Abstract
In the nematode Caenorhabditis elegans, the appropriate induction of dauer larvae development within growing populations is likely to be a primary determinant of genotypic fitness. The underlying genetic architecture of natural genetic variation in dauer formation has, however, not been thoroughly investigated. Here, we report extensive natural genetic variation in dauer larvae development within growing populations across multiple wild isolates. Moreover, bin mapping of introgression lines (ILs) derived from the genetically divergent isolates N2 and CB4856 reveals 10 quantitative trait loci (QTLs) affecting dauer formation. Comparison of individual ILs to N2 identifies an additional eight QTLs, and sequential IL analysis reveals six more QTLs. Our results also show that a behavioural, laboratory-derived, mutation controlled by the neuropeptide Y receptor homolog npr-1 can affect dauer larvae development in growing populations. These findings illustrate the complex genetic architecture of variation in dauer larvae formation in C. elegans and may help to understand how the control of variation in dauer larvae development has evolved.
Keywords: Caenorhabditis elegans, dauer larvae, npr-1, QTL mapping, complex traits, introgression lines
Introduction
In the nematode Caenorhabditis elegans, dauer larvae are formed when conditions are unsuitable for growth and reproduction, with worms developing as dauer larvae in response to low food availability, high conspecific population density and high temperatures (Golden and Riddle, 1984; Hu, 2007). Dauer pheromone, a complex mix of structurally related ascarosides (Jeong et al., 2005; Butcher et al., 2007; Pungaliya et al., 2009; Park et al., 2012), is used by C. elegans to assess population density, with these ascarosides also acting to regulate aggregation, mate recognition and dispersal (Srinivasan et al., 2008; Harvey, 2009; Pungaliya et al., 2009; Izrayelit et al., 2012; Jang et al., 2012). Although there is an extensive and detailed understanding of the genetic pathways that specify dauer and non-dauer larval development in C. elegans (see Hu (2007) for a review), the ecology of the species is still poorly understood. Indeed, the natural habitat of C. elegans, rotting fruit, has been proposed only recently (Kiontke et al., 2011; Félix and Duveau, 2012). Under such conditions, populations grow to large sizes (Félix and Duveau, 2012) and the principle cues for dauer larvae development, pheromone level and food availability will change as a consequence of the growth and consumption of bacterial food, and of worm population growth and pheromone production. Given this, the ecological relevance of the extensive variation between C. elegans isolates in their sensitivity to dauer-inducing conditions as assessed by analysing age-matched individuals at high pheromone concentrations and limiting amounts of food (Viney et al., 2003; Harvey et al., 2008) is unclear. As methods to allow the analysis of dauer larvae development in growing populations have now been validated (Green and Harvey, 2012), we have investigated variation in dauer larvae formation in growing populations, conditions that, because of the dynamic nature of the food and pheromone levels in such assays, will more closely approximate the natural conditions experienced by the species.
Dauer larvae are generally resistant to environmental stress and long-lived, and are often crucial for dispersal. There are also clear phenotypic links between the dauer larvae of free-living species and the infective stages of many parasitic nematodes (for example, Stasiuk et al. (2012); and see Sudhaus (2010) for a recent discussion of this topic). Understanding how dauer larvae development is regulated is therefore important in determining how nematode parasitism evolved, in targeting intervention in parasite life histories, and in predicting how any such interventions will affect selection on parasites. To investigate the genetic architecture of natural variation in dauer larvae formation, we analysed a panel of introgression lines (ILs) derived from the genetically divergent strains CB4856 and N2 (Doroszuk et al., 2009). The genome of an IL is composed of a recipient genome contributed by one of the parental strains (Bristol N2) and a short, homozygous segment of the donor genome contributed by CB4856. We first conducted a straightforward bin mapping approach, followed by comparisons to a common reference (N2), and then a sequential pairwise analysis (Shao et al., 2010). These analyses indicate that the genetic architecture of dauer traits is highly complex, with a large number of quantitative trait loci (QTLs) affecting dauer larvae development in growing populations. We further demonstrate that a laboratory-derived polymorphism controlled by npr-1, a G-protein-coupled receptor related to the mammalian neuropeptide Y receptor (de Bono and Bargmann, 1998), can act as a major determinate of dauer larvae formation in growing populations.
Materials and methods
Worms
N2, DA650 npr-1 (ad609), DA609 npr-1 (n1353) and DA508 npr-1 (g320) were obtained from the Caenorhabditis Genetics Centre. Wild isolates were obtained from Marie-Anne Félix (IBENS, Paris, France). The CB4856/N2 ILs used are described in Doroszuk et al. (2009). They were derived from recombinant inbred lines obtained from crosses between CB4856 and N2 (see, for details, Li et al., 2006; Kammenga et al, 2007, 2008; Li et al., 2010; Viñuela et al., 2010, 2012; Elvin et al., 2011; Rodriquez et al., 2012). Isolates were maintained using standard methods on nematode growth media (NGM) plates (Stiernagle, 1999), with the OP50 strain of Escherichia coli as a food source. All assays were performed at 20 °C and were initiated with fourth larval stage worms (L4s) grown from synchronised, arrested, L1s. Within each experiment, plates were blind-coded and treatments (genotypes) were randomised, with plates that became contaminated or on which the population had failed to grow discarded.
