Abstract
Baculoviruses are insect-specific viruses commonly found in nature. They are not able to replicate in mammalian cells but can transduce them when equipped with an appropriate mammalian cell active expression cassette. Although the viruses have been studied in several types of mammalian cells from different origins, the receptor that baculovirus uses to enter or interact with mammalian cells has not yet been identified. Due to the wide tropism of the virus, the receptor has been suggested to be a generally found cell surface molecule. In this article, we investigated the interaction of baculovirus and mammalian cell surface heparan sulfate proteoglycans (HSPG) in more detail. Our data show that baculovirus requires HSPG sulfation, particularly N- and 6-O-sulfation, to bind to and transduce mammalian cells. According to our results, baculovirus binds specifically to syndecan-1 (SDC-1) but does not interact with SDC-2 to SDC-4 or with glypicans. Competition experiments performed with SDC-1 antibody or recombinant SDC-1 protein inhibited baculovirus binding, and SDC-1 overexpression enhanced baculovirus-mediated transduction. In conclusion, we show that SDC-1, a commonly found cell surface HSPG molecule, has a role in the binding and entry of baculovirus in vertebrate cells. The results presented here reveal important aspects of baculovirus entry and can serve as a basis for next-generation baculovirus vector development for gene delivery.
INTRODUCTION
Baculoviruses are enveloped insect viruses belonging to the family Baculoviridae, and they have a large DNA genome of approximately 80 to 180 kbp in size (1). They are commonly found in nature and have been widely used as biological control agents (2). Within the past 30 years, however, the baculoviruses have also been utilized and developed for the broad purposes of biotechnology and as viral vectors to deliver transgenes into mammalian cells (3–5). As gene delivery vehicles, baculoviruses have a wide range of advantages, such as their large transgene capacity, easy production, and high gene expression in transduced cells (6). One of the biggest advantages, however, is their inability to replicate in mammalian cells, making them safe to use for gene transfer in cells outside their natural tropism (7, 8). The most widely studied and used baculovirus, AcMNPV (Autographa californica multiple nucleopolyhedrovirus), belonging to the genus Alphabaculovirus, can transduce a wide variety of mammalian cells and tissues (4, 5, 9, 10). The most permissive mammalian cells for AcMNPV have been found to be the ones from hepatic and osteosarcoma origin, and the poorest ones are from hematopoietic origin (5, 10–15).
The trafficking route which AcMNPV uses to travel from the cell surface to the nucleus has been investigated yet still remains largely unknown (16–21). At the cell surface, the virus has been suggested to take advantage of nonspecific electrostatic interactions (21, 22) and attach to a general cell surface molecule, such as a phospholipid or heparan sulfate proteoglycan (HSPG) (19, 21–25). No specific receptor has been identified so far. Baculovirus trafficking and clathrin-independent entry have been shown to be regulated by RhoA and Arf6 (16). Following entry, the virus is vesicularly transported until the pH-dependent fusion of the viral envelope with the endosome facilitates the release of the capsid to the cytoplasm (16, 19, 26). The escaped nucleocapsid enters the nucleus via nuclear pores with the help of actin filaments (27, 28). When the nucleus is reached, the nucleocapsid disassembles and releases the viral DNA (17, 18, 20, 29).
HSPGs are molecules located on the plasma membrane of all animal cells and constitute the major components of extracellular matrices. They are divided into two major subfamilies, syndecans and glypicans (30). The syndecans in particular have been shown to serve as receptors for several viruses, including HIV-1 (31, 32), herpes simplex virus (33), and human papillomavirus (34). Syndecans are highly anionic and linear HSPGs displayed at the cell surface as transmembrane receptors (35–37). The syndecan family is divided into 4 members, syndecan-1 (SDC-1) to SDC-4, all of which have a short cytoplasmic domain, a single-span transmembrane domain, and an extracellular domain containing attachment sites for 3 to 5 heparan sulfate or chondroitin sulfate chains. Transmembrane and cytoplasmic domains are highly conserved among the members, and the expression of the syndecans is cell and tissue specific (38). SDC-1 and SDC-3 have two distinct glycosaminoglycan attachment (GAG) sites near the N terminus and the membrane attachment site. SDC-2 and SDC-4 have the GAG attachment sites in the distal part of the ectodomain (38). SDC-1 is mainly found in epithelial and plasma cells; SDC-2 in fibroblasts, endothelial cells, neurons and smooth muscle cells; and SDC-3 in the nervous system. SDC-4 is the most ubiquitously expressed (35, 39). In general, syndecans have a role in cell proliferation, differentiation, adhesion, and migration. They are able to bind a wide variety of different molecules, including heparin-binding growth factors, such as fibroblast and vascular endothelial growth factors, transforming growth factor-β, and platelet-derived growth factors (40). In the extracellular space, in addition to binding viruses and growth factors, syndecans bind to bacteria, lipoproteins, proteases, and extracellular matrix proteins via their HS chains (38, 41). They also facilitate the formation of signaling complexes by acting as coreceptors by concentrating and presenting ligands to the cell surface receptors (36). Syndecans have the ability to internalize their ligands via endocytosis (36) and have strongly conserved cytoplasmic domains that establish connections with signaling and cytoskeletal molecules (42).
HS has previously been shown to be involved in glycoprotein 64 (gp64)-mediated baculovirus binding into mammalian cells (21, 22). Heparin as well as heparinases I and II were also shown to have an effect on the binding of baculovirus to mammalian cells, further indicating the role of HSPGs in virus entry (21, 22). However, the role of HSPG sulfation status and the exact member of the HSPG family acting as a receptor in the binding and entry of AcMNPV remains unsolved. In this study, we investigated the role of HSPGs in detail and show that N- and 6-O-sulfation is important for SDC-1 mediated internalization and binding of AcMNPV into mammalian cells.
MATERIALS AND METHODS
Cells.
HepG2 (human liver carcinoma cells; CRL-11997; ATCC), 293T (human embryonic kidney cells; CRL-11268; ATCC), EA.hy926 (hybridoma of human umbilical vein endothelial cells and A549 human alveolar basal epithelial cells) (64), and MG-63 cells (human osteosarcoma cells; CRL-1427; ATCC) were used in the experiments. 293T, EA.hy926, and MG-63 cells were cultured in Dulbecco's modified Eagle's medium (D6429; Sigma) and HepG2 cells in minimum essential medium Eagle (M2279; Sigma). Both media were supplemented with 10% fetal bovine serum (FBS; CNH0003; HyClone) and antibiotics (100 U/ml of penicillin and 100 μg/ml of streptomycin; 15070-022; Gibco). EA.hy926 cell culture medium was supplemented with 1× HAT (5 mM hypoxanthine, 20 μM aminopterin, and 0.8 mM thymidine; 21060-017; Gibco) and HepG2 cell medium with an additional 2 mM l-glutamine (25030-024; Gibco), 0.1 mM nonessential amino acids (111360-035; Gibco), and 1.0 mM Na-pyruvate (11360-039; Gibco). All of the cells were grown in humidified 5% (vol/vol) CO2 at 37°C.
Viruses, binding/entry assays, and transduction experiments.
Baculovirus Ba-CAG-EGFP, with woodchuck hepatitis posttranscriptional regulatory element (WPRE), and baculovirus p24Cherry, without WPRE, were used for the binding, entry, and transduction experiments. Sucrose gradient-purified viruses were produced as described earlier (16, 43). Different multiplicities of infection (MOIs; 200, 400, 500, and 800) were used depending on experimental setup. In baculovirus binding assays, the cells were incubated with the virus for 1 or 2 h on ice with gentle shaking in cell culture medium containing 1% FBS. The unbound virus was removed by washing the cells three times with 0.5% bovine serum albumin-phosphate-buffered saline (BSA-PBS) and fixed with 4% PFA-PBS. In antibody-mediated inhibition experiments and colocalization studies, the virus was allowed to enter the cells for 30 min, 1 h, or 4 h at 37°C, and the cells were further processed as described for the binding assays.
In transduction experiments, the virus was allowed to transduce the cells in full cell culture medium. After transduction (2, 4, or 48 h), the culture medium was renewed. The percentage of enhanced green fluorescent protein (EGFP)-expressing cells and their mean fluorescence intensity was analyzed at 48 h posttransduction by a FACSCanto II and FACSDiva software (10,000 gated cells; BD Biosciences). All fluorescence-activated cell sorter (FACS) samples were prepared by washing the cells with PBS following trypsinization and suspension into 1% FBS-PBS.
Antibodies, immunofluorescence staining, and confocal microscopy.
