Abstract
Antigen-presenting cells are a heterogeneous group of cells that are characterized by their functional specialization. Consequently, targeting specific antigen-presenting cell subsets offers opportunities to induce distinct T cell responses. Here we report on the generation and use of nanobodies (Nbs) to target lentivectors specifically to human lymph node-resident myeloid dendritic cells, demonstrating that Nbs represent a powerful tool to redirect lentivectors to human antigen-presenting cell subsets.
TEXT
Since the discovery of dendritic cells (DCs) in 1973, these antigen-presenting cells have been at the center of attention in vaccine development (1). In the past decade, an extra dimension of complexity was introduced by revealing an unforeseen diversity of DC subsets with distinct functions and locations (2–4). Since DC subsets induce distinct immune responses, it might be beneficial to develop strategies to target particular DC subsets and as such exploit the immune system to its full potential (5).
Numerous animal studies have proven the efficiency and safety of lentivectors (LVs) as vaccination moieties (6, 7). As lentivectors are intrinsically immunogenic, they deliver both antigens and activation signals to antigen-presenting cells (8). Furthermore, the lentivectors' envelopes are well suited for engineering, enabling the design of targeted lentivectors. Several methods have been described to redirect lentivectors to specific antigen-presenting cells (9–12). However, to our knowledge subset-specific delivery of transgenes has not been described.
Targeting myeloid DCs could be advantageous as they are considered to be important mediators of antigen-specific immunity. They are able to induce proper and oriented stimulation of CD4+ T helper 1 and CD8+ cytotoxic T cells. In addition, targeting may reduce the risk of adverse reactions such as autoimmune responses or induction of tolerance due to transgene expression and presentation by non-antigen-presenting cells or tolerogenic DC subtypes. Finally, as myeloid DCs have a limited life span, their targeting should result in a natural clearance of the lentivector and as such in a reduction of the risk of insertional mutagenesis.
We recently delivered a proof of concept on the use of nanobodies (Nbs) to target lentivectors to antigen-presenting cells (10). Nbs or VHH fragments are antibody fragments of about 12 to 25 kDa that are engineered from heavy-chain-only antibodies found in Camelidae. Because of their size and target affinity, they are of particular interest as targeting moieties. In the present study, we further refined the transduction profile of lentivectors, targeting them to human myeloid DCs using Nbs.
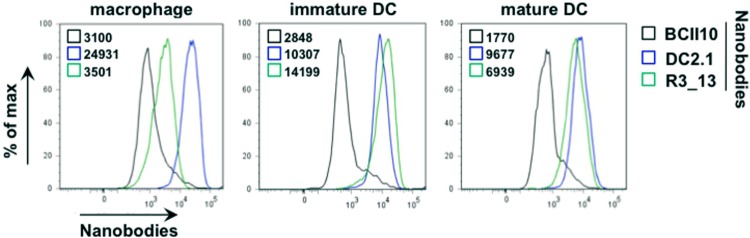
Two Nb libraries, derived from peripheral blood lymphocytes of llamas that were immunized with immature or lipopolysaccharide-stimulated murine bone marrow-derived DCs, were at our disposal (13). These were screened for cross-reactivity with human immature DCs. We selected two Nbs, DC2.1 and R3_13, after three consecutive rounds of cellular panning and extensive flow cytometry-based characterization on in vitro-generated human DCs and macrophages (Fig. 1). Nb BCII10, specific for subunit 10 of β-lactamase, served as a negative control.
Fig 1.
Screening of antigen-presenting cell binding nanobodies. Two libraries of phage-displaying Nbs obtained from peripheral blood lymphocytes of llamas immunized with immature or lipopolysaccharide-stimulated murine DCs were subjected to three consecutive rounds of cellular panning on human DCs. The gene segments encoding antigen-presenting cell binding Nbs were recloned into an expression vector introducing a hexahistidine tag-coding sequence at the carboxy terminus. Binding to human in vitro-generated monocyte-derived macrophages and DCs (immature or stimulated with a maturation cocktail composed of the Toll-like receptor 7 [TLR7]/8 agonist R848, gamma interferon [IFN-γ], tumor necrosis factor alpha [TNF-α], and prostaglandin E2 for 24 h) was visualized by flow cytometry, using fluorescein isothiocyanate (FITC)-tagged anti-histidine antibodies as detecting reagents. The black graphs represent the binding profiles of the Nb BCII10 (control). The blue and green graphs represent the binding profiles of the antigen-presenting cell binding Nbs DC2.1 and R3_13. The numbers next to the colored boxes represent the mean fluorescence intensities. The results of one representative experiment are shown (n = 2).
