Synopsis
Dysfunctional bioenergetics has emerged as a key feature in many chronic pathologies such as diabetes and cardiovascular disease. This has led to the mitochondrial paradigm in which it has been proposed that mitochondrial DNA (mtDNA) sequence variation contributes to disease susceptibility. In this study we present a novel animal model of mtDNA polymorphisms, the mitochondrial nuclear exchange mouse (MNX), in which the mtDNA from C3H/HeN mouse has been inserted onto the C57/BL6 nuclear background and vice versa to test this concept. Our data show a major contribution of the C57/BL6 mtDNA to the susceptibility to the pathological stress of cardiac volume overload which is independent of the nuclear background. Mitochondria harboring the C57/BL6J mtDNA generate more reactive oxygen species (ROS) and have a higher mitochondrial membrane potential relative to those having the C3H/HeN mtDNA, independent of nuclear background. We propose this is the primary mechanism associated with increased bioenergetic dysfunction in response to volume overload. In summary, these studies support the “mitochondrial paradigm” for the development of disease susceptibility, and show that the mtDNA modulates, cellular bioenergetics, mitochondrial reactive oxygen species generation and susceptibility to cardiac stress.
Keywords: mitochondria, disease susceptibility, mtDNA, bioenergetics, oxidative stress
Introduction
While significant progress has been made in understanding the pathobiology underlying heart disease development and progression, it is still not clear why some individuals with identical risk factors develop disease while others do not. It is widely thought that disease development is influenced by a combination of genetic, environmental, and behavioral factors that influence an individual’s biological response to disease risk factors. However, studies have not yet provided a clear genetic or mechanistic basis for “common” disease susceptibility. In this regard, the potential contribution of mitochondrial genetics and function is a consideration that has largely been lacking from prior susceptibility analyses. This “mitochondrial paradigm” for disease has been difficult to critically assess due the lack of animal models in which it can be tested. In this study we describe a novel approach to developing such models and illustrate its application in a model of bioenergetic dysfunction in the heart.
Mitochondria are ancient bacterial symbionts with their own DNA, RNA, and protein synthesis systems [1]. Each cell contains hundreds to thousands of mitochondria and every mitochondrion contains 5 – 10 copies of maternally inherited mtDNA. The mammalian mtDNA encodes two rRNAs (12S and 16S) and 22 tRNAs that are necessary for the proper translation of 13 mtDNA genes that encode polypeptides essential for electron transport and oxidative phosphorylation (OXPHOS). While the nucleus encodes all of the remaining mitochondrial proteins for the organelle, the mtDNA retains key structural subunits required for the catalytic activity of four of the five OXPHOS enzyme complexes (I, III, IV and V). It is clear that mutations in these mtDNA subunits alter the basic features of mitochondrial metabolism as has been shown in a broad range of mitochondrial diseases in which substantial loss of respiratory function occurs [2]. Recently, the contribution of functional but distinct mitochondrial DNA polymorphisms present in different human populations, to the susceptibility disease has been proposed [2]. While mtDNA sequence variation has been investigated in certain animal models of disease [3] few experimental models are available to test this hypothesis which is supported by human epidemiological data.
The mitochondrial paradigm has been proposed as a mechanism of how prehistoric humans were able to successfully establish populations out of Africa through exploiting mtDNA mutations which subtly alter mitochondrial bioenergetics. The key concept is that mtDNA haplotypes that promote heat generation conveyed greater reproductive and survival potential in our prehistoric ancestors even at some cost to energetic efficiency [4]. Today, in the presence of excessive dietary intake and sedentary lifestyle it is hypothesized that these mtDNA mutations will be linked to differences in bioenergetics and oxidant production, and thus, influence susceptibility to age-related diseases associated with increased oxidative stress or decreased energetic capacity [2;5]. Several studies have shown that specific mtDNA polymorphisms and haplotypes are associated with increased risk for diseases believed to have an environmental component. Since these studies have been epidemiologic the distinct and interactive contributions of the mtDNA and nuclear backgrounds on the molecular mechanisms of bioenergetic function in human populations has remained uncertain [6–10]. One of the central ideas from this body of work is that mtDNA polymorphisms code for respiratory chains with different bioenergetic characteristics and generation of reactive oxygen species.
To test this hypothesis we developed Mitochondrial – Nuclear eXchange (MNX) mice in which nuclear and mitochondrial genomes have been exchanged from different M. musculus strains. This approach is distinct from conplastic [11] and xenomitochondrial [12] animals in that MNX mice are generated directly with 100% of the desired nuclear and mtDNA complements from respective donor strains through nuclear transfer and thus do not require repeated back-crossings (as do conplastics) to generate animals having the desired genotype. Furthermore, MNX mice allow direct, unambiguous assessment of mtDNA contributions to disease since there is no complexity introduced by potential nuclear cross-over and combinational effects in the filial generations associated with standard backcrossing methods used to generate conplastic mice. By using C57BL/6 and C3H/HeN mice which have been previously shown to have distinct susceptibilities to dietary induced atherosclerosis and insulin resistance [13–18], we generated MNX mice having either the nucleus of a C57BL/6 or C3H/HeN mouse and the mtDNA of either the C3H/HeN or C57BL/6 animal, respectively. Mitochondrial bioenergetics, cellular oxidant production, and susceptibility to a model of cardiac volume overload of aortocaval fistula (ACF), a model which recapitulates the problem of valvular regurgitation [19] associated with increased oxidative stress [20;21] were examined in these mice compared to control animals (wild-type C57BL/6 and C3H/HeN). We show that mtDNA genetic background significantly modulated bioenergetics, oxygen utilization, and oxidant production in cardiac tissue. In addition, mtDNA background was a major determinant of ACF induced mitochondrial damage in the left ventricle independent of nuclear background – the typically sensitive C57BL/6 mouse was rescued from ACF induced mitochondrial damage by replacing the C57BL/6 mtDNA with the C3H/HeN mitochondrial genome, whereas the previously ACF resistant C3H/HeN mouse became sensitive when the C3H/HeN mtDNA was replaced with the C57BL/6 mtDNA in the MNX mouse. These findings provide direct evidence and support the concept that mitochondrial genetic background significantly influences cellular bioenergetics and disease susceptibility.
Experimental Procedures
Generation of Mitochondrial – Nuclear eXchange (MNX) Mice
C57BL/6J and C3H/HeN mice were purchased from Jackson Laboratories (Bar Harbor, ME) and Harlan Laboratories (Indianapolis, IN), respectively. Female donor mice (3–4 weeks of age) were super-ovulated hormonally and paired with males of the same nuclear genotype. Their cumulus masses were harvested as previously described to produce single-celled embryos [22]. Pronuclear embryos were placed in M2 medium containing cytochalasin B (5 µg/mL) and colcemid (0.1µg/mL) at 37°C for five minutes, and remained in a microinjection drop of the same medium at room temperature to prevent lysis during manipulation. A micropipette similar in size and shape to a beveled pipet used for embryonic stem cell injection injections was used to remove both pronuclei of each embryo. First, the pipet was positioned so that it put slight pressure on the zona pellucida. Then a high-intensity piezo pulse was applied until the zona was ruptured. The pipet was slowly advanced to each pronucleus, and with gentle suction applied to the needle, the two pronuclei were aspirated and removed as a single unit (karyoplast). The isolated pronuclei from one strain were then implanted into the enucleated embryo of the other strain. Ten centimeter tissue culture dishes were loaded with microdrops of M2 medium and covered in mineral oil and each embryo was placed into its own drop of media. An electrode was placed into the drop positioning the embryo between the two poles. A single 90 V pulse was applied to each re-constructed zygote and all zygotes were cultured overnight [22]. Two-cell embryos were transferred to the ostia of oviducts of 0.5 day CD-1 pseudopregnant mice to term. Nuclear genotype and mtDNA haplotype were verified in offspring via nuclear SNP and complete mtDNA genome sequence analysis.
MNX mouse colonies were generated by crossing female MNX mice (mtDNA haplotype confirmed by sequencing) with males of the matching nuclear background. MtDNA haplotypes were confirmed in F1 progeny by sequencing and subsequent generations by restriction fragment length polymorphism (RFLP) analysis of PCR products screening for diagnostic mtDNA SNP’s that delineate between the C57 and C3H mtDNAs (Supplemental Figure S1). C57BL/6 and C3H/HeN mtDNA were distinguished based on an A to G mutation at bp9348 that results in the loss of an AspI site in the C3H mtDNA but not the C57 mtDNA. Animals used in these studies were F7 or higher generation.
Euthanasia
Mice were paralyzed by interperitoneal injection of Ketastat/Xylazine (60mg/kg Ketastat/10mg/kg Xylazine) as previously described [23].
Cell-free Mitochondrial Preparations
Mitochondria were isolated following a modification of the method of Rasmussen et al. using a custom-built mitochondria isolation station/homogenizer (CL Krumdieck, DR Moellering) as previously described [24].
High Resolution Respirometry
Mitochondrial respiration assays using freshly isolated heart mitochondria (100 µg) were performed by measuring oxygen consumption in a two-channel respirometer (Oroboros Oxygraph with DatLab software; Oroboros, Innsbruck, Austria) as previously described [25]. Respiration rates were measured with 15 mM glutamate/2 mM malate as a substrate followed by addition of inhibitors. Determination of state 2, 3, and induced state 4 respiration rates were made in the presence of excess substrate alone (state 2), after the addition of 100 µM ADP (state 3), and after addition of 2 µg/ml oligomycin (induced state 4), an inhibitor of the ATP synthase. Respiration through complex IV only was performed in the presence of ascorbate and tetramethylphenylenediamine (TMPD) as previously described [26].
