Abstract
GABAA receptors play a crucial role in the actions of general anesthetics. The recently published crystal structure of the general anesthetic propofol bound to Gloeobacter violaceus ligand-gated ion channel (GLIC), a bacterial homolog of GABAA receptors, provided an opportunity to explore structure-based ligand discovery for pentameric ligand-gated ion channels (pLGICs). We used molecular docking of 153,000 commercially available compounds to identify molecules that interact with the propofol binding site in GLIC. In total, 29 compounds were selected for functional testing on recombinant GLIC, and 16 of these compounds modulated GLIC function. Active compounds were also tested on recombinant GABAA receptors, and point mutations around the presumed binding pocket were introduced into GLIC and GABAA receptors to test for binding specificity. The potency of active compounds was only weakly correlated with properties such as lipophilicity or molecular weight. One compound was found to mimic the actions of propofol on GLIC and GABAA, and to be sensitive to mutations that reduce the action of propofol in both receptors. Mutant receptors also provided insight about the position of the binding sites and the relevance of the receptor’s conformation for anesthetic actions. Overall, the findings support the feasibility of the use of virtual screening to discover allosteric modulators of pLGICs, and suggest that GLIC is a valid model system to identify novel GABAA receptor ligands.
Introduction
GABAA receptors are targets of many therapeutically important agents. In particular, clinically used general anesthetics including propofol, isoflurane, and midazolam are positive allosteric modulators of most GABAA receptors (Garcia et al., 2010). At clinically relevant concentrations, general anesthetics typically increase receptor sensitivity to GABA and thereby prolong inhibition of postsynaptic neuronal excitability (Garcia et al., 2010). However, the precise sites and mechanisms of action of these drugs have not been defined, in part because of the lack of high-resolution structural data for the GABAA receptors. A recent advance in this area has been the discovery of bacterial pentameric ligand-gated ion channels (pLGICs) that are homologous to GABAA receptors and may serve as valuable models for structure and function of their human homologs (Tasneem et al., 2005). The validity of prokaryotic pLGICs as models for GABAA receptor structure and function has been supported by sequence homology (Tasneem et al., 2005) and subsequent crystal structures of the Erwinia chrysanthemi ligand-gated ion channel (Hilf and Dutzler, 2008) and Gloeobacter violaceus ligand-gated ion channel (GLIC) (Bocquet et al., 2009; Hilf and Dutzler, 2009). These two receptors shared predicted structure properties of pLGICs, including five subunits each containing an extracellular domain and four transmembrane helices (TM1–4) with TM2 facing the ion-conducting pore. Although E. chrysanthemi ligand-gated ion channel and GLIC lack the extensive intracellular loop and characteristic extracellular disulfide bridge of GABAA receptors, sequence alignments of the TM2 domains show over 60% similarity with eukaryotic pLGICs (Thompson et al., 2012). Similar to GABAA receptors, GLIC is modulated by n-alcohols. Indeed, the potency of n-alcohols increases with chain-length up to nine carbons (cutoff) for GLIC, which agrees well with the behaviors observed for GABAA, glycine, and nicotinic acetylcholine (nACh) receptors (Howard et al., 2011). Furthermore, GLIC is modulated by several anesthetics, including propofol (Weng et al., 2010), and was recently cocrystalized with propofol and desflurane bound in a lipophilic intrasubunit cavity in the TM domain (Nury et al., 2011). The conservation of this crystallographic binding site in eukaryotic pLGICs was recently supported by photolabeling studies of the Torpedo marmorata nACh receptor (Jayakar et al., 2013), indicating this site of action may be a general feature of a variety of receptor family members.
One limitation of high-throughput drug discovery for pLGICs is that functional screening commonly requires time-consuming electrophysiological recordings from transfected cells. The determination of pLGIC structures opens the possibility to use virtual screening of chemical libraries to identify molecules with the ability to modulate the behavior of these channels via allosteric pockets (Jorgensen, 2009). The high cost and the low hit-rates of functional screening methods have further encouraged use of these cheaper and faster computational alternatives. However, the success of molecular docking has been found to vary widely depending on the target protein and algorithm used (Warren et al., 2006), and these approaches remain to be fully validated for targets such as pLGICs. The recently determined crystal structure of GLIC has been used in docking calculations of six general anesthetics to the protein. AutoDock (http://autodock.scripps.edu) determined several binding sites for the anesthetic agents in the GLIC crystal structure, among them the anesthetic binding site identified through cocrystallography, and the anesthetics’ predicted affinities correlated significantly with their known EC50 values (Liu et al., 2012). Because GLIC is sensitive to several clinically used drugs known to act on GABAA receptors, this raised the possibility that compounds acting at this site might modulate GABAA receptor function in a similar fashion. In this study, we used molecular docking against the propofol binding site in GLIC to screen more than 150,000 small-sized compounds and select a small number of candidates suitable for electrophysiological testing on GLIC expressed in Xenopus laevis oocytes. We asked whether this approach could identify novel modulators of GLIC, and whether any of these compounds would also be effective on GABAA receptors. We also tested the specificity of active compounds through site-directed mutagenesis of GLIC and GABAA receptors. Our results demonstrate that molecular docking screening can be used to discover allosteric modulators of pLGICs and that prokaryotic homologs may be generalized to human members of this receptor family in designing novel pharmaceutic agents.
Materials and Methods
Preparation of the Molecular Docking Screen.
Preparation of the molecular docking screen was performed as several steps. First, a molecular dynamics (MD) simulation was performed to obtain coordinates for the phospholipids in the vicinity of the propofol binding site. Second, docking was performed against 302 snapshots from the MD simulation to identify a structure that was suitable for virtual screening. In the last step, a molecular docking screen of commercially available molecules was performed against one of these structures.
As the majority of lipids were absent in crystal structures of GLIC, an MD simulation was performed to obtain conformations of phospholipids in the opening of the propofol binding site. If the membrane was not included in the virtual screening step, molecules would occasionally be docked on the outside of the propofol binding cavity. The presence of the membrane constrained the docking search algorithm to the site occupied by propofol, which was hypothesized to be necessary for allosteric modulation of GLIC. For the MD simulations, the crystal structure of GLIC with propofol bound (PDB ID 3P50) was used for initial coordinates (Nury et al., 2011). The system was embedded into a dioleoylphosphatidylcholine lipid bilayer, and solvated and ionized as described previously elsewhere (Murail et al., 2012). The simulation was run in GROMACS 4.5.5 (http://www.gromacs.org/) (Hess et al., 2008), using position restraints of 10,000 and 1000 kJ/mol/nm2 on all non-H atoms of propofol and the protein, respectively. The simulation was run with a time step of 2.0 femtoseconds and the temperature was controlled to 310 K, applying simulation parameters as in previous work (Murail et al., 2012). Parameters for the protein were taken from the Amber99sb-ILDN force field (Sorin and Pande, 2005), and the Berger force field was used for dioleoylphosphatidylcholine lipids (Berger et al., 1997), combined with TIP3P water (Jorgensen et al., 1983). The ligand atom and bond types were taken from the general Amber Force Field (http://ambermd.org/) (Duan et al., 2003) and assigned with Antechamber; the restrained electrostatic potential point charges were derived from a density functional theory calculation as previously described elsewhere (Murail et al., 2012).
