Abstract
As we are confronted with an increasing number of emerging and reemerging viral pathogens, the identification of novel pathogen-specific and broad-spectrum antivirals has become a major developmental objective. Targeting of host factors required for virus replication presents a tangible approach toward obtaining novel hits with a broadened indication range. However, the identification of developable host-directed antiviral candidates remains challenging. We describe a novel screening protocol that interrogates the myxovirus host-pathogen interactome for broad-spectrum drug candidates and simultaneously probes for conventional, pathogen-directed hits. With resource efficiency and pan-myxovirus activity as the central developmental parameters, we explored coscreening against two distinct, independently traceable myxoviruses in a single-well setting. Having identified a pair of unrelated pathogenic myxoviruses (influenza A virus and measles virus) with comparable replication kinetics, we observed unimpaired coreplication of both viruses, generated suitable firefly and Renilla luciferase reporter constructs, respectively, and validated the protocol for up to a 384-well plate format. Combined with an independent counterscreen using a recombinant respiratory syncytial virus luciferase reporter, implementation of the protocol identified candidates with a broadened antimyxovirus profile, in addition to pathogen-specific hits. Mechanistic characterization revealed a newly discovered broad-spectrum lead that does not block viral entry but stimulates effector pathways of the innate cellular antiviral response. In summary, we provide proof of concept for the efficient discovery of broad-spectrum myxovirus inhibitors in parallel to para- and orthomyxovirus-specific hit candidates in a single screening campaign. The newly identified compound provides a basis for the development of a novel broad-spectrum small-molecule antiviral class.
INTRODUCTION
In recent decades, small-molecule therapeutics have revolutionized the treatment of a variety of viral infections. Despite this success, the reach of licensed antivirals is frequently limited by a single pathogen indication profile and preexisting or rapidly emerging viral resistance. Furthermore, the significant resources required for clinical drug development prohibit attempts to generate individual antivirals against all clinically relevant viral pathogens or to flexibly respond to newly emerging virological threats. New antiviral leads and novel discovery strategies are therefore needed to expand the portfolio of treatable viral diseases within the realms of presently available manufacturing technologies.
Therapeutic targeting of host factors required for virus replication has emerged as a novel concept of antiviral therapy that has high promise to advance beyond some of these limitations (1–3). Different viruses, in particular those representing related viral families, frequently rely on an overlapping set of host cell factors and pathways for their replication (1, 2). As substantiated by recent evidence (4, 5), many host requirements are conserved among related viruses, increasing the possibility for broad-spectrum antiviral activity. Likewise, the host-directed approach may contribute to reducing the frequency of viral escape from inhibition (6–8), since individual viral mutations are less likely to compensate for functional loss of a host factor or pathway required for viral replication.
Recognizing that these advantages will be offset by a heightened risk of inducing drug-related side effects, viral pathogens associated predominantly with acute disease appear particularly suitable for this therapeutic approach, because treatment time, and hence host exposure to the drug, can be kept limited. Myxoviruses such as influenza viruses, in the Orthomyxoviridae family, and respiratory syncytial virus (RSV), human parainfluenza viruses (HPIVs), mumps virus (MuV), and measles virus (MeV), in the Paramyxoviridae family, are collectively responsible for major human morbidity and mortality due to acute viral respiratory disease (9–14).
Influenza virus in particular remains the leading cause of death from respiratory disease in the United States, despite the existence of vaccine prophylaxis. The licensed influenza virus neuraminidase inhibitors zanamivir and oseltamivir carboxylate are beneficial when treatment is initiated early, but this is increasingly offset by viral resistance (15–17). Despite extensive research, no vaccines are currently available for paramyxoviruses such as RSV and the HPIVs, and declining mumps vaccine uptake in several developed countries has, in conjunction with primary and secondary vaccine failures, resulted in a recent resurgence of mumps (14). Ribavirin is approved for RSV treatment, but its utility is limited due to efficacy and toxicity issues (18). RSV prophylaxis using antibody therapies (19, 20) is reserved for high-risk pediatric patients. Considering their clinical significance, unmet medical need, and predominant association with acute disease, myxovirus family members are viable targets for novel pathogen- and host-directed antiviral campaigns.
Discovery paths toward pathogen-directed drug candidates are well established, but diverse strategies are currently used to identify druggable host targets. For influenza virus in particular, several target-driven approaches have recently been employed, originating from either knowledge-based host target selection (21–23) or systemwide genetic screens for host factors that are involved in pathogen replication (24–26). Chosen targets can then be pursued through narrow drug screens or, if available, use of existing inhibitors. Repurposing of the MEK kinase inhibitor U0126, blocking the Raf/MEK/ERK cascade (21, 27), and the CDC-like kinase 1 inhibitor TG003 (24) for influenza virus inhibition serves as a case in point. While these examples hold some promise, the limited pool of attractive knowledge-based druggable targets, the low reproducibility between comparable RNA interference (RNAi) screens (28), misjudgment of the druggability of target candidates (29), and the challenges associated with bioinformatics-driven triaging of systemwide gene data sets based on differently curated pathway databases (8) are major obstacles in the path toward developing applicable host-directed therapeutics.
In search of an alternative, compound-driven approach, we propose a resource-efficient drug discovery protocol that allows the interrogation of the full host-pathogen interactome for druggable host targets with broad-spectrum antiviral effects in parallel to the discovery of conventional, pathogen-directed hits. In this study, we examined the hypothesis that the anticipated broadened pathogen indication spectrum of host-directed antivirals itself can be employed as a viable selector for host-directed hits. Having identified representatives of the Ortho- and Paramyxoviridae with compatible replication kinetics, we examined independent virus replication after coinfection of cells with both viruses in a single-well setting. Usage of independently quantifiable luciferase reporters set the stage for a high-throughput screening (HTS) protocol design that affords the identification of paramyxovirus-specific, orthomyxovirus-specific, and broadly myxovirus-specific, likely host-directed, compounds in a single assay (visualized conceptually in Fig. 1A). Implementation of the protocol against a 10,000-entry diversity set identified, among others, a novel chemical class of broad-spectrum myxovirus inhibitors.
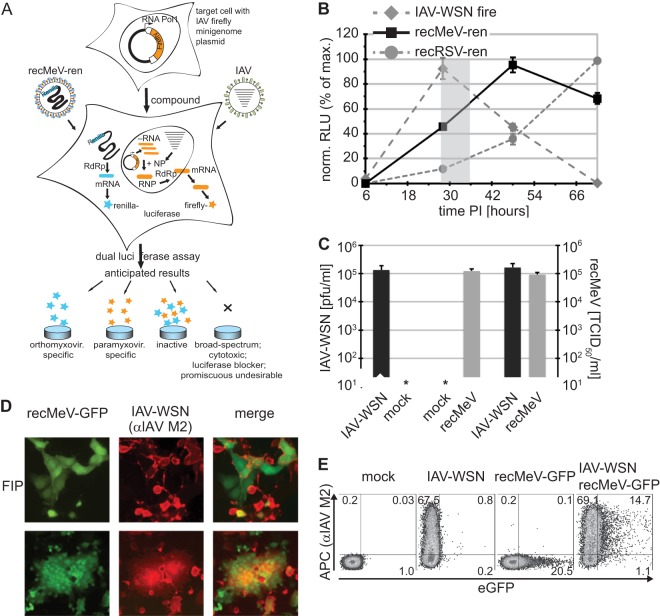
Fig 1.
Coinfection assay to identify host-directed pan-myxovirus inhibitors. (A) Schematic of the dual-target antiviral screen. Anticipated outcomes are specified. (B) Expression profiles of the different luciferase reporter constructs. Vero cells were independently infected with recMeV-ren and recRSV-ren (MOI = 0.2) or transfected with the IAV-firefly luciferase reporter and superinfected with IAV-WSN (MOI = 0.1). At the indicated times postinfection (PI), relative luciferase units (RLU) were determined. The shaded area shows the experimental window (28 to 34 hpi). Values were normalized to peak activities of the respective data series and represent means ± SD for three experiments. (C) Progeny titers of released (IAV-WSN) or cell-associated (recMeV) particles after infection of Vero cells singly or in combination. Values represent means and SD for three experiments. (D) Microscopic examination of Vero cells coinfected with recMeV-GFP and IAV-WSN. Where indicated, cells were incubated in the presence of 100 μM fusion inhibitory peptide (FIP). Recording was done at a magnification of ×200. (E) Cytometric analysis of doubly infected cells as described for panel B, with cells incubated in the presence of 100 μM FIP until harvest. Cells were immunostained with specific anti-IAV M2 antibodies (with APC conjugate), and APC fluorescence and EGFP autofluorescence were determined. Numbers show % distributions of singly and doubly fluorescent cells.
MATERIALS AND METHODS
Cells, viruses, and cloning.