ATP Quantification
Steady – state ATP levels were quantified in isolated cardiac mitochondria (100 µg) under state 3 respiration using a luciferase based kit (Molecular Probes, Eugene, OR) following manufacturer’s instructions. Briefly, ATP levels were determined in isolated mitochondria (heart) after a 1 minute incubation in the absence or presence of glutamate/malate and ADP, followed by addition of oligomycin. Negative controls including (i) mitochondria in the presence of oligomycin prior to substrate and ADP addition, (ii) mitochondria with glutamate/malate only, (iii) and ADP only (no mitochondria) were used. ATP levels produced by mitochondria following ADP addition were calculated using standard curves of known ATP concentrations.
NADH dehydrogenase, Succinate dehydrogenase, and Cytochrome c Oxidase Activities
Enzymatic activites for complexes I, II and IV were measured as previously described [27]. Isolated heart mitochondria were used for all assays. For complex I, rotenone was added as negative control and also subsequently to account for any rotenone insensitive activity. The complex II assay measured the reduction of 2,6-cichlorophenolindophenol (DCPIP) coupled to complex II catalyzed reduction of decylubiquinone. Complex IV activity was measured by following the oxidation of reduced cytochrome c.
Blue Native Proteomics
Composition and quantification of mitochondrial electron transport chain proteins were determined in isolated cardiac mitochondria as previously described [28].
Cardiomyocyte Isolation
Primary adult cardiac myocytes were isolated from sham and ACF mice from each of the different strains, as described previously [29]. Briefly, hearts were perfused with perfusion buffer (120 mM NaCl, 15 mM KCl, 0.5 mM KH2PO45 M NaHCO310 mM HEPES, and 5 mM glucose, pH 7.0) for 5 min and digested with perfusion buffer containing 2% collagenase II (Invitrogen, Carlsbad, CA) for 30 min at 37°C. The right ventricle, atria and apex were removed before the perfused-heart was minced. The digestion was filtered and washed, and the cells were pelleted. Only samples with purity > 95% and viability (rod-shaped) > 80% were used.
Cellular Bioenergetics
To determine the effects of mitochondrial haplotype on cellular bioenergetics, an XF24 analyzer (Seahorse Bioscience, Billerica, MA) was used to measure O2 consumption by live cells in culture [30]. This instrument allows for high throughput, real-time, non-invasive measurements of O2 consumption by using fluorescent probes adhered to disposable assay cartridges. These measurements can then be used to correlate O2 consumption rate (OCR) to mitochondrial function. Primary adult cardiac myocytes were attached to specialized V28 plates (Seahorse Bioscience, Billerica, MA) coated with laminin at 7,500 cells/well. The cells were then allowed to attach for 2 hr, after which time the culture media was changed to MEM supplemented with 1% FBS (Atlanta Biologicals, Lawrenceville, GA) and 4 mM l-glutamine (Invitrogen) for the XF24 assays.
Various parameters of mitochondrial function were measured utilizing the ability of the XF24 to inject compounds into the wells through an assay as described previously [30]. Briefly, basal O2 consumption of the primary adult cardiac myocytes was determined by measuring the OCR of the cells over time without any treatment. Next, FCCP (1 µM) was injected to stimulate the maximal OCR of the cells. This maximal OCR also allows for the calculation of the reserve capacity, which is the difference between the maximal and basal OCR, and represents the cells spare mitochondrial function that is available to be utilized during an increased work load or stress. Finally, antimycin A (10 µM) and rotenone (1 µM) were injected simultaneously to completely inhibit the mitochondrial electron transport chain, thus yielding the non-mitochondrial OCR of the myocytes.
Mitochondrial membrane potential (Δψ) and ROS generation in cardiomyocytes
The mitochondrial inner membrane electrochemical Δψ was assessed using epifluorescence microscopy with 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolcarbocyanine iodide (JC-1). Briefly, adult cardiomyocytes were seeded in a four-well chambered coverglass. Live cells were then incubated in culture medium supplemented with JC-1 (5 µM) for 30 min or DCF-DA (20 µM) for 20 min at 37°C. Next, cells were washed with serum complete medium, and images were acquired from three or more randomly chosen fields using an inverted epifluorescence microscope with suitable filter setup (model IX70; Olympus). The levels of JC-1 (red and green) and DCFH-DA (green for ROS generation) fluorescence were measured. The authors appreciate the considerations related to the use of DCFH-DA for the detection of ROS [31], and under the current circumstances (changing only the mtDNA), we believe its use to be reasonable. Quantitative data were expressed as total red channel pixel intensity per number of cell. Results are mean ± SEM from 35–50 cardiomyocytes.
Aortocaval Fistula Surgery
Control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) male mice (18–24 weeks) were used for volume overload studies. An aortocaval fistula (ACF) was surgically created as described previously [20]. Briefly, ACF and sham operations were performed under sterile conditions on all four strains of mice at 18 to 24 wk of age. With mice under isoflurane anesthesia, the abdominal cavity was opened via a midline incision, and a fistula was created between the abdominal aorta and the inferior vena cava. Induced sham and ACF mice were sacrificed 3 days after surgery, at which point the hearts were excised and used for the isolation of cardiomyocytes for bioenergetic measurements, or for LV sections for transmission electron microscopy. All groups had a decrease in mean arterial pressure (MAP) indicating successful ACF.
Transmission Electron Microscopy
Heart LV tissue was fixed in Karnovsky’s solution (2% paraformaldehyde/2/5% glutaraldehyde in 0.1 M phosphate buffer), and treated as previously described for TEM [21]. For analysis, n = 2 animals were used for the controls (C57n:C57mt and C3Hn:C3Hmt) and C57n:C3Hmt MNX, per group (sham and ACF); for the C3Hn:C57mt MNX animals 3 ACF and 4 shams were used. A minimum of 8 different fields were analyzed per mouse. For the calculation of average mitochondrial number per field, samples were blinded; a total of 78 fields were analyzed with a minimum of 8 fields per group.
Statistical Analyses
Results are expressed as means ± SE. ANOVA was used to test the null hypothesis that all samples were drawn from a single population and if the significant differences existed (P < 0.05), a Student-Newman-Keuls test was used for group comparisons. All analyses were carried out using SigmaStat statistical software.
Results
Generation of Mitochondrial – Nuclear eXchange (MNX) mice
C57BL/6 and C3H/HeN mouse strains were used to produce Mitochondrial – Nuclear eXchange (MNX) mice because these strains show significant differences in their susceptibility to dietary induced atherosclerosis, insulin resistance and obesity, and have distinct mtDNA sequences [13–18;32]. MNX mice were generated by enucleating fertilized oocytes from one strain and reconstituting with the nucleus from the other (Supplemental Figure S1). MNX embryos were then implanted into a surrogate mother and resulting offspring had the nuclear genome of one strain and the mitochondrial genome of the other; MNX nomenclature is indicated by strainnuclear:strainmtDNA (strainn:strainmt), e.g. mice with C3H nuclear DNA and C57 mtDNA are indicated by C3Hn:C57mt (Supplemental Figure S1). Genetic identity was initially confirmed via SNP analysis of a panel of 38 distinguishing nuclear SNPs (Supplemental Table S1) and mtDNA sequence analysis of F0 founders and F1 offspring reveal that C57 and C3H mouse mitochondrial haplotypes distinct at three locations in terms of missense and tRNA mutations, consistent with previous reports [32]. MtDNA haplogroup was determined in subsequent generations of MNX mice from tail clips (at weaning) and heart tissue (subsequent to sacrifice) by restriction fragment length polymorphism analysis using the restriction enzyme AspI which cleaves a 385 base pair (bp) PCR product into 274 bp + 111 bp product for the C57 mtDNA whereas the C3H mtDNA remains uncut (Supplemental Figure S1). Similarly, BclI cleaves a 204 bp PCR product into 166 + 38 bp products for the C3H mtDNA while the C57 mtDNA is not cleaved (Supplemental Figure S1). All MNX mice were healthy and fertile with no overt physical or behavioral phenotypes.