Assays
Population assays were performed as described by Green and Harvey (2012), except that agar concentration was varied, with normal, dauer agar, plates containing 20 g l−1 and sloppy agar plates containing 4 g l−1. This corrected an initial issue with the population assays where the wild isolates tended to burrow into the agar, making recovery and counting impossible. Sloppy agar is solid enough to hold a pellet of bacterial food, allows worms to burrow freely and allows recovery of all worms for assay. Comparison of population growth and of dauer larvae formation on normal and sloppy agar plates indicated that there are differences in population growth rate and in the population size at food exhaustion and that dauer larvae formation is much higher in sloppy agar (Supplementary Figure 1). These data suggest, as would be expected given the sensitivity of the C. elegans life history to its environment, that the reproductive schedule is affected by the difference between the two environments. The higher dauer larvae formation observed in sloppy agar also suggests that under these conditions, either access to food signals is reduced or access to dauer pheromone is increased.
For the analyses of the wild isolates and of the ILs, populations were initiated with 100 μl of a 20% (w v−1) suspension of E. coli in water and monitored daily until food exhaustion, when the population size and the number of dauer larvae were determined as described by Green and Harvey (2012). Wild isolates were analysed in two experimental blocks with an N2 control in each block and 10 plates per isolate. Testing the N2 controls of these two batches showed that they were not different (F1,17=4.37, P=0.052 and F1,17=0.05, P=0.82, for population size and dauer larvae formation, respectively). The two batches were therefore grouped and investigated as a combined set. Differences between the different wild isolates and N2 were tested by a two-sided Student's t-test, assuming unequal variance. The wild isolates and N2 were then grouped by significance (see Figure 1). These data were also used to estimate the heritability of the analysed traits. This was calculated by taking the adjusted sum of squares of the components in an analysis of variance, with genotype and batch fitted as factors. The adjusted sum of squares of the genotype term was then used as the among genotype variance, which was divided by the total variance to obtain the heritability. The CB4856/N2 ILs were analysed in six experimental blocks, one per chromosome, with all ILs from that chromosome and both CB4856 and N2 controls analysed in each block and between 10 and 15 plates initiated per isolate (this varied depending on the number of ILs per chromosome and was required to limit the total assay size). To further investigate individual QTLs, specific ILs, and the effect of variation in npr-1 genotype, additional assays were undertaken as described above. To investigate the effect of variation in npr-1 genotype, individual isolates were compared, as described above, with isolates compared by a two-sided t-test, assuming unequal variance. To address the significance and effect of any mutation in npr-1 on dauer formation, we fitted linear models to investigate the effects of genotype, genotype and batch, and genotype, batch and population size.
Figure 1.
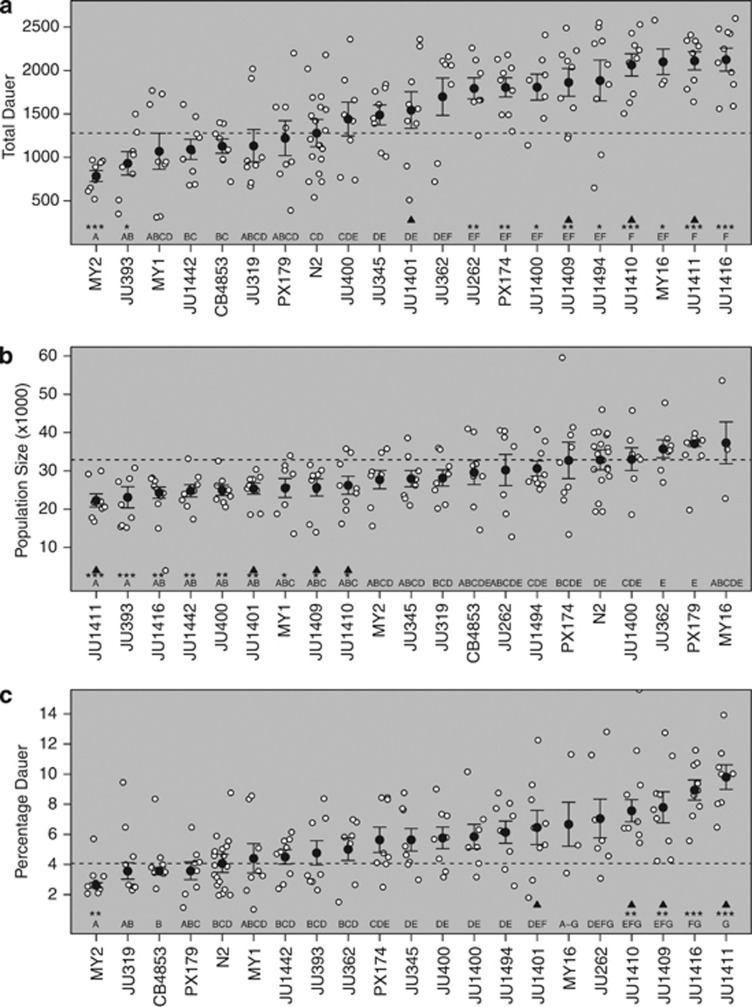
Natural variation in growing populations. The number of dauer larvae (a), the population size (b) and the percentage dauer larvae (c) at food exhaustion for N2 and 20 wild isolates. Dotted black horizontal lines show mean N2 values. Means per line are shown by the solid circle. Error bars indicate standard errors. Significance is shown by the asterisks on the x axis (*P<0.05, **P<0.01, ***P<0.001), denoting isolates that differ from N2. The letters show the significantly different groups. Black triangles show isolates identified as a clonal set by Andersen et al. (2012).
Mapping
For all mapping procedures, the chromosomes are treated as separate experiments. This can be carried out because all the ILs for one chromosome and N2 and CB4856 controls are measured within one experiment. Thresholds can be found in Supplementary Tables 1 and 2. This design maximises our ability to detect variation between ILs as comparisons are made between ILs within a chromosome and between individual ILs and the N2 controls for that block. However, as ILs on the same chromosome were assayed together, linkage is conflated with growth condition variation and other potential block effects. This approach therefore limits the value of estimates of QTL effect size and means that comparisons of effect sizes between QTLs on different chromosomes should be carried out with caution.
