Abstract
Previous studies indicate that the processing of hepatitis C virus (HCV) E2-p7-NS2 precursor mediated by host signal peptidase is relatively inefficient, resulting in the accumulation of E2-p7-NS2 and E2-p7 precursors in addition to E2 in mammalian cells. In this study, we discovered that a significant inhibition of the processing at an E2-p7 junction site is detrimental for HCV production, whether it was caused by the mutations in p7 or by the strategic introduction of a mutation at a terminal residue of E2 to block the signal peptidase-mediated cleavage of this junction site. However, complete separation of E2 and p7 by inserting an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) between these two proteins also moderately inhibited virus production. These results indicate that optimal processing of the E2-p7 junction site is critical for efficient HCV production. We further demonstrated that disrupting E2-p7 processing inhibits both NS2 localization to the putative virus assembly sites near lipid droplets (LD) and NS2 interaction with NS3 and E2. However, the impact, if any, of the p7-NS2 processing efficiency on HCV production seems relatively minor. In conclusion, these results imply that effective release of E2 and p7 from the precursor E2-p7 promotes HCV production by enhancing NS2-associated virus assembly complex formation near LD.
INTRODUCTION
Hepatitis C virus (HCV) is one of the major agents responsible for causing severe liver diseases, including chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (1, 2). It is an enveloped, positive-stranded RNA virus belonging to the Hepacivirus genus in the Flaviviridae family of viruses (3, 4). HCV encodes a single, large open reading frame encoding an ∼3,000-amino-acid polyprotein flanked by 5′ and 3′ noncoding sequences. The internal ribosome entry site (IRES) located at the 5′ noncoding region mediates translation of a polyprotein that is subsequently processed by both host- and virus-encoded proteases to yield 10 different proteins. The N-terminal one-third of the polyprotein encoding the structural proteins, including core and envelope proteins (E1 and E2), followed by p7 and NS2, is processed by host signal peptidase (5–7). Core protein is further processed by signal peptide peptidase into a mature form (8). The junction site between NS2 and NS3 is cleaved by an NS2-NS3 autoprotease (9, 10), and the remaining nonstructural proteins (NS3, NS4A, NS4B, NS5A, and NS5B) are processed by NS3 protease with its cofactor NS4A (11–13). Proteins NS3 to NS5B, along with the 5′- and 3′-terminal noncoding regions, were shown to be sufficient for the autonomous replication of HCV RNA (14).
Recent advances in the infectious HCV cell culture model allowed the investigation of HCV virus particle assembly processes (15–18). We have learned that the association of HCV core protein with lipid droplets (LD) is critical for virus assembly since disrupting the core protein localization to LD inhibited infectious virus production (19, 20). We also learned that the association of the C-terminal domain of NS5A with the core protein on LD is important for virus production (19, 21–24). It was suggested that NS5A is responsible for recruiting a replication complex to the vicinity of the LD since the localization of other nonstructural proteins in close proximity of LD depended on the localization of NS5A to LD (19). Several independent studies, including ours, showed that NS2 is involved in virus particle assembly by interacting with both the structural proteins E1 and E2 and also the nonstructural proteins NS3 and NS5A (25–28). Interestingly, these NS2-mediated interactions are associated with colocalization of E1, E2, NS2, NS3, and NS5A at dot-like structures near LD, and the relative percentages of HCV replicating cells exhibiting these dot-like structures positively correlated with the infectious virus titer (25, 26, 28). Thus, it is likely that these dot-like structures in the vicinity of LD represent virus particle assembly sites.
p7 is a small hydrophobic protein with two transmembrane domains connected by a short stretch of basic residues in the cytoplasm (29, 30). Previous studies showed that p7 forms a cation-selective ion channel as a hexameric or heptameric complex (31, 32). The ion channeling function of p7 is important for infectious HCV production since the mutations that inhibited the ion channel function of p7 also inhibited infectious virus production. Furthermore, the ectopic expression of the influenza virus M2 ion channel protein or treatment with bafilomycin partially rescued the virus production defect caused by some p7 mutations (33, 34). However, Brohm and colleagues showed that p7 or its precursors, but not M2, was capable of rescuing the virus production defect of the Δp7half (lacking residues 1 to 32) deletion mutant (35). These results suggest that p7 is involved in HCV production in both ion channel-dependent and -independent manners (34–36).
Several previous studies demonstrated that the processing by signal peptidase between the junction sites of E2 and p7 and also of p7 and NS2 is relatively inefficient, as evidenced by the detection of E2-p7 and E2-p7-NS2 precursors, despite the fact that the signal peptides of p7 and NS2 can function efficiently when they are fused to reporters (6, 37, 38). Subsequently, Carrère-Kremer and colleagues demonstrated that the incomplete cleavage at these junction sites is due to the structural determinants at the p7 junctions (37). The influence of the p7 sequence on the processing of p7 precursors was additionally supported by recent studies in which investigators showed that some mutations introduced to p7 affected the processing at these two junction sites (33–35). The negative impact of p7 mutations on the processing of the p7 precursors makes it difficult to interpret the p7 mutation-mediated viral phenotypes since it is unclear whether the effects of some p7 mutations on HCV replication are due to a functional defect of p7 or an aberrant processing of p7 precursors. Importantly, the significance of the delayed processing of p7 junction sites on HCV replication is currently unknown.
In this report we describe results suggesting that the optimal processing of the E2-p7 junction site is critical for efficient virus production.
MATERIALS AND METHODS
Cells.
Clonal derivatives of Huh-7, Huh-7.5 (39), and FT3-7 (40) were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum at 37°C in a 5% CO2 environment.
Plasmids.
HJ3-5, HJ3-5/p7(KRAA) (where KRAA indicates the mutations K33A R35A in p7), HJ3-5/p7HA/IRES (where p7HA is p7 with a C-terminal hemagglutinin [HA] tag), HJ3-5/p7HA/IRES/p7(KRAA), and HJ3-5/p7/IRES were described previously (25, 41). The HA epitope tag (YPYDVPDYA) followed by triple glycine residues was fused in frame to the first residue of p7 within pHJ3-5, HJ3-5/p7(KRAA), and HJ3-5/p7/IRES by using overlapping primers and a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) to make HJ3-5/HAp7 (p7 with an N-terminal HA tag), HJ3-5/HAp7(KRAA), and HJ3-5/HAp7/IRES. The mutation at the last residue of E2 A384R was introduced to HJ3-5 to make HJ3-5/E2(AR) by using a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). An encephalomyocarditis virus (EMCV) IRES was introduced to HJ3-5 to make HJ3-5/E2/IRES and HJ3-5/E2/IRES/p7(KRAA) by conducting the following steps. First, we introduced a unique PmeI restriction site between E2 and the p7 region of HJ3-5. Second, overlapping PCR was carried out to generate an EMCV IRES-NS2 fragment with an N-terminal PmeI site. Third, this fragment was digested with PmeI and BglII (located within NS2) restriction enzymes and ligated to PmeI/BglII fragments derived from pHJ3-5 with the PmeI site indicated above to make HJ3-5/E2/IRES. p7(KRAA) mutations were introduced to HJ3-5/E2/IRES to make HJ3-5/E2/IRES/p7(KRAA) by the PCR-based mutagenesis methods described above. The sequences of the regions manipulated within each plasmid were verified by DNA sequencing.
In vitro HCV RNA synthesis and transfection.
HCV cDNA (1 μg) was linearized with XbaI (NEB, Hitchin, United Kingdom), followed by transcription to RNA by using a T7 Megascript kit (Ambion, Austin, TX). DNA-free RNA was purified with an RNeasy RNA isolation kit (Qiagen, Valencia, CA). RNA integrity and concentration were determined by agarose gel electrophoresis and absorbance at 260 nm, respectively. In vitro-transcribed HCV RNA was transfected by electroporation into FT3-7 cells. In brief, 5 × 106 cells were mixed with 10 μg of in vitro-transcribed HCV RNA in a 4-mm cuvette and pulsed once at 270 V and 950 μF in a Gene Pulser System (Bio-Rad, Hercules, CA). Electroporated cells were seeded into 12-well plates for HCV RNA analysis and six-well plates for virus titration and HCV protein analysis.