Mitochondrial Bioenergetic Assessment
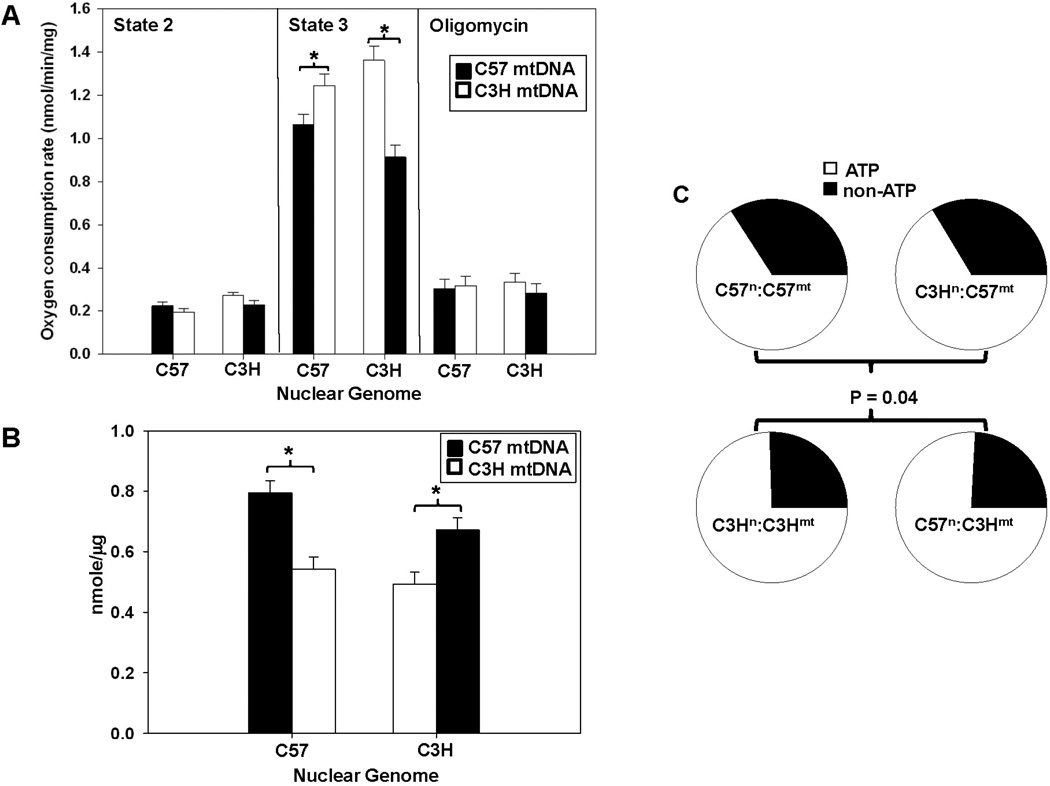
Cardiac mitochondrial oxygen consumption experiments were performed with control and MNX mice. No differences were observed in the presence of the complex I substrates glutamate and malate alone (state 2 respiration). State 3 respiration rates were significantly different (P < 0.05) between mitochondria isolated from control C57 and C3H mice and moreover, these rates were significantly changed by mtDNA background, as shown in the MNX animals (Figure 1A). No significant differences were observed between groups in the presence of oligomycin, an inhibitor of the ATP synthase (Figure 1A). Consequently, under conditions of ATP generation (State 3 respiration), the mtDNA significantly influenced mitochondrial oxygen consumption. Next, ATP generation was measured in heart mitochondrial isolates under state 3 conditions. Figure 1B shows that mitochondria harboring the C57 mtDNA generated more ATP compared to those having the C3H mtDNA, irrespective of the nuclear background. These data suggest that the relative proportion of oxygen utilized for generating ATP (State 3) versus non-ATP oxygen consumption (oligomycin resistant) in the mitochondrion was dominated by mitochondrial genetic background. This is illustrated in Figure 1C which shows that mitochondria with the C57 mtDNA (C57n:C57mt and C3Hn:C57mt) utilize a significantly (P < 0.05) lower proportion of oxygen for ATP production compared to those with C3H mtDNA (C3Hn:C3Hmt and C57n:C3Hmt).
Figure 1. Mitochondrial function and utilization segregates with mitochondrial haplotype.
Respiration and ATP levels were determined in mitochondria isolated from control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) mice (N ≥ 5 per group). For the bar graphs, the nuclear genome for each group is indicated on the X-axis, whereas the filled or open bars indicate the presence of either the C57 mtDNA or C3H mtDNA, respectively. (A) Respiration under state 2 (glutamate/malate), state 3 (glutamate/malate + ADP), and induced state 4 (glutamate/malate + ADP, followed by oligomycin addition). An asterisk (*) indicates a significant difference (p < 0.05) exists compared to nuclear genome matched control. (B) ATP levels determined under state 3 respiration conditions. An asterisk (*) indicates a significant difference (p < 0.05) exists compared to nuclear genome matched control. (C) Pie charts illustrating oxygen utilization profiles (percent oxygen consumption for ATP or non-ATP purposes) for controls (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) mice determined in heart mitochondrial isolates. Percent oxygen utilization for ATP production was determined by: ATP = (State 3 – Oligomycin)/State 3, whereas the proportion of oxygen utilized for non-ATP purposes was calculated by non-ATP = Oligomycin/State 3. The pie charts show that overall oxygen utilization was significantly influenced by the mtDNA (P = 0.04).
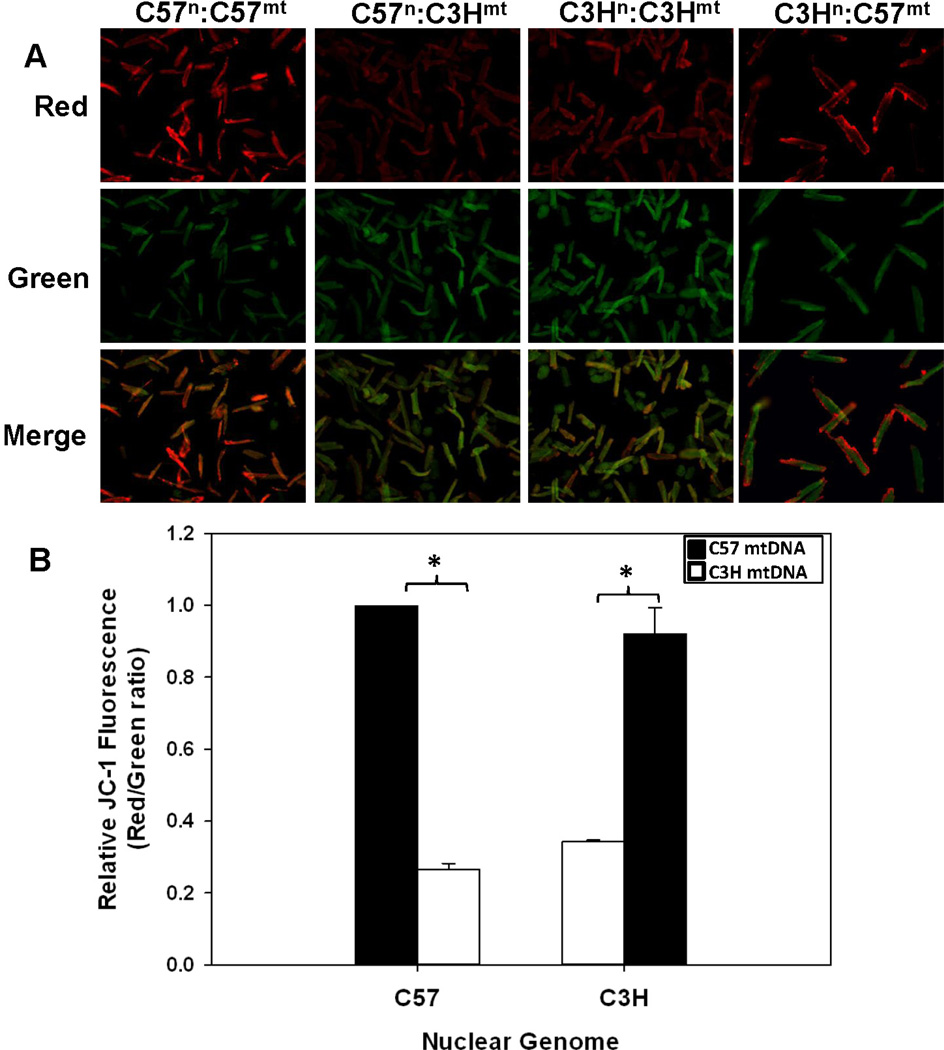
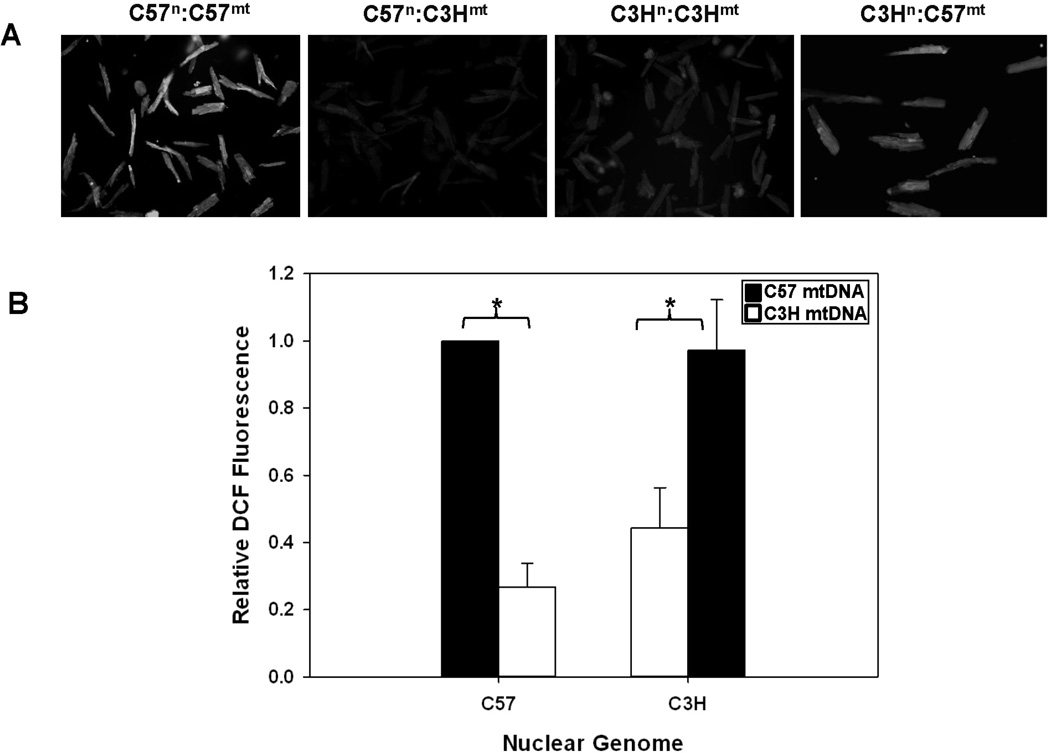
To determine the impact of mtDNA background on mitochondrial membrane potential and cellular ROS production, JC-1 and DCF-DA fluorescence was measured in isolated cardiomyocytes (Figures 2 and 3). Cardiomyocytes sharing the same mtDNA background had more similar measures of both mitochondrial membrane potential (Figure 2A and B) and cellular ROS production (Figure 3A and B), independent of nuclear background; cells harboring the C3H mtDNA (C3Hn:C3Hmt and C57n:C3Hmt) had the lowest membrane potentials and cellular ROS levels compared to those sharing the C57 mtDNA (C57n:C57mt and C3Hn:C57mt) which had significantly (P < 0.05) higher membrane potentials and cellular ROS (Figures 2 and 3). These findings strongly suggest that mtDNA background significantly influences cellular bioenergetics and ROS formation.