The molecular docking calculations were performed with the program DOCK3.6 (http://dock.compbio.ucsf.edu/DOCK3.6/) (Lorber and Shoichet, 1998, 2005; Irwin et al., 2009). All water molecules and ions were removed from the simulation snapshots. The docking was performed against one subunit of GLIC, and all protein residues within ∼20 Å of the propofol binding pocket were explicitly included in the calculations. The phospholipids in the opening of the cavity were maintained to favor posing of docked molecules in this pocket. The protonation states of ionizable residues of all Asp, Glu, Arg, and Lys residues were set to their charged states. The protonation states of His residues were set by manual inspection of the local hydrogen bonding network. Only His235 was close to the propofol binding pocket, and it was protonated on the epsilon-nitrogen.
The flexible ligand-sampling algorithm in DOCK3.6 superimposes atoms of the docked molecule onto binding site matching spheres, which indicate putative ligand atom positions (Lorber and Shoichet, 1998, 2005). Sixty matching spheres were used, and these were based on the atoms of the cocrystallized ligand (propofol). The spheres were also labeled for chemical matching based on the local receptor environment (Shoichet and Kuntz, 1993).
The degree of ligand sampling is determined by the bin size, bin size overlap, and distance tolerance. These three parameters were set to 0.3, 0.1, and 1.4 Å, respectively, for both the binding site matching spheres and the docked molecules. For ligand conformations passing an initial steric filter, a physics-based scoring function was used to evaluate the fit to the binding site. For the best scoring conformation of each docked molecule, 100 steps of rigid-body minimization were performed (Lorber and Shoichet, 1998, 2005). The score for each conformation was calculated as the sum of the receptor–ligand electrostatic and van der Waals interaction energy, corrected for ligand desolvation (Shoichet et al., 1999). These three terms are evaluated from precalculated grids. The three-dimensional map of the electrostatic potential in the binding site was prepared using the program DelPhi (http://wiki.c2b2.columbia.edu/honiglab_public/index.php/Software:DelPhi) (Nicholls and Honig, 1991). In this calculation, partial charges from the united atom AMBER force field (Weiner et al., 1984) were used for all receptor atoms except the side chain hydroxyls of Y254 and T255, for which the dipole moment was increased by +0.4 on the polar hydrogens and decreased by −0.4 on the oxygens to favor hydrogen bonding to these residues as described previously elsewhere (Carlsson et al., 2010). The program CHEMGRID (a DOCK tool) was used to generate a van der Waals grid, which is based on a united atom version of the AMBER force field (Meng et al., 1992). The desolvation penalty for a ligand conformation is estimated from a precalculated transfer free energy of the molecule between solvents of dielectrics 78 and 2. The docked molecules were assumed to be completely desolvated upon binding to the propofol binding pocket (Shoichet et al., 1999). Before the DOCK3.6 calculation, each molecule was prepared for docking by pregenerating up to 600 conformations using the program OMEGA (OpenEye Scientific Software, Santa Fe, NM). Partial atomic charges and transfer free energies for each molecule were calculated using AMSOL (http://comp.chem.umn.edu/amsol/) (Chambers et al., 1996; Li et al., 1998) and van der Waals parameters were derived from an all-atom AMBER potential (Weiner et al., 1986).
The molecular docking screen was performed in two steps. Docking was first performed against 302 snapshots from the MD simulation. In this step, two criteria were used to identify a structure (of the protein and membrane) that was suitable for virtual screening. For each MD snapshot, the ability of the snapshots to identify propofol among a set of decoys (nonbinders) and to reproduce the binding mode of propofol were assessed. As few experimentally verified nonbinders were available, 100 molecules with similar chemical properties but different chemical structures were selected as decoys according to the DUDE protocol (Mysinger et al., 2012). Propofol and the 100 decoys were docked to each of the 302 structures and were ranked based on their predicted energy scores. The 10 MD snapshots with the best abilities to identify propofol among the decoys were analyzed visually. From these, a snapshot that both reproduced the binding mode of propofol and had phospholipids that tightly enclosed the binding pocket was selected for the screening of commercially available compounds. All molecules with 13 or fewer non-H atoms in the ZINC fragment-like library (Irwin et al., 2012) (mol. wt. <250, logP <3.5, and rotatable bonds <5) were screened against the selected MD snapshot. In total, 153,000 compounds were docked and ranked based on their predicted binding energy.
Oocyte Preparation.
Oocytes from female X. laevis frogs were extracted, isolated, and stored as described previously elsewhere (McCracken et al., 2010; Howard et al., 2011). For the expression of GLIC, 32 nl cDNA solution was injected into the oocyte nucleus via the animal pole using a microdispenser (Drummond Scientific, Broomall, PA) with a 20–30 µm outer diameter glass capillary tip pulled with a Flaming/Brown Micropipette Puller (Sutter Instrument Co., Novato, CA). We used 1 ng GLIC wild-type cDNA, 9 ng GLIC M205W cDNA, and 3 ng of all other GLIC mutant cDNA per oocyte. For the expression of GABAA receptors, cRNA encoding wild-type or mutant α1, β2, and γ2s subunits in a 1:1:3 ng ratio was injected into the oocyte cytoplasm. The capped cRNAs encoding GABAA receptor subunits were made using mMessage mMachine (Life Technologies, Grand Island, NY). After injection, oocytes were incubated individually in 96-well plates filled with incubation media consisting of 88 mM NaCl, 10 mM HEPES, 2.4 mM NaHCO3, 1 mM KCl, 0.91 mM CaCl2, 0.82 mM MgSO4, 0.33 mM Ca(NO3)2, 2 mM sodium pyruvate, 0.5 mM theophylline, 0.1 mM gentamicin, 17 µM streptomycin, and 10,000 U/l penicillin. Oocytes expressing GABAA receptors were incubated for 2–5 days, and GLIC-expressing oocytes were stored until the signal reached between −0.8 and −20 µA when activated with an EC10 (concentration at which the channels conduct 10% of their maximal currents) of protons. An exception was made for GLIC M205W mutants, where we accepted signals as low as −0.3 µA.