Bin mapping
Data from the ILs were analysed by bin mapping using a linear model as described by Doroszuk et al. (2009). ILs were grouped per marker per chromosome, and tested against the N2 controls from the same block (Supplementary Figure 2). The chromosome-wide significance threshold was determined by 1000 permutations by randomly distributing the phenotypic values over the ILs and N2 per test. So for marker X, the individual phenotypic scores of N2 and the ILs containing an introgression on marker X were taken and randomly re-distributed (Supplementary Figure 3). In this way, the genetic structure of the IL population was kept intact. For all analyses, outliers per genotype (outside the mean±2 s.d.) were removed per group before testing. No genotypes were removed completely and the maximum number of outliers per genotype was 2 (out of ca. 15) (see Supplementary Figure 2 for an example of this). In each permutation run, the maximum of each chromosome/experiment-wide permutation profile was taken. These 1000 maximum values were ordered and the 950th value used as the 0.05 false-positive rate threshold (Supplementary Table 1).
Single IL mapping
Genomic regions affecting variation in population size, the total number of dauer larvae and the percentage of dauer larvae at food exhaustion were also mapped by comparing each IL against N2 and testing for a significant difference by a two-sided t-test, assuming unequal variance. The P-values were Bonferroni adjusted to correct for multiple testing. The ILs with Bonferroni-corrected P-values ⩽0.05 were determined to be significantly different from N2. QTLs were then defined by comparing the results of overlapping and bordering ILs as is standard for the mapping of QTLs by ILs.
In addition, all ILs per chromosome were tested for significant differences. Groups were made when lines were not statistically different (P>0.01). In this way, the genomic limits the various QTLs were placed. Closely linked QTLs were tested by a linear model in which the phenotypic scores of all the ILs involved were explained by the two QTLs (Supplementary Figure 4).
Sequential IL analysis
This method was proposed by Shao et al. (2010) and adjusted to the population used in this study. ILs that share one break-point were selected and used in sequential (two by two) analyses to identify QTLs. A two-sided t-test, assuming unequal variance, was used to determine significance, with a Bonferroni adjustment of the P-values used to correct for multiple testing. The pairs of ILs with genome-wide Bonferroni-corrected P-values ⩽0.05 were considered to be significantly different and to indicate the presence of a QTL.
Results
Comparison of wild isolates
The population size and the number of dauer larvae at food exhaustion were determined for 20 wild isolates and N2. These data indicate that many of the wild isolates have significantly more dauer larvae at food exhaustion than N2, with only two isolates showing a lower number of dauer larvae (Figure 1a). In contrast, the observed pattern for population size was reversed, with more isolates showing a lower population size than N2 (Figure 1b). Across the wild isolates, there was no indication that the number of dauer larvae was correlated with the population size (Pearson's product–moment correlation: r=0.08, P=0.73), a situation that would be expected if variation in dauer larvae development was purely a consequence variation in traits affecting population growth rates. This conclusion is supported by the observed pattern of variation in the percentage of dauer larvae (Figure 1c). Estimates of heritability indicate a higher heritability for the dauer larvae development traits (0.86 and 0.71 for the total number of dauer larvae and the percentage of dauer larvae, respectively) than for population size at food exhaustion (0.38). These analyses demonstrate that the differences between the wild isolates have a genetic basis. Data from Andersen et al. (2012) identifies four of these wild isolates, JU1401, JU1409, JU1410 and JU1411, as belonging to a clonal set, defined as having fewer single nucleotide polymorphisms between isolates than expected given the false-positive rate in sequence data from ca. 8% of the genome. Here, our analyses identify no differences between these lines in population size, but indicate that the number of dauer larvae at food exhaustion is higher than in N2 for JU1409, JU1410 and JU1411, but not for JU1401 (Figure 1a).
IL analysis
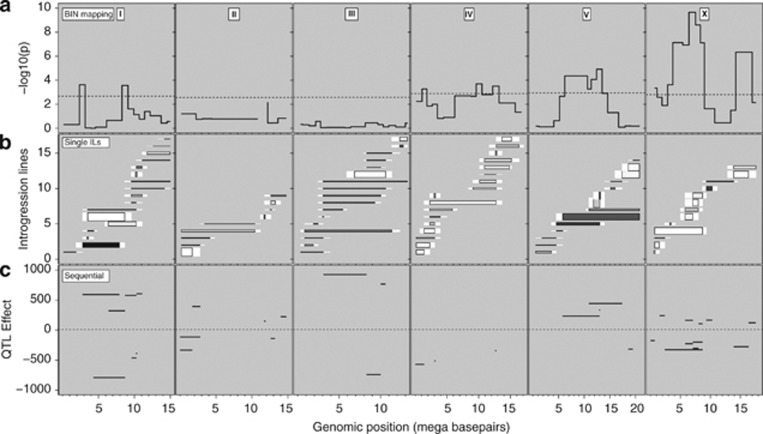
Population size and the number of dauer larvae present were determined at food exhaustion for 85 ILs, representing >96% of the CB4856 genome introgressed into the N2 genome (Doroszuk et al., 2009). Both of the traits were found to be variable between ILs, with the number of dauer larvae varying far more than the population size (Supplementary Figures 5 and 6) and the variation for percentage dauer larvae closely mirroring that observed for dauer larvae numbers (Supplementary Figure 7). The parental strains N2 and CB4856 showed a different number of dauer larvae at food exhaustion. In N2 dauer larvae are always formed, whereas in CB4856 only a very low number of dauer larvae are formed. Bin mapping in the ILs identified 10 dauer larvae formation QTLs on four chromosomes (Figure 2a), eight where the CB4856 allele decreases the number of dauer larvae and two that increase the number of dauer larvae (Table 1). So, even though CB4856 hardly forms any dauer larvae under these circumstances, the isolate still contains alleles that, in an N2 genetic background, have a positive effect on dauer larvae formation. Bin mapping of population size in the ILs did not identify any QTLs, while mapping of the percentage dauer larvae identified six QTLs (Supplementary Figure 8 and Supplementary Table 3), five of which colocalise with QTLs identified in the mapping of the number of dauer larvae (Table 1).
