Abstract
Sample displacement chromatography (SDC) in reversed-phase and ion-exchange modes was introduced approximately twenty years ago. This method was first used for the preparative purification of peptides and proteins. Recently, SDC in ion-exchange mode was also successfully used for enrichment of low abundance proteins from human plasma. In this paper, the use of SDC for the separation of plasma proteins in hydrophobic interaction mode is demonstrated. By use of two or more columns coupled in series during sample application, and subsequent elution of detached columns in parallel, additional separation of bound proteins was achieved. Further low-abundance, physiologically active proteins could be highly enriched and detected by ESI-MS/MS.
Keywords: Sample displacement chromatography, Hydrophobic interaction, Human plasma, Therapeutic proteins
1. Introduction
Hodges’ group [1, 2] introduced sample displacement chromatography (SDC) at the end of the 1980s for preparative purification of peptides in reversed-phase mode. This group demonstrated that, under overloading conditions, there is competition among the sample components for the binding sites on the surface of the stationary phase. The more peptides with higher affinity compete for the hydrophobic sites of the support, the more components with lower affinity to the surface will be displaced from the column and replaced with high affinity binding ones. A few years later, Veeraragavan et al. [3] demonstrated that SDC can be also used for the separation of some standard proteins in ion-exchange mode. After further development, Hodges’ group used SDC for simple and cost-effective purification of synthetic peptides [4], as well as preparative isolation of proteins from troponin, a skeletal muscle multi-protein complex [5]. Manseth et al. [6] applied SDC in affinity mode – Heparin Sepharose was used for the purification of thrombin from plasma of Atlantic salmon. This was the first time that the separation of a physiologically active protein was achieved by use of SDC. We recently demonstrated the application of ion-exchange monolithic supports for separation of proteins from human plasma. When SDC in anion- and cation-exchange mode was used, several physiologically active middle- and low-abundance proteins could be highly enriched by use of this chromatographic procedure [7].
Hydrophobic interaction chromatography (HIC) is now widely used as a method for the purification of monoclonal antibodies [8, 9] and other physiologically active proteins produced by use of recombinant technology [10, 11]. However, after early experiments, the use of this chromatographic technique for both analytical and preparative separation of proteins from human serum and plasma is relatively uncommon [12, 13]. Moreover, despite its high potential, HIC is hardly ever used for sample preparation in order to reduce the complexity of biological materials prior to proteomic analyses [14, 15].
Displacement chromatography was discovered by Tiselius in 1943 [16]. It was initially used for separation of amino acids [17] and further developed by Horváth’s group in the early 1980s [18]. In displacement chromatography, the sample is introduced onto the column, and then displaced by a constant infusion of a displacer solution. The affinity of the displacer for the stationary phase has to be higher than the affinity of any feed components [19]. Displacement chromatography of proteins and peptides is mainly performed in ion-exchange [19], followed by hydrophobic interaction mode [20].
In this paper we demonstrate that SDC in HIC mode is an effective method for the separation of complex biological mixtures and the concentration of low abundance proteins in both micro-preparative and preparative scale.
2. Materials and methods
2.1. Starting material
The starting material for HIC was cryopoor, single donor human plasma (Rhode Island Blood Center, Providence, RI, USA). After donation, plasma samples were screened in order to exclude the presence of blood-borne viruses (hepatitis A, B and C and HIV). Cryoglobulins were removed from plasma by precipitation at 4°C as described previously [21]. The cryopoor human plasma was diluted two-fold with 10 mM Tris-HCl, pH 7.4 (Buffer B). If 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4 was used as Buffer A, the same volume of 3.4 M (NH4)2SO4 in Buffer B was added to diluted plasma, in order to adjust the starting salt concentration to 1.7 M ammonium sulfate. When chromatography was performed by use of 4M NaCl in Buffer B as mobile phase (Buffer A), the proper concentration was reached by direct addition of this salt to diluted plasma. After storage at 4°C for 1 hour, precipitated proteins were removed by centrifugation at 3000 rpm (Centrifuge 5804 R, Eppendorf, Hamburg, Germany). Protein was determined in both precipitate and supernatant (sample for further chromatographic separation) and both samples were also analyzed by SDS-PAGE.
2.2. Hydrophobic interaction chromatography
For hydrophobic interaction chromatography, glass columns (Tosoh Bioscience, Stuttgart, Germany) with an I.D. of 6.5 mm packed with Toyopearl Phenyl 650 S support (Tosoh Bioscience) was used. The bed volume was 0.35 mL. After washing with HPLC water, the column was equilibrated with the high ionic strength buffer (Buffer A), containing either 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4, or 4M NaCl, and the sample was loaded. Unbound proteins were collected and subsequently analyzed. After sample application, the column was washed with at least five column volumes of Buffer A, and bound proteins were eluted with the low ionic strength buffer (Buffer B, 10 mM Tris-HCl, pH 7.4 ). The flow rates for all chromatographic separations were between 0.5 and 1 mL/min. In some runs under overloading conditions, three identical columns coupled in series were used. After sample loading and wash with Buffer A, columns were detached, and bound proteins were eluted in three parallel, separate steps. All runs were performed at 4–8°C, and a BioLogic Duo Flow chromatographic system (BioRad, Hercules, CA, USA) was used. Proteins were detected by UV absorption at 280, 260 and 210 nm. Protein amounts in collected fractions and starting material were determined with the Bicinchoninic Acid Protein Assay kit (Pierce, Rockford, IL, USA) according to the manufacturer’s procedure. Each experiment was performed, at least, in triplicate.
