Abstract
Natural hosts of simian immunodeficiency virus (SIV), African green monkeys (AGMs), rarely transmit SIV via breast-feeding. In order to examine the genetic diversity of breast milk SIV variants in this limited-transmission setting, we performed phylogenetic analysis on envelope sequences of milk and plasma SIV variants of AGMs. Low-diversity milk virus populations were compartmentalized from that in plasma. However, this compartmentalization was transient, as the milk virus lineages did not persist longitudinally.
TEXT
Postnatal human immunodeficiency virus type 1 (HIV-1) transmission via breast-feeding results in nearly half of the 330,000 infant infections occurring annually (1). Interruption of this mode of infant HIV-1 transmission is key to achieving an HIV-free generation. Interestingly, natural primate hosts of simian immunodeficiency virus (SIV), who coevolved with the virus for thousands of years and sustain a nonpathogenic infection, appear to transmit the virus to their infants postnatally only rarely (2–6). This negligible rate of SIV transmission via breast-feeding in natural SIV hosts contrasts with that of nonnatural SIV hosts, rhesus macaques, who transmit SIV via breast-feeding at a high rate (7). Understanding the mechanisms underlying the rarity of breast milk transmission in natural hosts of SIV could guide the development of interventions to impede postnatal HIV-1 transmission.
In humans, high plasma and milk viral loads are associated with the risk of breast milk transmission of HIV (8). However, in natural SIV hosts, African green monkeys (AGMs) (Chlorocebus sabaeus), breast milk viral loads are similar to that of rhesus monkeys, suggesting that the low rate of postnatal transmission of SIV in AGMs is not a result of low milk virus content (9). A low number of CCR5-expressing CD4+ target cells in infant AGMs (5) and sooty mangabeys (A. Chahroudi, E. Cartwright, D. G. Carnathan, S. T. Lee, B. Lawson, P. M. Carnathan, T. Hashempoor, M. K. Murphy, T. Meeker, S. Ehnert, C. Souder, J. G. Else, J. Cohen, T. H. Vanderford, S. R. Permar, C. A. Derdeyn, F. Villinger, and G. Silvestri, submitted for publication), another natural host species, has also been suggested as a key factor in the low infant infection rate. On the other hand, the strong neutralizing-antibody response observed in breast milk of AGMs (9) is a possible mechanism for the prevention of postnatal virus transmission in the natural hosts of SIV as it contrasts with the limited virus neutralization response detected in the milk of rhesus monkeys (9) and humans (10). Neutralizing antibodies are directed toward and exert selective pressure on the virus Envelope (Env) glycoprotein (11), the key determinant of host-cell interactions and neutralization sensitivity (12, 13). However, the role of neutralizing-antibody and neutralization escape mutants in postnatal HIV/SIV transmission remains unclear (14, 15). Thus, the role of local humoral immune responses and virus env evolution in the breast milk of the natural SIV hosts warrants further investigation.
Breast milk is separated from the systemic compartment by the mammary epithelium, which tightly regulates breast milk content (16). Therefore, it is possible that breast milk represents a distinct virologic compartment, which is characterized by the restriction of viral gene flow between tissues (17). Our previous studies in humans and rhesus monkeys, nonnatural hosts of HIV/SIV, revealed that milk virus variants exhibit limited compartmentalization from plasma virus variants (18, 19). However, whereas the breast milk viral load in humans and rhesus monkeys is 1 to 2 logs lower than that of plasma (19, 20), milk and plasma viral loads in AGMs are similar (9). This finding suggests distinct virus origins and replication mechanisms in breast milk of natural and nonnatural SIV hosts. Experimental infection of AGMs with an endemic SIV strain is a robust model for studying virus populations and immune responses in natural hosts of SIV (21). To study the milk virus population in natural SIV hosts, we inoculated pharmacologically induced lactating AGMs (20) and evaluated their systemic and breast milk virus populations to determine if natural SIV hosts harbor a distinct virus population in breast milk. Analysis of the virus population in milk of AGMs may provide insight on mechanisms that evolved to limit infant acquisition of SIV in natural hosts and has implications for the design of immunologic strategies to prevent postnatal transmission of HIV.
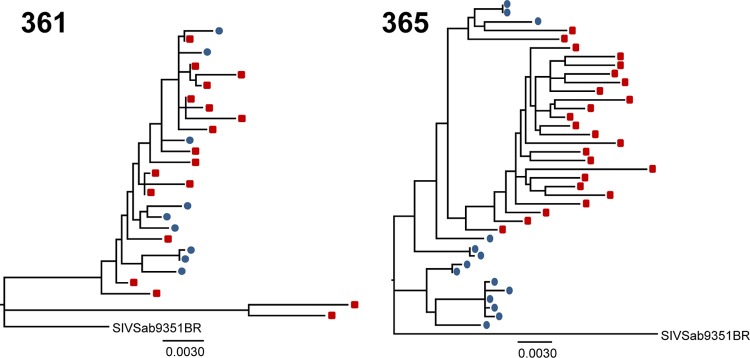
Compartmentalization of breast milk virus variants from the plasma virus population in AGMs chronically infected with a swarm SIVsab virus stock.