All cell lines were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 7.5% fetal bovine serum. Vero (African green monkey kidney epithelial) cells (ATCC CCL-81) stably expressing human signaling lymphocytic activation molecule (Vero-SLAM cells) (30) and baby hamster kidney (BHK-21) cells stably expressing T7 polymerase (BSR-T7/5 [BHK-T7] cells) (31) were incubated at every third passage in the presence of 500 μg/ml G-418 (Geneticin). Human peripheral blood mononuclear cells (PBMCs) (obtained under Emory University Institutional Review Board approval IRB00045690) were prepared and stimulated as previously described (4). Lipofectamine 2000 (Invitrogen) was used for cell transfections. The QuikChange protocol (Stratagene) was used for all standard site-directed mutagenesis assays. Virus strains used in this study were recombinant MeV-Edmonston (recMeV) (32), MuV strain F, recombinant RSV A2 (recRSV) (33), and influenza A virus strains IAV/New York/55/2004 (H3N2) (IAV-New York), IAV/Aichi/2/1968 (H3N2) (IAV-Aichi), IAV/Mexico/INDRE4489/2009 (H1N1) (IAV-Mexico), IAV/WSN/1933 (H1N1) (IAV-WSN), IAV/Brisbane/59/2007 (H1N1) (IAV-Brisbane), IAV/Pennsylvania/08/2008 (H1N1) (IAV-Pennsylvania), and IAV/Texas/15/2009 (H1N1) (IAV-Texas). MeV stocks were grown and titrated by 50% tissue culture infective dose (TCID50) titration on Vero-SLAM cells. MuV was grown and plaque assay titrated on Vero cells. RSV was grown and immuno-plaque assay titrated on HEp-2 cells (ATCC HB-8065), as described previously (33). IAV strains were grown and plaque assay titrated on Madin-Darby canine kidney (MDCK) cells or grown on MDCK cells and titrated by TaqMan real-time PCR-based quantification of progeny genome copy numbers, as described previously (4). recMeV-ren and recRSV-ren stocks were subjected to ultracentrifugation through a 20%-60% one-step sucrose gradient (90 min, 100,000 × g, 4°C), with recovery of viral particles concentrated at the gradient interphase to reduce contamination with free luciferase protein synthesized during virus amplification.
Compounds.
All compounds were dissolved in dimethyl sulfoxide (DMSO) and stored at −80°C. The previously characterized pan-myxovirus inhibitor JMN3-003 (4), MeV RNA-dependent RNA polymerase (RdRp) inhibitor AS-136A (34), and MeV entry inhibitor AS-48 (35) were synthesized in-house, and their purity was confirmed to be >95% by liquid chromatography-mass spectrometry (LC-MS) and nuclear magnetic resonance (NMR) analysis. The screening library (ChemDiv) was designed to cover a broad chemical space within the boundaries of drug-like physical-chemical properties, a molecular weight range of 120 to 500, and best adherence to the Lipinski rule of 5 (36). Individual hit candidates were sourced from ChemDiv, MolPort, or Ambienter.
Generation of luciferase reporter systems.
The basis for the generation of recMeV-ren was a plasmid harboring a complete cDNA copy of the recMeV-eGFP genome (37), which contains the enhanced green fluorescent protein (EGFP) open reading frame in the pre-MeV N position. EGFP and Renilla luciferase open reading frames were exchanged using standard cloning techniques, and viral recombinants were recovered after sequence confirmation, as described previously (38). The generation and recovery of the equivalently designed recRSV-ren recombinant were described previously (33). For construction of an IAV-firefly luciferase minireplicon reporter plasmid under the control of the RNA polymerase I (Pol I) promoter, the firefly luciferase gene was amplified with appropriate primers and transferred into the pHH21 vector, restoring the noncoding flanking regions of IAV gene segment 5 as specified previously (39), with the exception of an A-to-G exchange at position 8 in the 5′-noncoding region.
Virus-driven luciferase reporter assays.
Luciferase enzymatic activity was measured to quantify reporter protein expression. Unless otherwise specified, 293T cells were transfected with 1.0 μg of IAV-firefly luciferase minigenome reporter plasmid/105 cells and then cryopreserved at 28 h posttransfection. Thawed cells were seeded at a density of 3 × 104 cells/well in a 96-well plate and infected with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin-activated IAV strains (multiplicity of infection [MOI] = 0.1 PFU/cell), recMeV-ren (MOI = 0.2 TCID50/cell), and/or recRSV-ren (MOI = 0.2 PFU/cell) after a 20-hour incubation, unless stated otherwise for individual experiments. Twenty-eight hours after infection with IAV strains or recMeV-ren, Bright-Glo, Renilla-Glo, or Dual-Glo substrate (all from Promega) was added as specified in the manufacturer's instructions, and bioluminescence intensities were determined using an Envision Multilabel microplate reader (PerkinElmer). For experiments involving recRSV-ren, HEp-2 cells were seeded at a density of 1.5 × 104 cells/well and bioluminescence was assessed at 40 h postinfection (hpi). Where specified, the previously characterized pan-myxovirus inhibitor JMN3-003 (4), MeV RdRp inhibitor AS-136A (34), or ERK2 inhibitor 5-iodotubercidin (40) was added as a control.
Fluorescence microscopy.
Vero-SLAM cells seeded in multiwell slides were infected with recMeV-eGFP (MOI = 0.2 TCID50/cell) and IAV-WSN (MOI = 0.1 PFU/cell), followed by a 40-hour incubation. Where indicated, 100 μM fusion inhibitory peptide (FIP) was added to the cultures at 1 h postinfection. Slides with 4% paraformaldehyde-fixed cells were blocked with bovine serum albumin (BSA), washed, and stained with specific anti-IAV M2 protein antibodies (Thermo Scientific) and allophycocyanin (APC)-labeled anti-mouse secondary antibodies (Jackson). Images were taken on a Nikon Diaphot 200 fluorescence microscope at a magnification of ×200.
Flow cytometry.
Vero cells infected with recMeV-eGFP (MOI = 0.8 TCID50/cell) and IAV-WSN (MOI = 0.1 PFU/cell) were incubated in the presence of 100 μM FIP for 40 h, stained with anti-IAV M2 protein antibodies and APC-conjugated secondary antibodies, fixed, and subjected to cytometric analysis in a FACSCanto II instrument as previously described (38).
HTS.
Cryopreserved cells transfected with the IAV-firefly luciferase minigenome plasmid were seeded as described above in white 96-well plates or, at a density of 104 cells/well, in 384-well plates, followed by a 20-hour incubation. Test articles dissolved in DMSO were added at a 5 μM final concentration (final DMSO concentration, <0.2%). As a control, the pan-myxovirus inhibitor JMN3-003 (final concentration, 1 μM) and vehicle (DMSO)-only wells were added to each plate in four (96-well plate format) or eight (384-well plate format) replicates each. Cells were then infected with a mixture of TPCK-trypsin-activated IAV-Texas (MOI = 0.1 PFU/cell) and recMeV-ren (MOI = 0.2 TCID50/cell). In the time window of 28 to 32 h postinfection, Dual-Glo luciferase substrate was added, and firefly and Renilla luciferase activities were quantified in an Envision Multilabel or Synergy H1 (BioTek) microplate reader.
HTS data analysis and IP search.
Complete plate reader raw data sets were automatically reformatted into a three-column layout by use of an in-house program, followed by import into the cellHTS2 application package (41, 42). For analysis according to the plate median method, each value was normalized to the median value for all compound wells, and normalized values were scaled to the median absolute deviation of the plate. Stronger inhibition (a reduction in signal) is represented by larger (positive) Z scores. For data analysis of confirmatory screens after cherry picking of hits, the normalized percent inhibition (NPI) method was applied, and relative values were calculated by subtracting each compound value from the average for the plate vehicle controls, followed by dividing the results by the difference between the means for the plate vehicle and JMN3-003 controls. The SciFinder database package (American Chemical Society) was used to query chemical databases with hit candidate structures to evaluate known bioactivities of analogs, commercial availability, and free intellectual property (IP) space. Z′ values were calculated based on the following formula: Z′ = 1 − [(3 SDC + 3 SDB)/(meanC − meanB)], where SD is the standard deviation, C is the control, and B is the background (43). The coefficient of variation (CV) was calculated as follows: CV = SDC/meanC.
Assessment of compound cytotoxicity.
The CytoTox 96 nonradioactive cytotoxicity assay (Promega) was used to quantify compound toxicity. In 96-well plates, cells were exposed to candidates for 28 hours at 2-fold the screening concentration (10 μM). Substrate was then added, and color development was recorded at 490 nm (specific value) and 650 nm (reference value) in a Synergy H1 microplate reader. Values were normalized to vehicle controls according to % toxicity as follows: % toxicity = 100 − {[(specific value − reference value)/(vehicle value − reference value)] × 100}. To calculate 50% cytotoxic concentrations (CC50s), the compound was added in a 3-fold serial dilution range from 30 to 0.1 μM, and mean values for three replicates were subjected to three-parameter nonlinear regression fitting.
Dose-response curves and EC50 calculation.