2.3. SDS-PAGE
For SDS-PAGE, approximately 15 µg protein of each sample was solubilized in NuPAGE sample buffer (Invitrogen, Carlsbad, CA, USA) and separated as described previously [22]. The gels were stained with GelCodeBlue dye (Pierce) and visualized by a VersaDoc Imaging System (BioRad).
2.4. Sample preparation for MS analysis
For “in-solution” digestion, 50 µg of the acetone-precipitated and denatured protein pellet was resolubilized in 100 µl of NH4HCO3 (pH 8.0) / 8 M urea. The resolubilized proteins were reduced with 20 mM dithiothreitol (37°C, 45 min) and then alkylated with 50 mM iodoacetamide at room temperature for 30 min in the dark. Before tryptic digestion, 100 mM ammonium bicarbonate buffer was added to reduce the concentration of urea. Trypsin was added to the protein mixture at an enzyme to substrate ratio of 1: 60 (w/w). After incubating at 37°C overnight, the tryptic peptides were dried in a vacuum centrifuge (Vacufuge, Eppendorf). The pellet was then redissolved in a solution of 0.5% (v/v) formic acid and 20% (v/v) acetonitrile, with vacuum drying again. Subsequently, the peptides were isolated using a strong cation exchange TipTop™ (PolyLC, Inc., Columbia, MD, USA) according to the manufacturer’s instructions after resuspending in the same solvent and confirming the pH value. The resulting tryptic peptides were dried once more and were subject to the LC-MS/MS analysis after being redissolved in formic acid:water:acetonitrile:trifluoroacetic acid mixture (0.1:95:5:0.01).
2.5. Identification of proteins with LC-MS/MS
Tryptic digests of whole fractions obtained by ion-exchange chromatography (“in-solution” digestion) were separated with a reversed-phase column (C-18 PepMap 100, LC Packings/Dionex, Synnyvale, CA, USA) as previously described [23]. Briefly: The column eluate was introduced directly onto a QSTAR XL mass spectrometer (Applied Biosystems and Sciex, Concord, Ontario, Canada) via electrospray ionization (ESI). Half-second MS scans (300–1500 Thompson, Thompson (Th) = Da/z) were used to identify candidates for fragmentation during MS/MS scans. Up to five 1.5 s MS/MS scans (65–1500 Th) were collected after each scan. An ion had to be assigned a charge in the range of +2 to +4. The dynamic exclusion was 40 s. Protein identifications were completed with ProteinPilot (Applied Biosystems and Sciex), using the human and “RefSeq” databases from NCBI (http://www.ncbi.nlm.nih.gov/RefSeq/). ProteinPilot is the successor to ProID and ProGroup, and uses the same peptide and protein scoring method. Scores above 2.0 require that at least two sequence-independent peptides have been identified [23].
In parallel experiments, an additional LC-MS/MS system was used (Agilent Technologies, Paolo Alto, CA, USA, and Thermo Electron Corporation, San Jose, CA, USA). When this system was used, tryptic peptides were separated on a 12 cm (75 µm I.D.) analytical column with 3 µm Monitor C18 resin (Orochem Technologies, Inc., Lombard, IL, USA) and containing an integrated 10 µm ESI emitter tip (PicoTip; New Objective, Woburn, MA, USA). Solvent A was 0.1 M acetic acid in water and solvent B was 0.1 M acetic acid in acetonitrile. Peptides were eluted using a linear acetonitrile gradient (0–70% solvent B over 30 min).
Eluting peptides were introduced onto an LTQ Orbitrap Velos hybrid mass spectrometer (Thermo Scientific, San Jose, CA) with a 1.8 kV electrospray voltage. Full MS scans in the m/z range of 300–1700 were followed by data-dependent acquisition of MS/MS spectra for the ten most abundant ions, using a 30-second dynamic exclusion time.
Protein identification was performed in, at least, two independent experiments.
Peak list files were created from the mass spectrometer file by the program extract_msn.exe, using the following settings: The mass had to fall in the range of 600 to 4500 Daltons. The minimum total ion current for the scan had to be over 1000. The precursor tolerance for grouping was 0.005 Daltons, with no differing intermediate scans allowed and only a single scan required to create a peak file. The minimum signal-to-noise for a peak to be written to the peak file was 3, and 5 such peaks had to be found for a peak file to be created. The program determined charge states.
A program developed in-house was used to concatenate the peak files into a Mascot Generic Format (MGF). Database searching using a human IPI database (v. 3.79; downloaded January 23, 2011) was performed by MASCOT [27]. The precursor-ion tolerance was 7 ppm and the fragment-ion tolerance was 0.5 Daltons. Enzymatic digestion was specified as trypsin, with up to 2 missed cleavages allowed. The search database contained concatenated real (“target”) and sequence-reversed (“decoy”) proteins. The identifications were filtered on MOWSE score to yield a group of peptide assignments with a 1% false discovery rate [24].
3. Results
3.1. Protein precipitation
After treatment with 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4, roughly 22–24% of plasma proteins were precipitated and removed from the sample. The ten most abundant proteins in the precipitate are shown in Table 1A. The SDS-PAGE of precipitated proteins and the supernatant (starting material), and the complete list of identified proteins in the precipitate are shown in the Supplement (Figure S1A and Table S1A).
