of 7.8% of total counts per minute in L929 TK1+ cells and 6.8% of total counts per minute in L929 TK1– cells (data not shown). The intracellular concentrations of [3H]N5-2OH and [3H]N5-2OH-MP in TK1+ cells were 4.6 ± 0.5 and 1.5 ± 0.12 pmol/106 cells, respectively (Table 2). Retention of [3H]N5-2OH-MP in TK1– cells was approximately 5-fold lower than in TK1+ cells, whereas the level of [3H]N5-2OH was similar in both cell lines (Table 2).
A control experiment was performed with [3H]dThd, i.e., a 2-hour labeling period followed by 24 hours incubation in compound-free medium and, in this case, 9.6% of total counts per minute was retained in L929 TK1+ cells and 0.3% of total counts per minute in L929 TK1– cells (data not shown). Intracellular concentrations of dThd, dTMP, and dTDP in L929 TK1+ were 1.5 ± 0.16, 5.8 ± 0.56, and 0.5 ± 0.03 pmol/106 cells, respectively, and 1.4 ± 0.09, 0.039 ± 0.004, and 0 pmol/106cells, respectively, in L929 TK1– cells (Table 2).
The obtained data showed that N5-2OH could be taken up and retained in a TK1-independent fashion in two pairs of TK1-positive and -negative cell lines. In the case of L929 TK1+ cells, [3H]N5-2OH-MP was retained significantly longer than in L929 TK1– cells, indicating that formation of N5-2OH-MP was catalyzed by TK1. There was no indication for further phosphorylation of this metabolite in L929 cells (Supplemental Figs. 1 and 2).
Phosphorylation Studies with N5 and N5-2OH Monophosphates Using Recombinant Kinases.
It is well established that N5, N5-2OH, and numerous other 3CTAs are good substrates of TK1 (Khalil et al., 2013). To study the further anabolism of N5 and N5-2OH, a series of experiments with human nucleoside mono- and diphosphate kinases were performed using appropriate phosphorylated substrates. Two recombinant human pyrimidine nucleoside monophosphate kinases, UMP-CMPK and TMPK, were tested for their capacity to phosphorylate N5- and N5-2OH-MP. Monophosphates and diphosphates (DP) of N5 and N5-2OH (Fig. 1) were synthesized as described in Supplemental Schemes 1 and 2. N5-MP and N5-DP were used in these studies as surrogate systems for the corresponding phosphates of N5-2OH because of anticipated stability issues during synthesis of the latter ones, in particular N5-2OH-diphosphate (DP) (Supplemental Data). This approach was warranted since N5 and N5-2OH appeared to have generally similar enzymatic properties (Al-Madhoun et al., 2004; Hasabelnaby et al., 2012).
Both recombinant UMP-CMPK and TMPK showed high activities with their endogenous substrates dCMP and dTMP, whereas N5-MP or N5-2OH-MP substrates did not produce detectable amounts of diphosphate products (Fig. 5A), even at extended reaction times (Supplemental Fig. 4). Computational docking studies reinforced our experimental finding that N5-2OH-MP is not a substrate for TMPK (Supplemental Fig. 3). Docking of dTMP into the crystal structure of TMPK reproduced the pose of cocrystallized dTMP accurately [lowest root mean squared deviation (RMSD) = 1.03 Å], whereas N5-2OH-MP docked in marked distance to the original dTMP binding site (lowest RMSD = 6.43 Å).
It was demonstrated by TLC that human NDPK phosphorylated N5-DP to N5-TP with a phosphorylation efficiency of approximately 30% relative to that of dTDP (Fig. 5B). Increasing the reaction time from 30 to 60 minutes led to an increase of N5-TP formation by a factor of 2, whereas the rate of dTTP formation using dTDP as the substrate remained unchanged, indicating a substantially lower efficiency of N5-DP to serve as substrate for NDPK.
DNA Incorporation Studies.