Cells were infected with TPCK-trypsin-activated IAV (MOI = 0.002 PFU/cell) in the presence of 3-fold serial dilutions of compound (the highest concentration assessed was 10 μM) or vehicle. At 1 h postinfection, virus inoculum was removed and cells were incubated in the presence of compound and 3 μg/ml TPCK-trypsin for 40 to 44 h. Progeny virions in culture supernatants were quantified as described above. For all paramyxovirus inhibition curves, infected cells (MuV MOI = 0.1 PFU/cell, recMeV MOI = 0.4 TCID50/cell, and recRSV MOI = 0.05 PFU/cell) were incubated in the presence of serial dilutions of compound as described above, for 40 (recMeV) to 72 (MuV and recRSV) hours, followed by titration of cell-associated progeny particles. Fifty percent effective concentrations (EC50s) were calculated based on four-parameter variable-slope nonlinear regression fitting of mean values for three experiments.
Minireplicon reporter assay.
293T cells were transfected with plasmid DNA encoding the IAV (0.5 μg)- or MeV (1 μg) (44)-luciferase minigenome reporter and plasmids encoding the RdRp components MeV-L (1.1 μg), MeV-N (0.4 μg), and MeV-P (0.3 μg), for MeV replicon assays, or IAV-NP, -PA, -PB1, and -PB2 (0.5 μg each), for IAV replicon assays. In the case of MeV replicons, cells were infected with modified vaccinia virus Ankara expressing T7 polymerase (MVA-T7) (45) at 16 h pretransfection. Compound 09167 was added at 4 h posttransfection, and luciferase reporter activities were determined using Bright-Glo substrate as described above.
Fusion-from-without cell-to-cell fusion assay.
A dual split-protein cell content mixing assay was employed to quantify MeV envelope glycoprotein-mediated membrane fusion in the presence of compound. NP2-DSP1–7 and NP2-DSP8–11 cells (46), stably transfected with EGFP-Renilla luciferase dual split fusion proteins DSP1–7 and DSP8–11, respectively, were coseeded in black 96-well microtiter plates, preloaded with EnduRen luciferase substrate (Promega) at 1 h preinfection, and then spin inoculated with recMeV (1,000 × g, 30 min, 4°C; MOI = 10 TCID50/ml). Plates were transferred to 37°C, and luciferase activity was recorded in an Envision Multilabel microplate reader (PerkinElmer) at the specified time points. As a control, the MeV entry inhibitor AS-48 was added to a 50 μM final concentration.
Time-of-addition variation (TOAV) assays.
293T cells were incubated in the presence of compound 09167 at a final concentration of 1.0 or 0.25 μM at 37°C for up to 6 h preinfection, followed by infection with recMeV (MOI = 0.8 TCID50/ml) in the presence of equal compound concentrations. Where indicated, the compound was added to infected cells at the specified time points postinfection. Cell-associated progeny particles were titrated at 24 hpi. Reference samples received volume equivalents of vehicle (DMSO) only.
Quantitation of cellular mRNA levels.
293T cells (9 × 105) were incubated in the presence of compound 09167 (final concentration, 1.0 μM) or the volume equivalent of vehicle (DMSO) for 20 h at 37°C, followed by preparation of total RNA by use of a QIAcube automated extractor (Qiagen) and an RNeasy minikit (Qiagen) as described above. Quantitative TaqMan reverse transcription-PCR (RT-PCR) was performed using TaqMan Fast master mix (Applied Biosystems) combined with proprietary primer and probe sets specifically detecting spliced mRNAs, but not genomic DNA, for IL28A, IFNB1, IL3RA, IRF3, IRGM, ISG15, MDA5, RIG-I, and IFIT1. To calculate ΔΔCT values, threshold cycle (CT) values obtained for each sample were standardized to expression levels of the 60S ribosomal protein L30 (RPL30) as a reference, and then ΔCT values of compound 09167-treated samples were normalized to the vehicle controls. Final quantification was based on three independent experiments, each conducted in duplicate.
Immunoblotting.
Cells (approximately 1 × 106 per treatment condition) were lysed in RIPA buffer (1% sodium deoxycholate, 1% NP-40, 150 mM NaCl, 50 mM Tris-Cl, pH 7.2, 10 mM EDTA, 50 mM NaF, 0.05% SDS, protease inhibitors [Roche], 1 mM phenylmethylsulfonyl fluoride). Cleared lysates (20,000 × g, 30 min, 4°C) were mixed with urea buffer (200 mM Tris, pH 6.8, 8 M urea, 5% SDS, 0.1 mM EDTA, 0.03% bromophenol blue, 1.5% dithiothreitol) for 30 min at 50°C, fractionated by SDS-PAGE, and blotted onto polyvinylidene difluoride (PVDF) membranes. Immunoblots were decorated with anti-RIG-I (Cell Signaling), anti-IFIT1 (Pierce), and anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (Calbiochem) monoclonal antibodies and developed using a species-specific IgG light chain conjugate and a ChemiDoc XRS digital imaging system (Bio-Rad).
RESULTS
Productive coinfection of cells in a high-throughput drug screen mandates the following: (i) the replication profiles of the selected myxovirus representatives must be compatible with each other to allow synchronized infection and analysis, and (ii) infection and protein expression from either viral genome must be unaffected by the presence of the other virus in the same cell population. Members of both the orthomyxovirus (47) and paramyxovirus (48, 49) families employ different strategies to block the cellular antiviral response, including the suppression of host cell protein expression in infected cells (47, 50). However, neither myxovirus family induces rapid host cell lysis or apoptosis, and genome transcription and replication of the Paramyxoviridae occur in the cytosol, while the orthomyxoviruses adhere to nuclear transcription and replication of their genetic information. We therefore hypothesized that myxovirus family members may be suitable for productive coinfection of cells.
A myxovirus reporter pair with compatible expression profiles.
In search of a clinically relevant myxovirus pair meeting the above requirements, we focused on RSV, MeV, and IAV, and we first generated reporter constructs that allowed independent quantification of para- and orthomyxovirus replication. In the case of RSV and MeV, recombinant reporter viruses were generated by inserting an additional transcription unit encoding Renilla luciferase in the primary position into cDNA copies of the viral genomes, followed by recovery of the corresponding viral recombinants, recRSV-ren (33) and recMeV-ren, respectively. For IAV, we generated a minigenome reporter plasmid on the basis of gene segment 5 (NP) through insertion of a firefly luciferase transcription unit. Expression of the resulting IAV-firefly luciferase replicon reporter is driven through superinfection of transfected cells with IAV, which provides the required viral NP and polymerase proteins.
When we compared relative luciferase activity profiles after individual infections of cells with these reporter systems, we found a broad overlap of MeV- and IAV-driven reporter activity peaks at approximately 28 to 36 h postinfection. In contrast, substantial luciferase activity in cells infected with recRSV-ren emerged only after approximately 50 h postinfection (Fig. 1B). The highest paramyxovirus MOI assessed in this assay was 0.2 infectious unit/cell, since limited virus titers achievable in stock preparations restrict the maximal MOI that can be realized in 96- and 384-well plate formats. Based on these expression profiles, we selected recMeV-ren as the better-suited paramyxovirus representative for coinfection with IAV, and we chose a harvesting time of 28 to 32 hpi as the target window for subsequent experiments.
Myxovirus replication after coinfection.
To assess the level of viral replication after coinfection, we infected cells with recMeV and IAV-WSN, either individually or in combination, and quantified yields of progeny virus. The resulting viral titers were essentially identical, regardless of whether they originated from individual or coinfections (Fig. 1C). Microscopic examination of cells infected with both viruses revealed a mixture of individually and doubly infected cells, provided that MeV glycoprotein-induced syncytium formation was chemically suppressed (Fig. 1D, top panels). Under standard conditions, however, the high cell-to-cell fusion activity of MeV resulted in the rapid formation of large, multinucleated syncytia harboring both viruses (Fig. 1D, bottom panels). Quantitative analysis of doubly infected cells by flow cytometry confirmed that the majority of cells expressing MeV proteins also stained positive for IAV-WSN antigen (Fig. 1E). Taken together, these findings indicate unrestricted coreplication of both IAV and MeV in a single-well setting.
Optimization of coinfection conditions for HTS.
To prepare the single-well coinfection–luciferase reporter system for automated screening, we interrogated the assay in a 96-well plate format for host cell type, effect of MOI on reporter expression after coinfection, and IAV strain used. When a panel of human cell lines were infected individually with either virus, we found that all lines supported efficient recMeV-ren replication, returning high luciferase activity levels (Fig. 2A). However, IAV-driven IAV-firefly luciferase replicon expression was highest in 293T cells (Fig. 2A) and remained stable over a spectrum of 0.5 to 1.5 μg plasmid DNA/105 cells transfected. Using this robust 293T cell host system and 1.0 μg of replicon DNA/105 cells, we explored the effects of coinfection with increasing amounts of recMeV-ren particles on firefly and Renilla luciferase activities. Over a recMeV-ren MOI range of 0.05 to 0.2 infectious unit/cell, activity levels of both luciferases remained largely unchanged compared to those found in individually infected controls (Fig. 2B). Since slightly higher Renilla luciferase activity levels were observed after infection of cells with 0.2 infectious unit of recMeV-ren/cell, this MOI was chosen for all subsequent screens.