Table 1.
| A Major plasma proteins in the 1.7 M (NH4)2SO4. precipitate identified by LC-MS/MS. | |||
|---|---|---|---|
| Acc. No. | Protein name | Score | Uniq. pept. |
| IPI007839 | Complement C3 | 3892.14 | 78 |
| IPI22513100.3 | Apolipoprotein B | 3471.07 | 72 |
| IPI00745872.2 | Human serum albumin | 2822.99 | 56 |
| IPI00478003.3 | Alpha-2-macroglobulin | 2762.14 | 49 |
| IPI00887154.2 | Complement component 4B | 1852.10 | 35 |
| IPI00022463.2 | Serrotransferrin | 1841.48 | 37 |
| IPI00029739.5 | Complement factor H | 1541.70 | 31 |
| IPI00029717.1 | Fibrinogen | 1489.83 | 28 |
| IPI00867588.1 | Fibronectin | 1375.11 | 27 |
| IPI00017601.1 | Ceruloplasmin | 1321.44 | 24 |
| B Major plasma proteins in the 4M NaCl precipitate identified by LC-MS/MS. | |||
|---|---|---|---|
| Acc. No. | Protein name | Score | Uniq. pept. |
| IPI00745872.2 | Human serum albumin | 2887.13 | 55 |
| IPI00783987.2 | Complement C3 | 2879.83 | 56 |
| IPI00022229.2 | Apolipoprotein B | 2437.76 | 50 |
| IPI00022463.2 | Serrotransferrin | 2298.05 | 41 |
| IPI00478003.3 | Alpha-2-macroglobulin | 2064.90 | 37 |
| IPI00298497.3 | Fibrinogen | 1610.39 | 32 |
| IPI00845263.1 | Fibronectin | 1463.43 | 29 |
| IPI00019580.1 | Plasminogen | 1428.37 | 30 |
| IPI00019580.1 | Complement component 4B | 1317.76 | 25 |
| IPI00641737.1 | Haptoglobin | 1093.51 | 22 |
| IPI00947307.1 | Ceruloplasmin | 642.2 | 14 |
- Plasminogen
- Alpha-1-antitrypsin
- lal, heavy chain 4
- lalp
- Alpha-1-antichymotrypsin
- Prothrombin
- Alpha-1-antitrypsin
- Prothrombin
- lalp
- Alpha-1-antichymotrypsin
- Antithrombin-lll
- lal heavy chain 4
By use of 4M NaCl in 10 mM Tris-HCl, pH 7.4, only about half as much (11–12%) protein is precipitated from plasma (compared to the 22–24% that is precipitated with 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4). The ten most abundant proteins in the NaCl precipitate are also listed in the Table 1B. Again, the complete list of identified proteins and the SDS-PAGE of the NaCl precipitate and supernatant are shown in the Supplement (Figure S1B and Table S1B).
3.2.1. Separation with 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4 as a mobile phase
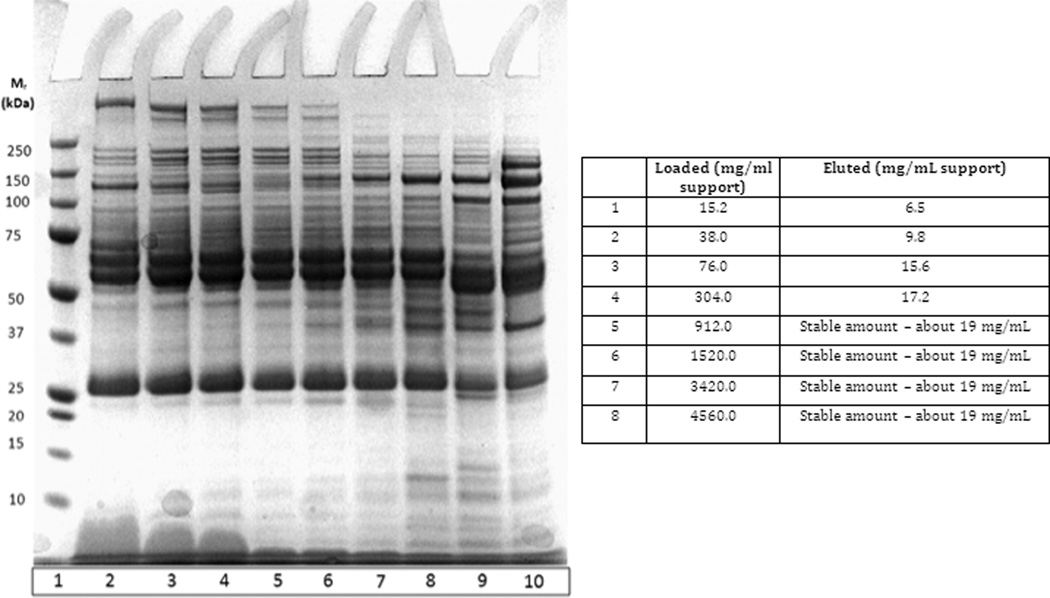
After precipitation with 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4 (Buffer A, see Materials and methods), different amounts of human plasma proteins (between 15 and 4560 mg protein/mL support) were applied to a hydrophobic interaction column by adjusting the volume of the supernatant that was applied. In these experiments, the column volume was 0.34 mL. After washing with 5 column volumes of Buffer A, bound proteins were eluted with 100% Buffer B (10 mM Tris-HCl, pH 7.4) by use of a step gradient. A typical chromatogram for this kind of separation is shown in Figures 1A and B. The corresponding SDS-PAGE of eluted proteins after loading different amounts of sample to the column packed with Toyopearl 650S Phenyl is shown in Figure 2.
