Abstract
Context
Epidermal growth factor receptor (EGFR) is overexpressed in up to 80% of colorectal and endometrial carcinomas. Deletions of the polyA tract in the 3′ untranslated region (3′ UTR) have been reported in microsatellite instability–high (MSI-H) colonic carcinomas, but their impacts on EGFR expression and downstream pathways are unclear. This phenomenon has not been reported in other MSI-H tumors.
Objective
To assess the 3′ UTR polyA tract of EGFR in both endometrial and colorectal carcinomas and the mutational status of EGFR downstream pathways.
Design
Ninety-eight colorectal carcinomas and 47 endometrial carcinomas were included. EGFR 3′ UTR polyA status was detected by capillary electrophoresis and Sanger sequencing. EGFR gene expression, EGFR copy numbers, and KRAS and BRAF mutation status were analyzed accordingly.
Results
The 3′ UTR polyA tract was deleted in 18 of 23 (78%) MSI-H versus 0 of 24 microsatellite-stable endometrial carcinomas (P < .001). Similar observations were seen in colorectal carcinomas, in which 29 of 36 (81%) MSI-H, 1 of 62 (1.6%) microsatellite instability–low, and none of the microsatellite-stable tumors harbored the deletion (P < .001). A moderate increase in EGFR mRNA level was observed in endometrial carcinomas with 3′ UTR polyA deletions versus those with wild-type polyA tract. Amplification of the EGFR gene was not observed. Deletions in polyA tract do not seem to affect the frequency of KRAS and BRAF mutations.
Conclusions
Deletions of EGFR 3′ UTR polyA are frequent in endometrial and colorectal carcinomas, are confined almost exclusively to MSI-H tumors, and do not affect KRAS and BRAF mutations.
The epidermal growth factor receptor (EGFR) is a receptor tyrosine kinase that belongs to the erbB proto-oncogene family.1 Binding of ligands to EGFR causes receptor dimerization and phosphorylation, leading to subsequent activation of its downstream targets, the phosphatidylinositol-3-kinase and mitogen-activated protein kinase pathways.1-3 Overexpression of EGFR has been observed in multiple types of solid tumors, including more than 80% of colorectal and endometrial carcinomas, 4-6 and correlates with tumor progression and prognosis.7-10
Recent studies have reported a possible correlation between EGFR mRNA level and the length of CA dinucleotide repeat or the length of a polyA tract in the 5′ and 3′ untranslated regions (UTRs) of the gene, respectively.11-13 However, the results are controversial. Yuan et al11 showed that the EGFR gene contained an A13/A14 polymorphism in the 3′ UTR, and mononucleotide or dinucleotide deletions in this polyA tract could stabilize EGFR mRNA and result in overexpression of EGFR protein in microsatellite instability–high (MSI-H) colon cancer cell lines. Baranovskaya and colleagues,13 in contrast, reported that deletions in the 3′ UTR polyA of EGFR had no effect on EGFR expression. Instead, the expansion of the 5′ UTR (CA)n repeats correlated with down-regulation of the EGFR gene.
Microsatellite instability (MSI) is a hallmark of tumors arising in hereditary nonpolyposis colon cancer syndrome and is present in 15% to 20% of sporadic colorectal carcinomas. A high frequency of deletion in the 3′ UTR polyA sequence of EGFR has been reported in MSI-H colon cancers. To our knowledge, the EGFR 3′ UTR polyA mutational status in endometrial carcinomas has never been reported.
In this study, we assessed for deletions of the 3′ UTR polyA sequence of the EGFR gene in a group of endometrial and colorectal carcinomas that were subdivided according to the MSI status as high (MSI-H), low (MSI-L), or stable (MSS). For the first time, we reported frequent deletions of the 3′ UTR polyA in MSI-H endometrial carcinomas, and this observation was further confirmed in colorectal carcinomas.
MATERIALS AND METHODS
Sample Selection
Cases included in this study were selected from approximately 500 formalin-fixed, paraffin-embedded colorectal and endometrial carcinoma samples submitted for MSI testing to the Molecular Diagnostics Laboratory at the University of Texas M. D. Anderson Cancer Center (Houston, Texas) from 2003 to 2009. Genomic DNA was available in 98 colorectal carcinoma (36 MSIH, 22 MSI-L, and 40 MSS) and 17 endometrial carcinoma (7 MSI-H and 10 MSS) cases. Because of the limited number of cases in the latter group, frozen tissues from 15 MSI-H and 15 MSS endometrial cancers were included. All the cases selected for this study had more than 50% tumors. The areas with the tumor were manually dissected to minimize contamination from adjacent stromal or inflammatory cells. A subgroup of the cases was from patients with Lynch syndrome.