We performed single-genome amplification, sequencing, and maximum-likelihood phylogenetic analysis on milk and plasma env virus sequences from two lactating AGMs 3 years following intravenous inoculation with the heterogeneous uncloned SIVsab9351BR virus stock (Fig. 1) (9). Viral RNA was extracted from plasma and milk, reverse transcribed into cDNA, and amplified by single-genome amplification, a method which reduces the likelihood of Taq-induced recombination and template resampling (18, 19, 22, 23). A nested PCR was performed on endpoint diluted-cDNA using the following primers: outer forward (5′-GTGGGTGTTGGAGCTTAGTGTGG-3′), outer reverse (5′-CCATCTCTCAAGCTCTTGGCGC-3′), inner forward (5′-GTTGGAGCTTAGTGTGGTTAGTCC-3′), and inner reverse (5′-GGCGCATTCTTCTTGGATGTG-3′). Amino acid sequences were aligned using MUSCLE (24), and the resulting amino acid alignment was back-translated into the corresponding DNA sequence alignment. Maximum-likelihood trees of plasma and milk virus env sequences were constructed through PAUP* (25), applying the general time-reversible (GTR) substitution model with fixed gamma-distributed rate variation across sites.
Fig 1.
Maximum-likelihood trees of plasma and breast milk virus variants of AGMs 3 years after infection with an uncloned SIVsab virus stock. The trees show full-length SIV env sequences amplified by single-genome amplification from plasma (red squares) and milk (blue circles). There is evidence of compartmentalization between breast milk and plasma virus variants. The P values were generated by the Slatkin-Maddison test of compartmentalization. The scale bars represent 0.003 substitutions per site.
Interestingly, the sequenced milk env variants generally clustered on branches distinct from those in plasma on phylogenetic analysis (Fig. 1). We applied the Slatkin-Maddison test of compartmentalization, a test that compares the minimum possible number of intercompartment migration events to the distribution of migration events in 10,000 randomized trees. Using this statistical test, there was statistical evidence of milk and plasma virus compartmentalization in both monkeys 361 (P = 0.01) and 365 (P < 0.0004).
Acute infection of lactating AGMs with an infectious molecular clone T/F SIVsab variant.
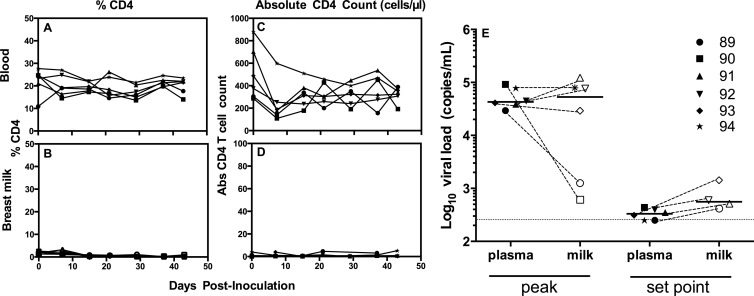
While we observed compartmentalization of breast milk and plasma virus variants in two AGMs 3 years after intravenous infection with an SIV stock consisting of a swarm of viral quasispecies, this model is not representative of the typical mucosal HIV/SIV infection that is initiated by a single or small number of transmitter/founder (T/F) variants (23). Thus, we utilized a previously defined species-specific infectious molecular clone T/F SIV variant (SIVsab92018ivTF) to intravenously infect six female hormone-induced lactating AGMs as previously described (26) (500 ng of p27; TZM-bl 50% tissue culture infectious dose, 2.6 × 106). This model of cloned T/F SIV infection is advantageous for defining viral sequence evolution but also has its limitations, as intravenous infection is not representative of the likely more common mucosal route of SIV infection in this species. As CD4+ T lymphocytes in breast milk are likely to facilitate replication of SIV within the breast milk compartment, we first sought to quantify and compare percentages and absolute numbers of CD4+ T cells in the blood and breast milk following infection. The proportion of CD4+ T lymphocytes was considerably lower in breast milk cells (range, 0 to 3.2%; Fig. 2B) than in blood (range, 10.9 to 26.7%; Fig. 2A) of SIV-infected AGMs. Similarly, the absolute CD4+ T cell count was considerably lower in breast milk (range, 0 to 5 cells/μl; Fig. 2D) than in blood (range, 109 to 881 cells/μl; Fig. 2C). The low level of CD4+ T cells present in breast milk of these AGMs may be a result of the downregulation of CD4 on naive CD4+ T cells upon transition to memory cells (27). However, despite the low number of CD4+ T lymphocyte target cells in breast milk of all SIV-infected AGMs, the peak measured milk viral load at 14 days postinfection (median, 5.3 × 104 copies/ml; range, 6.1 × 102 to 12.2 × 104 copies/ml) was similar to that in plasma (median, 4.3 × 104 copies/ml; range, 2.9 × 104 to 9.2 × 104 copies/ml; P = 0.93; Fig. 2E), with only two of six monkeys having considerably lower milk virus load than plasma virus load. As samples were collected only at 7 days and 14 days postinfection, the peak of viremia may have been missed in the two monkeys with lower-than-expected peak viral loads. The set point viral load in breast milk at 50 days postinfection in animals with adequate milk sample for virus load assessment (n = 4) (median, 5.6 × 102 copies/ml; range, 4.2 × 102 to 1.4 × 103 copies/ml) even trended toward slightly higher values than that of plasma (median, 3.3 × 102 copies/ml; range, 2.5 × 102 to 4.4 × 102 copies/ml; P = 0.09). Although the viral loads observed in this study were low compared to those determined in prior studies in AGMs (9, 26), the kinetics of viremia followed an expected course, and the finding that plasma and breast milk viral loads are of similar magnitudes throughout infection is consistent with results from our previously reported study (9).
