Abstract
Rabies virus (RABV) causes a fatal zoonotic encephalitis. Disease symptoms require replication and spread of the virus within neuronal cells; however, in infected animals as well as in cell culture the virus replicates in a broad range of cell types. Here we use a single-cycle RABV and a recombinant vesicular stomatitis virus (rVSV) in which the glycoprotein (G) was replaced with that of RABV (rVSV RABV G) to examine RABV uptake into the African green monkey kidney cell line BS-C-1. Combining biochemical studies and real-time spinning-disk confocal fluorescence microscopy, we show that the predominant entry pathway of RABV particles into BS-C-1 cells is clathrin dependent. Viral particles enter cells in pits with elongated structures and incomplete clathrin coats which depend upon actin to complete the internalization process. By measuring the time of internalization and the abundance of the clathrin adaptor protein AP2, we further show that the pits that internalize RABV particles are similar to those that internalize VSV particles. Pharmacological perturbations of dynamin or of actin polymerization inhibit productive infection, linking our observations on particle uptake with viral infectivity. This work extends to RABV particles the finding that clathrin-mediated endocytosis of rhabdoviruses proceeds through incompletely coated pits which depend upon actin.
INTRODUCTION
Rabies virus (RABV) is the prototypical member of the zoonotic lyssavirus genus responsible for fatal encephalitis in animals and humans. A single-stranded negative-sense RNA virus, RABV encases its RNA genome in a bullet-shaped, enveloped particle that incorporates a single surface glycoprotein (G). RABV G mediates all internalization steps from cell binding to membrane fusion. In addition, G is a major determinant of RABV neurotropism (1). Conjugation or pseudotyping with the ectodomain of pathogenic RABV G or peptides derived from receptor-binding regions allows retargeting of biologically active molecules to the central nervous system (CNS) for drug delivery or as neurotracers (2, 3). Since a large part of RABV pathogenesis is reliant on the virus garnering access to neurons and the CNS, G is also a determining factor in RABV virulence. The pathogenicity of attenuated strains can be effectively increased by replacing the glycoprotein with one from a neurotropic, virulent strain (4).
Like other rhabdoviruses, RABV gains access to the cellular interior by endocytosis and subsequent low pH-dependent fusion (5–7). Electron micrographs of viral particles in vesicles with electron-dense coats suggest that clathrin-coated pits mediate the uptake of RABV in both neuronal and nonneuronal cells (8, 9). However, static images cannot inform on the fate of such particles or the relevance of these interactions for subsequent infection. High-resolution live-imaging techniques permit tracking of viral uptake into coated pits (10–15). Fluorescence tagging of coated-pit components and quantitative analysis methods have revealed differences for the pits engaging fluorescently tagged viral particles (11, 12). In particular, vesicular stomatitis virus (VSV) particles are internalized through partially coated clathrin pits that require actin for the completion of envelopment (11). The morphology of the particle is a key determinant of actin dependence, since a truncated, defective interfering particle of VSV, DI-T, does not require actin polymerization (12). This observation also extends to other viruses with dimensions compatible with canonical coated pits, as clathrin-dependent uptake of the 60-nm-diameter parvovirus is also actin independent (10, 12).
In the present study, we employed biochemical and high-resolution imaging approaches to study RABV internalization. For this purpose, we used a recombinant VSV (rVSV) expressing RABV G (rVSV RABV G) that mimics the morphology of authentic RABV and allows experimentation at biosafety level 2. To establish rVSV RABV G as a surrogate virus, we compared its entry behavior with that of a “single-cycle” version of RABV which lacks a copy of the G gene and is amplified in cells that express RABV G (rRABV ΔG). We show that, like that of VSV, the predominant internalization route of RABV into BS-C-1 cells is through pits that are partially coated with clathrin and depend upon actin for internalization. The kinetics of internalization of RABV particles are also indistinguishable from those of VSV in the time between particle attachment to cells and association of the particles with the clathrin machinery and in the time of clathrin-dependent uptake.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney BS-C-1 cells (ATCC CCL-26; American Type Culture Collection, Manassas, VA), either the wild type or a line stably expressing the σ2 subunit of AP2 fused to enhanced green fluorescent protein (eGFP) (AP2-eGFP) (16), were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Tissue Culture Biologicals, Tulare, CA). Recombinant G deletion RABV strain SAD L16 expressing eGFP (rRABV ΔG) was a kind gift of E. M. Callaway (17). rRABV ΔG stocks were produced by propagation of the virus in 293T cells stably expressing SAD B19 RABV G (also a gift of E. M. Callaway). Once amplified, rRABV ΔG was purified using the same protocol as for recombinant VSV. Titer was estimated by infecting noncomplementing 293T cells with serial dilutions of purified rRABV ΔG and counting eGFP-expressing infected cells at 30 hours postinfection (hpi) by flow cytometry. rVSV eGFP, rVSV RABV G eGFP (rVSV RABV G) (18), and a recombinant VSV expressing the Ebola virus glycoprotein, rVSV EboV GP eGFP (rVSV EboV GP) (19), were amplified, purified, and maintained as previously described (11). rVSV eGFP was generated by amplification of the eGFP open reading frame (ORF) and insertion into the Xho and MscI sites of pVSV1(+)41 (20). The resulting virus was recovered as described previously (21).
Protein composition of purified virions.
To separate the viral proteins, we subjected purified virions to SDS-PAGE using 10% polyacrylamide (wt/vol) and 0.13% (wt/vol) bis-acrylamide and visualized them by staining with Coomassie blue. Relative amounts of N or G protein were established using ImageJ (U.S. National Institutes of Health, Bethesda, Maryland; http://rsb.info.nih.gov/ij/).
Inhibitors.