Isolation of Genomic DNA and Determination of MSI Status
Following manual microdissection, genomic DNA was isolated from formalin-fixed, paraffin-embedded tissue sections of endometrial and colorectal carcinomas using the DNA Mini Extraction Kit (Qiagen, Valencia, California) or the PicoPure DNA Extraction Kit (MDS Analytical Technologies, Toronto, Ontario, Canada). No microdissection was performed on frozen endometrial tissue samples prior to DNA isolation. Microsatellite instability status was evaluated by fluorescence-labeled microsatellite marker polymerase chain reaction (PCR) followed by capillary electrophoresis fragment size analysis using an ABI 3130 sequencer and Genescan software (Applied Biosystems, Foster City, California). The microsatellite markers included the National Cancer Institute panel (BAT25, BAT26, D2S123, D5S346, D17S250) with or without BAT40 and/or TGFBRII. Carcinomas were classified as MSI-H if 2 or more markers showed altered allelic size in tumor DNA, MSI-L if one marker showed allelic shift, and MSS if no markers showed an allelic shift.
Detection of EGFR 3′ UTR PolyA Status by Capillary Electrophoresis and Sanger Sequencing
The polyA region of EGFR 3′ UTR was amplified by PCR using a 6-carboxyfluorescein–labeled forward primer (5′-ACAGAA ACGCATCCAGCAA-3′) and a reverse primer tagged with the 7mer fragment analysis research group consensus sequence (5′-GTTTCTTACTTGTGGCTTGTGCTCCTT-3′).11,14 Polymerase chain reaction amplification was performed using AmpliTaq Gold (Applied Biosystems) and 20 ng genomic DNA per the manufacturer’s instructions. The PCR thermocycling conditions were as follows: 95°C for 5 minutes, followed by 20 cycles of 95°C for 30 seconds, 55°C for 2 minutes, and 72°C for 1 minute. The annealing temperature was increased 0.5°C after each cycle; 20 cycles of 95°C for 30 seconds, 65°C for 1 minute, and 72°C for 3 minutes, and extension at 72°C for 20 minutes. Fragment analysis was performed using a 3100 Genetic Analyzer (Applied Biosystems). An M13-tagged primer was used in the sequencing reaction. Applied Biosystems’ 3700 DNA analyzer was used for sequencing.
Determination of EGFR Gene Expression by Real-Time Quantitative PCR
Total RNA was extracted from frozen endometrial tumors (15 MSI-H and 15 MSS) using an RNeasy Mini kit (Qiagen) following the manufacturer’s protocol. For cDNA synthesis, 100 ng total RNA was reverse transcribed using SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, California) in a final volume of 20 μL. The TaqMan gene expression assay for EGFR and the endogenous control glucuronidase beta (GUSB) were obtained from Applied Biosystems. The PCR reactions were performed as follows: 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The mRNA level of EGFR was calculated by relative quantitation using the comparative threshold cycle method.15
Determination of EGFR Copy Number Changes by Real-Time Quantitative PCR
EGFR copy numbers were determined by real-time quantitative PCR using TaqMan Copy Number Assays (Applied Biosystems) according to the manufacturer’s instructions in 26 endometrial and colorectal tumors with polyA deletions. The single copy gene/allele RNase P was used as a copy number reference gene. Polymerase chain reactions were performed using 20 ng genomic DNA, 1× TaqMan Universal PCR master mix, 1× TaqMan Copy Number Assay for EGFR, and 1× TaqMan Reference Assay for RNase P on a 7900HT Sequence Detection System (Applied Biosystems) in a 96-well format. The thermocycling conditions were as follows: 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. Copy number analysis was performed by Applied Biosystems CopyCaller Software.
Detection of KRAS and BRAF Mutation Status
KRAS mutations at codons 12, 13, and 61 and BRAF mutations at codons 11 and 15 were analyzed by pyrosequencing as described previously.16
Statistical Analysis
Statistical analyses were performed with 2-tailed t test. P values < .05 were considered significant.
