Abstract
Identifying factors and the extent of their roles in the differentiation of populations is of great importance for understanding the evolutionary process in which a species is involved. Ctenomys minutus is a highly karyotype–polymorphic subterranean rodent, with diploid numbers ranging from 42 to 50 and autosomal arm numbers (ANs) ranging from 68 to 80, comprising a total of 45 karyotypes described so far. This species inhabits the southern Brazilian coastal plain, which has a complex geological history, with several potential geographical barriers acting on different time scales. We assessed the geographical genetic structure of C. minutus, examining 340 individuals over the entire distributional range and using information from chromosomal rearrangements, mitochondrial DNA (mtDNA) sequences and 14 microsatellite loci. The mtDNA results revealed seven main haplogroups, with the most recent common ancestors dating from the Pleistocene, whereas clustering methods defined 12 populations. Some boundaries of mtDNA haplogroups and population clusters can be associated with potential geographical barriers to gene flow. The isolation-by-distance pattern also has an important role in fine-scale genetic differentiation, which is strengthened by the narrowness of the coastal plain and by common features of subterranean rodents (that is, small fragmented populations and low dispersal rates), which limit gene flow among populations. A step-by-step mechanism of chromosomal evolution can be suggested for this species, mainly associated with the metapopulation structure, genetic drift and the geographical features of the southern Brazilian coastal plain. However, chromosomal variations have no or very little role in the diversification of C. minutus populations.
Keywords: allopatry, isolation by distance, phylogeography, metapopulation, Pleistocene, Holocene
Introduction
The role of chromosomal rearrangements in the speciation process has been widely discussed over several decades. Traditional chromosomal speciation models propose that the fitness of heterozygous carriers is diminished owing to meiotic abnormalities, leading to reductions in gene flow between chromosomally divergent populations and thus facilitating reproductive isolation (White, 1978). However, recent models suggest that chromosomal changes reduce gene flow and accelerate the divergence between populations by reducing recombination in heterokaryotypes and extending the effects of linking isolated genes or locally adapted alleles, and not because of underdominance (Rieseberg, 2001). However, it is questionable whether all the kinds of chromosomal rearrangements could be involved in the speciation process (Baker and Bickham, 1986).
On the other hand, the role of geographical factors in the process of biological diversification is increasingly understood, and its contribution as an evolutionary force shaping the genetic structure of the species is broadly accepted (Barraclough and Vogler, 2000). The geographical distribution of a species depends on, among other intrinsic and extrinsic factors, when and where it originated and what kinds of geographical barriers it encountered. Mountain chains, deserts, habitat fragmentation or watercourses can sometimes be insuperable geographical barriers for individuals that reach them, reducing or preventing gene flow between populations located on opposite sides, and thus accelerating the process of divergence (Slatkin, 1987). Steinberg and Patton (2000) suggested that the geographical context on the process of speciation in subterranean rodents is trivial, in which the vicariance or peripheral isolation may account for the allopatric divergence.
The burrowing rodents of the genus Ctenomys, popularly known as tuco-tucos, comprise approximately 60 living species, showing impressive intra- and interspecific karyotypic variation (Reig et al., 1990; Woods and Kilpatrick, 2005). Some of their characteristics, such as low rates of adult dispersal (Busch et al., 2000) and their distribution in relatively small and fragmented populations, foster the establishment of small genetic units where genetic variation is low and interpopulation divergence is high (Wlasiuk et al., 2003). These characteristics make the species more vulnerable to stochastic demographic factors and changes in their habitats (Patton et al., 1996), and also favor the fixation of new chromosomal rearrangements, which have been considered to have an important role in the diversification of ctenomyids (Reig et al., 1990; Lessa and Cook, 1998). This genus provides an excellent study system for exploring the role of discontinuities in the environment and chromosomal rearrangements in the speciation process (Mora et al., 2006, 2007; Fernández-Stolz, 2007; Tomasco and Lessa, 2007) and ultimately for identifying the important genes involved in reproductive isolation of these taxa. Thus, the investigation of population genetic structure presented in this manuscript is a first step in the process of exploring these large questions.
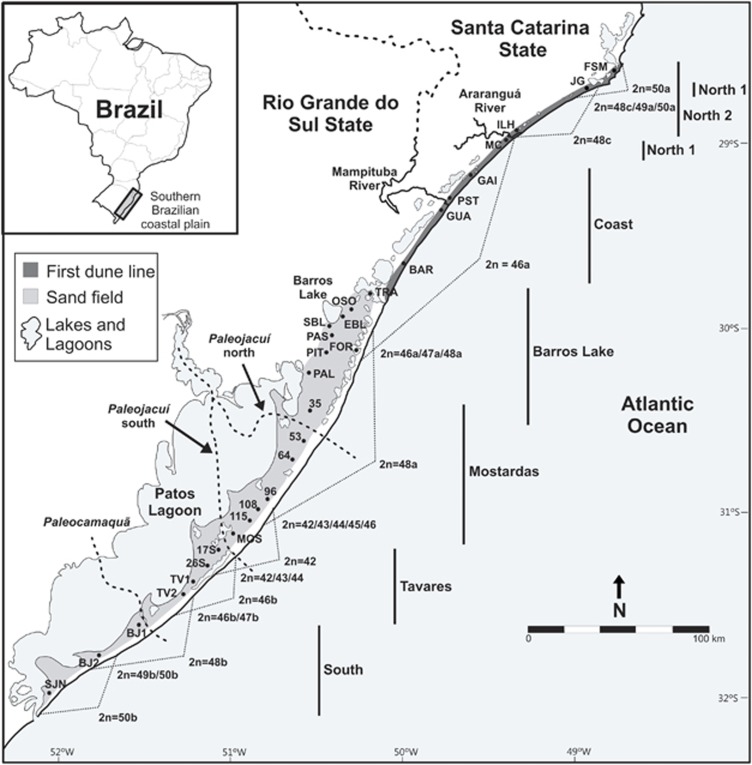
Ctenomys minutus Nehring, 1887 has a narrow distribution on the southern Brazilian coastal plain. The landscape of the region is a mosaic of innumerable lakes and lagoons, rivers, swamps, sand fields and dunes. During the Quaternary Period, fluctuations in the Atlantic Ocean sea level produced large lateral displacements of the shoreline, originating four barrier–lagoon systems that shaped the present coastline. The first three barrier–lagoon systems originated during the Pleistocene Period (hereafter called Barriers I, II and III), and the fourth system was formed in the Holocene Period (Barrier IV; Supplementary Figure S1; Tomazelli et al., 2000).
C. minutus inhabits only the sand fields (corresponding to Barriers II and III) in the southern portion of its distribution; and in the north, these tuco-tucos preferentially occupy the first dune line (Barrier IV; Figure 1 and Supplementary Figure S1; Freygang et al., 2004). The range includes many watercourses that have existed in different periods, affecting the local rodent populations over different time scales. According to studies conducted by Weschenfelder et al. (2008) and Baitelli (2012), three major paleochannels existed on the coastal plain (Paleojacuí north, Paleojacuí south and Paleocamaquã), but they were completely closed by sediments during the Pleistocene and Holocene Periods. Nowadays, the Patos Lagoon and the Araranguá and Mampituba Rivers form major discontinuities in the landscape of the region, and all of them flow into the Atlantic Ocean (Figure 1).
Figure 1.
Sampling localities of Ctenomys minutus, and the distribution of the parental karyotypes and intraspecific hybrid zones. The seven mtDNA haplogroups are highlighted in the figure. BAR, Barco Beach; BJ1, Bujuru 1; BJ2, Bujuru 2; EBL, East Barros Lake; FOR, Fortaleza Lake; FSM, Farol de Santa Marta; GAI, Gaivota Beach; GUA, Guarita Beach; ILH, Ilhas; JG, Jaguaruna; MC, Morro dos Conventos; MOS, Mostardas; PAL, Palmares do Sul; PAS, Passinhos; PIT, Pitangueira; PST, Passo de Torres; OSO, Osório; 35, road km 35; 53, road km 53; 64, road km 64; 96, road km 96; 108, road km 108; 115, road km 115; SBL, South Barros Lake; 17S, 17 km south of Mostardas; 26S, 26 km south of Mostardas; SJN, São José do Norte; TRA, Tramandaí TV1, Tavares 1; TV2, Tavares 2.
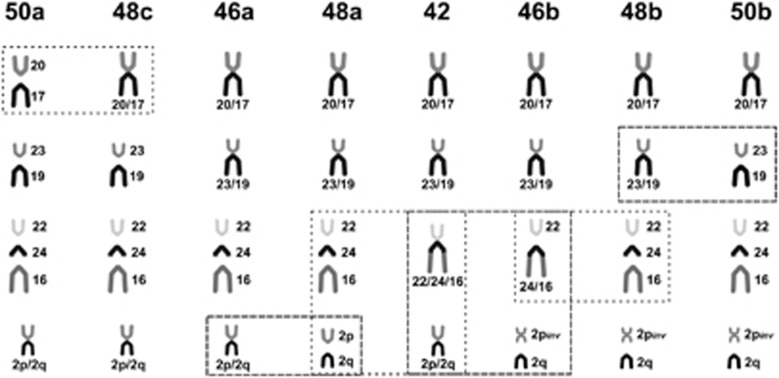
C. minutus is a highly karyotype–polymorphic species, with diploid numbers ranging from 42 to 50 and autosomal ANs from 68 to 80. The chromosomal variation consist of eight parapatrically distributed parental karyotypes (2n=50a, 48c, 46a, 48a, 42, 46b, 48b and 50b) and six intraspecific hybrid zones, among them: (i) 50a × 48c=49a; (ii)46a × 48a=47a; (iii)48a × 42=43, 44, 45, 46; (iv) 42 × 46b=43, 44, 45; (v) 46b × 48b=47b; and (vi) 48b × 50b=49b. The intraspecific hybrid zones were determined based on diploid numbers and chromosomal rearrangements observed through G-band patterns compared with the adjacent parental karyotypes (Freitas, 1997; Gava and Freitas, 2002, 2003; Freygang et al., 2004; Castilho et al., 2012; this study). The distribution of these karyotypes shows 2n=50 at both ends of the geographical range, whereas toward the middle of the distribution, it is progressively reduced to 2n=42 through Robertsonian rearrangements, tandem fusions/fissions and a pericentric inversion, which originated the distinct karyotypic systems ‘a/c' and ‘b' see Figures 1 and 2 (Freitas, 1997; Freygang et al., 2004).