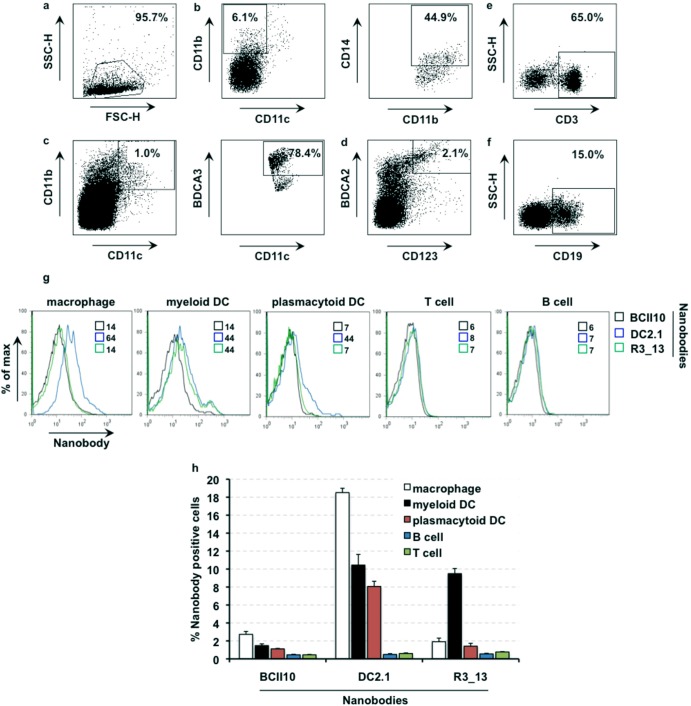
We next evaluated their specificity on single-cell suspensions derived from human lymph nodes. These cells were stained for 1 h with 1 μg/ml of the respective Nbs as well as antibodies that are directed against markers that allow discrimination of macrophages (CD11b, CD11c, CD14), myeloid DCs (CD11b, CD11c, BDCA3), plasmacytoid DCs (CD123, BDCA2), and T (CD3) and B (CD19) cells (Fig. 2a to f). We observed that Nb DC2.1 bound to human lymph node-resident macrophages and myeloid and plasmacytoid DCs, whereas binding of Nb R3_13 was restricted to myeloid DCs (Fig. 2g and h). The latter is contradictory to the binding pattern observed using in vitro-generated human macrophages. This might be explained by a difference in the levels of expression of the antigen recognized by Nb R3_13 on primary versus in vitro-generated macrophages. Importantly, this observation highlights the necessity of evaluating targeting moieties on primary cell types.
Fig 2.
Binding of nanobodies DC2.1 and R3_13 to human lymph node-derived single-cell suspensions. Panels a to f depict the gating strategy to define macrophages and myeloid and plasmacytoid DCs as well as T and B cells. (a) Dead and contaminating cells were excluded by gating on forward scatter (height) (FSC-H)/side scatter (height) (SSC-H) characteristics. (b) To define macrophages, the cells were first gated as CD11b+ and CD11c− cells. Within this population, the cells were further defined as CD14+. (c) To define myeloid DCs, cells were gated as CD11b/CD11c+ cells and further gated as BDCA3+ cells. (d) Plasmacytoid DCs were defined as CD123/BDCA2+ cells. (e and f) T cells (e) and B cells (f) were defined based on the expression of CD3 and CD19, respectively. (g) Binding of the respective Nbs on human lymph node-derived cells was evaluated by flow cytometry (n = 2). The numbers next to the colored boxes represent the mean fluorescence intensities. (h) The column graph depicts the percentages of Nb+ cells (y axis) in the macrophage, myeloid DC, plasmacytoid DC, B cell, and T cell populations. The percentages are shown as means and standard errors of the means of the results of two independent experiments.