HCV infectivity assays.
For virus titration, serial 10-fold dilutions of clarified cell culture supernatant or cell lysates that had been subjected to freezing and thawing were inoculated in 100-μl aliquots onto naive Huh-7.5 cells seeded at 1 × 105 cells/well in 48-well plates. Cells were incubated at 37°C in a 5% CO2 environment and fed with 200 μl of medium 24 h later. At 3 days postinoculation, the cells were fixed with methanol-acetone (1:1) for 10 min at room temperature and then immunostained for core protein expression by using monoclonal antibody C7-50 (1:600 dilution; Thermo Scientific, Rockford, IL,), followed by AlexaFluor 488-conjugated goat anti-mouse IgG (1:1,000 dilution; Invitrogen, Carlsbad, CA). Infectivity was determined by counting the clusters of infected cells staining for core protein, which were considered to be single, infectious focus-forming units (FFU).
Western blot analysis.
At day 2 postelectroporation, cells were lysed with ice-cold lysis buffer [phosphate-buffered saline (PBS) supplemented with 1% CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate) and complete protease inhibitor cocktail (Roche, Indianapolis, IN)]). Aliquots of cell lysates containing 20 μg of proteins were separated by performing SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were probed with monoclonal antibodies to core protein (C7-50, 1:2,000 dilution; Thermo Scientific, Rockford, IL), polyclonal rabbit anti-NS2 antibody (25) (1:10,000 dilution), NS3 (9-G2, 1:1,000 dilution; ViroGen, Watertown, MA), polyclonal goat anti-E2 antibody (1:2,000 dilution; Virostat, Inc., Portland, ME), and monoclonal anti-HA antibody (1:1,000 dilution; Sigma, St. Louis, MO). Proteins were visualized by subsequently probing the membranes with IRdye 800CW goat anti-mouse, IRdye 680 goat anti-rabbit, and IRdye 680 donkey anti-goat secondary antibodies (Li-Cor Biosciences, Lincoln, NE), followed by imaging with an Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE).
Coimmunoprecipitation.
Cell lysates were incubated with anti-NS2 antibody overnight at 4°C, followed by incubation for an additional 30 min on ice after the addition of protein G MicroBeads (Miltenyi Biotech, Auburn, CA). These solutions were applied to μ columns that had been set up in the magnetic field of a μMACS separator (Miltenyi Biotech, Auburn, CA). The columns were washed four times using wash buffer 1 (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl, pH 8.0), followed by one wash with buffer 2 (20 mM Tris-HCl, pH 7.5). Immune complexes were eluted by applying a preheated elution solution supplied by the manufacturer (Miltenyi Biotech, Auburn, CA).
Quantitative real-time RT-PCR.
Viral RNA was detected by a quantitative TaqMan reverse transcription-PCR (RT-PCR) assay (25). Total RNA was isolated from cell lysates by using an RNeasy kit (Qiagen, Valencia, CA) in accordance with the manufacturer's instructions. Quantitative real-time TaqMan RT-PCR analysis was carried out in a Bio-Rad iQ5 Real-time PCR Detection System by using the following primer pairs and a probe targeting a conserved 221-base sequence within the 5′ nontranslated RNA segment of the genome: HCV84FP, GCCATGGCGTTAGTATGAGTGT; HCV JFH_303RP, CGCCCTATCAGGCAGTACCACAA; and HCV146BHQ, FAM-TCTGCGGAACCGGTGAGTACACC-DBH1 (where FAM is 6-carboxyfluorescein). Reaction mixtures were incubated at 50°C for 2 min, 60°C for 45 min, and 95°C for 2 min, followed by 40 cycles of 95°C for 20 s and 60°C for 1 min.
Deglycosylation with endoglycosidase H.
Endoglycosidase H (Endo-H; NEB, Hitchin, United Kingdom) treatment was performed according to the manufacturer's recommendations. Briefly, 20 μg of protein was added to 4× denaturing SDS sample buffer and heated at 100°C for 10 min. Subsequently, 2,000 units of Endo-H (2 μl) and 2 μl of 50 mM sodium citrate (pH 5.5) were added to samples in a final volume of 20 μl, and then these reaction mixtures were incubated for 90 min at 37°C, followed by inactivation of Endo-H at 75°C for 10 min.
Confocal microscopy.
Electroporated cells were plated on eight-well chamber slides (BD Biosciences, Bedford, MA) at a density of 1 × 104 cells per well. Two days later, slides were washed with PBS, fixed with 4% formaldehyde for 20 min at room temperature, and permeabilized with 0.2% Triton X-100 in PBS for 10 min. Fixed and permeabilized cells were incubated with NS2 antibody (1:1,000 dilution) for 2 h at room temperature, followed by incubation with AlexaFluor 488-conjugated, goat anti-rabbit antibody (1:1,000; Invitrogen, Carlsbad, CA). Lipid droplets were stained with HCS LipidTOX deep red neutral lipid stain (1:1,000) (Molecular Probes, Inc., Eugene, OR) for 30 min at room temperature. Nuclei were labeled with Hoechst stain (Anaspec, Inc., Fremont, CA). Slides were examined with an Olympus FV1000-Fluoview confocal laser scanning biological microscope.
Statistical analyses.
Student's t test (unpaired, with Welch's correction) was performed by using GraphPad Prism, version 6, software (GraphPad, La Jolla, CA) to determine the significance in differences between two paired values.
RESULTS
HA epitope tag introduced to the N terminus of p7 rescues the virus production defect of a p7 mutant.
In a previous study, we introduced either a yellow fluorescence protein (p7YFP) or an HA (p7HA) epitope tag in frame to the C terminus of p7 in an effort to detect p7 since p7 antibody was not readily available (25). However, while YFP tagging of p7 allowed the detection of YFP-fused p7, this modification prevented the production of infectious HCV (25). On the other hand, while the introduction of the HA epitope tag to the C terminus of p7 (previously described as HJ3-5/p7HA/IRES) only slightly reduced virus production, we failed to detect HA-tagged p7, other than E2-p7HA precursor (25). In this study, we introduced an HA epitope tag at the N terminus of p7 followed by a 3-amino-acid linker sequence (Gly-Gly-Gly) to a genotype 1a/2a (gt1a/2a) chimeric HCV HJ3-5 (HJ3-5/HAp7) (Fig. 1A). This modification not only allowed the detection of HA-tagged p7 (HAp7) (Fig. 1C, lane 3) but also permitted the production of infectious virus, although at a moderately reduced level (∼1 log) compared to that from HJ3-5 at day 2 postelectroporation of this RNA (Fig. 1B, compare lanes 1 and 3). These results confirmed the recent report by Vieyres and colleagues who showed that introducing an N-terminal double HA tag to the p7 of the gt2a/2a chimera Jc1 allowed the production of virus, albeit at a reduced level compared to that from Jc1 (42). Since p7 in HJ3-5 is derived from gt1a, these results also indicate that p7 with an N-terminal epitope tag was functional in virus production in both gt1a and gt2a genetic backgrounds.
Fig 1.
The effect of the HA epitope tag at the N terminus of p7 on infectious HCV production and p7-NS2 processing. (A) Organization of the gt1a/gt2a HCV chimera HJ3-5 modified to encode the HA epitope tag at the N terminus of p7 with or without the insertion of a stop codon followed by the EMCV IRES at the junction site of p7 and NS2. The location of p7(KRAA) mutations is also indicated. The gt1a-based sequence is shaded. C, core protein. (B) Intracellular and extracellular virus titers expressed as infectious focus-forming units (FFU) (see Materials and Methods). The dotted line indicates infectious virus detection limit. Mean titers ± standard deviations from four different experiments are shown. (C) The expression of NS2, NS3, HAp7, and HAp7-NS2 determined by Western blot analysis by using anti-NS2 (α-NS2), anti-NS3 (α-NS3), and anti-HA (α-HA) antibodies, at day 2 postelectroporation of the indicated HCV RNAs.