Fig 2.
CD4+ T cell counts and viral loads in blood and breast milk of AGMs acutely infected with a T/F SIVsab variant. (A and B) Percentages of CD4+ T lymphocytes in blood (A) and breast milk (B) from six AGMs during acute infection with SIVsab92018ivTF. (C and D) Absolute CD4 T cell counts in blood (C) and breast milk (D) during acute infection with SIVSab92018ivTF. (E) The peak SIV viral load in plasma (closed symbols) is similar to that in breast milk (open symbols) of four SIV-infected AGMs (P = 0.93), whereas the set point milk viral load trends toward a magnitude higher than that in plasma (P = 0.09). Dashed lines are drawn between plasma and milk viral loads from the same monkey, and solid horizontal lines display the medians of each data set. The dotted horizontal line represents the minimum of detection in the virus load assay.
Compartmentalization of breast milk virus variants from the plasma virus population at 5 months post-T/F SIV infection in AGMs.
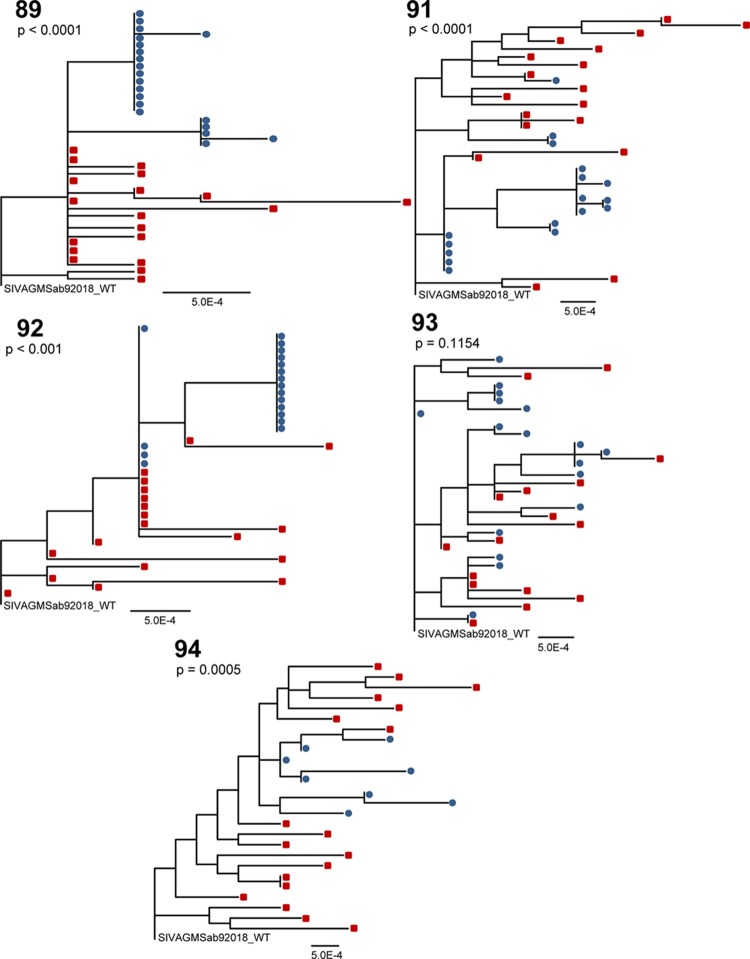
We next investigated SIVsab env sequences of virus variants in milk and blood of five of the six AGMs infected with the cloned T/F variant. Monkey 90 was excluded from analysis as there was insufficient breast milk to extract viral RNA. Primers for SIVsab92018ivTF single-genome amplification included the following: outer forward (5′-CCGCATATGGCCCAGGAAGAG-3′), outer reverse (5′-CACAATTCCCCACTCATTAAGAG-3′), inner forward (5′-ATGCTGCTATCATTGTCCGCTT-3′), and inner reverse (5′-ATTACCCTCAGAGGTACCCTG-3′). Sequences with nonsense mutations were removed from statistical analysis, and a region corresponding to amino acids 123 to 131 (ASKTSKTT) in the SIVSab92018 T/F env clone was removed from the multiple-sequence alignment of the monkey 92 data because substitutions and multiple amino acid insertions in that region generated an ambiguous alignment. The maximum-likelihood trees of plasma and milk virus env sequences revealed clustering of milk virus variants separately from those in plasma in four of five monkeys (Fig. 3). In contrast to the other four monkeys, the plasma and milk virus variants in monkey 93 were largely interspersed throughout the phylogenetic tree. The four monkeys that displayed little to no intermingling of the plasma and breast milk virus variants (monkeys 89, 91, 92, and 94) showed statistical evidence of phylogenetic compartmentalization between blood and milk virus variants by the Slatkin-Maddison test (P value range, P < 0.0001 to P < 0.001). However, the low genetic distance from the breast milk virus variants to their most recent common ancestor compared to that of plasma variants in monkeys 89, 92, and 94 suggested limited evolution of milk virus variants within the mammary gland (data not shown). Furthermore, there did not appear to be differences in the pairwise distances of the plasma and breast milk virus variants from the challenge virus sequence within any of the five AGMs (data not shown), indicating that the breast milk and plasma virus variants evolved at similar rates.