Changes in current as a response to receptor activation were measured by two electrode voltage clamp electrophysiology (Wagner et al., 2000). During recordings, each oocyte was kept in a ∼100 µl perfusion chamber connected to a peristaltic pump (Cole-Palmer, Vernon Hills, IL) flowing at 2 ml/min via 18-gauge inert tubing. Expressing oocytes were clamped at −70 mV using an oocyte clamp (Warner Instruments, Hamden, CT), and continuously recorded using a digitizer controlled by LabChart Pro software (AD Instruments, Colorado Springs, CO).
Oocyte Recordings.
As the effect of allosteric modulators on pLGICs has been shown to be greater at lower levels of activation, we used an activating concentration of protons (GLIC) or GABA (GABAA receptors) of EC10. As mutations introduced to a channel can alter its sensitivity to protons, agonist response curves for each GLIC mutant were fit to find its EC10. Activation buffer for GLIC experiments consisted of 123 mM NaCl, 10 mM sodium citrate, 2 mM KCl, 2 mM MgSO4 and 2 mM CaCl2, adjusted to each channel’s EC10 pH. GLIC running buffer was identical to activation buffer, but with 10 mM HEPES in place of citrate; the pH was adjusted either to pH 8.0 for GLIC I201F and I202W/I258W, or to pH 7.4 for all other clones. Running buffers for GABAA receptor experiments consisted of 96 mM NaCl, 2 mM KCl, 1 mM CaCl2, 1 mM MgCl2, and 5 mM HEPES adjusted to pH 7.5. In GABAA receptor experiments, the EC10 GABA concentration was determined for each oocyte after application of a maximal GABA concentration. Propofol and all novel compounds were prepared as 50–100 mM stocks in dimethylsulfoxide (DMSO). Less than 2 hours before use, stock solutions were diluted to the appropriate concentration in running or activation buffer and sonicated in an Ultrasonic Cleaner Model 450, E/MC (RAI Research, Long Island, NY) for at least 2 minutes before application.
To measure each drug response, wild-type or mutant receptors were first activated for 2 minutes (GLIC) or 30 seconds (GABAA receptors) with EC10 activation buffer (containing the appropriate concentration of protons or GABA, respectively), followed by a 5–15-minute washout with running buffer. Then, the test compound was preapplied in running buffer for 1 minute. Immediately after preapplication, the compound was coapplied with activation buffer for 2 minutes (GLIC) or 30 seconds (GABAA receptors), followed by another 5–15-minute washout. Finally, activation buffer was applied again for 2 minutes (GLIC) or 30 seconds (GABAA receptors). The percentage of modulation was calculated as the ratio of the peak coapplication current to the average of the initial and final activations in the absence of compound.
Statistics.
Prism 6.0 (GraphPad Software, La Jolla, CA) was used for statistical analysis of all data. Pooled data were calculated as value mean ± S.E.M. Initial validation was performed using one-way analysis of variance (ANOVA) to determine the statistically significant differences between each compound and vehicle (0.1–0.2% DMSO). All other statistical analyses were performed using either a Student’s t test, one-way ANOVA, or two-way ANOVA, as indicated.
Mutagenesis.
We used molecular visualization in UCSF Chimera 1.6.1 (http://www.cgl.ucsf.edu/chimera/) (Pettersen et al., 2004) to choose amino acids in the GLIC-propofol cocrystal structure (Nury et al., 2011) that would most likely block propofol binding if mutated to phenylalanine or tryptophan. For mutagenesis of both GLIC and GABAA receptors, we used commercially made mutagenic primers (Integrated DNA Technologies, Coralville, IA) and the QuikChange II site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA).
Results
Molecular Docking Screen and Compound Selection.
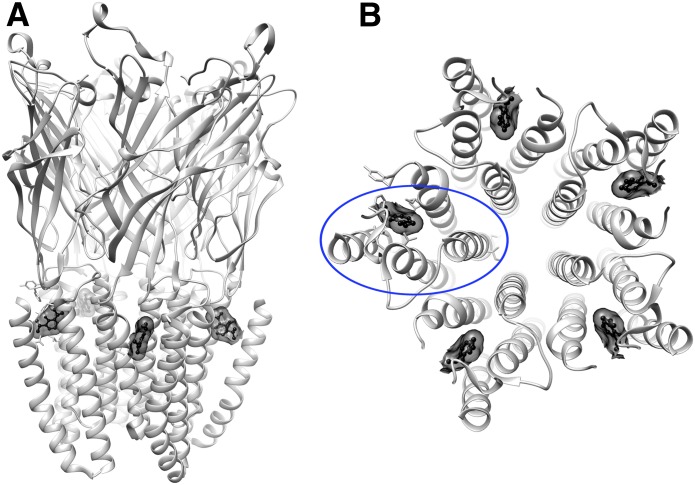
The molecular docking screen was performed against the structure of GLIC that has propofol bound to an allosteric, intrasubunit TM site (PDB ID 3P50). The binding pocket is completely occluded from aqueous solvent but accessible from the lipid membrane (Fig. 1).
Fig. 1.
GLIC propofol structure (PDB ID 3P50) showing propofol in ball-and-stick and transparent surface representation (black), bound near the top of the helical TM domains within each subunit. (A) Side view. (B) View from the top of the TM domains, with one of the subunits circled in blue.
In the first step, propofol was (re)docked to the intrasubunit pocket. Docking algorithms have largely been developed for soluble proteins, creating two challenges in these initial membrane-embedded screens. First, because the phospholipids surrounding the binding site were not resolved in the crystal structure, the docking algorithm occasionally produced ligand orientations outside the intrasubunit pocket. Second, as the region surrounding the intrasubunit pocket was treated as a medium with a dielectric representative of aqueous solution, polar groups of docked molecules were sometimes positioned toward the membrane. To achieve a more physically correct description of ligand binding to the pocket, an MD simulation of GLIC with propofol bound was performed in a hydrated lipid-bilayer. In the simulation, GLIC and propofol were tightly restrained to their initial coordinates while the membrane and solvent were allowed to equilibrate for 25 nanoseconds. A snapshot of the equilibrated membrane, which shows the location of the propofol binding pocket, is shown in Supplemental Fig. 1. From this simulation, 302 snapshots of GLIC with equilibrated phospholipids surrounding the intrasubunit pocket were extracted. Propofol together with 100 decoys (nonbinders) were then docked to each structure using DOCK3.6. One of the MD snapshots that had the best ability to identify propofol among the decoys was chosen for the prospective virtual screen. For this structure, propofol was ranked as number 17 of the 101 docked molecules.