We reported that double mutations located at the cytoplasmic loop region of p7 (K33A R35A, noted as KRAA) disrupted the ion channel activity of p7 and completely blocked infectious HJ3-5 virus production (Fig. 1B, lane 2) (25, 34). Corresponding p7 mutations were also lethal for gt2a JFH1 and gt1b/2a chimeric Con1/C3 virus production (33). Also, these mutations severely decreased the virus production from gt2a/2a chimeras, including J6 and J6/JFH1 (33, 43). Thus, we were surprised to find that HJ3-5/HAp7 with KRAA mutations [HJ3-5/HAp7(KRAA)] is capable of infectious virus production at a significant level (Fig. 1B, lane 4). These results suggested that the N-terminal HA tag in p7 compensated for the virus assembly defect caused by KRAA mutations within p7. Thus, we further investigated the mechanisms of this unexpected compensation by the N-terminal HA tag at p7 for the virus assembly defect caused by p7 mutations.
HAp7-NS2 precursor is not responsible for the promotion of virus production from a p7 mutant.
One of our first findings upon introducing the N-terminal HA epitope tag to p7 in HJ3-5 to make HJ3-5/HAp7 was the marked appearance of an HAp7-NS2 precursor detectable by both anti-NS2 and anti-HA antibodies (Fig. 1C, lane 3). We also detected the corresponding HAp7-NS2 precursor from HJ3-5/HAp7(KRAA), although in reduced quantity compared to that from HJ3-5/HAp7 (Fig. 1C, compare lanes 3 and 4). This could be due to a lower stability of p7 caused by p7(KRAA) mutations since the level of HAp7(KRAA) from HJ3-5/HAp7(KRAA) was significantly lower than that of HAp7 from HJ3-5/HAp7 (Fig. 1C, compare lanes 3 and 4). It is also possible that the processing of the HAp7(KRAA)-NS2 precursor is more efficient than that of the wild-type (wt) counterpart, resulting in a lesser accumulation of this precursor from the KRAA mutant (Fig. 1C, compare lanes 3 and 4). The third possibility is the preferential accumulation of the E2-HAp7(KRAA)-NS2 precursor from the KRAA mutant, as described below, which would consequently reduce the level of HAp7(KRAA)-NS2 precursor (Fig. 2A, arrowhead in lane 4).
Fig 2.
E2-p7 processing efficiency. (A) E2 and its precursors were detected by using Western blot analyses of cell lysates collected at day 2 postelectroporation of the indicated HCV RNAs to Huh-7 cells by using anti-E2 (α-E2) and anti-HA (α-HA) antibodies. The arrowheads indicate the locations of E2-HAp7-NS2. The Western blots of endoglycosidase H (Endo-H)-treated samples to better separate the E2 and p7 are shown in the bottom panel. E2 is detected in doublet bands upon Endo-H treatment, similar to results from previous studies (49–51). GND represents replication-defective HCV RNA. (B) The percentage of E2 in unprocessed E2-p7 precursor forms, including E2-p7, E2-HAp7, and E2-p7HA, was calculated from Western blot analyses of Endo-H-treated samples following quantification of E2-related protein bands by using an Odyssey Infrared Imaging system (Li-Cor) after background correction. Mean percentages of E2-p7 precursors and the standard deviations from three to five experiments are shown. Statistical analyses were performed by using GraphPad Prism, version 6, software (see Materials and Methods). Asterisks indicate statistically significant differences between two paired values: ***, P < 0.0005; **, P < 0.005; *, P < 0.05. The differences with a P value of >0.05 were considered not significant (ns).
To investigate whether the increased level of HAp7(KRAA)-NS2 precursor could have accounted for the significant level of virus production observed from the HJ3-5/HAp7(KRAA) mutant, we inserted the stop codon followed by an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) at the junction site between p7 and NS2 to eliminate this precursor formation (Fig. 1A). This modification effectively eliminated the protein band detectable by both anti-NS2 and anti-HA antibodies, as expected, thus confirming that this protein band was indeed the p7-NS2 precursor (Fig. 1C, compare lanes 3 and 4 with lanes 5 and 6).
The RNA replication levels of the bicistronic genomes [HJ3-5/HAp7/IRES and HJ3-5/HAp7(KRAA)/IRES] were comparable to those of their monocistronic counterparts [HJ3-5/HAp7 and HJ3-5/HAp7(KRAA)] (Fig. 3A, compare lanes 6 and 8 and lanes 7 and 9). However, both the intracellular and extracellular virus titers were moderately decreased from HJ3-5/HAp7/IRES compared to those from HJ3-5/HAp7 by about 1 log (Fig. 1B, compare lanes 3 and 5). Also, the virus titer from HJ3-5/HAp7(KRAA)/IRES was increased, rather than decreased, compared to that from HJ3-5/HAp7(KRAA) (Fig. 1B, compare lanes 4 and 6). Based on these results, it is unclear whether the delayed processing of p7-NS2 leading to the accumulation of p7-NS2 may play a role during HCV replication or not (see Discussion). Importantly, these results suggested that aberrant processing between p7 and NS2 caused by the N-terminal HA tagging of p7, which led to the accumulation of the p7-NS2 precursor, is not responsible for the restoration of the virus production defect caused by p7 KRAA mutations.
Fig 3.
Time course of HCV RNA replication and virus production. (A) The results of the quantitative TaqMan RT-PCR assays for HCV RNAs in lysates of electroporated Huh-7 cells at the indicated time points. The relative HCV RNA represents the copy number for each construct relative to the value present at 4 h after electroporation.) Extracellular (B) and intracellular (C) virus titers were determined at the indicated time points. Mean titers ± standard deviations from four different experiments are shown. The dotted line is the limit of virus titration assays.
HA epitope at the N-terminal region of p7 restores the E2-p7 processing defect caused by the p7 KRAA mutations.
During our previous study, we detected significantly higher levels of E2-p7 precursor from HJ3-5/p7(KRAA) than from HJ3-5 (25). It was described previously by Carrère-Kremer and colleagues that modifying the sequence within the N-terminal region of p7, including the insertion of an HA epitope sequence, enhanced the processing between E2 and p7 when they were expressed by using a vaccinia virus expression system (37). The location of HA epitope insertion was slightly different between previous and our current studies, being between the P3′ and P4′ positions within p7 and at the N terminus of p7, respectively. However, we considered the possibility that E2-p7 processing might have been enhanced in our case as well following HA insertion to the p7 N terminus. To investigate this, we electroporated HCV RNAs to Huh-7 cells and detected E2 and E2-p7 proteins by performing Western blot analysis 48 h later. To better distinguish the protein bands corresponding to the E2-p7 precursors (E2-p7, E2-HAp7, or E2-p7HA) and E2, we also analyzed the cell lysates following treatment with endoglycosidase H (Endo-H) to remove the glycans from the E2 (Fig. 2A, bottom panel). The protein band intensities corresponding to the E2-p7 precursors and E2 from three to five independent experiments were quantified by using an Odyssey Infrared Imaging System (see Materials and Methods for details), and the mean percentages of E2 in E2-p7 precursor forms from such analyses along with standard deviations are shown in Fig. 2B.
First, we confirmed our previous finding that p7(KRAA) mutations in HJ3-5 significantly inhibit processing between E2 and p7 compared to the wt (25), as shown in the Western blot analysis of the cell lysates from HJ3-5 and HJ3-5/p7(KRAA) by using an anti-E2 antibody (Fig. 2A and B, compare lanes 1 and 2). We detected ∼60% of total E2 from the p7(KRAA) mutant in the E2-p7 precursor form compared to ∼15% of that from HJ3-5 (Fig. 2B, lanes 1 and 2). Second, the HA tag at the C terminus of p7 did not significantly affect the E2-p7 processing in either HJ3-5/p7HA/IRES or HJ3-5/p7HA(KRAA)/IRES compared to that in HJ3-5 or HJ3-5/p7(KRAA) (Fig. 2A and B, compare lanes 1 and 7 and lanes 2 and 8). Third, these analyses clearly indicate that the N-terminal insertion of an HA tag to p7 significantly enhanced the processing of E2-p7 derived from both HJ3-5/HAp7 and HJ3-5/HAp7(KRAA), decreasing the detectable level of E2-HAp7 precursor from HJ3-5/HAp7 to ∼4% and that from HJ3-5/HAp7(KRAA) to ∼9%, compared to that from HJ3-5 and HJ3-5/p7(KRAA) (Fig. 2B, compare lanes 3 and 4 to lanes 1 and 2). Thus, the E2-p7 processing defect caused by the p7(KRAA) mutations was corrected by the HA tag at the N terminus of p7.