Fig 3.
Compartmentalization of SIVsab92018ivTF env variants in breast milk of four of five AGMs at 5 months postinfection. Maximum-likelihood trees of full-length SIV env sequences amplified by single-genome amplification from plasma (red squares) and milk (blue circles) are shown. The P values were generated by the Slatkin-Maddison test of compartmentalization. The scale bars represent 0.0005 nuclear substitutions per site. WT, wild type.
Local replication of virus variants in the breast milk compartment of SIV-infected AGMs.
In humans and rhesus monkeys, groups of identical HIV/SIV env variants were more common in milk than in blood, indicating the local production of virus (18, 19). In this study, three (monkeys 89, 91, and 92) of the four monkeys that displayed statistical evidence of compartmentalization by the Slatkin-Maddison test demonstrated milk viruses that clustered in large groups of identical or nearly identical sequences, suggesting clonal amplification of virus variants that seeded the breast milk compartment from blood. These large numbers of identical sequences can inflate statistical measures of compartmentalization. However, the compartmentalization observed in the two AGMs infected with an uncloned SIVsab virus stock (Fig. 1) and in monkey 94 (Fig. 3) did not include identical breast milk env sequences. Furthermore, we repeated the Slatkin-Maddison test on the data sets of monkeys 89, 91, and 92 after removing identical sequences within the same compartment. Monkeys 89 and 91 retained statistical evidence of compartmentalization (P < 0.01 and P = 0.003), and yet statistical evidence of compartmentalization was lost in monkey 92 (P = 1), who had only two clonal virus species amplified from milk. Thus, after monotypic sequences were removed, 3 of 5 (60%) monkeys exhibited evidence of compartmentalization, and yet the power of this Slatkin-Maddison test was reduced because so few breast milk sequences remained after removal of the identical sequences.
Fisher's exact test revealed that the number of identical sequences in breast milk was significantly greater than that in plasma for two (monkeys 91 and 92) of the five AGMs (Table 1). Overall, the frequency of identical plasma sequences observed in these monkeys is consistent with those expected based on the number of identical sequences observed in chronically infected HIV subjects (19, 28, 29). However, as there were deletions and insertions among the virus variants in a region that was removed for sequence alignment in monkey 92, the number of identical plasma sequences and the number of identical breast milk sequences may be inflated in this animal (Fig. 3). Nevertheless, the large number of identical sequences in the breast milk compartment of these monkeys suggests that a restricted number of plasma viruses traffic to the breast milk compartment, where they are clonally amplified. If this is correct, then compartmentalization is not a consequence of an independent, evolving subpopulation of viruses in the breast but is rather an effect of a short-lived expansion of migratory viruses or infected cells.
Table 1.
Number of clonally amplified virus variants in plasma and breast milk virus variants of SIV-infected AGMs
| Monkey | Sample type | No. of amplicons that are part of a group of identical sequences | Total no. of Env sequences | % of Env sequences that are part of a group of identical sequences | P value from Fisher's exact test |
|---|---|---|---|---|---|
| 89 | Plasma | 13 | 19 | 68.4 | 0.1274 |
| Breast milk | 18 | 20 | 90.0 | ||
| 91 | Plasma | 2 | 18 | 11.1 | <0.0001 |
| Breast milk | 15 | 17 | 88.2 | ||
| 92 | Plasma | 7 | 19 | 36.8 | <0.0001 |
| Breast milk | 18 | 18 | 100 | ||
| 93 | Plasma | 2 | 17 | 11.8 | 0.2245 |
| Breast milk | 5 | 16 | 31.3 | ||
| 94 | Plasma | 2 | 18 | 11.1 | 1 |
| Breast milk | 0 | 8 | 0 |
Persistent seeding of the breast milk compartment from the systemic virus population and transient compartmentalization of breast milk virus variants in SIV-infected AGMs.
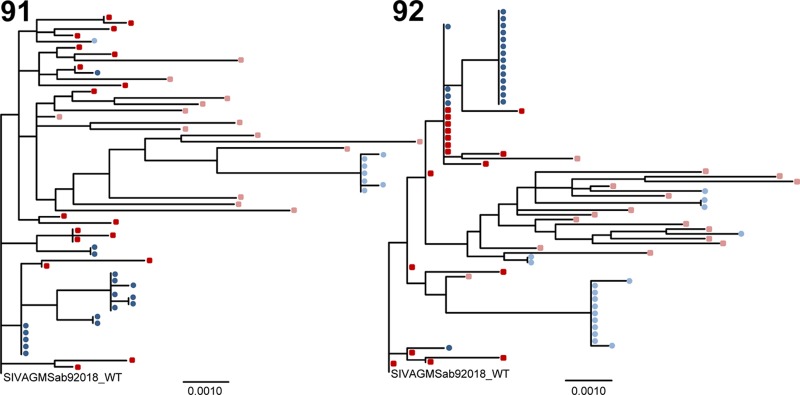
To assess whether there is independent evolution of breast milk virus lineages as a result of selective pressure in the breast milk compartment or whether compartmentalization of breast milk virus variants is transient, we amplified and sequenced env variants from two AGMs infected with the cloned T/F variant 1 year after infection with SIVsab92018ivTF. Compartmentalization of milk virus variants was again demonstrated via the Slatkin-Maddison test at 1 year postinfection in these two monkeys (P < 0.0004 for both). Maximum-likelihood trees of plasma and milk SIV env variants isolated from 5 months and 1 year postinfection show that milk virus variants were most closely related to the concurrent plasma virus population (Fig. 4). As the breast milk virus lineages observed at 5 months postinfection did not persist at 1 year postinfection, the milk virus variants did not evolve as a distinct viral population over time. Moreover, groups of identical breast milk virus variants present at 1 year postinfection were distinct from those present at 5 months postinfection. Thus, virus variants from the periphery are likely continually trafficking to the breast milk compartment and undergoing clonal amplification, producing transient breast milk virus lineages.