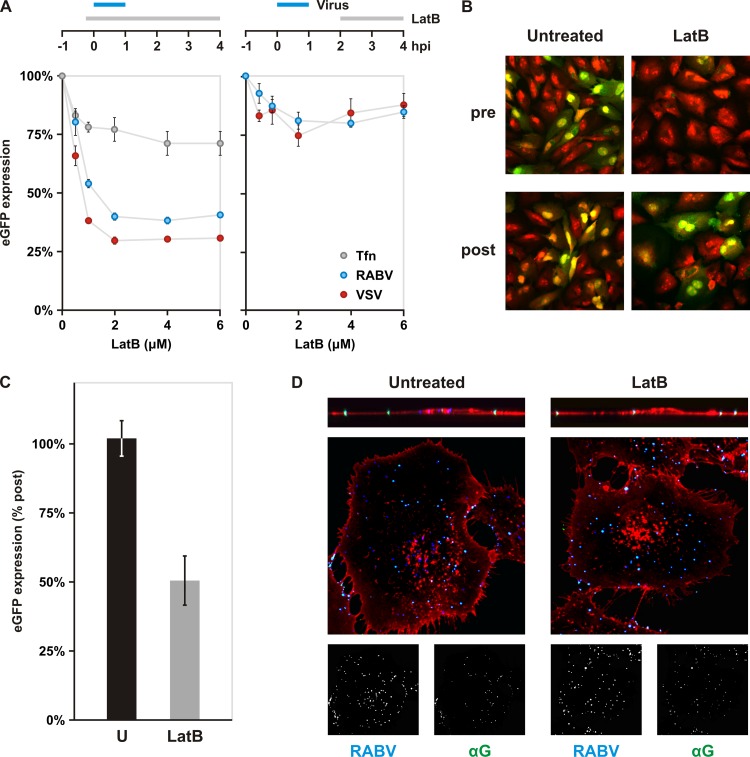
The following chemicals were administered at the listed concentrations: 0.1 μM bafilomycin A1 (BAF A1; Calbiochem, Millipore); 100 μM dynasore; and 25 μM 5-(N-ethyl-N-isopropyl)amiloride (EIPA; Sigma-Aldrich). Latrunculin B (LatB; Sigma-Aldrich) concentrations ranged from 0.5 to 6 μM, as illustrated (see Fig. 7). One micromolar LatB was administered for internalization assays.
Fig 7.
Impact of actin depolymerization on rVSV RABV G internalization. (A) Impact of chemical depolymerization of actin on rVSV RABV G (RABV) and rVSV eGFP (VSV) infections. BS-C-1 cells were treated with various concentrations of latrunculin B (LatB) to inhibit polymerization of actin. As shown in the schematic, drug was added either 10 min prior to a 1-h inoculation with virus or at 2 hpi and maintained until analysis. eGFP expression was assayed by cytofluorimetry at 4 hpi. rVSV eGFP is shown in red and rVSV RABV G in blue. Uptake of fluorescent transferrin (Tfn, gray) is included as a control for general disruption of clathrin-dependent endocytosis. Cells were pretreated with LatB 10 min prior to Tfn addition. Cells were incubated with Tfn for 7 min. Following a wash with acid buffer to remove surface-bound Tfn, intracellular Tfn levels were measured by flow cytometry. (B) Effect of actin depolymerization on rRABV ΔG infection. BS-C-1 cells were treated with 6 μM LatB prior to inoculation (pre) or at 2 hpi (post). Expression of eGFP (green) from viral genomes was detected by confocal microscopy at 25 hpi. Cells were stained with propidium iodide (red). (C) Quantification of LatB effect on rRABV ΔG infection. BS-C-1 cells treated as for panel B were collected and analyzed for eGFP expression by flow cytometry. (D) Confocal microscopy of internalization assay in cells. Above the merged images are single-plane z-stack cross sections. Cells pretreated as indicated were inoculated with AF647-labeled rVSV RABV G (RABV), fixed, and stained with antibody against RABV G in the absence of permeabilization. The LatB concentration used was 1 μM. RABV (blue) and αG (green) signals are shown separately in inlays below the merged images. WGA is shown in red. Singly labeled, internalized particles appear blue; dually labeled, surface-bound particles appear cyan.
Nucleic acid transfection.
To visualize clathrin and actin dynamics simultaneously, we cotransfected rat mCherry-LCa (constructed as described for tomato-LCa [22]) with mouse actin-eGFP into 90%-confluent BS-C-1 cells in 6-well plates. Plasmid DNA was introduced into the cells using FuGene HD (Roche Diagnostics, Indianapolis, IN) according to the manufacturer's instructions, using a ratio of 3 μg DNA to 5 μl FuGENE HD in 100 μl Opti-MEM. Following overnight incubation at 37°C, cells were reseeded onto 25-mm coverslips at 25% confluence. Cells were imaged approximately 5 h later, after complete adherence to the coverslips was verified by visual inspection.
Electron microscopy (EM).
To visualize viral morphology, we deposited purified rVSV eGFP or rVSV RABV G particles onto carbon-coated copper grids and stained them with 2% phosphotungstic acid (wt/vol) in H2O (pH 7.5). To visualize viral particles in clathrin-coated pits, we inoculated BS-C-1 cells at a multiplicity of infection (MOI) of 1,000 for 15 min at 37°C. Samples were then processed for ultrathin sectioning as previously described (11, 23). Virus particles and ultrathin sections of cells were viewed using a Tecnai G2 Spirit BioTWIN transmission electron microscope (FEI, Hillsboro, OR).
Dye conjugation to virus particles.
Viral particles were labeled with 40 μg ml−1 Alexa Fluor (AF647) succinyl esters (Molecular Probes, Invitrogen, Eugene, OR) as previously described (11). Titration of virus preparations before and after labeling showed that dye conjugation had a negligible effect on infectivity.
Infectivity studies.
Cytofluorimetry experiments were carried out in 24-well plates and epifluorescence microscopy experiments in black, clear-bottomed 96-well microplates (Corning, Tewksbury, MA). BS-C-1 cells were treated either 15 min prior to infection or at 2 hpi with DMEM plus inhibitors as indicated on the figures and in the figure legends. rRABV ΔG inoculations were incubated for 2 h at 37°C with an estimated MOI of 0.5, whereas rVSV RABV G inoculations were incubated with an MOI of 0.5 for 1 h at 37°C. Following inoculation, cells were washed and media were replaced as indicated. Cells were processed for epifluorescence microscopy or cytofluorimetry at 4 to 6 hpi for VSV-RABV or 25 hpi for rRABV ΔG. LatB was removed at 12 hpi to prevent cytotoxicity. Cells were washed three times with DMEM and media was replaced with DMEM supplemented with 10% FBS. For epifluorescence microscopy, cells were washed twice with phosphate-buffered saline (PBS) and fixed for 15 min with 4% paraformaldehyde (PFA). The fixed cells were washed with PBS and stained with DAPI (4′,6-diamidino-2-phenylindole) in the presence of 0.1% Triton X-100 for 30 min at 37°C. rVSV eGFP-infected cells were imaged using a CellWorX automated microscope (Applied Precision, Issaquah, WA); rRABV SAD B19 ΔG-infected samples were imaged using an ImageXpress Micro automated microscope (Molecular Devices, Sunnyvale, CA). For cytofluorimetry, cells were washed with PBS and harvested using 0.5 mM EDTA in PBS. Mean fluorescence measurements were obtained using a modified FACSCalibur (Cytek Development, Fremont, CA) instrument and analyzed using FlowJo (Tree Star Industries, Ashland, OR).