RESULTS
Deletion of EGFR 3′ UTR PolyA Tract in Colorectal and Endometrial Carcinomas
To determine whether deletions in EGFR 3′ UTR polyA are present in endometrial carcinomas, we examined the 3′ UTR polyA length in 47 cases of endometrial carcinomas (23 MSI-H and 24 MSS cases). Eighteen of 23 (78%) MSI-H endometrial carcinomas had 3′ UTR polyA deletions, but no polyA deletion was detected in 24 MSS endometrial carcinomas (P < .001; Table 1). As shown in Table 1, 10 (43.5%), 4 (17.4%) and 4 (17.4%) of the 23 MSI-H endometrial carcinomas had the presence of a 1-, 2-, or 3-base deletion mutation within this element, resulting in an A12, A11, or A10 repeats, respectively. Four-base deletion A9 was not observed because of limited sample volume. Deletions identified by capillary electrophoresis were confirmed by Sanger sequencing.
Table 1.
Frequency and Size of EGFR 3′ UTR PolyA Deletion in Colorectal and Endometrial Carcinomas
| Carcinoma and size of deletion | MSI-H, % (No.) | MSI-L, % (No.) | MSS, % (No.) |
|---|---|---|---|
| Colorectal | |||
| 1 nucleotide | 38.8 (14/36) | 4 (1/22) | |
| 2 nucleotides | 25 (9/36) | 0 (0/22) | |
| 3 nucleotides | 13.9 (5/36) | 0 (0/22) | |
| 4 nucleotides | 2.8 (1/36) | 0 (0/22) | |
| Total | 81 (29/36) | 4 (1/22) | 0 (0/40) |
| Endometrial | |||
| 1 nucleotide | 43.5 (10/23) | 0 (0/24) | |
| 2 nucleotides | 17.4 (4/23) | 0 (0/24) | |
| 3 nucleotides | 17.4 (4/23) | 0 (0/24) | |
| 4 nucleotides | 0 (0/23) | 0 (0/24) | |
| Total | 78 (18/23) | 0 (0/24) |
Abbreviations: MSI-H, microsatellite instability–high; MSI-L, microsatellite instability–low; MSS, microsatellite-stable; UTR, untranslated region; EGFR, epidermal growth factor receptor.
Similar observations were made in colorectal carcinomas. Twenty-nine of 36 (81%) MSI-H colorectal carcinomas harbored deletions in the EGFR 3′ UTR polyA. The lengths of these deletions ranged from 1 to 4 nucleotides. In contrast, only 1 of 22 (4%) MSI-L tumors showed a single nucleotide deletion and none of the 40 MSS tumors had deletions (P < .001, Table 1). The mutations in the EGFR 3′ UTR is therefore significantly linked to the presence of MSI-H for both colorectal and endometrial carcinomas. In accordance with the observation in endometrial carcinomas, longer deletions had lower frequencies. The majority of the 3′ UTR polyA deletions identified were 1 or 2 nucleotides: 38.8% had 1 nucleotide deleted and 25% had 2 nucleotides deleted. Three- and 4- nucleotide deletions were present in 13.9% and 2.8% of cases, respectively (Table 1). Deletions identified by capillary electrophoresis were also confirmed by Sanger sequencing (representative cases shown in Figure 1).
Figure 1.
