Abstract
Recent evidence has identified the role of granzyme B- and perforin-expressing CD4+ T cells with cytotoxic potential in antiviral immunity. However, the in vivo cytokine cues and downstream pathways governing the differentiation of these cells are unclear. Here, we have identified that CD4+ T cells with cytotoxic potential are specifically induced at the site of infection during influenza virus infection. The development of CD4+ T cells with cytotoxic potential in vivo was dependent on the cooperation of the STAT2-dependent type I interferon signaling and the interleukin-2/interleukin-2 receptor alpha pathway for the induction of the transcription factors T-bet and Blimp-1. We showed that Blimp-1 promoted the binding of T-bet to the promoters of cytolytic genes in CD4+ T cells and was required for the cytolytic function of the in vitro- and in vivo-generated CD4+ T cells with cytotoxic potential. Thus, our data define the molecular basis of regulation of the in vivo development of this functionally cytotoxic Th subset during acute respiratory virus infection. The potential implications for the functions of these cells are discussed.
INTRODUCTION
Naive CD4+ T cells can differentiate into diverse Th effector subsets upon activation in response to environmental cues and often, the cytokine milieu (1). The appropriate differentiation of a variety of functionally specialized effector Th cells is required for generating productive immune responses against different types of pathogens. Classically, Th cells participate in the antiviral immune responses by providing essential “help” for the development of effector and memory CD8+ T and B cell responses. In contrast, direct effector functions, such as target cell killing or pathogen neutralization, are typically mediated by CD8+ cytotoxic T cells and/or via antibody production by B cells. However, the detection of CD4+ T cells with cytotoxic potential has challenged this conventional view (2, 3). Antigen-dependent cytolytic activity of CD4+ T cells was initially observed in murine and human T cell clones and T cell lines in vitro (4–7). Subsequently, CD4+ T cells with cytotoxic potential were detected in both mice and humans with chronic viral infections such as HIV, cytomegalovirus, Epstein-Barr virus, and lymphocytic choriomeningitis virus (LCMV) infection as well as acute viral infections, such as influenza virus infection (8, 9). CD4+ T cells with cytotoxic potential express cytolytic molecules, such as granzyme B (Gzmb) and perforin, and can undergo granule exocytosis upon antigen stimulation. Studies have established that the expression of these cytolytic molecules is crucial for the in vitro and in vivo killing activities of these CD4+ T cells (9, 10).
Recent advances have also begun to uncover the functions of CD4+ T cells with cytotoxic potential in antiviral and antitumor immunity in vivo (7, 10, 11). In a mouse model of ectromelia virus infection, CD4+ T cells with cytotoxic potential were shown to directly control virus replication in a perforin-dependent manner, demonstrating the physiological role of these cells during a primary viral infection (10). In a murine advanced model of melanoma, T cell receptor (TCR)-transgenic naive CD4+ T cells differentiated into Gzmb- and perforin-expressing cells capable of lysing melanoma cells in vitro and in vivo, suggesting the potential of these cells for cancer immunotherapy (11). Conversely, CD4+ T cells with cytotoxic potential may also exhibit adverse effects that trigger autoimmunity and tissue injury. CD4+ T cells with cytotoxic potential have been detected in rheumatoid arthritis and ankylosing spondylitis and were able to mediate transplant rejection through a perforin-dependent mechanism (2, 12). Thus, understanding the cellular and molecular mechanisms regulating the development of these cells is of considerable significance with respect to viral infections, cancer, and autoimmunity. Currently, interleukin-2 (IL-2) signaling, low antigen dose, as well as OX40 and 41BB stimulation (13–15), have been suggested to promote the development of CD4+ T cells with cytotoxic potential. However, the physiological signals in vivo and the downstream signaling pathways, as well as the underlying transcription factors regulating the development of these CD4+ T cells remain undefined.
Influenza virus is the leading cause of upper and lower respiratory infections and constitutes an ongoing threat to global health. B cells and CD4+ and CD8+ T cells all contribute to the clearance of influenza virus during primary and secondary infection. Besides their role in helping B cell and CD8+ T cell responses, evidence has suggested the direct effector activity of CD4+ T cells in anti-influenza virus immunity (3). CD4+ T cell clones derived from influenza virus-infected mice and the in vitro-generated influenza virus-specific primary CD4+ T cells exhibited cytotoxic activity and were capable of protecting against lethal influenza virus infection (5, 16). More recently, it was demonstrated that CD4+ T cells isolated from influenza virus-infected lungs expressed Gzmb and perforin and were able to kill virus-infected cells through perforin-dependent mechanisms (9). Furthermore, such perforin-dependent cytotoxic mechanisms were able to place evolutionary pressure on the selection of epitope-specific influenza virus escape mutants (17). Collectively, these data support the importance of CD4+ T cells with cytotoxic potential in promoting the clearance of influenza virus in vivo and highlight the need to better understand the cellular and molecular mechanisms guiding the development of these T cells in vivo during influenza virus infection.
We investigated the molecular cues in vivo that govern the development of CD4+ T cells with cytotoxic potential during influenza virus infection. We found that the innate antiviral type I interferons (IFNs) and the adaptive cytokine IL-2 coordinated the in vivo development of CD4+ T cells with cytotoxic potential. We further identified downstream signaling pathways and two transcription factors (T-bet and Blimp-1) that control the development of these CD4+ T cells during influenza virus infection. Thus, we have begun to unravel the complex molecular network that controls the development of CD4+ T cells with cytotoxic potential during an acute viral infection.
MATERIALS AND METHODS
Mice and infection.
Wild-type (WT) C57BL/6 mice were purchased from the Jackson Laboratory. T-bet-deficient mice (Tbx21−/−), IFNAR1-deficient (Ifnar1−/−), Blimp-1 control (Prdm-1fl/fl), Blimp-1 conditional knockout (cKO; CD4-Cre Prdm1fl/fl), STAT4-deficient (Stat4−/−), STAT2-deficient (Stat2−/−), IL-2 receptor α-deficient (Il2rα−/−), Rag1-deficient (Rag1−/−), OTII TCR-transgenic, perforin-deficient OTII (Prf1−/− OTII), Blimp-1 cKO OTII, and Thy1.1 congenic mice were bred in-house. All mice were housed in a specific-pathogen-free environment, and all animal experiments were performed in accordance with protocols approved by the University of Virginia Animal Care and Use Committee or the Indiana University Institutional Animal Care and Use Committee. For influenza virus infection, mice were infected with an ∼500 egg infectious units (EIU) dose of wild-type A/PR/8-34 or ∼2,500 (for Rag1-deficient mice), or ∼5,000 (for WT mice) EIU of A/PR/8/34 expressing ovalbumin (PR8-OVA) in serum-free Iscove's medium intranasally following anesthesia with ketamine and xylazine. In vivo IL-2 blockade was achieved through the injection of anti-IL-2 (clone S4B6; Bio-X-Cell) at day 3 and day 5 postinfection (p.i.) intraperitoneally (i.p.; 750 μg/mouse/injection).
Quantitative RT-PCR.
Lung single-cell suspensions were prepared as previously described (18). CD4+ cells pooled from 2 to 3 lungs per group were purified through magnetic-activated cell sorting (MACS) beads (Miltenyi Biotech) through positive selection. mRNA from in vivo-purified cells or in vitro-cultured cells, as indicated, was isolated by using an RNeasy kit (Qiagen) and treated with DNase I (Invitrogen). Random primers (Invitrogen) and SuperScript II (Invitrogen) were used to synthesize first-strand cDNAs from equivalent amounts of RNA from each sample. Reverse transcription-PCR (RT-PCR) was performed with SYBR green PCR master mix (Applied Biosystems). Data were generated with the comparative threshold cycle (ΔCT) method by normalizing the results to those for hypoxanthine phosphoribosyltransferase (HPRT). Prf1, IFNa6, and IFNb1 primers were bought from Qiagen. Sequences for the rest of the primers used in the studies were as follows: Hprt forward, 5′-CTCCGCCGGCTTCCTCCTCA-3′, and reverse, 5′-ACCTGGTTCATCATCGCTAATC-3′; Prdm1 forward, 5′-GAAGGGAACACGCTTTGGAC-3′, and reverse, 5′-GATTCACGTAGCGCATCCAG-3′. Il2 forward, 5′-CGGCATGTTCTGGATTTGAC-3′, and reverse, 5′-CATCATCGAATTGGCACTCAA-3′.