Figure 2.
Three strategies for mapping QTLs affecting dauer larvae development. Chromosomes are indicated by the Roman numeral at the top of the panels. (a) Bin mapping, with chromosome-specific threshold (false discovery rate=0.05) indicated by the horizontal line and significance (−log 10 (P)) per marker is shown in black. (b) Single IL analysis, with ILs shown as horizontal bars. The length indicates the introgression size, the colour the CB4856 allelic effect (darker than background is positive and lighter than background is negative) and the width indicates the significance. For exact values and IL names see Supplementary Figure 6. (c) Sequential IL analysis, with QTLs shown by the black horizontal bars. The y axis shows the effect of the CB4856 allele. For exact values, see Supplementary Table 2.
Table 1. Locations and effect of QTLs detected for the number of dauer larvae.
| Chr. | QTL | Limits | Effect | Sequential IL mapping | IL mapping | Bin mapping | Other traits |
|---|---|---|---|---|---|---|---|
| I | 1 | 1.9–3.5 | P | 1.9–8.7, (N2,02) | 1.9–3.5, (02) | X | +% Dauer |
| 2 | 4.3–9.6 | N | 4.3–9.6, (04,06) | 7.9–9.6, (05,06) | X | −% Dauer | |
| 3 | 10.3–11.1 | P | 8.7–11.1, (06,07) | ||||
| P | 10.3–11.8, (13,14) | ||||||
| 4 | 9.6–11.1 | N | 9.6–11.1, (13) | −% Dauer | |||
| II | 5 | 0–2.8 | N | 0–2.8, (N2,19) | 0–2.8, (19) | −% Dauer | |
| 6 | 1.7–3.4 | P | 1.7–3.4, (19,20) | ||||
| 7 | 12.6–14.0 | N | 12.6–14.0, (25,26) | 11.8–14.0, (26) | |||
| 8 | 11.2–12.6 | P | 11.2–12.6, (24) | −% Dauer | |||
| 9 | 13.2–17.5 | P | 13.2–17.5, (26,27) | ||||
| III | 10 | 2.5–8.3 | P | 2.5–8.3, (40,36) | |||
| 11 | 8.0–10.6 | N | 8.0–10.6, (N2,40) | 8.0–10.6, (40) | −% Dauer | ||
| 12 | 10.0–11.3 | P | 10.0–11.3, (40,41) | ||||
| IV | 13 | 0.8–2.3 | N | 0–2.3, (N2,45) | 0.8–2.3, (45,46,48) | X | −% Dauer |
| 14 | 2.3–3.9 | N | 2.3–3.9, (N2,53) | 2.3–3.9, (52,53) | −% Dauer | ||
| 15 | 9.1–10.9 | N | 9.1–10.9, (52) | X | |||
| 16 | 11.7–13.7 | N | 11.7–13.7, (52,58,59,60) | X | |||
| V | 17 | 11.8–14.0 | P | 10.4–17.4, (75,70) | 11.8–14.0 (69,71,72) | X | +% Dauer |
| P | 11.8–14.0, (N2,72) | ||||||
| 18 | 17.4–19.5 | N | 17.4–19.5, (77,76) | 17.4–19.5, (75,76) | −% Dauer | ||
| X | 19 | 0–1.5 | N | 0–1.5, (78,79,81) | X | −Popn. size | |
| −% Dauer | |||||||
| 20 | 1.5–3.3 | P | 1.5–3.3, (79,80) | ||||
| 21 | 3.3–5.8 | N | 2.4–9.3, (80,81) | 3.3–5.8, (81) | X | −Popn. size | |
| 22 | 5.8–8.0 | N | 5.0–8.0, (N2,83) | 5.8–8.0, (81,83,85,86) | X | −Popn. size | |
| −% Dauer | |||||||
| N | 5.8–8.7, (N2,85) | ||||||
| N | 5.8–9.3, (N2,86) | ||||||
| 23 | 8.7–11.1 | P | 8.7–11.1, (N2,87) | 8.7–11.1, (87) | |||
| 24 | 13.9–17.6 | N | 12.9–17.6, (N2,89) | 13.9–17.6, (89,90) | X | −Popn. size | |
| +% Dauer |
Abbreviations: IL, introgression line; QTL, quantitative trait loci; N, negative; P, positive.
Limits denote the maximum possible QTL region. For the sequential and IL mapping, the limits defined by those analyses and the ILs tested are given. Other traits mapping to these regions are also noted. QTLs defined only by sequential IL mapping are shown in bold.