The capacity of the Klenow DNA polymerase I enzyme to incorporate N5-TP, AZT-TP, and dTTP into oligonucleotide templates during processive DNA synthesis, i.e., using running start primer-templates, was investigated (Copeland et al., 1992; Eriksson et al., 1995). For these studies we used a coupled synthesis of dTTP and AZT-TP by adding a mixture of recombinant human TK1, TMPK, UMP-CMPK, and NDPK to either dThd or AZT. For the synthesis of N5-TP we used N5-DP as a substrate for NDPK as described above. As a control for each enzyme step, we carried out a series of assays using only TK1 or combinations of TK1 with TMPK or UMP-CMPK. A TLC analysis of mono- and diphosphate products in the enzyme reactions with dThd and AZT is shown in Supplemental Fig. 5. The running start primer reaction was performed as described in Materials and Methods, and a gel analysis of the reaction products with Klenow DNA polymerase I and dTTP, AZT-TP, and N5-TP using a running start primer-template DNA is shown Fig. 5, C and D. Incubation of the 16-mer primer template with T4 polynucleotide kinase and [γ−32P]ATP led to 32Pi-labeled 18-mer nucleotide product. Both dTTP and coupled synthesis dTTP product were incorporated by Klenow DNA polymerase I into the running start primer with the formation of 32Pi-labeled 19-mer nucleotide products. The coupled synthesis AZT-TP product was also a substrate for Klenow DNA polymerase I, whereas N5-TP apparently was not, since no incorporation of N5-TP into the oligonucleotide was detected (Fig. 5D).
Cellular Efflux of [3H]N5-2OH.
Efflux of N5-2OH was measured in CEM-TK1+ cells, which were incubated with 1 μM [3H]N5-2OH for 1 hour and then maintained in nucleoside-free growth medium for 150 minutes, as described in Materials and Methods. One milliliter of medium was removed at different time points for the determination of total radioactivity from [3H]N5-2OH and its metabolites (Fig. 6A) and for HPLC analysis of [3H]N5-2OH and [3H]N5-2OH-MP released from cells (Fig. 6B).
Removing [3H]N5-2OH from the media led to a rapid loss of labeled nucleoside from the cells. There was decrease in radioactivity (~30%) 60 minutes after the transfer of cells to nucleoside-free media. Following this, radioactivity levels reached a plateau (Fig. 6A). Time-dependent efflux of [3H]N5-2OH and [3H]N5-2OH-MP is shown in Fig. 6B. Approximately 86% of the total radioactivity in the media at 150 minutes was [3H]N5-2OH, with the remaining 14% being [3H]N5-2OH-MP. During the incubation period from 60 to 150 minutes, the ratio of extracellular to intracellular [3H]N5-2OH-MP was 15–85% (data not shown).
We also studied effects of specific efflux inhibitors—e.g., dipyridamole, indomethacin and verapamil—on extracellular N5-2OH-MP levels (Fig. 6C). Results showed that dipyridamole, an inhibitor of MRP4 and MRP5 (Ritter et al., 2005), and indomethacin, a relatively selective inhibitor of MRP4 (Ritter et al., 2005), decreased the efflux of N5-2OH-MP by approximately 80%. Verapamil, an inhibitor of P-gp (Summers et al., 2004), decreased efflux of N5-2OH-MP from CEM TK+ cells only by approximately 30%.
Discussion
Data from earlier studies suggested that phosphorylation by TK1 was required for uptake and retention of the N5-2OH in malignant cells both in vitro and in vivo (Al-Madhoun et al., 2004; Barth et al., 2004, 2008). However, little information is available regarding the complete intracellular metabolism of N5-2OH, including its potential incorporation into DNA. In addition, mechanisms of cellular membrane traversal of N5-2OH and its metabolites are largely unexplored.
Only hENT1-mediated Urd transport was inhibited by N5-2OH, both in yeast and CEM cells. In experiments with CEM cells, Urd uptake inhibition by N5-2OH was shown to be competitive. However, the high affinity hENT1 transport inhibitor NBMPR had minimal effects on uptake of N5-2OH, suggesting that hENT1 played no major role in the uptake of N5-2OH by CEM cells. Furthermore, the cytotoxicity of N5-2OH for CEM cells in the presence of 1 μM NBMPR was the same as in its absence, thus confirming that hENT1 was not important in mediating uptake of N5-2OH into CEM cells.