Figure 2. Mitochondrial membrane potential is influenced by the mtDNA in cardiomyocytes.
A. Mitochondrial membrane potential was determined using the JC-1 fluorescence method (representative images); cardiomyocytes isolated from control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) mouse hearts (emission-green 529 nm/emission-red 590 nm) B) Bar graph illustrating the quantification of fluorescence relative to the C57n:C57mt controls. The nuclear genome for each group is indicated on the X-axis, whereas the filled or open bars indicate the presence of either the C57 mtDNA or C3H mtDNA, respectively. Data = mean ± SEM from 35–50 cardiomyocytes. An asterisk (*) indicates a significant difference (p < 0.001) exists compared to nuclear genome matched control.
Figure 3. Cellular ROS generation is influenced by the mtDNA in cardiomyocytes.
A) Cardiomyocytes isolated from control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) mouse hearts were treated with DCF-DA and cellular fluorescence measured (485 nm/530 nm - excitation/emission) B) Bar graph illustrating fluorescence relative to the C57n:C57mt controls. The nuclear genome for each group is indicated on the X-axis, whereas the filled or open bars indicate the presence of either the C57 mtDNA or C3H mtDNA, respectively. Data = mean ± SEM from 35–50 cardiomyocytes. An asterisk (*) indicates a significant difference (p < 0.001) exists compared to nuclear genome matched control.
NADH dehydrogenase and Cytochrome c oxidase (CO) mtDNA missense mutations
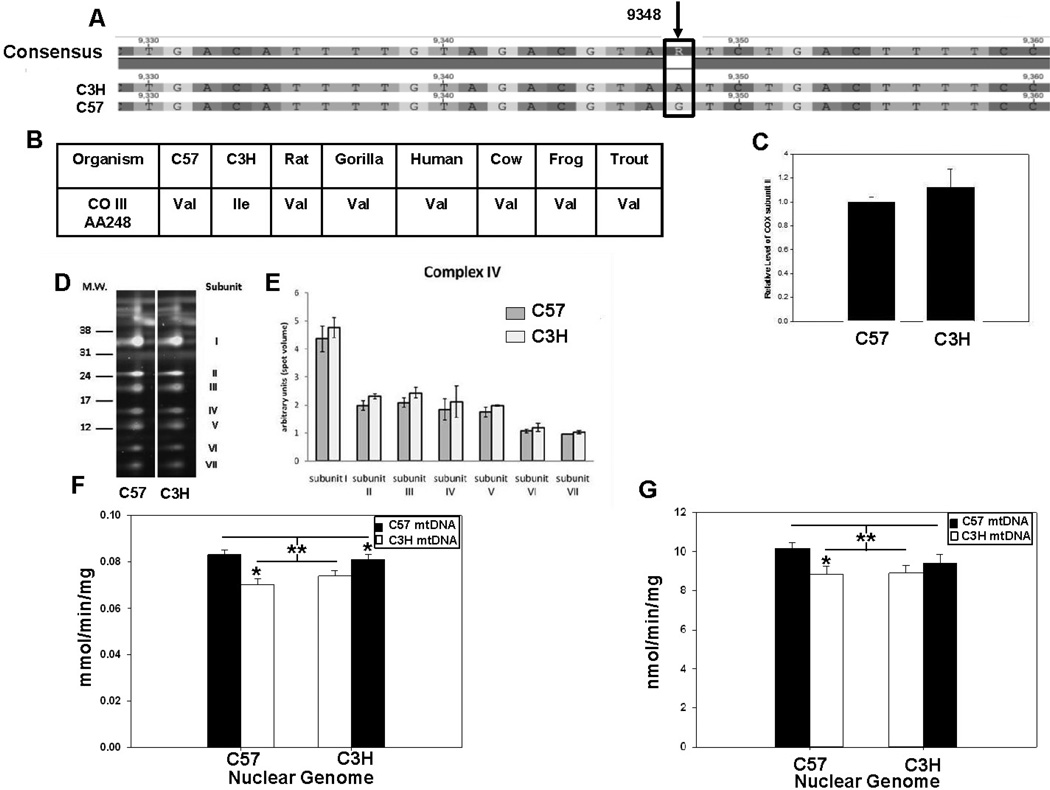
MtDNA sequence results revealed that C57 and C3H mouse mitochondrial haplotypes had two distinct missense mutations, an A to C transversion at nucleotide 9461 in subunit III of NADH dehydrogenase (ND3), and a G to A transition at nucleotide 9348 in subunit III of cytochrome c oxidase (CO3), consistent with previous reports [32]. The ND3 mutation results in the replacement of a methionine start codon in the C57 with an isoleucine codon in the C3H which could potentially influence ND3 levels in complex I. However proteomic analysis revealed no differences in detectable complex I subunit levels between mouse strains, and no differences in complex I enzyme activity were observed between mitochondrial isolates of C57 and C3H hearts (Supplementary Figure S2). Similarly, activity of complex II (succinate dehydrogenase) which contains no mtDNA encoded subunits, showed no differences between mouse strains (Supplementary Figure S3). In contrast, the G to A mutation in CO3 replaces a highly conserved valine to isoleucine at AA248 in subunit III between the C57 and C3H mouse, respectively (Figure 4A and B). While this change does not appear to affect either CO subunit II levels (Figure 4C) nor the levels of the CO holoenzyme (Figure 4D and E), quantification of CO activity in heart mitochondria by following the oxidation of reduced cytochrome c revealed those with the C57 mtDNA had a slightly higher activity than those having the C3H mtDNA, suggesting that CO activity segregated with mtDNA background (Figure 4F). Complementary analysis of complex IV respiration using ascorbate + TMPD showed that respiration rates between C57n:C57mt and C57n:C3Hmt mitochondrial isolates were significantly different. But the activity of the C3Hn:C3Hmt and C3Hn:C57mt mice was not statistically different (Figure 4G).
Figure 4. Complex IV (cytochrome oxidase) protein levels and activity in C57 and C3H mitochondria.
Proteomic and enzymatic analyses of heart mitochondrial isolates for cytochrome c oxidase. (A) MtDNA sequence of C3H and C57 mice showing a G to A transitional missense mutation in subunit III of complex IV. (B) Conservation of valine at AA248 throughout vertebrates. (C) Levels of cytochrome oxidase (COX) subunit II and (D) blue native 2D proteomics of additional cytochrome oxidase subunits. (E) Quantification of subunits, indicating no differences in the expression of isolated complex IV subunits. (F) Cytochrome c oxidase activity following the oxidation of reduced cytochrome c at 550 nm in cardiac mitochondria from control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) mice. The nuclear genome for each group is indicated on the X-axis, whereas the filled or open bars indicate the presence of either the C57 mtDNA or C3H mtDNA, respectively. An asterisk indicates a significant difference (*p < 0.05) exists between the nuclear matched control and MNX animal; the double asterisk (**) with lines indicates a significant difference exists between groups having the C57 versus C3H mtDNA. (G) Cytochrome c oxidase oxygen consumption was determined in cardiac mitochondria using ascorbate + TMPD from control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) mice. The nuclear genome for each group is indicated on the X-axis, whereas the filled or open bars indicate the presence of either the C57 mtDNA or C3H mtDNA, respectively. An asterisk indicates a significant difference (*p < 0.05) exists between the nuclear matched control and MNX animal; the double asterisk with lines indicates a significant difference exists between groups having the C57 versus C3H mtDNA.
MtDNA background affects susceptibility to cardiac volume overload (VO)
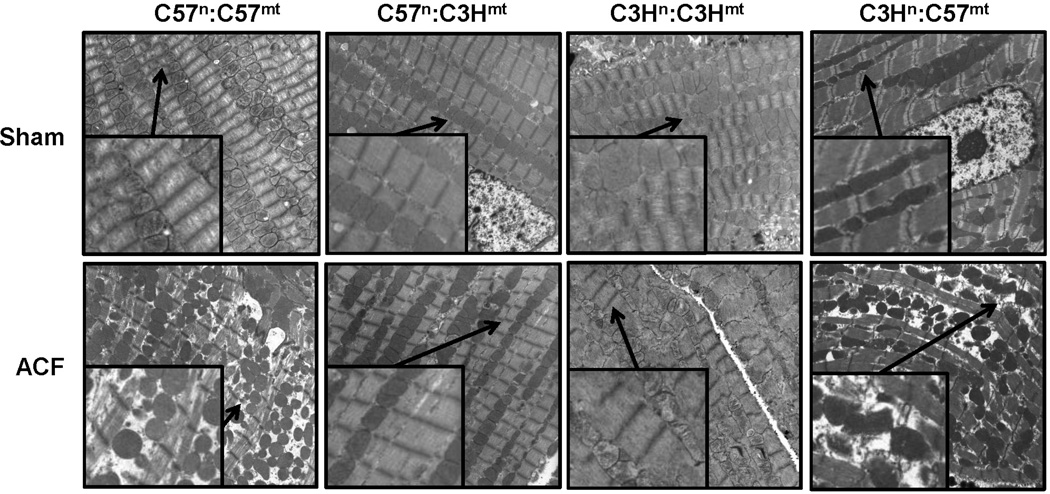
To determine whether these changes associated with mitochondrial genetic background and bioenergetics also influenced susceptibility to cardiac stress, a heart volume overload (VO) model was performed using the aortocaval fistula (ACF) mouse model [19]. Control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt and C3Hn:C57mt) mice underwent sham surgery or 3 days of ACF and heart left ventricle (LV) tissue was excised and examined using transmission electron microscopy (TEM). Figure 5 shows dramatic ultrastructural changes in the LV from ACF treated C57 control mice (C57n:C57mt), indicating substantial myofibrillar and mitochondrial degeneration, whereas C3H control mice (C3Hn:C3Hmt) showed very little disruption of the cytoskeleton and myofibrils, only a minor degree of mitochondrial swelling and thus are minimally impacted by 3 days of ACF. Remarkably, susceptibility to ultrastructural changes was inherently linked to the mitochondrial genetic origin and not the nuclear background; C57n:C3Hmt mouse hearts had little or no evidence of the pathological structural changes observed in the C57n:C57mt animals in response to ACF (Figure 5), whereas the C3Hn:C57mt had marked pathological changes compared to C3Hn:C3Hmt mice in response to ACF treatment.