Figure 2.
Comparison of chromosomal rearrangements found among parental karyotypes based on the G-band patterns. The karyotype 2n=50a, AN=76 was used as standard to compare the chromosomal rearrangements in the other karyotypes. Squares highlight the main chromosomal rearrangements involved in the intraspecific hybrid zones. Modified from Freitas (1997), Gava and Freitas (2003) and Freygang et al. (2004).
Considering the several potential water barriers acting on different time scales along the southern Brazilian coastal plain, and since tuco-tucos seem to be very poor swimmers (Reig et al., 1990), we investigated here if perennial streams could lead to a process of genetic differentiation between C. minutus populations in allopatry and also if geographical features could have influenced the fixation of chromosomal differences. We also explored the possibility that the chromosomal rearrangements found in this species could represent incipient genetic barriers to reduce gene flow between chromosomally divergent populations. In this contribution, we attempted to examine both historical and contemporary processes that shaped and continue to shape the geographical genetic structure of C. minutus, regarding the coastal plain dynamics, by means of the mitochondrial DNA (mtDNA) control region (CR) and cytochrome c oxidase subunit I sequences (COI), 14 microsatellite loci, using specimens sampled and karyotyped on a fine scale.
Materials and methods
Sample collection and cytogenetics
The samples analyzed included 3 to 25 specimens of C. minutus from each of 30 localities, totaling 276 individuals analyzed for mtDNA, and 340 individuals analyzed for microsatellite loci, across the species' entire distributional range (see Table 1 and Figure 1). Most of the specimens were collected and karyotyped previously in studies by Freitas (1997), Gava and Freitas (2002, 2003), Freygang et al. (2004) and Castilho et al. (2012) (Table 1). The tissue samples were preserved in 95% ethanol and stored at −20 °C in the collection of the Laboratório de Citogenética e Evolução of the Departamento de Genética of the Universidade Federal do Rio Grande do Sul, Brazil. Additional specimens were collected and karyotyped for this study and are highlighted in Table 1. For these specimens, the mitotic preparations were obtained from bone marrow, according to Ford and Hamerton (1956). The diploid and autosomal numbers were determined by analyses of at least 20 metaphase spread cells stained with Giemsa.
Table 1. Genetic structure and variability observed among the localities (abbreviations within parentheses) sampled for Ctenomys minutus for both mitochondrial DNA sequences and nuclear microsatellite markers.
| Localities | 2n(AN) | mtDNA haplogroup | No. S | CC | Structure clusters | Tess clusters | No. G | A | F IS |
|---|---|---|---|---|---|---|---|---|---|
| Farol de Santa Marta (FSM) | 50a (76) | North 2 | 15a | CC48(2), CC49(8), CC50(3), CC51(1), CC52(1) | I | A | 15a | 2.57 | 0.038 |
| Jaguaruna (JG) | 48c (76), 49a (76), 50a (76) | North 1, North 2 | 12b+5a | CC46(14), CC47(3) | II | B | 12b+5a | 4.14 | 0.087 |
| Ilhas (ILH) | 48c (76) | North 2 | 15a | CC45(15) | II | B | 15a | 2.93 | 0.200c |
| Morro dos Conventos (MC) | 46a (76) | North 1 | 12b | CC43(6), CC44(6) | II, III | B | 14b | 4.36 | 0.467 |
| Gaivota Beach (GAI) | 46a (76) | Coast | 10b | CC42(10) | III | C | 19 | 5.21 | 0.218 |
| Passo de Torres (PST) | 46a (76) | Coast | 3b | CC39(1), CC40(1), CC41(1) | III, IV | C,D,H | 3b | 3.57 | 0.404 |
| Guarita Beach (GUA) | 46a (76) | Coast | 12b | CC38(12) | IV | D | 18b | 3.71 | 0.443 |
| Barco Beach (BAR) | 46a (76) | Coast | 7b | CC35(3), CC36(1), CC37(3) | IV | D | 7b | 3.71 | 0.071 |
| Tramandaí (TRA) | 46a (76) | Barros Lake | 2b | CC31(1), CC34(1) | V | E | 4b | 3.29 | 0.464 |
| Osório (OSO) | 46a (76) | Barros Lake | 9b | CC25(3), CC31(2), CC32(3), CC33(1) | V | E | 25b | 6.36 | 0.198c |
| East Barros Lake (EBL) | 46a (76), 47a (76), 48a (76) | Barros Lake | 5b | CC27(5) | V | E | 13b | 3.64 | 0.295c |
| South Barros Lake (SBL) | 47a (76), 48a (76) | Barros Lake | 11b | CC26(9), CC28(2) | VI | F | 13b | 3.50 | 0.447 |
| Passinhos (PAS) | 48a (76) | Barros Lake | 4b | CC26(4) | VI, VII | F | 9b | 3.71 | 0.224 |
| Pitangueira (PIT) | 48a (76) | Barros Lake | 5b | CC24(2), CC25(3) | VI, VII | F | 5b | 2.79 | -0.041 |
| Fortaleza Lake (FOR) | 46a (76), 47a (76), 48a (76) | Barros Lake | 12b | CC26(1), CC29(1), CC30(10) | VII | G | 10b | 4.00 | 0.130 |
| Palmares do Sul (PAL) | 48a (76) | Barros Lake | 7b | CC21(5), CC23(1), CC24(1) | VII | G | 7b | 3.64 | 0.029 |
| Road km 35 (35) | 48a (76) | Barros Lake Mostardas | 4b | CC20(3), CC22(1) | VII | G | 4b | 3.07 | -0.137 |
| Road km 53 (53) | 48a (76) | Mostardas | 5b | CC17(2), CC18(2), CC19(1) | VIII | H | 5b | 3.21 | -0.131 |
| Road km 64 (64) | 48a (76) | Mostardas | 3b | CC16(2), CC17(1) | VIII | H | 3b | 2.86 | 0.424 |
| Road km 96 (96) | 48a (76) | Mostardas | 6b | CC13(4), CC15(2) | VIII | H | 7b | 4.71 | 0.287c |
| Road km 108 (108) | 42 (68,69,70,71,72,73,74), 43 (70,72,73,74,75), 44 (72,73,74,75,76), 45 (74,76,78,80), 46 (71,74,76,77,78) | Mostardas | 15b | CC11(15) | VIII, IX | H/I | 17b | 5.07 | 0.149 |
| Road km 115 (115) | 42 (74) | Mostardas | 3b | CC11(2), CC13(1) | IX | I | 3b | 3.21 | 0.244 |
| Mostardas (MOS) | 42 (74) | Mostardas | 16b | CC11(5), CC12(1), CC13(6), CC14(4) | IX | I | 21b | 6.21 | 0.177c |
| 17 km south of Mostardas (17S) | 42 (74), 43 (74) | Tavares | 10a | CC9(8), CC10(2) | X | J | 11a | 5.43 | 0.087 |
| 26 km south of Mostardas (26S) | 42 (74), 43 (70,72,74), 44 (74), 45 (76), 46b (76) | Tavares | 4b+11a | CC4(10), CC8(3), CC9(2) | X | J | 7b+11a | 5.64 | 0.023 |
| Tavares 1 (TV1) | 46b (76) | Tavares | 14b | CC4(10), CC5(1), CC6(3) | X | J | 14b | 4.93 | 0.645 |
| Tavares 2 (TV2) | 46b (76), 47b(76) | Tavares | 6b | CC4(2), CC6(3), CC7(1) | X | J | 6b | 3.07 | -0.031 |
| Bujuru 1 (BJ1) | 48b (76,78) | South | 12b | CC3(12) | XI | K | 15b | 3.29 | 0.001 |
| Bujuru 2 (BJ2) | 49b (76,77), 50b(76) | South | 9b | CC2(9) | XI | K | 9b | 2.50 | 0.174 |
| São José do Norte (SJN) | 50b (76,77) | South | 12b | CC1(12) | XII | L | 13b | 2.07 | 0.134 |
| 30 localities | 45 karyotypes | 7 haplogroups | 276 | 52 haplotypes | 12 clusters | 12 clusters | 340 | 3.88 |
Abbreviations: A, mean number of alleles per locality; CC, number of specimens per haplotype of concatenated mtDNA in parentheses; FIS, within-population structure; 2n(AN), diploid and autosomal arm numbers found within the localities, in this and other studies; mtDNA, mitochondrial DNA; No. G, number of samples genotyped; No. S, number of samples sequenced.
Only most representative clusters of Structure and Tess were cited in the localities showing specimens with admixed ancestries.
Number of samples collected for this study.
Number of samples collected and karyotyped previously by Freitas (1997), Gava and Freitas (2002, 2003), Freygang et al. (2004) and Castilho et al. (2012).
Statistical significance (P<0.05).
DNA amplification, sequencing and genotyping
Total DNA was extracted using a standard phenol:chloroform protocol (Sambrook and Russel, 2001). Two fragments of mtDNA were amplified by polymerase chain reaction: Part of HVS1 from the CR was amplified using the primers TucoPro (5′-TTCTAATTAAACTATTTCTTG-3′ Tomasco and Lessa, 2007) and TDKD (5′-CCTGAAGTAGGAACCAGATG-3′ Kocher et al., 1989), following a protocol modified from Tomasco and Lessa (2007). The cytochrome c oxidase subunit I (COI) followed the protocols suggested in http://www.barcoding.si.edu/DNABarCoding.htm, using the primers LCO-1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO-2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′ Folmer et al., 1994).
The polymerase chain reaction products were purified using Exonuclease I and Shrimp Alkaline Phosphatase (GIBCO-BRL Life Sciences/Invitrogen, Carlsbad, CA, USA), following the guidelines of the suppliers, and sequenced in an ABI 3730 (Applied Biosystems, Foster City, CA, USA) automated sequencer, using the forward primers TucoPro and LCO-1490.