Fig 4.
Maximum-likelihood trees of plasma and breast milk virus variants from 5 months postinfection and 1 year postinfection. Trees show full-length SIV env sequences from plasma (red squares) and milk (blue circles) at 5 months postinfection and plasma (pink squares) and milk (light blue circles) at 1 year postinfection. There was transient compartmentalization of virus variants in the breast milk of SIV-infected AGMs, but milk virus variants do not evolve distinctly from that in plasma over time. There were large groups of identical or nearly identical sequences among the breast milk virus variants. The scale bars represent 0.001 nuclear substitutions per site.
This phenomenon of persistent seeding of the breast milk compartment and subsequent clonal replication of virus variants is similar to that found in SIV-infected rhesus monkeys and HIV-infected women (18, 19, 29). However, whereas in this study, there was evidence of compartmentalization in the majority of AGMs both before and after removing monotypic sequences from analysis, in previous studies in rhesus monkeys (18) and humans (19, 29), only limited compartmentalization was observed. The AGM breast milk compartment may be more permeable to viral lineages than that of humans and rhesus monkeys, where there is a greater degree of intermingling between the peripheral and breast milk compartment. Therefore, the transient compartmentalization of milk virus variants in AGMs could reflect the clonal expansion of a more restricted diversity of milk virus variants in the low-level CD4+ T lymphocyte target cell environment (Fig. 2).
Lack of a signature SIV env sequence or unique SIV Env phenotype among AGM breast milk virus variants.
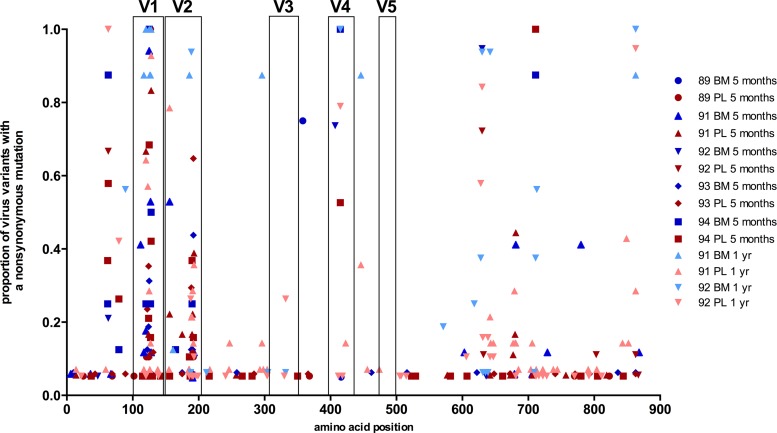
The anatomic compartmentalization of milk virus variants in AGMs warrants an analysis of the differential genotypic and phenotypic characteristics of AGM milk virus variants. We analyzed env gene sequence alignments for each AGM at 5 months (see Fig. S1 in the supplemental material) and 1 year (see Fig. S2) postinfection using Los Alamos National Laboratory's highlighter plot analysis tool to observe sequence attributes common to milk viruses among distinct animals. Furthermore, we mapped the location and calculated the frequency of nonsynonymous substitutions throughout the Env protein (Fig. 5). Using Los Alamos National Laboratory's sequence locator tool (30), the Env variable regions of SIVsab92018ivTF were located according to those previously defined in SIV Mne CL8, a molecular clone of SIV (31). There was a high frequency of nonsynonymous substitutions located in the V1 and V2 loop regions in both plasma and milk env variants. However, there was no single env gene signature sequence that was shared among the milk virus variants across the monkeys and was not found in plasma virus variants. Interestingly, at 5 months postinfection there were a few substitutions that appeared relatively consistently among the milk virus variants within each animal that were absent in plasma virus variants (N358S in monkey 89, S127P in monkey 91, Q407R in monkey 92). However, these substitutions did not persist at 1 year postinfection. Therefore, the distinct nonsynomymous substitutions of milk virus variants at a single time point may reflect selection of virus variants in milk that evolve from a limited number of plasma virus variants crossing the mammary gland epithelial cell barrier.
Fig 5.
Nonsynonymous amino acid substitutions in breast milk (BM) and plasma (PL) Env variants of SIV-infected AGMs. The frequencies of substitutions in plasma virus variants and breast milk virus variants at each amino acid position in the SIVsab92018ivTF Env are shown. There is minimal clustering of substitutions in breast milk virus variants to a specific region of the Env that is distinct from those in plasma across the five monkeys. High-frequency substitutions in both plasma and milk virus variants are clustered in the V1 and V2 loop regions.