Uptake of 1 μM [3H]N5-2OH into CEM cells was further evaluated in presence or absence of several transport inhibitors, including dilazep, NBMPR, probenecid, and cimetidine as well as excess nonradioactive Urd and N5-2OH. Excess nonradioactive N5-2OH inhibited [3H]N5-2OH uptake to a low extent, suggesting the possible presence of an unidentified uptake mechanism. All other transport inhibitors had little or no effect on uptake of [3H]N5-2OH into cells. In contrast, [3H]Urd uptake in CEM cells was inhibited by both dilazep and excess Urd, which is consistent with mediated uptake by hENT1, a transporter known to be selectively expressed in CEM cells (Belt et al., 1993). The effect of excess nonradioactive N5-2OH on uptake of 1 μM [3H]Urd by recombinant hENT1 in yeast suggested that N5-2OH was binding to the uridine binding site of hENT1 and competitively blocked [3H]Urd uptake. Overall, these results strongly suggest that cellular uptake of N5-2OH by CEM cells occurred mainly through passive diffusion, a conclusion supported by the minimal effect of NBMPR on the moderate cytotoxicity observed for N5-2OH.
In our metabolic studies with CEM TK1+/TK1– and L929 TK+/TK1– cells, important similarities and differences between [3H]dThd and [3H]N5-2OH became apparent. These studies confirmed that the presence of TK1 was crucial for accumulation of dThd nucleotides (dTMP and dTDP) in cells. In the absence of TK1 activity, uptake and retention of [3H]dThd was drastically reduced and only minor quantities of dTMP were observed. In the case of [3H]N5-2OH, however, substantial uptake and retention of the parental nucleoside was also observed in TK1– cells. Significant formation of N5-2OH-MP by TK1 takes place in TK1+ cells. However, even in these cells the parental nucleoside was the major intracellular component reaching concentration levels comparable to that of TK– cells. There was no indication for further phosphorylation of N5-2OH-MP in TK1+ L929 cells (Supplemental Data). Thus, we cannot exclude the possibility that hitherto unidentified mechanisms specific to TK1+ wild-type cells and their TK1– variants used in our studies contributed to the observed cellular uptake pattern of N5-2OH. It is also conceivable that this 3CTA binds nonspecifically to lipophilic cellular components, as the agent itself is very lipophilic (Al-Madhoun et al., 2004).
To substantiate the metabolic profile found for N5-2OH in cell culture experiments, a series of experiments with recombinant human pyrimidine nucleoside monophosphate kinases, UMP-CMPK and TMPK, were performed to test their capacity to phosphorylate N5-MP and N5-2OH-MP. Both monophosphate kinases showed high activity with their endogenous substrates dCMP and dTMP, respectively. In contrast, when N5-MP and N5-2OH-MP were used as substrates, no diphosphate products were observed. This finding correlated with the metabolite analysis of N5-2OH in our cell culture studies. Surprisingly, however, human NDPK did catalyze the phosphorylation of N5-DP to N5-TP although with a substantially lower efficiency compared with endogenous dTDP.
Both efficacy as boron delivery agents for NCT and cytotoxicity of 3CTAs may be related to their potential incorporation into DNA, as has been discussed by us previously (Barth et al., 2004; Tjarks et al., 2007). Thus, we studied the capacity of Klenow DNA polymerase I to incorporate N5-TP, AZT-TP, and dTTP into oligonucleotide templates during processive DNA synthesis, i.e., using a running start primer-template system. Both AZ-TP and dTTP, generated via coupled enzymatic synthesis, were good substrates for Klenow DNA polymerase I. In contrast, N5-TP, synthesized enzymatically from NDPK/N5-DP, apparently was not incorporated into template oligonucleotide by this polymerase. This result is consistent with data from a previous study indicating that 3-methyl-dTTP could not be incorporated into DNA (Huff and Topal, 1987). The apparent lack of DNA incorporation of N5-TP supports the moderate in vitro and in vivo toxicity observed for 3CTAs (see also Fig. 3B) (Al-Madhoun et al., 2004; Barth et al., 2004, 2008). On the other hand, the absence of DNA incorporation did not appear to hamper the efficacy of N5-2OH in preclinical NCT studies, as previously reported (Barth et al., 2008).