In addition, 14 microsatellite loci were amplified, using fluorescently labeled primers, isolated from the Argentinean species C. haigi (Hai2, Hai3, Hai4, Hai5, Hai6, Hai9, Hai10, Hai12; Lacey et al., 1999) and C. sociabilis (Soc1, Soc2, Soc3, Soc4, Soc5, Soc6; Lacey, 2001). Polymerase chain reaction amplifications were carried out following the protocols described by Lacey et al. (1999) and Lacey (2001). The genotypes were obtained in the sequencer ABI 3730, mixing two primer pairs with different labels, per reaction. To define the allele sizes, we used the program PeakScanner 1.0 (http://www.appliedbiosystems.com).
Data analyses
Genetic diversity
Sequence electropherograms were visually inspected using Chromas 2.33 (http://www.technelysium.com.au/chromas.html), and aligned using the Clustal W algorithm with default options, implemented in Mega 4.0.2 (Tamura et al., 2007). All analyses were performed with the three data sets: CR and COI separately, and a concatenated data set of CR+COI (CC); however, mainly the CC results will be presented here. Measures of mtDNA diversity, such as average number of nucleotide differences (k), average number of nucleotide differences per site between all pairs of sequences (π), definitions of haplotypes (H) and haplotype diversity (Hd), were calculated with DnaSP 5.00.03 (Librado and Rozas, 2009).
We used Arlequin 3.5.1.2 (Excoffier and Schneider, 2005) and Genepop 4.0 (Rousset, 2008) to test deviations from the Hardy–Weinberg equilibrium, to calculate the observed and expected heterozygosis (Ho and He) and to perform the analyses of linkage equilibrium (LE) for microsatellite loci. The significance levels (α=0.05) of Hardy–Weinberg equilibrium and LE were adjusted using sequential Bonferroni corrections. Genotyping errors were tested with Micro-Checker (van Oosterhout et al., 2004), under a 95% confidence interval. The measurements of microsatellite diversity were assessed using Arlequin 3.5.1.2.
Genetic differentiation
Wright's F-statistics were applied to analyze the within- (FIS) and between- (FST) population structures for microsatellite data, using Arlequin. Pairwise RST population estimates (Slatkin, 1995) was also calculated, using Genepop 4.0. Localities with fewer than five specimens were excluded from pairwise comparisons, and sequential Bonferroni corrections were applied to adjust the significance levels (α=0.05).
The program Arlequin 3.5.1.2 was used to perform analyses of molecular variance (AMOVA) to investigate two different scenarios of hypothesized population subdivisions, regarding the karyotypic variation and the potential geographical barriers. All the specimens collected in intraspecific karyotypic hybrid zones were excluded from both analyses. First, the samples were subdivided into eight groups according to parental karyotypes: (i) only specimens with 2n=50a, from Farol de Santa Marta (FSM) (the complete names of the localities and the corresponding abbreviations are listed in Table 1); (ii) specimens with 2n=48c, from Ilhas (ILH); (iii) 2n=46a, all specimens from Morro dos Conventos (MC), Gaivota Beach (GAI), Passo de Torres (PST), Guarita Beach (GUA), Barco Beach (BAR), Tramandaí (TRA) and Osório (OSO); (iv) 2n=48a, from Passinhos (PAS), Pitangueira (PIT), Palmares do Sul (PAL), road km 35 (35), road km 53 (53), road km 64 (64) and road km 96 (96); (v) 2n=42, from road km 115 (115) and Mostardas (MOS); (vi) 2n=46b, from Tavares 1 (TV1); (vii) 2n=48b, from Bujuru 1 (BJ1); and (viii) specimens with 2n=50b, from São José do Norte (SJN). The same data set was used in a second test, but the samples were separated into seven groups, considering present and possible former geographical barriers: (i) specimens from FSM and ILH, isolated to the south by the Araranguá River; (ii) specimens from MC, GAI and PST, isolated to the south by the Mampituba River; (iii) specimens collected in GUA and BAR, delimited to the south by the transition between sand fields and dunes; (iv) all specimens from TRA, OSO, PAS, PIT, PAL and 35, isolated in the past to the south by the Paleojacuí north; (v) samples from 53, 64, 96, 115 and MOS, isolated in the past to the south by the Paleojacuí south; (vi) TV1, isolated in the past to the south by the Paleocamaquã; and (vii) specimens sampled in BJ1 and SJN, isolated in the southernmost part of the C. minutus distribution.
The FST and RST estimates, and the AMOVA tests were also performed using reduced data sets, excluding loci showing evidence of potential null alleles and/or linkage disequilibrium, to ensure that these loci do not bias the estimations of gene flow and the genetic structure results observed for C. minutus.
Isolation-by-distance analysis
To search for positive correlations between genetic and geographical distances, a simple Mantel test was implemented, based on pairwise estimates of FST and linear geographical distances between localities. Partial Mantel tests were also computed to observe whether genetic and chromosomal variations are positively correlated, while controlling for the spatial influence. These analyses were computed in Arlequin 3.5.1.2.
The Robertsonian rearrangements and the pericentric inversion shown in Figure 2 were coded as shown in Supplementary Table S1. As we do not have the G-band patterns of some specimens from locality road km 108 (108), and all specimens from 17 km south of Mostardas (17S) and 26 km south of Mostardas (26S), we excluded these individuals from the analyses. The chromosome rearrangements indicated for karyotype 48c were inferred based on its diploid and autosomal arm numbers (ANs), and also on the G-band patterns of karyotypes 50a and heterozygotes 49a. The pairwise karyotypic distances between sampling sites were calculated based on Nei's genetic distance (Nei, 1978), implemented in Popdist 1.2.4 (Guldbrandtsen et al., 2000).
Moreover, a spatial autocorrelation analysis was performed in the program Alleles In Space 1.0 (Miller, 2005), assuming 30 equal distance classes with unequal sample sizes. We also checked whether the chromosomal rearrangements follow a pattern of isolation by distance. The matrix of chromosomal rearrangements was applied in the program Alleles In Space as codominant diploid data, together with the geographical coordinates of the specimens, to implement the spatial autocorrelation analysis and Mantel test analyses. The statistical significances of the tests were assessed assuming 1000 random permutations
Phylogenetic analysis
An incongruence length difference test was implemented in the program PAUP* 4.0 (Swofford, 2002), to ensure that both CR and COI data do not have significantly different phylogenetic signals, and can be combined. We applied a heuristic search, under 1000 random permutations. We assumed a significance level of 0.01, as the incongruence length difference test is prone to the type I error (Cunningham, 1997). A phylogenetic analysis using Bayesian inference was performed with MrBayes 3.1.2 (Huelsenbeck and Ronquist, 2001). The CR and COI sequences were combined into a single data set (incongruence length difference, P=0.015), with the data partitioned by fragment, for which the appropriate model of nucleotide sequence evolution, determined in MrModeltest 2.2 (http://www.abc.se/~nylander), was applied (CR: HKY+I+G; and COI: HKY+G). Two independent replicates of the Markov chain Monte Carlo search were performed with 1 000 000 simulations, each with four chains, sampling trees every 100 generations. After the convergence test, the first 500 trees were discarded. Homologous sequences of C. torquatus Lichtenstein, 1830 and C. flamarioni Travi, 1981 were used as the outgroup. The topological relationship between the haplotypes was estimated using the program Network 4.5.1.0 (http://www.fluxux-engineering.com) with the median-joining approach, for the concatenated mtDNA data set.
To better visualize the relationships among parental karyotypes represented by a dendrogram, we implemented the neighbor-joining method in the program PAUP* 4.0, based on a matrix of presence/absence of chromosomal rearrangements, assuming 1000 random replicates.
Demographic history and divergence time estimates
The program Arlequin 3.5.1.2 was used to assess the demographic history of population expansion, employing Tajima's D and Fu's FS neutrality tests, and mismatch distribution analysis. To test the goodness of fit of the observed mismatch distribution, we used the sum of squared deviations (SSD) and the Harpending's raggedness index (RI) (Harpending, 1994). In addition, to examine past population dynamics and the time of the most recent common ancestor (tMRCA) for the seven main haplogroups identified in phylogenetic analysis, we applied the Bayesian skyline approach implemented in the program Beast 1.7.2 (Drummond and Rambaut, 2007), using the molecular models of evolution provided by MrModeltest 2.2. We applied a strict molecular clock with the substitution rates of 0.0213 (95% confidence interval: 0.0142–0.0287) and 0.0296 (95% confidence interval: 0.0165–0.0442) substitutions per site per million years for COI and CR, respectively, estimated for the genus Ctenomys by Roratto (2012) as normal priors, with 50 000 000 of Markov chain Monte Carlo simulations sampled every 1000 chains. Runs were summarized in TreeAnnotator 1.5.0 (Drummond and Rambaut, 2007), with a burn-in of the first 10% of the trees.
Bayesian clustering of populations
To determine the pattern of population genetic structuring and to assign individuals to populations, we used the Bayesian Markov chain Monte Carlo approach with and without spatial data, implemented in the programs Tess 2.3.1 (François et al., 2006) and Structure 2.3.3 (Pritchard et al., 2000), respectively. The number of clusters (K) was estimated in Tess 2.3.1 using the CAR admixture model, a burn-in of 50 000 sweeps followed by 50 000 iterations. We applied 10 independent runs for each K, ranging from 2 to 30. The values of deviance information criterion (DIC) of each run were plotted against the Kmax, and the most likely number of clusters was considered when the DIC curve approaches a plateau. The Structure 2.3.3 analyses also followed 10 independent runs from K=2–30, 2 000 000 Markov chain Monte Carlo iterations, and a burn-in period of 1 500 000 steps, assuming an admixture model, correlated allele frequencies and a priori location information for each sample. The most likely number of clusters was based on the second-order rate of change in the log probability of the data (ΔK; Evanno et al., 2005).