Figure 1.
Separation of plasma proteins by HIC. After treatment with 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4 (Buffer A) and removal of precipitated proteins by centrifugation (see Materials and methods) 26 mg of plasma protein from the supernatant were applied to the Toyopearl Phenyl 650 S column (I.D. 6.5 mL, column volume 0.34 mL). After washing with 5 mL of Buffer A, bound proteins were eluted with 10 mM Tris-HCl, pH 7.4 (Buffer B). A – Sample application step; B – elution step. Optical density was determined at 280, 260 and 210 nm. Chromatographic conditions – Flow rate 0.5 mL/min, temperature 4–8°C, pressure 1 bar. Collected fractions were analyzed by SDS-PAGE, and LC-MS/MS – see Table 1A.
Figure 2.
Separation of proteins from human plasma on the hydrophobic interaction support Toyopearl Phenyl 650 S (column volume 0.34 mL). 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4, was used as Buffer A. Increasing amount of human plasma was applied. Bound proteins were eluted with 10 mM Tris-HCl, pH 7.4. SDS-PAGE of eluted proteins is shown. For other conditions – see Materials and methods and Figure 1.
3.2.1. Separation with 4M NaCl in 10 mM Tris-HCl, pH 7.4 as a mobile phase
In a parallel experiment, proteins were precipitated with 4M NaCl in a solution that was buffered with 10 mM Tris-HCl (pH 7.4). Again, different amounts of sample (between 15 and 4560 mg protein/mL support) were applied to the same Toyopearl 650S Phenyl column. The SDS-PAGE of eluted proteins after loading increasing amounts of sample is shown in Figure 3.
Figure 3.
Separation of proteins from human plasma on the hydrophobic interaction support Toyopearl Phenyl 650 S (column volume 0.34 mL). 4 M NaCl in 10 mM Tris-HCl, pH 7.4 was used as Buffer A. Increasing amount of human plasma was applied. Bound proteins were eluted with 10 mM Tris-HCl, pH 7.4. SDS-PAGE of eluted proteins is shown. For other conditions – see Materials and methods and Figure 1.
3.3. Use of a three-column system for SDC in HIC mode
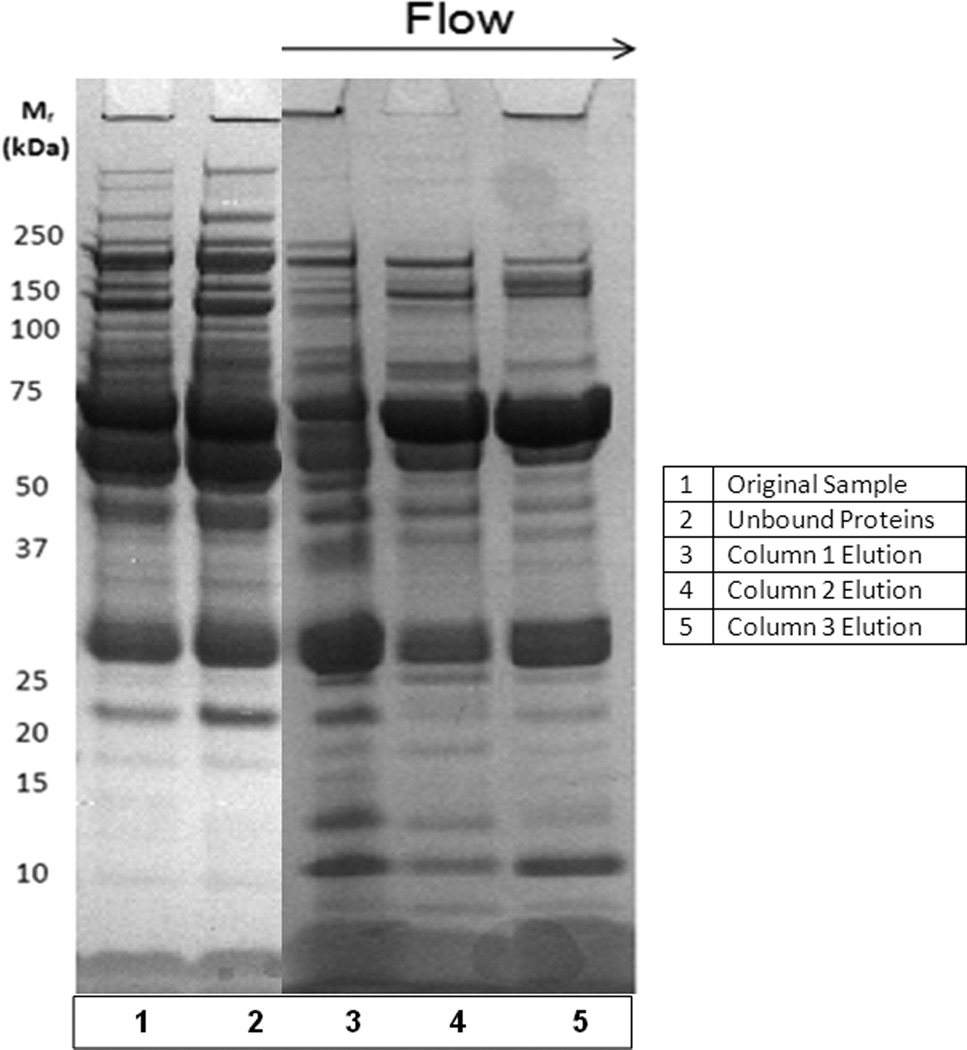
Three columns (shown in Figure 4A) were coupled in series during the sample application and columns were subsequently washed with the Buffer A. The elution was performed in parallel, after detachment of columns. As in previous experiments, Buffer B was used for elution of bound proteins (see also Materials and methods).