Uncoating assay.
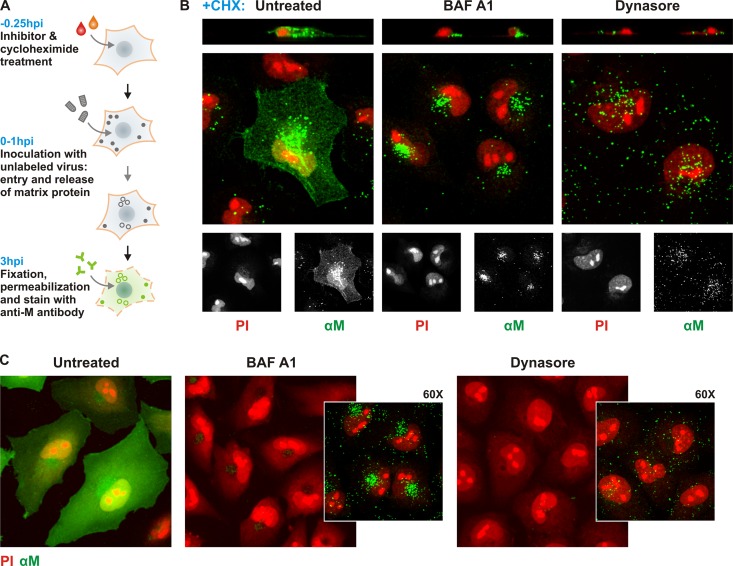
Confluent BS-C-1 cells seeded onto 12-mm coverslips were exposed for 15 min to DMEM containing 5 μg ml−1 cycloheximide (CHX) alone or with an entry inhibitor as indicated (see Fig. 3). Cells were then inoculated with rVSV RABV G at an MOI of 1,000. Following 1 h of incubation at 37°C, unabsorbed virus was removed by washing with fresh media. The treatment media were replaced, and cells were incubated for 2 h at 37°C. Note that CHX and the indicated entry inhibitors were maintained on cells throughout the inoculation as well as during the subsequent 2 h of incubation. Cells were washed with PBS and fixed with 4% PFA for 15 min. Following permeabilization with 0.1% Triton X-100, cells were exposed to mouse 23H12 anti-M antibody (gift of D. Lyles) (24, 25) and 1:3,000 propidium iodide (PI; Sigma-Aldrich, St. Louis, MO) and an AF488-labeled goat anti-mouse secondary antibody. Coverslips were mounted onto slides using ProLong Gold antifade mounting media (Molecular Probes). z stacks were collected at 0.3-μm intervals. Images were analyzed and processed using ImageJ, maintaining contrast adjustments constant across samples for the AF488-conjugated secondary-antibody signal.
Fig 3.
Absence of M protein release and expression in cells treated with BAF A1 or dynasore. (A) Schematic of the uncoating assay protocol. Release of M protein is monitored as an indication of viral fusion. Fifteen minutes prior to infection, cells were treated with 5 uM cycloheximide (CHX) to prevent de novo synthesis of viral proteins. CHX was administered alone or in conjuction with entry inhibitor, 0.1 uM BAF A1, or 100 uM dynasore, as indicated. Cells were inoculated with rVSV RABV G virus (MOI = 500) for 1 h. At 3 hpi, cells were fixed, permeabilized (indicated by dashed cell outline), and stained with antibody against VSV M (αM; green) and propidium iodide (PI; red). External, surface-bound virus is shown as filled circles, internalized virus as open circles. Anti-M antibody stained both diffuse M in the cell and tightly packed M in individual particles. (B) Confocal microscopy of M release in cells treated with entry inhibitors and CHX. Above the merged images are single-plane z-stack cross sections. PI (red) and αM (green) signals are shown separately below the merged images. (C) Confocal microscopy of M localization in the absence of CHX treatment. Cells were stained with PI (red) and αM (green). Large panels were imaged with a 40× lens objective; insets for BAF A1- and dynasore-treated conditions were imaged with a 60× lens objective, allowing visualization of aggregates of unfused viral particles.
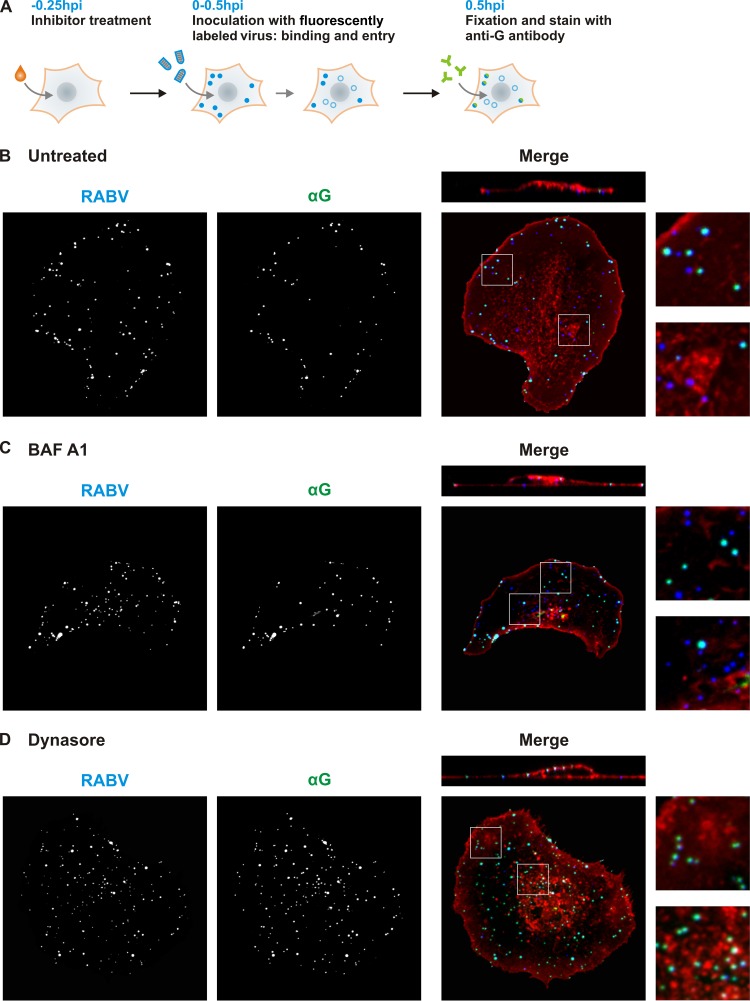
Internalization assay.