We performed 50 additional runs for the most likely K found previously in Tess 2.3.1 and Structure 2.3.3, following the same parameters described above. We showed the results for the run that retrieved the highest mean of the estimated logarithm of probability of the data (ln Pr (X/K)) for Structure 2.3.3, and the lowest DIC value for Tess 2.3.1. We assumed an arbitrary threshold of membership probability q⩾0.80, to ensure that specimens with more than 80% of their genomic content represented in one cluster will be assigned to it.
Results
Cytogenetics
Of the 21 specimens collected at sampling sites 17S and 26S, 12 individuals were successfully karyotyped, revealing three specimens with a known karyotype (2n=42, AN=74), described by Freitas (1997), and nine specimens with four new karyotypes, intermediate between the forms 2n=42 and 46b, described in Supplementary Table S2 and Supplementary Figure S2A–D.
All specimens recovered in FSM had the karyotype 2n=50a, AN=76, previously described by Freitas (1997); the specimens from ILH had unknown karyotype, 2n=48c, AN=76 (Supplementary Table S2 and Supplementary Figure S2E); and at locality JG, we found specimens with 2n=48c and 50a, and heterozygotes with 2n=49a, AN=76.
Considering all cytogenetic data available for C. minutus, there is evidence that this species has nine diploid numbers (2n=42, 43, 44, 45, 46, 47, 48, 49, and 50), eight parental karyotypes (2n=42, 46a, 46b, 48a, 48b, 48c, 50a, and 50b) and six hybrid zones (between: 50a × 48c; 46a × 48a; 48a × 42; 42 × 46b; 46b × 48b; and 48b × 50b), comprising a total of 45 karyotypes known so far (Table 1).
Genetic diversity
From 398 bp of the CR, 43 polymorphic sites were obtained, resulting in a total of 40 haplotypes. The COI sequences were 620 bp in length, resulting in 44 variable sites and 35 haplotypes, and the concatenated data comprised 1018 bp, generating a total of 52 haplotypes. The levels of diversity in the three data sets were mostly moderate to high compared with indices of mtDNA diversity for other ctenomyids (overall estimates, π=0.0100–0.0236; k=6.20–15.59; Hd=0.947–0.965; Table 2).
Table 2. Mitochondrial genetic variability estimates for the seven main haplogroups identified for C. minutus and the total variation.
| North 1 | North 2 | Coast | Barros Lake | Mostardas | Tavares | South | Total | |
|---|---|---|---|---|---|---|---|---|
| N | 26 | 33 | 32 | 56 | 51 | 45 | 33 | 276 |
| π (CR) | 0.0026 | 0.0069 | 0.0066 | 0.0049 | 0.0054 | 0.0035 | 0.0024 | 0.0236 |
| π (COI) | 0.0006 | 0.0047 | 0.0029 | 0.0040 | 0.0017 | 0.0029 | 0.0007 | 0.0100 |
| π (CC) | 0.0014 | 0.0055 | 0.0044 | 0.0044 | 0.0032 | 0.0031 | 0.0014 | 0.0153 |
| k (CR) | 1.03 | 2.75 | 2.64 | 1.95 | 2.14 | 1.41 | 0.95 | 9.39 |
| k (COI) | 0.37 | 2.88 | 1.81 | 2.49 | 1.07 | 1.80 | 0.48 | 6.20 |
| k (CC) | 1.40 | 5.63 | 4.46 | 4.44 | 3.21 | 3.20 | 1.43 | 15.59 |
| Hd (CR) | 0.517 | 0.699 | 0.758 | 0.845 | 0.729 | 0.702 | 0.682 | 0.961 |
| Hd (COI) | 0.369 | 0.722 | 0.758 | 0.864 | 0.462 | 0.643 | 0.477 | 0.947 |
| Hd (CC) | 0.628 | 0.735 | 0.764 | 0.882 | 0.764 | 0.702 | 0.682 | 0.965 |
| D (CR) | 2.07 | 0.44 | 0.20 | 0.32 | −0.58 | −0.64 | 1.87 | 1.01 |
| D (COI) | 0.67 | 1.37 | 0.13 | −0.13 | −1.07 | 1.42 | 1.42 | −0.36 |
| D (CC) | 1.93 | 1.73 | 0.19 | 0.06 | −0.88 | 0.24 | 2.15 | 0.33 |
| FS (CR) | 3.21 | 2.84 | 0.56 | −0.34 | 0.08 | −0.97 | 1.58 | −2.46 |
| FS (COI) | 1.00 | 2.85 | −0.65 | −0.83 | −0.79 | 2.66 | 1.60 | −4.67 |
| FS (CC) | 2.43 | 3.89 | 1.68 | −5.30* | −4.17 | −0.94 | 2.80 | −1.59 |
Abbreviations: CC, concatenated data; COI, cytochrome c oxidase subunit I; CR, control region; D, Tajima's D neutrality test; FS, Fu's FS neutrality test; Hd, haplotype diversity; N, number of specimens; k, average nucleotide differences; π, nucleotide diversity.
*P<0.05 Fu's FS significance.
For microsatellite data, the total number of alleles per locus among the 14 loci analyzed ranged from 7 to 16, and from 1 to 12 per locus per locality. Sampling sites FSM, JG, ILH, BAR, EBL, 35 and 96 showed one monomorphic locus; TV2, BJ1 and BJ2 showed two monomorphic loci; and SJN had six monomorphic loci. The monomorphic loci differed among the localities. The mean numbers of alleles per locality are presented in Table 1.
Departures from Hardy–Weinberg equilibrium were found in six sampling sites: OSO (Hai12 and Soc3); EBL (Hai4); SBL (Soc5); 96 (Soc3); MOS (Hai6); and BJ2 (Hai4 and Soc6). These deviations could be explained by (i) null alleles detected with Micro-Checker; (ii) as a consequence of reduced effective population sizes, which together with low rates of dispersal can lead to deviations from random mating, as evidenced in estimates of significant local FIS in ILH, OSO, EBL, 96 and MOS (Table 1); or even (iii) the departures from Hardy–Weinberg equilibrium detected at EBL, SBL and BJ2 may be a consequence of hybridization events, as three of six loci that were not in equilibrium were from these hybrid localities. Pairwise comparisons of LE within localities revealed significant results among five pairs of loci: Hai3 × Soc5 (OSO), Hai10 × Soc2 (JG, OSO, 108), Hai12 × Soc1 and Soc4 × Soc5 (GAI) and Soc3 × Soc6 (MOS). However, these loci were not excluded from the subsequent analysis because there were no significant deviations from LE, for the same loci, in other pairwise comparisons for C. minutus. The influence of potential null alleles and loci under linkage disequilibrium seems to be negligible regarding the FST and RST estimates, and the AMOVA tests performed with reduced data sets, without these loci (data not show).
Phylogeographic pattern of haplotype distributions
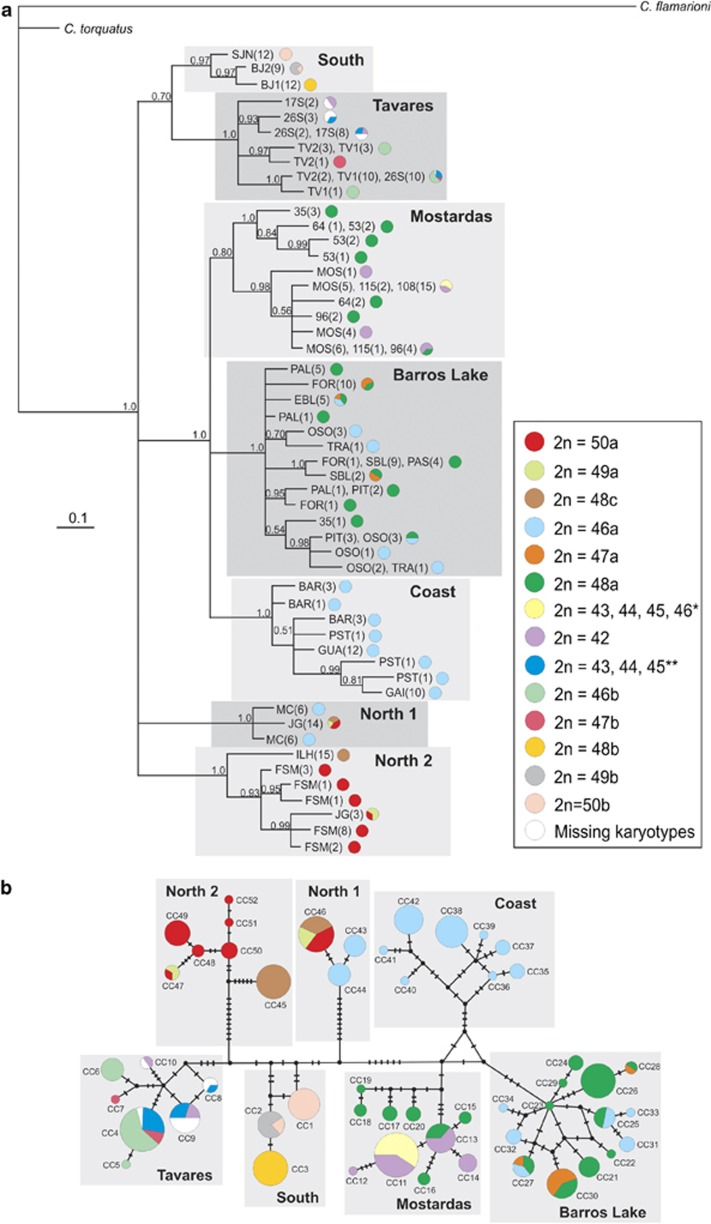
Seven main clades were highlighted in the Bayesian phylogenetic tree (Figure 3a), all of them showing a strong relationship between specimen clustering and the geographical distribution. This same pattern could also be observed in the seven haplogroups retrieved by the median-joining haplotype network (Figure 3b). The northern localities were subdivided into two separate clades: North 1 was formed by all specimens from MC and 14 specimens from JG (six specimens with 2n=50a, three with 2n=49a and five with 2n=48c); and North 2 included the specimens from FSM, ILH and three specimens from JG (two with 2n=49a and one with 2n=50a). The middle of the C. minutus distribution was subdivided into three clades, among them: Coast, formed by specimens inhabiting only the shoreline (GAI, PST, GUA and BAR); Barros Lake, comprising sampling sites TRA, OSO, EBL, SBL, PAS, FOR, PIT and PAL, and one specimen from 35; and Mostardas, represented by the specimens from 35, 53, 64, 96, 108, 115 and MOS. The Tavares clade was represented by specimens from 17S, 26S, TV1 and TV2. Finally, the South clade, comprising specimens from BJ1, BJ2 and SJN.