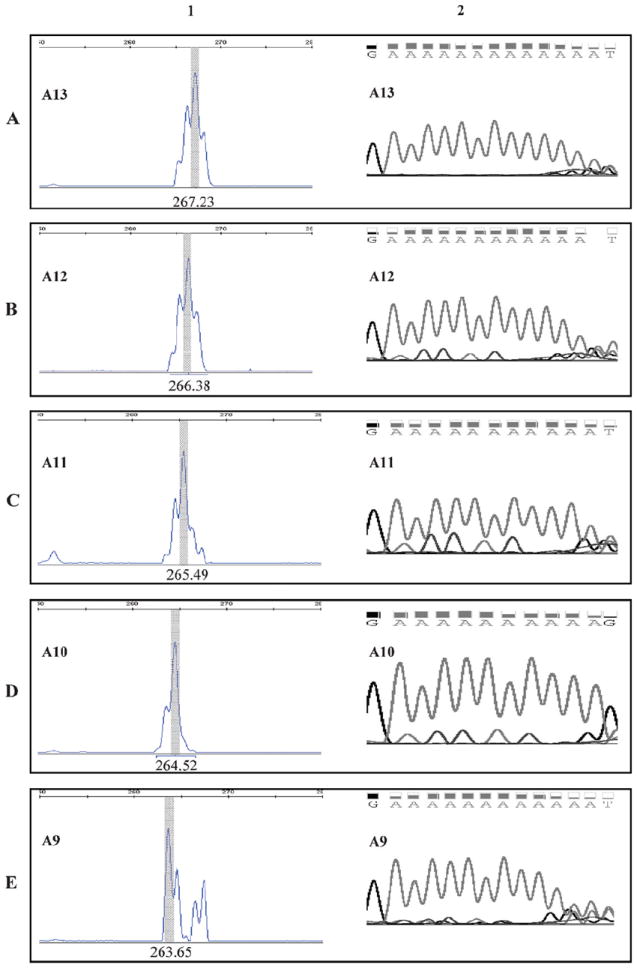
Deletion of polyA tract in EGFR 3′ untranslated region (3′ UTR) determined by capillary electrophoreses (CE) and Sanger sequencing. The length of EGFR 3′ UTR polyA was measured by CE and confirmed by Sanger sequencing as described in “Materials and Methods.” The size of the polymerase chain reaction fragment with wild-type (WT) 3′ UTR polyA is 267 bases. Column 1, fragment analysis by CE. Column 2, chromatograms of the 3′ UTR polyA regions by Sanger sequencing. A, A13: WT polyA tract with 13 nucleotides. B, A1: polyA tract with 1 nucleotide deletion. C, A11: polyA tract with 2-nucleotide deletion. D, A10: polyA tract with 3-nucleotide deletion. E, A9: polyA tract with 4-nucleotide deletion.
The EGFR 3′ UTR polyA has been reported to be polymorphic, with either 13 or 14 repeats in normal individuals.11 We therefore tested adjacent normal tissues from 24 MSI-H tumor cases that harbored 3′ UTR polyA deletions. The A13 repeat was present in 23 (96%) of the normal tissues whereas the A14 repeat was present in 1 case (4%) (data not shown). None of the normal tissues harbored a deletion mutation in the polyA tract, suggesting that the 3′ UTR polyA deletions identified are specific for carcinomas.
Correlation of EGFR 3′ UTR PolyA Deletion and EGFR mRNA Expression
Because Yuan et al11 showed that the EGFR gene polyA tract mutation could stabilize EGFR mRNA and result in expression of EGFR protein in cell line studies, we quantified the mRNA level of EGFR in 30 fresh-frozen endometrial carcinomas, including 15 MSI-H tumors and 15 MSS tumors. Overall, MSI-H tumors had a 1.5-fold higher EGFR level compared with MSS tumors (Figure 2, A). There was a moderate (1.5-fold) increase in EGFR mRNA in tumors with 3′ UTR polyA deletions compared with those that had wild-type (WT) 3′ UTR polyA (P = .29) (Figure 2, B). In MSI-H tumors, a 1.6-fold increase in EGFR mRNA was observed in tumors with 3′ UTR polyA deletion versus tumors without polyA deletion (Figure 2, C). This trend did not reach statistical significance, which is possibly because of the limitation of case volume.
Figure 2.
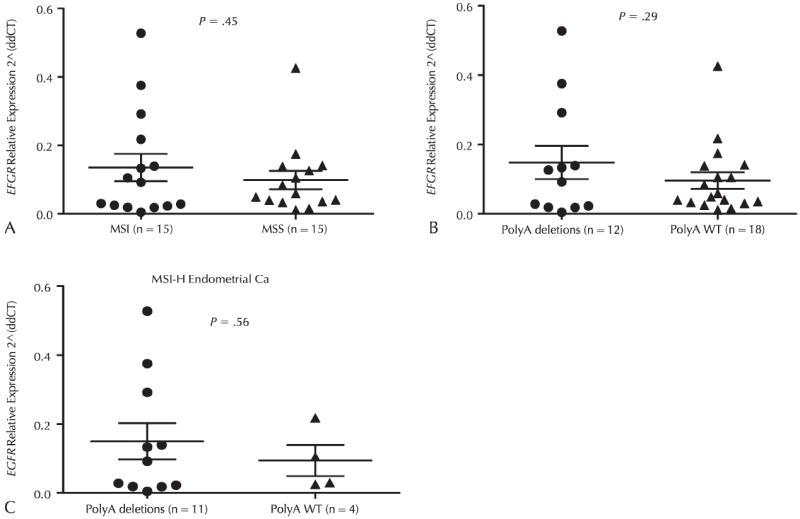
Expression of EGFR mRNA in endometrial carcinomas. The expression of EGFR was measured in fresh-frozen microsatellite instability–high (MSI-H) endometrial carcinomas (Ca) by real-time quantitative polymerase chain reaction as described in “Materials and Methods.” A, EGFR expression in MSI-H versus microsatellite-stable (MSS) tumors. (B) EGFR expression in tumors with and without polyA deletions. C, EGFR expression in MSI-H tumors with and without polyA deletions. Abbreviation, WT, wild-type; 2 (ddCT), relative expression level; EGFR, epidermal growth factor receptor.