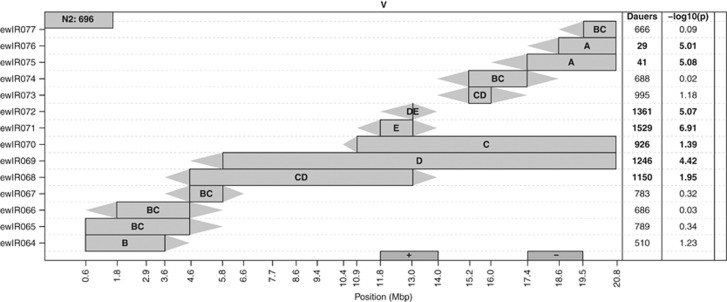
Analysis of individual ILs vs N2 indicated that bin mapping underestimates the number of loci affecting variation in all traits, identifying a further eight QTLs affecting the number of dauer larvae (Figure 2b, Supplementary Figure 9 and Table 1). For example, analysis of the chromosome V ILs identifies an additional QTL affecting dauer larvae development, where the CB4856 genotype acts to decrease the number of dauer larvae (Figure 3). It is particularly noteworthy that several of the QTLs detected individually switch the phenotype from one parental form to another (Supplementary Figures 6 and 7). Similar analyses identify four QTLs affecting population size and an additional 10 QTLs affecting the percentage of dauer larvae (Supplementary Table 3 and Supplementary Figure 9). Again QTLs affecting the percentage of dauer larvae predominantly colocalise with QTLs identified in the mapping of the number of dauer larvae (Table 1). It should be mentioned here that comparison to a common reference N2 does not attempt to predict trait architecture, but merely to localise QTLs that act in the same direction as the parental difference. Sequential IL analysis of the number of dauer larvae identifies, at a genome-wide significance threshold, 20 QTLs (Figure 2c, Table 1 and also see Supplementary Table 4). The bulk of these QTLs (14/20) match those identified by the other methods, but this approach does identify six additional QTLs (Figure 2c and Table 1). As would be expected given the nature of the comparisons made in this analysis, the QTLs found only by this approach are all ones where the CB4856 allele increases the number of dauer larvae (Table 1).
Figure 3.
ILs of chromosome V. The CB4856 introgression per IL is shown by the coloured rectangle. Triangles join adjacent CB4856 and N2 markers. The letters in the introgression box indicate to which of the five significantly different groups an IL belongs. IL names are shown on the left y axis. N2 dauer larvae number is shown in the grey box in the upper left corner. On the right side, the number of dauer larvae is shown, and in bold, if significantly different from N2. Also shown is the significance in −log 10 (P). QTLs are indicated at the x axis by the box marked with ‘+' (positive CB4856) and the box marked with ‘−' (negative CB4856).
QTL confirmation
To test the QTLs detected here, ILs spanning the X chromosome were retested. These data indicated that QTLs 19, 21, 22 and 24, which were all detected via the bin mapping, could be detected in both data sets (Table 2). QTL 22, which was only detected in the sequential mapping via comparison of ewIR079 and ewIR080, was also significant in the retest, with a similar effect size observed in both data sets (Table 2). QTL 23 was not detected in the retest.
Table 2. Retest of the ILs of the X chromosome.
| IL | Batch | Total dauer difference with N2 | Batch | Total dauer difference with N2 | Same sign | Significant in both |
|---|---|---|---|---|---|---|
| ewIR078 | A | −352.5 | B | −442.1 | Y | N |
| ewIR079 | A | −445 | B | −1086.8 | Y | Y |
| ewIR080 | A | 31.7 | B | −511.8 | N | N |
| ewIR081 | A | −623.75 | B | −1193.8 | Y | Y |
| ewIR082 | A | −190.7 | B | −465.2 | Y | N |
| ewIR083 | A | −455 | B | −882.1 | Y | Y |
| ewIR084 | A | −289.4 | B | −893.8 | Y | Y |
| ewIR085 | A | −611.7 | B | −1177.8 | Y | Y |
| ewIR086 | A | −402.5 | B | −156 | Y | N |
| ewIR087 | A | 326.3 | C | −200 | N | N |
| ewIR088 | A | 95 | C | 200 | Y | N |
| ewIR089 | A | −556.1 | C | −355.6 | Y | Y |
| ewIR090 | A | −316.7 | C | −477.8 | Y | Y |
Abbreviations: IL, introgression line; N, no; Y, yes.
Total dauer difference with N2 per batch. Values in bold are significantly different from N2 (from the same batch). Same sign column shows if the difference is in the same direction in both tests. Significant in both shows if the difference is significant in both tests.