In all immunofluorescence and confocal microscopy studies, the cells were grown on coverslips or in cell culture chambers and fixed with 4% PFA-PBS. Permeabilization, when needed, was performed with 0.1 to 0.2% Triton X-100-PBS. All antibodies used were diluted in 3% BSA-PBS, and cells were stained by using a standard protocol for immunofluorescence staining. Mouse monoclonal gp64 (B12D5), mouse monoclonal vp39 (p10C6), and rabbit polyclonal anti-baculovirus primary antibodies were a kind gift from L. Volkman. Other primary antibodies were SDC-1 (sc-5632), SDC-2 (sc-15348), SDC-3 (sc-15349), SDC-4 (sc-15350; all from Santa Cruz Biotechnology), and CD59 (ab18237; Abcam). Fluorescence-conjugated goat secondary antibodies against mouse or rabbit antibodies were Alexa 488, 555, and 594 (Life Technologies). The coverslips were mounted with ProLong gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen) or with Vectashield hard set with DAPI (Vector Laboratories). Slides were imaged with an Olympus FV1000-IX81 or Zeiss LSM700 confocal microscope. Appropriate excitation and emission settings were used (488-nm argon laser and 543-nm HeNe laser). An UPLSAPO objective (60×; 1.35 numeric aperture) and 20×, 0.5-numeric-aperture EC Plan-Neofluar objective with a resolution of 512 by 512 pixels/image were used.
Desulfated heparins.
Differentially desulfated, commercially available heparins (Iduron) were studied to investigate the possible preference of baculovirus to utilize a certain sulfation group on HSPGs. In 2-O-desulfated heparin sulfate, groups of C2 of iduronate were removed (De2SHep). In 6-O-desulfated heparin sulfate, groups of C6 glucosamine were removed (De6SHep), and in N-desulfated heparin, N-sulfates of glucosamine were removed (DeNS). The N-desulfated heparin also contains a free amino group (NH+3). Heparin and N-desulfated, 2-O-desulfated, and 6-O-desulfated heparins were preincubated with baculoviruses for 1 h at 37°C in PBS at a concentration of 2 mg/ml, and the viruses (MOI, 500) were then added to cells. After 4 h, the virus inoculum was removed and cell culture medium was renewed. The percentage of EGFP-expressing cells and their fluorescence intensity were analyzed 48 h later by FACS.
Inhibition assays.
Antibody inhibition assays with antibody against SDC-1 were performed in HepG2 cells. One day after plating the cells, medium containing 1% FBS, including syndecan antibodies (0 to 20 μg/ml), was added and incubated for 30 min on ice with gentle shaking. In transduction assays, baculovirus (MOI, 400) was added and incubated at 37°C until the percentage of EGFP-expressing cells and their fluorescent intensity was analyzed 48 h later by FACS. In the baculovirus entry assay, the baculovirus (MOI, 800) was added for 30 min at 37°C and the samples were fixed and stained. IgG antibody (I-5000; Vector Laboratories) was used as a control.
Recombinant SDC-1 protein (ab83609; Abcam) competition/inhibition assay was performed in HepG2 cells. The protein was added to the cells at different concentrations (0, 1, 2.5, and 5 μg/ml) and incubated for 1 h at 37°C. Baculovirus (MOI, 200) was added and kept on the cells for 2 h at 37°C. The virus inoculum was removed and cell culture medium was renewed. The percentage of EGFP-expressing cells and their fluorescent intensity were analyzed 48 h later by FACS.
Sodium chlorate (NaClO3; 244147; Sigma-Aldrich) was added to the cell culture medium with 0, 25, 50, and 75 mM per day prior to baculovirus (MOI, 200) addition and kept along with the virus on the cells until the transduction data were analyzed by FACS 48 h later. The effect of sodium chlorate on the binding of baculovirus to the surface of HepG2 and EA.hy926 cells was studied by treating the cells with 0, 25, 50, and 75 mM sodium chlorate before the addition of the virus (MOI, 400). After 24 h of treatment, the cells were washed and the virus was added to the cells in cell culture medium containing 1% FBS. MTS assay (CellTiter 96 aqueous one solution cell proliferation assay; Promega) was performed on sodium chlorate-treated cells in order to ensure that sodium chlorate did not affect the cell viability and influence the virus binding or transduction. The assay was performed according to the manufacturer's instructions.
To study the role of glypicans, phosphatidylinositol-specific phospholipase C (PI-PLC; P5542; Sigma-Aldrich) was used to enzymatically remove glycosylphosphatidylinositol (GPI)-anchored proteins from the cell membrane. After a wash with PBS, the cells were incubated in the absence or presence of 80 mU/ml PI-PLC for 40 min at 37°C. To confirm the effectiveness of the enzyme treatment, the amount of GPI-anchored CD59 membrane glycoprotein on the cell membrane was studied by staining the cells with anti-CD59 and analyzed with a confocal microscope. To study the baculovirus binding on the cells after treatment, the cells were incubated with the virus (MOI, 500) for 2 h on ice in cell culture medium containing 1% FBS with gentle shaking.
SDC-1 transfection.
HepG2 cells were transfected with plasmid coding for SDC-1 (Source BioScience imaGenes) to detect whether overexpression of SDC-1 had an effect on baculovirus transduction. Transfection was performed according to the manufacturer's instructions by JetPei hepatocyte (HepG2) transfection reagent (Polyplus transfection). Twenty-four h after transfection, baculovirus (MOI, 400) transduction was performed, and the percentage of EGFP-expressing cells and their mean fluorescent intensity were analyzed 48 h later by FACS. The increase of expressed SDC-1 at the time of virus addition was verified by immunolabeling. The transfection efficiency of the HepG2 cells was determined with the aid of EGFP-encoding plasmid from the control cells. The transfection percentage, which was estimated at the time of transfection (data not shown), was relatively low (17.2% ± 1.2%).
Analysis of the microscopic data.
Quantification of virus binding and the amounts of various proteins were performed using a free, open-source software package, BioImageXD (44), by intensity threshold segmentation. To quantify the level of labeled antigen, 30 images from three independent experiments were taken. Each image contained 1 to 5 cells, which were randomly selected and imaged. The threshold for each channel was manually adjusted to separate the signal from noise. The total intensity from immunofluorescence was divided by the DAPI signal from the nucleus (as a measure of the nuclear volume) to determine the intensity-per-nuclear-volume value.
To analyze the colocalization, BioImageXD was used (45). Colocalization was evaluated from the center slice of the cells. Thresholds were adjusted manually to eliminate fluorescence originating from the background and from diffuse staining. The colocalization percentage shown represents the signal derived from baculovirus overlapping with signal derived from different syndecans. Statistical significance of observed colocalization was calculated by the Costes algorithm (45). Only colocalization with zero coincidence probability was taken into account (i.e., P = 1.00).
Statistical analysis.
Statistical analysis was performed with GraphPad Prism software. Statistical significance of pair-wise differences was determined by Student's t test (*, P < 0.05; **, P < 0.01; and ***, P < 0.001). All data are presented as means ± standard errors of the means (SEM).
RESULTS
HSPG sulfation is essential for baculovirus binding and transduction.
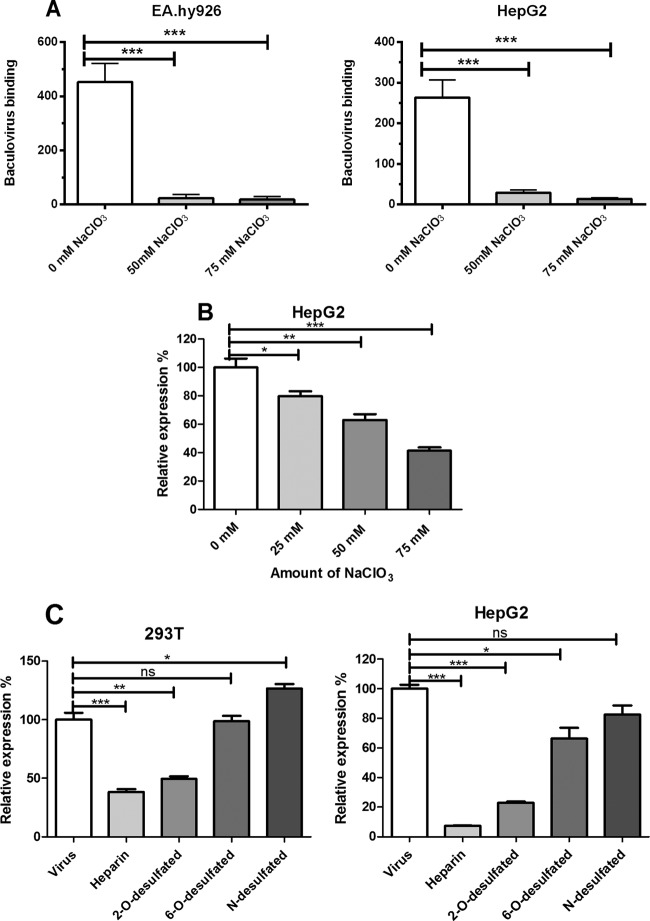
Neutralization of negatively charged epitopes on cell surfaces or heparinase treatment has previously been shown to inhibit baculovirus binding onto mammalian cells (21, 22). In this study, we investigated in more detail the role of different subfamilies of HSPGs and HSPG sulfate groups in both baculovirus binding and transduction in mammalian cells. Previously, NaClO3 has been shown to have an effect on the sulfation degree of cell surface GAG by preventing sulfate donation to newly synthesized polysaccharide chains (Fig. 1B) (46). This results in undersulfated GAGs but has no effect on protein synthesis or other posttranslational modifications (46–48). To study the role of HSPG sulfate groups in baculovirus binding, HepG2 and EA.hy926 cells were treated with various concentrations of NaClO3 (0, 25, 50, and 75 mM). The removal of HSPG sulfation with NaClO3 concentrations of 50 to 75 mM was shown to decrease significantly the amount of bound baculovirus on the surface of both cell lines as detected by confocal microscopy (Fig. 2A). This indicates that baculovirus requires sulfated HSPGs to bind to the surface of mammalian cells. In order to see whether the effect of NaClO3 on virus binding is also reflected in baculovirus transduction efficiency, permissive HepG2 cells were transduced with EGFP/WPRE-bearing baculovirus in medium containing NaClO3 (0, 25, 50, and 75 mM) and analyzed 48 h later by FACS. In line with the viral binding studies, the removal of sulfation had a clear dose-dependent effect on the baculovirus transduction rate. Compared to control cells (100.0% ± 6.2%), the relative EGFP expression in HepG2 cells decreased significantly, with NaClO3 treatments being 79.7% ± 3.3% (25 mM), 63.0% ± 4.0% (50 mM), and 41.3% ± 2.3% (75 mM), respectively (Fig. 2B). 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay performed on NaClO3-treated cells revealed no cytotoxicity for the concentrations used (data not shown).