Figure 3.
(a) Bayesian phylogenetic tree; and (b) median-joining haplotype network topology obtained for concatenated mtDNA data. The seven main genetic haplogroups are highlighted by squares. Shading represents diploid numbers of parental karyotypes and intraspecific hybrids, as indicated in the legend. The abbreviations of the localities and the number of specimens per sampling site (within parentheses) are shown in the tree, and correspond to those in Table 1. The node posterior probabilities are given on the branches. Small black circles represent extinct or unsampled haplotypes in the network topology, and circle areas are proportional to the haplotype frequencies. *Diploid numbers of hybrids from crossing between 2n=42 × 48a; **diploid numbers of hybrids from crossing between 2n=42 × 46b. The color reproduction of this figure is available on the Hereditary journal online.
The mean tMRCAs calculated for each haplogroup were as follows: North 1—34 719 years (95% highest posterior density (HPD): 1613–82 431 years); North 2—179 390 years (95% HPD: 66 141–310 040 years); Coast—152 430 years (95% HPD: 58 048–259 190 years); Barros—122 130 years (95% HPD: 49 026–212 930 years); Mostardas—162 350 years (95% HPD: 65 339–276 080 years); Tavares—109 440 years (95% HPD: 37 531–194 680 years); and South—33 032 years (95% HPD: 1424–78 114 years).
In general, most haplotypes were separated by several mutational steps, and were limited to single localities, that is, private alleles. The shared haplotypes were mainly among neighboring sampling sites (Figures 1 and 3a and Table 1). Concerning chromosomal polymorphisms, only specimens from parapatric karyotypes and their hybrid forms shared haplotypes in the three mtDNA data sets (Supplementary Table S3).
The dendrogram of chromosomal rearrangements demonstrated that the karyotypes distributed at the north and south extremes of the geographical distribution of C. minutus (that is, 2n=50a, 48c and 50b) are more similar to each other than with the karyotypes observed in the middle of the distribution (Supplementary Figure S3).
Demographic history and isolation by distance
Although the plots of mismatch distributions were not clearly unimodal (Supplementary Figure S4), the tests of goodness of fit indicated that the observed and expected distributions of pairwise differences among haplotypes did not differ significantly under a model of recent population expansion, for COI (SSD=0.002, P=0.77; RI=0.006, P=0.88) and CC data sets (SSD=0.004, P=0.28; RI=0.004, P=0.24). In contrast, the observed and expected mismatch distributions differed significantly for the CR data set (SSD=0.012, P=0.001; RI=0.016, P=0.001).
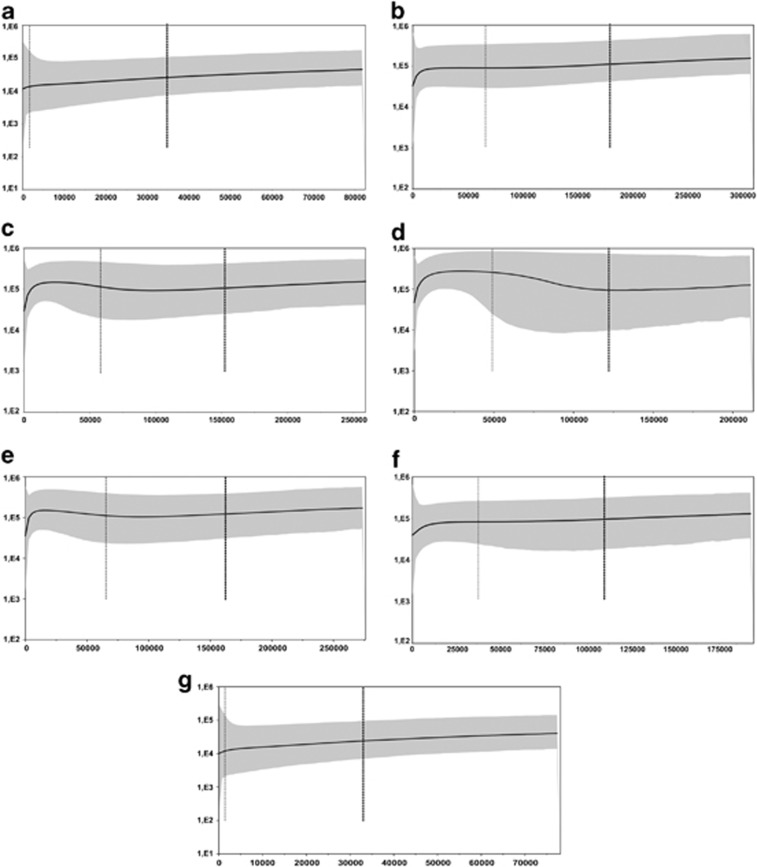
There was no statistical support to accept a recent history of population expansion for C. minutus, according to the results of neutrality tests. This hypothesis is reinforced by high average number of nucleotide differences among haplotypes (ranging from k=6.20 to 15.59; Table 2), the limited distribution of haplotypes, and also by the absence of a star-like topology in the median-joining network, which is expected under a scenario of recent population expansion. The overall results of Tajima's D showed positive values for CR and CC data and a negative D for COI, although none of the values was significant (Table 2). For the global Fu's FS test, all values were negative but not significant. With respect to the neutrality test results for the seven haplogroups of mtDNA, only Barros Lake showed negative and significant Fu's FS values, for the CC data set (Table 2). The effective population sizes were mostly constant among all the seven haplogroups, according to the Bayesian skyline analyses, and only some minor sudden retractions were observed from the last 13 000 years (Figure 4).
Figure 4.
Bayesian skyline plots of concatenated mtDNA data for the seven main haplogroups of C. minutus: (a) North 1; (b) North 2; (c) Coast; (d) Barros Lake; (e) Mostardas; (f) Tavares; and (g) South. The gray area corresponds to the 95% highest posterior density limits for the effective population size. The thin and thick dotted lines are the lowest and the mean estimated tMRCA. Below the plot is the time represented in years, and in the left side are the effective population sizes.
The Mantel's test detected a positive and significant correlation between genetic and geographical distances, for the overall geographical range in CC mtDNA (r=0.38, P=0.00), microsatellite data (r=0.53, P=0.00) and for chromosomal rearrangements (r=0.60, P=0.00). The partial Mantel tests do not detect significant correlations between genetic and chromosomal variations, while partial out the geographical distance, in both CC mtDNA (r=−0.07, P=0.73) and microsatellite data sets (r=0.10, P=0.20).
Clear positive phylogeographic structures were also observed through the spatial autocorrelation analyses (Supplementary Figure S5). The mean genetic distances (Ay) estimated for CC mtDNA, microsatellites and chromosomal rearrangements were 0.015, 0.770 and 0.363, respectively. For CC mtDNA data, highly positive correlations were evident among the first three distance classes (from 0 to 42 km). For microsatellite data, the positive structure persisted up to around 56 km, and for chromosomal rearrangements, the strongest correlations were observed up to around 70 km.
Population structure
The microsatellite pairwise estimates of FST and RST revealed moderate to high levels of genetic differentiation among sampling sites, ranging from 0.09 to 0.61 for FST, and from 0.05 to 0.79 for RST. Most pairwise comparisons showed values higher than 0.3 (Supplementary Table S4).
The AMOVA results for the three mtDNA data sets, using eight groups of parental karyotypes showed most of the genetic variation distributed among populations within karyotypes (49.75–62.16%). On the other hand, the test in which the populations were subdivided into seven groups, considering present and past geographical barriers, revealed higher percentages of genetic variation at the group level, ranging from 55.58 to 58.28% (Table 3). For microsatellite data, the two AMOVA tests showed more than 67% of the genetic variation within populations; however, the percentage of variation among karyotypic groups was slightly higher when compared with geographical groups, 10.49% versus 8.77%, respectively (Table 3).
Table 3. Results of analyses of molecular variance for mitochondrial DNA data sets and microsatellites.
|
CR
|
COI
|
CC
|
Microsatellites
|
|||||
|---|---|---|---|---|---|---|---|---|
| PK | GB | PK | GB | PK | GB | PK | GB | |
| Among groups (FCT) | 30.30% (0.30) | 55.58% (0.56) | 41.81% (0.42) | 58.28% (0.58) | 35.00% (0.35) | 56.67% (0.57) | 10.49% (0.10) | 8.77% (0.09) |
| Among populations within groups (FSC) | 62.16% (0.89) | 37.00% (0.83) | 49.75% (0.85) | 33.33% (0.80) | 57.09% (0.88) | 35.51% (0.82) | 22.09% (0.25) | 23.21% (0.25) |
| Within populations (FST) | 7.54% (0.92) | 7.41% (0.93) | 8.44% (0.92) | 8.40% (0.92) | 7.91% (0.92) | 7.81% (0.92) | 67.43% (0.33) | 68.01% (0.32) |
Abbreviations: CC, concatenated data between CR+COI; COI, cytochrome c oxidase subunit I; CR, control region; GB, seven groups, considering potential present and former geographical barriers; PK, eight groups, considering parental karyotypes.
Values in bold are significant (P=0.00).
The percentage of variation for each hierarchical level and the fixation indices (within parentheses) are shown.