Assessment of EGFR Copy Numbers by Real-Time Quantitative PCR
In order to eliminate the possibility that the observed EGFR overexpression in MSI endometrial carcinomas was due to EGFR gene amplification, we examined EGFR copy numbers by real-time quantitative PCR. No amplification of EGFR was observed in all cases tested, including 26 carcinomas with 3′ UTR polyA deletions (12 endometrial and 14 colorectal carcinomas) and 37 carcinomas with wild-type 3′ UTR polyA sequences (14 endometrial and 23 colorectal carcinomas) (Table 2).
Table 2.
Assessment of EGFR Copy Numbers by Real-Time qPCR
| Colorectal Carcinoma
|
Endometrial Carcinoma
|
|||
|---|---|---|---|---|
| ΔPolyA | WT PolyA | ΔPolyA | WT PolyA | |
| Case number | 14 | 23 | 12 | 14 |
| Copy number | 2 | 2 | 2 | 2 |
Abbreviations: Δ, deletion; qPCR, quantitative polymerase chain reaction; WT, wild-type; EGFR, epidermal growth factor receptor.
Evaluation of KRAS and BRAF Mutational Status
To assess for a possible link between EGFR 3′ UTR polyA deletion and the mutational status of KRAS and BRAF, which are downstream regulators of the EGFR signaling pathway, we tested samples with available DNA for KRAS mutations in codons 12, 13, and 61, and BRAF mutations in exons 11 and 15 (Table 3). In endometrial carcinomas, no significant difference was found in KRAS mutation between endometrial carcinomas with or without deletions in the polyA tract (Figure 3, A), but a significant correlation between the polyA deletion and microsatellite status was found in tumors with or without KRAS mutations (P < .001 and P = .02, respectively) (Figure 3, B and C). In MSI-H endometrial carcinomas, tumors with polyA deletions are more likely to have KRAS mutations (Table 3). Only 1 of 47 endometrial carcinomas harbored a BRAF V600E mutation, which is a MSI-H case with a 3-nucleotide 3′ UTR polyA deletion. In colorectal carcinomas, the frequencies of KRAS and BRAF mutations in tumors with polyA deletions were higher compared with tumors with WT polyA tract in both MSI-H and MSS tumors (Table 3).
Table 3.
KRAS and BRAF Mutation in Colorectal and Endometrial Carcinomas
| Genes | Colorectal Carcinoma
|
Endometrial Carcinoma
|
||||
|---|---|---|---|---|---|---|
| MSI-H
|
MSS
|
MSI-H
|
MSS
|
|||
| ΔPolyA, % (No./Total) | WT PolyA, % (No./Total) | WT PolyA, % (No./Total) | ΔPolyA, % (No./Total) | WT PolyA, % (No./Total) | WT PolyA, % (No./Total) | |
| KRAS | 11 (3/26) | 8 (2/26) | 38 (8/21) | 13 (3/23) | 9 (2/23) | 22 (5/23) |
| BRAF | 17.2 (5/29) | 6.9 (2/29) | 12 (2/17) | 4 (1/26) | 0 (0/26) | 0 (0/26) |
Abbreviations: Δ, deletion; MSI-H, microsatellite instability–high; MSS, microsatellite-stable; WT, wild-type.
Figure 3.