In vitro DC/T cell coculture.
Bone marrow-derived DC (BMDC) were generated as described previously (18). CD4+ T cells were isolated from spleen and lymph nodes of the indicated mice by using MACS beads (Miltenyi Biotech). Then, we mixed DC with CD4+ T cells at the ratio of 1:10, DC:T cells, in round-bottom 96-well plates (5 ×104 T cells/well) in the presence of 0.1 μg/ml anti-CD3. The conditions of the culture are indicated below. For RNA isolation and RT-PCR, T cells were harvested at day 3 of culture. For Gzmb and T-bet intracellular staining, T cells were harvested at day 4 of culture. For generating large numbers of T cells for use in the chromatin immunoprecipitation (ChIP) assay, T cells were cultured in 6-well tissue culture plate. T-bet and Gzmb staining was performed using the Foxp3 staining buffer set (eBioscience) according to the manufacturer's protocols. The concentrations of the cytokines used were as follows: human IL-2, 30 U/ml (low dose) or 300 U/ml (high dose); IFN-α4 (eBioscience), 50 ng/ml.
Retroviral transduction.
CD4+ T cells were stimulated with plate-bound anti-CD3 and soluble anti-CD28 (both 2 μg/ml) to ensure optimal T cell activation for retroviral transduction. At days 1 and 2 of culture, cells were transduced with bicistronic retrovirus expressing enhanced green fluorescent protein (EGFP) only (control), T-bet and EGFP (T-bet), hCD2 only (control), or hCD2–Blimp-1 (Blimp-1) through spin infection (2,500 rpm; 90 min). After transduction, cells were cultured for an additional 2 days prior to direct flow cytometry or cell sorting (for GFP-expressing virus), followed by intracellular staining of Gzmb as described above.
ChIP.
The ChIP assay was performed as previously described (19). In brief, 6 × 106 cells were cross-linked for 10 min with 1% formaldehyde, lysed by sonication, and precleared with salmon sperm DNA, bovine serum albumin, and a protein A-agarose bead slurry (50%). Cell extracts were incubated with either rabbit polyclonal T-bet (4B10) or normal rabbit IgG (Milipore) overnight at 4°C. The immunocomplexes were precipitated with protein A-agarose beads at 4°C for 2 h, washed, eluted, and reverse cross-linked at 65°C overnight. DNA was purified, resuspended in H2O, and analyzed by quantitative PCR. The following primer pairs were used for T-bet binding: Gzmb forward, 5′-ATGCTCCTGATTACCCTCAC-3′, and reverse, 5′-CAGAGAACCACCACTTACAG-3′; Prf1 forward, 5′-GTACTAGCCTGCTCAAACCT-3′, and reverse, 5′-CTAATCACAGTGTCCCATGAG-3′. ChIP results are represented as percentages of input, and the amount of immunoprecipitated DNA from the IgG control was subtracted from the amount of immunoprecipitated DNA from the specific antibody ChIP, followed by normalization against the amount of input DNA.
BMDC and PMA-ionomycin stimulation.
On day 6 to 7 of BMDC culture, BMDC were harvested and infected with influenza virus at an approximate multiplicity of infection (MOI) of 100 for 6 h. Then, BMDC were counted and mixed with total lung cells at a 1.5-to-1 ratio in the presence of Golgi-Stop (BD Biosciences, 1 μl/ml) and human IL-2 (hIL-2; 40 U/ml) for an additional 6 h. The surface staining of cell surface markers and intracellular staining of cytokines were performed using the Foxp3 staining buffer set (eBioscience) according to the manufacturer's instructions. For CD107α staining, lung cells were stimulated with influenza virus-infected BMDC or phorbol myristate acetate (PMA;100 ng/ml) and ionomycin (1 μg/ml; all from Sigma) in the presence of fluorescein isothiocyanate–anti-CD107α, hIL-2, and Golgi-Stop (BD Biosciences) for 5 to 6 h. The surface staining of cell surface markers and intracellular staining of cytokines as well as CD107α staining were performed as described before (20). For perforin staining, lung cells were restimulated with PMA and ionomycin as described previously (18), and then the surface staining of cell surface markers and intracellular staining of perforin were performed with the Foxp3 staining buffer set (eBioscience).
Generation of mixed bone marrow chimeras.
To generate mixed bone marrow chimeras, we lethally irradiated (1,100 rads) WT mice and then intravenously injected the 1:1 mixed BM cells from WT (Thy1.1+) mice and BM cells from IL-2Rα-deficient mice. After 12 weeks for reconstitution, the chimeric mice were then infected with influenza A/PR8 virus.
OTII cell sorting.
Eight million splenocytes from WT OTII or Blimp-1 cKO OTII mice were transferred into Thy1.1 congenic mice. Then, the mice were infected with PR8-OVA virus. At day 7 p.i., infected lungs were collected and CD4+ T cells were first enriched with MACS beads. Then, the cells were stained with Thy1.2, and Thy1.2+ cells were sorted using an I-cyt Reflection cell sorter (Sony Biotechnology). The sorted OTII cells were then mixed with peptide-pulsed target cells to determine the cytotoxicity of the OTII cells.
Calcein-AM release cytotoxicity assay.
LB27.4 target cells expressing I-Ab were pulsed with OVA323–339 peptide at a concentration of 2 μg/ml for 2 h. Calcein AM (Invitrogen) was added to target cells for the second hour of pulsing at a concentration of 30 μM. LB27.4 target cells were then washed twice and adjusted to a concentration of 2 × 105 cells/ml. Effector OTII cells were cultured as described and added to LB27.4 cells at the indicated effector-to-target (E/T) ratios. After 3 to 4 h of incubation, supernatants were collected, and cytotoxicity (based on absorbance) was measured using a Spectramax Gemini dual-scanning microplate spectrofluorimeter (Molecular Devices) with an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Specific cytotoxicity was calculated as follows: percent specific cytotoxic killing = (experimental release − spontaneous release) × 100/(maximum release − spontaneous release). Spontaneous release represents calcein release from target cells in medium alone, and maximum release is the calcein release from target cells lysed in medium plus 5% Triton X-100.
Cell transfer and infection.
To generate sufficient numbers of T cells for in vivo transfer, we modified our initial culture by first stimulating WT OTII cells or Blimp-1-deficient OTII cells with BMDC and anti-CD3 for 3 days. Then, we expanded and polarized the activated cells with IL-2 plus IFN-α. We confirmed that the expression of Gzmb by OTII cells is Blimp-1 dependent in culture (data not shown). After an additional 2 days in culture, equal numbers (∼3.5 million) of WT OTII effector cells or Blimp-1-deficient OTII effector cells were transferred into Rag1-deficient mice. Two hours later, the mice that received the transferred cells were infected with PR8-OVA virus. Weight loss was monitored daily.
Airway cytokine determination.
Bronchoalveolar lavage (BAL) fluid was obtained by flushing the airway multiple times with a single volume of 600 μl sterile phosphate-buffered saline. Cells in the BAL fluid were spun down, and supernatants were collected for multiplex analysis (Millipore) according to the manufacturer's instructions.
FACS analysis.
All fluorescence-activated cell sorting (FACS) antibodies were purchased from Biolegend, BD Biosciences, or eBioscience. Cells were acquired through a FACSCalibur, FACSCanto, or LSR II apparatus (BD Biosciences). Data were then analyzed by using FlowJo software (Treestar).
Statistical analyses.
Data are reported as means ± standard errors of the means. A paired or unpaired two-tailed Student's t test was used. We considered P values of <0.05 significant.
RESULTS
T-bet- and Blimp-1-dependent development of CD4+ T cells with cytotoxic potential in vivo during influenza virus infection.