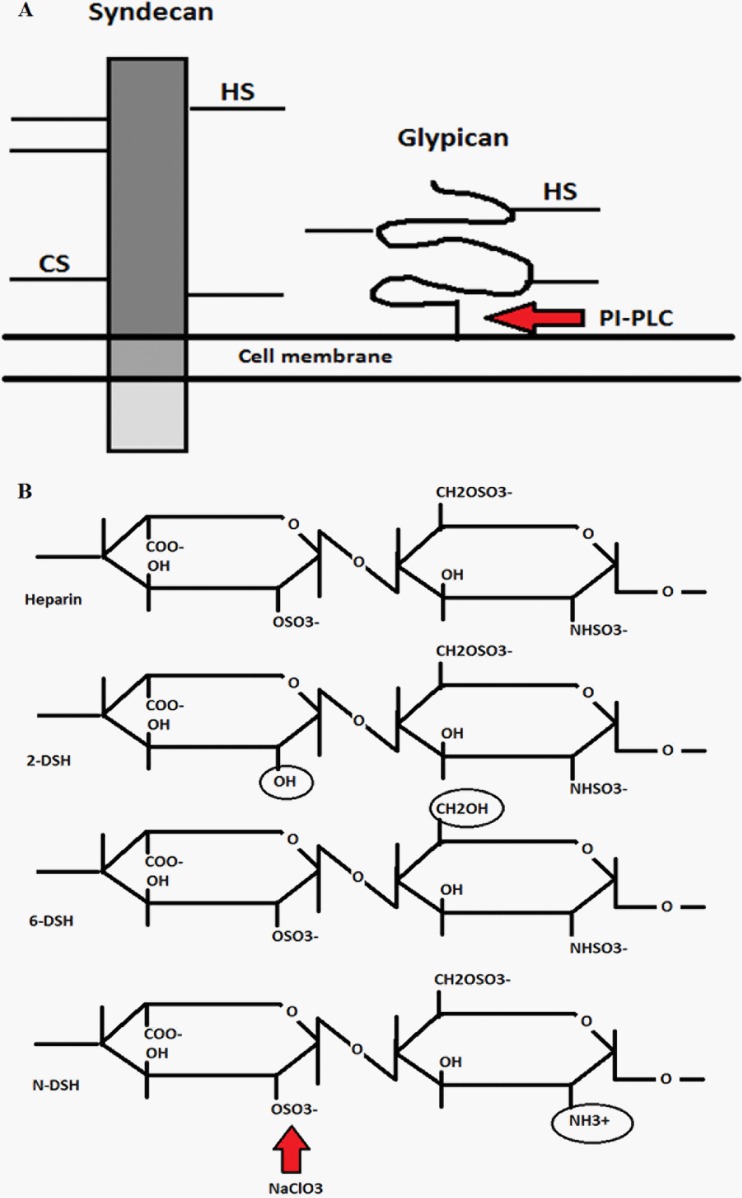
Fig 1.
Schematic of syndecan and glypican at the plasma membrane and the effect of treatments. (A) Syndecans are extracellular transmembrane proteins which have heparan (HS) and chondroitin sulfate (CS) side chains attached to the extracellular core protein (ectodomain). These glycosaminoglycan chains consist of repetitive differentially sulfated polysaccharides. Glypicans have the same type of side chains but are attached to the plasma membrane by a GPI anchor. Treatment with PI-PLC cuts the GPI anchor and releases the glypicans from the cell surface. (B) Schematic showing differentially desulfated heparan sulfate/heparins (2-DSH, 2-O-desulfated; 6-DSH, 6-O-desulfated; N-DSH, N-desulfated). Different desulfation positions have been marked with circles. An example where NaClO3 removes the sulfation on heparan sulfate is indicated by an arrow.
Fig 2.
Role of HSPG sulfation on baculovirus binding and transduction. (A) Quantification of cell surface-bound baculovirus on EA.hy926 and HepG2 cells treated with NaClO3 (0 to 75 mM). Baculovirus (MOI, 400) was allowed to bind to the surface of NaClO3-treated cells (1 h). The bound virus was stained with mouse anti-gp64 and anti-mouse Alexa 488-conjugated secondary antibody and imaged with confocal microscopy (60× magnification). Image analysis was performed as described in Materials and Methods. (B) HepG2 cells treated with different concentrations of NaClO3 (0 to 75 mM) and transduced with baculovirus (MOI of 200) for 48 h. The virus-mediated transgene (EGFP) expression percentages were analyzed by FACS. (C) HepG2 and 293T cells transduced with baculoviruses (MOI, 500) pretreated with basic and differentially 2-O-, 6-O-, and N-desulfated heparins (2 mg/ml). The percentage of EGFP-positive cells was analyzed 48 h later by FACS. In all experiments, EGFP/WPRE-bearing baculovirus was used. Mean fluorescence values and standard deviations are shown.
Since the sulfation of HSPGs was shown to be important in baculovirus binding and transduction, we next investigated if a certain sulfation residue on the HSPGs could be involved in baculovirus and mammalian cell surface interaction. Differentially desulfated heparins (Fig. 1B) were used in a competition assay to study the role of sulfation. Basic heparin and N-desulfated, 2-O-desulfated, and 6-O-desulfated heparins (1 to 2 mg/ml) were preincubated with EGFP/WPRE-bearing baculoviruses (MOI, 500) for 1 h at 37°C, and the viruses then were added to the cells. The number of EGFP-positive cells was analyzed 48 h later by FACS. Pretreatment of baculoviruses with both heparin and 2-O-desulfated heparin had a clear inhibitory effect on the transduction rate in both studied permissive cell lines (Fig. 2C). Compared to heparin (100.0% ± 5.8%), the relative EGFP expression in 293T cells with 2-O-, 6-O-, and N-desulfated heparins was 38.2% ± 2.4%, 49.5% ± 2.0%, 98.5% ± 4.8%, and 126.5% ± 4.0%, respectively. In HepG2 cells, the corresponding values were 100.0% ± 2.8%, 7.1% ± 0.3%, 22.8% ± 0.8%, 66.3% ± 7.3%, and 82.5% ± 6.0%, respectively. In conclusion, heparins with 6-O-desulfation and N-desulfation did not negatively affect transduction efficiency. This suggests that baculovirus probably utilizes 6-O- and N-sulfated residues for its interaction with heparin. Thus, sulfation has an important role in baculovirus binding and transduction efficiency in mammalian cells.
Glypicans do not mediate baculovirus binding on the plasma membrane.