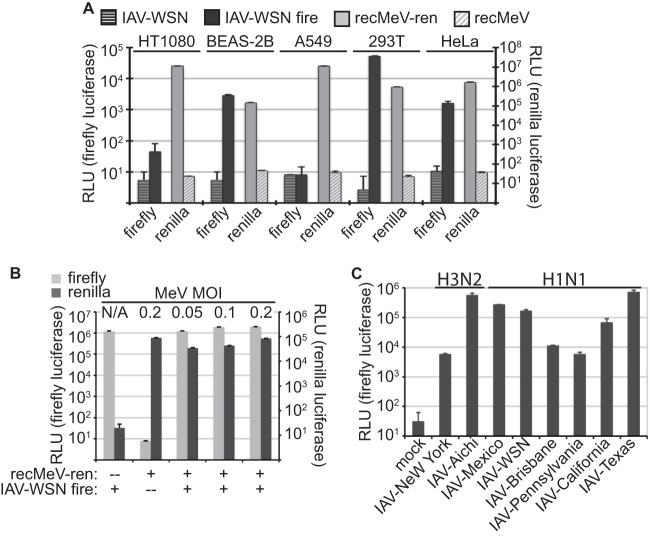
Fig 2.
Assay optimization. (A) 293T cells returned the highest activity levels for both reporter constructs. Cells were infected individually with recMeV-ren or transfected with the IAV minigenome reporter and superinfected with IAV-WSN. RLU values represent means and SD for four experiments. (B) Efficient expression of both luciferase reporters in coinfected 293T cells. Cells transfected with the IAV-firefly luciferase plasmid were superinfected with IAV-WSN and increasing amounts of recMeV-ren, ranging from 0.05 to 0.2 infectious unit/cell. Relative activities of either reporter were determined at 30 hpi. Controls lacked either recMeV-ren or IAV-WSN. Values represent means and SD for three experiments. (C) The highest IAV-firefly luciferase reporter expression levels were achieved with swine-origin IAV-Texas. Cells transfected with IAV-firefly luciferase minigenome plasmid DNA were infected with different IAV strains at an MOI of 0.1 infectious unit/cell. Luciferase activities were determined at 30 hpi. Values represent means and SD for three experiments.
In search of the most competent driver of the IAV-firefly luciferase replicon, we scanned a panel of different IAV strains representing H1N1 and H3N2 genotypes. Under the experimental conditions established above, infection of minireplicon-transfected cells with swine-origin IFA/Texas/2009 (H1N1) (IAV-Texas) resulted in the highest luciferase activities overall, which were approximately 4-fold higher than those observed for IAV-WSN (Fig. 2C). We therefore selected IAV-Texas as the orthomyxovirus representative of choice for subsequent validation and screening campaigns.
Assay validation and miniaturization.
Having established the basic infection parameters, we first tested the robustness of the protocol in a 96-well plate format, and the assay was then miniaturized to a 384-well scale. For positive controls with distinct antiviral profiles, we chose the previously developed small-molecule compounds AS-136A, an MeV-specific RdRp inhibitor (34, 51); JMN3-003, a broadly acting pan-myxovirus inhibitor (4); and 5-iodotubercidin, a potent ERK2 inhibitor (40) that we found to block IAV, but not MeV, replication. Using the coinfection protocol, the effect of each of these control compounds was assessed in dose-response assays in independent replicate plates. This approach yielded dose-dependent paramyxovirus-specific, orthomyxovirus-specific, and pan-myxovirus antiviral profiles with little plate-to-plate variation (Fig. 3A), confirming the capacity of the assay to reliably detect representatives of each desired inhibitor class in a single screen.
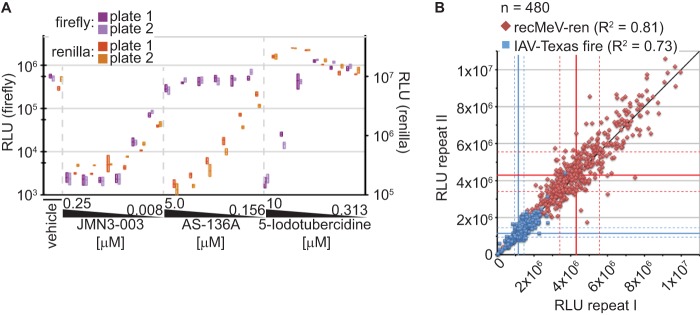
Fig 3.
Assay validation for high-throughput inhibitor screening. (A) Control inhibitors with distinct antimyxovirus profiles were reproducibly identified in single-well coinfection assays. IAV-firefly luciferase-transfected cells were infected with IAV-Texas and recMeV-ren in the presence of JMN3-003 (pan-myxovirus inhibitor), AS-136A (MeV RdRp inhibitor), 5-iodotubercidin (IAV inhibitor), or vehicle control (DMSO). Relative luciferase activities in the wells were determined at 30 hpi. Each concentration was assessed in three replicates; two independent plates were prepared and analyzed. Symbols show means (lines) and minimum and maximum values (floating bars). (B) Plate-to-plate variation assessment using a random test set of 480 compounds. Cells were treated at a final concentration of 5 μM and then coinfected, and luciferase activities were determined at 30 hpi. Symbols represent values for each compound obtained from two independent replicate sets. A direct linear correlation (black line), median RLU values for each reporter data set (solid blue and red lines), and 25th and 75th percentiles (dashed blue and red lines) are shown.
Using a random test set of 480 small-molecule compounds (six plates) with unknown antiviral activity, we next quantified plate-to-plate variability for each reporter under screening conditions. Plotting of relative luciferase activities obtained for each compound in replicate experiments for both target viruses resulted in linear correlations with R2 values of 0.73 and 0.81 (Fig. 3B). To quantitatively assess the suitability of the assay for automated screening, we selected the myxovirus inhibitor JMN3-003 as a pan-myxovirus positive control and calculated Z′ values (43), signal-to-background ratios, and coefficients of variation for the 96-well and, after miniaturization, 384-well plate sizes (Table 1). For both formats, the values were within the acceptable range for automated screening (43, 52).
Table 1.
Comparison of assay formats used in this study
| Plate format | Target virusa | Z′ valueb | S/B ratioc | % CV |
|---|---|---|---|---|
| 96 wells | recMeV-ren | 0.74 | 31 | 8.0 |
| IAV-firefly luciferase | 0.79 | 109 | 6.7 | |
| IAV-Texas | ||||
| 384 wells | recMeV-ren | 0.74 | 41 | 8.2 |
| IAV-firefly luciferase | 0.67 | 89 | 10.9 | |
| IAV-Texas |
Cryopreserved 293T cells transfected with IAV-firefly luciferase were plated and coinfected with recMeV-ren and IAV-Texas in the presence of the pan-myxovirus inhibitor JMN3-003 (final concentration, 1 μM) or an equivalent amount of vehicle (DMSO). Relative luciferase unit values were determined at 30 hpi.
Z factor (43). Statistical analyses are based on means for four independent experiments.
Ratio of signal to background.
HTS of a 10,000-entry diversity set.
To conceptually test the assay in HTS mode, we screened a 10,000-entry diversity set of drug-like small molecules following the filter strategy depicted in Fig. 4A. All compounds were tested at a concentration of 5 μM, and each plate contained vehicle controls and the pan-myxovirus reference inhibitor JMN3-003 in quadruplicate. Primary HTS data were normalized by plate to the control inhibitor, Z score scaled, and organized by increasing score values (Fig. 4B). The top-scoring candidates, with Z scores of ≥2 standard deviations above the mean for IAV-Texas (238 entries) or ≥2.5 standard deviations above the mean for MeV (246 entries), were cherry picked for further analysis. Of these, 124 showed broad antiviral activity against both myxovirus targets in the primary screen (Fig. 4C) and were thus considered pan-myxovirus inhibitor candidates.
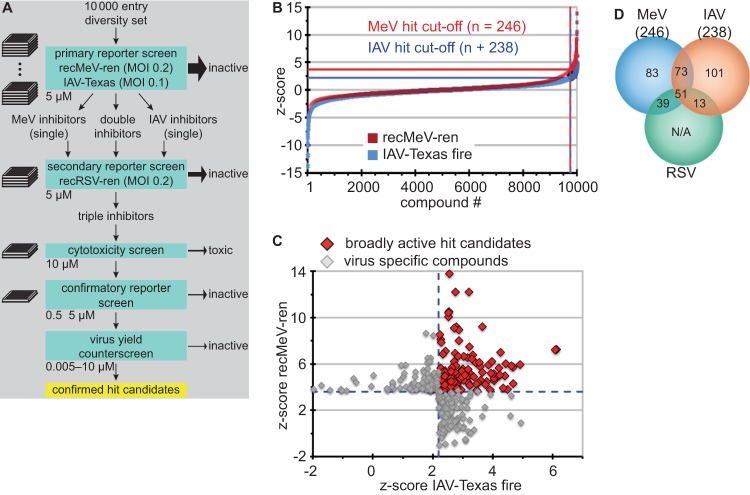
Fig 4.