Over 153,000 commercially available molecules from the ZINC fragment-like library were docked to the intrasubunit pocket of GLIC. The 700 top-ranked compounds, corresponding to 0.5% of the screened library, were inspected for properties that are not taken into account in the docking scoring function, such as similarity to known ligands, availability from vendors, and physical properties. In total, 22 compounds among the top-ranked molecules were selected for experimental testing (Supplemental Table 1). A majority of the compounds had a six-membered aromatic ring but varied in the composition, polarity, and number of substituents.
Functional Validation of Molecular Docking Screen.
We divided the experimental testing of the predicted ligands into three phases. First, we asked whether molecular docking could provide novel ligands that modulate GLIC. Second, we mutated amino acids critical for propofol action on GLIC to answer whether the screened compounds bind specifically to the site targeted by computational docking. Third, we asked if novel GLIC modulators also altered the function of GABAA receptors, and if these effects were specific to the GABAA receptor propofol site.
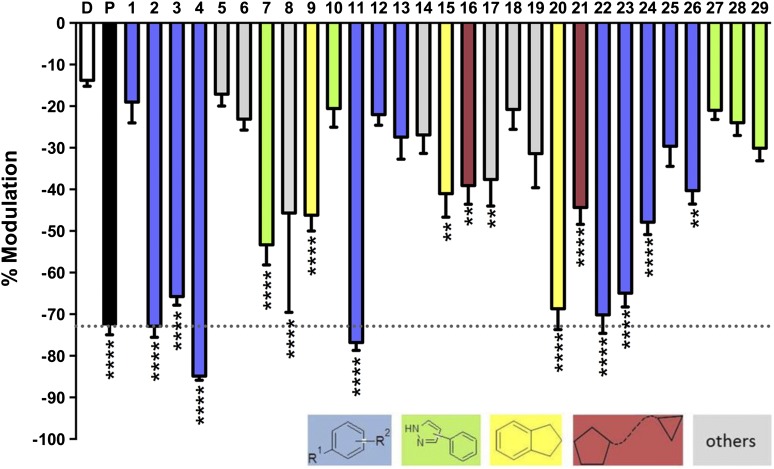
To test the ability of virtual screening to find compounds that modulate GLIC function, the 22 compounds that were selected from the docking screen (Supplemental Table 1) were tested at 50 µM for their ability to alter proton-activated GLIC currents (pH 5.5, ∼EC10) and compared with vehicle (0.1% DMSO) by ANOVA (Fig. 2). The concentration of 50 µM is within the range reported to be lethal in 50% of the subjects (30–100 µM) (Krasowski et al., 2001) but it produced a non-maximal effect on GLIC, which is not as sensitive as the GABAA receptors. A total of 13 compounds (59%) were positive hits, defined by significantly stronger inhibition than vehicle (P ≤ 0.05), and are shown in Table 1. Five of the most significant hits (P ≤ 0.0001) were unbranched disubstituted benzenes, and, similar to many anesthetics, these compounds were all halogenated. Three of the strongest inhibitors, compounds 2–4, were distinguished only by the position of the methyl group on the benzene ring (ortho, meta, and para, respectively). The para-substituted compounds, 4 and 11, led to the greatest inhibition, but all potencies were within ±15% of the propofol response.
Fig. 2.
Modulation of wild-type GLIC by novel compounds. Modulation of proton-activated GLIC currents (pH 5.5, ∼EC10) by vehicle (0.1% DMSO, D), 50 μM propofol (P, black, dotted line), or the predicted ligand (50 µM) were determined from 4–15 measurements as indicated at the base of every column. Statistical significance is relative to vehicle, Dunnet’s multiple comparison test, ANOVA (**P ≤ 0.01; ****P ≤ 0.0001).
TABLE 1.
Predicted ligands from the docking screen and analogs of positive hits that significantly inhibited GLIC
Activity is the percentage of inhibition of the proton-induced current in GLIC (see Fig. 2 for details).
| Compound # | Structure | Activity (%) | Docking Rank | Compound # | Structure | Activity (%) | Docking Rank |
|---|---|---|---|---|---|---|---|
| 2 | −77 | 698 | 16 | −39 | 335 | ||
| 3 | −66 | 351 | 17 | −38 | 111 | ||
| 4 | −85 | 350 | 20 | −64 | 280 | ||
| 7 | −55 | 9 | 21 | −43 | 497 | ||
| 8 | −46 | 208 | 22 | −63 | 399 | ||
| 9 | −43 | 179 | 23 | −65 | A | ||
| 11 | −77 | 313 | 24 | −48 | A | ||
| 15 | −41 | 67 | 26 | −40 | A |
A, analog of a positive hit from the docking screen.
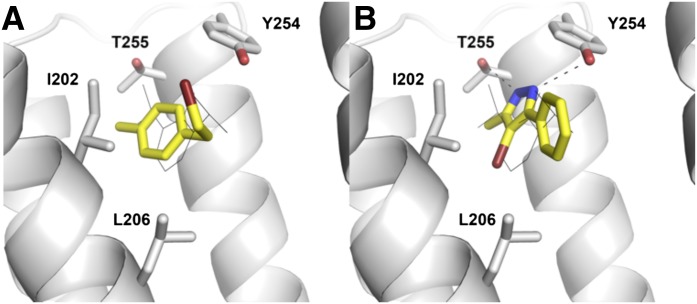
To further investigate the structure–activity relationship for the discovered ligands, seven additional analogs were purchased (23–29; Supplemental Table 2). A potential concern with screening molecules containing alkylhalide moieties is that such compounds may be reactive and be a source of false-positives in high-throughput screening (Rishton, 1997). To verify that compounds 2, 3, 4, and 20 were not acting via such a mechanism, analog 23 was tested against GLIC. In compound 23, the methyl and ethylbromide substituents of compound 4 were replaced with a bromo and propyl group, respectively, and these rearrangements only changed the extent of GLIC modulation slightly. Analogs of compounds 7 and 11 were also tested. Two of the purchased analogs of compounds 11, 24, and 26 were still active, but the inhibition was reduced by ∼10 and 30%, respectively. The rather small change of the thioether in compound 11 to an ether moiety in compound 25 decreased inhibition by 50%, producing an effect not significantly different from vehicle. Only one out of five tested pyrazole compounds led to a significant inhibition of activation. The presence of a methyl rather than an amine group in active compound 7 versus inactive compound 10 (Supplemental Table 1) increased inhibition by 30%. Similarly, replacing the bromine in compound 7 with a hydrogen in compound 28 also led to a significant reduction of activity. None of the three analogs of compound 7 (27–29; Supplemental Table 2) modulated GLIC to a significantly greater extent than the vehicle. Considering all 29 tested compounds, a total of 16 (55%) produced a significant inhibition compared with the vehicle (P ≤ 0.05). The predicted binding modes for compounds 4 and 7 in the GLIC crystal structure are shown in Fig. 3.
Fig. 3.