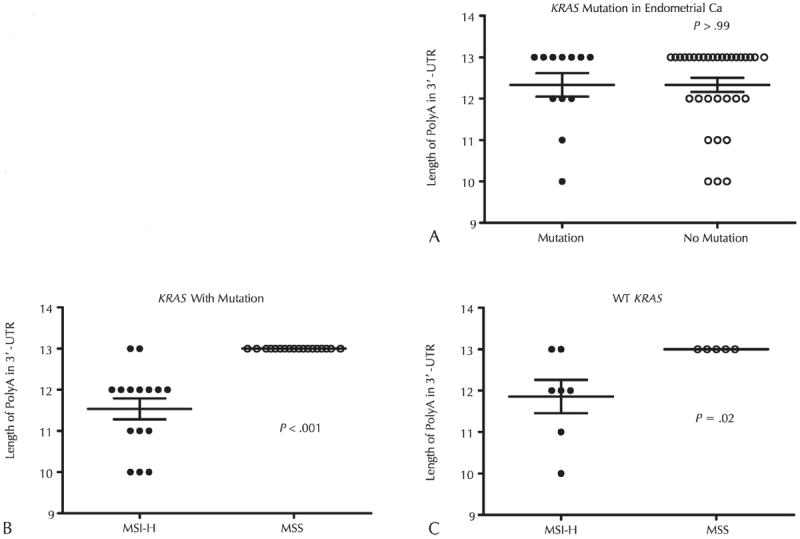
Association analysis between deletions in the length of EGFR 3′ untranslated region (3′ UTR) polyA tract and KRAS mutation in endometrial carcinoma (Ca). Statistical analyses were performed with 2-tailed t test. P values < .05 were considered significant. A, The association of the length of the polyA and microsatellite status in endometrial Ca cases with KRAS mutation. B, The association of polyA tract length and microsatellite status in endometrial carcinoma cases with KRAS mutation. C, The correlation of polyA tract length and microsatellite status in endometrial Ca with wild-type (WT) KRAS. Abbreviations: UTR, untranslated region.
COMMENT
Previous publications had shown that the EGFR gene contains a 3′ UTR polyA tract that commonly carries deletions that affect EGFR expression in colorectal carcinoma cell lines and limited cases of patient samples. The polyA deletion was also found to be related to MSI status. The 3′ UTR polyA tract had not been assessed in endometrial carcinomas, another tumor type in which a substantial subset of cases exhibit microsatellite instability. Here, we showed for the first time that these deletions were very common, present in almost 80% of cases, and were confined almost exclusively to MSI-H tumors.
The 3′ UTR was reported to modulate gene expression by altering RNA stability.11,17-19 We observed that in endometrial carcinomas, there was a slightly higher EGFR mRNA level in MSI-H tumors with deletions in 3′ UTR polyA compared with MSI-H tumors without polyA deletion and MSS tumors. Although not statistically significant, the trend is consistent with the observation on colorectal carcinoma cell lines.11 A recent study using chromogenic in situ hybridization showed that immunohistochemical expression of EGFR in colorectal carcinoma correlated with a high level of EGFR gene amplification, contrary to most of the reports in the literature.20,21 In our cohort, gene amplification was not observed in endometrial and colorectal carcinomas with deletions in polyA tract. Although EGFR is overexpressed in several solid tumors,21,22 reduced EGFR expression in tumors compared with normal tissues has been reported.20 Similar results were also obtained in MSI-H colorectal carcinomas in comparison with normal tissue.13 One possible explanation for this paradox is that the activation of apoptotic pathways by EGFR was through other ligands, such as tumor growth factors.23,24
Previous studies have shown polymorphism of EGFR 3′ UTR (A13/A14) in normal tissues.11 In our study, only 1 out of 24 cases showed this polymorphism in normal tissues. In addition to the 1- and 2-nucleotide deletions reported in the same study,11 we identified subgroups of MSI-H colorectal tumors with 3- or 4-nucleotide deletions. The high incidence of polyA deletions in MSI-H tumors suggested that deletion of 3′ UTR polyA is characteristic of microsatellite unstable carcinomas.
It is well documented that the clinical response in endometrial cancers treated with EGFR inhibitors does not correlate with EGFR immunohistochemical expression and the mutational status of KRAS and BRAF predicts the response to anti-EGFR therapy.25-29 Also, it has been proposed that MSI tumors are likely to have a lower incidence of RAS mutations.30 Although no direct correlations between deletions in polyA tract and the incidence of KRAS and BRAF mutations were observed, we found that in MSI-H endometrial and colorectal carcinomas, tumors with polyA deletions were more likely to have BRAF or KRAS mutations.
