Abstract
Minor group human rhinoviruses bind low-density lipoprotein (LDL) receptors for endocytosis. Once they are inside endosomes, the acidic pH triggers their dissociation from the receptors and conversion into hydrophobic subviral A particles; these attach to the membrane and transfer their single-strand, positive-sense RNA genome into the cytosol. Here, we allowed human rhinovirus 2 (HRV2) A particles, produced in vitro by incubation at pH 5.4, to attach to liposomes; cryo-electron microscopy 3-dimensional single-particle image reconstruction revealed that they bind to the membrane around a 2-fold icosahedral symmetry axis.
TEXT
HRV2, produced and purified as described previously (1), was incubated at pH 5.4 for 15 min at room temperature. This results in complete conversion of native virus into subviral (expanded full) A particles with a sedimentation constant of 135S, compared to 150S for native virus. We noticed that our viral preparations also contained variable proportions of natural empty capsids (NEC), also called natural top component. At a pH of ≤5.6, these are converted into expanded empty shells with diameters and sedimentation rates identical to those of the empty particles remaining after uncoating of native virions, except that they contain VP0 instead of VP2 and VP4 (reference 2 and our unpublished observations).
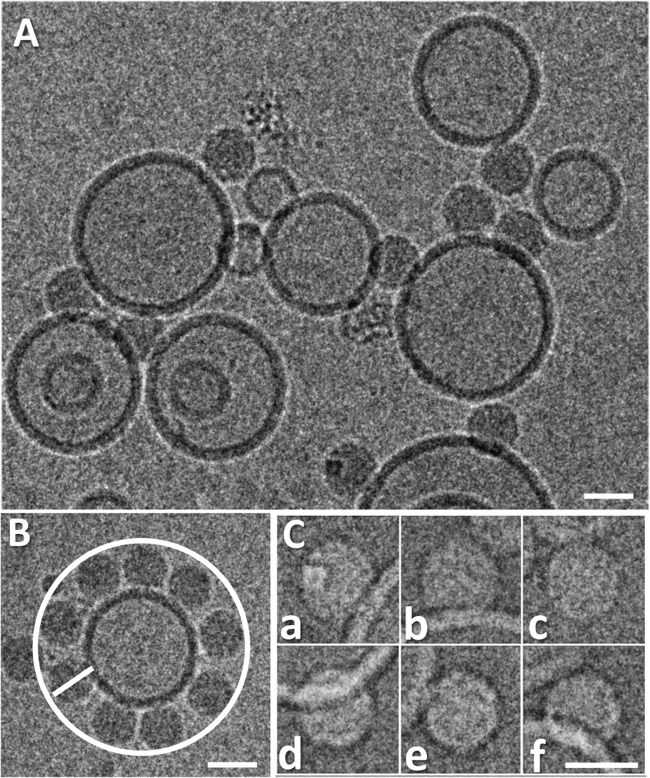
The material resulting from acidification was incubated with unilamellar liposomes (∼100 nm in diameter) (3) for 30 min, applied to holey carbon grids, flash frozen, and viewed in a Polara cryo-electron microscope equipped with a charge-coupled device (CCD) camera at a detector magnification of ×79,372 (sampling rate, 1.89 Å/pixel [px]). Micrographs (example shown in Fig. 1A) were acquired at an underfocus between 1.5 and 5.4 μm, and 239 single particles were manually picked by using Xmipp 2.4 (4) and boxed (256 by 256 px) with RELION (5). The high defocus was used to increase the visibility of the contacts between virus and membrane while accepting the ensuing fringe-related artifacts. We found it impossible to ascertain that a particle entirely below or above a vesicle is definitely membrane attached; therefore, only images with a clear side view of the lipid bilayer (that was about 5 nm in thickness) were selected. These included some particles seen to be attached to the circumference of the liposome (like those in Fig. 1B and C, panels c and e) and above or below it (like those in Fig. 1C, panels a, b, d, and f). Our preliminary electron cryo-tomography results also ascertained that such particles were definitely in contact with the membrane and had not just accidentally become positioned close to it (our unpublished results). The contrast transfer function (CTF) was estimated with ctffind3 (6); although this software is able to correct major and minor defocus values, micrographs with grossly astigmatic Thon rings were discarded. Because of the low number of individual particles per micrograph, those with similar CTFs were combined in 10 groups for better noise estimation (7). CTF-corrected, contrast-inverted, and normalized particle images were subjected to three consecutive rounds of two-dimensional classification (5). During this process, 15 particle images with obvious distortions segregated into separate groups and were discarded.