Predicted binding modes of compounds 4 and 7 in the propofol binding pocket of GLIC. The carbon atoms of the discovered ligands are shown as yellow sticks, and black dotted lines indicate hydrogen bonds to the protein. The GLIC backbone is shown in white ribbons with selected amino acids shown as sticks. Propofol is shown in black lines. This figure was generated with PyMOL (version 1.4.1).
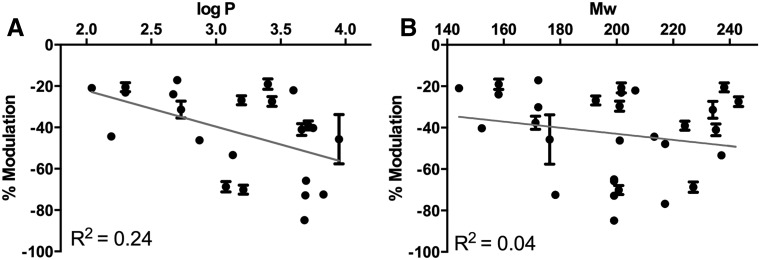
Multiple studies have shown that the potency of anesthetics correlates with simple physicochemical properties. For this reason, we next asked if the physicochemical properties of the discovered compounds correlated with differences in GLIC modulation. The extent of modulation was plotted against the partition coefficient (logP; Fig. 4A) and the molecular weight of each compound (Fig. 4B). Among the compounds tested, potency did not correlate with lipophilicity (R2 = 0.24) or molecular weight (R2 = 0.04).
Fig. 4.
Correlation between potency and physicochemical properties. (A) Percentage of modulation versus lipophilicity (logP), with value for the correlation coefficient (R2) of a linear regression fit (gray line). (B) Percentage of modulation versus molecular weight (Mw), reported as in A.
GLIC Binding Site Specificity.
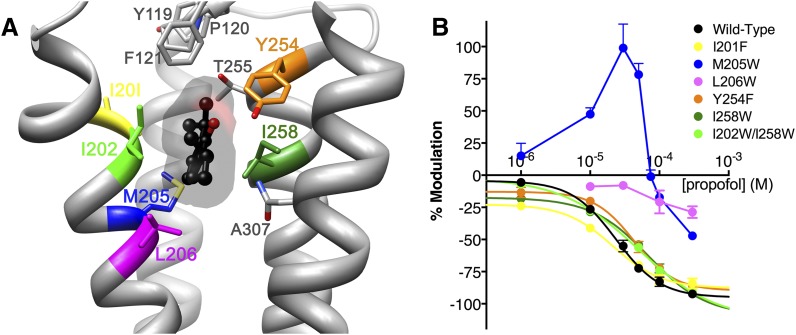
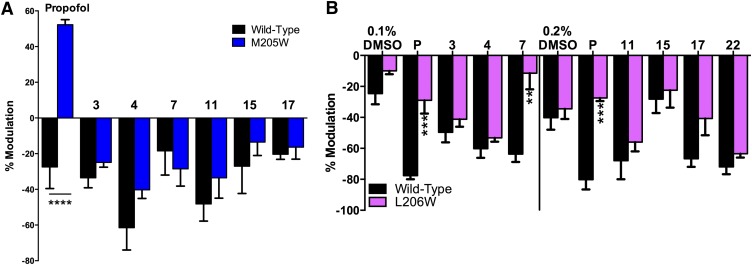
In the second part of the validation process, we explored whether virtual screening yielded compounds that bound to the same site as propofol on GLIC. We tested a series of mutations in the propofol binding pocket for reduced propofol inhibition, as such mutations should also block novel compounds from binding if they modulated GLIC via the same site and mechanism (Fig. 5A). Mutation of leucine to tryptophan at position 206 in TM1 reduced apparent propofol binding, as propofol modulation was drastically reduced over a wide concentration range (Fig. 5B). Conversely, propofol inhibition of GLIC mutants I201F, Y245F, I258W, and the double mutant I202W/I258W followed sigmoidal concentration dependence with inhibitory concentration 50 (IC50) values similar to wild-type GLIC. An interesting effect was observed in the M205W mutant, for which low concentrations of propofol (1 to 50 µM) potentiated proton activation, while higher concentrations (100 µM and higher) inhibited channel function. When tested in the M205W mutant, six of the hit compounds (3, 4, 7, 11, 15, and 17) produced an inhibitory modulation that did not show significant differences compared with wild-type (Fig. 6A). Due to its apparent influence on propofol binding, the L206W mutant was tested with the same hit compounds plus 22. Inhibition by compound 7 was markedly reduced for L206W compared with wild-type GLIC (Fig. 6B).
Fig. 5.
Disruption of modulation by propofol site mutagenesis. (A) GLIC propofol structure (PDB ID 3P50) showing propofol in ball-and-stick and transparent surface representation (black), with residues within 5 Å of propofol shown as sticks. Residues mutated to probe propofol specificity are colored. (B) Propofol concentration-response curves for wild-type and mutant GLIC. Curves represent nonlinear regression fits for wild-type GLIC and all mutants except M205W and L206W, which did not follow standard inhibition profiles; for these two mutants, data points were connected through straight lines. Propofol inhibitory concentration 50 (IC50) values (in µM): wild-type 24, I201F 21, I258W 70, I202W/I258W 50, Y254F 43. Number of oocytes tested: 2–15.
Fig. 6.
Effect of propofol and selected compounds from the virtual screen on wild-type (WT) and mutated GLIC receptors activated at the 10% level. (A) Effects of vehicle (DMSO), propofol (P), and selected hit compounds at 10 μM on WT and M205W. (B) Effect of propofol (P) and virtually screened compounds (100 µM) on EC10 responses in GLIC receptors. The results are grouped according to the DMSO concentration present (0.1 or 0.2%), determined by the concentration of the stock used. Statistical significance for mutant versus WT determined by unpaired t test (**P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001), n = 2–9.
Modulation of GABAA Receptors.