In summary, we found that the deletion in the 3′ UTR polyA tract of EGFR is a common somatic mutation that occurs in MSI-H colorectal and endometrial carcinomas. No significant changes of EGFR mRNA levels were found in association with specific mutations. Our findings indicate that the deletions in the EGFR 3′ UTR polyA tract represent a marker of DNA mismatch repair deficiency, similar to microsatellite markers routinely used for MSI testing. The pathogenetic and clinical significance of deletions in the 3′ UTR polyA tract of EGFR on tumor progression and therapeutic response warrant further analysis.
Acknowledgments
This work was funded in part by NIH Uterine Cancer Spore grant 1P50CA098258-01 to Dr Broaddus.
Footnotes
The authors have no relevant financial interest in the products or companies described in this article.
References
- 1.Yarden Y. The EGFR family and its ligands in human cancer: signalling mechanisms and therapeutic opportunities. Eur J Cancer. 2001;37(suppl 4):S3–S8. doi: 10.1016/s0959-8049(01)00230-1. [DOI] [PubMed] [Google Scholar]
- 2.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110(6):669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon MA. Ligand-induced ErbB receptor dimerization. Exp Cell Res. 2009;315(4):638–648. doi: 10.1016/j.yexcr.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oza AM, Eisenhauer EA, Elit L, et al. Phase II study of erlotinib in recurrent or metastatic endometrial cancer: NCIC IND-148. J Clin Oncol. 2008;26(26):4319–4325. doi: 10.1200/JCO.2007.15.8808. [DOI] [PubMed] [Google Scholar]
- 5.Vallbohmer D, Lenz HJ. Epidermal growth factor receptor as a target for chemotherapy. Clin Colorectal Cancer. 2005;5(suppl 1):S19–S27. [PubMed] [Google Scholar]
- 6.Cunningham MP, Essapen S, Thomas H, et al. Coexpression of the IGF-IR, EGFR and HER-2 is common in colorectal cancer patients. Int J Oncol. 2006;28(2):329–335. [PubMed] [Google Scholar]
- 7.Inada S, Koto T, Futami K, Arima S, Iwashita A. Evaluation of malignancy and the prognosis of esophageal cancer based on an immunohistochemical study (p53, E-cadherin, epidermal growth factor receptor) Surg Today. 1999;29(6):493–503. doi: 10.1007/BF02482343. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. Eur J Cancer. 2001;37(suppl 4):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 9.Roberts RB, Min L, Washington MK, et al. Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis. Proc Natl Acad Sci U S A. 2002;99(3):1521–1526. doi: 10.1073/pnas.032678499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasei M, Modjtahedi H, Ale-Booyeh O, et al. Amplification and expression of EGFR and ERBB2 in Wilms tumor. Cancer Genet Cytogenet. 2009;194(2):88–95. doi: 10.1016/j.cancergencyto.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Z, Shin J, Wilson A, et al. An A13 repeat within the 3′-untranslated region of epidermal growth factor receptor (EGFR) is frequently mutated in microsatellite instability colon cancers and is associated with increased EGFR expression. Cancer Res. 2009;69(19):7811–7818. doi: 10.1158/0008-5472.CAN-09-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rego RL, Foster NR, Smyrk TC, et al. Prognostic effect of activated EGFR expression in human colon carcinomas: comparison with EGFR status. Br J Cancer. 2010;102(1):165–172. doi: 10.1038/sj.bjc.6605473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranovskaya S, Martin Y, Alonso S, et al. Down-regulation of epidermal growth factor receptor by selective expansion of a 5′-end regulatory dinucleotide repeat in colon cancer with microsatellite instability. Clin Cancer Res. 2009;15(14):4531–4537. doi: 10.1158/1078-0432.CCR-08-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ballard L, Adams P, Bao Y, et al. Strategies for genotyping: effectiveness of tailing primers to increase accuracy in short tandem repeat determinations. J Biomol Tech. 2002;13(1):20–29. [PMC free article] [PubMed] [Google Scholar]