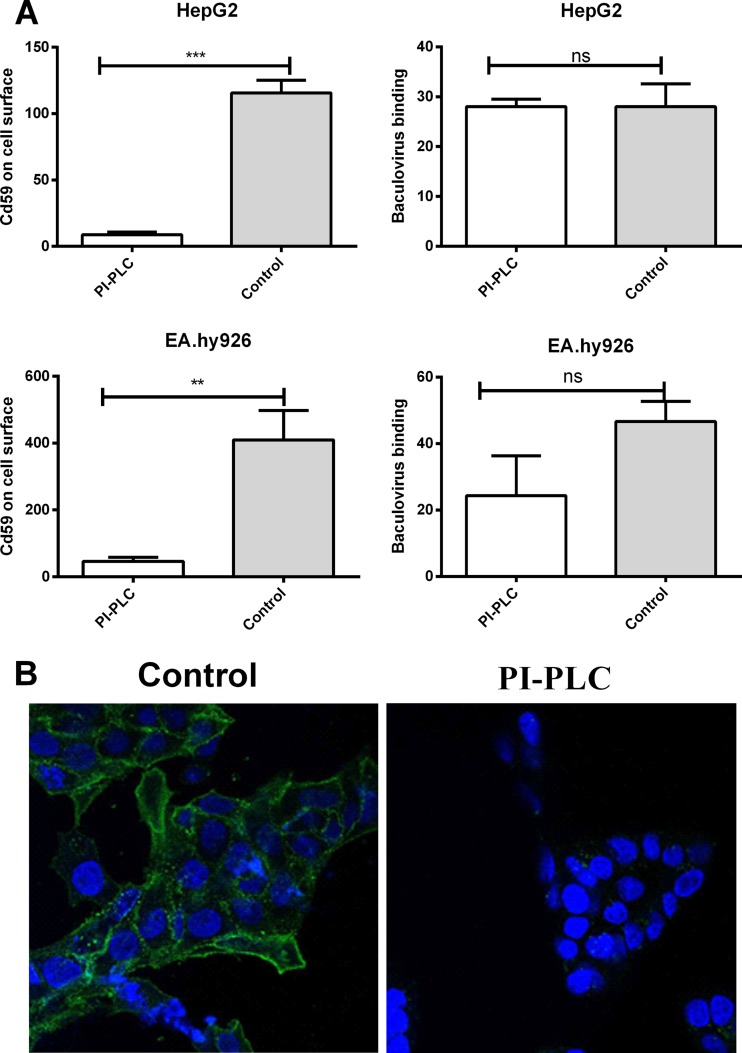
Main subfamilies of the cell surface HSPGs are transmembrane syndecans and GPI-anchored glypicans. To distinguish the role of baculovirus binding between these two families, PI-PLC, an enzyme that cleaves the GPI anchor of GPI-anchored proteins, was used to remove the glypicans from the cell surface. In practice, HepG2 and EA.hy926 cells were treated with PI-PLC (80 mU/ml), and the virus was then allowed to bind to the surface of the cells. The amount of attached virus was quantified by confocal microscopy. The successful removal of GPI-anchored proteins was detected with an antibody against CD59, a member of the group of GPI proteins to which glypicans also belong. Within the PI-PLC-treated cells, only a small amount of CD59 was detected on the cell surface, indicating the effective cleavage of the cell surface GPI proteins (Fig. 3A and B). After the PI-PLC treatment, statistically nonsignificant effects on baculovirus binding were observed (Fig. 3A), implying that the HSPG glypicans are not important for binding of baculovirus to the mammalian cell surface. Accordingly, no change in the EGFP transgene expression was detected in permissive HepG2 cells transduced with EGFP/WPRE-bearing baculovirus after PI-PLC treatment (data not shown).
Fig 3.
Effect of PI-PLC treatment on glypican removal and baculovirus binding. (A) HepG2 and EA.hy926 cells versus cells treated with PI-PLC. Baculovirus (MOI, 500) was allowed to bind to the surface of the cells. The virus was stained with rabbit anti-baculovirus antibody together with anti-rabbit Alexa-555-conjugated secondary antibody. The amount of GPI proteins was quantified by staining the cell surface with mouse anti-CD59 and anti-mouse Alexa 488 and imaged with confocal microscopy (60× magnification). Image analysis was performed as described in Materials and Methods. Mean fluorescence values and standard deviations are shown. (B) Representative images of CD59-stained, PI-PLC-treated versus control HepG2 cells. Staining was performed as described for panel A. CD59 is seen in green, and DAPI-stained nuclei are seen in blue.
SDC-1 expression levels and baculovirus binding in different cell types.
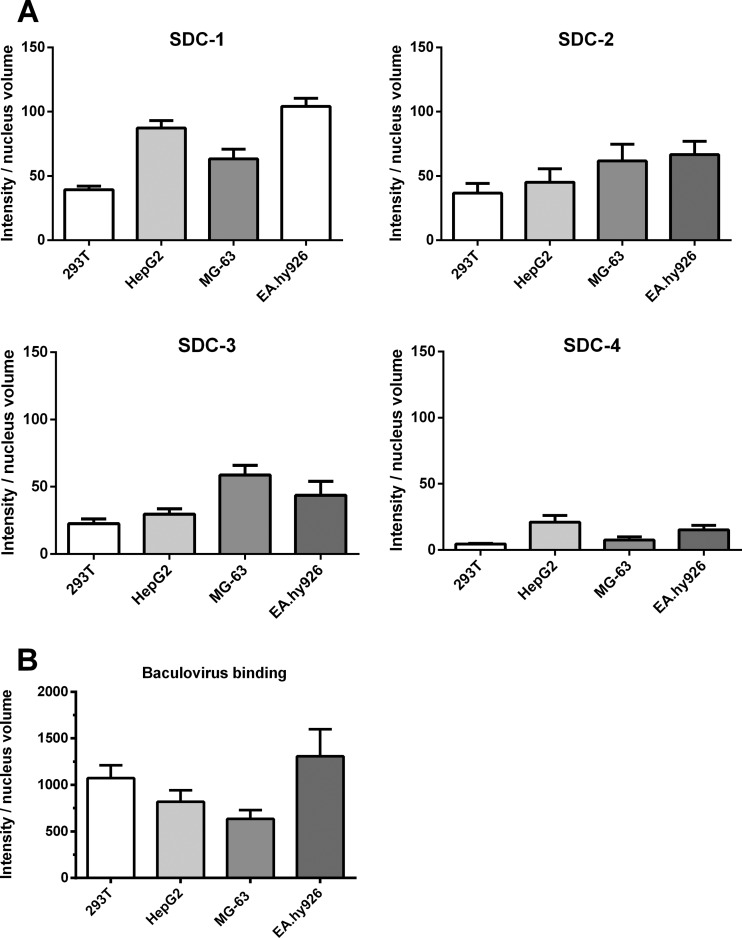
Since the glypicans apparently did not mediate the binding of the baculovirus, we studied further the other main members of the HSPGs, the syndecans. To observe SDC-1 to SDC-4 at the surface of different types of cells, HepG2, 293T, EA.hy926, and MG-63 cells were first stained with specific antibodies against each member of the syndecan family, and antibody binding was detected with confocal microscopy. The expression levels of SDC-1 seemed to be slightly higher, on average, in EA.hy926 and HepG2 cells, whereas the expression levels of SDC-2, SDC-3, and SDC-4 were quite uniform in all studied cells (Fig. 4A). In order to observe if there was any relation between the amount of syndecans expressed at the cell surface and the level of baculovirus binding among different cell types, baculovirus was allowed to attach to the surface of the cells and the amount of bound virus was quantified. The detected amount of bound baculovirus was highest in EA.hy926 cells (Fig. 4B), which also expressed the highest levels of SDC-1. The binding of the baculovirus to MG-63 and HepG2 cells seemed to have a similar trend with the detected amount of SDC-1. However, in 293T cells, the trend between SDC-1 expression and baculovirus binding was not evident.
Fig 4.
Syndecan expression levels and baculovirus binding in various mammalian cell types. (A) 293T, MG-63, HepG2, and EA.hy926 cells were immunostained with rabbit anti-SDC-1 to -4 antibodies and anti-rabbit Alexa 488 antibody and imaged by confocal microscopy (60× magnification). Image analysis was performed as described in Materials and Methods. Mean fluorescence values and standard deviations are shown. (B) The amount of baculovirus binding at the surface of 293T, MG-63, HepG2, and EA.hy926 cells. The virus (MOI, 500) was allowed to attach on the surface of the cells. The bound virus was stained with rabbit anti-baculovirus antibody and anti-rabbit Alexa 488 antibody and imaged by confocal microscopy (60× magnification). Image analysis was performed as described in Materials and Methods. Mean fluorescence values and standard deviations are shown.