Figure 5. Transmission Electron Microscopy (TEM) of left ventricular (LV) tissue in sham and aortacaval fistula (ACF) treated mice.
Representative images from control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmt, and C3Hn:C57mt) mice that underwent either sham or ACF surgery and sacrificed 3 days thereafter, and LV tissue prepared for TEM. Mitochondrial swelling and disorganization along with myofibrillar degeneration was observed in the ACF treated mice having the C57 mtDNA (C57n:C57mt and C3Hn:C57mt animals), which was absent in the ACF treated mice having the C3H mtDNA (C3Hn:C3Hmt and C57n:C3Hmt mice). Mag = 4500x (insets are 9000x, arrows indicate enlarged area). Imaging by Emlabs, Inc., Birmingham, AL.
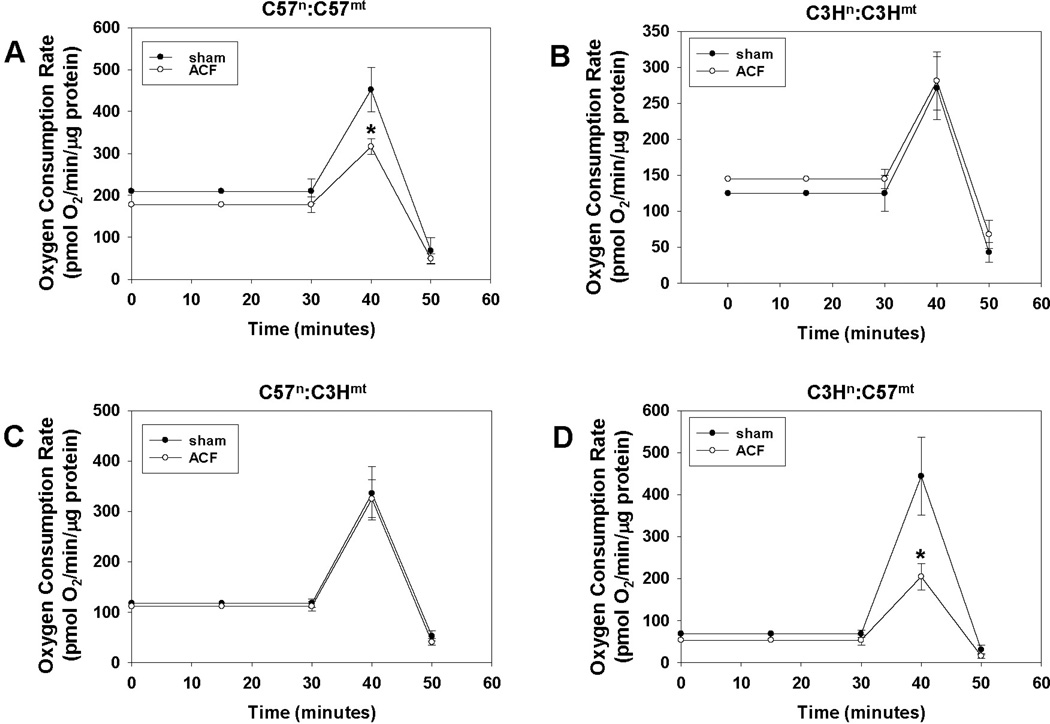
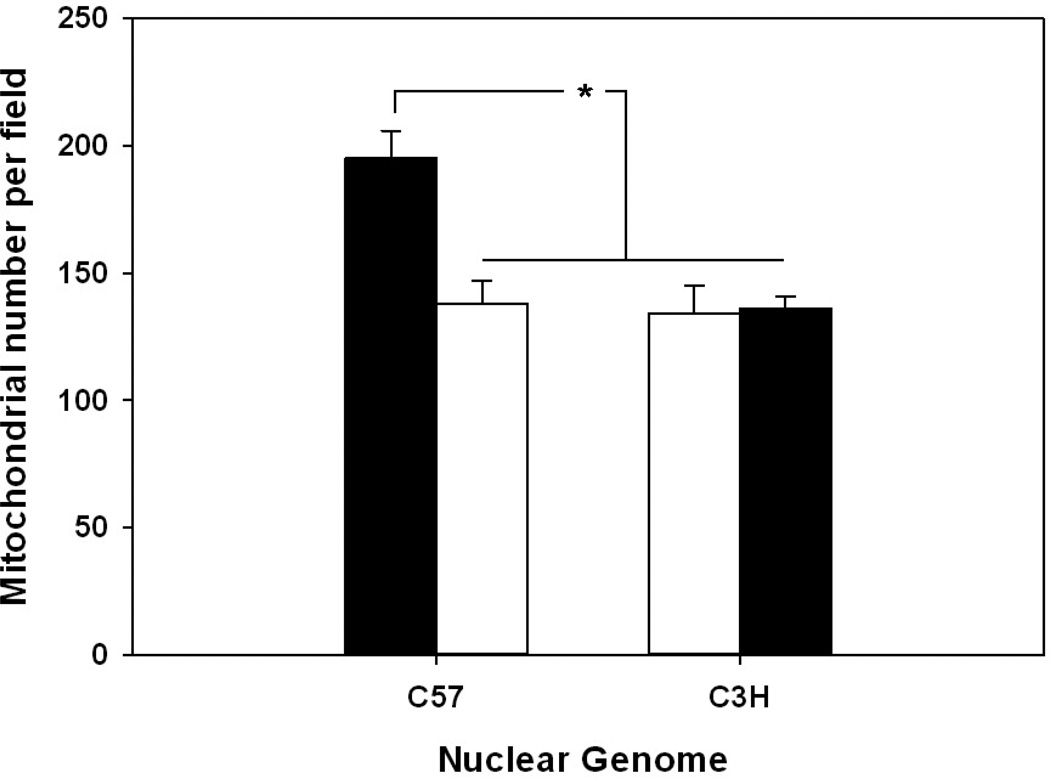
Finally, to assess the ACF effects on cardiomyocyte bioenergetics, oxygen consumption rates (OCRs) were determined in isolated cardiomyocytes from both sham and ACF treated mice (Figure 6). ACF treatment significantly decreased the maximal OCR in cardiomyocytes compared to cells derived from sham treated animals which harbored the C57 mtDNA (C57n:C57mt and C3Hn:C57mt), independent of nuclear background, whereas no differences were seen in those sharing the C3H mtDNA (C3Hn:C3Hmt and C57n:C3Hmt). Interestingly, the maximal OCR in the sham treated animals was lowest in those having the C3H mtDNA (271 + 43.4 and 336 + 52.8 pmol O2/min/µg protein, in C3Hn:C3Hmt and C57n:C3Hmt mice respectively), versus those harboring the C57 mtDNA (452 + 53.3 and 444 + 92.4 pmol O2/min/µg protein in C57n:C57mt and C3Hn:C57mt mice, respectively) consistent with the findings that complex IV activity was decreased in mitochondria having the C3H mtDNA. Interestingly, it was also observed that the basal respiration rate was higher in the C57n:C57mt sham animals compared to the other groups; to determine a potential basis for this difference mitochondrial numbers per cell were estimated by calculating the mean number of mitochondria observed per TEM field in the sham animals. Figure 7 reveals that the C57n:C57mt mice had significantly higher numbers of mitochondria compared to all remaining groups, which had similar amounts of mitochondria, potentially providing an explanation for the observed differences in basal rates. No differences in mitochondrial number were observed between ACF versus sham animals within each strain (data not shown).
Figure 6. Cardiomyocyte bioenergetics in sham and aortacaval fistula (ACF) treated hearts.
Graphs illustrating basal, maximum, and non-mitochondrial oxygen consumption rates in cardiomyocytes isolated from sham and ACF treated hearts for (A) C57n:C57mt(B) C3Hn:C3Hmt(C) C57n:C3Hmtand (D) C3Hn:C57mt (see methods, N > 5 per group). Asterisks (*) indicate a significant (P < 0.05) difference exists between ACF and sham treated control.
Figure 7. Average number of mitochondria in left ventricular tissue in control and MNX mice.
Graph illustrating mean number of mitochondria per TEM field from control (C57n:C57mt and C3Hn:C3Hmt) and MNX (C57n:C3Hmtand C3Hn:C57mt) mice. Values were calculated from a total of 78 fields with a minimum of 8 fields per mouse strain. Asterisks (*) indicate a significant (P < 0.05) difference exists between C57n:C57mt versus C3Hn:C3HmtC57n:C3Hmt and C3Hn:C57mt left ventricular tissue.