Dual myxovirus screen of a 10,000-entry diversity set. (A) Schematic of primary, confirmatory, and counterscreens for hit identification. (B) Normalized, scaled, and ordered screening scores (Z scores) obtained for each target virus. Lines mark hit selection cutoffs; selected hit candidates are shown to the right of the dashed lines. (C) Selected hit candidates plotted by the Z score obtained for each target virus. Dashed blue lines mark the cutoffs for MeV (approximately 2.5 SD above the mean) and IAV-Texas (approximately 2 SD above the mean). Red symbols mark compounds with activity against both myxovirus targets. (D) Venn diagram summarizing the second-tier confirmatory screen of all 360 primary hit candidates shown in panel C against recRSV-ren (hit cutoff, 2 SD above the mean).
For second-tier hit filtration, we tested the primary candidates individually against recRSV-ren, which served as an independent confirmatory paramyxovirus target. Of 360 distinct compounds examined in this assay, 13 candidates exclusively blocked IAV-Texas and RSV, a 3-fold larger contingent (39 entries) inhibited MeV and RSV—and were thus considered pan-paramyxovirus inhibitor candidates—and 51 suppressed reporter expression by all three viral targets (Fig. 4D). Since the library was not pretested for cytotoxic compounds, the last group was anticipated to comprise cytotoxic compounds, promiscuous hits (53, 54), and pan-myxovirus inhibitor candidates.
Hit confirmation and counterscreening.
To distinguish between these alternatives and provide further insight into the inhibitory potential of individual hits, all 51 candidates that suppressed MeV, RSV, and IAV were subjected to quantitative cytotoxicity assessment at twice the screening concentration (final concentration, 10 μM) and to an independent, two-concentration (5 and 0.5 μM) third-tier confirmatory screen against MeV and IAV-Texas. For quantitative comparisons after cherry picking, we calculated toxicity and virus inhibition relative to those of vehicle-treated controls. Antiviral effects of the hit candidates with the highest Z scores against all targets were due to strong toxicity, defined as <75% of cellular metabolic activity remaining after 28 h of exposure (Fig. 5A). However, 15 candidates returned >85% inhibition against both MeV and IAV-Texas at 5 μM, and in the case of 4 candidates, this also extended to ≥50% inhibition of viral titers at 0.5 μM (Fig. 5A).
Fig 5.
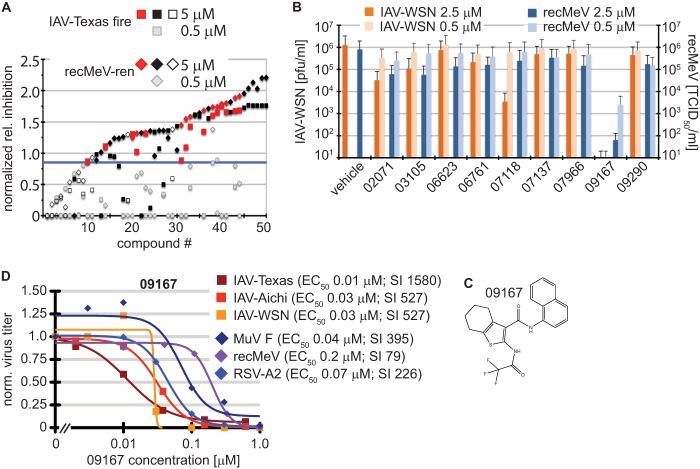
Identification of a nanomolar pan-myxovirus inhibitor class. (A) Two-concentration confirmatory screen of the selected 51 hit candidates. Values were normalized to those for plate controls and represent inhibition relative to that of JMN3-003. Compounds are ordered by increasing relative inhibition values. Black filled symbols mark cytotoxic compounds (reducing cell metabolic activity by >25%), open symbols represent compounds that were inactive against at least one of the target viruses at 5 μM, and red symbols highlight the remaining hit candidates. Gray symbols show the corresponding normalized scores obtained for each entry at 0.5 μM. The blue line marks the hit cutoff (0.85) relative to JMN3-003 for all candidates at 5.0 μM. (B) Virus yield-based counterscreen of nine candidates at 2.5 and 0.5 μM against IAV-WSN and recMeV. Values represent averages for two independent experiments, and error bars show data ranges. (C) Chemical scaffold of lead candidate 09167 {2,2,2-trifluoro-N-[3-(N-naphthylcarbamoyl)(4,5,6,7-tetrahydrobenzo [β]thiophen-2-yl)]acetamide}. (D) Virus yield-based dose-response assays. Virus titers were determined through plaque assay (IAV-WSN, MuV F, and RSV-A2), TaqMan genome copy number quantification (IAV-Texas and IAV-Aichi), or TCID50 titration (recMeV) and then normalized to vehicle (DMSO) controls. Data points represent means for three experiments. EC50s were calculated based on four-parameter variable-slope nonlinear regression models. Specificity indices (SI) represent CC50/EC50.
Visual inspection of the chemical scaffolds of all 15 candidates with confirmed bioactivity identified three compounds with undesirable properties, such as highly reactive substructures or other common features of promiscuous small-molecule screening hits (53, 54), and another five compounds represented analogs of only two distinct chemical scaffolds. Excluding these readily undesirable and structurally redundant candidates, we subjected the remaining nine compounds to counterscreening, assessing their ability to reduce yields of progeny recMeV and IAV-WSN viral particles at concentrations of 2.5 and 0.5 μM, respectively (Fig. 5B). Three candidates reduced titers of both target viruses by >90% at 2.5 μM, and one compound (09167) induced >99% inhibition at submicromolar concentrations.
The lead candidate, compound 09167 (Fig. 5C), was sourced for further characterization. Database queries with the chemical scaffold did not return close (≥90% similarity) chemical analogs with defined bioactivity, arguing against multitarget promiscuity. Virus yield-based dose-response curves furthermore confirmed nanomolar EC50s of the compound against a set of three different IAV strains (Fig. 5D). Potent antiviral activity was not restricted to orthomyxovirus family members but extended equally to a group of clinically relevant paramyxoviruses, including MeV, MuV, and RSV. Based on these results, we selected the 09167 hit for initial characterization of the mechanism of action (MOA).
MOA of first-generation lead 09167.
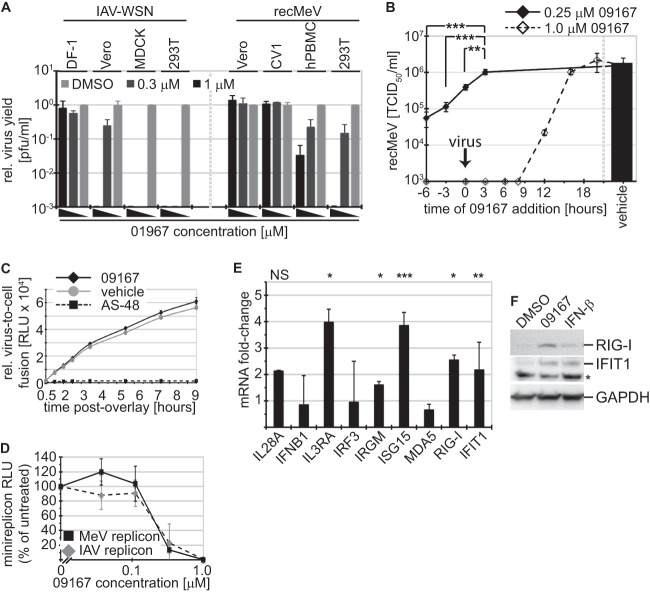
A host-directed antiviral mechanism frequently coincides with host cell species dependence of the inhibitory activity (4). When we examined the bioactivity of 09167 in a variety of cell lines of different species origins, we observed the most potent inhibition in human and canine cell lines and primary human PBMCs (Fig. 6A). IAV-WSN was more sensitive to inhibition than recMeV-Edm in both 293T and Vero cell lines in this assay. In contrast, antiviral activity was minimal or absent in cell lines of nonhuman primate and avian origins. These data exclude a direct virucidal effect of the compound and confirm a host-directed mechanism of action.
Fig 6.