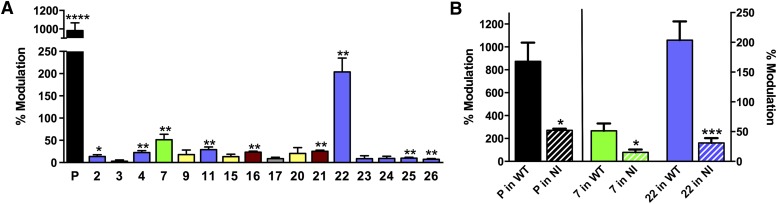
The sedative and anesthetic actions of propofol have been attributed primarily to modulation of GABAA receptors (Jurd et al., 2003; Garcia et al., 2010). Therefore, we asked if compounds which modulated GLIC would also allosterically modulate recombinant α1β2γ2 GABAA receptors. We tested these compounds on a submaximal GABA concentration (EC10) at a concentration of 2 µM, and compared them with propofol at the same concentration. Propofol produced a potentiation of 137% (n = 4), while the rest of the compounds had minimal effects (−2 to 6%, data not shown). We then tested propofol and the other compounds at the concentration (50 µM) used on GLIC, to be able to measure and compare all effects. At 50 μM, propofol potentiated α1β2γ2 GABAA receptors activated by EC10 GABA ∼11-fold (Fig. 7A). Compounds 7 and 22 also produced substantial potentiation of GABA responses, enhancing currents ∼0.5- and ∼2-fold at 50 μM, respectively. The effect of compound 7 was concentration dependent, increasing to ∼1.5-fold potentiation at 100 μM (data not shown). Most of the other novel compounds produced a small potentiation between 3 and 29% (Fig. 7A).
Fig. 7.
Effect of propofol and compounds identified in the virtual screen on GABA responses. (A) Modulation of EC10 GABA responses in recombinant α1β2γ2 GABAA receptors by 50 μM propofol (P) and selected hit compounds. Statistical significance is versus a hypothetical value of 0 (*P ≤ 0.05; **P ≤ 0.01; ****P ≤ 0.0001), one-sample t test, n = 4–12. (B) Effect of mutation N265I in the β2 subunit (NI) on responses to 50 μM propofol and compounds 7 and 22. Statistical significance is versus wild-type (WT) (*P ≤ 0.05; ***P ≤ 0.001), unpaired t test, n = 4–6.
Mutations at residue 265 in TM2 of the GABAA receptor β subunit have been previously shown to reduce actions of propofol (Jurd et al., 2003). We confirmed that β2(N265I) reduced the effects of 50 μM propofol (Fig. 7B), and asked if this mutation would also reduce the effects of our most potent novel compounds. For both compounds 7 and 22, the β2(N265I) mutation caused a significant decrease in potentiation, similar to that of propofol (Fig. 7B).
We considered the possibility that the compounds that did not have effect per se could be binding to the same site as propofol without producing any effect. If that were the case, they could act as competitive antagonists and block propofol effects. When 14 compounds (50 µM) were individually coapplied with propofol (2 µM), most did not have any effect, and six showed only a very small antagonistic effect on propofol potentiation, even with a concentration 25 times higher (Supplemental Fig. 2). Thus, none of the 14 compounds appeared to have clear antagonistic effects.
Discussion
The aim of this study was to investigate whether high-resolution crystal structures could be used to identify novel modulators of pLGICs. Docking against the crystallographic propofol binding pocket, as recently demonstrated by other groups (Liu et al., 2012), revealed challenges in targeting allosteric sites of membrane proteins with virtual screening approaches. A combined MD and molecular docking approach enabled us to explicitly account for interactions of potential ligands with both the protein and surrounding membrane environment. With this protocol, it was possible to build on previous docking studies (Liu et al., 2012) to perform a screen of commercially available chemicals for those that complemented the propofol binding pocket of GLIC. Of the 29 compounds selected for functional testing on GLIC, 55% showed significantly higher inhibition than vehicle, including three compounds (10%) that produced equal or greater modulation than propofol. This hit-rate compares favorably with previous virtual screening studies against membrane receptors. For example, virtual screens against the dopamine D3 receptor (Carlsson et al., 2011) and adenosine A2A receptor (Carlsson et al., 2010) crystal structures produced hit rates of 23 and 35%, respectively. As in the case of these GPCRs, the deep and enclosed intrasubunit pocket of GLIC is likely to interact favorably with many small organic compounds.
The level of activity of the hit compounds did not correlate with simple physicochemical properties, supporting that these allosteric modulators act via specific interactions with GLIC rather than changing the properties of the membrane as was historically suggested for anesthetics (McCreery and Hunt, 1978). Because of the small size of the site, compounds that were selected for virtual screening were restricted to molecules with less or equal to 13 non-H atoms; with this restriction, functional experiments on GLIC revealed no correlation between potency and molecular weight. Similarly, while all compounds identified by virtual screening fell in a logP range of 2.2 to 4.0, the potency of compounds in this range did not depend purely on lipophilicity, as the correlation between modulation and logP was low.
Given that physicochemical properties did not explain differences in GLIC modulation, we examined possible associations between potency and structural features. Several structural classes were capable of inhibiting GLIC activation. Identification of a single pharmacophore for modulation was not possible, but two-thirds of the compounds with disubstituted benzenes showed significant modulation. It should be noted that a majority of all discovered ligands were halogenated, which is also a characteristic of many anesthetics, particularly volatile agents as desflurane. Among the 12 most potent hit compounds (P ≤ 0.0001 in Fig. 2), nine share the feature of an aromatic ring structure connected to an unbranched side chain with at least three non-H atoms. However, the position of the side chains seems to be of marginal importance for methylbenzenes, as compounds 2, 3, and 4 all led to high inhibition. Intriguingly, even minor structural differences in the longer side chain led to differences in potency.
We used mutagenesis around the presumed propofol binding site in GLIC to test the specificity of hit compounds for the site targeted by virtual screening. One mutation, L206W, strongly reduced propofol inhibition and also inhibited the action of one of the seven compounds that we tested (compound 7). Notably, this L206W-sensitive compound was ranked among the top 10 molecules in the docking screen of over 150,000 compounds against the GLIC crystal structure. It was also an effective potentiator of GABAA receptors, and its modulation appeared to be specifically decreased by the propofol-blocking GABAA receptor mutation β2(N265I). These findings are consistent with the presence of a conserved binding site for propofol and compound 7 in GLIC and GABAA receptors. Compared with propofol, compound 7 is less lipophilic and could be valuable as a chemical tool for further understanding of pLGIC.
Conversely, most of the active compounds from the docking screen were not significantly affected by the L206W mutation. This result could suggest that many of our discovered ligands do not bind at the same site as propofol, and that the true docking hit rate—the percentage of the tested compounds that actually target the intrasubunit pocket—is lower than 55%. However, it is also possible that these compounds, as well as propofol, act via multiple sites, particularly given their small size and amphiphilic nature. It may also be that our model of anesthetic binding based on the GLIC crystal structure is imperfect, particularly given that this structure represents a presumed open state (Nury et al., 2011) that may differ dramatically from the conformation stabilized by allosteric inhibitors.