Figure 4.

Sample displacement chromatography of plasma proteins by use of three identical columns packed with Toyopearl Phenyl 650 S (column volume 0.34 mL each). A – all three columns were connected in series during sample loading and washing. B – for the elution step, the columns were detached and bound proteins were eluted by a step gradient of 100% Buffer B (10 mM Tris-HCl, pH 7.4). Different grades of yellow color express the different relative concentration of serum albumin, which carries yellow colored pigment from human plasma.
3.3.1. SDC in HIC mode with 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4, as a mobile phase
If 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4 was used as a mobile phase, sample displacement can also be monitored visually when severe overloading conditions are used, e.g., by application of more than 1000 mg protein/mL HIC support (see Figure 4B). As shown here, highly concentrated serum albumin (yellow because it binds some hydrophobic pigments from human plasma) has been displaced from the first column by other, more strongly binding proteins. After detachment, this protein is subsequently eluted from the second (partially enriched) and third column (highly enriched, see also Figure 5A and Tables 2A–C). The corresponding SDS-PAGE of proteins eluted from each column is shown in Figure 5A. The lists of the major proteins eluted from each column are given in Tables 2A-C. The complete list of identified proteins is attached in the Supplement (Table S 2A–C).
Figure 5.
Sample displacement chromatography of plasma proteins by use of three identical columns packed with Toyopearl Phenyl 650 S (column volume 0.34 mL each). SDS-PAGE of proteins eluted from each column after detachment (see Figure 4 and Materials and methods). A - 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4 was used as Buffer A; B - 4 M NaCl in 10 mM Tris-HCl, pH 7.4 was used during sample application and washing. For other conditions – see Figure 4.
Table 2.
| A Major plasma proteins eluted from the first column after detachment from the three-column system (see Figure 4A) identified by LC-MS/MS. 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4 was used as Buffer A. | |||
|---|---|---|---|
| Acc. No. | Protein name | Score | Uniq. pept. |
| IPI00022229.1 | Apolipoprotein A1 | 1538.56 | 34 |
| IPI00745872.2 | Human serum albumin | 1428.25 | 30 |
| IPI00783987.2 | Complement C3 | 1327.91 | 26 |
| IPI00021841.1 | Apolipoprotein A1 | 941.18 | 17 |
| IPI00022463.1 | Serrotransferrin | 793.18 | 17 |
| IPI00304273.2 | Apolipoprotein A-IV | 706.79 | 17 |
| IPI00947307.1 | Ceruloplasmin | 679.00 | 14 |
| IPI00478003.3 | Alpha-2-macroglobulin | 580.1 | 15 |
| IPI00019568.1 | Prothrombin | 549.3 | 12 |
| IPI00910625.1 | Beta-2-glycoprotein | 394.8 | 9 |
| B Major plasma proteins eluted from the second column after detachment from the three-column system (see Figure 4A) identified by LC-MS/MS. 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4 was used as Buffer A. | |||
|---|---|---|---|
| Acc. No. | Protein name | Score | Uniq. pept. |
| IPI00022229.2 | Apolipoprotein B | 4847.00 | 92 |
| IPI000222463.2 | Serotransferrin | 2696.04 | 46 |
| IPI00745872.2 | Human serum albumin | 2237.30 | 44 |
| IPI00783987.2 | Complement C3 | 1905.52 | 37 |
| IPI00478003.3 | Alpha-2-macroglobulin | 1834.15 | 35 |
| IPI00021841.1 | Apolipoprotein A1 | 1232.86 | 23 |
| IPI00017601.1 | Ceruloplasmin | 1141.65 | 22 |
| IPI00641737.1 | Haptoglobin | 1064.82 | 21 |
| IPI00022395.1 | Complement component C9 | 738.77 | 13 |
| IPI00019568.1 | Prothrombin | 636.6 | 14 |
| C Major plasma proteins eluted from the third column after detachment from the three-column system (see Figure 4A) identified by LC-MS/MS. 1.7 M (NH4)2SO4 in 10 mM Tris-HCl, pH 7.4 was used as Buffer A. | |||
|---|---|---|---|
| Acc. No. | Protein name | Score | Uniq. pept. |
| 225131003 | Apolipoprotein B | 4109.81 | 82 |
| IPI00745872.2 | Human serum albumin | 3749.71 | 67 |
| IPI00021841.1 | Apolipoprotein A-1 | 1530.40 | 27 |
| IPI00478003.3 | Alpha-2-macroglobulin | 548.2 | 9 |
| IPI00553177.1 | Alpha-1-antitrypsin | 486.1 | 9 |
| IPI00742696.2 | Vitamin D-binding protein | 431.5 | 9 |
| 619383 | Apolipoprotein D | 403.32 | 8 |
| IPI00021842.1 | Apolipoprotein E | 397.0 | 8 |
| IPI00032220.3 | Angiotensinogen | 273.58 | 4 |
| IPI00847635.1 | Alpha-1-antichymotrypsin | 259.87 | 4 |
- Clotting factor IX
3.3.2. SDC in HIC mode with 4M NaCl in 10 mM Tris-HCl, pH 7.4, as a mobile phase
The SDS-PAGE profiles of eluted proteins after binding to the Toyopearl Phenyl 650 S column under overloading conditions are shown in Figure 5B. Again, three columns were coupled in series, and 4M NaCl in 10 mM Tris-HCl, pH 7.4, was used as mobile phase (Buffer A, see Materials and methods). After loading of 3500 mg protein/mL support and washing with Buffer A, columns were detached and separately eluted with Buffer B. The predominant proteins eluted from each column are listed in Tables 3A–C. The complete list of eluted proteins is attached as a Supplement (Table S 3A–C).