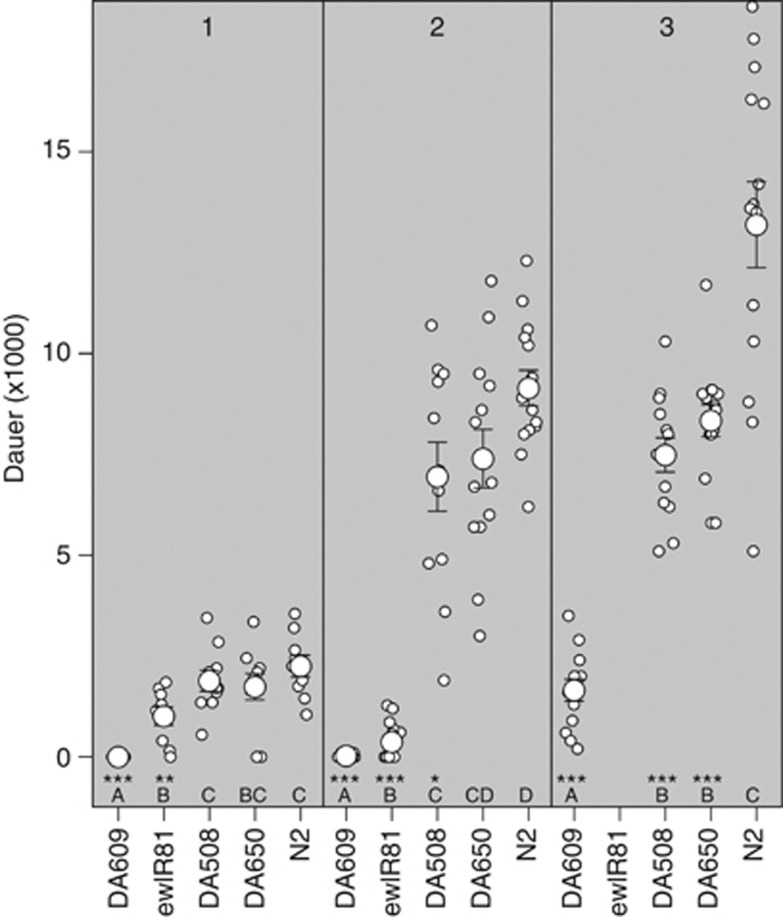
Separate assays were also undertaken to investigate the neuropeptide Y receptor homolog npr-1. This locus is of interest as the lab-derived (McGrath et al., 2009), N2 allele of npr-1 has been found to be important in a number of phenotypes (for example, Doroszuk et al., 2009; McGrath et al., 2009; Reddy et al., 2009; Gaertner et al., 2012). This locus was therefore investigated using the isolate DA650, which contains the g320 npr-1 allele from RC301 backcrossed 10 times into an N2 background (Gloria-Soria and Azevedo, 2008), and isolates DA609 and DA508, which contain different npr-1 loss-of-function alleles (de Bono and Bargmann, 1998). Analysis of these isolates indicates that npr-1 affects dauer larvae formation in growing populations (see Supplementary Table 5 for full model), with alleles that produce a bordering phenotype lowering the number of dauer larvae produced (Figure 4). Interestingly, these analyses indicate that the effect of variation in npr-1 on dauer larvae formation is allele-specific, with a much stronger effect on dauer larvae development observed in DA609 (Figure 4 and Supplementary Table 5).
Figure 4.
Candidate gene analysis. The number of dauer larvae in three npr-1 mutants (alleles DA508, DA609 and DA650), an IL (ewIR081) containing the CB4856 npr-1 allele and N2 in three experiments with increasing dauer numbers are shown (1, 2 and 3). Individual plates are indicated by small white dots, means by large white dots and standard errors by the error bars. Line names shown on the x axis. Letters just above the x axis show the significance group of the IL (A is significantly different from B and B from C, and so on). Significance is shown by the asterisks on the x axis (*P<0.05, **P<0.01, ***P<0.001), denoting isolates that differ from N2.
Discussion
Here, we have demonstrated extensive variation in both dauer larvae formation and population size in growing populations of C. elegans (Figure 1). Interestingly, dauer larvae formation is significantly higher in many of the wild isolates studied than it is in N2. This contrasts with previous observations that, in standard dauer larvae development assays, N2 had higher rates of dauer larvae formation than all tested wild isolates (Viney et al., 2003; Harvey et al., 2008). This may imply that wild isolates are more sensitive to their own pheromone than they are to N2 pheromone (as used in Viney et al. (2003) and Harvey et al. (2008)). Alternatively, this may reflect variation between the isolates in aspects of their biology that are not captured by standard dauer larvae development assays. For example, dauer larvae formation in growing populations would be affected by variation between isolates in population growth rates and by differences in rates of pheromone production, traits that would not affect the results of standard dauer larvae development assays. This result also makes an interesting comparison to the findings of Mayer and Sommer (2011), who showed that 13 out of 16 tested strains of Pristionchus pacificus produced a pheromone that induced higher dauer larvae formation in different genotypes. This suggests that P. pacificus is attempting to induce precocious dauer larvae formation in other strains (Mayer and Sommer, 2011). Given these differences, it would be interesting to investigate how growing populations of C. elegans respond to pheromone from different isolates and to investigate the properties of mixed-genotype growing populations in both C. elegans and P. pacificus.
Mapping of dauer larvae development and of population size in the ILs identified a total of 24 QTLs for dauer larvae development (Table 1), with these QTLs found to be robust and reproducible (Table 2). This represents an unprecedented number of QTLs for a single trait from one mapping panel in C. elegans. The identification of these QTLs in the ILs provides a simplified and tractable approach to further genetic analysis of these loci. The developmental choice between dauer and non-dauer larvae development is based on assessment of the levels of various ascarosides (Jeong et al., 2005; Butcher et al., 2007; Pungaliya et al., 2009; Park et al., 2012), on food availability and temperature (Golden and Riddle, 1984) and potentially on an assessment of individual quality (Harvey and Orbidans, 2011). Within growing populations, variation between genotypes in the number of dauer larvae could therefore be a consequence of variation in traits that affect the likelihood of dauer larvae development, the perception of the environment, or the way in which the population grows.
One potentially confounding factor is that ILs can reveal the presence of Dobzhansky–Muller incompatibilities. In this case, IL regions displaying Dobzhansky–Muller incompatibilities might reduce population growth rates and hence affect dauer larvae development via effects on pheromone accumulation. This would be manifested as additional QTLs where the CB4856 allele negatively affected both population size and dauer larvae development. Most QTLs do not show such effects, but QTLs displaying these characteristics are detected on the X chromosome (Table 1) and a large number of negative effect QTLs are also seen on chromosome IV.