Twenty-five to fifty percent-confluent BS-C-1 cells seeded onto 12-mm coverslips were treated with chemicals as indicated (see Fig. 4). AF647-labeled rVSV RABV G was added at an MOI of 100 and incubated for 30 min at 37°C. Cells were washed with PBS, fixed with 4% PFA, and exposed to anti-RABV G antibody (1:500; Millipore, Billerica, MA), which was detected by a secondary goat anti-mouse AF488 (1:500; Molecular Probes). To stain the cell membrane, we also exposed cells to 10 μg ml−1 AF594-labeled wheat germ agglutinin (WGA; Molecular Probes). Coverslips were mounted onto slides using ProLong Gold antifade mounting media (Molecular Probes). Samples were imaged by spinning-disk confocal microscopy with z stacks collected at 0.3-μm intervals. Stacks were processed using ImageJ (National Institutes of Health, Bethesda, MD; http://imagej.nih.gov/ij/). For increased clarity, z-stack maximal projections were processed to exclude coverslip-bound viruses. The WGA signal was used to generate a mask matching the outline of the cell(s). The area outside the border of the cell was replaced with a black background. Exposure and contrast adjustments were kept constant across samples throughout image acquisition and processing; WGA contrast and brightness were optimized to aid visualization of the cell boundaries.
Fig 4.
Dynasore blocks internalization at the plasma membrane. (A) Schematic of internalization assay. Fifteen minutes prior to infection, cells were treated with DMEM only (B), 0.1 uM BAF A1 (C), or 100 uM dynasore (D). AF647-labeled rVSV RABV G virus (MOI = 50) was administered to cells for 30 min. During incubation, surface-bound virus (filled blue circles) internalizes and travels to the perinuclear region (open blue circles). Cells were fixed and stained with antibody against RABV G (αG) in the absence of permeabilization, which exclusively dually labels external particles (blue-green filled circles). Internal particles are protected from αG staining and remain singly labeled (open blue circles). WGA staining was used to detect cell surfaces. (B, C, and D) Confocal microscopy of internalization assay in cells treated as listed above. AF647-labeled rVSV RABV G (RABV) and αG signals are shown individually. In the merged panels, the RABV signal is shown in blue, αG in green, and WGA in red. Singly labeled internal particles appear blue and dually labeled external particles appear cyan. Above the merged images are single-plane z-stack cross sections. Smaller panels (right) show magnified portions of the cells corresponding to the white squares in the merged panels.
Transferrin uptake.
For cytofluorimetry experiments, BS-C-1 cells were seeded onto 24-well plates to be confluent the next day. For fixed confocal microscopy, BS-C-1 cells were seeded onto 12-mm coverslips to be 50% confluent the next day for ease of imaging. BS-C-1 cells were pretreated with DMEM plus LatB or DMEM alone for 5 to 10 min. AF488-labeled transferrin (Tfn; Molecular Probes) was added to the media at a final concentration of 20 μg ml−1, and the cultures were incubated for 7 min at 37°C. On ice, cells were rapidly washed with acid solution (PBS, 0.5 M NaCl, 0.2 M acetic acid) and 3 times with PBS. Tfn uptake was assessed by spinning-disk confocal cytofluorimetry. Cells intended for analysis by flow cytometry were harvested and analyzed as described above.
Fixed-cell imaging.
Slides were imaged using a Marianas system (Intelligent Imaging Innovations) based on a Zeiss observer microscope (Carl Zeiss MicroImaging, Thornwood, NY) outfitted with a CSU-22 spinning-disk confocal unit (Yokogawa Electric Corporation, Tokyo, Japan) and a 40× (Plan-Neofluar, NA 1.3; Carl Zeiss MicroImaging) or 63× (Plan-Apochromat, NA 1.4; Carl Zeiss Microimaging) objective lens. Excitation wavelengths were 491 nm for AF488, 561 nm for AF594, and 660 nm for AF647. For three-dimensional acquisitions, the vertical position was manipulated in 0.3-μm increments using a PZ-2000 automated stage (Applied Scientific Instrumentation, Eugene, OR). Images were collected using a Photometrics Cascade II electron multiplication camera (Photometrics, Tucson, AZ). SlideBook versions 4 and 5 (Intelligent Imaging Innovations, Denver, CO) were used to command the hardware devices and visualize the acquired data.
Live-cell imaging.
Live single-particle tracking on AP2-eGFP-expressing BS-C-1 cells was conducted as described previously, collecting images at 3-s intervals over a period of 10 min per cell (11). Slidebook 4.2.13 and 5.0.0.2 (Intelligent Imaging Innovations) were used to command the hardware devices and visualize the acquired data. Prior to imaging, Alexa-labeled virus was centrifuged briefly on a tabletop microcentrifuge to remove aggregates. Cells were inoculated with a volume of virus calibrated to result in attachment and internalization of approximately 100 particles per cell within 20 min (∼1 × 109 PFU of rVSV RABV G). Movies were generated by exporting time-lapse TIFF files from Slidebook and compiling the images into a single AVI file using ImageJ. All contrast editing, cropping, and scaling were performed using ImageJ.
Analysis of time series.