Table 3.
| A Major plasma proteins eluted from the first column after detachment from the three-column system (see Figure 4A) identified by LC-MS/MS. 4M NaCl in 10 mM Tris-HCl, pH 7.4 was used as Buffer A. | |||
|---|---|---|---|
| Acc. No. | Protein name | Score | Uniq. pept. |
| IPI00783987.2 | Complement C3 | 2825.45 | 54 |
| 62739186 | Complement factor H | 2424.82 | 46 |
| IPI00019580.1 | Plasminogen | 1927.69 | 42 |
| IPI00478003.3 | Alpha-2-macroglobulin | 1806.31 | 37 |
| IPI00298497.3 | Fibrinogen | 1677.13 | 32 |
| IPI00022229.2 | Apolipoprotein B | 1661.45 | 36 |
| IPI00745872.2 | Human serum albumin | 1546.10 | 29 |
| IPI00892547.1 | Complement component 4A | 1444.19 | 26 |
| IPI00797833.3 | LMW Kininogen=1 | 1365.61 | 24 |
| IPI00867588.1 | Fibronectin | 1211.96 | 23 |
| B Major plasma proteins eluted from the second column after detachment from the three-column system (see Figure 4A) identified by LC-MS/MS. 4 M NaCl in 10 mM Tris-HCl, pH 7.4 was used as Buffer A. | |||
|---|---|---|---|
| Acc. No. | Protein name | Score | Uniq. pept. |
| IPI00783987.2 | Complement C3 | 3951.21 | 73 |
| IPI00022229.2 | Apolipoprotein B | 2765.35 | 59 |
| IPI00892547.1 | Complement component 4A | 2079.37 | 38 |
| IPI00478003.3 | Alpha-2-macroglobulin | 2061.22 | 40 |
| IPI00029739.5 | Complement factor H | 1955.98 | 37 |
| IPI00797833.3 | LMW Kininogen=1 | 1744.34 | 32 |
| IPI00745872.2 | Human serum albumin | 1463.63 | 30 |
| IPI00298497.3 | Fibrinogen | 1369.93 | 25 |
| IPI00019580.1 | Plasminogen | 1234.44 | 26 |
| IPI00021841.1 | Apolipoprotein A-1 | 1177.26 | 21 |
| C Major plasma proteins eluted from the third column after detachment from the three-column system (see Figure 4A) identified by LC-MS/MS. 4M NaCl in 10 mM Tris-HCl, pH 7.4 was used as Buffer A. | |||
|---|---|---|---|
| Acc. No. | Protein name | Score | Uniq. pept. |
| I225131003 | Apolipoprotein B | 4801.35 | 97 |
| IPI00783987.2 | Complement C3 | 4688.66 | 89 |
| IPI00892547.1 | Complement component 4A | 2357.27 | 46 |
| IPI00478003.3 | Alpha-2-macroglobulin | 2010.13 | 39 |
| IPI00215894.1 | LMW Kininogen-1 | 1777.46 | 33 |
| IPI00029739.1 | Complement factor H | 1713.16 | 34 |
| IPI00745872.2 | Human serum albumin | 1571.07 | 32 |
| IPI00032291.2 | Complement C5 | 1380.07 | 30 |
| IPI00021841.1 | Apolipoprotein A-1 | 1310.37 | 25 |
| IPI00654888.4 | Plasma kallikrein | 1174.02 | 21 |
- Clotting factor XI (highly enriched)
- Hyaluronan-binding protein 2 (highly enriched)
- lalp heavy chain 4
- Clotting factor V
- Clotting factor XII
- Protein S
- Clotting factor V
- Clotting factor XII
- lalp heavy chain 4
- Prothrombin
- Clotting factor V
- Clotting factor XI
- Clotting factor IX
- Protein S
- Clotting factor XII
4. Discussion
In early experiments, the Hodges group showed that SDC can be used for very effective purification of synthetic peptides in reversed-phase mode [1, 2]. Veeraragavan et al. [3] extended this purification strategy to the separation of proteins in ion-exchange mode. However, at that time, only standard proteins were used. The real potential of sample displacement chromatography was recognized more than ten years later, when Husband et al. [4] used a combination of a hydrophobic and a hydrophilic column for purification of peptide mixtures. In this system, hydrophilic peptides as impurities were bound to the first, hydrophilic column. They displaced the hydrophobic peptides that then bind to the second, hydrophobic column, and were subsequently eluted by use of a solvent gradient. A similar strategy was subsequently used for the purification of rabbit skeletal troponin complex, a relatively simple protein mixture containing four single polypeptides [4]. Hodges’ group usually combined SDC with gradient elution chromatography. Consequently, the advantageous potential of displacement chromatography for yielding relatively high concentration product was not completely explored [5, 25]. In order to optimize the SDC separation, Agner suggested the use of a multicolumn system in series followed by parallel elution from individual columns in order to optimize the separation [25, 26]. After moderate success in the purification of Atlantic salmon thrombin for nutritional use by employing “conventional” heparin affinity chromatography [27], Manseth et al. [6] significantly improved the production process and product purity by use of SDC on the same matrix, namely Heparin Sepharose. A multicolumn system was successfully used for the first time for the separation of a physiologically active protein, thrombin from the plasma of Atlantic salmon [6].