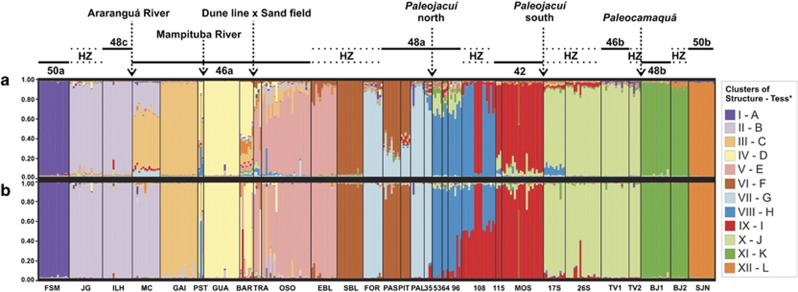
The most likely number of clusters in the Structure analysis, according to the ΔK approach of Evanno was 12 (Supplementary Figure S6). The major divergences among the 60 runs at K=12 were regarding the estimated membership coefficients (q) and the assignment of specimens from the localities of MC and SJN. The samples from MC showed a private cluster in some runs, whereas in other runs, the specimens showed mixed ancestries (q<0.8) between the clusters representing the neighboring localities of JG/ILH and GAI. The locality of SJN kept all the individuals in a private cluster in some runs, and in other runs, the specimens were grouped in the same cluster of the neighboring localities of BJ1 and BJ2. All the other sampling sites showed consistent results among different runs. The plot of population clustering retrieved in the run with the highest ln Pr (X/K)=−13 404.3 is represented in Figure 5a. In the Tess analysis, the plot of DIC × Kmax began to approach a plateau around K=12 (Supplementary Figure S7). We analyzed the best runs for Ks from 12 to 16. All these Ks retrieved the same pattern of grouping localities in 12 main clusters, and the cluster retrieved above 12 comprised only minor subdivisions in the sampling sites LLB, FOR, 53, 64, TV1 or TV2 (data not shown).
Figure 5.
Bayesian clustering and specimen assignments for the 12 clusters identified by the (a) Structure and (b) Tess programs. Each specimen is represented by a single bar, and each cluster by a color. Sampling sites are plotted geographically in the North–South direction, from left to right. Above plot: parental parapatric karyotypes, hybrid zones among them (HZ) and main possible geographical barriers. Below plot: BAR, Barco Beach; BJ1, Bujuru 1; BJ2, Bujuru 2; EBL, East Barros Lake; FOR, Fortaleza Lake; FSM, Farol de Santa Marta; GAI, Gaivota Beach; GUA, Guarita Beach; ILH, Ilhas; JG, Jaguaruna; MC, Morro dos Conventos; MOS, Mostardas; PAL, Palmares do Sul; PAS, Passinhos; PIT, Pitangueira; PST, Passo de Torres; OSO, Osório; 35, road km 35; 53, road km 53; 64, road km 64; 96, road km 96; 108, road km 108; 115, road km 115; SBL, South Barros Lake; 17S, 17 km south of Mostardas; 26S, 26 km south of Mostardas; SJN, São José do Norte; TRA, Tramandaí TV1, Tavares 1; TV2, Tavares 2. *Roman numbers correspond to Structure clusters, and the uppercase letters correspond to Tess clusters.
The 12 clusters retrieved by both Structure and Tess tended to group together specimens from the same sampling site and nearby localities, in a similar pattern. One of the main discrepancies was with respect to MC, which was completely admixed for Structure among several clusters, but in Tess its ancestry came from a single cluster (B; Figure 5b). Moreover, Tess showed a smaller proportion of individuals with admixed ancestries (q⩽0.8) compared with Structure (∼10% versus 27%, respectively). This difference is mainly associated with localities MC, BAR, TRA, OSO, PAS, PIT, 35, 53, 64, 96 and 115, in which most specimens could be classified as non-admixed by Tess, but had mixed ancestries in the Structure results. On the other hand, sampling site 108 showed more admixed specimens in Tess than in Structure.
Discussion
Isolation by distance
C. minutus has a nearly linear geographical distribution along the southern Brazilian coastal plain. In general, when species of small mammals have a linear distribution, dispersal and gene flow will be geographically limited to neighboring populations, mainly because of their low ability to disperse over significant distances relative to the species' range, which is a common feature in subterranean rodents (Busch et al., 2000) and may result in a pattern of isolation by distance. Phylogeographical studies based on control region, COI and/or cytochrome b mtDNA data revealed that species of tuco-tucos such as C. pearsoni, C. flamarioni, C. australis and C. lami have low levels of gene flow among populations, and a pattern of isolation by distance associated with their narrow geographical distribution (Mora et al., 2006; Fernández-Stolz, 2007; Tomasco and Lessa, 2007; Lopes and Freitas, 2012).
C. minutus showed a geographical limitation on the distribution of mtDNA haplotypes, as most of them were limited to single sampling sites, and shared haplotypes were mainly among neighboring localities (Figures 1 and 3a and Table 1). The most widely separated localities that shared haplotypes, for CR, COI and CC data, respectively, were FOR and 35 (41.5 km apart, haplotype CR20), SBL and MOS (140 km apart, haplotype COI8) and 96 and MOS (32 km apart, CC13; Supplementary Table S3). Bayesian clustering of populations based on microsatellite data tended to group together specimens from the same sampling site and nearby localities (Figures 5a and b). Likewise, the distribution of parental karyotypes was parapatric, and no specimen was found outside the limits of its source karyotypic population (Table 1).
Analyses of the variation in the skull shape of C. minutus found a weak but significant association between morphometric and geographical distances, and the authors suggested that differences among populations follow a subtle pattern of isolation by distance (Fornel et al., 2010).
The Mantel tests and the spatial autocorrelation analyses showed overall positive and significant correlations between genetic and geographical distances, for the mitochondrial, nuclear and chromosomal data sets, and the strongest positive structures were observed up to around 70 km. However, the values of Ay did not increase linearly with increasing geographical distance, showing an unstable pattern of variation for all data sets, mainly from the 10th distance class, corresponding to 140 km onwards (Supplementary Figures S5A–C). This pattern was described by Diniz-Filho and Telles (2002) as a ‘stabilizing profile', which may reflect influences of positive genetic/geographical associations among groups of closely distributed populations, and phylogeographical groups of populations primarily structured by other historical events, for example, the genetic structures revealed by the seven haplogroups for mtDNA data or the 12 clusters of localities retrieved in the Bayesian clustering analyses. The sharp increase in genetic and geographic distance correlations, until it reaches an irregular pattern, is expected under a model of isolation with asymmetric gene flow in a metapopulation. A similar pattern was observed in the perrensi group of ctenomyids (Fernández et al., 2012).
Geographical barriers
The phylogenetic tree generated from concatenated data (Figure 3a) showed a basal polytomy between the C. minutus branches and the outgroup. Polytomies are commonly found among and within species of the genus Ctenomys, and have been suggested to be the result of an early simultaneous cladogenesis event (Lessa and Cook, 1998). Nevertheless, we were able to recover a pattern of genetic structure of the sampling sites subdivided in seven main haplogroups.
Time estimates and calibration of molecular clock is often a hard task because of the scarcity and uncertainty of the fossil records for some taxa, sometimes resulting in substitution rates with large confidence intervals, which may account for the largely overlapping 95% HPD of tMRCA calculated for the seven mtDNA haplogroups of C. minutus. Nevertheless, we can observe a general trend of older mean estimates of tMRCA for the haplogroups of C. minutus came from its northern range, with exception of North 1, and toward the south the mean tMRCA decreased, reaching its minimum in the South haplogroup (Figure 4). Moreover, the southern locality of SJN has the lowest levels of microsatellite variability, showing a total of six monomorphic loci. This pattern may result from a more ancient genetic structure achieved in the northern geographical distribution, while the southern sampling sites may account to a founder effect of more recent occurrence.
Around 200 ka, the sea level started a regressive cycle, and in 141 ka, the southern Brazilian coastal plain became wider than nowadays. During this period, both Paleojacuí north and Paleocamaquã were present in the region. The subsequent transgressive cycle, around 133 ka, closed completely the Palejacuí north outlet to the sea. A new glacial cycle led to a long and oscillating regressive phase of the Atlantic sea level, which persisted for around 100 000 years and reached its maximum at 18 ka. The Paleojacuí south arose on the coastal plain in the beginning of this regressive cycle. Both the Paleojacuí south and the Paleocamaquã were completely closed by sediments 6 ka, during the last great transgressive cycle of the Atlantic sea level (Tomazelli et al., 2000; Weschenfelder et al., 2008; Baitelli, 2012).
These three paleochannels may have reduced the gene flow between populations from opposite sides of the rivers during the Pleistocene. As a consequence, we detected associated patterns of genetic structure that have persisted to the present, which can be observed in Bayesian clustering analyses (Figure 5) and in mtDNA haplogroups (Figures 1 and 3a and b). The persistence of these genetic structures may have been reinforced by the metapopulation structure, common to ctenomyids, in which the effects of low levels of gene flow hamper the gene exchange between the subpopulations, and the genetic drift associated with the small subpopulation sizes maintain the low levels of genetic diversity.
The transition between the sand fields and dune line represent another environmental discontinuity along the range of C. minutus, which may inhibit free dispersal among specimens from different habitats. For mtDNA data, we recovered the Coastal haplogroup (corresponding to specimens from the dune line) and Barros Lake (sand fields; Figure 1). For microsatellite data, a pattern of genetic structuring following the transition between these two environments was less pronounced. The tuco-tucos living near the transition region showed admixed ancestries (q⩽0.8) among one main cluster (IV–D for specimens from the dune line, and V–E for specimens from sand fields), and several other less-representative clusters (Figure 5).
Environmental characteristics such as soil hardness lead to morphological adaptations in tuco-tucos, mainly in the anterior and posterior limbs and in the cranium, as they are burrowing rodents and use their teeth, paws and legs to dig tunnels. Fornel et al. (2010) found significant differences in skull morphology between specimens of C. minutus that live in sand fields or dunes. Therefore, these different environments may also be associated with the genetic discontinuities observed in our data.
The role of the Mampituba and Araranguá Rivers as effective geographical barriers to gene flow were conflicting regarding mitochondrial and nuclear molecular markers. One possible explanation for the absence of clear genetic structuring caused by the Mampituba and Araranguá Rivers may lie in the past transient status of the river mouths (Tomazelli and Villwock, 2000; S Dillenburg, personal communication), which may have often allowed gene flow between populations from opposite banks, which have not reached reproductive isolation. Nowadays, the banks of both rivers have stabilized, hampering the contact between specimens from opposite sides.