However, although the processing of the E2-p7 junction site from HJ3-5/HAp7(KRAA), as judged by the relative level of this precursor, was more augmented than that from parental HJ3-5 (∼9% versus ∼15%) (Fig. 2B, compare lanes 1 and 4), it was still relatively low in this mutant compared to that in HJ3-5/HAp7 (∼9% versus ∼4%) (Fig. 2B, compare lanes 3 and 3). These data may indicate that p7(KRAA) mutations still exert a negative influence on E2-p7 processing even when the processing of this junction site was augmented by the HA tag at the p7 N terminus. We also detected an E2-HAp7-NS2 precursor that was detectable by anti-E2, anti-HA, and anti-NS2 antibodies from an HJ3-5/HAp7(KRAA) mutant (Fig. 2A, arrowhead in lane 4; also data not shown) that is likely generated, in part, due to the relatively inefficient processing of E2-p7 and p7-NS2 junction sites in this mutant, as described above (Fig. 1C and 2A, lane 4). This E2-HAp7-NS2 precursor was hardly detectable from HJ3-5/HAp7, probably due to the efficient processing between E2 and p7 in this HCV clone (Fig. 2A, lane 3). Interestingly, we observed a small, but significant, increase in the level of E2-HAp7 precursor from HJ3-5/HAp7/IRES compared to that from HJ3-5/HAp7 (Fig. 2B, compare lanes 3 and 5). This result suggests the possibility that efficient processing of the p7-NS2 junction site may have a negative impact on E2-p7 processing. However, we did not observe any significant difference in the relative amounts of this precursor between HJ3-5/HAp7(KRAA)/IRES and HJ3-5/HAp7(KRAA) (Fig. 2B, compare lanes 4 and 6), probably due to the inherent adverse effect of KRAA mutations on E2-p7 cleavage.
Our attempt to directly measure the E2-p7 processing kinetics by using a pulse-chase experiment did not yield conclusive data due to the difficulty in separating the E2-p7 and E2 from each other and/or from host proteins that coseparated with them (data not shown). Therefore, we could not completely eliminate the possibility that differences in the relative abundance of E2 in unprocessed E2-p7 precursor forms that we detected in this study (Fig. 2B) may have been derived from the relative difference in E2-p7 and/or E2 stabilities rather than from the difference in protein processing efficiencies. However, the probability of the former is low, based on the following information. First, the average levels of total E2 (E2 plus E2-p7) from the different HCV derivatives used in this study were similar between them (Fig. 2; also data not shown). Second, most of the HCV constructs encode exactly the same E2 sequences.
Optimal processing of E2-p7 precursor plays a critical role in HCV production.
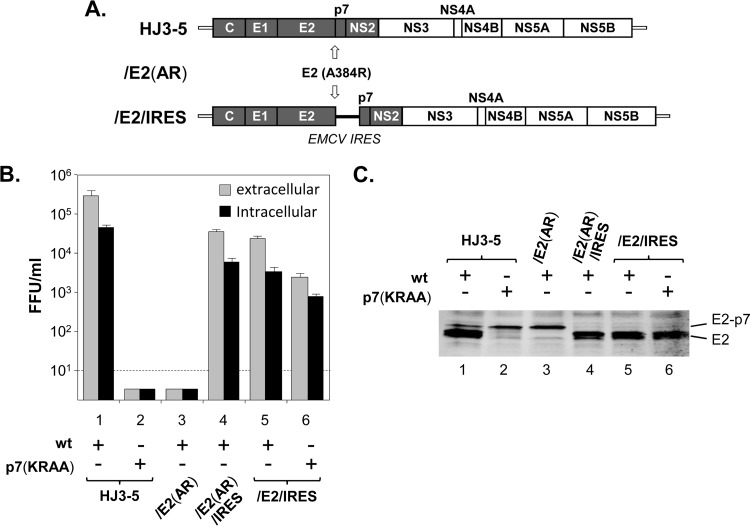
To formally demonstrate that enhanced processing of the E2-p7 junction in HJ3-5/HAp7(KRAA) is responsible for the production of infectious virus from this mutant and also to determine the importance of E2-p7 processing on HCV production, we performed the following experiments. First, we introduced a termination codon, followed by an EMCV IRES, at the junction site of E2 and p7 to separate these two proteins, independent of the signal peptidase-mediated cleavage (Fig. 4A). This modification caused a more than 1-log reduction in virus production, for both intracellular and extracellular virus, from the resulting HJ3-5/E2/IRES compared to that from HJ3-5 (Fig. 4B, compare lanes 1 and 5) although viral RNA replication was not affected by this change (Fig. 3A, compare lanes 1 and 4). However, the same modification led to infectious virus production from the resulting HJ3-5/E2/IRES/p7(KRAA), despite the fact that this clone lacked an HA tag at the N terminus of p7 (Fig. 4B, lane 6). These results imply that the HA tag at the N terminus of p7 per se was not responsible for causing the HJ3-5/HAp7(KRAA) to produce infectious virus. Rather, these results support the notion that improved processing between E2 and p7 caused by the HA insertion at the N terminus of p7 in HJ3-5/HAp7(KRAA) led to the production of infectious virus from this mutant. To further understand the importance of E2-p7 processing on virus production, we introduced a mutation at the last residue of E2 (A384R) in HJ3-5 [HJ3-5/E2(AR)] to block the signal peptidase-mediated cleavage of the E2-p7 junction site (44). As shown in Fig. 4C, lane 3, the processing of E2-p7 precursor was severely impaired in HJ3-5/E2(AR), and nearly ∼86% of E2 was detected as an E2-p7 precursor form (Fig. 2B, lane 9). Importantly, similar to the case of HJ3-5/p7(KRAA), this mutation completely blocked virus production (Fig. 4B, lane 3), despite the fact that the p7 sequence in this mutant is wt.
Fig 4.
The effect of E2-p7 processing on HCV production. (A) The organization of HJ3-5 with and without E2 terminal residue mutation A384R (AR) to block E2-p7 processing by the signal peptidase and HJ3-5/E2/IRES, which is modified to encode a stop codon followed by an EMCV IRES at the junction of E2 and p7 to separate E2 and p7. (B) Intracellular and extracellular virus titers determined at day 2 postelectroporation of indicated HCV RNAs. Mean titers ± standard deviations from four different experiments are shown. The dotted line is the limit of virus titration. (C) Western blot analysis of cell lysates collected at the time point indicated above following Endo-H treatment to detect E2 and E2-p7 by using an anti-E2 antibody.
To verify that the defect in E2-p7 processing, and not an AR mutation-mediated functional defect in E2, was responsible for the defective virus production from HJ3-5/E2(AR), we also generated HJ3-5/E2(AR)/IRES, in which we introduced a stop codon followed by an IRES sequence between E2 and p7 of HJ3-5/E2(AR) to separate these two proteins. As expected, E2(AR)-p7 precursor was no longer detected in cells replicating HJ3-5/E2(AR)/IRES (Fig. 4C, lane 4). Importantly, we detected virus production from HJ3-5/E2(AR)/IRES at a level similar to that from HJ3-5/E2/IRES, despite the fact that the former encoded E2(AR) and the latter encoded wt E2 (Fig. 4B, lanes 4 and 5). These results indicate that the AR mutation in E2 did not affect E2 functions involved in virus production.
In aggregate, these results indicate that complete separation of E2-p7, as in the case of HJ3-5/E2/IRES, and significantly reduced processing of this site, as in the case of HJ3-5/E2(AR) and HJ3-5/p7(KRAA), are both detrimental for HCV production. These data support the notion that optimal processing of E2-p7 is critical for efficient production of infectious HCV. It is also worth mentioning that the defective processing of E2-p7 in HJ3-5/p7HA(KRAA)/IRES, as shown in Fig. 2 (lane 8), due to the lack of an apparent positive effect from a C-terminal HA tag at p7 on E2-p7 processing, correlates nicely with the virus production null phenotype of this mutant that we demonstrated before (25).
p7(KRAA) mutations delay the kinetics of virus assembly independent of their effect on viral protein processing.