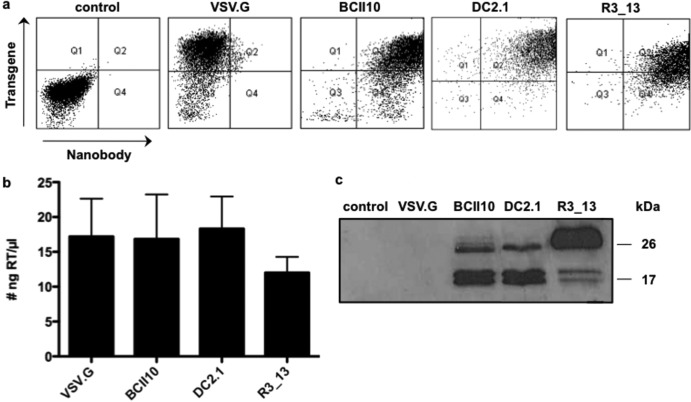
We next applied the Nb-display technology to generate targeted lentivectors. This innovative approach exploits the budding mechanism of lentivectors to incorporate both Nbs and the binding-defective but fusion-competent vesicular stomatitis virus glycoprotein (VSV.Gs) on the viral surface (10, 14). Producer cells that were modified to express Nb BCII10, Nb DC2.1 or Nb R3_13 on their cell membrane were generated by transducing HEK293T cells with lentivectors encoding the membrane-bound form of these Nbs (10). These cells were used to generate Thy1.1-encoding Nb-displaying lentivectors (referred to as BCII10-, DC2.1-, and R3_13-LVs), while nonmodified HEK293T cells were used to generate broad-tropism lentivectors (VSV.G-LVs). Three days after transfection, flow cytometry revealed high Thy1.1 expression on all producer cells as well as high expression of the Nbs (myc tag) on Nb-modified HEK293T cells (Fig. 3a). A commercially available colorimetric reverse transcriptase (RT) assay was used to determine the amount of RT in the 1,000-fold-concentrated lentivector preparations. We demonstrated high levels of RT in the different lentivector preparations (Fig. 3b). Comparison of the RT content with the titer of VSV.G-LVs determined in flow cytometry revealed that 1 ng RT correlates with 2.5 × 104 infectious particles. Western blot analysis (using 5 ng RT per lentivector) was performed to confirm the presence of VSV.Gs and Nbs on the targeted lentivectors. To that end, we used an anti-hemagglutinin (HA) antibody, which binds to the HA tag present within both VSV.Gs and Nbs. Lentivectors pseudotyped with VSV.G served as a negative control, as this envelope does not contain an HA tag and as these vectors are devoid of Nbs (Fig. 3c). Of note, we observed that R3_13-LVs had incorporated an amount of Nbs on their surface that was larger than the amounts seen with BCII10- and DC2.1-LVs. This is explained by the level of Nbs that are expressed on the producer cell lines, which, based on the mean fluorescence intensity, was higher in Nb R3_13-modified producer cells (data not shown).
Fig 3.
Production of DC2.1- and R3_13-displaying lentivectors. (a) Nonmodified HEK293T cells or HEK293T cells stably expressing Nb BCII10, DC2.1, or R3_13 were used to produce VSV.G-LVs or Nb-displaying lentivectors, respectively. Three days after transfection of the producer cells, flow cytometry was applied to evaluate the expression of the transgene (Thy1.1) and the Nbs (myc tag) on the cell membrane. Nontransfected HEK293T cells served as a control. The results of one representative experiment are shown (n = 7). (b) To characterize the lentivectors, we determined their RT content. The graph depicts the amounts of RT as means and standard errors of the means (n = 7). (c) Western blot analysis of the lentivectors was performed as a quality control. After separation on a 15% sodium dodecyl sulfate-polyacrylamide gel and transfer to a nitrocellulose membrane, the Nbs (± 25 kDa) and VSV.Gs (± 15 kDa), which both contained an HA tag, were detected with an anti-HA antibody. The results of one representative experiment are shown (n = 3).