Our studies indicated that N5-2OH-MP was subject to cellular efflux from CCRF-CEM wild-type cells and that both dipyridamole and indomethacin inhibited the efflux of N5-2OH-MP, whereas verapamil had minimal effects on N5-2OH-MP efflux. Previous studies have shown that MRP4 is a major efflux pump in CCRF-CEM wild-type cells (Peng et al., 2008). Thus, the strong inhibition of N5-2OH-MP efflux by the selective MRP4 inhibitor indomethacin suggests a key role for this pump as a regulator of the intracellular accumulation of N5-2OH-MP.
In conclusion, N5-2OH traverses cell membranes via passive diffusion and its monophosphate appears to be a substrate of MRP4 in CCRF-CEM wild-type cells. In addition, 3CTAs, such as N5 and N5-2OH, appear to possess a unique metabolic profile among established biomedical/clinical nucleoside analogs because they are metabolized exclusively by TK1 within the range of enzymes that can bind the dThd scaffold intracellularly. Previous studies have shown that N5-2OH is not a substrate of thymidine kinase 2 (TK2) and catabolizing thymidine phosphorylase (TPase) and deoxynucleotidase-1 (dNT-1) did not catalyze dephosphorylation of N5-2OH-MP (Al-Madhoun et al., 2004; Tjarks et al., 2007).
Supplementary Material
Acknowledgments
The authors thank Dr. Michael V. Darby for supervising work in the radiochemistry laboratory at The Ohio State University.
Abbreviations
- ATCC
American Type Culture Collection
- AZT-TP
AZT triphosphate
- AZT
zidovudine
- CMM
complete minimal medium
- CNT
concentrative nucleoside transporter
- 3CTA
3-carboranyl thymidine analog
- dThd
thymidine
- DTT
dithiothreitol
- dTTP
thymidine triphosphate
- ENT
equilibrative nucleoside transporter
- HPLC
high-performance liquid chromatography
- MRP
multidrug resistance-associated proteins
- MTS
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- N5
3-[5-(o-carboran-1-yl)pentan-1-yl]thymidine
- N5-2OH-MP
N5-2OH monophosphate
- N5-2OH
3-[5-{2-(2,3-dihydroxyprop-1-yl)-o-carboran-1-yl}pentan-1-yl]thymidine
- N5-MP
N5-monophosphate
- N5-TP
N5-triphosphate
- NBMPR
nitrobenzylmercaptopurine ribonucleoside
- NCT
neutron capture therapy
- NDPK
nucleoside diphosphate kinase
- OAT
organic anion transporter
- OCT
organic cation transporter
- P-gp
P-glycoprotein
- PBS
phosphate-buffered saline
- PeSt
penicillin/streptomycin mixture
- TK1
thymidine kinase 1
- TK2
thymidine kinase 2
- TLC
thin-layer chromatography
- TMPK
thymidine monophosphate kinase
- UMP-CMPK
uridine monophosphate–cytidine monophosphate kinase
- Urd
uridine
Authorship Contributions
Participated in research design: Sjuvarsson, Damaraju, Nakkula, Barth, Cass, Eriksson, Tjarks.
Conducted experiments: Sjuvarsson, Mowles, Tiwari, Agarwal, Khalil, Nakkula.
Contributed new reagents or analytic tools: Tiwari, Agarwal, Khalil, Hasabelnaby, Goudah.
Performed data analysis: Sjuvarsson, Damaraju, Tiwari, Agarwal, Khalil.
Wrote or contributed to the writing of the manuscript: Sjuvarsson, Damaraju, Sawyer, Tiwari, Khalil, Barth, Cass, Eriksson, Tjarks.
Footnotes
This study was supported in part by the National Institutes of Health National Cancer Institute [Grant 5R01 CA127935]; The Ohio State University College of Pharmacy; the Swedish Research Council; and the Canadian Cancer Society Research Institute.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the mentioned organizations/institutions.