Most of mtDNA genetic variation is apportioned among groups geographically delimited, according to AMOVA (Table 3), but microsatellite data showed the lowest proportions of genetic variation among groups, suggesting the involvement of factors other than geographical barriers or chromosomal rearrangements affecting the partition of genetic variation in nuclear genome. The potential homoplasy and high levels of polymorphism in microsatellites, and the lower effective population sizes of mtDNA, may account for different results observed in AMOVA tests.
Cytonuclear genome discordance has been described for some vertebrates at both intra- and interspecific levels. Genome components, with distinct modes of inheritance, commonly show differences in phylogeographic patterns and levels of introgression between hybridizing species/populations. These differences can result from distinct mutation rates and effective population sizes among molecular markers, and also from sex-biased dispersal (Petit and Excoffier, 2009). The mtDNA has non-recombinant maternal inheritance, which allows the retention of haplotypes shared between populations recently isolated, or to keep the differentiation between populations in recent secondary contact that had previously differentiated in allopatry. Whereas, male-biased dispersal, that is characteristic of the species of the genus (Malizia and Busch, 1991; Cutrera et al., 2006), can lead to faster and farther dissemination of alleles of nuclear molecular markers relative to mtDNA, and associated with the nuclear genome recombination and high mutation rates of microsatellite loci, it can mask the signals of historical evolutionary processes in the nuclear genome (Petit and Excoffier, 2009; Yang and Kenagy, 2009).
The skull morphometric data for C. minutus indicate that there was no direct relationship between Robertsonian fusions/fissions and skull morphology, and these characters are independent, although both vary with the geographical distribution (Fornel et al., 2010). A phenotypic structuring into three morphological groups was suggested for C. minutus: (i) the populations in the northern part of the geographical range, isolated from the other localities by the Araranguá River; (ii) populations in the southern part of the distribution, isolated by the Paleojacuí south, which might be reinforced by the pericentric inversion; (iii) and all other populations associated with lakes and lagoons, in the middle of the distribution.
The geographical features of the southern Brazilian coastal plain may also be associated with the fixation of new chromosomal rearrangements in C. minutus, and two different scenarios of a step-by-step mechanism of chromosomal evolution can be suggested for this species. The first hypothesis relies on the metapopulation structure, with small subpopulations distributed in patches of suitable habitats (Fernández et al., 2012). Whether a new chromosomal rearrangement causes little or no negative meiotic effects, it can be more easily fixed in peripheral and/or small isolated populations (Baker and Bickham, 1986) by means of genetic drift, spreading to new, recently colonized areas. The second hypothesis is an extension of the first, also based on the fixation of chromosomal rearrangements by means of genetic drift, but in this case the chromosomal rearrangements remain isolated from each other in allopatry because of effective geographical barriers to gene flow. An example from this study is the transition between karyotypes 2n=48c and 46a, which are delimited by the Araranguá River (Figure 1). As these barriers no longer exist, the gene flow between specimens from parapatric karyotypes, which were not reproductively isolated, could give rise to secondary-contact hybrid zones, for example, between karyotypes 2n=42 × 46b, overlapping the region corresponding to Paleojacuí south (Figure 1).
Chromosomal rearrangements
Subterranean rodents show some of the highest rates of chromosomal evolution known in mammals (Reig et al., 1990). The cytogenetic data available for C. minutus reveal a total of 45 distinct karyotypes (Freitas, 1997; Gava and Freitas, 2002, 2003; Freygang et al., 2004; Castilho et al., 2012; this study), and most of this polymorphism was retrieved in the intraspecific karyotypic hybrid zones.
The six hybrid zones identified for C. minutus arose even between parapatric karyotypes differing by one Robertsonian rearrangement: (i) 2n=50a × 48c; (ii) 46a × 48a; (iii) 46b × 48b; (iv) and 48b × 50b; or more than one: (v) 2n=48a × 42; and between karyotypes differing by Robertsonian rearrangements associated with a pericentric inversion: (vi) 2n=42 × 46b (Figure 2). Adaptability and fertility in chromosomally heterozygous carriers can be maintained by mechanisms that partially or totally suppress recombinations with harmful effects (White, 1978; Rieseberg, 2001). Heterozygotes for simple Robertsonian centric fusion/fission rearrangements, for example, can produce balanced meiotic systems, by means of trivalents, which often show normal segregation during meiosis (Baker and Bickham, 1986).
Different patterns of genetic structures and levels of introgression were observed among the hybrid zones of C. minutus. These patterns may vary depending on the molecular marker analyzed, which include differences in mutation rates, recombination and effective population sizes; the kind of chromosomal rearrangements involved and whether they are implicated or not in reductions in gene flow between chromosomally divergent populations; whether there are different patterns of crossings, or both sexes from both parental karyotypes are able to mate and breed equally; or whether the hybrid zone is associated or not with a geographical barrier and the effectiveness and persistence of this geographical feature as a barrier to gene flow between populations.
The recent literature about ctenomyids provides some examples of karyotypic polymorphic species that fail to show genetic structure following a pattern of chromosomal rearrangements (Lopes and Freitas, 2012). For C. pearsoni, it is suggested that new chromosomal rearrangements, besides being a common feature, seem not to be followed by sterility, reductions in fitness or negative heterosis of heterozygote carriers (Tomasco and Lessa, 2007). Studies by Gava and Freitas (2002, 2003), using cytogenetic data for the C. minutus hybrid zones between 2n=46a × 48a and 2n=48a × 42 showed no evidence of underdominance, and demonstrated that the chromosomal polymorphisms fail to cause sterility in heterotypes. The chromosomal rearrangements in the ‘perrensi' group were considered to be the result of an ongoing isolation with a migration process, rather than representing a barrier to gene flow (Caraballo et al., 2012; Fernández et al., 2012). Some highly chromosomally polymorphic mammals, such as Mus musculus domesticus and Sorex araneus, show a step-by-step mechanism of karyotype evolution, similar to that of C. minutus. However, although the presence of karyotypic populations is well defined, molecular markers do not match this subdivision, and the karyotypic populations are still considered as part of a single species (Wójcik et al., 2002; Piálek et al., 2005).
C. minutus shows no chromosomal polymorphism within the parental karyotypic populations (outside hybrid zones), and differences among neighboring parental karyotypes, although minor, are fixed. However, chromosomal rearrangements are not strongly associated with the patterns of nuclear genetic structure observed, as suggested by AMOVA, partial Mantel test and Bayesian clustering analyses, and thus probably has no or very little role in the diversification of C. minutus populations. As suggested by Lukhtanova et al. (2011) ‘we cannot exclude that geographically distant and chromosomally divergent populations would display reduced fertility if crossed, although they are connected by a chain of compatible populations that should allow gene flow'.
In summary, the effects of genetic drift, low levels of gene flow relative to the geographical range, the metapopulation structure and the geographical features of the southern Brazilian coastal plain may account for the patterns of genetic/geographical structure and the step-by-step mechanism of chromosomal evolution observed in this study for C. minutus. Further researches regarding the chromosomal rearrangements and their effects on heterozygote carriers will allow to better understand the evolutionary process in which C. minutus is involved.
Data archiving
Sequence data have been submitted to GenBank. Ctenomys minutus accession numbers: HM236969–HM237008 and HM237009–HM237043; Ctenomys torquatus: HM443438 and HM443439; Ctenomys flamarioni: JQ341041 and JQ341052. Microsatellite data have been submitted to Dryad repository, doi:10.5061/dryad.d12j8.
Acknowledgments
We thank Capes, CNPQ, FAPERGS and the Departamento de Genética/UFRGS for supporting this project. We are grateful to Camila S Castilho, Lígia Tchaicka and Cristina C Freygang for providing samples and cytogenetic data, to Paula A Roratto and Gabriela de P Fernández for providing the outgroup sequences, Florian Alberto for suggestions and discussions and three anonymous reviewers who highly improved this manuscript. We also thank colleagues from the Laboratório de Citogenética e Evolução and Departamento de Genética—UFRGS for support at various stages of this research.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Baitelli R.2012. Evolução Paleogeográfica do Sistema de Paleodrenagem do Rio Jacuí na Planície Costeira do Rio Grande do Sul. PhD thesis, Universidade Federal do Rio Grande do Sul.
- Baker RJ, Bickham JW. Speciation by monobrachial centric fusions. Proc Natl Acad Sci USA. 1986;83:8245–8248. doi: 10.1073/pnas.83.21.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraclough TG, Vogler AP. Detecting the geographical pattern of speciation from species-level phylogenies. Am Nat. 2000;155:419–434. doi: 10.1086/303332. [DOI] [PubMed] [Google Scholar]
- Busch C, Antinuchi CD, del Valle JC, Kittlein MJ, Malizia AI, Vassallo AI, et al. 2000Population ecology of subterranean rodentsIn: Lacey EA, Patton JL, Cameron GN, (eds)Life Underground: The Biology of Subterranean Rodents The University of Chicago Press: Chicago, IL, USA; 183–226. [Google Scholar]
- Caraballo DA, Abruzzese GA, Rossi MS. Diversity of tuco-tucos (Ctenomys, Rodentia) in the Northeastern wetlands from Argentina: mitochondrial phylogeny and chromosomal evolution. Genetica. 2012;140:125–136. doi: 10.1007/s10709-012-9664-7. [DOI] [PubMed] [Google Scholar]
- Castilho CS, Gava A, Freitas TRO. Hybrid zone in genus Ctenomys: a study case in southern Brazil. Genet Mol Biol. 2012;35:990–997. doi: 10.1590/s1415-47572012000600014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CW. Can the incongruence test predict when data should be combined. Mol Biol Evol. 1997;14:733–740. doi: 10.1093/oxfordjournals.molbev.a025813. [DOI] [PubMed] [Google Scholar]
- Cutrera AP, Antinuchi CD, Mora MS, Vasallo AI. Home range and activity patterns of the South American subterranean rodent Ctenomys talarum. J Mammal. 2006;87:1183–1191. [Google Scholar]
- Diniz-Filho JAF, Telles MPDC. Spatial autocorrelation analysis and the identification of operational units for conservation in continuous populations. Conserv Biol. 2002;16:924–935. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Schneider S. Arlequin version 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fernández-Stolz GP.2007. Estudos Evolutivos, Filogeográficos e de Conservação em uma Espécie Endêmica do Ecossistema de Dunas Costeiras do Sul do Brasil, Ctenomys flamarioni (Rodentia—Ctenomyidae), Através de Marcadores Moleculares Microssatélites e DNA Mitocondrial. PhD thesis, Universidade Federal do Rio Grande do Sul.