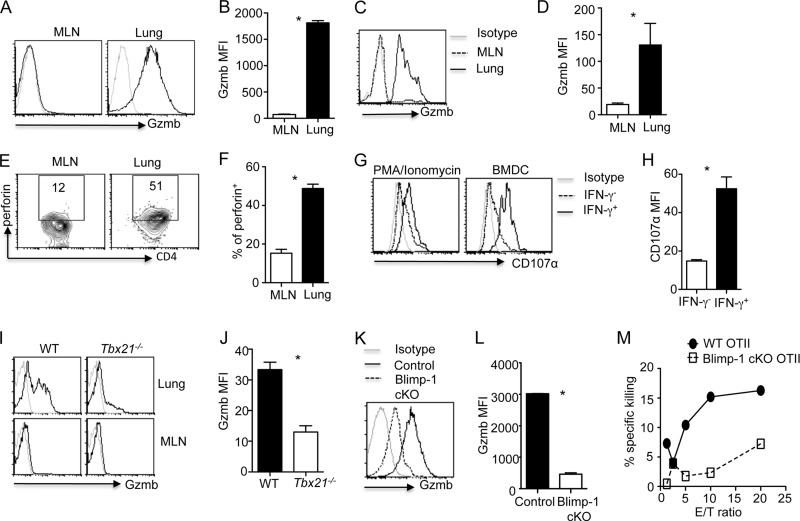
Previous studies had demonstrated that effector CD4+ T cells can directly kill target cells by employing the cytolytic molecules Gzmb and perforin (13, 16). We examined the expression of Gzmb in CD4+ T cells during influenza A/PR8 virus infection. We found that lung CD4+ T cells, but not CD4+ T cells in the draining mediastinal lymph nodes (MLN), expressed Gzmb at day 7 p.i. (Fig. 1A and B). We also examined Gzmb expression in antigen-specific MLN and lung CD4+ T cells during influenza virus infection by transferring OVA-specific CD4+ OTII cells into Thy1-mismatched congenic mice and infecting the mice with OVA epitope-expressing influenza PR8 virus (PR8-OVA). We found that lung OTII T cells but not MLN or spleen OTII cells expressed Gzmb (Fig. 1C and D and data not shown), demonstrating that Gzmb expression in antigen-specific CD4+ T cells was also restricted to the site of infection. We also found that lung OTII cells expressed more perforin than MLN or spleen OTII cells (Fig. 1E and F and data not shown). Notably, MLN and spleen OTII cells expressed significant levels of IFN-γ, suggesting they indeed were Th1 effector cells (data not shown). A hallmark of cytotoxic T cells is that they undergo granule exocytosis following antigenic stimulation. Therefore, we examined whether lung CD4+ T cells could transiently upregulate CD107α as a marker for T cell cytolytic granule exocytosis (21). We found that lung antigen-specific CD4+ T cells (IFN-γ+) underwent the degranulation process in response to mitogen (PMA and ionomycin) or antigen (virus-infected BMDC) stimulation (Fig. 1G and H). Collectively, these data, in agreement with a recent report (9), demonstrated that influenza virus infection induces the development of CD4+ T cells with a cytotoxic potential at the site of infection. Importantly, Gzmb- and perforin-expressing CD4+ T cells have recently been demonstrated to directly kill virus-infected major histocompatibility complex II (MHC-II)-expressing cells and confered protection against influenza virus infection (9). However, the underlying molecular and cellular cues required for the development of these CD4+ T cells during viral infection remain elusive.
Fig 1.
T-bet- and Blimp-1-dependent induction of CD4+ T cells with cytotoxic potential in vivo. (A and B) Gzmb expression (A) and MFI (B) in WT lung and MLN CD4+ T cells at day 7 post-PR8 infection (n = 3). (C to F) OTII cells were transferred into Thy1-mismatched WT mice, and then the mice were infected with PR8-OVA (n = 3). Gzmb expression (C) and MFI (D), perforin expression (E), and the percentage of perforin+ cells (F) in lung and MLN OTII cells at day 7 p.i. are illustrated. (G) Lung CD4+ T cell degranulation (CD107α expression) in IFN-γ+ cells (indication of antigen-specific CD4+ cells) and IFN-γ− cells following stimulation with PMA-ionomycin or virus-infected BMDC. (H) Lung CD107α MFI in IFN-γ+ and IFN-γ− cells following stimulation with virus-infected BMDC (n = 3). (I and J) Gzmb expression (I) and MFI (J) of MLN and lung CD4+ T cells from WT or T-bet-deficient mice at day 7 post-PR8 infection (n = 2 to 3). (K and L) Gzmb expression (K) and MFI (L) of lung CD4+ T cells from WT or Blimp-1-deficient (Blimp-1 cKO) mice at day 7 post-PR8 infection (n = 3). (M) Cytolytic activity of WT or Blimp-1-deficient lung OTII cells (day 7 post-PR8-OVA infection) was measured by calcein AM release of peptide-pulsed LB27.4 target cells. Data are representative of two to four separate experiments. *, P < 0.05.
We next sought to investigate the molecular mechanisms underlying the development of CD4+ T cells with cytotoxic potential during influenza virus infection. T-bet is a transcription factor that plays important roles in the development of CD8+ cytotoxic T cells (22). We found that lung polyclonal or OTII TCR-transgenic CD4+ T cells expressed higher levels of T-bet than their counterparts in MLN or spleen, and lung CD4+ T-bet+ cells expressed higher levels of Gzmb than lung CD4+ T-bet− cells (data not shown), suggesting that T-bet may positively regulate the development of CD4+ T cells with cytotoxic potential. Consistent with this idea, we found that T-bet deficiency impaired the expression of Gzmb in CD4+ T cells in vivo (Fig. 1I and J). Thus, these data indicate that T-bet is required for the generation of CD4+ T cells with cytotoxic potential in vivo.
Although T-bet is required for the induction of CD4+ T cells with cytotoxic potential in the lung, T-bet expression alone was not sufficient to induce Gzmb expression in MLN CD4+ T cells (data not shown), suggesting that additional factors play a role in regulating the development of CD4+ T cells with cytotoxic potential in vivo. We found that lung CD4+ T cells expressed higher levels of Blimp-1 mRNA (Prdm1) than MLN CD4+ T cells (data not shown), indicating a possible role of Blimp-1 in regulating Gzmb and perforin expression in lung CD4+ T cells in vivo. To explore this idea, we infected control (Prdm1fl/fl) or T cell-specific Blimp-1 conditional mutant mice (CD4-Cre Prdm1fl/fl) with influenza virus and examined the Gzmb expression by CD4+ T cells. We found that Blimp-1 deficiency in T cells drastically impaired the expression of Gzmb in CD4+ T cells (Fig. 1K and L). We also examined whether CD4+ T cells isolated in vivo were able to kill peptide-pulsed MHC-II-expressing target cells in a Blimp-1-dependent manner. To this end, we transferred naive WT OTII cells or Blimp-1-deficient OTII cells (by crossing OTII transgenic mice to conditional Blimp-1 mutant mice) into Thy1-mismatched congenic mice and infected the mice with PR8-OVA. We then sorted WT and Blimp-1-deficient OTII cells from the infected lungs and examined their ability to kill target cells in vitro. We found that Blimp-1 deficiency impaired the ability of the in vivo-generated CD4+ T cells with cytotoxic potential to kill target cells (Fig. 1M). Together, these data indicated that the generation of CD4+ T cells with cytotoxic potential in vivo is dependent on the expression of Blimp-1 in CD4+ T cells.
STAT4-independent, STAT2-dependent development of CD4+ T cells with cytotoxic potential during influenza virus infection.