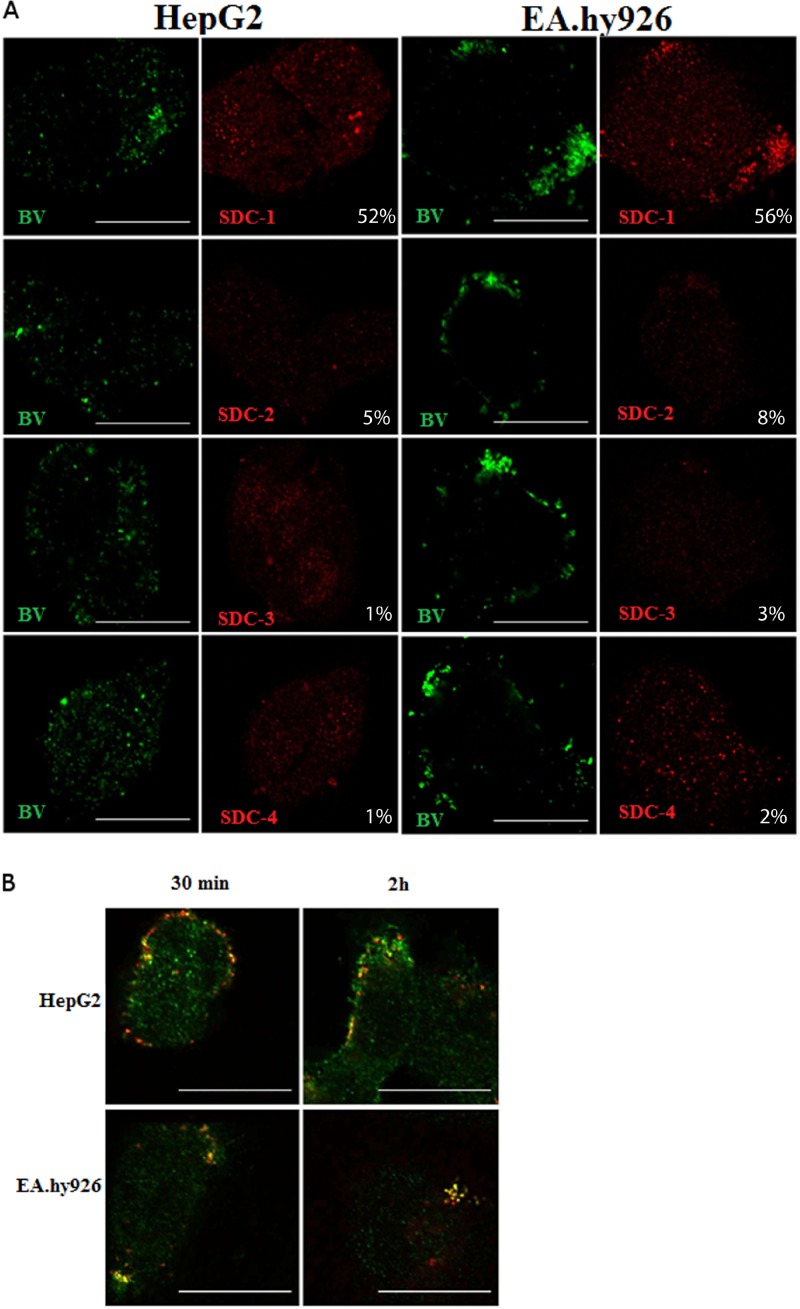
Baculovirus colocalizes with SDC-1.
Since the experiments suggested a role for syndecans in baculovirus binding and transduction, we next studied further the colocalization of baculovirus with different syndecans in HepG2 and EA.hy926 cells. The baculovirus was allowed to bind and enter the cells for 4 h, and the syndecans and the viruses were stained with specific antibodies and imaged. Colocalization of baculovirus in EA.hy926 and HepG2 cells was evident and statistically significant only with SDC-1 (Fig. 5A). Only minimal colocalization was detected with other members of the syndecan family, further highlighting the role of SDC-1 in the interaction of baculovirus on the mammalian cell surface and within the cells. The colocalization was also evident when virus internalization was monitored at different time points postinternalization (30 min, 2 h, and 4 h). Only SDC-1 showed colocalization with the virus (Fig. 5B), whereas no colocalization was observed with other members of the syndecan family (data not shown).
Fig 5.
Localization of baculovirus with different syndecans in EA.hy926 and HepG2 cells. (A) Baculovirus (MOI, 500) was allowed to bind to and enter the cells and was immunostained with mouse anti-vp39 and anti-mouse Alexa 488. Syndecans were stained with rabbit anti-SDC-1 to -4 antibodies and anti-rabbit Alexa 555 antibody. Baculoviruses are seen in green and syndecans in red. Imaging was performed by confocal microscopy (60× magnification). Colocalization percentages represent the percent overlap of baculovirus with SDC-1 to -4 signals. Scale bars, 20 μm. (B) SDC-1 and baculovirus colocalization at different time points (30 min and 2 h) postinternalization in HepG2 and EA.hy926 cells. Baculovirus (MOI, 500) was allowed to bind to and enter the cells, and the virus was immunostained with mouse anti-vp39 and anti-mouse Alexa 555. SDC-1 was stained with rabbit anti-SDC-1 antibody and anti-rabbit Alexa 488 antibody. SDC-1 is seen in green, and baculoviruses are seen in red. Imaging was performed by confocal microscopy (60× magnification). Scale bars, 20 μm.
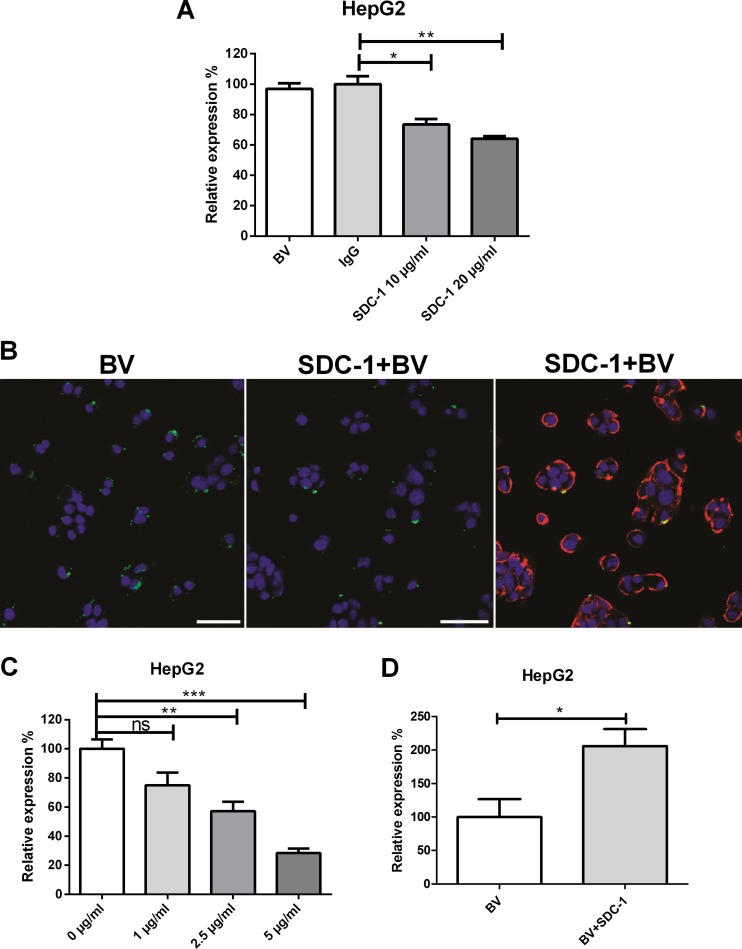
Since the colocalization studies suggested a role for SDC-1 in the interaction of baculovirus with mammalian cells, we next tested whether the masking of the cell surface with the SDC-1 molecule had an effect on baculovirus transduction. Permissive HepG2 cells were first pretreated with SDC-1 antibody (0, 10, and 20 μg/ml), followed by baculovirus transduction for 48 h and detection of the virus-mediated EGFP expression by FACS (Fig. 6A). The antibody-mediated masking of the cell surface SDC-1 showed that SDC-1 antibody was able to decrease baculovirus transduction efficiency in HepG2 cells compared to that in control antibody-treated cells. The negative effect of SDC-1 antibody on baculovirus uptake was also seen with confocal microscopy at 30 min posttransduction (Fig. 6B). To further study the role of SDC-1 in baculovirus transduction, a competition assay with recombinant SDC-1 protein was performed. SDC-1 protein was added to the cells 1 h before the addition of the virus, and the detection of virus-mediated EGFP expression by FACS was performed again 48 h later. The increase in the amount of SDC-1 recombinant protein was shown to decrease the baculovirus transduction levels dose dependently (Fig. 6C). Altogether, these data show that masking of the cell surface SDC-1 with the aid of antibody or the competition provided by recombinant SDC-1 protein can prevent both baculovirus uptake and transduction in HepG2 cells.
Fig 6.