Fig 1.
Examples of cryo-electron micrographs of A particles attached to liposomes. HRV2 was incubated for 15 min in 50 mM sodium acetate (pH 5.4), resulting in its conversion into A particles, which were subsequently incubated with a suspension of liposomes (∼100-nm diameter) at pH 7.4 for 30 min. The sample was then applied onto holey carbon grids and vitrified with a Leica EM GP automatic grid plunger, and images were collected at a magnification of ×79,372 in an FEI Tecnai F30 Polara electron microscope with a Gatan Ultrascan 4000 4k CCD camera as described in reference 19. (A) Section of one of the micrographs (5.1-μm underfocus). (B) HRV2 A particles attached to a liposome within the same plane, as seen from an equal distance from their centers from the membrane contacts. (C) Six examples of extracted (contrast-inversed) particle images (of the 224 used in the 3DR). Particles are attached at the circumference of the vesicle (as in panel B) (panels c and e) or below or above it (panels a, b, d, and f). Note the strong negative density at the contact site between the A particle and the membrane, which is most probably an artifact resulting from fringes occurring at the high defocus. It might give rise to the cleft seen in the reconstructions (Fig. 2E, arrow). Scale bars, 30 nm.
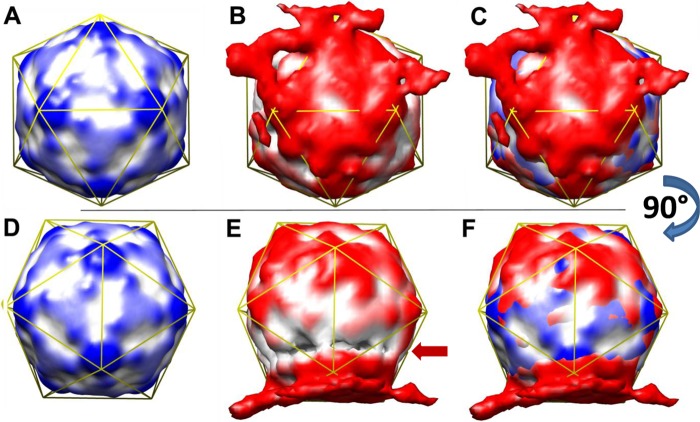
A 3D reconstruction (3DR) was then achieved with RELION run to convergence on the final data set of 224 single particle images, covering the defocus values indicated above, using a cryo-electron microscopy 3DR model of the HRV2 A particle (A. Pickl-Herk, D. Luque, L. Vives-Adrián, J. Querol-Audí, D. Garriga, B. L. Trus, N. Verdaguer, D. Blaas, and J. R. Castón, submitted for publication) filtered to 60 Å as a starting map. In the first round, icosahedral (I2) symmetry was enforced using particle images masked with an (apodized) circular mask of diameter 330 Å, i.e., 175 px, leading to a 3DR with about a 20-Å final resolution (Fig. 2A). This model (filtered to 60 Å) was then used as a starting map for 3DR without imposing symmetry. Several 3DRs were run using values for the particle mask diameter between 350 and 390 Å to vary the contribution of the (asymmetric) liposomal membrane and the (symmetric subviral) particle to the final model. A mask diameter of 380 Å appeared to best reveal the icosahedral symmetry of the particle together with a distinct adhering membrane patch; it converged at a resolution of about 37 Å (Fig. 2B). Despite substantial distortion with respect to a perfect icosahedron and a cleft at the membrane-proximal side of the particle, the superposition of the symmetric (I2) 3DR and the asymmetric (C1) 3DR allowed unequivocal identification of the symmetry axes of the icosahedral virion shell (Fig. 2C) and established their position with respect to the lipid bilayer. The membrane extended over one of the 2-fold axes, including two star-like mesas at the 5-fold axes. This was confirmed by difference mapping, arithmetically subtracting the I2 3DR from the C1 3DR; the most important difference was again manifest just above one of the 2-fold axes (data not shown). The cleft (Fig. 2E, arrow) is most probably an artifact resulting from fringes appearing at the high defocus values chosen for better visibility of the membrane contacts and/or from an unequal angular distribution of the (small number of) particle images.