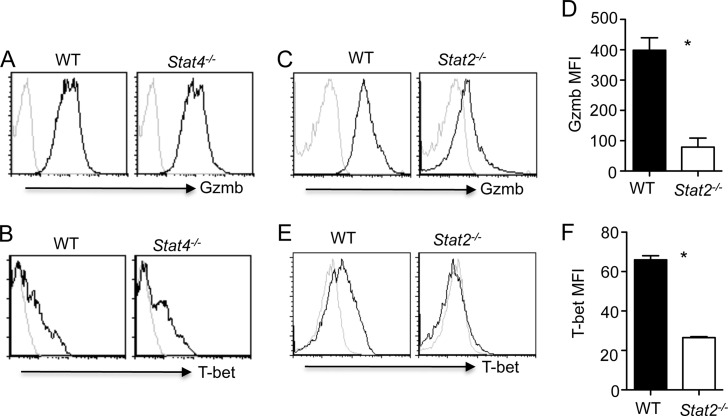
We next examined the upstream signaling pathways required for the generation of CD4+ T cells with cytotoxic potential during influenza virus infection. Since STAT4 is a major factor that controls T-bet expression in CD4+ T cells (23), we examined whether STAT4 is required for the development of CD4+ T cells with cytotoxic potential during influenza virus infection. Remarkably, STAT4-deficient CD4+ T cells expressed equivalent levels of Gzmb and T-bet as WT CD4+ T cells upon viral challenge (Fig. 2A and B and data not shown), suggesting that STAT4 is dispensable for the induction of CD4+ T cells with cytotoxic potential in vivo during influenza virus infection. We also infected WT and STAT2-deficient mice with influenza virus and examined Gzmb expression in lung CD4+ T cells at day 7 p.i. We found that STAT2 deficiency significantly abrogated Gzmb expression in lung CD4+ T cells (Fig. 2C and D). The diminished expression of Gzmb in CD4+ T cells was correlated with lower T-bet expression in STAT2-deficient CD4+ T cells (Fig. 2E and F). Together, these data suggested that STAT2 rather than STAT4 is required for the development of CD4+ T cells with cytotoxic potential in vivo during influenza virus infection.
Fig 2.
STAT4-independent, STAT2-dependent development of CD4+ T cells with cytotoxic potential in vivo. WT, STAT4-deficient, or STAT2-deficient mice were infected with PR8. (A and B) Gzmb (A) and T-bet (B) expression in lung CD4+ T cells from WT or STAT4-deficient mice at day 7 p.i. (n = 2 to 3). (C to F) Gzmb expression (C) and MFI (D), T-bet expression (E), and MFI (F) of lung CD4+ T cells from WT or STAT2-deficient mice at day 7 p.i. (n = 2 to 3). Data are representative of three separate experiments. *, P < 0.05.
IL-2 and type I IFNs cooperate to induce the development of CD4+ T cells with cytotoxic potential in vitro.
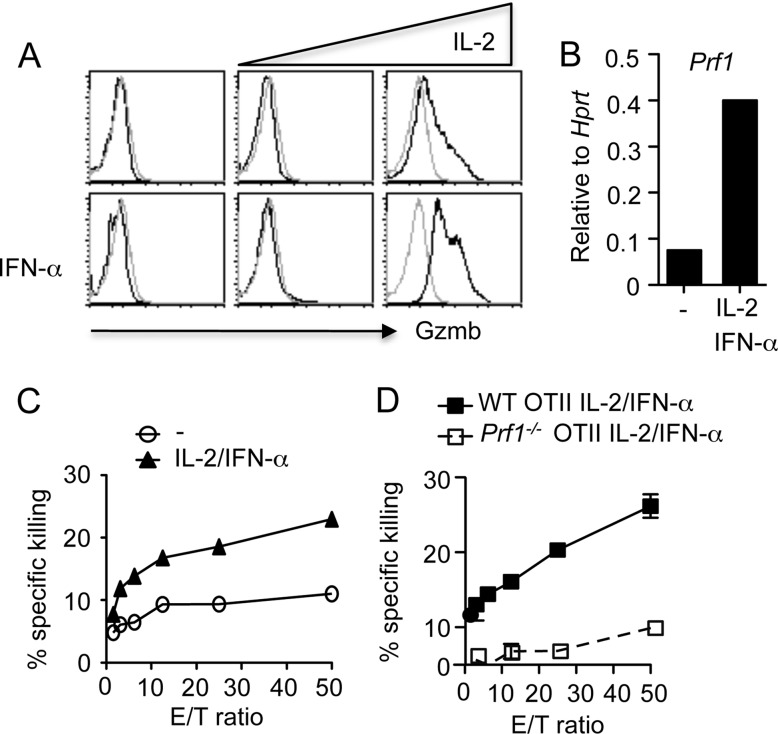
The fact that STAT2 is required for the development of CD4+ T cells with cytotoxic potential in vivo suggests that type I IFNs, which signal through STAT2, may be required for the generation of CD4+ T cells with cytotoxic potential. In addition, a role for IL-2 has also been indicated in the induction of cytotolytic molecule expression in CD8+ T cells (24). To investigate the role of IL-2 and type I IFNs in the development of CD4+ T cells with cytotoxic potential, we developed an in vitro coculture system in which we stimulated CD4+ T cells with BMDC plus soluble anti-CD3 in the presence or absence of IFN-α and increasing concentrations of IL-2. We then examined Gzmb expression in CD4+ T cells. We found that addition of IFN-α or IL-2 alone to the culture modestly increased Gzmb expression in CD4+ T cells (Fig. 3A). However, IFN-α and a high concentration of IL-2 (300 U) had drastic synergistic effects in the induction of Gzmb expression in CD4+ T cells (Fig. 3A). IL-2 plus IFN-α also induced perforin mRNA expression in CD4+ T cells (Fig. 3B). These data suggest that IL-2 cooperates with type I IFNs to induce cytolytic gene expression in CD4+ T cells in vitro. We next examined whether OTII cells cultured in the presence of IL-2 and IFN-α could kill peptide-pulsed MHC-II-expressing target cells. Consistent with the enhanced Gzmb and perforin expression in CD4+ T cells cultured with IL-2 plus IFN-α, we found that OTII cells cultured under this condition exhibited an enhanced ability to kill target cells (Fig. 3C). Importantly, the killing of the target cells by these OTII cells was perforin dependent (Fig. 3D), further supporting the idea that they have cytotoxic potential. Taken together, these data suggest that a high concentration of IL-2 in combination with type I IFNs induces the development of CD4+ T cells with cytotoxic potential in vitro.
Fig 3.
IL-2 and type I IFNs cooperate to induce the development of CD4+ T cells with cytotoxic potential in vitro. (A and B) CD4+ T cells were cultured in the absence or presence of IFN-α plus increasing concentrations of hIL-2 (low concentration, 30 U/ml; high concentration, 300 U/ml). Gzmb protein (A) and perforin mRNA (B) expression in CD4+ T cells was measured. (C) Cytolytic activity of WT OTII cells cultured under neutral (no cytokine added) or in the presence of IL-2 plus IFN-α was measured based on calcein AM release by peptide-pulsed LB27.4 target cells. (D) Cytolytic activity of WT or perforin-deficient OTII cells cultured in the presence of IL-2 plus IFN-α was measured based on calcein AM release by peptide-pulsed LB27.4 target cells. Data are representative of at least three separate experiments.
T-bet and Blimp-1 are required for the induction of CD4+ T cells with cytotoxic potential by IL-2 plus type I IFNs.