Given the large number of QTLs detected (Table 1) and the large number of loci known to be able to, either singly or in combination, affect dauer larvae development, potential candidate genes underlying most QTLs were not investigated in detail (cf. Terpstra et al., 2010). It is however clear that variation at the npr-1 loci does affect dauer larvae development in growing populations (Figure 4) and it is likely that it is variation in npr-1 that underlies QTL 21. This effect of social behaviour on dauer larvae formation is a clear example of an indirect effect on dauer larvae development, particularly given that it has previously been shown that there is no effect of bordering on dauer larvae development in standard dauer assays (Viney et al., 2003). As lower numbers of dauer larvae are observed in the npr-1 mutants, the opposite result to what one might predict if clumping behaviour produces high local pheromone concentrations and low local food levels, the reduction of dauer larvae development may be similar to the effects on dauer larvae development seen as a consequence of reduced maternal food availability (Harvey and Orbidans, 2011), that is, a harsher maternal environment resulting in the production of progeny that are less likely to develop as dauer larvae. The reduced numbers of dauer larvae formed in the npr-1 mutants also suggests that there is significant genetic variation which would act to increase dauer larvae development in many wild isolates (Figure 1), as the majority of these lines display the clumping phenotype.
There is a strong possibility that variation between isolates in dauer larvae formation is a consequence of variation in the chemoreceptors required to sense both pheromone and food signals. As ascaroside signalling is known to be complex, with some receptors showing excessive specificity and others able to respond to a range of ascarosides (Kim et al., 2009; McGrath et al., 2011; Park et al., 2012), variation in receptor sensitivity would allow fine-tuning of the dauer larvae development decision. Interestingly, QTL 24 on the X chromosome colocalises with individual markers identified as being associated with variation in dauer larvae formation between N2 and DR1350 (Harvey et al., 2008) and contains srg-36 and srg-37, genes known to encode receptors for ascaroside C3, one of the components of C. elegans pheromone (McGrath et al., 2011).
Currently available isolates of C. elegans are characterised by a limited number of large and relatively common shared haplotypes on four chromosomes (Andersen et al., 2012), with population genetic modelling suggesting that this pattern of variation is a consequence of recent chromosome-scale selective sweeps (Andersen et al., 2012). Given the association between C. elegans and rotting fruit (Kiontke et al., 2011), it is tempting to speculate that human-induced changes in the prevalence and morphology of various fruits may be related to these selective sweeps. If this is the case, then it is possible that dauer larvae development in growing populations is one of the traits under selection. The large number of QTLs identified here also contrasts with the three dauer larvae development QTLs identified by Harvey et al. (2008) in an analysis of recombinant inbred lines produced from crosses between N2 and DR1350. Given the genetic isolation of CB4856 from DR1350 and N2 it would therefore be very informative to determine if the more complex architecture revealed here between CB4856 and N2 is a consequence of the differences in methodologies (analysis of ILs and of growing populations) or reflects differences due to selective history. It is, however, clear that it would be very worthwhile developing and analysing additional genome-wide IL sets in C. elegans.
Data archiving
Data are available at WormQTL (http://www.wormqtl.org) (Snoek et al., 2013) with matrix IDs 44–49.
Acknowledgments
We thank Helen Orbidans for assistance with the IL assays, the Caenorhabditis Genetics Centre and Marie-Anne Félix for strains, K Joeri Van der Velde and Morris A Swertz from the Groningen Bioinformatics Centre, University of Groningen for assistance with WormQTL and WormBase (www.wormbase.org) for being an easily accessible and versatile source of information. JG and SH were supported by Canterbury Christ Church University. LBS and JK were funded by the ERASysbio-plus ZonMW project GRAPPLE—Iterative modelling of gene regulatory interactions underlying stress, disease and ageing in C. elegans (project no. 90201066), and NEMADAPT, NWO-ALW (project no. 855.01.151).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, et al. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44:285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher RA, Fujita M, Schroeder FC, Clardy J. Small-molecule pheromones that control dauer development in Caenorhabditis elegans. Nat Chem Biol. 2007;3:420–422. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]
- De Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- Doroszuk A, Snoek LB, Fradin E, Riksen J, Kammenga J. A genome-wide library of CB4856/N2 introgression lines of Caenorhabditis elegans. Nucleic Acids Res. 2009;37:e110. doi: 10.1093/nar/gkp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvin M, Snoek LB, Frejno M, Klemstein U, Kammenga JE, Poulin GB. A fitness assay for comparing RNAi effects across multiple C. elegans genotypes. BMC Genom. 2011;12:510. doi: 10.1186/1471-2164-12-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix MA, Duveau F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 2012;10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertner BE, Parmenter MD, Rockman MV, Kruglyak L, Phillips PC. More than the sum of its parts: a complex epistatic network underlies natural variation in thermal preference behavior in Caenorhabditis elegans. Genetics. 2012;192:1533–1542. doi: 10.1534/genetics.112.142877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloria-Soria A, Azevedo RB. npr-1 regulates foraging and dispersal strategies in Caenorhabditis elegans. Curr Biol. 2008;18:1694–1699. doi: 10.1016/j.cub.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Golden JW, Riddle DL. The Caenorhabditis elegans dauer larva: developmental effects of pheromone, food, and temperature. Dev Biol. 1984;102:368–378. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- Green JWM, Harvey SC. Caenorhabditis elegans dauer larvae development in growing populations. Nematology. 2012;14:165–173. [Google Scholar]