Fig 2.
HRV2 attaches to a lipid bilayer over a 2-fold axis, including two star-like mesas at the 5-fold axes. Rendered with Chimera (20). (A) Image computed by imposing I2 symmetry, viewed down a 2-fold axis. (B) Image computed without imposing symmetry; same view as in panel A). (C) Superposition of panels A and B. (D to F) As in A to C but rotated 90° around the x axis. Images were rendered at a contour level of ∼2 sigma (I2 3DR) and ∼1.7 sigma (C1 3DR). Color gradients (distance from the particle center) are 140 Å (white) to 150 Å (dark colors). For better orientation, an icosahedral mesh is displayed in yellow. The arrow points to a cleft that is probably the product of fringes resulting from the high defocus values and the strong local changes in density at the contact points between the viral protein and the lipid membrane.
Strauss and colleagues recently reported the 3D structure of membrane-bound poliovirus A particles solved by cryo-tomography (8). They triggered receptor-catalyzed conversion of virus prebound to nickel-nitrilotriacetic acid (Ni-NTA)-lipid-containing liposomes carrying His-tagged soluble poliovirus receptor by incubation at 37°C (9). The ensuing A particles are presumably transferred to the membrane, where they were seen to interact with the lipid bilayer via elongated extensions close to a 2-fold axis.
We show here that single-particle reconstruction from 224 particles can reliably localize the site of interaction between the A particle of HRV2, another member of the genus Enterovirus, and a lipid membrane. Mixing A particles, prepared by incubation of native HRV2 at acidic pH to mimic the physiologic conditions within the endosome during infection, with liposomes resulted in direct attachment of the lipophilic particles to the lipid membrane (10). By analogy to poliovirus, attachment likely occurs via the externalized amphiphilic N-terminal segments of VP1 (11) and can lead to RNA transfer through channels in the lipid bilayer (12). Poliovirus particles were shown to be infectious, although with low efficiency, indicating the capacity to transfer their RNA through a cellular membrane (3). The same might apply to other members of the genus Enterovirus, such as HRV2. Our reconstruction of the membrane-bound HRV2 A particles does not show the “umbilical” connections reported for poliovirus A particles (8). Rather, the protein shell was found to be in intimate contact with the membrane. This is clearly seen in the 3DRs, despite the presence of the artifactual cleft most probably resulting from the high defocus. Why our data and those obtained with poliovirus A particles bound to liposomes differ is unclear; as stated by the authors of reference 8, the “umbilici” might be aggregates composed of extruded VP4, the N-terminal extensions of VP1 together with RNA, and/or poliovirus receptor molecules. In our case, no receptors were present, making the system cleaner and more easily interpretable. Our present data on HRV2 suggest that the N-terminal amphipathic segments of VP1 which become exposed between the tips of the three-bladed propellers at the 3-fold symmetry axes and the ascent of the star-like mesa at the 5-fold axes (13) and that mediate membrane attachment (11, 14) might “pull” the protein shell toward the membrane. This would result in the establishment of a tight seal around the hole at the 2-fold axis, where the RNA exits.
When cryo-EM images were taken from native HRV2 that had been preincubated with recombinant 6His-tagged soluble very-low-density lipoprotein receptor fragments (1) and exposed to Ni-NTA-lipid-containing liposomes (15), 3DR demonstrated more prominent domes at the 5-fold axes because of receptor-derived additional density (16). Furthermore, density stemming from the membrane above one to three of the domes could be discerned (our unpublished results); however, the distance from the virion center to the membrane surface was longer than that in subviral particles directly attached to the membrane. Thus, like in poliovirus (17), RNA passage from within receptor-bound virions through the lipid bilayer into the cytosol is unlikely. This agrees with older data (18) in that the virus has to leave the receptor and be converted into A particles that subsequently (or concomitantly) attach directly to the lipid bilayer of the endosome for RNA passage into the cytoplasm.