Lead candidate 09167 primes the host antiviral response. (A) Host cell species dependence of 09167 antiviral activity. Avian DF-1, canine MDCK, primate Vero and CV1, and human 293T cells were infected with IAV-WSN or MeV in the presence of 1.0 and 0.33 μM 09167 or vehicle control. Progeny virus yields were normalized to titers in vehicle controls. (B) Priming of target cells with 09167 enhances antiviral activity. The compound was added to cells at 0.25 and 1.0 μM at the specified time points before or after infection with MeV, and progeny virus titers were determined at 24 hpi. Values represent means ± SD for three experiments (**, P <0.01; ***, P < 0.001). (C) Quantitative cell content mixing assay assessing MeV glycoprotein-induced virus-to-cell fusion kinetics, carried out in the presence of 1 μM 09167, vehicle (DMSO), or 75 μM fusion inhibitor AS-48 (control). At the indicated time points, reconstitution of double GFP-Renilla split-luciferase proteins, indicating cell content mixing, was determined. Values represent averages ± SD for five replicates/time point. (D) Plasmid-based IAV and MeV minigenome reporter assays to determine viral RdRp activity in the presence of increasing 09167 concentrations. Relative luciferase reporter activities were determined after 24 hours of exposure. Values represent means ± SD for at least three experiments. (E) Treatment of 293T cells with 09167 stimulates expression of several ISGs. Cells were exposed to 1.0 μM 09167 or vehicle (DMSO) for 20 h, followed by TaqMan RT-PCR quantitation of relative expression levels of a panel of genes associated with the host innate immune system. IL28A, interleukin-28; IFNB1, IFN-β; IL3RA, interleukin-3 receptor α; IRF3, interferon regulatory factor 3; IRGM, interferon-inducible protein 1; ISG15, interferon-induced 17-kDa protein; MDA5, melanoma differentiation-associated protein 5; RIG-I, retinoic acid-inducible gene 1; IFIT1, interferon-induced protein with tetratricopeptide repeats 1. Values represent averages and SD for three independent experiments; each sample was analyzed in duplicate. Symbols: *, P < 0.05; **, P < 0.01; ***, P < 0.001; NS, not significant. (F) Total lysates of cells treated as described for panel E or exposed to 50 U IFN-β were subjected to gel fractionation and immunoblotting using specific antibodies directed against RIG-I and IFIT1. As a control, blots were decorated with anti-GAPDH antibodies. *, nonspecific crossreaction.
To narrow the range of possible host-pathogen interactome targets of compound 09167, we examined the effect of time-of-addition variation (TOAV) on antiviral potency. MeV served as the viral reporter in these experiments, and the compound was administered at concentrations equivalent to 1.25- and 5-fold higher than the EC50 at distinct time points pre- or postinfection. Virus replication was completely suppressed over a wide addition time frame at the higher concentration, indicating a fast-acting host effect of the compound. Importantly, dosing at lower levels revealed significantly increased antiviral potency when cells were pretreated with the compound prior to infection (Fig. 6B), suggesting priming of a host cell antiviral stage. Supporting the TOAV profile, we found the kinetics of virus-to-cell fusion to be unimpaired by 09167 (Fig. 6C), but we observed a dose-dependent inhibition of the viral RNA-dependent RNA polymerase activity (Fig. 6D), which would be expected if the compound stimulates cellular antiviral defense pathways (55, 56).
To test this hypothesis, we determined whether 09167 triggers traditional RNA virus pattern recognition receptor (PRR) signaling pathways, such as RIG-I/MDA5 and Toll-like receptor 3 (TLR3), resulting in activation of the type I interferon (IFN) response and IFN-β secretion. TaqMan RT-PCR revealed that IFNB1 and IRF3 mRNA levels were unaffected by the compound (Fig. 6E), and enzyme-linked immunosorbent assay (ELISA)-based quantification of IFN-β in culture supernatants showed no compound 09167-induced increase in interferon release. In contrast, expression of several IFN-stimulated downstream effector genes (ISGs), including the ISG15, RIG-I, and IFIT1 genes, was significantly increased after treatment of cells with 09167 (Fig. 6E). Immunodetection of RIG-I and IFIT1 after exposure of cells to 09167 confirmed that changes in relative mRNA contents translate into increased effector protein steady-state levels. Taken together, these results advocate that screening lead 09167 represents a novel small-molecule activator of the host cell antiviral ISG system.
DISCUSSION
Emerging and reemerging viral pathogens mandate the development of novel therapeutic strategies. Broad-spectrum antivirals have become a major goal of drug discovery campaigns, but selecting druggable host targets and identifying viable leads remain challenging.
Recent genomewide RNA interference screens have expanded our insight into the host-pathogen interactome for several viruses, including influenza virus (24–26). Genetically identified host target candidates could be pursued through repurposing known drugs or can become the objective of target-based HTS activities. However, large-scale implementation of such a strategy with an antibacterial endpoint has yielded sobering results (29), demonstrating that the genomics-based deliberate selection of fruitful drug targets remains challenging. These experiences have resulted in the realization that “following the compound” rather than the target may in fact be the more rewarding path toward drug discovery (57).
Guided by these considerations, we describe an HTS protocol that supports identification of pathogen-specific and broad-spectrum myxovirus hit candidates through parallel interrogation of possible pathogen targets and the host-pathogen interactome. Based on the approximately 30-hour (to harvest) timeline of the protocol and the nature of the readout, we anticipate such screens to reveal predominantly inhibitors of virus attachment, entry, and/or polymerase activity, while late-stage blockers of particle assembly and egress are less likely to be discovered. Simultaneous screening against two viral targets representing related but distinct viral families does not guarantee per se that broad-spectrum hits will follow a host-directed activity profile. Broadened activity could, alternatively, ensue from interference with conserved pathogen structures; for instance, inhibition of viral glycoprotein-mediated virus entry through lectins (58) in a topical application or the use of lipid-active compounds suggested to interfere with membrane function of enveloped viruses (59) serve as cases in point. A mechanistic analysis of successful small-molecule antivirals (60) reveals, however, that the majority of pathogen-targeting compounds are highly virus specific, whereas broad-spectrum hits engage predominantly host factors required for virus replication or host pathogen control.
In primary HTS assays, broad-spectrum and pathogen-specific antiviral activities can be assessed in silico after distinct serial screens of a single library against individual viral targets or by simultaneous probing against both viruses in a single-well format. Provided that replication of either target virus is not affected by the presence of the other, a parallel screening campaign is substantially more effective in regard to equipment, reagents and supplies, and time, resulting in significant resource advantages when larger diversity sets are assessed. In addition, this approach adds confidence to the identification of pathogen-specific inhibitor candidates, since highly divergent screening scores of a candidate compound for either viral target will argue against common cell-based HTS liabilities such as general cytotoxicity, compound promiscuity, or technical issues. Independent of serial or parallel screening strategies, however, broad-spectrum hit candidates must be considered at risk of being enriched for undesirable compounds (61).
Although coinfection with ortho- and paramyxoviruses has been observed clinically (62), downregulation of host protein expression by pathogens of either family could prevent successful coreplication in a single-well format. For instance, the influenza virus NS1 protein has been demonstrated to block correct processing of cellular mRNAs (47), while the MeV N protein has been implicated in interference with host mRNA translation through interaction with the translation initiation factor eIF3-p40 (63). Moreover, the high cell-to-cell fusion activity associated with MeV infection (64) results in the rapid formation of very large syncytia that can comprise entire cell monolayers. One can therefore assume that both pathogens will rapidly be located in the same cellular environment after coinfection, even when originally added at lower multiplicities of infection. However, protein expression shutoff by MeV is inefficient (65), and cytosolic transcription and replication may shield paramyxovirus genomes from the nuclear functions of NS1. Our results demonstrating efficient IAV and MeV coreplication and protein expression are consistent with this view, and they identify ortho- and paramyxovirus family members as suitable target pairs for parallel drug screens.
Quantitative validation of the firefly and Renilla luciferase reporter-based screen returned the anticipated distinct hit profiles for positive-control compounds with defined pathogen-specific or broad antimyxovirus patterns with a high reproducibility. Application to a 10,000-entry test set demonstrated the general feasibility of the strategy for simultaneous identification of pathogen-specific and broad-spectrum hit candidates. We noted some bias toward doubly false-positive results after coinfection, but we found a nearly 2-fold higher count of MeV-IAV double inhibitor candidates than, for instance, MeV-RSV blockers after primary and confirmatory screening. The screen also confirmed that the pool of primary broad-spectrum candidates was enriched for cytotoxic, promiscuous, and/or assay-interfering compounds.
Triangular confirmatory screening of all MeV and IAV hit candidates against RSV provided an attractive avenue toward reducing the rate of assay false-positive results and potentially identifying pan-paramyxovirus blockers. Although they represent distinct paramyxovirus subfamilies, the phylogenetic proximity of RSV and MeV to each other is substantially closer than that of either to IAV. Indeed, our assay returned a 3-fold higher hit rate for MeV-RSV than IAV-RSV inhibitor candidates. While the molecular characterization of these compounds is pending, we consider it unlikely that this pattern reflects pathogen-directed inhibition of conserved paramyxovirus targets. Rather, we would expect members of different paramyxovirus subfamilies to rely on a more homogenous host factor pool than those for ortho- and paramyxovirus representatives. Follow-up testing in search of candidates with an antiparamyxovirus as opposed to pan-myxovirus profile will center on those compounds that combine the highest Z values against both MeV and RSV with the lowest scores against IAV as the point of entry.