Effect of SDC-1 plasmid transfection, antibody-mediated SDC-1 inhibition, and SDC-1 recombinant protein competition on baculovirus transduction and binding. (A) HepG2 cells were pretreated with SDC-1-specific (0, 10, and 20 mg/ml) and control antibodies (20 μg/ml; IgG). Baculovirus (MOI, 800) was added, and the percentage of EGFP-positive cells was analyzed 48 h later by FACS. (B) HepG2 cells were pretreated with 20 μg/ml of SDC-1 antibody. Baculovirus (MOI, 800) was added and allowed to enter the cells for 30 min at 37°C. Baculovirus was stained with anti-vp39 and anti-mouse Alexa 488 antibodies, and SDC-1 was stained with anti-rabbit Alexa 594 antibody. Baculovirus is seen in green and SDC-1 in red. Imaging was performed by confocal microscopy (20× magnification). (C) HepG2 cells were pretreated with SDC-1 recombinant protein (0, 1, 2.5, and 5 μg/ml) and the baculovirus (MOI, 200) was added. The percentage of EGFP-positive cells was analyzed 48 h later with FACS. (D) HepG2 cells were transfected with SDC-1-encoding plasmid, and baculovirus (MOI, 400) was added 24 h later. The percentage of EGFP-positive cells was analyzed 48 h later by FACS. In all transduction experiments, EGFP/WPRE-bearing baculovirus was used. Mean fluorescence values and standard deviations are shown.
In order to see if the level of baculovirus transduction could be enhanced by increasing the expression of SDC-1 at the surface of the cells, HepG2 cells were transfected with a plasmid encoding SDC-1 and transduced with baculovirus. As a result, enhanced baculovirus transduction efficiency in SDC-1-transfected cells was detected by FACS (Fig. 6D). The functionality of the plasmid was checked by immunolabeling the cell surface SDC-1 of transfected cells (data not shown). In conclusion, the overexpression of SDC-1 was able to enhance the baculovirus transduction in HepG2 cells. Altogether, our results suggest that baculovirus requires HSPG sulfation, particularly N- and 6-O-sulfation, to bind to and transduce mammalian cells. Also, our colocalization studies suggest that baculovirus binds to SDC-1 but does not interact with SDC-2 to -4 or with glypicans. In addition, competition experiments performed with SDC-1 antibody or recombinant SDC-1 protein, shown to inhibit baculovirus binding and SDC-1 overexpression, lead to enhanced baculovirus-mediated transduction.
DISCUSSION
The determination of cellular factors influencing the susceptibility of cells to virus transduction is crucial in understanding virus-cell interactions and for development of next-generation viral vectors for gene therapy. Baculoviruses hold great promise as gene therapy vectors, since they are able to transduce a wide variety of cells (5). Additionally, they have already been used as vaccines (49). In this respect, it is remarkable that the receptor(s) which baculovirus interacts with in insect or mammalian cells is not yet known. So far, general cell surface molecules, such as phospholipids or heparan sulfate proteoglycans (HSPGs), have been suggested to be responsible for the interaction (19, 21, 22); however, this interaction has not been studied in detail. A wide variety of pathogens utilize the HSPGs, especially the members of the syndecan family (41). In this study, we show that baculovirus interacts with specifically sulfated HSPGs in mammalian cells and that it uses SDC-1 for its binding and entry.
HSPGs are widely expressed on the surface of adherent cells and in the extracellular matrix. They are composed of a core protein into which one or several HS glycosaminoglycan chains bearing N- and O-sulfated linear polysaccharides are attached (Fig. 1). The expression and the sulfation degree of cell surface HSPGs differ depending on the cell type (38). The majority of pathogens have been shown to interact particularly with the HS moieties of HSPGs (41). Our desulfation experiments performed with NaClO3 showed a clear dose-dependent effect on both baculovirus transduction and binding, indicating the crucial importance of HS sulfation in baculovirus-cell surface interaction. It is known that several viruses utilize specific sulfation residues over others when binding to the HSPGs. N- and 6-O-sulfations have been reported to be crucial for coxsackie B virus internalization (50), whereas 6-O-sulfation is important for HIV-1 binding (51). To determine if the binding of baculovirus was based solely on random electrostatic interactions or if there was a specific sulfation residue which the baculovirus interacts with, we used differentially desulfated heparins. The experiments showed that the interaction was not unspecific, as both N- and 6-O-sulfations had an effect in transduction while 2-O-sulfation played no role. Interestingly, the baculovirus gp64 peptides responsible for the binding of the virus to the mammalian cell surface HSPGs do not seem to function in a similar fashion in Sf9 insect cells (21).
HSPGs are composed of two main families, syndecans and glypicans. To discriminate the binding of baculovirus between the two families, we removed glypicans from the cell surface with PI-PLC and allowed the virus to bind. The removal of GPI-anchored proteins with PI-PLC did not have a significant impact on baculovirus binding in EA.hy926 and HepG2 cells. Although the removal was detected to be successful, the virus was still able to bind to the surface of the cells. This is in line with studies showing that most of the identified HSPG-mediated interactions with different viruses have been seen to occur with members of the syndecan family (41). Our previous study also suggested that GPI-anchored proteins were not involved in baculovirus entry into mammalian cells (16). Based on these results, the role of the glypican family was ruled out, and our further studies focused on investigating the role of the syndecan family in baculovirus binding and entry.
Syndecan HSPGs are ubiquitously expressed in different kinds of mammalian cells, but the expression patterns of the four members vary and are highly dependent on the cell and tissue type (40). To estimate the expression patterns in different cells types, we immunostained all four cell surface syndecans in 293T, EA.hy926, HepG2, and MG-63 cells. As a result, SDC-1 was found to be more expressed in HepG2 and EA.hy926 cells than in 293T and MG-63 cells. Additionally, the binding of baculovirus was detected to be most efficient in the same HepG2 and EA.hy926 cell types. Interestingly, despite the lower level of SDC-1 expression in 293T cells, baculovirus was able to bind to the cells efficiently. This could be due to significant differences in the disaccharide composition of the HS chains of SDC-1 in fibroblastic, endothelioid, and epithelial cells (37). The number and size of the GAG chains also can vary in a tissue- and cell type-specific manner (37). These differences may reflect the ligand binding properties and syndecan function. Since HS functions primarily as a coreceptor that catalyzes ligand encounters with other signaling receptors on the cell surface (31, 32), it is possible that other molecules influence the baculovirus binding as well. Although EA.hy926 and MG-63 cells express relatively large amounts of SDC-1 at their cell surface, the transduction rate of baculovirus in these cells is very low (14, 18). However, this does not exclude the role of SDC-1 in baculovirus interaction on the surface of the cells, as shown by our results. Baculovirus is able to bind to and interact with SDC-1 of EA.hy926 cells, but in this nonpermissive cell line it cannot reach the nucleus where the transgene expression takes place (18). Additionally, we showed recently that the dynamics of vimentin expression and the phosphorylation status of protein kinase C alpha (PKC-α) and -ε regulate the nonpermissive phenotype of EA.hy926 and MG-63 cells, affecting baculovirus transduction (52). The phenomenon of the virus getting trapped and aggregated in EA.hy926 cells has also been reported in the case of AAV-2, a virus known to apply HSPG as a receptor (53).
From our studies, several lines of evidence demonstrated the importance of SDC-1 over the other syndecans. First, the binding of baculovirus seems to follow a similar trend in the estimated levels of SDC-1 expression in various cell types, suggesting that SDC-1 is a key member in baculovirus binding. Second, colocalization studies, which were carried out with all syndecans and baculovirus, showed no colocalization with SDC-2 to -4. The only evident colocalization detected was with baculovirus and SDC-1, indicating that baculovirus uses SDC-1 for its binding to and entry into the cells. Third, when antibody inhibition studies with SDC-1 antibody were performed, SDC-1 antibody was able to prevent baculovirus binding and entry. Fourth, a decrease in baculovirus transduction was seen in a competition assay with recombinant SDC-1 protein. Finally, as the expression levels of SDC-1 were upregulated with SDC-1-encoding plasmid, an increase in the transduction levels of baculovirus was detected. Together, these data suggest that SDC-1 has a role in the binding and transduction of baculovirus in mammalian cells.
The binding and entry of baculoviruses into mammalian cells has been considered controversial. Some studies have identified cell surface phospholipids important for entry (23–25, 54), whereas others suggest roles of HSPGs (21, 22). Recently, Wu and Wang (21) suggested that baculoviruses utilize two different methods, nonspecific electrostatic interactions and more specific receptors in attachment. Kataoka et al. (24) concluded that baculovirus internalization is mediated by lipid rafts and is based on receptor-mediated endocytosis, whereas O'Flynn et al. (54) suggested that baculovirus takes advantage of a lipid-based receptor-independent route. Although previous reports show controversy between receptor-independent and receptor-mediated entry (21, 24, 54), there is a consensus for a requirement of gp64 in the internalization process. Gp64 has been shown to interact with cell surface phospholipids (23–25, 54) and heparins (21), suggesting that these two factors act in cooperation or that two distinct internalization routes exist for baculovirus. The cooperation hypothesis is supported by the fact that in several baculovirus-phospholipid studies, baculovirus internalization has been specifically associated with cholesterol-rich raft areas (24, 54). Upon ligand-induced clustering, SDC-1 also has been shown to move to and internalize from the raft areas (55–60). Furthermore, we have shown earlier that clustering of the cell surface cholesterol by filipin inhibits baculovirus internalization (16). Kataoka et al. (24) also observed that methyl-β-cyclodextrin, which removes cell surface cholesterol, inhibits baculovirus uptake. Methyl-β-cyclodextrin treatment has previously been shown to remove raft-associated SDC-1 from the cell surface (58).