Image analysis was performed as previously described (11, 12) with the following modifications. Images were exported from Slidebook 4.2.13 and 5.0.0.2, cropped using ImageJ, and analyzed using a previously described automated image analysis application (IMAB) within MATLAB (MathWorks, Natick, MA) (22). As previously described (11, 12, 22), only events where single particles docked onto cells and were seen to undergo internalization were included in our analysis. Virus aggregates and predocked particles were excluded from the analysis. Internalization events were confirmed by observation of rapid directional movement of viral particles toward the perinuclear region. For comparison, 50 pits lacking virus particles within the time frame of virus uptake were also analyzed to provide baseline measurements of coat lifetime and intensity.
RESULTS
Recombinant VSV expressing RABV G.
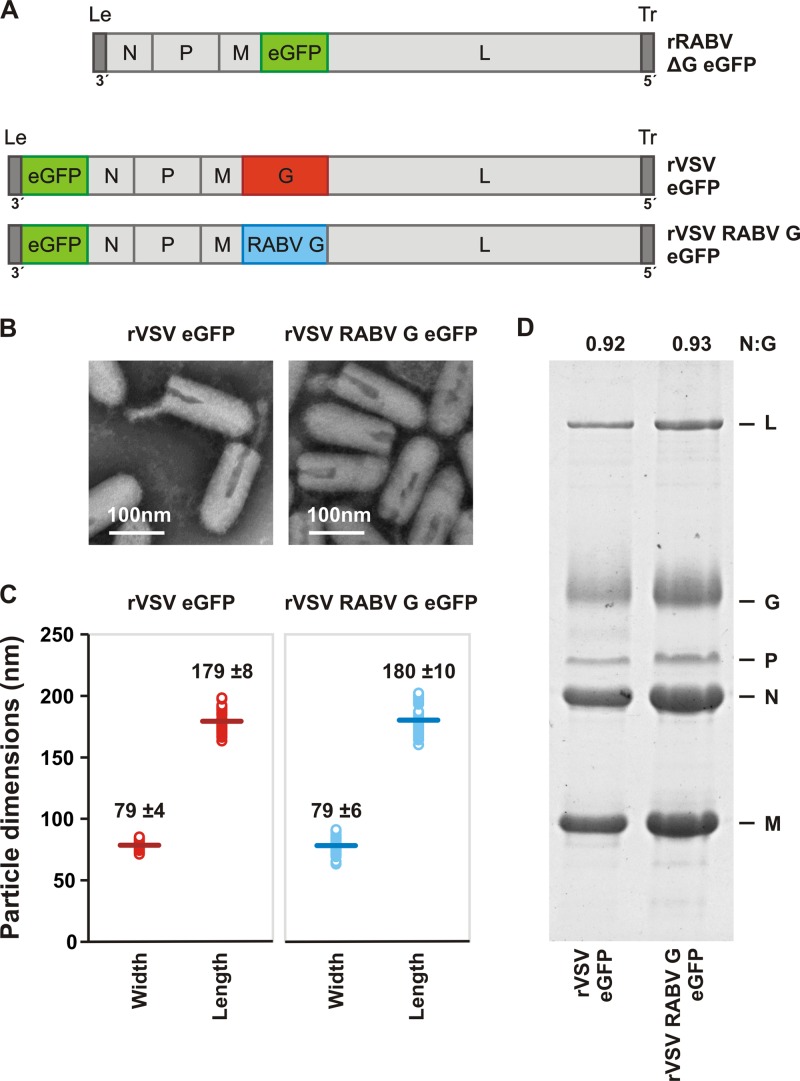
Recombinant VSVs in which the glycoproteins from Ebola, Marburg, and Lassa fever viruses replace the endogenous glycoprotein (G) serve as useful surrogates to study the uptake pathways of these viruses (18, 19, 26) under biosafety level 2 conditions. We adapted such a strategy to permit the study of RABV infection by replacing VSV G with that of RABV SAD B19 (rVSV RABV G) (Fig. 1A) (18). As expected, rVSV RABV G retains the morphological features of authentic RABV: rVSV RABV G virions are bullet shaped and measure on average 180 nm in length and 80 nm in width (Fig. 1B and C). These dimensions are typical for RABV and also match those of the parental VSV strain rVSV eGFP (Fig. 1B and C) (27, 28). Purified rVSV RABV G virions incorporate RABV G to an extent comparable to VSV G incorporation into particles (Fig. 1D). Collectively, these data show that the rVSV RABV G particles are structurally indistinct from those of VSV except that they now contain RABV G on their surfaces and thus appear externally as RABV.
Fig 1.
Characterization of recombinant VSV expressing RABV G. (A) Genomic structures of rRABV ΔG eGFP, rVSV eGFP, and rVSV RABV G eGFP. The single-stranded, negative-sense RNA genomes are shown in a 3′ to 5′ orientation. N, nucleocapsid gene; P, phosphoprotein gene; M, matrix gene; G, glycoprotein gene; L, large polymerase gene. Noncoding genomic leader (Le) and trailer (Tr) regions serve as promoters for RNA synthesis and genomic RNA encapsidation, respectively. All viruses express the eGFP gene (green) as a marker for infection. The glycoprotein open reading frame of rRABV ΔG eGFP was replaced with that of eGFP. To produce infectious virus, rRABV ΔG was grown in complementing cells expressing SAD B19 G. In rVSV RABV G eGFP, the wild-type VSV G (1,535 nt) was replaced with RABV G (1,574 nt) from the SAD B19 strain. (B) Electron micrographs of rVSV eGFP and rVSV RABV G eGFP viral particles demonstrate morphological homogeneity. Particles were negatively stained with 1% PTA. (C) Dimensions of individual viral particles measured from micrographs like those shown in panel B. Each open circle represents the measurement for a single virion. A line denotes the mean (± the standard deviation [SD]; n = 50) for each population, the value of which is provided. (D) SDS-PAGE analysis of purified virions. Viral proteins were stained with Coomassie blue. The ratio of N to G was quantified using ImageJ to estimate the average glycoprotein density in each particle population.
Productive infection of rVSV RABV G occurs through a clathrin-dependent endocytic pathway and requires endosomal acidification.