ACKNOWLEDGMENTS
We thank I. Gösler for preparing virus, José Caston and Daniel Luque for help with Xmipp, G. Resch (Electron Microscopy Facility of the VBC) for assistance with handling the Polara microscope, Sjorn Scheres for aid with RELION, Holland Cheng for valuable suggestions, and the Vienna Scientific Cluster Team for installation of software.
This study was funded by the Austrian Science Fund (FWF), P 20915-B13. Part of the computations were run on the Vienna Scientific Cluster, P 70172. M.K. was supported by the DK ‘Structure and Interaction of Macromolecules' funded by the FWF.
Footnotes
Published ahead of print 14 August 2013
REFERENCES
- 1.Hewat EA, Neumann E, Conway JF, Moser R, Ronacher B, Marlovits TC, Blaas D. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19:6317–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korant BD, Lonberg Holm K, Noble J, Stasny JT. 1972. Naturally occurring and artificially produced components of three rhinoviruses. Virology 48:71–86. [DOI] [PubMed] [Google Scholar]
- 3.Bilek G, Matscheko NM, Pickl-Herk A, Weiss VU, Subirats X, Kenndler E, Blaas D. 2011. Liposomal nanocontainers as models for viral infection: monitoring viral genomic RNA transfer through lipid membranes. J. Virol. 85:8368–8375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marabini R, Masegosa IM, San Martin MC, Marco S, Fernandez JJ, de la Fraga LG, Vaquerizo C, Carazo JM. 1996. Xmipp: An image processing package for electron microscopy. J. Struct. Biol. 116:237–240. [DOI] [PubMed] [Google Scholar]
- 5.Scheres SH. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180:519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mindell JA, Grigorieff N. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347. [DOI] [PubMed] [Google Scholar]
- 7.Scheres SH. 2012. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415:406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM. 2013. RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J. Virol. 87:3903–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsang SK, McDermott BM, Racaniello VR, Hogle JM. 2001. Kinetic analysis of the effect of poliovirus receptor on viral uncoating: the receptor as a catalyst. J. Virol. 75:4984–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lonberg Holm K, Gosser LB, Shimshick EJ. 1976. Interaction of liposomes with subviral particles of poliovirus type 2 and rhinovirus type 2. J. Virol. 19:746–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fricks CE, Hogle JM. 1990. Cell-induced conformational change in poliovirus—externalization of the amino terminus of Vp1 is responsible for liposome binding. J. Virol. 64:1934–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosteson MT, Chow M. 1997. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J. Virol. 71:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745–7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Cheng N, Chow M, Filman DJ, Steven AC, Hogle JM, Belnap DM. 2011. An externalized polypeptide partitions between two distinct sites on genome-released poliovirus particles. J. Virol. 85:9974–9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bilek G, Kremser L, Wruss J, Blaas D, Kenndler E. 2007. Mimicking early events of virus infection: capillary electrophoretic analysis of virus attachment to receptor-decorated liposomes. Anal. Chem. 79:1620–1625. [DOI] [PubMed] [Google Scholar]
- 16.Verdaguer N, Fita I, Reithmayer M, Moser R, Blaas D. 2004. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11:429–434. [DOI] [PubMed] [Google Scholar]
- 17.Bubeck D, Filman DJ, Hogle JM. 2005. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat. Struct. Mol. Biol. 12:615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konecsni T, Berka U, Pickl-Herk A, Bilek G, Khan AG, Gajdzig L, Fuchs R, Blaas D. 2009. Low pH-triggered beta-propeller switch of the low-density lipoprotein receptor assists rhinovirus infection. J. Virol. 83:10922–10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harutyunyan S, Kumar M, Sedivy A, Subirats X, Kowalski H, Kohler G, Blaas D. 2013. Viral uncoating is directional: exit of the genomic RNA in a common cold virus starts with the poly-(A) tail at the 3′-end. PLoS Pathog. 9:e1003270. 10.1371/journal.ppat.1003270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. [DOI] [PubMed] [Google Scholar]