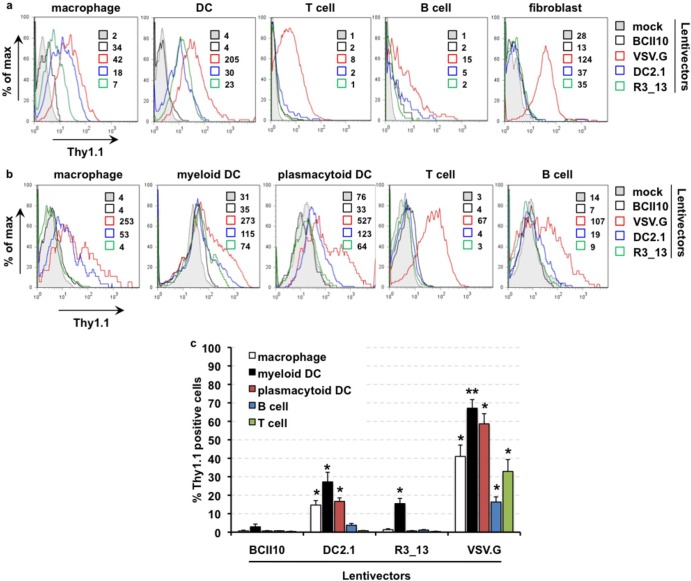
To assess the specificity of the lentivectors, we transduced in vitro-generated human macrophages and DCs as well as thawed peripheral blood lymphocytes with the respective Thy1.1-encoding lentivectors at 60 ng RT per 105 cells. Human in vitro-cultured fibroblasts were transduced at 20 ng RT per 105 cells. Three days later, flow cytometry showed Thy1.1 expression in all cell types upon transduction with VSV.G-LVs whereas none of the cell types were transduced when incubated with BCII10-LVs. More importantly, upon transduction with DC2.1- and R3_13-LVs, we observed Thy1.1 expression only in macrophages and DCs, except for a very low level of B cell transduction by DC2.1-LVs (Fig. 4a). We next evaluated the transduction profile of the lentivectors on the stated human lymph node-derived single-cell suspensions. Three days after transduction, we evaluated the expression of Thy1.1 on the different cell subsets by flow cytometry (Fig. 4b and c). These experiments demonstrated that VSV.G-LVs transduced all cell types evaluated, whereas BCII10-LVs did not. Transduction of macrophages and of myeloid and plasmacytoid DCs, as well as a low level transduction of B cells was observed upon transduction with DC2.1-LVs. Importantly, only myeloid DCs were transduced when R3_13-LVs were used, which is in line with the binding profile of the Nbs on human lymph node-derived cells.
Fig 4.
Transduction of human cells by broad tropism using DC2.1- or R3_13-displaying lentivectors. (a) Human in vitro-generated monocyte-derived macrophages and DCs (day 3 of culture) and thawed peripheral blood lymphocytes were transduced with the respective lentivectors (60 ng RT per 105 cells), while human in vitro-cultured fibroblasts were transduced at 20 ng RT per 105 cells (n = 2). (b and c) Human lymph nodes were reduced to single-cell suspensions. These were transduced with lentivectors at 60 ng RT per 105 cells. Analysis of transgene expression (Thy1.1) was performed 3 days after transduction by flow cytometry (n = 3). (c) The numbers next to the colored boxes represent the mean fluorescence intensities. (d) The column graph depicts the percentages of Thy1.1+ (hence, transduced) cells (y axis) in the respective cell populations. The percentages are shown as means and standard errors of the means (n = 3). A Student t test comparing transduction of the respective populations with DC2.1-, R3_13-, or VSV.G-LVs to the results obtained with BCII10-LVs was performed to evaluate the statistical significance. Numbers of asterisks in the figures indicate the levels of statistical significance as follows: *, P < 0.05; **, P < 0.01.