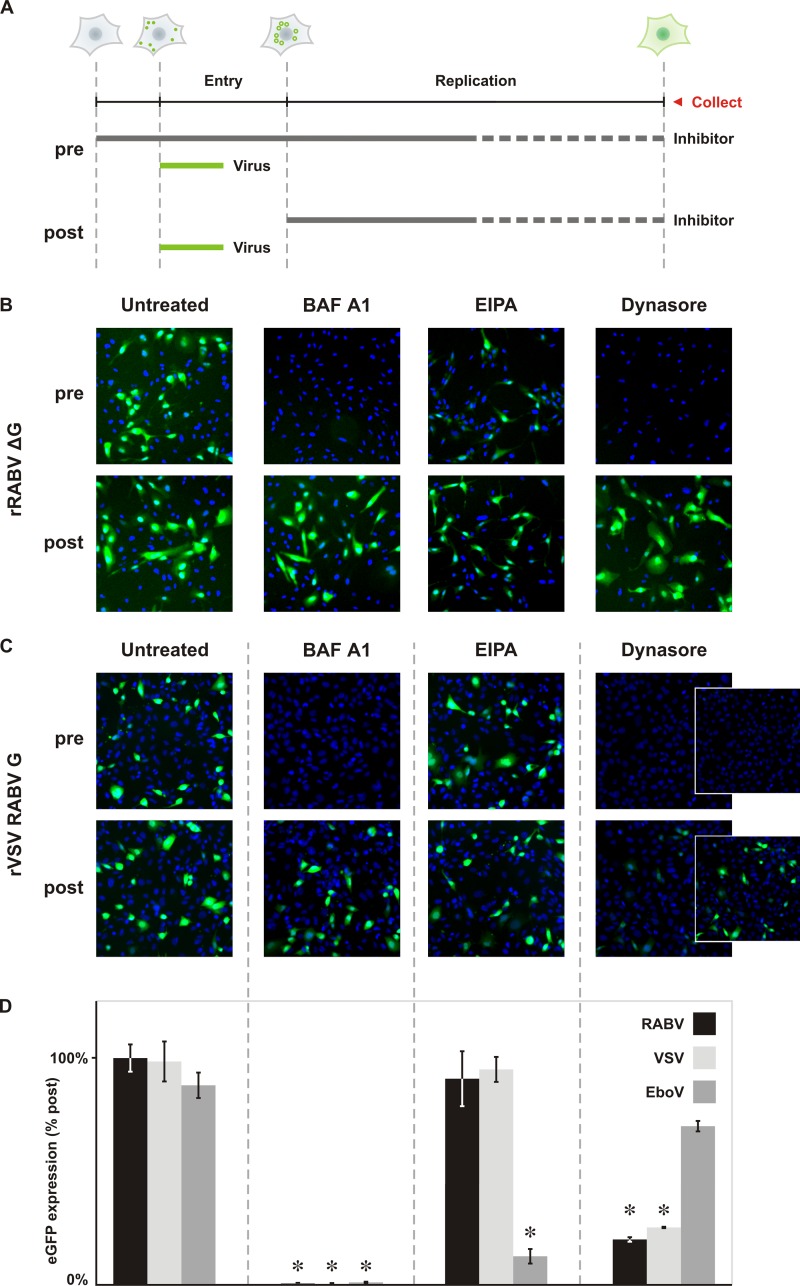
To determine whether rVSV RABV G serves as a useful surrogate to study RABV entry and define the route of infection, we compared its sensitivity to inhibitors of different endocytic mechanisms with that of a recombinant single-cycle RABV virus, rRABV ΔG. We tested bafilomycin A1 (BAF A1), an inhibitor of the H+ ATPase pump which by blocking endosomal acidification eliminates the trigger of conformational rearrangements in G necessary for membrane fusion; amiloride (EIPA), an inhibitor of macropinocytosis (29); and dynasore, an inhibitor of dynamin required for the scission step of clathrin-mediated endocytosis (30). To discriminate between the effect of inhibitors on entry versus postentry steps of viral replication, we compared their effects when added prior to or 2 h following inoculation with virus (Fig. 2A). For controls, we monitored the impact of these inhibitors on infection with rVSV eGFP, a virus that depends upon clathrin-dependent endocytosis and endosome acidification, and a recombinant VSV expressing the Ebola virus glycoprotein (rVSV EboV GP), which requires macropinocytic uptake (Fig. 2D). For each virus, infection was measured by the expression of eGFP detected by fluorescence microscopy (Fig. 2B and C) or cytofluorimetry (Fig. 2D).
Fig 2.
Dynamin and endosomal acidification are required for productive RABV G-dependent infection. (A) Schematic of the methods employed for panels B and C. A graphical representation of a cell at each time point indicates the stage of infection. Circles represent viral particles, surface bound (filled) or internalized (hollow). Expression of the reporter gene, eGFP, is shown as a diffuse green in the cytoplasm. BS-C-1 cells were inoculated with virus (MOI = 0.5) and assayed for infection by detection of the eGFP reporter gene either by autoscope microscopy or cytofluorimetry at the collection point (25 hpi for rRABV ΔG; 4 to 6 hpi for rVSV RABV G). The effect of inhibitor on viral infection was assayed by treating cells with drug prior (pre) or following (post) inoculation with virus. Comparison of pretreated and posttreated samples enables determination of the effect of each inhibitor on viral entry. (B) Fluorescence microscopy of single-cycle rRABV ΔG infection following treatment with inhibitor. The H+ pump inhibitor bafilomycin A1 (BAF A1; 0.1 μM), macropinocytosis inhibitor amiloride (EIPA; 25 μM), and dynamin inhibitor dynasore (100 μM) were tested for their abilities to impact RABV infection. rRABV ΔG infection was detected via expression of the eGFP reporter gene (green). Cell nuclei were labeled with DAPI (blue). (C) Fluorescence microscopy of rVSV RABV G infection following treatment with inhibitors. rVSV RABV G infection is also detected as expression of the eGFP reporter gene (green). Cell nuclei were labeled with DAPI (blue). Images were collected using the same exposure and processed to the same brightness and contrast settings. Dynasore has a moderate effect on the rate of viral-gene expression; as a result, the brightness and contrast were readjusted in the dynasore insets to highlight lower-intensity infected cells. (D) Cytofluorimetric analysis of infection. Total eGFP expression following pretreatment with drug was calculated from mean fluorescence intensites using FlowJo software and expressed as percentages relative to that of controls treated following inoculation (% post). rVSV eGFP (VSV) and rVSV EboV GP (EboV) served as prototypical clathrin-dependent and macropinosome-dependent pathogens, respectively. Statistically significant differences are demarcated by asterisks (Student's t test; P < 0.01).