We observed a delay in virus production kinetics from p7(KRAA) mutants, including HJ3-5/E2/IRES/p7(KRAA), HJ3-5/HAp7(KRAA), and HJ3-5/HAp7(KRAA)/IRES, compared to their p7 wt counterparts (Fig. 3B, compare lanes 4 and 5, 6 and 7, and 8 and 9). It is unlikely that the difference in RNA replication efficiency between the wt and p7(KRAA) mutants resulted in this phenotype since the HCV RNA replication levels were comparable between HJ3-5/HAp7(KRAA) and HJ3-5/HAp7(KRAA)/IRES and their p7 wt counterparts (Fig. 3A, compare lanes 6 and 7 and lanes 8 and 9). Also, although we detected a reduction in HCV RNA levels from the HJ3-5/E2/IRES/p7(KRAA) mutant compared to its p7 wt counterpart, a significant difference in HCV RNA level was detected at and after the 48-h time point and not at the 24-h time point when we detected a virus production delay from this mutant (Fig. 3A and B, compare lanes 4 and 5). The inherent virus production efficiency (as judged by steady-state virus titer) alone also could not explain the delayed kinetics of virus production observed from all of these virus-producing KRAA mutants that we investigated, since the maximum virus titer from HJ3-5/HAp7(KRAA)/IRES was comparable to that from its p7 wt counterpart (Fig. 3B, compare lanes 8 and 9), although the virus titers from HJ3-5/E2/IRES/p7(KRAA) and HJ3-5/HAp7(KRAA) were reduced compared to those from their p7 wt counterparts (Fig. 3B, compare lanes 4 and 5 and lanes 6 and 7).
The delayed kinetics of virus production from KRAA mutants is also independent from the processing efficiency of the junction site between E2 and p7 or between p7 and NS2 since we observed this particular phenotype in the backgrounds of both HJ3-5/E2/IRES and HJ3-5/HAp7/IRES, in which the processing efficiency of these two junction sites is no longer an issue. We observed similarly delayed kinetics of intracellular virus production from HJ3-5/E2/IRES/p7(KRAA) and HJ3-5/HAp7(KRAA)/IRES compared to their p7 wt counterparts (Fig. 3C, compare lanes 1 and 2 and lanes 3 and 4). Thus, the delayed virus production from these p7 mutants is due to the delay in the intracellular virus assembly rather than the secretion of virus to the extracellular medium. According to the study by StGelais and colleagues, KRAA mutations in p7 impaired not only its ion channel function but also its insertion into membranes although they did not substantially affect the folding and oligomerizing properties of p7 (45). It is likely that some of the disrupted functions of p7 in the p7(KRAA) mutant may be responsible for the delay in virus production. Alternatively, the potentially decreased stability of p7 due to the KRAA mutations, as evidenced by the lower level of HAp7 detected from HJ3-5/HAp7(KRAA) and HJ3-5/HAp7(KRAA)/IRES than that from HJ3-5/HAp7 and HJ3-5/HAp7/IRES (Fig. 1C, compare lanes 3 and 4 and lanes 5 and 6), may have played a role in delaying the kinetics of HCV assembly by reducing the level of p7 involved in virus assembly.
Both E2-p7 processing and functional p7 are required for NS2 localization to the dot-like complexes adjacent to LD.
In a previous study, we showed that p7(KRAA) mutations disrupted the localization of NS2 to the putative virus assembly site near LD and inhibited NS2-mediated viral protein interaction (25). However, since p7(KRAA) mutations disrupted both the p7 function and E2-p7 processing, it was unclear which of these defects led to these phenotypes. To understand the determinants involved in NS2 subcellular localization and NS2-mediated viral protein complex formation, we electroporated HCV RNAs, including HJ3-5/E2(AR), which is defective in E2-p7 processing but encodes wt p7, and HJ3-5/E2/IRES/p7(KRAA), which is defective in p7 function but guarantees the complete separation of E2-p7 precursor, to Huh-7 cells. Next, we determined the localization of NS2 and the interaction between NS2 and NS3 or E2 by carrying out immunofluorescence analyses and coimmunoprecipitation assays, respectively.
First, confirming our previous findings (25), NS2 from HJ3-5 was localized to the distinct dot-like complexes near LD (punctate LD distribution) at 48 h postelectroporation of this RNA in the majority of NS2-positive cells that we analyzed (∼ 80%) (Fig. 5A and B, lane a). NS2 from HJ3-5/p7(KRAA) showed endoplasmic reticulum (ER)-like diffused distribution in the absence of strong colocalization with LD (ER-like distribution) in most of the cells replicating this RNA (∼90%) and, in the remaining cells, less distinct, occasional distribution near LD (intermediate distribution) (Fig. 5A, frame e, inset 2, and B, lane b). Second, the NS2 from HJ3-5/E2(AR), which is defective in E2-p7 processing, displayed exactly the same NS2 distribution phenotype as that from HJ3-5/p7(KRAA) (Fig. 5A and B, compare lanes b and c). These results suggest that NS2 localization to virus assembly sites near LD depends on the processing between E2 and p7. Third, we also detected punctate LD localization of NS2 from HJ3-5/E2/IRES in about ∼ 50% of cells replicating this RNA and that from HJ3-5/HAp7 in about ∼60% of cells, which are lower percentages than observed from HJ3-5 replicating cells (Fig. 5A and B, compare lane a with lanes d and f). These results suggest that the enhanced separation between E2 and p7 also may have a negative impact on NS2 localization to virus assembly sites near LD. It is likely that this reduced localization of NS2 to the virus assembly sites is at least partly responsible for reduced virus production from HJ3-5/E2/IRES and HJ3-5/HAp7 compared to that from HJ3-5 (Fig. 3B, compare lane 1 with lanes 4 and 6). Fourth, about 25% of cells replicating HJ3-5/E2/IRES/p7(KRAA) or HJ3-5/HAp7(KRAA) displayed non-ER-like distributions of NS2, in comparison to about 10% of cells replicating HJ3-5/p7(KRAA) (Fig. 5A and B, compare lane b with lanes e and g). Interestingly, NS2 from HJ3-5/E2/IRES/p7(KRAA) in this non-ER-like distribution category showed intermediate distribution and that from HJ3-5/HAp7(KRAA) showed punctate LD localization (Fig. 5A and B, compare lanes e and g). Since E2 and p7 are completely separated in HJ3-5/E2/IRES/p7(KRAA) and incompletely in HJ3-5/HAp7(KRAA), these data provide additional support of our hypothesis that incomplete separation of E2-p7 is important for efficient localization of NS2 to the punctate virus assembly sites near LD. It is likely that the difference in NS2 distributions between these two HCVs might have caused an ∼3-fold reduction in titer from HJ3-5/E2/IRES/p7(KRAA) compared to that from HJ3-5/HAp7(KRAA) (Fig. 3B, compare lanes 5 and 7). Importantly, these results suggest that separation of E2-p7 could enhance the localization of NS2 to the virus assembly sites even in the presence of p7(KRAA) mutations and explain the detectable levels of virus production from HJ3-5/E2/IRES/p7(KRAA) and HJ3-5/HAp7(KRAA) (Fig. 3B, lanes 5 and 7). Fifth, p7(KRAA) mutations reduced NS2 localization to punctate LD even when the E2-p7 processing defect caused by these mutations is no longer an issue, as in the case of HJ3-5/E2/IRES and its p7(KRAA) mutant pair and HJ3-5/HAp7 and its HAp7(KRAA) mutants pair (Fig. 5B, compare lanes d and e and lanes f and g). In summary, these results suggest that the disruption of normal E2-p7 processing (both the inhibition and enhancement) and the defect in p7 function caused by p7(KRAA) mutations negatively regulate NS2 localization to the punctate LD, which is a putative virus assembly site.
Fig 5.