- 15.Luthra R, Singh RR, Luthra MG, et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27(52):6667–6678. doi: 10.1038/onc.2008.256. [DOI] [PubMed] [Google Scholar]
- 16.Zuo Z, Chen SS, Chandra PK, et al. Application of COLD-PCR for improved detection of KRAS mutations in clinical samples. Mod Pathol. 2009;22(8):1023–1031. doi: 10.1038/modpathol.2009.59. [DOI] [PubMed] [Google Scholar]
- 17.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’ for pathology? Nat Med. 2000;6(6):637–641. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 18.Kondo S, Kubota S, Eguchi T, et al. Characterization of a mouse ctgf 3′-UTR segment that mediates repressive regulation of gene expression. Biochem Biophys Res Commun. 2000;278(1):119–124. doi: 10.1006/bbrc.2000.3780. [DOI] [PubMed] [Google Scholar]
- 19.Puga I, Lainez B, Fernandez-Real JM, et al. A polymorphism in the 3′ untranslated region of the gene for tumor necrosis factor receptor 2 modulates reporter gene expression. Endocrinology. 2005;146(5):2210–2220. doi: 10.1210/en.2004-1366. [DOI] [PubMed] [Google Scholar]
- 20.Koenders PG, Peters WH, Wobbes T, Beex LV, Nagengast FM, Benraad TJ. Epidermal growth factor receptor levels are lower in carcinomatous than in normal colorectal tissue. Br J Cancer. 1992;65(2):189–192. doi: 10.1038/bjc.1992.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koenders PG, Beex LV, Kienhuis CB, Kloppenborg PW, Benraad TJ. Epidermal growth factor receptor and prognosis in human breast cancer: a prospective study. Breast Cancer Res Treat. 1993;25(1):21–27. doi: 10.1007/BF00662397. [DOI] [PubMed] [Google Scholar]
- 22.de Wit PE, Moretti S, Koenders PG, et al. Increasing epidermal growth factor receptor expression in human melanocytic tumor progression. J Invest Dermatol. 1992;99(2):168–173. doi: 10.1111/1523-1747.ep12616793. [DOI] [PubMed] [Google Scholar]
- 23.Hemmings C, Broomfield A, Bean E, Whitehead M, Yip D. Immunohistochemical expression of EGFR in colorectal carcinoma correlates with high but not low level gene amplification, as demonstrated by CISH. Pathology. 2009;41(4):356–360. doi: 10.1080/00313020902884477. [DOI] [PubMed] [Google Scholar]
- 24.Scartozzi M, Bearzi I, Mandolesi A, et al. Epidermal growth factor receptor (EGFR) gene copy number (GCN) correlates with clinical activity of irinotecancetuximab in K-RAS wild-type colorectal cancer: a fluorescence in situ (FISH) and chromogenic in situ hybridization (CISH) analysis. BMC Cancer. 2009;9:303. doi: 10.1186/1471-2407-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fakih MM. KRAS mutation screening in colorectal cancer: from paper to practice. Clin Colorectal Cancer. 2010;9(1):22–30. doi: 10.3816/CCC.2010.n.003. [DOI] [PubMed] [Google Scholar]
- 26.Italiano A, Hostein I, Soubeyran I, et al. KRAS and BRAF mutational status in primary colorectal tumors and related metastatic sites: biological and clinical implications. Ann Surg Oncol. 2010;17(5):1429–1434. doi: 10.1245/s10434-009-0864-z. [DOI] [PubMed] [Google Scholar]
- 27.Lievre A, Blons H, Laurent-Puig P. Oncogenic mutations as predictive factors in colorectal cancer. Oncogene. 2010;29:3033–3043. doi: 10.1038/onc.2010.89. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Crapez E, Mineur L, Emptas H, Lamy PJ. KRAS status analysis and anti-EGFR therapies: is comprehensiveness a biologist’s fancy or a clinical necessity? Br J Cancer. 2010;102(6):1074–1075. doi: 10.1038/sj.bjc.6605582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Modjtahedi H, Essapen S. Epidermal growth factor receptor inhibitors in cancer treatment: advances, challenges and opportunities. Anticancer Drugs. 2009;20(10):851–855. doi: 10.1097/CAD.0b013e3283330590. [DOI] [PubMed] [Google Scholar]
- 30.Ishii S, Xu YH, Stratton RH, Roe BA, Merlino GT, Pastan I. Characterization and sequence of the promoter region of the human epidermal growth factor receptor gene. Proc Natl Acad Sci U S A. 1985;82:4290–4294. doi: 10.1073/pnas.82.15.4920. [DOI] [PMC free article] [PubMed] [Google Scholar]