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Al-Madhoun AS, Johnsamuel J, Barth RF, Tjarks W, Eriksson S. (2004) Evaluation of human thymidine kinase 1 substrates as new candidates for boron neutron capture therapy. Cancer Res 64:6280–6286 [DOI] [PubMed] [Google Scholar]
- Arnér ES, Eriksson S. (1995) Mammalian deoxyribonucleoside kinases. Pharmacol Ther 67:155–186 [DOI] [PubMed] [Google Scholar]
- Barth RF, Yang W, Al-Madhoun AS, Johnsamuel J, Byun Y, Chandra S, Smith DR, Tjarks W, Eriksson S. (2004) Boron-containing nucleosides as potential delivery agents for neutron capture therapy of brain tumors. Cancer Res 64:6287–6295 [DOI] [PubMed] [Google Scholar]
- Barth RF, Yang W, Wu G, Swindall M, Byun Y, Narayanasamy S, Tjarks W, Tordoff K, Moeschberger ML, Eriksson S, et al. (2008) Thymidine kinase 1 as a molecular target for boron neutron capture therapy of brain tumors. Proc Natl Acad Sci USA 105:17493–17497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belt JA, Marina NM, Phelps DA, Crawford CR. (1993) Nucleoside transport in normal and neoplastic cells. Adv Enzyme Regul 33:235–252 [DOI] [PubMed] [Google Scholar]
- Borst P, Balzarini J, Ono N, Reid G, de Vries H, Wielinga P, Wijnholds J, Zelcer N. (2004) The potential impact of drug transporters on nucleoside-analog-based antiviral chemotherapy. Antiviral Res 62:1–7 [DOI] [PubMed] [Google Scholar]
- Carnrot C, Wang L, Topalis D, Eriksson S. (2008) Mechanisms of substrate selectivity for Bacillus anthracis thymidylate kinase. Protein Sci 17:1486–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass CE, Young JD, Baldwin SA. (1998) Recent advances in the molecular biology of nucleoside transporters of mammalian cells. Biochem Cell Biol 76:761–770 [DOI] [PubMed] [Google Scholar]
- Copeland WC, Chen MS, Wang TS. (1992) Human DNA polymerases α and β are able to incorporate anti-HIV deoxynucleotides into DNA. J Biol Chem 267:21459–21464 [PubMed] [Google Scholar]
- Damaraju VL, Damaraju S, Young JD, Baldwin SA, Mackey J, Sawyer MB, Cass CE. (2003) Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene 22:7524–7536 [DOI] [PubMed] [Google Scholar]
- Deville-Bonne D, El Amri C, Meyer P, Chen Y, Agrofoglio LA, Janin J. (2010) Human and viral nucleoside/nucleotide kinases involved in antiviral drug activation: structural and catalytic properties. Antiviral Res 86:101–120 [DOI] [PubMed] [Google Scholar]
- Eriksson S, Xu B, Clayton DA. (1995) Efficient incorporation of anti-HIV deoxynucleotides by recombinant yeast mitochondrial DNA polymerase. J Biol Chem 270:18929–18934 [DOI] [PubMed] [Google Scholar]
- Eriksson S, Munch-Petersen B, Johansson K, Eklund H. (2002) Structure and function of cellular deoxyribonucleoside kinases. Cell Mol Life Sci 59:1327–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmarini CM, Mackey JR, Dumontet C. (2001) Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia 15:875–890 [DOI] [PubMed] [Google Scholar]
- Harley ER, Paterson AR, Cass CE. (1982) Initial rate kinetics of the transport of adenosine and 4-amino-7-(beta-D-ribofuranosyl)pyrrolo[2,3-d]pyrimidine (tubercidin) in cultured cells. Cancer Res 42:1289–1295 [PubMed] [Google Scholar]
- Hasabelnaby S, Goudah A, Agarwal HK, abd Alla MS, Tjarks W. (2012) Synthesis, chemical and enzymatic hydrolysis, and aqueous solubility of amino acid ester prodrugs of 3-carboranyl thymidine analogs for boron neutron capture therapy of brain tumors. Eur J Med Chem 55:325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff AC, Topal MD. (1987) DNA damage at thymine N-3 abolishes base-pairing capacity during DNA synthesis. J Biol Chem 262:12843–12850 [PubMed] [Google Scholar]