- Harvey SC. Non-dauer larval dispersal in Caenorhabditis elegans. J Exp Zool B. 2009;312:224–230. doi: 10.1002/jez.b.21287. [DOI] [PubMed] [Google Scholar]
- Harvey SC, Orbidans HE. All eggs are not equal: the maternal environment affects progeny reproduction and developmental fate in Caenorhabditis elegans. PLoS ONE. 2011;6:e25840. doi: 10.1371/journal.pone.0025840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey SC, Shorto A, Viney ME. Quantitative genetic analysis of life-history traits of Caenorhabditis elegans in stressful environments. BMC Evol Biol. 2008;8:15. doi: 10.1186/1471-2148-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu PJ.2007DauerIn: The C. elegans Research Community edsWormBook 1–19. . http://www.wormbook.org/ . 17988074
- Izrayelit Y, Srinivasan J, Campbell SL, Jo Y, von Reuss SH, Genoff MC, et al. Targeted metabolomics reveals a male pheromone and sex-specific ascaroside biosynthesis in Caenorhabditis elegans. ACS Chem Biol. 2012;7:1321–1325. doi: 10.1021/cb300169c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Kim K, Neal SJ, Macosko E, Kim D, Butcher RA, et al. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron. 2012;75:585–592. doi: 10.1016/j.neuron.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong PY, Jung M, Yim YH, Kim H, Park M, Hong E, et al. Chemical structure and biological activity of the Caenorhabditis elegans dauer-inducing pheromone. Nature. 2005;433:541–545. doi: 10.1038/nature03201. [DOI] [PubMed] [Google Scholar]
- Kammenga JE, Doroszuk A, Riksen JA, Hazendonk E, Spiridon L, Petrescu AJ, et al. A Caenorhabditis elegans wild type defies the temperature-size rule owing to a single nucleotide polymorphism in tra-3. PLoS Genet. 2007;3:e34. doi: 10.1371/journal.pgen.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga JE, Phillips PC, De Bono M, Doroszuk A. Beyond induced mutants: using worms to study natural variation in genetic pathways. Trends Genet. 2008;24:178–185. doi: 10.1016/j.tig.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Kim K, Sato K, Shibuya M, Zeiger DM, Butcher RA, Ragains JR, et al. Two chemoreceptors mediate developmental effects of dauer pheromone in C. elegans. Science. 2009;326:994–998. doi: 10.1126/science.1176331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke KC, Félix MA, Ailion M, Rockman MV, Braendle C, Pénigault JB, et al. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011;11:339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JA, et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2:e222. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Breitling R, Snoek LB, van der Velde KJ, Swertz MA, Riksen JAG, et al. Global genetic robustness of the alternative splicing machinery in Caenorhabditis elegans. Genetics. 2010;186:405–410. doi: 10.1534/genetics.110.119677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MG, Sommer RJ. Natural variation in Pristionchus pacificus dauer formation reveals cross-preference rather than self-preference of nematode dauer pheromones. Proc Biol Sci. 2011;278:2784–2790. doi: 10.1098/rspb.2010.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, et al. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath PT, Xu Y, Ailion M, Garrison JL, Butcher RA, Bargmann CI. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature. 2011;477:321–325. doi: 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, O'Doherty I, Somvanshi RK, Bethke A, Schroeder FC, Kumar U, et al. Interaction of structure-specific and promiscuous G-protein-coupled receptors mediates small-molecule signaling in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109:9917–9922. doi: 10.1073/pnas.1202216109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pungaliya C, Srinivasan J, Fox BW, Malik RU, Ludewig AH, Sternberg PW, et al. A shortcut to identifying small molecule signals that regulate behavior and development in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106:7708–7713. doi: 10.1073/pnas.0811918106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Snoek LB, Riksen JA, Bevers RP, Kammenga JE. Genetic variation for stress-response hormesis in C. elegans lifespan. Exp Gerontol. 2012;47:581–487. doi: 10.1016/j.exger.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Shao H, Sinasac DS, Burrage LC, Hodges CA, Supelak PJ, Palmert MR, et al. Analyzing complex traits with congenic strains. Mamm Genome. 2010;21:276–286. doi: 10.1007/s00335-010-9267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snoek LB, Van der Velde KJ, Arends D, Li Y, Beyer A, Elvin M, et al. WormQTL—public archive and analysis web portal for natural variation data in Caenorhabditis spp. Nucleic Acids Res. 2013;41:D738–D743. doi: 10.1093/nar/gks1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PE, et al. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiuk SJ, Scott MJ, Grant WN. Developmental plasticity and the evolution of parasitism in an unusual nematode, Parastrongyloides trichosuri. EvoDevo. 2012;3:1. doi: 10.1186/2041-9139-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle T.1999MethodsIn: Hope IA (ed)C. elegans: A Practical Approach Oxford University Press: Oxford, UK; 51–67. [Google Scholar]
- Sudhaus W. Preadaptive plateau in Rhabditida (Nematoda) allowed the repeated evolution of zooparasites, with an outlook on evolution of life cycles within Spiroascarida. Palaeodiversity. 2010;3:117–130. [Google Scholar]
- Terpstra IR, Snoek LB, Keurentjes JJ, Peeters AJ, van den Ackerveken G. Regulatory network identification by genetical genomics: signalling downstream of the Arabidopsis receptor-like kinase ERECTA. Plant Physiol. 2010;154:1067–1078. doi: 10.1104/pp.110.159996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viney ME, Gardner MP, Jackson JA. Variation in Caenorhabditis elegans dauer larva formation. Dev Growth Differ. 2003;45:389–396. doi: 10.1046/j.1440-169x.2003.00703.x. [DOI] [PubMed] [Google Scholar]
- Viñuela A, Snoek LB, Riksen JA, Kammenga JE. Genome-wide gene expression regulation as a function of genotype and age in C. elegans. Genome Res. 2010;20:929–937. doi: 10.1101/gr.102160.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viñuela A, Snoek LB, Riksen JAG, Kammenga JE. Aging uncouples heritability and expression-QTL in Caenorhabditis elegans. G3. 2012;2:597–605. doi: 10.1534/g3.112.002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.