To predict potential phenotypic differences in the Env glycoprotein of breast milk and plasma viruses, we investigated whether there were differences in the number of N-linked glycosylation sites or the length of the env gene between the plasma and milk virus variants. Neutralization escape of HIV/SIV has been previously associated with a high number of glycosylation sites and deletions in the external regions of the Env protein which come in contact with Env-specific antibodies (32). We used Los Alamos National Laboratory's N-GlycoSite tool (32) to tally the number of predicted N-linked glycosylation sites within each env sequence. The median numbers of predicted N-linked glycoslyation sites of plasma and milk virus variants were not different between the two compartments (Table 2) (P = 0.32). Similarly, there was no difference in the lengths of the Env amino acid sequences of plasma and milk virus variants (Table 2) (P = 0.5).
Table 2.
Number of N-linked glycosylation sites and the amino acid length of the Env in plasma and breast milk virus variants of SIV-infected AGMsa
| Monkey | Mo postinfection | Sample type | No. of N-linked glycosylation sites |
Env amino acid length |
||
|---|---|---|---|---|---|---|
| Median | Range | Median | Range | |||
| 89 | 5 | Plasma | 23 | 23–24 | 868 | 867–869 |
| 5 | Breast milk | 23 | 23–24 | 868 | 868 | |
| 91 | 5 | Plasma | 24 | 23–25 | 868 | 864–872 |
| 5 | Breast milk | 23 | 22–24 | 868 | 867–870 | |
| 12 | Plasma | 22 | 21–24 | 868 | 863–870 | |
| 12 | Breast milk | 23 | 23 | 865 | 863–865 | |
| 92 | 5 | Plasma | 24 | 24 | 867 | 866–869 |
| 5 | Breast milk | 23 | 22–24 | 867 | 867–869 | |
| 12 | Plasma | 23 | 22–24 | 868 | 865–870 | |
| 12 | Breast milk | 24 | 22–24 | 868 | 866–869 | |
| 93 | 5 | Plasma | 23 | 22–24 | 868 | 866–868 |
| 5 | Breast milk | 23.5 | 23–24 | 866.5 | 866–870 | |
| 94 | 5 | Plasma | 23 | 23–24 | 868 | 867–873 |
| 5 | Breast milk | 24 | 23–24 | 868 | 868 | |
P = 0.32 and P = 0.5 (Wilcoxon signed-rank test) for N-linked glycosylation site and Env amino acid length data, respectively.
We next assessed whether there was evidence of positive selection acting on the breast milk sequences compared to the plasma sequences. The Codeml program implemented in PAML (33) was used to calculate the percentage of codons in the SIV env that were under negative, neutral, or positive selection. The ratio of the nonsynonymous substitution rate to the synonymous substitution rate (ω) indicates the presence of negative (ω < 0), neutral (ω = 0), or positive (ω > 1) selection. There was evidence of positive selection on milk env sequences in two of five of the monkeys, with 2.3% of the env codons in monkey 91 (ω = 28.3) and 2.2% of the env codons in monkey 92 (ω = 11.1) exhibiting positive selection. However, the sites of positive selection did not overlap in the two monkeys and did not appear to locate to particular regions of the Env, further suggesting that there is no distinct evolution of breast milk variants.
Finally, in order to assess whether evolutionary rates differed between the plasma and breast milk lineages, we applied the Bayesian Markov chain Monte Carlo (MCMC) method, implemented by BEAST v1.7.5 (34), to analyze the evolutionary rate of the SIV env in the two monkeys for which samples were collected at both 5 months and 1 year postinfection.
BEAST was run with the following parameters: no coalescent model; uncorrelated lognormal relaxed clock; GTR substitution model; root height = 7 to 12.5 months; 50,000,000 MCMC iterations; sampling every 5,000 steps. We manually inspected the trace to ensure convergence. The burn-in was set at 10% of the total number of iterations, and all parameters had an effective sample size (ESS) greater than 300. The substitution rate for the SIVsab env of milk and plasma virus variants ranged between 2.81 × 10−3 and 4.07 × 10−3 nucleotide substitutions per site per year, which is consistent with previously reported SIVagm substitution rates (35). Although the substitution rate on the branch leading to the major breast milk lineage at 1 year postinfection is slightly higher than that on branches leading to plasma virus variants, there was no consistent pattern to suggest evidence that the breast milk virus lineage evolved at a higher rate (see Fig. S3 in the supplemental material). Together, these observations suggest that plasma and milk viruses likely evolve under the same selective pressure.
In summary, the breast milk virus variants in SIV-infected AGMs are largely compartmentalized from that in blood, which is in contrast to the limited compartmentalization previously described in the human and rhesus monkey HIV/SIV hosts (18, 19, 29). However, the lack of distinct breast milk substitutions that persist longitudinally and the lack of phenotypic differences between the plasma and breast milk sequences support the model of stochastic migration and subsequent local replication of plasma virus variants in the mammary gland. While this model has been previously observed in humans and rhesus monkeys as well (18, 19), the low number of CD4+ T lymphocyte target cells in the breast milk compartment of AGMs may contribute to a more limited diversity of milk virus variants and subsequently a greater degree of compartmentalization. As only a limited number of maternal HIV variants are postnatally transmitted to infants (7, 36, 37), it is possible that the restricted diversity of AGM milk virus variants limits the number of virus variants fit for transmission across the genetic bottleneck introduced by the mucosal barrier of the infant gastrointestinal tract. The restricted diversity of milk variants and the limited number of CCR5-expressing CD4+ target cells in infant AGMs (5, 38) may together contribute to the rarity of breast milk transmission in natural hosts. Future investigations should delve into the fitness, neutralization sensitivity, and infectivity of these milk viruses in low-level CCR5-expressing target cell populations to determine the contribution of the restricted virus population in breast milk of AGMs to the rarity of infant virus transmission in natural SIV hosts.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brandon Keele for provision of assistance with SGA and primer sequences and Beatrice Hahn and Nicholas Parrish for provision of the SIVsab92018ivTF clone.