The M205W mutation was unique in that propofol acted as a potentiator at lower concentrations, but turned into an inhibitor at concentrations 100 µM and higher. These results could reflect a change in the functional outcome of binding, such that the targeted cavity is converted from an inhibitory into a potentiating site; however, this hypothesis does not easily explain why propofol acts as an inhibitor at higher concentrations. Alternately, the M205W mutation could create or favor a second binding site, at which propofol acts to enhance function. This two-site hypothesis was recently supported by work with n-alcohols (Howard et al., 2011), where mutations that altered the potentiating effects of short-chain alcohols on GLIC did not affect inhibition by long-chain alcohols, which suggests that they bind in a distinct site. Indeed, recent cocrystal structures with the volatile anesthetic bromoform associated binding in a novel inter-subunit cavity with GLIC potentiation, whereas channels inhibited by bromoform showed only binding in the intrasubunit cavity (Sauguet et al., 2013). Our virtual screening hit compound 7 appears to bind more specifically to the inhibitory binding site than propofol, as it was not affected by the potentiation-enhancing M205W mutation.
A third hypothesis is that propofol binding to the crystallographic site is actually not the important mechanism for channel inhibition. Interestingly, the four residues in closest proximity to the crystallographic propofol binding site in GLIC showed almost no effect on propofol modulation when mutated. However, tryptophan substitutions at M205 and L206, which are located deeper within the subunit than most of the predicted binding site residues, led to a significantly different propofol response than in the wild-type. These data suggest that propofol may bind deeper within the channel than predicted in the crystal structure, and/or may bind in multiple sites as suggested above. Indeed, recent MD studies have suggested a variety of alternative sites of anesthetics binding to pLGICs, including intersubunit cavities (Bondarenko et al., 2013; Spurny et al., 2013) and the channel pore (LeBard et al., 2012). This distinction may reflect the determination of the GLIC crystal structure with propofol in a presumed open state, whereas it is assumed that the inhibitory action of propofol stabilizes the closed state (Nury et al., 2011). Our data suggest that the functionally relevant inhibitory site for propofol may be more deeply buried in the intrasubunit cavity than indicated in the crystal structure.
Many of our hit compounds proved to modulate GABAA receptors in addition to GLIC, although none approached the high potency and effectiveness of propofol, and effects on behavioral pharmacology phenotypes such as loss of righting reflex are beyond the scope of this study. On one hand, the identification of GABAA modulators by virtual screening of GLIC suggests that these receptors share a conserved binding site for anesthetics. On the other hand, the low potency of these compounds indicates that the binding sites are different enough for virtual screening to select compounds that bind much more specifically to GLIC than to GABAA receptors. Although studies of the Torpedo nACh receptor have supported conservation of the crystallographic intrasubunit propofol site from GLIC in eukaryotic receptors (Jayakar et al., 2013), previous studies suggested a neighboring intersubunit site for propofol potentiation of GABAA receptors (Chiara et al., 2012). Our identification of hit compounds for GABAA receptors suggests the binding sites for propofol on both channels are similar; however, it remains plausible that the precise binding site on GABAA receptors differs from that on GLIC (Chiara et al., 2012), or that multiple sites of action contribute to the net pharmacologic profile (Murail et al., 2012). In either case, identification of novel anesthetics will be facilitated by new GABAA receptor homology models; our results suggest that the GLIC structure is a valid starting point for such modeling.
In this study, we demonstrate that computational screening of commercially available chemical libraries can prioritize candidates for new allosteric modulators of GLIC with a high hit rate, and identify novel compounds with propofol-like actions on GABAA receptors. In addition, our mutational studies on GLIC suggest the presence of a propofol site slightly deeper within the protein than suggested by the crystal structure. These findings emphasize the value of combining mutational, functional, and crystallographic data in understanding the structure and function of brain receptors, and in the development of better methods for drug discovery. In particular, the high hit rate of our approach in finding new ligands for GLIC, along with our more limited success with GABAA receptors, emphasizes the need for better GABAA receptor models in the search for new anesthetics.
Supplementary Material
Acknowledgments
The authors thank Dr. James R. Trudell for many insightful and valuable discussions; and R.J.H., C.M.B., M.A.C., U.S.L., and R.A.H. thank Mendy Black and Aanika Das for excellent technical assistance.
Abbreviations
- ANOVA
one-way analysis of variance
- DMSO
dimethylsulfoxide
- EC10
effective concentration 10
- GLIC
Gloeobacter violaceus ligand-gated ion channel
- pLGICs
pentameric ligand-gated ion channels
- MD
molecular dynamics
- nACh
nicotinic acetylcholine
- TM
transmembrane helices
Authorship Contributions
Participated in research design: Lindahl, Carlsson, Harris.
Conducted experiments: Heusser, Howard, Borghese, Cullins, Broemstrup, Lee, Lindahl, Carlsson.
Performed data analysis: Heusser, Howard, Borghese, Cullins, Broemstrup, Lee.
Wrote or contributed to the writing of the manuscript: Heusser, Howard, Borghese, Lindahl, Carlsson, Harris.
Footnotes
This work was supported by the National Institutes of Health National Institute on Alcohol Abuse and Alcoholism [Grants AA06399 and AA19875]; the Institute of Pharmaceutical Sciences, Swiss Federal Institute of Technology; the Swedish Foundation for Strategic Research and the Swedish e-Science Research Center; and the Knut and Alice Wallenberg Foundation. Computational resources were provided by the Swedish National Infrastructure for Computing.