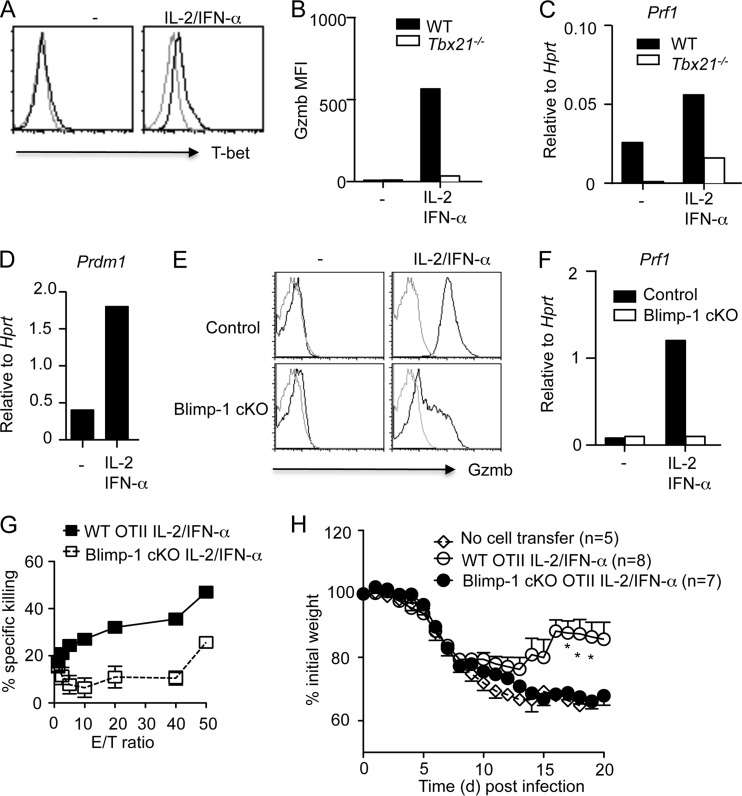
We next examined whether the induction of CD4+ T cells with cytotoxic potential by IL-2 and IFN-α was dependent on T-bet and/or Blimp-1. To this end, we found that IL-2 plus IFN-α induced T-bet expression in CD4+ T cells in vitro (Fig. 4A). Furthermore, T-bet-deficient CD4+ T cells displayed greatly reduced levels of Gzmb and perforin mRNA when cultured in the presence of IL-2 plus IFN-α (Fig. 4B and C). Thus, T-bet is required for the induction by IL-2 plus IFN-α of CD4+ T cells with cytotoxic potential in vitro. IL-2 plus IFN-α also induced Blimp-1 expression in CD4+ T cells (Fig. 4D), suggesting that Blimp-1 is required for the induction by IL-2 and IFN-α of CD4+ T cells with cytotoxic potential. To explore this idea, we cultured WT or Blimp-1-deficient CD4+ T cells in the presence of IL-2 plus IFN-α to induce CD4+ T cells with cytotoxic potential. We observed that Blimp-1 deficiency greatly impaired the expression of Gzmb in CD4+ T cells (Fig. 4E). We also found that Blimp-1 deficiency impaired perforin mRNA expression in CD4+ T cells (Fig. 4F). To determine the role of Blimp-1 in regulating the function of CD4+ T cells with cytotoxic potential, we cultured WT OTII or Blimp-1-deficient OTII cells with IL-2 plus IFN-α and examined their ability to kill target cells. We found that Blimp-1 deficiency impaired the ability of the in vitro-generated CD4+ T cells with cytotoxic potential to kill target cells (Fig. 4G). Recent evidence demonstrated that, when transferred into naive hosts, in vitro- and in vivo-generated CD4+ T cells with cytotoxic potential are able to provide protection against influenza virus infection (9, 13, 16). We next examined whether the protection from influenza virus infection by CD4+ T cells with cytotoxic potential was Blimp-1 dependent. To do so, we cultured WT and Blimp-1-deficient OTII cells with IL-2 plus IFN-α and transferred the cultured cells into Rag1-deficient mice. We then infected Rag1-deficient mice with PR8-OVA and monitored the weight loss of the mice following infection. Consistent with previous findings (16), we found that CD4+ T cells with cytotoxic potential generated in vitro were able to provide protection to the host (Fig. 4H). Importantly, Blimp-1 deficiency in CD4+ T cells impaired the ability of these cells to provide protection against influenza virus-induced host morbidity (Fig. 4H). These data thus demonstrated that Blimp-1 is essential for the function of CD4+ T cells with cytotoxic potential in vivo.
Fig 4.
Induction of CD4+ T cells with cytotoxic potential by IL-2 plus type I IFNs is dependent on T-bet and Blimp-1. (A) T-bet expression in CD4+ T cells cultured under the indicated conditions. (B and C) WT or T-bet-deficient CD4+ T cells were cultured under the indicated conditions. Gzmb protein (B) and perforin mRNA (C) expression in WT or T-bet-deficient CD4+ T cells are shown. (D) Blimp-1 mRNA expression in CD4+ T cells cultured under the indicated conditions. (E and F) Control or Blimp-1-deficient CD4+ T cells were cultured under the indicated conditions. Gzmb protein (E) and perforin mRNA (F) expression in control or Blimp-1-deficient CD4+ T cells are shown. (G) Cytolytic activity of WT or Blimp-1-deficient OTII cells cultured under the indicated conditions was measured based on calcein AM release of peptide-pulsed LB27.4 target cells. (H) Weight loss of PR8-OVA-infected Rag1-deficient mice which received IL-2/IFN-α-cultured WT or Blimp-1-deficient (cKO) OTII cells. Data are representative of two to three separate experiments. Data were pooled from two independent experiments (n = 5 to 8). *, P < 0.05.
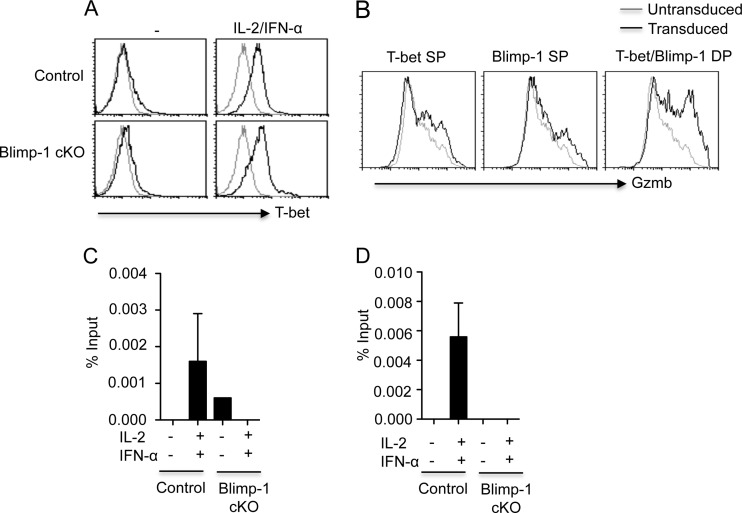
Blimp-1 controls T-bet binding to cytolytic genes in CD4+ T cells.
Blimp-1 is a transcriptional repressor, and it is unclear how Blimp-1 positively controls Gzmb and perforin expression in T cells. We found that Blimp-1 deficiency enhanced T-bet expression in CD4+ T cells (Fig. 5A), suggesting that the diminished Gzmb expression in Blimp-1-deficient T cells was not due to the absence of T-bet. We next examined whether T-bet or Blimp-1 expression was sufficient to induce Gzmb expression in CD4+ T cells. To do so, we ectopically expressed T-bet and Blimp-1 in CD4+ T cells by transducing CD4+ T cells with T-bet- and Blimp-1-expressing retrovirus. We found that ectopic T-bet or Blimp-1 expression alone modestly enhanced Gzmb expression in CD4+ T cells (Fig. 5B). However, coexpression of T-bet and Blimp-1 cooperatively induced higher levels of Gzmb expression in CD4+ T cells (Fig. 5B). T-bet can directly bind to the Gzmb and Prf1 promoters in CD8+ T cells to promote the expression of these cytotoxic molecules (25). We next examined whether T-bet was able to bind to the Gzmb and Prf1 promoters in CD4+ T cells in ChIP experiments. We found that T-bet bound directly to the Gzmb and Prf1 promoters in CD4+ T cells cultured under the conditions to induce CD4+ T cells with cytotoxic potential (Fig. 5C and D). Interestingly, Blimp-1 deficiency impaired the binding of T-bet to the Gzmb and Prf1 promoters (Fig. 5C and D). These data suggest that Blimp-1 controls the development of CD4+ T cells with cytotoxic potential by regulating the binding of T-bet to the promoters of cytolytic molecules. These data also established that T-bet and Blimp-1 act cooperatively during the development of CD4+ T cells with cytotoxic potential.
Fig 5.
Blimp-1 controls T-bet binding to cytolytic genes in CD4+ T cells. (A) T-bet expression in control or Blimp-1-deficient CD4+ T cells cultured under the indicated conditions. (B) CD4+ T cells were cotransduced with T-bet- plus Blimp-1-expressing retroviruses. Gzmb expression in untransduced cells, T-bet single-transduced cells (T-bet SP), Blimp-1 single-transduced cells (Blimp-1 SP), or T-bet and Blimp-1 double-transduced cells (T-bet and Blimp-1 DP) is depicted. (C and D) Control or Blimp-1 cKO CD4+ T cells were cultured under the indicated conditions. The binding of T-bet to the Gzmb (C) or Prf1 (D) promoters in cultured CD4+ T cells was assessed by ChIP. ChIP data are presented as the percentages of input, as described in Materials and Methods. Data are representative of three separate experiments.