Support for this work was provided by National Institutes of Health grants R21AI100760 and K08AI087992 (S.R.P.), the Center For HIV/AIDS Vaccine Immunology (CHAVI; grant U19 AI067854), and the Duke University Center for AIDS Research (CFAR; P30 AI064518).
Footnotes
Published ahead of print 7 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01643-13.
REFERENCES
- 1.Nduati R, John G, Mbori-Ngacha D, Richardson B, Overbaugh J, Mwatha A, Ndinya-Achola J, Bwayo J, Onyango FE, Hughes J. 2000. Effect of breastfeeding and formula feeding on transmission of HIV-1. JAMA 283:1167–1174. [DOI] [PubMed] [Google Scholar]
- 2.Chahroudi A, Meeker T, Lawson B, Ratcliffe S, Else J, Silvestri G. 2011. Mother-to-infant transmission of simian immunodeficiency virus is rare in sooty mangabeys and is associated with low viremia. J. Virol. 85:5757–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otsyula M, Gettie A, Suleman M, Tarara R, Mohamed I, Marx P. 1995. Apparent lack of vertical transmission of simian immunodeficiency virus (SIV) in naturally infected African green monkeys, Cercopithecus aethiops. Ann. Trop. Med. Parasitol. 89:573. [DOI] [PubMed] [Google Scholar]
- 4.Pandrea I, Sodora DL, Silvestri G, Apetrei C. 2008. Into the wild: simian immunodeficiency virus (SIV) infection in natural hosts. Trends Immunol. 29:419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pandrea I, Onanga R, Souquiere S, Mouinga-Ondéme A, Bourry O, Makuwa M, Rouquet P, Silvestri G, Simon F, Roques P. 2008. Paucity of CD4+ CCR5+ T cells may prevent transmission of simian immunodeficiency virus in natural nonhuman primate hosts by breast-feeding. J. Virol. 82:5501–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fultz PN, Gordon TP, Anderson DC, McClure HM. 1990. Prevalence of natural infection with simian immunodeficiency virus and simian T-cell leukemia virus type I in a breeding colony of sooty mangabey monkeys. AIDS 4:619–626. [DOI] [PubMed] [Google Scholar]
- 7.Amedee AM, Lacour N, Ratterree M. 2003. Mother-to-infant transmission of SIV via breast-feeding in rhesus macaques. J. Med. Primatol. 32:187–193. [DOI] [PubMed] [Google Scholar]
- 8.Rousseau CM, Nduati RW, Richardson BA, John-Stewart GC, Mbori-Ngacha DA, Kreiss JK, Overbaugh J. 2004. Association of levels of HIV-1-infected breast milk cells and risk of mother-to-child transmission. J. Infect. Dis. 190:1880–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilks AB, Perry JR, Ehlinger EP, Zahn RC, White R, Gauduin M-C, Carville A, Seaman MS, Schmitz JE, Permar SR. 2011. High cell-free virus load and robust autologous humoral immune responses in breast milk of simian immunodeficiency virus-infected African green monkeys. J. Virol. 85:9517–9526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fouda GG, Yates NL, Pollara J, Shen X, Overman GR, Mahlokozera T, Wilks AB, Kang HH, Salazar-Gonzalez JF, Salazar MG. 2011. HIV-specific functional antibody responses in breast milk mirror those in plasma and are primarily mediated by IgG antibodies. J. Virol. 85:9555–9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richman DD, Wrin T, Little SJ, Petropoulos CJ. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144–4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffin JM. 1986. Genetic variation in AIDS viruses. Cell 46:1–4. [DOI] [PubMed] [Google Scholar]
- 13.Overbaugh J, Rudensey LM. 1992. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J. Virol. 66:5937–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouda GG, Mahlokozera T, Salazar-Gonzalez JF, Salazar MG, Learn G, Kumar SB, Dennison SM, Russell E, Rizzolo K, Jaeger F, Cai F, Vandergrift NA, Gao F, Hahn B, Shaw GM, Ochsenbauer C, Swanstrom R, Meshnick S, Mwapasa V, Kalilani L, Fiscus S, Montefiori D, Haynes B, Kwiek J, Alam SM, Permar SR. 2013. Postnatally-transmitted HIV-1 envelope variants have similar neutralization-sensitivity and function to that of nontransmitted breast milk variants. Retrovirology 10:3. 10.1186/1742-4690-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch JB, Nduati R, Blish CA, Richardson BA, Mabuka JM, Jalalian-Lechak Z, John-Stewart G, Overbaugh J. 2011. The breadth and potency of passively acquired human immunodeficiency virus type 1-specific neutralizing antibodies do not correlate with the risk of infant infection. J. Virol. 85:5252–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgeson JC, Filteau SM. 2000. Physiology, immunology, and disease transmission in human breast milk. AIDS Patient Care STDs 14:533–539. [DOI] [PubMed] [Google Scholar]
- 17.Nickle DC, Shriner D, Mittler JE, Frenkel LM, Mullins JI. 2003. Importance and detection of virus reservoirs and compartments of HIV infection. Curr. Opin. Microbiol. 6:410–416. [DOI] [PubMed] [Google Scholar]