Taken together, the present data show that immunization of llama with murine DCs elicits Nbs that cross-react with human myeloid cells. Importantly, one of these Nbs (R3_13) can target lentivectors specifically to lymph node-resident myeloid DCs. This finding opens the perspective of a vaccination strategy that induces very particular immune responses by targeting a specific DC subset, which enables full exploitation of the immune system.
ACKNOWLEDGMENTS
This research was performed with the financial support of the Research Foundation Flanders (FWO-V), the Agency of Innovation by Science and Technology (IWT-SBO), the Interuniversity Attraction Poles Program (IUAP), the Belgian State-Belgian Science Policy, and the research committee of the VUB (OZR). C.G. and K.B. are funded by the FWO-V.
Footnotes
Published ahead of print 17 July 2013
REFERENCES
- 1.Steinman RM, Cohn ZA. 1973. Identification of a novel cell type in peripheral lymphoid organs of mice. I. Morphology, quantitation, tissue distribution. J. Exp. Med. 137:1142–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coquerelle C, Moser M. 2010. DC subsets in positive and negative regulation of immunity. Immunol. Rev. 234:317–334. [DOI] [PubMed] [Google Scholar]
- 3.Liu YJ. 2001. Dendritic cell subsets and lineages, and their functions in innate and adaptive immunity. Cell 106:259–262. [DOI] [PubMed] [Google Scholar]
- 4.Ueno H, Klechevsky E, Morita R, Aspord C, Cao T, Matsui T, Di Pucchio T, Connolly J, Fay JW, Pascual V, Palucka AK, Banchereau J. 2007. Dendritic cell subsets in health and disease. Immunol. Rev. 219:118–142. [DOI] [PubMed] [Google Scholar]
- 5.Palucka K, Banchereau J, Mellman I. 2010. Designing vaccines based on biology of human dendritic cell subsets. Immunity 33:464–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Escors D, Breckpot K. 2010. Lentiviral vectors in gene therapy: their current status and future potential. Arch. Immunol. Ther. Exp. (Warsz) 58:107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu B, Tai A, Wang P. 2011. Immunization delivered by lentiviral vectors for cancer and infectious diseases. Immunol. Rev. 239:45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breckpot K, Escors D, Arce F, Lopes L, Karwacz K, Van Lint S, Keyaerts M, Collins M. 2010. HIV-1 lentiviral vector immunogenicity is mediated by Toll-like receptor 3 (TLR3) and TLR7. J. Virol. 84:5627–5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ageichik A, Buchholz CJ, Collins MK. 2011. Lentiviral vectors targeted to MHC II are effective in immunization. Hum. Gene Ther. 22:1249–1254. [DOI] [PubMed] [Google Scholar]
- 10.Goyvaerts C, De Groeve K, Dingemans J, Van Lint S, Robays L, Heirman C, Reiser J, Zhang XY, Thielemans K, De Baetselier P, Raes G, Breckpot K. 2012. Development of the Nanobody display technology to target lentiviral vectors to antigen-presenting cells. Gene Ther. 19:1133–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morizono K, Ku A, Xie Y, Harui A, Kung SK, Roth MD, Lee B, Chen IS. 2010. Redirecting lentiviral vectors pseudotyped with Sindbis virus-derived envelope proteins to DC-SIGN by modification of N-linked glycans of envelope proteins. J. Virol. 84:6923–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang L, Yang H, Rideout K, Cho T, Joo KI, Ziegler L, Elliot A, Walls A, Yu D, Baltimore D, Wang P. 2008. Engineered lentivector targeting of dendritic cells for in vivo immunization. Nat. Biotechnol. 26:326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Groeve K, Deschacht N, De Koninck C, Caveliers V, Lahoutte T, Devoogdt N, Muyldermans S, De Baetselier P, Raes G. 2010. Nanobodies as tools for in vivo imaging of specific immune cell types. J. Nucl. Med. 51:782–789. [DOI] [PubMed] [Google Scholar]
- 14.Cronin J, Zhang XY, Reiser J. 2005. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 5:387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]