- Khalil A, Ishita K, Ali T, Tjarks W. (2013) N3-substituted thymidine bioconjugates for cancer therapy and imaging. Future Med Chem 5:677–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsell H, Endou H. (2004) The SLC22 drug transporter family. Pflugers Arch 447:666–676 [DOI] [PubMed] [Google Scholar]
- Kreimeyer A, Schneider B, Sarfati R, Faraj A, Sommadossi JP, Veron M, Deville-Bonne D. (2001) NDP kinase reactivity towards 3TC nucleotides. Antiviral Res 50:147–156 [DOI] [PubMed] [Google Scholar]
- Lunato AJ, Wang J, Woollard JE, Anisuzzaman AKM, Ji W, Rong F-G, Ikeda S, Soloway AH, Eriksson S, Ives DH, et al. (1999) Synthesis of 5-(carboranylalkylmercapto)-2′-deoxyuridines and 3-(carboranylalkyl)thymidines and their evaluation as substrates for human thymidine kinases 1 and 2. J Med Chem 42:3378–3389 [DOI] [PubMed] [Google Scholar]
- Parks RE, Jr, Agarwal RP. (1973) Nucleoside diphosphokinases, in The Enzymes, 3rd ed (Boyer PD. ed) vol. 8, pp 307–333, Academic Press, New York [Google Scholar]
- Pasti C, Gallois-Montbrun S, Munier-Lehmann H, Veron M, Gilles AM, Deville-Bonne D. (2003) Reaction of human UMP-CMP kinase with natural and analog substrates. Eur J Biochem 270:1784–1790 [DOI] [PubMed] [Google Scholar]
- Pastor-Anglada M, Cano-Soldado P, Molina-Arcas M, Lostao MP, Larráyoz I, Martínez-Picado J, Casado FJ. (2005) Cell entry and export of nucleoside analogues. Virus Res 107:151–164 [DOI] [PubMed] [Google Scholar]
- Peng XX, Shi Z, Damaraju VL, Huang XC, Kruh GD, Wu HC, Zhou Y, Tiwari A, Fu L, Cass CE, et al. (2008) Up-regulation of MRP4 and down-regulation of influx transporters in human leukemic cells with acquired resistance to 6-mercaptopurine. Leuk Res 32:799–809 [DOI] [PubMed] [Google Scholar]
- Plunkett W, Saunders PP. (1991) Metabolism and action of purine nucleoside analogs. Pharmacol Ther 49:239–268 [DOI] [PubMed] [Google Scholar]
- Ritter CA, Jedlitschky G, Meyer zu Schwabedissen H, Grube M, Köck K, Kroemer HK. (2005) Cellular export of drugs and signaling molecules by the ATP-binding cassette transporters MRP4 (ABCC4) and MRP5 (ABCC5). Drug Metab Rev 37:253–278 [DOI] [PubMed] [Google Scholar]
- Summers MA, Moore JL, McAuley JW. (2004) Use of verapamil as a potential P-glycoprotein inhibitor in a patient with refractory epilepsy. Ann Pharmacother 38:1631–1634 [DOI] [PubMed] [Google Scholar]
- Tjarks W, Tiwari R, Byun Y, Narayanasamy S, Barth RF. (2007) Carboranyl thymidine analogues for neutron capture therapy. Chem Commun (Camb) (47):4978–4991 [DOI] [PubMed] [Google Scholar]
- Vickers MF, Kumar R, Visser F, Zhang J, Charania J, Raborn RT, Baldwin SA, Young JD, Cass CE. (2002) Comparison of the interaction of uridine, cytidine, and other pyrimidine nucleoside analogues with recombinant human equilibrative nucleoside transporter 2 (hENT2) produced in Saccharomyces cerevisiae. Biochem Cell Biol 80:639–644 [DOI] [PubMed] [Google Scholar]
- Zhang J, Visser F, Vickers MF, Lang T, Robins MJ, Nielsen LP, Nowak I, Baldwin SA, Young JD, Cass CE. (2003) Uridine binding motifs of human concentrative nucleoside transporters 1 and 3 produced in Saccharomyces cerevisiae. Mol Pharmacol 64:1512–1520 [DOI] [PubMed] [Google Scholar]
- Zhang J, Smith KM, Tackaberry T, Visser F, Robins MJ, Nielsen LP, Nowak I, Karpinski E, Baldwin SA, Young JD, et al. (2005) Uridine binding and transportability determinants of human concentrative nucleoside transporters. Mol Pharmacol 68:830–839 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.