Part of this work was presented as follows: Heusser SA (2012) Functional Validation of Rationally Designed Allosteric Ion Channel Modulators. Master’s thesis, Institute of Pharmaceutical Sciences, Swiss Federal Institute of Technology, Zurich, Switzerland.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Berger O, Edholm O, Jähnig F. (1997) Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys J 72:2002–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. (2009) X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature 457:111–114 [DOI] [PubMed] [Google Scholar]
- Bondarenko V, Mowrey D, Liu LT, Xu Y, Tang P. (2013) NMR resolved multiple anesthetic binding sites in the TM domains of the α4β2 nAChR. Biochim Biophys Acta 1828:398–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J, Coleman RG, Setola V, Irwin JJ, Fan H, Schlessinger A, Sali A, Roth BL, Shoichet BK. (2011) Ligand discovery from a dopamine D3 receptor homology model and crystal structure. Nat Chem Biol 7:769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson J, Yoo L, Gao ZG, Irwin JJ, Shoichet BK, Jacobson KA. (2010) Structure-based discovery of A2A adenosine receptor ligands. J Med Chem 53:3748–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers CC, Hawkins GD, Cramer CJ, Truhlar DG. (1996) Model for aqueous solvation based on class IV atomic charges and first solvation shell effects. J Phys Chem 100:16385–16398 [Google Scholar]
- Chiara DC, Dostalova Z, Jayakar SS, Zhou X, Miller KW, Cohen JB. (2012) Mapping general anesthetic binding site(s) in human α1β3 γ-aminobutyric acid type A receptors with [³H]TDBzl-etomidate, a photoreactive etomidate analogue. Biochemistry 51:836–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012 [DOI] [PubMed] [Google Scholar]
- Garcia PS, Kolesky SE, Jenkins A. (2010) General anesthetic actions on GABA(A) receptors. Curr Neuropharmacol 8:2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447 [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. (2009) Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature 457:115–118 [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. (2008) X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452:375–379 [DOI] [PubMed] [Google Scholar]
- Howard RJ, Murail S, Ondricek KE, Corringer PJ, Lindahl E, Trudell JR, Harris RA. (2011) Structural basis for alcohol modulation of a pentameric ligand-gated ion channel. Proc Natl Acad Sci USA 108:12149–12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, Cao Y. (2009) Automated docking screens: a feasibility study. J Med Chem 52:5712–5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. (2012) ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52:1757–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayakar SS, Dailey WP, Eckenhoff RG, Cohen JB. (2013) Identification of propofol binding sites in a nicotinic acetylcholine receptor with a photoreactive propofol analog. J Biol Chem 288:6178–6189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL. (2009) Efficient drug lead discovery and optimization. Acc Chem Res 42:724–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935 [Google Scholar]
- Jurd R, Arras M, Lambert S, Drexler B, Siegwart R, Crestani F, Zaugg M, Vogt KE, Ledermann B, Antkowiak B, et al. (2003) General anesthetic actions in vivo strongly attenuated by a point mutation in the GABA(A) receptor β3 subunit. FASEB J 17:250–252 [DOI] [PubMed] [Google Scholar]
- Krasowski MD, Jenkins A, Flood P, Kung AY, Hopfinger AJ, Harrison NL. (2001) General anesthetic potencies of a series of propofol analogs correlate with potency for potentiation of γ-aminobutyric acid (GABA) current at the GABA(A) receptor but not with lipid solubility. J Pharmacol Exp Ther 297:338–351 [PubMed] [Google Scholar]
- LeBard DN, Hénin J, Eckenhoff RG, Klein ML, Brannigan G. (2012) General anesthetics predicted to block the GLIC pore with micromolar affinity. PLOS Comput Biol 8:e1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhu T, Cramer CJ, Truhlar DG. (1998) New class IV charge model for extracting accurate partial charges from wave functions. J Phys Chem A 102:1820–1831 [Google Scholar]
- Liu R, Perez-Aguilar JM, Liang D, Saven JG. (2012) Binding site and affinity prediction of general anesthetics to protein targets using docking. Anesth Analg 114:947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorber DM, Shoichet BK. (1998) Flexible ligand docking using conformational ensembles. Protein Sci 7:938–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorber DM, Shoichet BK. (2005) Hierarchical docking of databases of multiple ligand conformations. Curr Top Med Chem 5:739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken ML, Borghese CM, Trudell JR, Harris RA. (2010) A transmembrane amino acid in the GABA(A) receptor β2 subunit critical for the actions of alcohols and anesthetics. J Pharmacol Exp Ther 335:600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCreery MJ, Hunt WA. (1978) Physico-chemical correlates of alcohol intoxication. Neuropharmacology 17:451–461 [DOI] [PubMed] [Google Scholar]
- Meng EC, Shoichet BK, Kuntz ID. (1992) Automated docking with grid-based energy evaluation. J Comput Chem 13:505–524 [Google Scholar]
- Murail S, Howard RJ, Broemstrup T, Bertaccini EJ, Harris RA, Trudell JR, Lindahl E. (2012) Molecular mechanism for the dual alcohol modulation of Cys-loop receptors. PLOS Comput Biol 8:e1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysinger MM, Carchia M, Irwin JJ, Shoichet BK. (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55:6582–6594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A, Honig B. (1991) A rapid finite difference algorithm, utilizing successive over-relaxation to solve the Poisson–Boltzmann equation. J Comput Chem 12:435–445 [Google Scholar]
- Nury H, Van Renterghem C, Weng Y, Tran A, Baaden M, Dufresne V, Changeux J-P, Sonner JM, Delarue M, Corringer P-J. (2011) X-ray structures of general anaesthetics bound to a pentameric ligand-gated ion channel. Nature 469:428–431 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612 [DOI] [PubMed] [Google Scholar]
- Rishton GM. (1997) Reactive compounds and in vitro false positives in HTS. Drug Discov Today 2:382–384 [Google Scholar]
- Sauguet L, Howard RJ, Malherbe L, Lee US, Corringer PJ, Harris RA, Delarue M. (2013) Structural basis for potentiation by alcohols and anaesthetics in a ligand-gated ion channel. Nat Commun 4:1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet BK, Kuntz ID. (1993) Matching chemistry and shape in molecular docking. Protein Eng 6:723–732 [DOI] [PubMed] [Google Scholar]
- Shoichet BK, Leach AR, Kuntz ID. (1999) Ligand solvation in molecular docking. Proteins 34:4–16 [DOI] [PubMed] [Google Scholar]
- Sorin EJ, Pande VS. (2005) Exploring the helix-coil transition via all-atom equilibrium ensemble simulations. Biophys J 88:2472–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurny R, Billen B, Howard RJ, Brams M, Debaveye S, Price KL, Weston DA, Strelkov SV, Tytgat J, Bertrand S, et al. (2013) Multisite binding of a general anesthetic to the prokaryotic pentameric Erwinia chrysanthemi ligand-gated ion channel (ELIC). J Biol Chem 288:8355–8364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasneem A, Iyer LM, Jakobsson E, Aravind L. (2005) Identification of the prokaryotic ligand-gated ion channels and their implications for the mechanisms and origins of animal Cys-loop ion channels. Genome Biol 6:R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Alqazzaz M, Ulens C, Lummis SC. (2012) The pharmacological profile of ELIC, a prokaryotic GABA-gated receptor. Neuropharmacology 63:761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner CA, Friedrich B, Setiawan I, Lang F, Bröer S. (2000) The use of Xenopus laevis oocytes for the functional characterization of heterologously expressed membrane proteins. Cell Physiol Biochem 10:1–12 [DOI] [PubMed] [Google Scholar]
- Warren GL, Andrews CW, Capelli A-M, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, et al. (2006) A critical assessment of docking programs and scoring functions. J Med Chem 49:5912–5931 [DOI] [PubMed] [Google Scholar]
- Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner P. (1984) A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc 106:765–784 [Google Scholar]
- Weiner SJ, Kollman PA, Nguyen DT, Case DA. (1986) An all atom force field for simulations of proteins and nucleic acids. J Comput Chem 7:230–252 [DOI] [PubMed] [Google Scholar]
- Weng Y, Yang L, Corringer PJ, Sonner JM. (2010) Anesthetic sensitivity of the Gloeobacter violaceus proton-gated ion channel. Anesth Analg 110:59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.