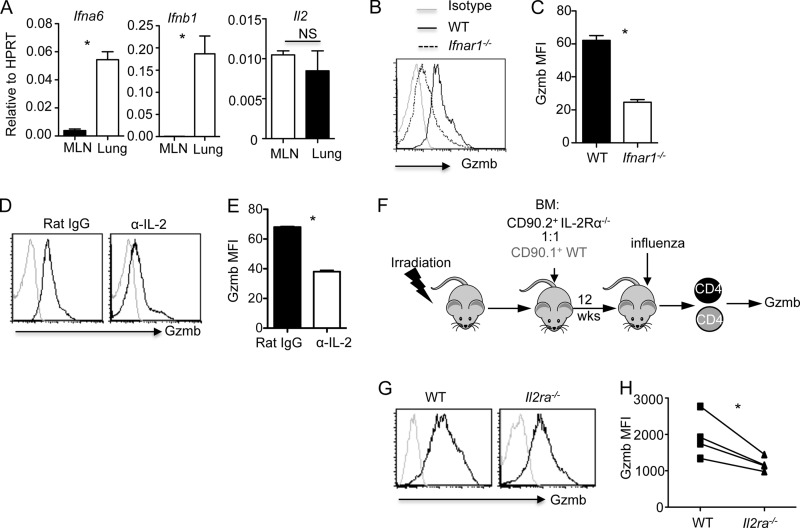
IL-2 and type I IFNs cooperate to induce the development of CD4+ T cells with cytotoxic potential in vivo. The fact that IL-2 and IFN-α induced cytolytic molecule expression in CD4+ T cells in vitro strongly suggests that they are required for the generation of CD4+ T cells with cytotoxic potential in vivo during influenza virus infection. Indeed, type I IFNs are produced at the time of effector T cell infiltration into the lungs during influenza virus infection (26, 27). Furthermore, we found that type I IFNs, but not IL-2, gene expression was highly expressed in the lung compared to MLN at the time of T cell infiltration (day 5 p.i.) (Fig. 6A), which is coincident with the specific expression of Gzmb in the lung but not MLN CD4+ T cells during influenza virus infection (Fig. 1). Therefore, we hypothesized that type I IFN signaling is required for the development of CD4+ T cells with cytotoxic potential in vivo during influenza virus infection. Consistent with this hypothesis, we found that the deficiency of IFNAR1 impaired Gzmb and T-bet expression in CD4+ T cells (Fig. 6B and C and data not shown). These data thus established that type I IFN signaling is required for optimal development of CD4+ T cells with cytotoxic potential during influenza virus infection. We next examined whether IL-2 is required for cytolytic molecule expression in CD4+ T cells in vivo. To this end, we infected WT mice with influenza virus and then blocked IL-2 signaling via injection of an IL-2-neutralizing antibody. We found that IL-2 neutralization in vivo partially, but significantly, diminished Gzmb expression in CD4+ T cells (Fig. 6D and E). To investigate whether IL-2 signaling is intrinsically required in CD4+ T cells, we infected WT and IL-2Rα-deficient mixed BM chimeric mice with influenza virus and examined Gzmb expression in CD4+ T cells (Fig. 6F). We found that the deficiency of IL-2Rα on CD4+ T cells diminished Gzmb expression (Fig. 6G and H), suggesting that the intrinsic IL-2Rα signaling in CD4+ T cells is required for the optimal expression of Gzmb in CD4+ T cells in vivo.
Fig 6.
IL-2 and type I IFN signaling are required for the development of CD4+ T cells with cytotoxic potential in vivo. (A) IFN-α6, IFN-β1, and IL-2 gene expression in the lung and MLN at day 5 p.i. (n = 3). (B and C) Gzmb expression (B) and MFI (C) of lung CD4+ T cells isolated from influenza virus-infected WT or IFNAR1-deficient mice at day 7 p.i. (n = 3). (D and E) WT mice were infected with influenza virus PR8 and treated under the indicated conditions (n = 3). Gzmb expression (D) and MFI (E) of lung CD4+ T cells at day 7 p.i. are shown. (F) Schematic of BM chimera construction and influenza virus infection. (G and H) WT and IL-2Rα-deficient mixed BM chimeras were infected with influenza virus (n = 4). Gzmb expression (G) and MFI (H) of WT (CD90.1+) or IL-2Rα-deficient (CD90.2+) lung CD4+ T cells at day 7 p.i. are shown. Data are representative of at least two separate experiments. *, P < 0.05.
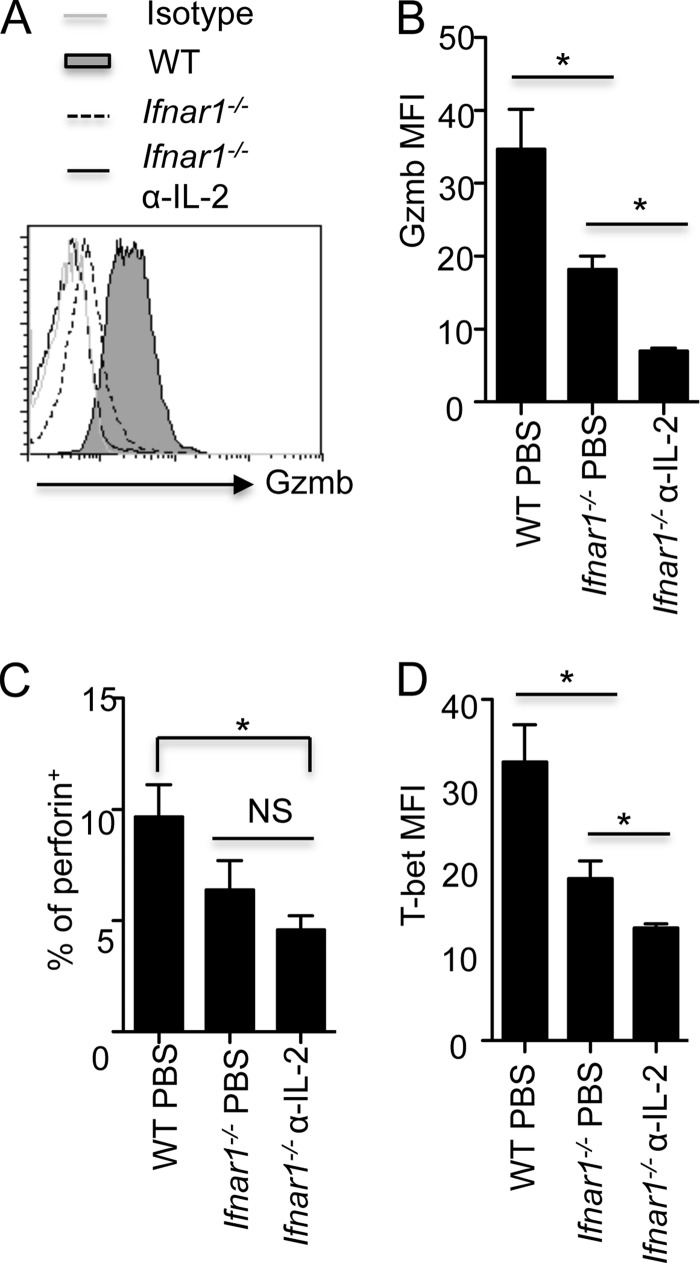
Notably, the single deficiency of IL-2Rα or IFNAR1 in vivo only partially impaired Gzmb expression in CD4+ T cells. Therefore, we blocked IL-2 and type I IFN signaling simultaneously during influenza virus infection by administering anti-IL-2 to influenza virus-infected IFNAR1-deficient mice. We found that the blockade of type I IFN and IL-2 signaling together completely abrogated Gzmb expression in CD4+ T cells (Fig. 7A and B). The combined blockade of type I IFN and IL-2 signaling also significantly impaired perforin expression in lung CD4+ T cells (Fig. 7C). The impaired expression of Gzmb and perforin in CD4+ T cells was associated with diminished T-bet expression in CD4+ T cells following the double blockade of type I IFN and IL-2 signaling in vivo (Fig. 7D). Taken together, these data suggest that the development of CD4+ T cells with cytotoxic potential in vivo is dependent on the cooperation between IL-2 and type I IFN signaling pathways during influenza virus infection. Notably, the blockade of IL-2 and type I IFN signaling together also disrupted the expression of Gzmb in CD8+ T cells (data not shown), suggesting that IL-2 and type IFNs coordinate the development of both cytotoxic CD4+ and CD8+ T cells in vivo during influenza virus infection.