We used monolithic, polyacrylamide-based convective interaction media (CIM) ion-exchange supports for SDC fractionation of human plasma [7]. In those experiments, displacement of two highly abundant proteins, human serum albumin (HSA) in anion-exchange mode and IgG (and other weakly binding proteins) in cation-exchange mode, by proteins that strongly bind to the support was achieved. It could be also demonstrated that SDC in ion-exchange mode can be successfully used for concentration of strongly binding low-abundance proteins when this very complex biological mixture was applied [7].
Body fluids, especially human blood plasma and serum, are the most important sources for discovery of disease biomarkers, and are therefore the topic of intensive investigations in order to find new biomarker candidates [14]. There are a plethora of papers about plasma fractionation as a sample preparation step for further LC-MS/MS investigations [28], but there is still a need for the development of new methods in order to enrich and detect further low abundance biomarker candidates [14, 28]. On the other hand, there is also a need for new methods in plasma fractionation, especially for the isolation of new therapeutic proteins, and in order to optimally use this important and valuable starting material [14, 28, 29].
Neither hydrophobic interaction chromatography nor displacement chromatography have been routinely applied to the separation of proteins from human plasma. Elution-modified displacement chromatography as an effective method for reversed-phase separation of tryptic peptides as sample preparation for ESI MS was developed almost ten years ago [30, 31], but this method did not find widespread acceptance. Ahrends et al. [32] recently compared displacement chromatography and gradient elution chromatography as the cation-exchange step for sample preparation for ESI-MS/MS. They demonstrated that displacement chromatography is well suited for this application. The use of displacement chromatography as an alternative for gradient elution, SDS-PAGE, or 2D-electrophoresis for separation of protein mixtures before tryptic digestion in proteomics is even more scarce [14].
The SDC method presented here was used for fractionation of plasma protein in hydrophobic interaction mode. This method offers an additional, very effective alternative for separation of this complex biological mixture and both isolation and detection of low abundant proteins. As shown here, some high abundance proteins, such as human serum albumin and IgG, can be eliminated by precipitation in the presence of high salt concentrations during sample preparation (see Tables 1A and B), or by sample displacement during chromatographic separation (see Figures 2 and 3). Low abundance plasma proteins such as alkaline phosphatase, the vitamin E-binding protein afamin, plasminogen and clotting factors and inhibitors could be highly enriched by use of this strategy (see Tables 2A–C and 3A–C).
The above-presented multicolumn system enables the elution by only one step gradient and it is sufficient to achieve a highly efficient separation (see Figures 4B and 5A and B and Tables 2A–C and 3A–C) and additional removal of high-abundance proteins and enrichment of proteins present in trace amounts in the sample. The multicolumn system can be applied for both large and small-scale separations.
The experiments presented in both our recent [7] and in the present paper demonstrate only the proof of principle, and further investigations of this kind of chromatography are necessary.
Finally, the use of different salts as mobile phases in HIC enables different selectivity of the HIC column, and can be utilized for modulation of the separation and isolation and detection of additional low-abundance proteins.
5. Conclusions
-
-
Sample displacement chromatography (SDC) in hydrophobic interaction mode is an additional, very effective method for separation of complex biological mixtures.
-
-
SDC in hydrophobic interaction mode can be applied to the separation of human plasma and the concentration of low abundance proteins.
-
-
Use of different salts (and eventually salt mixtures) yields in different selectivity of the HIC column.
-
-
By use of two or more columns coupled in series during sample application, and subsequent elution of detached columns in parallel, additional separation of bound proteins can be achieved.
Supplementary Material
Figure S1
SDS-PAGE of proteins after precipitation with Buffer A.
A – plasma proteins were precipitated with 1.7 M (NH4)2SO4.;
B – plasma proteins were precipitated with 4M NaCl.
Acknowledgements
This work was supported by National Institutes of Health (NIH), Centers for Biochemical Research Excellence (COBRE), grant No. P20RR017695 and NIH grant No. 1S10RR025623-01 (Dj. Josic), the National Science Foundation (NSF) Experimental Program to Stimulate Competitive Research (EPSCoR), grant No. 1004057 and NIH grant No. 1S10RR020923 (J. Clifton), and the National Science Foundation of Republic of Croatia (Dj. Josic, M. Srajer Gajdosikand D. Gaso-Sokac). M. Srajer Gajdosik was also supported by Fulbright Scholarship.