The addition of BAF A1 or dynasore prior to inoculation blocked rRABV ΔG infection, whereas EIPA had no effect. None of the inhibitors had a significant effect following postentry addition. This result indicates that rRABV ΔG entry is dependent on dynamin and is likely clathrin mediated. As expected, productive infection also requires endosome acidification. rVSV RABV G (Fig. 2C) shared the same sensitivity profile as rRABV ΔG (Fig. 2B), supporting that this recombinant virus serves as a useful surrogate to study rabies virus entry. To quantitate the reduction in infection, we used cytofluorimetry (Fig. 2D). Pretreatment of cells with BAF A1 reduced rVSV RABV G infection to 1% of that of postinoculation-treated controls. Pretreatment with dynasore reduced rVSV RABV G infection, like VSV infection, to 20% of that of postinoculation-treated controls, and pretreatment with EIPA had a negligible effect on eGFP expression (Fig. 2C and D). By contrast, infection of rVSV EboV GP was strongly inhibited by EIPA but not by dynasore (Fig. 2D). These results support the hypothesis that infection with rVSV RABV G requires dynamin and that macropinocytosis does not play a role in infection. Furthermore, the similarity in sensitivity profiles for rVSV RABV G and VSV is suggestive of a clathrin-dependent route of entry.
BAF A1 and dynasore block RABV infection at separate steps in internalization.
To identify the steps at which BAF A1 and dynasore block infection, we employed separate assays that directly monitor uncoating (Fig. 3) and internalization (Fig. 4) of the virus (31). For uncoating, we detected the release of internal virion matrix (M) protein following successful membrane fusion. Briefly, cells were infected in the presence of the protein synthesis inhibitor cycloheximide so that only the incoming virion proteins were detected. Successful membrane fusion releases the virion contents into the cytoplasm, a step that is visualized by monitoring the diffuse distribution of the input M protein by immunofluorescence (Fig. 3A and B), and in the absence of CHX, infection was established in almost every cell (Fig. 3C). By contrast, intact particles result in high-intensity perinuclear punctate staining, as shown by treatment of cells with BAF A1 in the presence (Fig. 3B) or absence (Fig. 3C) of CHX. This is consistent with the fact that inhibition of the vacuolar ATPase prevents the drop in endosomal pH required for RABV G fusion, leading to the accumulation of intracellular particles. In the presence of dynasore, there was no diffuse cytoplasmic M or perinuclear accumulation of intact particles. Intact particles appeared uniformly dispersed over the presumed cell surface and absent from the cell interior (Fig. 3B and C).
To confirm that dynasore prevents particle internalization at the plasma membrane, we inoculated cells with Alexa Fluor-labeled virions for 30 min and, following fixation, exposed cells to an antibody directed against RABV G in the absence of membrane permeabilization (Fig. 4A). This approach results in dually labeled surface-bound particles and permits their discrimination from singly labeled internalized particles. Following 30 min of inoculation, approximately half of cell-associated particles were internal and singly labeled, resulting in 42% colocalization of the Alexa Fluor signal with the anti-RABV G stain (Fig. 4B). Internal particles were distributed between the perinuclear region and the cell periphery. In the presence of BAF A1, rVSV RABV G particles were internalized into endocytic compartments, resulting in a percentage of colocalization (44%) similar to that of untreated controls (Fig. 4C). By contrast, treatment of cells with dynasore resulted in the accumulation of dually labeled particles distributed evenly on the cell surface (Fig. 4D, 78% colocalization). Those particles remained at the plasma membrane, as detected by WGA staining (Fig. 4D, z cross-section). We conclude that inhibition of dynamin-dependent processes prevents rVSV RABV G uptake at the cell surface.
Kinetics of rVSV RABV G uptake in clathrin-coated pits.
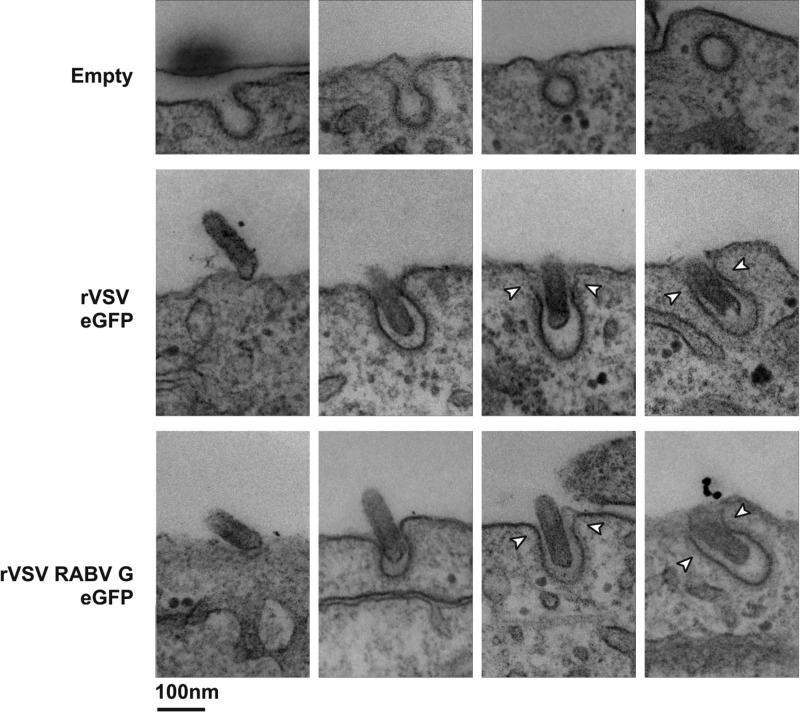
To further understand the steps of clathrin-mediated uptake of rVSV RABV G particles, we visualized single viral particles undergoing internalization by electron microscopy. Clathrin-coated pits are readily identifiable in electron micrographs as invaginations of the plasma membrane surrounded by an electron-dense coat (Fig. 5, first row). Vesicles arising from clathrin-mediated endocytosis are largely spherical and have an average interior diameter of 60 nm. As expected for VSV (11) and as shown here for rVSV RABV G, viral particles are found within elongated vesicles with electron-dense material visible on the spherical end (Fig. 5).