Fig 7.
IL-2 and type I IFN signaling cooperate in the development of CD4+ T cells with cytotoxic potential in vivo. (A to D) WT or IFNAR1-deficient mice were infected with influenza virus PR8 and treated under the indicated conditions (n = 3 to 5). Gzmb expression (A) and MFI (B), the percentages of perforin+ cells (C), and also the T-bet MFI (D) of lung CD4+ T cells at day 7 p.i. are shown. Data are representative of at least two experiments. *, P < 0.05; NS, nonsignificant.
DISCUSSION
Emerging evidence has highlighted the role of CD4+ T cells with cytotoxic potential in antiviral and antitumor immunity. In this report, we demonstrated that IL-2 and type I IFNs cooperatively induce the development of CD4+ T cells with cytotoxic potential in vitro and in vivo during acute respiratory virus infection. Mechanistically, IL-2 and type I IFNs induced the expression of T-bet and Blimp-1, two transcription factors that cooperate to induce the expression of cytolytic molecules in CD4+ T cells. Notably, we showed that the antiviral function of CD4+ T cells with cytotoxic potential in vivo is critically dependent on the expression of Blimp-1. We thus have identified the molecular mechanisms regulating the development of CD4+ T cells with cytotoxic potential during acute viral infection.
Recent analyses have begun to elucidate the function of CD4+ T cells with cytotoxic potential during viral infections. By using mice with a selective deficiency of perforin in CD4+ T cells, it was demonstrated that CD4+ T cells with cytotoxic potential directly controlled ectromelia virus replication in vivo (10). In influenza virus infection, antiviral effector CD4+ T cells generated in vitro or in vivo, upon transfer, were able to attenuate influenza virus-induced host morbidity and mortality in WT and B cell-deficient mice, in part through a perforin-dependent cytotoxic mechanism (9, 16). We likewise have reported here that antiviral CD4+ T cells exhibit Blimp-1-dependent protection against influenza virus infection in adaptive immune (B and T cell)-deficient recipients. These data suggest that CD4+ T cells with cytotoxic potential may contribute to the elimination of virus-infected cells in vivo during a primary or secondary influenza virus infection. Strikingly, a recent clinical study implicated preexisting CD4+ T cells in directly killing target cells through perforin-dependent mechanisms and contributing to the heterologous influenza virus immunity in humans (28). Notably, lung alveolar type II cells and some airway epithelial cells, the critical cell targets of influenza virus (29), constitutively or inducibly express MHC-II (30), and this raises the possibility that CD4+ T cells may directly control influenza virus replication in vivo. Furthermore, tumor necrosis factor-inducible nitric oxide synthase DC (TipDCs), the major injury-inducing cells during influenza virus infection (31–33), harbor influenza virus antigen and express high levels of MHC-II (31). Thus, it is also possible that CD4+ T cells with cytotoxic potential may indirectly protect the host from influenza virus-induced diseases through the killing of TipDCs. Although the exact physiological function of CD4+ T cells with cytotoxic potential during influenza virus infection warrants further studies, the existence of such cells presumably enlarges the pool of cytotoxic T cells to enable host clearance of virus-infected cells faster and/or to avert virus escape mutants. Potentially, the functionality of these CD4+ T cells with cytotoxic potential during a viral infection may be determined by virus tropism. CD4+ T cells with cytotoxic potential may play more significant roles when MHC-II+ cells are the prominent target of virus infection.
Various cytokines, including IL-2, IL-12, IL-27, type I IFNs, and type II IFN (IFN-γ), have been shown to promote Gzmb and/or perforin expression in CD8+ T cells and NK cells. However, the in vivo significance and relative contribution of these cytokines in regulating the expression of cytolytic molecules are currently poorly defined. IL-2 is critical in promoting Gzmb expression in CD8+ T cells in vitro and in vivo during LCMV infection (24, 34). With a murine influenza virus model, we recently showed that in an inflammatory milieu, the ablation of IL-2 signaling alone in T cells only partially affected Gzmb expression in effector CD8+ T cells (18). Similarly, we found here that IL-2 was partially required for Gzmb expression in CD4+ T cells in vivo during influenza virus infection. This is in contrast to a recent report that IL-2 ablation completely abrogated Gzmb expression in vivo in CD4+ T cells stimulated with OX40 plus 41BB (15). The presence of strong inflammation (i.e., abundant type I IFNs) at the site of infection may partially compensate for the requirement of IL-2 in Gzmb expression within both CD4+ and CD8+ T cells. In accordance with this idea, the combined blockade of IL-2 plus type I IFNs completely abrogated the expression of Gzmb in both CD4+ and CD8+ T cells (data not shown).
Blimp-1 is required for Gzmb expression in effector CD8+ T cells and NK cells (35, 36). However, the underlying mechanisms by which Blimp-1 controls the expression of Gzmb in CD8+ and NK cells remain to be determined. Here we have demonstrated that the expression of both Gzmb and perforin is dependent on the expression of Blimp-1 in CD4+ T cells as well. Blimp-1 was previously found to inhibit T-bet and IFN-γ expression during Th1 polarization (37). In agreement with this finding, we found here that Blimp-1-deficient T cells expressed higher levels of T-bet. However, T-bet was unable to bind to the Gzmb and Prf1 promoters in the absence of Blimp-1. Thus, Blimp-1 exhibits differential effects in regulating the expression of effector cytokine IFN-γ and cytolytic molecules (Gzmb and perforin) in CD4+ T cells (37). The exact mechanism underlying such differential effects of Blimp-1 in the expression of IFN-γ and cytolytic molecules is currently unknown. One possible reason is that Blimp-1 directly binds to Ifng and Tbx21 loci to repress their expression through its transcriptional repressor activity (37), while it works independently of its DNA binding/repressor activity to modulate the function of T-bet for Gzmb and Prf1 transcription. Whether Blimp-1 directly associates with T-bet or simply maintains the active chromatin structure of Gzmb and Prf1 loci to facilitate T-bet binding requires further investigation. In addition, Blimp-1 may also repress repressors of the Gzmb and Prf1 genes (such as microRNAs that target Gzmb and Prf1 genes) to promote the expression of cytolytic molecules during Th cell differentiation.
Recently, OX40-induced Gzmb expression in CD4+ T cells was shown to be Eomes-dependent (14). Furthermore, these CD4+ T cells, capable of producing both Th1 and Th2 cytokines, exhibit characteristics of both memory and terminal effector phenotypes (14). In contrast, we showed here that type I IFN- and IL-2-induced Gzmb expression requires T-bet expression but not Eomes (data not shown) and can be categorized as CD4+ effector cells under type 1 lineage. Interestingly, although antigen-specific CD4+ T cells in the MLN expressed T-bet and Th1 signature cytokine IFN-γ, they did not express Gzmb or perforin. The likely explanation is that a high threshold of T-bet is required for driving the transcription of cytolytic molecules in CD4+ T cells. However, ectopic expression of T-bet in CD4+ T cells is not sufficient to induce optimal expression of Gzmb in vitro, suggesting an additional factor, such as Blimp-1, plays an important role in the development of the cytotoxic potential of CD4+ T cells. Our data also suggest that the development of CD4+ T cells with cytotoxic potential passes through a conventional Th1 cell stage. Understanding the complex interaction of the cellular and molecular networks governing the differentiation of antiviral Th cells may provide the basis for better treatment and vaccine strategies for viral infections in the future.