Discussion
The factors that contribute to cellular bioenergetics and disease susceptibility are clearly complex. The mitochondrion is a multifunctional organelle which plays a vital role in energy production, cell growth, apoptosis, thermogenesis and redox signaling. Mitochondrial genetics has an “aging” mechanism in that there are thousands of copies of mtDNA per cell compared to two nuclear alleles per cell; this redundancy allows for the accumulation of mtDNA mutations and damage over time that contributes to an eventual decline in mitochondrial function with age [2;33–35]. Additionally, studies have shown that disease risk factors such as hypercholesterolemia and cigarette smoking increase mitochondrial damage and alter function in cardiovascular tissues [23;35–37]. Most ancient nuclear DNA polymorphisms are common to all global populations [38] whereas by virtue of its maternal transmission, the mtDNA displays marked regional variation. Maternal family history of certain diseases has also been suggested to convey greater risk than paternal history [39;40] and although it is thought that this association may be due to offspring – maternal behaviors, and/or nutritional effects that were experienced in utero [41], additional studies have shown that in utero exposure to disease risk factors increase mtDNA damage [37;42].
Wallace and co-workers originally proposed the mitochondrial paradigm in which environmental factors influenced prehistoric human radiation patterns and survival by selecting for aspects of mitochondrial function [2;4]. As humans radiated out of Africa, they encountered colder climates, a higher dependence upon dietary animal proteins and fats, and accumulated a greater frequency of mtDNA missense mutations [2;4]. It has been suggested that these mutations altered mitochondrial economy (the conversion of caloric energy into ATP, heat, and oxidants) enabling individuals harboring these mtDNAs to more effectively utilize caloric energy to meet local environmental challenges and establish populations northward from Africa. These findings have led to the hypothesis that in the present era, with the development of a more sedentary lifestyle, excessive caloric intake, and climate control, these variants in mitochondrial function influence cellular ROS formation and therefore disease susceptibility. Differences in the mtDNA have been linked to behavioral, cognitive, and age – related deafness in conplastic mice [11;43–45], these studies did not report mitochondrial or cellular bioenergetics. Other investigators using conplastic or cybrid approaches have examined the effects of mtDNA variation upon bioenergetics and ROS production [46–49]; and have concluded that ROS production can be influenced by mtDNA sequence, and that respiratory capacity, “per molecule of mtDNA” could be affected by mtDNA sequence [46]. Interpretation of these studies is complicated bv the possibility that Mendelian allelic selection events for “fitness” are also influenced by mtDNA – nuclear interactions. In this scenario selection for specific nuclear alleles may occur during the sequential backcrossings of hybrid conplastic animals resulting in a model with a unique combination of mitochondrial and nuclear backgrounds which cannot be controlled by the parent strains. MNX mice overcome this problem because they are produced directly without the generation of nuclear hybrids (thus circumventing any potential issue of nuclear allelic selection in hybrid animals), generating an animal having desired nuclear and mitochondrial genome in a single step. Consequently, MNX mice provide a means of unequivocally testing the influence of mtDNA background on cell function by circumventing a potential confounder of nuclear allelic selection that can occur with the generation of nuclear hybrids using conplastic methods.
In this regard, we now provide direct evidence that mtDNA plays a significant role in controlling the relationship between mitochondrial energetic and ROS formation, and this is associated with susceptibility to ACF induced pathological changes in the heart, a model of volume overload which ultimately leads to heart failure. While missense mutations exist between the C3H/HeN and C57BL/6 mtDNAs at both complex I (NADH dehydrogenase subunit 3) and complex IV (cytochrome c oxidase subunit III, COIII), no significant differences were observed in complex I protein or activity levels, whereas differences were noted at complex IV. While both spectrophotometic and polarographic assays for complex IV activity showed significant changes in mice harboring the C57 versus C3H mtDNA background, the spectrophotometic assay also showed significant differences between both controls and nuclear-matched MNX, whereas the polarographic measures showed only differences between the C57n:C57mt versus C57n:C3Hmt animals but not the C3Hn:C3Hmt versus C3Hn:C57mt mice. Because these assays utilize different mechanisms to quantify cytochrome c oxidase activity, e.g. by monitoring the rate of oxidation of exogenously available reduced cytochrome c (a single electron donor) via absorbance changes versus the reduction of oxygen (a four electron acceptor) dependent upon endogenous cytochrome c levels, using an oxygen electrode, these differences in sensitivity and mechanism probably underlie the discrepancy between the activity assays for complex IV. The COIII mutation replaces a highly conserved valine to an isoleucine. CO subunits I and II act as the catalytic subunits for redox reactions and proton pumping for cytochrome c oxidase, whereas subunit III is thought to play a role in the structural relationship between subunits I and II. It has also been shown that COIII plays a key role in proton pumping - absence of this subunit decreases proton pumping significantly by changing the pKa of Asp132 of subunit I [50]. Thus, the valine to isoleucine change in COIII may alter its conformation and in turn influence cytochrome c oxidase enzymatic activity and its ability to pump protons across the membrane, potentially manifesting as altered mitochondrial economy. This is consistent with the observation that under conditions of coupling, mitochondria with the C57BL/6 mtDNA have higher membrane potentials, consume less oxygen, and generate more ATP compared to those harboring the C3H/HeN mtDNA (Figures 1 and 2). While seeming contradictory with the higher CO enzyme activity results observed in with the C57BL/6 mtDNA (Figure 4), these measures were performed in absence of a membrane potential using uncoupled mitochondria. The higher membrane potential and economy associated with the C57BL/6 mtDNA under coupled conditions likely regulates mitochondrial oxygen consumption. Another feature not yet understood is whether the differences observed between the isolated mitochondrial experiments and cardiomyocyte respiration studies are due to: 1) the fact that cellular bioenergetics substrates are provided by the metabolic activity of the cells versus the saturating substrate conditions used for the isolated mitochondria experiments; 2) differences in mitochondrial number in hearts as shown in Figure 7; 3) both 1 and 2, or 4) as yet unidentified mechanisms. Regardless, the combination of mitochondrial and nuclear genetic background also may influence organelle biogenesis and regulation of mitochondrial number per cell. Future studies to further explore these observations are underway.
Several studies have noted differences in cardiovascular disease (CVD) susceptibility between certain strains of mice having specific nuclear gene differences [11;17;18;51–55]. For example, it has been suggested that the C3H/HeJ mouse (a descendant substrain of the C3H/HeN mouse used herein) is resistant to atherosclerosis by virtue of being Toll receptor- 4 (TLR4) null [56]. However, many TLR4 competent mice are also atherosclerosis resistant, such as the C3H/HeN and the A/J mouse, among others. While the loss of TLR4 may contribute to athero-resistance, it is not the only potential basis. Differences in nicotinamide nucleotide transhydrogenase (NNT) gene expression, a protein that regenerates NADPH within the mitochondrial inner membrane, have been suggested as the basis for the observed increase in dietary induced insulin resistance and obesity in C57 mice [17;52;53]. However, it remains unclear whether decreased or increased expression of NNT conveys these effects since studies suggesting both outcomes have been reported [52;55;57;58]. It is well established that other strains of mice which are NNT competent also display dietary induced insulin resistance and obesity [55;58] and hence differences at this locus cannot be the sole explanation for susceptibility. The findings that C57 mice are also more prone to obesity, insulin resistance, dietary induced atherogenesis, and are generally more vulnerable to the effects of environmental oxidants [17;53;56;58] compared to C3H mice are consistent with the notion of differences in mitochondrial function. These same traits would be predicted if differences in mitochondrial economy that influenced caloric utilization and oxidant generation existed between mouse strains.
Why some individuals with identical CVD risk factor profiles develop disease while others do not is unclear. In addition to endogenous and environmental risk factors (i.e. hypercholesterolemia and tobacco smoke exposure, respectively), CVD susceptibility is increased by age, family history [39;40], and ethnicity [59]. The “common disease, common variants” hypothesis suggests that CVD susceptibility has a multi-factorial, polygenic basis in that genetic variants present in many normal individuals, each with a relatively small effect, alone or in combination with modifier genes and environmental factors, contribute to overall CVD risk [60]. Numerous studies have sought associations between polymorphic gene mutations and CVD development. However, many original associations were lost in larger scale studies or were not as predictive for risk as plasma markers such as cholesterol levels [61–64]. Consequently, while important in advancing our understanding of gene groups that may be involved in influencing predilection to disease development, the underlying genetic and physiologic basis of why these differences exist are not well understood. A complementary possibility is that differences in mitochondrial function which are influenced by mtDNA affect cytoplasmic signaling to the nucleus thereby influencing aspects of nuclear gene expression. While similar to concepts of retrograde signaling, subtle changes in mitochondrial quality would play a role in the regulation of nuclear gene expression and hence, be distinct from current concepts of retrograde signaling that usually invoke significant mitochondrial stress or loss of function [65]. In this manner, changes in mitochondrial function, influenced by mtDNA mutations, would potentially have significant impact upon the expression of nuclear genes involved in disease susceptibility. In addition, it is likely that any change in mitochondrial function associated with mtDNA mutation would have distinct effects on different tissues. It is well established that a hierarchy exists in terms of tissue sensitivity to pathogenic mtDNA mutations [2;66], hence it is likely the same occurs with “normal” polymorphisms. Because the focus of this study was upon the heart the impact of different mtDNA backgrounds on other tissues or organs was not explored; however based upon what is known regarding pathogenic mtDNA mutations, it is possible that the same mtDNA background will have varied effects upon different tissues. It is also plausible that this interaction would be significantly affected by environmental factors and the combination of mtDNA and nuclear genomes. Consequently, we view the mitochondrial paradigm for common disease development as a significant yet complementary component of the “common disease, common variants” explanation for disease susceptibility. Future studies investigating this potential aspect of mitochondrial genetics in cellular bioenergetics and disease development and progression are now possible using MNX mice.