- 18.Permar S, Kang H, Wilks A, Mach L, Carville A, Mansfield K, Learn G, Hahn B, Letvin N. 2010. Local replication of simian immunodeficiency virus in the breast milk compartment of chronically-infected, lactating rhesus monkeys. Retrovirology 7:7. 10.1186/1742-4690-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salazar-Gonzalez JF, Salazar MG, Learn GH, Fouda GG, Kang HH, Mahlokozera T, Wilks AB, Lovingood RV, Stacey A, Kalilani L, Meshnick SR, Borrow P, Montefiori DC, Denny TN, Letvin NL, Shaw GM, Hahn BH, Permar SR; Center for HIV/AIDS Vaccine Immunology A0167854. 2011. Origin and evolution of HIV-1 in breast milk determined by single-genome amplification and sequencing. J. Virol. 85:2751–2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Permar SR, Kang HH, Carville A, Mansfield KG, Gelman RS, Rao SS, Whitney JB, Letvin NL. 2008. Potent simian immunodeficiency virus-specific cellular immune responses in the breast milk of simian immunodeficiency virus-infected, lactating rhesus monkeys. J. Immunol. 181:3643–3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pandrea I, Apetrei C, Dufour J, Dillon N, Barbercheck J, Metzger M, Jacquelin B, Bohm R, Marx PA, Barre-Sinoussi F, Hirsch VM, Müller-Trutwin MC, Lackner AA, Veazey RS. 2006. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J. Virol. 80:4858–4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BTM, Sharp PM, Shaw GM, Hahn BH. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping L-H, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar R. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods), v. 4 Sinauer Associates, Sunderland, MA. [Google Scholar]
- 26.Gnanadurai CW, Pandrea I, Parrish NF, Kraus MH, Learn GH, Salazar MG, Sauermann U, Töpfer K, Gautam R, Münch J, Stahl-Henning C, Apetrei C, Hahn BH, Kirchhoff F. 2010. Genetic identity and biological phenotype of a transmitted/founder virus representative of nonpathogenic simian immunodeficiency virus infection in African green monkeys. J. Virol. 84:12245–12254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beaumier CM, Harris LD, Goldstein S, Klatt NR, Whitted S, McGinty J, Apetrei C, Pandrea I, Hirsch VM, Brenchley JM. 2009. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat. Med. 15:879–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parrish NF, Wilen CB, Banks LB, Iyer SS, Pfaff JM, Salazar-Gonzalez JF, Salazar MG, Decker JM, Parrish EH, Berg A, Hopper J, Hora B, Kumar A, Mahlokozera T, Yuan S, Coleman C, Vermeulen M, Ding H, Ochsenbauer C, Tilton JC, Permar SR, Kappes JC, Betts MR, Busch MP, Gao F, Montefiori D, Haynes BF, Shaw GM, Hahn BH, Doms RW. 2012. Transmitted/founder and chronic subtype C HIV-1 use CD4 and CCR5 receptors with equal efficiency and are not inhibited by blocking the integrin α4β7. PLoS Pathog. 8:e1002686. 10.1371/journal.ppat.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gantt S, Carlsson J, Heath L, Bull ME, Shetty AK, Mutsvangwa J, Musingwini G, Woelk G, Zijenah LS, Katzenstein DA, Mullins JI, Frenkel LM. 2010. Genetic analyses of HIV-1 env sequences demonstrate limited compartmentalization in breast milk and suggest viral replication within the breast that increases with mastitis. J. Virol. 84:10812–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calef C, Mokili J, O'Connor DH, Watkins DI, Korber B. 2001. Numbering positions in SIV relative to SIVMM239 group 4:1. http://www.hiv.lanl.gov/content/sequence/HIV/REVIEWS/SIV_NUMBERING2001/SivNumbering.html.
- 31.Overbaugh J, Rudensey L, Papenhausen M, Benveniste R, Morton W. 1991. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J. Virol. 65:7025–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229–1246. [DOI] [PubMed] [Google Scholar]
- 33.Yang Z. 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- 34.Drummond A, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Müller-Trutwin MC, Corbet S, Tavares MD, Hervé VMA, Nerrienet E, Georges-Courbot M-C, Saurin W, Sonigo P, Barré-Sinoussio Fß. 1996. The evolutionary rate of nonpathogenic simian immunodeficiency virus (SIVagm) is in agreement with a rapid and continuous replication in vivo. Virology 223:89–102. [DOI] [PubMed] [Google Scholar]
- 36.Wolinsky S, Wike C, Korber B, Hutto C, Parks W, Rosenblum L, Kunstman K, Furtado M, Munoz J. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134–1137. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Tully DC, Hoffmann FG, He J, Kankasa C, Wood C. 2010. Restricted genetic diversity of HIV-1 subtype C envelope glycoprotein from perinatally infected Zambian infants. PLoS One 5:e9294. 10.1371/journal.pone.0009294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pandrea I, Parrish NF, Raehtz K, Gaufin T, Barbian HJ, Ma D, Kristoff J, Gautam R, Zhong F, Haret-Richter GS. 2012. Mucosal simian immunodeficiency virus transmission in African green monkeys: susceptibility to infection is proportional to target cell availability at mucosal sites. J. Virol. 86:4158–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.