ACKNOWLEDGMENTS
We thank J. S. Blum for critical reading of the manuscript. We thank E. J. Pearce and P. M. Allen (Washington University School of Medicine), C. Schindler (Columbia University), and T. R. Malek (University of Miami) for reagents. We thank S. Zhang and S. Farag for technical advice.
This work was supported by U.S. National Institutes of Health grants (AI099753 to J.S., AI045515 to M.H.K., AI083024 to T.J.B., and AI092212 to A.L.D.), AHA predoctoral fellowship 10PRE4620001 to D.S., NIH predoctoral training grant T32 HL007910 to D.P., and Showalter Trust funds to J.S.
Footnotes
Published ahead of print 28 August 2013
REFERENCES
- 1.O'Shea JJ, Paul WE. 2010. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327:1098–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appay V. 2004. The physiological role of cytotoxic CD4(+) T-cells: the holy grail? Clin. Exp. Immunol. 138:10–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swain SL, McKinstry KK, Strutt TM. 2012. Expanding roles for CD4(+) T cells in immunity to viruses. Nat. Rev. Immunol. 12:136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleischer B. 1984. Acquisition of specific cytotoxic activity by human T4+ T lymphocytes in culture. Nature 308:365–367 [DOI] [PubMed] [Google Scholar]
- 5.Graham MB, Braciale VL, Braciale TJ. 1994. Influenza virus-specific CD4+ T helper type 2 T lymphocytes do not promote recovery from experimental virus infection. J. Exp. Med. 180:1273–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maimone MM, Morrison LA, Braciale VL, Braciale TJ. 1986. Features of target cell lysis by class I and class II MHC-restricted cytolytic T lymphocytes. J. Immunol. 137:3639–3643 [PubMed] [Google Scholar]
- 7.Marshall NB, Swain SL. 2011. Cytotoxic CD4 T cells in antiviral immunity. J. Biomed. Biotechnol. 2011:954602. 10.1155/2011/954602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Appay V, Zaunders JJ, Papagno L, Sutton J, Jaramillo A, Waters A, Easterbrook P, Grey P, Smith D, McMichael AJ, Cooper DA, Rowland-Jones SL, Kelleher AD. 2002. Characterization of CD4(+) CTLs ex vivo. J. Immunol. 168:5954–5958 [DOI] [PubMed] [Google Scholar]
- 9.Brown DM, Lee S, Garcia-Hernandez Mde L, Swain SL. 2012. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J. Virol. 86:6792–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang M, Siciliano NA, Hersperger AR, Roscoe F, Hu A, Ma X, Shamsedeen AR, Eisenlohr LC, Sigal LJ. 2012. Perforin-dependent CD4+ T-cell cytotoxicity contributes to control a murine poxvirus infection. Proc. Natl. Acad. Sci. U. S. A. 109:9983–9988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA, Restifo NP, Allison JP. 2010. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J. Exp. Med. 207:637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grazia TJ, Plenter RJ, Weber SM, Lepper HM, Victorino F, Zamora MR, Pietra BA, Gill RG. 2010. Acute cardiac allograft rejection by directly cytotoxic CD4 T cells: parallel requirements for Fas and perforin. Transplantation 89:33–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown DM, Kamperschroer C, Dilzer AM, Roberts DM, Swain SL. 2009. IL-2 and antigen dose differentially regulate perforin- and FasL-mediated cytolytic activity in antigen specific CD4+ T cells. Cell. Immunol. 257:69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirschhorn-Cymerman D, Budhu S, Kitano S, Liu C, Zhao F, Zhong H, Lesokhin AM, Avogadri-Connors F, Yuan J, Li Y, Houghton AN, Merghoub T, Wolchok JD. 2012. Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J. Exp. Med. 209:2113–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qui HZ, Hagymasi AT, Bandyopadhyay S, St Rose MC, Ramanarasimhaiah R, Menoret A, Mittler RS, Gordon SM, Reiner SL, Vella AT, Adler AJ. 2011. CD134 plus CD137 dual costimulation induces Eomesodermin in CD4 T cells to program cytotoxic Th1 differentiation. J. Immunol. 187:3555–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown DM, Dilzer AM, Meents DL, Swain SL. 2006. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J. Immunol. 177:2888–2898 [DOI] [PubMed] [Google Scholar]
- 17.McKinstry KK, Strutt TM, Kuang Y, Brown DM, Sell S, Dutton RW, Swain SL. 2012. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J. Clin. Invest. 122:2847–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Dodd H, Moser EK, Sharma R, Braciale TJ. 2011. CD4+ T cell help and innate-derived IL-27 induce Blimp-1-dependent IL-10 production by antiviral CTLs. Nat. Immunol. 12:327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pham D, Vincentz JW, Firulli AB, Kaplan MH. 2012. Twist1 regulates Ifng expression in Th1 cells by interfering with Runx3 function. J. Immunol. 189:832–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun J, Madan R, Karp CL, Braciale TJ. 2009. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat. Med. 15:277–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. 2003. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods 281:65–78 [DOI] [PubMed] [Google Scholar]
- 22.Intlekofer AM, Banerjee A, Takemoto N, Gordon SM, Dejong CS, Shin H, Hunter CA, Wherry EJ, Lindsten T, Reiner SL. 2008. Anomalous type 17 response to viral infection by CD8+ T cells lacking T-bet and eomesodermin. Science 321:408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thieu VT, Yu Q, Chang HC, Yeh N, Nguyen ET, Sehra S, Kaplan MH. 2008. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity 29:679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. 2010. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity 32:91–103 [DOI] [PubMed] [Google Scholar]
- 25.Glimcher LH, Townsend MJ, Sullivan BM, Lord GM. 2004. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat. Rev. Immunol. 4:900–911 [DOI] [PubMed] [Google Scholar]
- 26.Jewell NA, Vaghefi N, Mertz SE, Akter P, Peebles RS, Jr, Bakaletz LO, Durbin RK, Flano E, Durbin JE. 2007. Differential type I interferon induction by respiratory syncytial virus and influenza A virus in vivo. J. Virol. 81:9790–9800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shahangian A, Chow EK, Tian X, Kang JR, Ghaffari A, Liu SY, Belperio JA, Cheng G, Deng JC. 2009. Type I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J. Clin. Invest. 119:1910–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, Xu XN. 2012. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 18:274–280 [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Nikrad MP, Phang T, Gao B, Alford T, Ito Y, Edeen K, Travanty EA, Kosmider B, Hartshorn K, Mason RJ. 2011. Innate immune response to influenza A virus in differentiated human alveolar type II cells. Am. J. Respir. Cell. Mol. Biol. 45:582–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gereke M, Jung S, Buer J, Bruder D. 2009. Alveolar type II epithelial cells present antigen to CD4(+) T cells and induce Foxp3(+) regulatory T cells. Am. J. Respir. Crit. Care Med. 179:344–355 [DOI] [PubMed] [Google Scholar]
- 31.Aldridge JR, Jr, Moseley CE, Boltz DA, Negovetich NJ, Reynolds C, Franks J, Brown SA, Doherty PC, Webster RG, Thomas PG. 2009. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc. Natl. Acad. Sci. U. S. A. 106:5306–5311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herold S, Steinmueller M, von Wulffen W, Cakarova L, Pinto R, Pleschka S, Mack M, Kuziel WA, Corazza N, Brunner T, Seeger W, Lohmeyer J. 2008. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J. Exp. Med. 205:3065–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD. 2008. CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. J. Immunol. 180:2562–2572 [DOI] [PubMed] [Google Scholar]
- 34.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 32:79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kallies A, Xin A, Belz GT, Nutt SL. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity 31:283–295 [DOI] [PubMed] [Google Scholar]
- 36.Shin H, Blackburn SD, Intlekofer AM, Kao C, Angelosanto JM, Reiner SL, Wherry EJ. 2009. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity 31:309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cimmino L, Martins GA, Liao J, Magnusdottir E, Grunig G, Perez RK, Calame KL. 2008. Blimp-1 attenuates Th1 differentiation by repression of ifng, tbx21, and bcl6 gene expression. J. Immunol. 181:2338–2347 [DOI] [PubMed] [Google Scholar]