The process of SDC-1 entry seems to be in accordance with the steps of baculovirus entry. Similar to baculovirus uptake (16), SDC-1 entry is clathrin and caveolin independent and actin dependent, and it occurs from the membrane raft areas (55, 56). Ligand binding causes syndecan clustering and subsequent internalization of the receptor and its cargo (55, 56). This is also in line with a previous study (19) showing clustering of the baculoviruses on the cell membrane prior to their internalization. In this study, we have suggested that heparin sulfate proteoglycan SDC-1 acts as a receptor for baculovirus in human cells. Evidence to support this includes the previous reports showing heparin dependency (21, 22) and similarities of internalization mechanisms of baculovirus and SDC-1 (16, 19, 55, 56). The data also fit the hypothesis that the seemingly contradictory results actually represent the several subsequent steps of baculovirus entry. Subsequent steps are required in attachment (HSPGs), signaling/trafficking to membrane raft areas/internalization (SDC-1), and gp64-mediated fusion (phospholipids) into the endosome, mediated by HSPGs, SDC-1, and phospholipids, respectively. Whether there are other receptors/coreceptors/molecules involved remains to be further studied.
In conclusion, we show that baculovirus interacts with 6-O- and N-sulfated HS chains of HSPGs. Most importantly, our results indicate that SDC-1 serves as a receptor for baculovirus in vertebrate cells. The binding and entry generally are complex processes which normally require the involvement of several molecules that are responsible for the different steps of virus binding, entry, and intracellular signaling, leading to the efficient internalization and transport to the nucleus. Thus, the involvement of other significant cellular factors cannot be excluded. However, the results presented here reveal important aspects of baculovirus entry which expand the current knowledge of virus-cell interaction and serve as a basis for the further development of baculovirus as a versatile vector for gene delivery into vertebrate cells. As SDC-1 is seen to be upregulated in many types of cancers (61), the use of baculovirus as a gene transfer vector to malignant cells is appealing and further justified (5, 62, 63).
ACKNOWLEDGMENTS
We thank Tarja Taskinen from Ark Therapeutics Oy, Anneli Miettinen and Joonas Malinen from University of Eastern Finland, and Laura Pitkänen and Arja Mansikkaviita from University of Jyväskylä for their excellent technical support. Loy Volkman (University of California, Berkley) is very much appreciated for the baculovirus antibodies.
This work was supported by the Academy of Finland and the Alfred Kordelin foundation.
Footnotes
Published ahead of print 7 August 2013
REFERENCES
- 1.Airenne KJ, Mahonen AJ, Laitinen OH, Yla-Herttuala S. 2009. Baculovirus-mediated gene transfer: an emerging universal concept, p 263–291. In Templeton N. (ed), Gene and cell therapy: therapeutic mechanisms and strategies, 3rd ed. CRC Press, Boca Raton, FL. [Google Scholar]
- 2.Hartig PC, Cardon MC, Kawanishi CY. 1992. Effect of baculovirus on selected vertebrate cells. Dev. Biol. Stand. 76:313–317. [PubMed] [Google Scholar]
- 3.Kost TA, Condreay JP, Jarvis DL. 2005. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 23:567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Airenne KJ, Makkonen K-E, Mähönen AJ, Ylä-Herttuala S. 2010. In vivo application and tracking of baculovirus. Curr. Gene Ther. 10:187–194. [DOI] [PubMed] [Google Scholar]
- 5.Airenne KJ, Hu Y-C, Kost TA, Smith RH, Kotin RM, Ono C, Matsuura Y, Wang S, Ylä-Herttuala S. 2013. Baculovirus: an insect-derived vector for diverse gene transfer applications. Mol. Ther. 21:739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O′Reilly DR, Miller LK, Luckov VA. 1994. Baculovirus expression vectors: a laboratory manual. Oxford University press, New York, NY. [Google Scholar]
- 7.Kost TA, Condreay JP. 2002. Innovations-biotechnology: baculovirus vectors as gene transfer vectors for mammalian cells: biosafety considerations. J. Am. Biol. Safety Assoc. 7:167–169. [Google Scholar]
- 8.Burges HD, Croizier G, Huger J. 1980. A review of safety tests on baculoviruses. Entomaphaga 25:329–339. [Google Scholar]
- 9.Hu Y-CC. 2008. Baculoviral vectors for gene delivery: a review. Curr. Gene Ther. 8:54–65. [DOI] [PubMed] [Google Scholar]
- 10.Airenne KJ, Laitinen OH, Mähönen AJ, Ylä-Herttuala S. 2009. Transduction of vertebrate cells with recombinant baculovirus. Cold Spring Harbor protocols 2009:pdb.prot5182. 10.1101/pdb.prot5182. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. 1995. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc. Natl. Acad. Sci. U. S. A. 92:10099–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyce FM, Bucher NL. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 93:2348–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condreay JP, Witherspoon SM, Clay WC, Kost TA. 1999. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. U. S. A. 96:127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song SU, Shin S-H, Kim S-K, Choi G-S, Kim W-C, Lee M-H, Kim S-J, Kim I-H, Choi M-S, Hong Y-J, Lee K-H. 2003. Effective transduction of osteogenic sarcoma cells by a baculovirus vector. J. Gen. Virol. 84:697–703. 10.1099/vir.0.18772-0. [DOI] [PubMed] [Google Scholar]
- 15.Chen C-Y, Lin C-Y, Chen G-Y, Hu Y-C. 2011. Baculovirus as a gene delivery vector: recent understandings of molecular alterations in transduced cells and latest applications. Biotechnol. Adv. 29:618–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laakkonen JP, Makela AR, Kakkonen E, Turkki P, Kukkonen S, Peranen J, Yla-Herttuala S, Airenne KJ, Oker-Blom C, Vihinen-Ranta M, Marjomaki V, Mäkelä AR, Peränen J, Ylä-Herttuala S, Marjomäki V. 2009. Clathrin-independent entry of baculovirus triggers uptake of E. coli in non-phagocytic human cells. PLoS One 4:e5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laakkonen JP, Kaikkonen MU, Ronkainen PHA, Ihalainen TO, Niskanen EA, Häkkinen M, Salminen M, Kulomaa MS, Ylä-Herttuala S, Airenne KJ, Vihinen-Ranta M. 2008. Baculovirus-mediated immediate-early gene expression and nuclear reorganization in human cells. Cell. Microbiol. 10:667–681. [DOI] [PubMed] [Google Scholar]
- 18.Kukkonen SP, Airenne KJ, Marjomaki V, Laitinen OH, Lehtolainen P, Kankaanpaa P, Mahonen AJ, Raty JK, Nordlund HR, Oker-Blom C, Kulomaa MS, Yla-Herttuala S, Marjomäki V, Kankaanpää P, Mähönen AJ, Räty JK, Ylä-Herttuala S. 2003. Baculovirus capsid display: a novel tool for transduction imaging. Mol. Ther. 8:853–862. [DOI] [PubMed] [Google Scholar]
- 19.Matilainen H, Rinne J, Gilbert L, Marjomaki V, Reunanen H, Oker-Blom C. 2005. Baculovirus entry into human hepatoma cells. J. Virol. 79:15452–15459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Loo ND, Fortunati E, Ehlert E, Rabelink M, Grosveld F, Scholte BJ. 2001. Baculovirus infection of nondividing mammalian cells: mechanisms of entry and nuclear transport of capsids. J. Virol. 75:961–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Wang S. 2012. A pH-sensitive heparin-binding sequence from baculovirus gp64 protein is important for binding to mammalian cells but not to Sf9. J. Virol. 86:484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duisit G, Saleun S, Douthe S, Barsoum J, Chadeuf G, Moullier P. 1999. Baculovirus vector requires electrostatic interactions including heparan sulfate for efficient gene transfer in mammalian cells. J. Gene Med. 1:93–102. [DOI] [PubMed] [Google Scholar]
- 23.Tani H, Nishijima M, Ushijima H, Miyamura T, Matsuura Y. 2001. Characterization of cell-surface determinants important for baculovirus infection. Virology 279:343–353. [DOI] [PubMed] [Google Scholar]
- 24.Kataoka C, Kaname Y, Taguwa S, Abe T, Fukuhara T, Tani H, Moriishi K, Matsuura Y. 2012. Baculovirus GP64-mediated entry into mammalian cells. J. Virol. 86:2610–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamiya K, Kobayashi J, Yoshimura T, Tsumoto K. 2010. Confocal microscopic observation of fusion between baculovirus budded virus envelopes and single giant unilamellar vesicles. Biochim. Biophys. Acta 1798:1625–1631. [DOI] [PubMed] [Google Scholar]