References
- 1.Hodges RS, Burke TWL, Mant CT. J. Chromatogr. A. 1988;444:349–362. doi: 10.1016/s0021-9673(01)94036-1. [DOI] [PubMed] [Google Scholar]
- 2.Lorne Burke TWL, Mant CT, Hodges RS. J. Liq. Chromatogr. 1988;11:1229–1247. [Google Scholar]
- 3.Veeraragavan K, Bernier A, Braendli E. J. Chromatogr. A. 1991;541:207–220. [Google Scholar]
- 4.Husband DL, Mant CT, Hodges RS. J. Chromatogr. A. 2000;893:81–94. doi: 10.1016/s0021-9673(00)00751-2. [DOI] [PubMed] [Google Scholar]
- 5.Mant CT, Hodges RS. J. Chromatogr. A. 2002;972:101–114. doi: 10.1016/s0021-9673(02)01079-8. [DOI] [PubMed] [Google Scholar]
- 6.Manseth E, Skjervold PO, Flengsrud R. J. Biochem. Biophys. Methods. 2004;60:39–47. doi: 10.1016/j.jbbm.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 7.Brgles M, Clifton J, Walsh R, Huang F, Rucevic M, Cao L, Hixson D, Müller E, Josic Dj. J. Chromatogr. A 1218. 2011;1218:2389–2395. doi: 10.1016/j.chroma.2010.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Tetrault J, Ley A. J. Chromatogr. A. 2008;1177:272–281. doi: 10.1016/j.chroma.2007.07.083. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez-Rey M, Lang DA. Biotechnol. Bioeng. 2011;108:1494–1508. doi: 10.1002/bit.23155. [DOI] [PubMed] [Google Scholar]
- 10.Zou W, Bi J, Zhao L, Wang Y, Li Y, Huamg Y, Ma G, Su Z. Process Biochem. 2007;42:751–756. [Google Scholar]
- 11.Passarinha LA, Bonifacio MJ, Soares-da-Silva P, Queiroz JA. J. Chromatogr. A 1177. 2008;1177:287–296. doi: 10.1016/j.chroma.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Aizawa P, Winge S, Karlsson G. Thromb. Res. 2008;122:560–567. doi: 10.1016/j.thromres.2007.12.027. [DOI] [PubMed] [Google Scholar]
- 13.Karlsson G. Protein Expr. Purif. 2003;27:171–174. doi: 10.1016/s1046-5928(02)00596-x. [DOI] [PubMed] [Google Scholar]
- 14.Surinova S, Schiess R, Hüttelhain R, Carciello F, Wollscheid B, Aebersold R. J. Proteome Res. 2011;10:5–16. doi: 10.1021/pr1008515. [DOI] [PubMed] [Google Scholar]
- 15.Josic Dj, Clifton JG. J. Chromatogr. A. 2007;1144:2–13. doi: 10.1016/j.chroma.2006.11.082. [DOI] [PubMed] [Google Scholar]
- 16.Tiselius A. Ark. Kemi. Mineral. Geol. 1943;16A:1–38. [Google Scholar]
- 17.Buchanan DC. J. Biol. Chem. 1957;229:211–229. [PubMed] [Google Scholar]
- 18.Horváth Cs, Nahum A, Frenz J. J. Chromatogr. 1981;218:365–393. [Google Scholar]
- 19.Kundu A, Barnthouse KA, Cramer SM. Biotechnol. Bioeng. 1997;56:119–129. doi: 10.1002/(SICI)1097-0290(19971020)56:2<119::AID-BIT1>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 20.Shukla AA, Sunasara KM, Rupp RP, Cramer SM. Biotechnol. Bioeng. 68. 2000;68:672–680. doi: 10.1002/(sici)1097-0290(20000620)68:6<672::aid-bit11>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 21.Josic Dj, Hoffer L, Buchacher A. J. Chromatogr. B. 2003;790:183–197. doi: 10.1016/s1570-0232(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 22.Josic Dj, Brown MK, Huang F, Lim Y-P, Rucevic M, Clifton JG, Hixson D. Proteomics. 2006;6:2874–2885. doi: 10.1002/pmic.200500563. [DOI] [PubMed] [Google Scholar]
- 23.Lawson E, Clifton JG, Huang F, Li X, Hixson D, Josic Dj. Electrophoresis. 2006;27:2747–2758. doi: 10.1002/elps.200600059. [DOI] [PubMed] [Google Scholar]
- 24.Elias JE, Gigy SP. Nature Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 25.Agner E. 6,576,134. U.S. Patent. 2003
- 26.Agner E. 6,245,238. U.S. Patent. 2001
- 27.Manseth EP, Skjervpld PO, Fjœra SO, Brosstad FR, Ødegaard R, Flengsrud R. J. Food Sci. 2003;68:1648–1652. [Google Scholar]
- 28.Cottingham K. J. Proteome Res. 2006;5:1298–1298. doi: 10.1021/pr062733+. [DOI] [PubMed] [Google Scholar]
- 29.Bandow E. Proteomics. 2010;10:1416–1425. doi: 10.1002/pmic.200900431. [DOI] [PubMed] [Google Scholar]
- 30.Gaso-Sokac D, Kovac S, Clifton J, Josic Dj. Electrophoresis. 2011;32:1104–1117. doi: 10.1002/elps.201000641. [DOI] [PubMed] [Google Scholar]
- 31.Over J. Presented at Plasma Protein Biotechnology 2011, Paphos, Cyprus, L 506; http://www.bo-conf.com/ppb11/ [Google Scholar]
- 32.Ahrends R, Lichtner B, Bertsch A, Kohlbacher O, Hildebrand D, Trusch M, Schlüter H. J. Chromatogr. A. 2010;1217:3321–3329. doi: 10.1016/j.chroma.2009.10.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
SDS-PAGE of proteins after precipitation with Buffer A.
A – plasma proteins were precipitated with 1.7 M (NH4)2SO4.;
B – plasma proteins were precipitated with 4M NaCl.