The impact of E2-p7 processing on NS2 localization and its interaction with NS3 from the following in panel A: frame a, HJ3-5; b, HJ3-5/p7(KRAA); c, HJ3-5/E2(AR); d, HJ3-5/E2/IRES; e, HJ3-5/E2/IRES/p7(KRAA); f, HJ3-5/HAp7; and g, HJ3-5/HAp7(KRAA). Confocal image analysis using an Olympus FluoView FV1000 laser scanning confocal microscope of cells at day 2 postelectroporation with HCV RNAs encoding indicated genomes. Anti-NS2 antibody (green) and LipidTOX deep red neutral lipid stain (red) were used to detect NS2 and lipid droplets. The numbered images at the bottom are enlargements of the corresponding areas from the images at the top. (B) The relative percentages of NS2-positive cells displaying different NS2 localization patterns NS2 localization patterns were analyzed from 50 NS2 immunostaining-positive cells from two independent (lanes a, b, c, d, and e) or single (lanes f and g) experiments. Punctate, NS2 localization similar to that detected from frames a, d, f, and g in panel A; intermediate, NS2 localization similar to that observed from frame e' in panel A; nonpunctate, NS2 localization similar to that detected from frames b and c in panel A.
E2-p7 processing determines the efficiency of NS2 association with NS3 and E2.
We have used NS2 antibody to pull down the NS2 and NS2-associated viral proteins. Interestingly, NS2 from HJ3-5/p7(KRAA), HJ3-5/E2(AR), and HJ3-5/E2/IRES/p7(KRAA) (that we designated group A) was less efficiently immunoprecipitated by this antibody than that from HJ3-5 and HJ3-5/E2/IRES (group B) although all of these HCVs encode exactly the same NS2 sequence (Fig. 6A, top panel, and B). Since functional p7 availability is decreased in group A [due to the disruption of E2-p7 processing and/or p7(KRAA) mutations] but not impaired in group B and since our previous findings suggested that p7 affects NS2 conformation (25), we speculate that the antibody-binding epitope in NS2 from group A is less accessible to NS2 antibody than that from group B. Due to this finding, the degree of NS2 interactions with NS3 and E2 was calculated by a three-step process. First, the relative level of immunoprecipitated NS2 (IP-NS2) was normalized with input NS2 (Fig. 6B, IP-NS2). Second, the relative level of coprecipitated NS3 (coIP-NS3) or E2 (coIP-E2) was normalized with input NS3 or E2 (Fig. 6C and E). Third, the degree of interaction between NS2 and NS3 or E2 was calculated by dividing the coIP-NS3 or coIP-E2 value by the IP-NS2 value (Fig. 6D and F). The relative amount of protein pulled down obtained from HJ3-5 was set at 100. The data were obtained from at least three different experiments, and the significance of the differences in NS2-mediated NS3 or E2 pulldown efficiencies between the different HCVs was calculated by performing an unpaired Student's t test (see Materials and Methods).
Fig 6.
The impact of E2-p7 processing on the interaction of NS2 with NS3 and E2. (A) The interaction of NS2 with NS3 and E2 was determined by Western blot analysis following NS2 pulldown assays. (B) The level of immunoprecipitated NS2 (IP-NS2) was normalized to that of NS2 present in input lysates (NS2-input). IP-NS2 (normalized with by NS2-input) from HJ3-5 was set to 100. The standard deviations from five different experiments are shown. Asterisks indicate statistically significant differences between a and b, c, d, or e paired values: ***, P < 0.0005; **, P < 0.005; *, P < 0.05. The differences with a P value of >0.05 were considered not significant (ns). (C) The same experiment as shown in panel B except that the level of immunoprecipitated NS3 (IP-NS3) was normalized to that of NS3 present in input lysates (NS3-input). Results from three different experiments are shown. (D) The data shown in panel C were normalized to those shown in panel B from three different experiments to determine the efficiency of NS2 and NS3 interaction. (E) The same experiment as shown in panel B except that the level of immunoprecipitated E2 (IP-E2) was normalized to that of E2 present in input lysates (E2-input). (F) The data shown in panel E were normalized to those shown in panel B.
We detected a significant decrease in interactions between NS2 and NS3 or E2 from HJ3-5/p7(KRAA) compared to those from HJ3-5 (Fig. 6A, D, and F, compare lanes a and b), which was similar to our previous findings when we used an NS2YFP (NS2 with an N-terminal YFP tag) pulldown assay (25). Importantly, NS2 interaction with NS3 and E2 was also significantly reduced in HJ3-5/E2(AR) (Fig. 6A, D, and F, lanes c). These results suggest that E2-p7 processing is critical for NS2-mediated complex formation with NS3 and E2.
Complete separation of E2 and p7 in HJ3-5/E2/IRES decreased the interaction between NS2 and E2 but not that between NS2 and NS3 (Fig. 6A, D, and F, lanes d). Similar phenotypes were described previously by using the gt2a chimera Jc1 modified to encode an EMCV IRES between E2 and p7 (27). These results suggest that the complete separation of E2 and p7 negatively regulates the interaction of NS2 with E2 in a genotype-independent manner. In addition, these findings provide mechanistic insights behind results showing a lower level of virus production from HJ3-5/E2/IRES than from HJ3-5 (Fig. 3B, compare lanes 1 and 4) and reinforce the notion that optimal separation of E2 and p7 is necessary for efficient NS2-mediated viral assembly complex formation. It is important to point out that separating NS2 and NS3 in HJ3-5 by inserting an IRES between these two proteins (HJ3-5/NS2/IRES) also resulted in an ∼5-fold reduction in virus production (data not shown), which is similar to the results from previous studies (27, 43). Interestingly, according to the results of Stapleford and Lindenbach, the interaction between NS2 and NS3, but not that between NS2 and E2, was defective in Jc1/NS2-IRES-NS3, which is a gt2a chimeric equivalent of HJ3-5/NS2/IRES (27). These results suggest that complete separation of E2 and p7 and of NS2 and NS3 due to IRES insertion between these proteins inhibited HCV production by specifically affecting the interaction between NS2 and E2 and between NS2 and NS3, respectively. Furthermore, these results imply that the reduced virus production from HJ3-5/E2/IRES and HJ3-5/NS2/IRES is unlikely to be caused by the indirect impact of IRES insertion per se.
Interestingly, p7(KRAA) mutations did not affect the interaction between NS2 and NS3 or E2 in an HJ3-5/E2/IRES background, in which E2-p7 processing is no longer an issue (Fig. 6A, D, and F, compare lanes d and e). These results suggest that p7 may not be involved in the interaction between NS2 and other viral proteins or, at least, that these particular mutations of p7 did not disrupt NS2-mediated protein interaction.
In aggregate, these results imply that inhibition of E2-p7 processing disrupts the interaction between NS2 with NS3 and E2, while the IRES insertion-mediated separation of E2 and p7 disrupts NS2 interaction with E2 but not with NS3. Therefore, optimal processing of E2-p7 is critical for NS2-mediated viral protein complex formation important for infectious virus production.
DISCUSSION
This study revealed that impairment of E2-p7 processing is detrimental for infectious HCV production without affecting viral RNA replication. We also showed that introducing a stop codon, followed by an EMCV IRES, between E2 and p7 to completely separate E2 and p7 in the gt1a-gt2a chimera HJ3-5 (HJ3-5/E2/IRES) resulted in a moderate reduction in virus production, similar to that observed by Jones and colleagues, who used a gt2a-gt2a chimera, J6/JFH1 (43). Combined with the findings that E2-p7 processing is naturally delayed due to structural determinants at p7 (6, 37, 38) (Fig. 2), these data suggest that delayed, but effective, processing of the E2-p7 junction site is critical for efficient HCV production.