Our test screen combined with virus titer-based counterscreens yielded a single pan-myxovirus inhibitor with nanomolar potency. This small hit number may well reflect the challenge of broad-spectrum inhibition and may be accentuated further by the stringent screening criteria applied. While a more relaxed screen may identify additional viable scaffolds, this potential gain must be offset against the likely discovery of undesirable promiscuous hits with essentially flat structure-activity relationships (SARs). Three lines of evidence support the hypothesis that the confirmed hit compound represents a novel class of agonists of the host innate immune response. First, compound 09167 shows host cell species dependence of the antiviral effect, supporting a host-directed mechanism of activity. Second, host cell preexposure substantially enhances the antiviral potency of the compound. This TOAV profile suggests priming of a host antiviral state as the basis for inhibitory activity. Lastly, quantitative analysis of ISG mRNA and protein levels demonstrated that the compound upregulates the expression of a subset of antiviral effector genes, including those for ISG15 and IFIT1, which recognize viral genomic and antigenomic RNAs bearing 5′-triphosphate groups (66, 67). Expression of multiple ISG products may be synergistically responsible for the potent overall antiviral effect of the compound. Current work is directed at the systematic characterization of the specific pathways triggered and the positive identification of the molecular target.
Despite the effective and well-documented stimulation of a broad antiviral state by type I IFN, small-molecule agonists of the innate immune system have not yet been exploited clinically for antiviral therapies. However, specific activation of innate immune signaling pathways for an antiviral endpoint has experienced a renaissance in recent drug screening campaigns (68–70). Several of these activities have identified fused heterocyclic compounds with DNA-binding (68) and/or DNA-intercalation activity (70, 71), which is inherently associated with high mutagenic potential (72), creating a developmental liability. Compound 09167 is structurally distinct from this set of DNA-active small molecules, but it features a thiophene substructure which is potentially reactive (61) and has appeared in low-potency hits (i.e., hits with EC50s of 13 to 300 μM) in previous screening campaigns (73, 74). This chemical scaffold therefore mandates careful examination. Based on consistent activity in our orthogonal counterscreening assays and the nanomolar, approximately 1,000-fold higher potency than that found for promiscuous thiophenes, 09167 shows all the features of a viable lead suitable for advanced synthetic hit-to-lead optimization.
In toto, we have demonstrated unrestricted coreplication of ortho- and paramyxovirus representatives after coinfection, which set the stage for a novel time- and resource-efficient HTS protocol that affords the identification of broad-spectrum myxovirus inhibitors in parallel to the discovery of conventional, pathogen-specific antivirals. This approach is readily transferable to nonmyxovirus pathogen-target pairs, provided that they share comparable replication kinetics capable of unimpaired coreplication. Application to a small-molecule test set emphasized the necessity for orthogonal counterscreening, in particular for the pursuit of broad-spectrum candidates, and identified a promising novel small-molecule inhibitor with nanomolar antimyxovirus activity.
ACKNOWLEDGMENTS
We thank M. Currier for software development, N. Kondo and Z. Matsuda for split-luciferase reporter plasmids, M. A. Brindley and R. Suter for help with computer graphics, and T. W. Moore, M. A. Brindley, and A. L. Hammond for discussions and comments on the manuscript.
This work was supported in part by Public Health Service grants AI087798 and AI095227 (to M.L.M.) and by grants AI071002, AI057157, and AI085328 from the NIH/NIAID (to R.K.P.).
Footnotes
Published ahead of print 7 August 2013
REFERENCES
- 1.Schwegmann A, Brombacher F. 2008. Host-directed drug targeting of factors hijacked by pathogens. Sci. Signal. 1:re8. [DOI] [PubMed] [Google Scholar]
- 2.Tan SL, Ganji G, Paeper B, Proll S, Katze MG. 2007. Systems biology and the host response to viral infection. Nat. Biotechnol. 25:1383–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller B, Krausslich HG. 2009. Antiviral strategies. Handb. Exp. Pharmacol. 2009:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krumm SA, Ndungu JM, Yoon JJ, Dochow M, Sun A, Natchus M, Snyder JP, Plemper RK. 2011. Potent host-directed small-molecule inhibitors of myxovirus RNA-dependent RNA-polymerases. PLoS One 6:e20069. 10.1371/journal.pone.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML. 2011. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 108:5777–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salerno D, Hasham MG, Marshall R, Garriga J, Tsygankov AY, Grana X. 2007. Direct inhibition of CDK9 blocks HIV-1 replication without preventing T-cell activation in primary human peripheral blood lymphocytes. Gene 405:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schang LM. 2006. First demonstration of the effectiveness of inhibitors of cellular protein kinases in antiviral therapy. Expert Rev. Anti Infect. Ther. 4:953–956. [DOI] [PubMed] [Google Scholar]
- 8.Prussia A, Thepchatri P, Snyder JP, Plemper RK. 2011. Systematic approaches towards the development of host-directed antiviral therapeutics. Int. J. Mol. Sci. 12:4027–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thompson WW, Comanor L, Shay DK. 2006. Epidemiology of seasonal influenza: use of surveillance data and statistical models to estimate the burden of disease. J. Infect. Dis. 194(Suppl 2):S82–S91. [DOI] [PubMed] [Google Scholar]
- 10.Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. 2004. Influenza-associated hospitalizations in the United States. JAMA 292:1333–1340. [DOI] [PubMed] [Google Scholar]
- 11.Johnson D. 2009. Croup. Clin. Evid. (Online) 2009:0321 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2907784/. [Google Scholar]
- 12.Bjornson CL, Johnson DW. 2008. Croup. Lancet 371:329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization. 2012. World Health Statistics 2012. WHO, Geneva, Switzerland: http://www.who.int/gho/publications/world_health_statistics/en/index.html. [Google Scholar]
- 14.MacDonald N, Hatchette T, Elkout L, Sarwal S. 2011. Mumps is back: why is mumps eradication not working? Adv. Exp. Med. Biol. 697:197–220. [DOI] [PubMed] [Google Scholar]
- 15.De Clercq E. 2006. Antiviral agents active against influenza A viruses. Nat. Rev. 5:1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiso M, Mitamura K, Sakai-Tagawa Y, Shiraishi K, Kawakami C, Kimura K, Hayden FG, Sugaya N, Kawaoka Y. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759–765. [DOI] [PubMed] [Google Scholar]
- 17.Sugaya N, Mitamura K, Yamazaki M, Tamura D, Ichikawa M, Kimura K, Kawakami C, Kiso M, Ito M, Hatakeyama S, Kawaoka Y. 2007. Lower clinical effectiveness of oseltamivir against influenza B contrasted with influenza A infection in children. Clin. Infect. Dis. 44:197–202. [DOI] [PubMed] [Google Scholar]
- 18.Anderson LJ, Parker RA, Strikas RL. 1990. Association between respiratory syncytial virus outbreaks and lower respiratory tract deaths of infants and young children. J. Infect. Dis. 161:640–646. [DOI] [PubMed] [Google Scholar]
- 19.Groothuis JR, Simoes EA, Levin MJ, Hall CB, Long CE, Rodriguez WJ, Arrobio J, Meissner HC, Fulton DR, Welliver RC, Tristram DA, Siber GR, Prince GA, Van Raden M, Hemming VG, Respiratory Syncytial Virus Immune Globulin Study Group. 1993. Prophylactic administration of respiratory syncytial virus immune globulin to high-risk infants and young children. N. Engl. J. Med. 329:1524–1530. [DOI] [PubMed] [Google Scholar]
- 20.Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, Dormitzer M, O'Grady J, Koenig S, Tamura JK, Woods R, Bansal G, Couchenour D, Tsao E, Hall WC, Young JF. 1997. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J. Infect. Dis. 176:1215–1224. [DOI] [PubMed] [Google Scholar]
- 21.Pleschka S, Wolff T, Ehrhardt C, Hobom G, Planz O, Rapp UR, Ludwig S. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3:301–305. [DOI] [PubMed] [Google Scholar]
- 22.Kujime K, Hashimoto S, Gon Y, Shimizu K, Horie T. 2000. p38 mitogen-activated protein kinase and c-jun-NH2-terminal kinase regulate RANTES production by influenza virus-infected human bronchial epithelial cells. J. Immunol. 164:3222–3228. [DOI] [PubMed] [Google Scholar]
- 23.Moss RB, Hansen C, Sanders RL, Hawley S, Li T, Steigbigel RT. 2012. A phase II study of DAS181, a novel host directed antiviral for the treatment of influenza infection. J. Infect. Dis. 206:1844–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, Maurer AP, Muller E, Wolff T, Rudel T, Meyer TF. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822. [DOI] [PubMed] [Google Scholar]
- 25.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ludwig S. 2009. Targeting cell signalling pathways to fight the flu: towards a paradigm change in anti-influenza therapy. J. Antimicrob. Chemother. 64:1–4. [DOI] [PubMed] [Google Scholar]
- 28.Barrows NJ, Le Sommer C, Garcia-Blanco MA, Pearson JL. 2010. Factors affecting reproducibility between genome-scale siRNA-based screens. J. Biomol. Screen. 15:735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. 6:29–40. [DOI] [PubMed] [Google Scholar]