Supplementary Material
Acknowledgements
Support was from NIH RO1 94518 (SWB) and RO1 103859 (SWB), the National Foundation for Cancer Research (DRW), Susan G. Komen for the Cure SAC111370 (DRW), U.S. Army Medical Research & Material Command W81XWH-07-1-0540d (SWB), and a pilot grant from the UAB Comprehensive Cancer Center CA013148 (DRW and SWB). Additional support was received from RO1 HL109785 (LJD); NIH predoctoral training program in cardiovascular pathophysiology T32 HL007918 (JLF); the American Heart Association predoctoral fellowships 09PRE2240046 (JLF) and 11PRE7650033 (KJD); the UAB Transgenic Mouse Facility (RAK) supported by awards NIH P30 CA13148, P30 AR048311, P30 DK074038, P30 DK05336, and P60 DK079626; and by the NIH-funded Diabetes Research Center Bioanalytical Redox Biology Core (P60 DK079626) located at the University of Alabama at Birmingham.
Footnotes
Author Contribution: JLF and BRZ performed the majority of the experimental work for this study. LWJ and JZ generated the original MNX founders for the mouse colonies. DRM, DGW, MJS, MJ, KJD, and BC assisted in the conduction experiments. WEB and CCW performed the TEM experiments. MP and XC provided assistance with MNX colony maintenance, haplotyping and breeding. TGS assisted JLF with the mtDNA sequencing experiments. RAK and LWJ assisted in the experimental design for developing the MNX model. LJD and VDU assisted in experimental design; DRW and SWB generated overall the experimental design. JLF, LJD, VDU, DRW and SWB wrote the manuscript.
Reference List
- 1.Margulis L. Origin of Mitochondria and Hydrogenosomes. History and Philosophy of the Life Sciences. 2008;30:473–477. [Google Scholar]
- 2.Wallace DC. A Mitochondrial Paradigm of Metabolic and Degenerative Diseases, Aging, and Cancer: A Dawn for Evolutionary Medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumarasamy S, Gopalakrishnan K, Shafton A, Nixon J, Thangavel J, Farms P, Joe B. Mitochondrial polymorphisms in rat genetic models of hypertension. Mamm.Genome. 2010;21:299–306. doi: 10.1007/s00335-010-9259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–226. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 5.Krzywanski DM, Moellering DR, Fetterman JL, Dunham-Snary KJ, Sammy MJ, Ballinger SW. The mitochondrial paradigm for cardiovascular disease susceptibility and cellular function: a complementary concept to Mendelian genetics. Lab Invest. 2011;91:1122–1135. doi: 10.1038/labinvest.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown MD, Starikovskaya E, Derbeneva O, Hosseini S, Allen JC, Mikhailovskaya IE, Sukernik RI, Wallace DC. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup. J Human Genetics. 2002;110:130–138. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- 7.Corral-Debrinski M, Shoffner JM, Lott MT, Wallace DC. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat.Res. 1992;275:169–180. doi: 10.1016/0921-8734(92)90021-g. [DOI] [PubMed] [Google Scholar]
- 8.Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. American Journal of Medical Genetics. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 9.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, Passarino G, Bonafe M, Monti D, Baggio G, Bertolini S, Mari D, Mattace R, Franceschi C. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. Faseb Journal. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 10.van den Ouweland JMW, Lemkes HHPJ, Ruitenbeek W. Mutation in mitochondrial tRNALeu(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nature Genetics. 1992;1:368–371. doi: 10.1038/ng0892-368. [DOI] [PubMed] [Google Scholar]
- 11.Yu X, Gimsa U, Wester-Rosenlof L, Kanitz E, Otten W, Kunz M, Ibrahim SM. Dissecting the effects of mtDNA variations on complex traits using mouse conplastic strains. Genome Research. 2009;19:159–165. doi: 10.1101/gr.078865.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenzie M, Trounce IA, Cassar CA, Pinkert CA. Production of homoplasmic xenomitochondrial mice. Proc.Natl.Acad.Sci.USA. 2004;101:1685–1690. doi: 10.1073/pnas.0303184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paigen B, Morrow A, Brandon C, Mitchell D, Holmes P. Variation in susceptibility to atherosclerosis among inbred starins of mice. Atherosclerosis. 1985;57:65–73. doi: 10.1016/0021-9150(85)90138-8. [DOI] [PubMed] [Google Scholar]
- 14.Kaku K, Fiedorek FT, Province M, Permutt MA. Genetic analysis of glucose tolerance in inbred mouse starins. Diabetes. 1988;37:707–713. doi: 10.2337/diab.37.6.707. [DOI] [PubMed] [Google Scholar]
- 15.Grimsditch DC, Penfold S, Latcham J, Vidgeon-Hart M, Groot PH, Benson GM. C3H apoE(−/−) mice have less atherosclerosis than C57BL apoE(−/−) mice despite having a more atherogenic serum lipid profile. Atherosclerosis. 2000;151:389–397. doi: 10.1016/s0021-9150(99)00400-1. [DOI] [PubMed] [Google Scholar]
- 16.Shi W, Wang NJ, Shih DM, Sun VZ, Wang X, Lusis AJ. Determinants of atherosclerosis susceptibility in the C3H and C57BL/6 mouse model: evidence for involvement of endothelial cells but not blood cells or cholesterol metabolism. Circ.Res. 2000;86:1078–1084. doi: 10.1161/01.res.86.10.1078. [DOI] [PubMed] [Google Scholar]
- 17.Freeman HC, Hugill A, Dear NT, Ashcroft FM, Cox RD. Deletion of nicotinamide nucleotide transhydrogenase: a new quantitive trait locus accounting for glucose intolerance in C57BL/6J mice. Diabetes. 2006;55:2153–2156. doi: 10.2337/db06-0358. [DOI] [PubMed] [Google Scholar]
- 18.Wang XS, Ria M, Kelmenson PM, Eriksson P, Higgins DC, Samnegard C, Petros C, Rollins J, Bennet AM, Wiman B, de Faire U, Wennberg C, Olsson PG, Ishii N, Sugamura K, Hamsten A, Forsman-Semb K, Lagercrantz J, Paigen B. Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nature Genetics. 2005;37:365–372. doi: 10.1038/ng1524. [DOI] [PubMed] [Google Scholar]
- 19.Scheuermann-Freestone M, Freestone NS, Langenickel T, Hohnel K, Dietz R, Willenbrock R. A new model of congestive heart failure in the mouse due to chronic volume overload. Eur.J.Heart Fail. 2001;3:535–543. doi: 10.1016/s1388-9842(01)00160-x. [DOI] [PubMed] [Google Scholar]
- 20.Gladden JD, Zelickson BR, Wei CC, Ulasova E, Zheng J, Ahmed MI, Chen Y, Bamman M, Ballinger S, Darley-Usmar V, Dell'Italia LJ. Novel insights into interactions between mitochondria and xanthine oxidase in acute cardiac volume overload. Free Radic.Biol.Med. 2011;51:1975–1984. doi: 10.1016/j.freeradbiomed.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ulasova E, Gladden JD, Chen Y, Zheng J, Pat B, Bradley W, Powell P, Zmijewski JW, Zelickson BR, Ballinger SW, Darley-Usmar V, Dell'Italia LJ. Loss of interstitial collagen causes structural and functional alterations of cardiomyocyte subsarcolemmal mitochondria in acute volume overload. J.Mol.Cell Cardiol. 2011;50:147–156. doi: 10.1016/j.yjmcc.2010.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Z, Cheng Y, Liang CG, Latham KE. Nuclear transfer in mouse oocytes and embryos. Methods Enzymol. 2010;476:171–184. doi: 10.1016/S0076-6879(10)76010-9. [DOI] [PubMed] [Google Scholar]
- 23.Knight-Lozano CA, Young CG, Burow DL, Hu Z, Uyeminami D, Pinkerton K, Ischiropoulos H, Ballinger SW. Cigarette Smoke Exposure and Hypercholesterolemia increase mitochondrial damage in cardiovascular tissues. Circulation. 2002;105:849–854. doi: 10.1161/hc0702.103977. [DOI] [PubMed] [Google Scholar]
- 24.Rasmussen HN, Andersen AJ, Rasmussen UF. Optimization of preparation of mitochondria from 25–100 mg skeletal muscle. Anal.Biochem. 1997;252:153–159. doi: 10.1006/abio.1997.2304. [DOI] [PubMed] [Google Scholar]
- 25.Lanza IR, Nair KS. Functional assessment of isolated mitochondria in vitro. Methods Enzymol. 2009;457:349–372. doi: 10.1016/S0076-6879(09)05020-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darley-Usmar V, Capaldi RA, Takamiya S, Millett F, Wilson MT, Malatesta F, Sarti P. Reconstitution and molecular analysis of the respiratory chain. In: Darley-Usmar V, Rickwood D, Wilson MT, editors. Mitochondria a practical approach. Oxford, England: IRL Press Limited; 1987. pp. 113–152. [Google Scholar]
- 27.Trounce I, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 28.Brookes PS, Pinner A, Ramachandran A, Coward L, Barnes S, Kim H, rley-Usmar VM. High throughput two-dimensional blue-native electrophoresis: a tool for functional proteomics of mitochondria and signaling complexes. Proteomics. 2002;2:969–977. doi: 10.1002/1615-9861(200208)2:8<969::AID-PROT969>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 29.Wei CC, Chen Y, Powell LC, Zheng J, Shi K, Bradley WE, Powell PC, Ahmad S, Ferrario CM, Dell'Italia LJ. Cardiac kallikrein-kinin system is upregulated in chronic volume overload and mediates an inflammatory induced collagen loss. PLoS.One. 2012;7:e40110. doi: 10.1371/journal.pone.0040110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill BG, Dranka BP, Zou LY, Chatham JC, Darley-Usmar VM. Importance of the bioenergetic reserve capacity in response to cardiomyocyte stress induced by 4-hydroxynonenal. Biochem.J. 2009;424:99–107. doi: 10.1042/BJ20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonini MG, Rota C, Tomasi A, Mason RP. The oxidation of 2',7'- dichlorofluorescin to reactive oxygen species: a self-fulfilling prophesy? Free Radic.Biol.Med. 2006;40:968–975. doi: 10.1016/j.freeradbiomed.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 32.Bayona-Bafaluy MP, Acin-Perez R, Mullikin JC, Park JS, Moreno-Loshuertos R, Hu PQ, Perez-Martos A, Fernandez-Silva P, Bai YD, Enriquez JA. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trounce I, Byrne E, Marzuki S. Decline in skeletal muscle mitochondrial respiratory chain function: possible factor in ageing. Lancet. 1989;1:44–45. doi: 10.1016/s0140-6736(89)92143-0. [DOI] [PubMed] [Google Scholar]