It is currently unknown how the delayed processing of the E2-p7 junction may affect HCV production. On one hand, it is possible that E2-p7 precursor may play a role during infectious virus production. However, if this is the case, the putative role played by E2-p7 precursor is not essential for virus production since removing this precursor completely, as in the case of HJ3-5/E2/IRES, allowed a reduced but significant level of virus production (Fig. 4B and C, lane 5) (43). Also, the defective processing of E2-p7 leading to a significant accumulation of E2-p7 precursor, as in the case of HJ3-5/p7(KRAA) and HJ3-5/E2(AR), prevented virus production (Fig. 4B and C, lanes 2 and 3). Thus, the E2-p7 precursor could not replace the functions of E2 and/or p7 during virus assembly. On the other hand, it is possible that the delayed processing of the E2-p7 precursor could ensure the delayed release of functional E2 and/or p7 until they are needed at late steps of HCV replication for virus particle assembly to prevent premature virus assembly. Our data support this possibility. First, the immediate availability of E2 and p7 without any delay during HCV replication, as in the case of HJ3-5/E2/IRES, reduced infectious virus production (Fig. 3B, compare lanes 1 and 4). Second, functional p7 is required for efficient virus production even when E2 and p7 are efficiently separated [Fig. 3B, compare lanes 4 and 5 and lanes 6 and 7 for the viral titers from HJ3-5/E2/IRES and HJ3-5/HAp7 in the presence and absence of p7(KRAA) mutations]. Third, the release of functional E2 allowed limited levels of virus production even in the absence of or with a reduced level of functional p7 since we observed the partial rescue of the virus production defect caused by p7(KRAA) mutations when E2 and p7 were efficiently separated, as in the case of HJ3-5/E2/IRES/p7(KRAA) and HJ3-5/HAp7(KRAA) (Fig. 3B, lanes 5 and 7). In aggregate, our data suggest that optimal availability of E2 and p7 is important for efficient HCV production.
The data shown in Fig. 5 and 6 revealed that E2-p7 processing is necessary for both NS2 localization to the putative virus assembly sites in close proximity of the LD and the interaction of NS2 with NS3 and E2 (Fig. 5 and 6, compare lanes a with lanes b and c). However, the separation of E2 and p7 alone was not sufficient for the localization of functional NS2 complexes near LD since p7(KRAA) mutations inhibited this localization even when the processing of E2-p7 precursor was no longer an issue (Fig. 5, compare lanes d and e and lanes f and g). These results suggest that, in addition to the E2-p7 processing, functional p7 is required for efficient localization of NS2 to the virus assembly sites. Supporting this notion, p7 was shown by Tedbury and colleagues to affect NS2 subcellular localization in an HCV subgenomic replicon system (46). On the other hand, the same p7(KRAA) mutations did not significantly affect the interaction of NS2 with NS3 and E2, as long as E2 and p7 were separated (Fig. 6D and F, compare lanes d and e). These results suggest that p7 may not be involved in NS2-mediated viral protein complex formation. However, further study is necessary to prove this notion. We also confirmed the previous findings by Stapleford and Lindenbach (27), who showed that separating E2 and p7 by inserting an EMCV IRES between these two proteins in a gt2a chimera inhibited the interaction between NS2 and E2 without affecting that between NS2 and NS3 by using the gt1a chimera HJ3-5/E2/IRES (Fig. 6D and F, compare lanes a and d). These results suggest that optimal processing of E2 and p7 is necessary to ensure an efficient interaction between the NS2 and E2 involved in the virus assembly process. Based on these results and the literature mentioned above, we propose that the efficiency of E2-p7 processing regulates HCV particle assembly by controlling the timely release of p7. Released p7 will then target the NS2 to the site of virus assembly to form virus assembly complexes by interacting with both the structural and nonstructural proteins. HCV may be actively delaying the release of p7 by placing structural constraints that make the cleavage of p7 junction sites relatively inefficient, as shown by Carrère-Kremer and colleagues (37), to delay the NS2 targeting to virus assembly sites by p7 until the time of optimal virus assembly to ensure an efficient interaction between NS2 and E2.
The significance of p7-NS2 processing on HCV replication is less clear. The substantial inhibition of p7-NS2 processing caused by HA epitope tagging at the N terminus of p7 in HJ3-5/HAp7 only moderately reduced infectious virus production (∼1 log or so) (Fig. 1, compare lanes 1 and 3). However, this modification also enhanced the processing between E2 and p7 (Fig. 2B, lane 3), and, thus, it is reasonable to expect that the enhanced E2-p7 processing could also have influenced the virus titer reduction in this HCV. Also, although the complete separation of HAp7 and NS2 by the insertion of an EMCV IRES in this HCV clone further decreased the virus titer (Fig. 1B and C, compare lanes 3 and 5), it is unclear whether this reduction is totally due to the absence of HAp7-NS2 precursor since this modification also affected E2-p7 processing (Fig. 2, compare lanes 3 and 5). In addition, we demonstrated previously that the insertion of an EMCV IRES to separate the C-terminal HA-tagged p7 and NS2 in HJ3-5/p7HA/EMCV resulted in only a 2- to 3-fold reduction in virus titer compared to that of the HJ3-5 (25). These data suggest that the impact of p7-NS2 processing efficiency on HCV production is relatively minor, if it occurs at all.
We observed that the efficient separation of E2 and p7, due to the insertion of either the EMCV IRES between them or of the N-terminal HA tag at p7, allowed moderate levels of virus production from HCV with p7(KRAA) mutations (Fig. 3B, lanes 5, 7, and 9). Interestingly, these p7 mutations caused delays of up to 12 h in virus production (Fig. 3B, compare lanes 4 and 5, 6 and 7, and 8 and 9). One explanation for this phenotype could be the reduced virus production efficiency of the p7(KRAA) mutants compared to that of their wt counterparts, as in the cases of wt and p7(KRAA) mutant pairs of HJ3-5/E2/IRES and HJ3-5/HAp7. However, the virus titers detected at different time points following the electroporation of HJ3-5/HAp7/IRES without and with p7(KRAA) mutations were strikingly similar starting from 36 h, despite the fact that we were able to detect virus production in the former from 24 h but not from the latter until 36 h (Fig. 3B, compare lanes 8 and 9, and C, compare lanes 3 and 4). As the P7(KRAA) mutations significantly diminished the NS2 localization to the putative virus assembly sites (Fig. 5A and B, compare lanes a and b, d and e, and f and g), we speculate that the p7-dependent localization of NS2 to virus assembly sites signaled the timely initiation of the HCV assembly step and that, in the absence or reduced level of functional p7, the slower, p7-independent localization of NS2 to the virus assembly site delayed the initiation of virus assembly.
Introducing the mutations to p7 often led to various degrees of E2-p7-NS2 processing defects, and, as a result, it had been difficult to understand the actual scope of p7-specific roles during HCV replication by p7 mutagenesis (33, 34). The p7(KRAA) mutations that we analyzed in this study were no exception, in that these mutations were shown to inhibit not only the p7 ion channeling function (34) but also E2-p7 processing when introduced to HJ3-5 (25). Interestingly, separating E2 and p7 in this p7-mutated HCV by introducing an EMCV IRES between them partly restored virus production defects, despite the fact that p7 ion channel activity was still defective (Fig. 4B, compare lanes 2 and 6). These results suggest to us that although the ion channel activity of p7 is required for efficient HCV production, it is not absolutely essential for virus production. The controversial, mostly negative, clinical efficacy of currently available p7 ion channel inhibitors could be due to the modest impact of p7 ion channel activity on HCV production (47, 48). However, it is likely that more potent p7 ion channel inhibitors in combination with other HCV inhibitors would benefit HCV therapy.
In conclusion, we showed that the efficiency of E2-p7 processing during HCV replication is the major determinant for optimal HCV assembly. Since the inhibition of this processing prevented HCV production, development of specific agents targeting this step may provide novel anti-HCV therapeutic options.
ACKNOWLEDGMENTS
We thank Leoncio Vergara for his help on confocal microscopic imaging analysis. We thank Arvind Patel, Charles Rice, and Takaji Wakita for providing AP33 antibody, Huh7.5 cells, and the JFH1 cDNA clone, respectively. We also thank Stanley M. Lemon, Robin Stephens, and Mardelle Susman for careful reading of the manuscript.