- Fernández MJG, Gaggiotti OE, Mirol P. The evolution of a highly speciose group in a changing environment: are we witnessing speciation in the Iberá wetlands. Mol Ecol. 2012;13:3266–3282. doi: 10.1111/j.1365-294X.2012.05598.x. [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–297. [PubMed] [Google Scholar]
- Ford CE, Hamerton JL. A colchicine, hypotonic citrate, squash sequence for mammalian chromosomes. Stain Technol. 1956;31:247–251. doi: 10.3109/10520295609113814. [DOI] [PubMed] [Google Scholar]
- Fornel R, Cordeiro-Estrela P, Freitas TRO. Skull shape and size variation in Ctenomys minutus (Rodentia: Ctenomyidae) in geographical, chromosomal polymorphism, and environmental contexts. Biol J Linn Soc Lond. 2010;101:705–720. [Google Scholar]
- François O, Ancelet S, Guillot G. Bayesian clustering using hidden Markov random fields in spatial population genetics. Genetics. 2006;174:805–816. doi: 10.1534/genetics.106.059923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas TRO. Chromosome polymorphism in Ctenomys minutus (Rodentia: Octodontidae) Genet Mol Biol. 1997;20:1–7. [Google Scholar]
- Freygang CC, Marinho JR, Freitas TRO. New karyotypes and some considerations of Ctenomys minutus (Rodentia: Ctenomyidae) on the coastal plain of the Brazilian State of Rio Grande do Sul. Genetica. 2004;121:125–132. doi: 10.1023/b:gene.0000040376.56321.be. [DOI] [PubMed] [Google Scholar]
- Gava A, Freitas TRO. Characterization of a hybrid zone between chromosomally divergent populations of Ctenomys minutus (Rodentia; Ctenomyidae) J Mammal. 2002;83:843–851. [Google Scholar]
- Gava A, Freitas TRO. Inter and intra-specific hybridization in tuco-tucos (Ctenomys) from Brazilian coastal plains (Rodentia: Ctenomyidae) Genetica. 2003;119:11–17. doi: 10.1023/a:1024465429856. [DOI] [PubMed] [Google Scholar]
- Guldbrandtsen B, Tokiuk J, Loeschcke V. POPDIST, version 1.1.1: a program to calculate population genetic distance and identity measures. J Hered. 2000;91:178–179. doi: 10.1093/jhered/91.2.178. [DOI] [PubMed] [Google Scholar]
- Harpending RC. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum Biol. 1994;66:591–600. [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, Villablanca FX, et al. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey EA. Microsatellite variation in solitary and social tuco-tucos: molecular properties and population dynamics. Heredity. 2001;86:628–637. doi: 10.1046/j.1365-2540.2001.00881.x. [DOI] [PubMed] [Google Scholar]
- Lacey EA, Maldonado JE, Clabaugh JP, Matocq MD. Interspecific variation in microsatellites isolated from tuco-tucos (Rodentia: Ctenomyidae) Mol Ecol. 1999;8:1753–1768. doi: 10.1046/j.1365-294x.1999.00723-1.x. [DOI] [PubMed] [Google Scholar]
- Lessa EP, Cook JA. The molecular phylogenetics of tuco-tucos (genus Ctenomys, Rodentia: Octodontidae) suggests an early burst of speciation. Mol Phylogenet Evol. 1998;9:88–99. doi: 10.1006/mpev.1997.0445. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lopes CM, Freitas TRO. Human impact in naturally patched small populations: genetic structure and conservation of the burrowing rodent, tuco-tuco (Ctenomys lami) J Hered. 2012;103:672–681. doi: 10.1093/jhered/ess027. [DOI] [PubMed] [Google Scholar]
- Lukhtanova VA, Dincã V, Tavalera G, Vila R. Unprecedented within-species chromosome number cline in the Wood White butterfly Leptidea sinapis and its significance for karyotype evolution and speciation. BMC Evol Biol. 2011;11:109. doi: 10.1186/1471-2148-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MP. Alleles In Space: computer software for the joint analysis of interindividual spatial and genetic information. J Hered. 2005;96:722–724. doi: 10.1093/jhered/esi119. [DOI] [PubMed] [Google Scholar]
- Mora MS, Lessa EP, Cutrera AP, Kittlein MJ, Vassalo AI. Phylogeographical structure in the subterranean tuco-tuco Ctenomys talarum (Rodentia: Ctenomyidae): contrasting the demographic consequences of regional and habitat-specific histories. Mol Ecol. 2007;16:3453–3465. doi: 10.1111/j.1365-294X.2007.03398.x. [DOI] [PubMed] [Google Scholar]
- Malizia AI, Busch C. Reproductive parameters and growth in the fossorial rodent Ctenomys talarum (Rodentia: Octodontidae) Mammalia. 1991;55:293–305. [Google Scholar]
- Mora MS, Lessa EP, Kittlein MJ, Vassallo AI. Phylogeography of the subterranean rodent Ctenomys australis (Rodentia: Ctenomyidae) in sand-dune habitats: evidence of recent population expansion. J Mammal. 2006;87:1192–1203. [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton JL, da Silva MN, Malcolm JR. Hierarchical genetic structure and gene flow in three sympatric species of Amazonian rodents. Mol Ecol. 1996;5:229–238. doi: 10.1111/j.1365-294x.1996.tb00310.x. [DOI] [PubMed] [Google Scholar]
- Petit RJ, Excoffier L. Gene flow and species delimitation. Trends Ecol Evol. 2009;24:386–393. doi: 10.1016/j.tree.2009.02.011. [DOI] [PubMed] [Google Scholar]
- Piálek J, Hauffe HC, Searle JB. Chromosomal variation in the house mouse. Biol J Linn Soc Lond. 2005;84:535–563. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reig OA, Busch C, Ortells MO, Contreras JL.1990An overview of evolution, systematics, population biology and molecular biology in CtenomysIn: Nevo E, Reig OA, (eds)Evolution of Subterranean Mammals at the Organismal and Molecular Levels Allan Liss: New York, NY, USA; 71–96. [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Roratto PA.2012. Tuco-Tucos do Pampa Rio-Grandense: a Filogeografia de Ctenomys torquatus (Rodentia: Ctenomyidae) e a Descrição de uma Nova Espécie. PhD thesis, Universidade Federal do Rio Grande do Sul.
- Rousset P. Genepop'007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russel DW.2001Rapid isolation of yeast DNAIn: Sambrook J, Russel DW, (eds)Molecular Cloning, A Laboratory Manual Cold Spring Harbor Laboratory: New York, NY, USA; 631–632. [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg EK, Patton JM.2000Genetic structure of subterranean rodentsIn: Lacey EA, Patton JL, Cameron GN, (eds)Life Underground: The Biology of Subterranean Rodents The University of Chicago Press: Chicago, IL, USA; 301–331. [Google Scholar]
- Swofford DL.2002PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods)Version 4Sinauer Associates: Sunderland, MA, USA [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tomasco I, Lessa EP.2007Phylogeography of the tuco-tuco Ctenomys pearsoni: mtDNA variation and its implication for chromosomal differentiationIn: Kelt DA, Lessa EP, Salazar-Bravo J, Patton JL, (eds)The Quintessential Naturalist: Honoring the Life and Legacy of Oliver P Pearson University of California Press: Berkeley, CA, USA; 859–882. [Google Scholar]
- Tomazelli LJ, Dillenburg SR, Villwock JA. Late Quaternary geological history of Rio Grande do Sul coastal plain, southern Brazil. Rev Bras Geociênc. 2000;30:474–476. [Google Scholar]
- Tomazelli LJ, Villwock JA.2000O Cenozóico no Rio Grande do Sul Geologia da Planície CosteiraIn: Holz M, de Ros LF, (eds)Geologia do Rio Grande do Sul Editora Universidade Federal do Rio Grande do Sul: Porto Alegre; 375–406. [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Weschenfelder J, Corrêa ICS, Junior EET, Baitelli R. Paleocanais como indicativo de eventos regressivos quaternários do nível do mar no sul do Brasil. Rev Bras Geofís. 2008;26:367–375. [Google Scholar]
- White MJD. Modes of Speciation. Freeman: San Francisco, CA, USA; 1978. [Google Scholar]
- Wlasiuk GJ, Garza C, Lessa EP. Genetic and geographic differentiation in the Rio Negro tuco-tuco (Ctenomys rionegrensis): inferring the roles of migration and drift from multiple genetic markers. Evolution. 2003;57:913–926. doi: 10.1111/j.0014-3820.2003.tb00302.x. [DOI] [PubMed] [Google Scholar]
- Wójcik JM, Ratkiewicz M, Searle JB. Evolution of the common shrew, Sorex araneus: chromosomal and molecular aspects. Acta Theriol. 2002;47:139–167. [Google Scholar]
- Woods C, Kilpatrick C.2005Infraorder Hystricognathi Brandt, 1855In: Wilson DE, Reeder DM, (eds)Mammal Species of the World: A Taxonomic and Geographic Reference3rd ednVol 2Johns Hopkins University Press: Baltimore, MD, USA; 1538–1600. [Google Scholar]
- Yang D-S, Kenagy GJ. Nuclear and mitochondrial DNA reveal contrasting evolutionary processes in populations of deer mice (Peromyscus maniculatus) Mol Ecol. 2009;18:5115–5125. doi: 10.1111/j.1365-294X.2009.04399.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.