- 30.Ono N, Tatsuo H, Hidaka Y, Aoki T, Minagawa H, Yanagi Y. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75:4399–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buchholz UJ, Finke S, Conzelmann KK. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter MA. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hotard AL, Shaikh FY, Lee S, Yan D, Teng MN, Plemper RK, Crowe JE, Jr, Moore ML. 2012. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology 434:129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoon JJ, Krumm SA, Ndungu JM, Hoffman V, Bankamp B, Rota PA, Sun A, Snyder JP, Plemper RK. 2009. Target analysis of the experimental measles therapeutic AS-136A. Antimicrob. Agents Chemother. 53:3860–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plemper RK, Erlandson KJ, Lakdawala AS, Sun A, Prussia A, Boonsombat J, Aki-Sener E, Yalcin I, Yildiz I, Temiz-Arpaci O, Tekiner B, Liotta DC, Snyder JP, Compans RW. 2004. A target site for template-based design of measles virus entry inhibitors. Proc. Natl. Acad. Sci. U. S. A. 101:5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. 2001. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 46:3–26. [DOI] [PubMed] [Google Scholar]
- 37.Ehrengruber MU, Hennou S, Bueler H, Naim HY, Deglon N, Lundstrom K. 2001. Gene transfer into neurons from hippocampal slices: comparison of recombinant Semliki Forest virus, adenovirus, adeno-associated virus, lentivirus, and measles virus. Mol. Cell. Neurosci. 17:855–871. [DOI] [PubMed] [Google Scholar]
- 38.Paal T, Brindley MA, St Clair C, Prussia A, Gaus D, Krumm SA, Snyder JP, Plemper RK. 2009. Probing the spatial organization of measles virus fusion complexes. J. Virol. 83:10480–10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neumann G, Watanabe T, Kawaoka Y. 2000. Plasmid-driven formation of influenza virus-like particles. J. Virol. 74:547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox T, Coll JT, Xie X, Ford PJ, Germann UA, Porter MD, Pazhanisamy S, Fleming MA, Galullo V, Su MS, Wilson KP. 1998. A single amino acid substitution makes ERK2 susceptible to pyridinyl imidazole inhibitors of p38 MAP kinase. Protein Sci. 7:2249–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boutros M, Bras LP, Huber W. 2006. Analysis of cell-based RNAi screens. Genome Biol. 7:R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pelz O, Gilsdorf M, Boutros M. 2010. web cellHTS2: a web-application for the analysis of high-throughput screening data. BMC Bioinformatics 11:185. 10.1186/1471-2105-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang JH, Chung TDY, Oldenburg KR. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67–73. [DOI] [PubMed] [Google Scholar]
- 44.Dochow M, Krumm SA, Crowe JE, Jr, Moore ML, Plemper RK. 2012. Independent structural domains in paramyxovirus polymerase protein. J. Biol. Chem. 287:6878–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutter G, Ohlmann M, Erfle V. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9–12. [DOI] [PubMed] [Google Scholar]
- 46.Kondo N, Miyauchi K, Matsuda Z. 2011. Monitoring viral-mediated membrane fusion using fluorescent reporter methods. Curr. Protoc. Cell Biol. Chapter 26:Unit 26.9. 10.1002/0471143030.cb2609s50. [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Sastre A. 2011. Induction and evasion of type I interferon responses by influenza viruses. Virus Res. 162:12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goodbourn S, Randall RE. 2009. The regulation of type I interferon production by paramyxoviruses. J. Interferon Cytokine Res. 29:539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramachandran A, Horvath CM. 2009. Paramyxovirus disruption of interferon signal transduction: STATus report. J. Interferon Cytokine Res. 29:531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vreede FT, Chan AY, Sharps J, Fodor E. 2010. Mechanisms and functional implications of the degradation of host RNA polymerase II in influenza virus infected cells. Virology 396:125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White LK, Yoon JJ, Lee JK, Sun A, Du Y, Fu H, Snyder JP, Plemper RK. 2007. Nonnucleoside inhibitor of measles virus RNA-dependent RNA polymerase complex activity. Antimicrob. Agents Chemother. 51:2293–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iversen PW, Beck B, Chen YF, Dere W, Devanarayan V, Eastwood BJ, Farmen MW, Iturria SJ, Montrose C, Moore RA, Weidner JR, Sittampalam GS. 2004. HTS assay validation, p 1–31. In Sittampalam GS, Gal-Edd N, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J, Lemmon V, Li Z, McGee J, McManus O, Minor L, Napper A, Riss T, Trask OJ, Weidner J. (ed), Assay guidance manual. Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, MD. [PubMed] [Google Scholar]
- 53.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. 2002. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 45:1712–1722. [DOI] [PubMed] [Google Scholar]
- 54.Coan KE, Maltby DA, Burlingame AL, Shoichet BK. 2009. Promiscuous aggregate-based inhibitors promote enzyme unfolding. J. Med. Chem. 52:2067–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. 2007. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. 6:975–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elliot RL. 2012. Four lessons from global health drug discovery: medicine for an ailing industry? Med. Chem. Lett. 3:688–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lifson J, Coutre S, Huang E, Engleman E. 1986. Role of envelope glycoprotein carbohydrate in human immunodeficiency virus (HIV) infectivity and virus-induced cell fusion. J. Exp. Med. 164:2101–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf MC, Freiberg AN, Zhang T, Akyol-Ataman Z, Grock A, Hong PW, Li J, Watson NF, Fang AQ, Aguilar HC, Porotto M, Honko AN, Damoiseaux R, Miller JP, Woodson SE, Chantasirivisal S, Fontanes V, Negrete OA, Krogstad P, Dasgupta A, Moscona A, Hensley LE, Whelan SP, Faull KF, Holbrook MR, Jung ME, Lee B. 2010. A broad-spectrum antiviral targeting entry of enveloped viruses. Proc. Natl. Acad. Sci. U. S. A. 107:3157–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Clercq E. 2012. Human viral diseases: what is next for antiviral drug discovery? Curr. Opin. Virol. 2:572–579. [DOI] [PubMed] [Google Scholar]
- 61.Baell JB, Holloway GA. 2010. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53:2719–2740. [DOI] [PubMed] [Google Scholar]
- 62.Esper FP, Spahlinger T, Zhou L. 2011. Rate and influence of respiratory virus co-infection on pandemic (H1N1) influenza disease. J. Infect. 63:260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato H, Masuda M, Kanai M, Tsukiyama-Kohara K, Yoneda M, Kai C. 2007. Measles virus N protein inhibits host translation by binding to eIF3-p40. J. Virol. 81:11569–11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Griffin DE. 2007. Measles virus, p 1551–1585. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 65.Wechsler SL, Fields BN. 1978. Intracellular synthesis of measles virus-specified polypeptides. J. Virol. 25:285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pichlmair A, Lassnig C, Eberle CA, Gorna MW, Baumann CL, Burkard TR, Burckstummer T, Stefanovic A, Krieger S, Bennett KL, Rulicke T, Weber F, Colinge J, Muller M, Superti-Furga G. 2011. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 12:624–630. [DOI] [PubMed] [Google Scholar]
- 67.Abbas YM, Pichlmair A, Gorna MW, Superti-Furga G, Nagar B. 2013. Structural basis for viral 5′-PPP-RNA recognition by human IFIT proteins. Nature 494:60–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martinez-Gil L, Ayllon J, Ortigoza MB, Garcia-Sastre A, Shaw ML, Palese P. 2012. Identification of small molecules with type I interferon inducing properties by high-throughput screening. PLoS One 7:e49049. 10.1371/journal.pone.0049049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bedard KM, Wang ML, Proll SC, Loo YM, Katze MG, Gale M, Jr, Iadonato SP. 2012. Isoflavone agonists of IRF-3 dependent signaling have antiviral activity against RNA viruses. J. Virol. 86:7334–7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patel DA, Patel AC, Nolan WC, Zhang Y, Holtzman MJ. 2012. High throughput screening for small molecule enhancers of the interferon signaling pathway to drive next-generation antiviral drug discovery. PLoS One 7:e36594. 10.1371/journal.pone.0036594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fornari FA, Randolph JK, Yalowich JC, Ritke MK, Gewirtz DA. 1994. Interference by doxorubicin with DNA unwinding in MCF-7 breast tumor cells. Mol. Pharmacol. 45:649–656. [PubMed] [Google Scholar]
- 72.Babudri N, Pani B, Tamaro M, Monti-Bragadin C, Zunino F. 1984. Mutagenic and cytotoxic activity of doxorubicin and daunorubicin derivatives on prokaryotic and eukaryotic cells. Br. J. Cancer 50:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Broom WJ, Auwarter KE, Ni J, Russel DE, Yeh LA, Maxwell MM, Glicksman M, Kazantsev AG, Brown RH., Jr 2006. Two approaches to drug discovery in SOD1-mediated ALS. J. Biomol. Screen. 11:729–735. [DOI] [PubMed] [Google Scholar]
- 74.Soelaiman S, Wei BQ, Bergson P, Lee YS, Shen Y, Mrksich M, Shoichet BK, Tang WJ. 2003. Structure-based inhibitor discovery against adenylyl cyclase toxins from pathogenic bacteria that cause anthrax and whooping cough. J. Biol. Chem. 278:25990–25997. [DOI] [PubMed] [Google Scholar]