- 26.Blissard GW. 1996. Baculovirus-insect cell interactions. Cytotechnology 20:73–93. [DOI] [PubMed] [Google Scholar]
- 27.Goley ED, Ohkawa T, Mancuso J, Woodruff JB, D'Alessio JA, Cande WZ, Volkman LE, Welch MD. 2006. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 314:464–467. [DOI] [PubMed] [Google Scholar]
- 28.Ohkawa T, Volkman LE, Welch MD. 2010. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J. Cell Biol. 190:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Au S, Panté N, Pante N. 2012. Nuclear transport of baculovirus: revealing the nuclear pore complex passage. J. Struct. Biol. 177:90–98. [DOI] [PubMed] [Google Scholar]
- 30.Bishop JR, Schuksz M, Esko JD. 2007. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446:1030–1037. [DOI] [PubMed] [Google Scholar]
- 31.Mondor I, Ugolini S, Sattentau QJ. 1998. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J. Virol. 72:3623–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gallay P. 2004. Syndecans and HIV-1 pathogenesis. Microbes Infect. 6:617–622. [DOI] [PubMed] [Google Scholar]
- 33.Cheshenko N, Liu W, Satlin LM, Herold BC. 2007. Multiple receptor interactions trigger release of membrane and intracellular calcium stores critical for herpes simplex virus entry. Mol. Biol. Cell 18:3119–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shafti-Keramat S, Handisurya A, Kriehuber E, Meneguzzi G, Slupetzky K, Kirnbauer R. 2003. Different heparan sulfate proteoglycans serve as cellular receptors for human papillomaviruses. J. Virol. 77:13125–13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park PW, Reizes O, Bernfield M. 2000. Cell surface heparan sulfate proteoglycans: selective regulators of ligand-receptor encounters. J. Biol. Chem. 275:29923–29926. [DOI] [PubMed] [Google Scholar]
- 36.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M, Götte M. 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68:729–777. [DOI] [PubMed] [Google Scholar]
- 37.Götte M, Gotte M. 2003. Syndecans in inflammation. FASEB J. 17:575–591. 10.1096/fj.02-0739rev. [DOI] [PubMed] [Google Scholar]
- 38.Carey DJ. 1997. Syndecans: multifunctional cell-surface co-receptors. Biochem. J. 327(Part 1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim CW, Goldberger OA, Gallo RL, Bernfield M. 1994. Members of the syndecan family of heparan sulfate proteoglycans are expressed in distinct cell-, tissue-, and development-specific patterns. Mol. Biol. Cell 5:797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tkachenko E, Rhodes JM, Simons M. 2005. Syndecans: new kids on the signaling block. Circ. Res. 96:488–500. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Gotte M, Liu J, Park PW, Götte M. 2008. Microbial subversion of heparan sulfate proteoglycans. Mol. Cells 26:415–426. [PubMed] [Google Scholar]
- 42.Lambaerts K, Wilcox-Adelman SA, Zimmermann P. 2009. The signaling mechanisms of syndecan heparan sulfate proteoglycans. Curr. Opin. Cell Biol. 21:662–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mähönen AJ, Airenne KJ, Purola S, Peltomaa E, Kaikkonen MU, Riekkinen MS, Heikura T, Kinnunen K, Roschier MM, Wirth T, Ylä-Herttuala S, Mahonen AJ, Yla-Herttuala S. 2007. Post-transcriptional regulatory element boosts baculovirus-mediated gene expression in vertebrate cells. J. Biotechnol. 131:1–8. [DOI] [PubMed] [Google Scholar]
- 44.Kankaanpaa P, Paavolainen L, Tiitta S, Karjalainen M, Paivarinne J, Nieminen J, Marjomaki V, Heino J, White DJ. 2012. BioImageXD: an open, general-purpose and high-throughput image-processing platform. Nat. Methods 9:683–689. [DOI] [PubMed] [Google Scholar]
- 45.Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 86:3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung CS, Hsiao JC, Chang YS, Chang W. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72:1577–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalia M, Chandra V, Rahman SA, Sehgal D, Jameel S. 2009. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 83:12714–12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keller KM, Brauer PR, Keller JM. 1989. Modulation of cell surface heparan sulfate structure by growth of cells in the presence of chlorate. Biochemistry 28:8100–8107. [DOI] [PubMed] [Google Scholar]
- 49.Harper DM, Franco EL, Wheeler C, Ferris DG, Jenkins D, Schuind A, Zahaf T, Innis B, Naud P, De Carvalho NS, Roteli-Martins CM, Teixeira J, Blatter MM, Korn AP, Quint W, Dubin G. 2004. Efficacy of a bivalent L1 virus-like particle vaccine in prevention of infection with human papillomavirus types 16 and 18 in young women: a randomised controlled trial. Lancet 364:1757–1765. [DOI] [PubMed] [Google Scholar]
- 50.Zautner AE, Jahn B, Hammerschmidt E, Wutzler P, Schmidtke M. 2006. N- and 6-O-sulfated heparan sulfates mediate internalization of coxsackievirus B3 variant PD into CHO-K1 cells. J. Virol. 80:6629–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Parseval A, Bobardt MD, Chatterji A, Chatterji U, Elder JH, David G, Zolla-Pazner S, Farzan M, Lee TH, Gallay PA. 2005. A highly conserved arginine in gp120 governs HIV-1 binding to both syndecans and CCR5 via sulfated motifs. J. Biol. Chem. 280:39493–39504. [DOI] [PubMed] [Google Scholar]
- 52.Turkki P, Makkonen K-E, Huttunen M, Laakkonen JP, Ylä-Herttuala S, Airenne KJ, Marjomäki V. 2013. Cell susceptibility to baculovirus transduction and echovirus infection is modified by PKC phosphorylation and vimentin organization. J. Virol. [Epub ahead of print.] 10.1128/JVI.01004-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pajusola K, Gruchala M, Joch H, Luscher TF, Yla-Herttuala S, Bueler H. 2002. Cell-type-specific characteristics modulate the transduction efficiency of adeno-associated virus type 2 and restrain infection of endothelial cells. J. Virol. 76:11530–11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Flynn NMJ, Patel A, Kadlec J, Jones IM. 2013. Improving promiscuous mammalian cell entry by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. Biosci. Rep. 33:23–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuki IV, Kuhn KM, Lomazov IR, Rothman VL, Tuszynski GP, Iozzo RV, Swenson TL, Fisher EA, Williams KJ. 1997. The syndecan family of proteoglycans. Novel receptors mediating internalization of atherogenic lipoproteins in vitro. J. Clin. Investig. 100:1611–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fuki IV, Meyer ME, Williams KJ. 2000. Transmembrane and cytoplasmic domains of syndecan mediate a multi-step endocytic pathway involving detergent-insoluble membrane rafts. Biochem. J. 351(Part 3):607–612. [PMC free article] [PubMed] [Google Scholar]
- 57.Wilsie LC, Gonzales AM, Orlando RA. 2006. Syndecan-1 mediates internalization of apoE-VLDL through a low density lipoprotein receptor-related protein (LRP)-independent, non-clathrin-mediated pathway. Lipids Health Dis. 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Podyma-Inoue KA, Hara-Yokoyama M, Shinomura T, Kimura T, Yanagishita M. 2012. Syndecans reside in sphingomyelin-enriched low-density fractions of the plasma membrane isolated from a parathyroid cell line. PLoS One 7:e32351. 10.1371/journal.pone.0032351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benmerah A, Lamaze C. 2007. Clathrin-coated pits: vive la différence? Traffic 8:970–982. [DOI] [PubMed] [Google Scholar]
- 60.Chen K, Williams KJ. 2013. Molecular mediators for raft-dependent endocytosis of syndecan-1, a highly conserved, multifunctional receptor. J. Biol. Chem. 288:13988–13999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teng YH, Aquino RS, Park PW. 2012. Molecular functions of syndecan-1 in disease. Matrix Biol. 31:3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo W-Y, Shih Y-S, Hung C-L, Lo K-W, Chiang C-S, Lo W-H, Huang S-F, Wang S-C, Yu C-F, Chien C-H, Hu Y-C. 2012. Development of the hybrid Sleeping Beauty: baculovirus vector for sustained gene expression and cancer therapy. Gene Ther. 19:844–851. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Balasundaram G. 2010. Potential cancer gene therapy by baculoviral transduction. Curr. Gene Ther. 10:214–225. [DOI] [PubMed] [Google Scholar]
- 64.Edgell CJ, McDonald CC, Graham JB. 1983. Permanent cell line expressing human factor VIII-related antigen established by hybridization. Proc. Natl. Acad. Sci. U. S. A. 80:3734–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]