This work is supported by grant number R01 AI075090 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print 14 August 2013
REFERENCES
- 1.Alter HJ. 2005. HCV natural history: the retrospective and prospective in perspective. J. Hepatol. 43:550–552. [DOI] [PubMed] [Google Scholar]
- 2.Leone N, Rizzetto M. 2005. Natural history of hepatitis C virus infection: from chronic hepatitis to cirrhosis, to hepatocellular carcinoma. Minerva Gastroenterol. Dietol. 51:31–46. [PubMed] [Google Scholar]
- 3.Houghton M, Weiner A, Han J, Kuo G, Choo QL. 1991. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology 14:381–388. [PubMed] [Google Scholar]
- 4.Robertson B, Myers G, Howard C, Brettin T, Bukh J, Gaschen B, Gojobori T, Maertens G, Mizokami M, Nainan O, Netesov S, Nishioka K, Shin i T, Simmonds P, Smith D, Stuyver L, Weiner A. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch. Virol. 143:2493–2503. [DOI] [PubMed] [Google Scholar]
- 5.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. U. S. A. 88:5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin C, Lindenbach BD, Pragai BM, McCourt DW, Rice CM. 1994. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J. Virol. 68:5063–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima H, Hijikata M, Asabe S, Hirota M, Kimura K, Shimotohno K. 1994. Two hepatitis C virus glycoprotein E2 products with different C termini. J. Virol. 68:6215–6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McLauchlan J, Lemberg MK, Hope G, Martoglio B. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reed KE, Rice CM. 1999. Expression and characterization of the HCV NS2 protease. Methods Mol. Med. 19:331–342. [DOI] [PubMed] [Google Scholar]
- 10.Lorenz IC, Marcotrigiano J, Dentzer TG, Rice CM. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442:831–835. [DOI] [PubMed] [Google Scholar]
- 11.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Failla C, Tomei L, De Francesco R. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68:3753–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin C, Pragai BM, Grakoui A, Xu J, Rice CM. 1994. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J. Virol. 68:8147–8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. [DOI] [PubMed] [Google Scholar]
- 15.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. [DOI] [PubMed] [Google Scholar]
- 18.Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:2310–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097. [DOI] [PubMed] [Google Scholar]
- 20.Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169. [DOI] [PubMed] [Google Scholar]
- 21.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim S, Welsch C, Yi M, Lemon SM. 2011. Regulation of the production of infectious genotype 1a hepatitis C virus by NS5A domain III. J. Virol. 85:6645–6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032. 10.1371/journal.ppat.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma Y, Anantpadma M, Timpe JM, Shanmugam S, Singh SM, Lemon SM, Yi M. 2011. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J. Virol. 85:86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jirasko V, Montserret R, Lee JY, Gouttenoire J, Moradpour D, Penin F, Bartenschlager R. 2010. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 6:e1001233. 10.1371/journal.ppat.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stapleford KA, Lindenbach BD. 2011. Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 85:1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popescu CI, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps V, Duverlie G, Penin F, Heliot L, Rouille Y, Dubuisson J. 2011. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 7:e1001278. 10.1371/journal.ppat.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffin SD, Harvey R, Clarke DS, Barclay WS, Harris M, Rowlands DJ. 2004. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J. Gen. Virol. 85:451–461. [DOI] [PubMed] [Google Scholar]
- 30.Carrère-Kremer S, Montpellier-Pala C, Cocquerel L, Wychowski C, Penin F, Dubuisson J. 2002. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 76:3720–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clarke D, Griffin S, Beales L, Gelais CS, Burgess S, Harris M, Rowlands D. 2006. Evidence for the formation of a heptameric ion channel complex by the hepatitis C virus p7 protein in vitro. J. Biol. Chem. 281:37057–37068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luik P, Chew C, Aittoniemi J, Chang J, Wentworth P, Jr, Dwek RA, Biggin PC, Venien-Bryan C, Zitzmann N. 2009. The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 106:12712–12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steinmann E, Penin F, Kallis S, Patel AH, Bartenschlager R, Pietschmann T. 2007. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 3:e103. 10.1371/journal.ppat.0030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wozniak AL, Griffin S, Rowlands D, Harris M, Yi M, Lemon SM, Weinman SA. 2010. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 6:e1001087. 10.1371/journal.ppat.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brohm C, Steinmann E, Friesland M, Lorenz IC, Patel A, Penin F, Bartenschlager R, Pietschmann T. 2009. Characterization of determinants important for hepatitis C virus p7 function in morphogenesis by using trans-complementation. J. Virol. 83:11682–11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steinmann E, Pietschmann T. 2010. Hepatitis C virus p7-a viroporin crucial for virus assembly and an emerging target for antiviral therapy. Viruses 2:2078–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrère-Kremer S, Montpellier C, Lorenzo L, Brulin B, Cocquerel L, Belouzard S, Penin F, Dubuisson J. 2004. Regulation of hepatitis C virus polyprotein processing by signal peptidase involves structural determinants at the p7 sequence junctions. J. Biol. Chem. 279:41384–41392. [DOI] [PubMed] [Google Scholar]
- 38.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. 1994. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 68:6147–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yi M, Ma Y, Yates J, Lemon SM. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi M, Ma Y, Yates J, Lemon SM. 2009. Trans-complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 5:e1000403. 10.1371/journal.ppat.1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vieyres G, Brohm C, Friesland M, Gentzsch J, Wölk B, Roingeard P, Steinmann E, Pietschmann T. 2013. Subcellular localization and function of an epitope-tagged p7 viroporin in hepatitis C virus-producing cells. J. Virol. 87:1664–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for infectious virus production. J. Virol. 81:8374–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cocquerel L, Op DB, Lambot M, Roussel J, Delgrange D, Pillez A, Wychowski C, Penin F, Dubuisson J. 2002. Topological changes in the transmembrane domains of hepatitis C virus envelope glycoproteins. EMBO J. 21:2893–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.StGelais C, Foster TL, Verow M, Atkins E, Fishwick CW, Rowlands D, Harris M, Griffin S. 2009. Determinants of hepatitis C virus p7 ion channel function and drug sensitivity identified in vitro. J. Virol. 83:7970–7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tedbury P, Welbourn S, Pause A, King B, Griffin S, Harris M. 2011. The subcellular localization of the hepatitis C virus non-structural protein NS2 is regulated by an ion channel-independent function of the p7 protein. J. Gen. Virol. 92:819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Soest H, van der Schaar PJ, Koek GH, de Vries RA, van Ooteghem NA, van Hoek B, Drenth JP, Vrolijk JM, Lieverse RJ, Houben P, van der Sluys Veer A, Siersema PD, Schipper ME, van Erpecum KJ, Boland GJ. 2010. No beneficial effects of amantadine in treatment of chronic hepatitis C patients. Dig. Liver Dis. 42:496–502. [DOI] [PubMed] [Google Scholar]
- 48.von Wagner M, Hofmann WP, Teuber G, Berg T, Goeser T, Spengler U, Hinrichsen H, Weidenbach H, Gerken G, Manns M, Buggisch P, Herrmann E, Zeuzem S. 2008. Placebo-controlled trial of 400 mg amantadine combined with peginterferon alfa-2a and ribavirin for 48 weeks in chronic hepatitis C virus-1 infection. Hepatology 48:1404–1411. [DOI] [PubMed] [Google Scholar]
- 49.Drummer HE, Maerz A, Poumbourios P. 2003. Cell surface expression of functional hepatitis C virus E1 and E2 glycoproteins. FEBS Lett. 546:385–390. [DOI] [PubMed] [Google Scholar]
- 50.Kien F, Abraham JD, Schuster C, Kieny MP. 2003. Analysis of the subcellular localization of hepatitis C virus E2 glycoprotein in live cells using EGFP fusion proteins. J. Gen. Virol. 84:561–566. [DOI] [PubMed] [Google Scholar]
- 51.Lucas M, Tsitoura E, Montoya M, Laliotou B, Aslanoglou E, Kouvatsis V, Entwisle C, Miller J, Klenerman P, Hadziyannis A, Hadziyannis S, Borrow P, Mavromara P. 2003. Characterization of secreted and intracellular forms of a truncated hepatitis C virus E2 protein expressed by a recombinant herpes simplex virus. J. Gen. Virol. 84:545–554. [DOI] [PubMed] [Google Scholar]