- 34.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 35.Ballinger SW, Patterson C, Knight-Lozano CA, Burow DL, Conklin CA, Hu Z, Reuf J, Horaist C, Lebovitz RM, Hunter G, McIntyre K, Runge MS. Mitochondrial integrity and function in atherogenesis. Circulation. 2002;106:544–549. doi: 10.1161/01.cir.0000023921.93743.89. [DOI] [PubMed] [Google Scholar]
- 36.Harrison CM, Pompilius M, Pinkerton KE, Ballinger SW. Mitochondrial oxidative stress significantly influences atherogenic risk and cytokineinduced oxidant production. Environ.Health Perspect. 2011;119:676–681. doi: 10.1289/ehp.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Z, Knight CA, Mamerow M, Vickers K, Penn A, Postlethwait E, Ballinger SW. Prenatal Environmental Tobacco Smoke Exposure Promotes Adult Atherogenesis and Mitochondrial Damage in apoE-/- mice fed a chow diet. Circulation. 2004;110:3715–3720. doi: 10.1161/01.CIR.0000149747.82157.01. [DOI] [PubMed] [Google Scholar]
- 38.Schurr TG. The peopling of the New World: Perspectives from molecular anthropology. Annu Rev Anthropol. 2004;33:551–583. [Google Scholar]
- 39.Sundquist K, Li XJ. Differences in maternal and paternal transmission of coronary heart disease. American Journal of Preventive Medicine. 2006;30:480–486. doi: 10.1016/j.amepre.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 40.Sesso HD, Lee IM, Gaziano JM, Rexrode KM, Glynn RJ, Buring JE. Maternal and paternal history of myocardial infarction and risk of cardiovascular disease in men and women. Circulation. 2001;104:393–398. doi: 10.1161/hc2901.093115. [DOI] [PubMed] [Google Scholar]
- 41.Barker DJ. Fetal origins of coronary disease. BMJ. 1995;311:171–174. doi: 10.1136/bmj.311.6998.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westbrook DG, Anderson PG, Pinkerton KE, Ballinger SW. Perinatal tobacco smoke exposure increases vascular oxidative stress and mitochondrial damage in non-human primates. Cardiovasc.Toxicol. 2010;10:216–226. doi: 10.1007/s12012-010-9085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gimsa U, Kanitz E, Otten W, Ibrahim SM. Behavior and Stress Reactivity in Mouse Strains with Mitochondrial DNA Variations. Neuroimmunomodulation: from Fundamental Biology to Therapy. 2009;1153:131–138. doi: 10.1111/j.1749-6632.2008.03960.x. [DOI] [PubMed] [Google Scholar]
- 44.Johnson KR, Zheng QY, Bykhovskaya Y, Spirina O, Fischel-Ghodsian N. A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nature Genetics. 2001;27:191–194. doi: 10.1038/84831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roubertoux PL, Sluyter F, Carlier M, Marcet B, Maarouf-Veray F, Cherif C, Marican C, Arrechi P, Godin F, Jamon M, Verrier B, Cohen-Salmon C. Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nature Genetics. 2003;35:65–69. doi: 10.1038/ng1230. [DOI] [PubMed] [Google Scholar]
- 46.Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez- Martos A, Rodriguez de CS, Gallardo ME, Enriquez JA. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat.Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- 47.Gusdon AM, Votyakova TV, Mathews CE. mt-Nd2a suppresses reactive oxygen species production by mitochondrial complexes I and III. J.Biol.Chem. 2008;283:10690–10697. doi: 10.1074/jbc.M708801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gusdon AM, Votyakova TV, Reynolds IJ, Mathews CE. Nuclear and mitochondrial interaction involving mt-Nd2 leads to increased mitochondrial reactive oxygen species production. J.Biol.Chem. 2007;282:5171–5179. doi: 10.1074/jbc.M609367200. [DOI] [PubMed] [Google Scholar]
- 49.Kumarasamy S, Gopalakrishnan K, bdul-Majeed S, Partow-Navid R, Farms P, Joe B. Construction of two novel reciprocal conplastic rat strains and characterization of cardiac mitochondria. Am.J.Physiol Heart Circ.Physiol. 2013;304:H22–H32. doi: 10.1152/ajpheart.00534.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adelroth P, Hosler J. Surface proton donors for the D-pathway of cytochrome c oxidase in the absence of subunit III. Biochemistry. 2006;45:8308–8318. doi: 10.1021/bi0605843. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Phelan SA, Petros C, Taylor EF, Ledinski G, Jurgens G, Forsman- Semb K, Paigen B. Peroxiredoxin 6 deficiency and atherosclerosis susceptibility in mice: significance of genetic background for assessing atherosclerosis. Atherosclerosis. 2004;177:61–70. doi: 10.1016/j.atherosclerosis.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Toye AA, Lippiat JD, Proks P, Shimomura K, Bentley L, Hugill A, Mijat V, Goldsworthy M, Moir L, Haynes A, Quarterman J, Freeman HC, Ashcroft FM, Cox RD. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia. 2005;48:675–686. doi: 10.1007/s00125-005-1680-z. [DOI] [PubMed] [Google Scholar]
- 53.Freeman H, Shimomura K, Horner E, Cox RD, Ashcroft FM. Nicotinamide nucleotide transhydrogenase: a key role in insulin secretion. Cell Metab. 2006;3:35–45. doi: 10.1016/j.cmet.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 54.ston-Mourney K, Wong N, Kebede M, Zraika S, Balmer L, McMahon JM, Fam BC, Favaloro J, Proietto J, Morahan G, Andrikopoulos S. Increased nicotinamide nucleotide transhydrogenase levels predispose to insulin hypersecretion in a mouse strain susceptible to diabetes. Diabetologia. 2007;50:2476–2485. doi: 10.1007/s00125-007-0814-x. [DOI] [PubMed] [Google Scholar]
- 55.Wong N, Blair AR, Morahan G, Andrikopoulos S. The deletion variant of nicotinamide nucleotide transhydrogenase (Nnt) does not affect insulin secretion or glucose tolerance. Endocrinology. 2010;151:96–102. doi: 10.1210/en.2009-0887. [DOI] [PubMed] [Google Scholar]
- 56.Shih DM, Welch C, Lusis AJ. New insights into atherosclerosis from studies with mouse models. Mol.Med.Today. 1995;1:364–372. doi: 10.1016/s1357-4310(95)93834-6. [DOI] [PubMed] [Google Scholar]
- 57.Lin SJ, Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Current Opinion in Cell Biology. 2003;15:241–246. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 58.Lin S, Thomas TC, Storlien LH, Huang XF. Development of high fat diet-induced obesity and leptin resistance in C57Bl/6J mice. Int.J.Obes.Relat Metab Disord. 2000;24:639–646. doi: 10.1038/sj.ijo.0801209. [DOI] [PubMed] [Google Scholar]
- 59.Forouhi NG, Sattar N. CVD risk factors and ethnicity - A homogeneous relationship? Atherosclerosis Supplements. 2006;7:11–19. doi: 10.1016/j.atherosclerosissup.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 60.American Heart Association. Relevance of genetics and genomics for prevention and treatment of cardiovascular disease: A scientific statement from the AHA council on epidemiology and prevention, the stroke council, and the functional genomics and translational biology interdisciplinary working group. Circulation. 2007;115:2878–2901. doi: 10.1161/CIRCULATIONAHA.107.183679. [DOI] [PubMed] [Google Scholar]
- 61.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 62.Connelly JJ, Wang T, Cox JE, Haynes C, Wang LY, Shah SH, Crosslin DR, Hale AB, Nelson S, Crossman DC, Granger CB, Haines JL, Jones CJH, Vance JM, Goldschmidt-Clermont PJ, Kraus WE, Hauser ER, Gregory SG. GATA2 is associated with familial early-onset coronary artery disease. Plos Genetics. 2006;2:1265–1273. doi: 10.1371/journal.pgen.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Humphries SE, Talmud PJ, Hawe E, Bolla M, Day INM, Miller GJ. Apolipoprotein E4 and coronary heart disease in middle-aged men who smoke: a prospective study. Lancet. 2001;358:115–119. doi: 10.1016/S0140-6736(01)05330-2. [DOI] [PubMed] [Google Scholar]
- 64.Keavney B, Parish S, Palmer A, Clark S, Youngman L, Danesh J, McKenzie C, Delepine M, Lathrop M, Peto R, Collins R. Large-scale evidence that the cardiotoxicity of smoking is not significantly modified by the apolipoprotein E epsilon 2/epsilon 3/epsilon 4 genotype. Lancet. 2003;361:396–398. doi: 10.1016/S0140-6736(03)12386-0. [DOI] [PubMed] [Google Scholar]
- 65.Hill BG, Benavides GA, Lancaster JR, Ballinger SW, Dell'italia L, Zhang J, Darley-Usmar VM. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol.Chem. 2012 doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ballinger SW, Shoffner JM, Wallace DC. Mitochondrial Myopathies - Genetic-Aspects. Current Topics in Bioenergetics. 1994;Vol 17(17):59–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.