Fig 5.
Visualization of clathrin-dependent uptake of rVSV RABV G by transmission electron microscopy. Electron micrographs show the presumed order of internalization for empty, rVSV eGFP, or rVSV RABV G clathrin-coated pits. BS-C-1 cells were exposed to virus at an MOI of 1,000 for 15 min prior to fixation and processing as outlined in Materials and Methods. Arrowheads highlight sections of the endocytic vesicle lacking electron-dense clathrin.
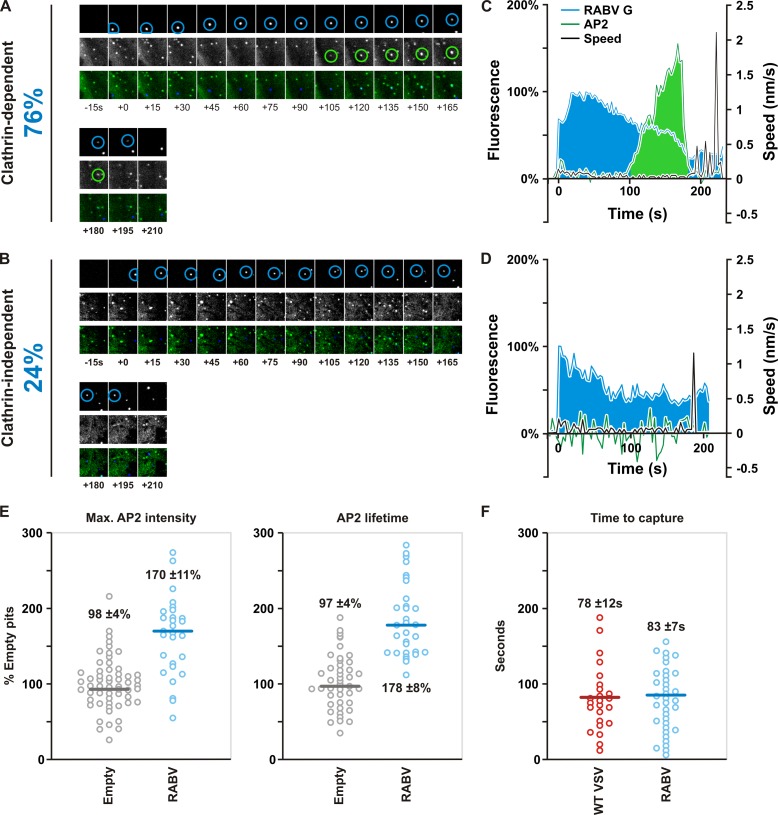
Previously we demonstrated that the clathrin-coated structures that internalize VSV take longer to form than pits lacking virus; they also lack the expected content of clathrin and its adaptor molecule, AP2 (11). To investigate whether pits internalizing rVSV RABV G share similar properties, we monitored the internalization of single viral particles into BS-C-1 cells stably expressing fluorescently tagged AP2 (11, 12). We analyzed particles that displayed the following properties: (i) docking of the particle on the cell surface; (ii) a period of surface association with the cell membrane characterized by limited motility and potential association with AP2; and finally, (iii) sustained, rapid, directed movement toward the perinuclear region. A representative internalization event shows docking and a period of short-range, low-speed movement at the cell surface followed by a steady increase in AP2 (Fig. 6A and C). After reaching peak intensity, the AP2 signal abruptly disappears, and the virus moves rapidly toward the perinuclear region (Fig. 6A and C; see Movie S1 in the supplemental material). This loss of the AP2 signal is indicative of the uncoating of clathrin following scission of the coated pit from the plasma membrane (32). We found that the majority (76%) of rVSV RABV G particles were seen within AP2-containing structures during entry (Fig. 6A). The remaining particles internalized in the absence of a detectable AP2 signal (Fig. 6B and D; Movie S2).
Fig 6.
Live-cell imaging and kinetics analysis of rVSV RABV G internalization. (A) Tile view of images taken from a 10-min time-lapse movie (see Movie S1 in the supplemental material) of rVSV RABV G labeled with AF647 (top, blue) entering a BS-C-1 cell stably expressing AP2-eGFP (middle, green) via clathrin-mediated endocytosis. Particles were imaged at a rate of 20 images min−1. Circles highlight the presence of detectable virus and AP2, respectively. The percentage (at left) refers to the fraction of recorded events that were clathrin dependent (76%; n = 66). (B) Tile view of images taken from a 10-min time-lapse movie (Movie S2) of AF647-labeled rVSV RABV G (top, blue) entering a BS-C-1 cell stably expressing AP2-eGFP (middle, green) in an AP2-independent manner. The percentage (at left) refers to the fraction of recorded events that were clathrin independent (24%; n = 66). (C and D) Graphical representations of the particle behaviors illustrated in panels A and B. AF647-labeled rVSV RABV G fluorescence (RABV G) was plotted as a percentage of its maximum. AP2 was plotted as a percentage of the average maximal fluorescence of empty pits. Particle speed is also plotted on the right y axes. (E) Plots of maximal AP2 intensity and duration during AP2-dependent internalization of rVSV RABV eGFP plotted as a percentage of the average for all measured empty pits. Empty and viral pits are compared. Empty circles refer to individual measurements; bars indicate the median value, which is shown on the plot. For both parameters, the difference between empty and viral pits is statistically significant as determined by Student's t test (P < 10−12). (F) Plot comparing the time between docking and AP2 capture for WT VSV and rVSV RABV G. Empty circles refer to individual measurements; bars indicate the median value, which is shown on the plot. The difference between time to capture of WT VSV and rVSV RABV G is not statistically significant as determined by Student's t test (P > 0.1).
To compare internalization of rVSV RABV G to that of the related rhabdovirus, VSV, we measured the following parameters: maximum AP2 intensity and duration relative to those of empty pits (Fig. 6E) and the time from docking to a cell to association with AP2 relative to the time for WT VSV (Fig. 6F). rVSV RABV G-internalizing pits recruit on average 1.7 times more AP2 than their empty counterparts (median value; n = 66) (Fig. 6E). In addition, the duration of AP2 recruitment was approximately 1.8 times that of a pit lacking virus. These changes mirror those previously measured for the internalization of WT VSV into pits (11, 12). We also found that the time to capture of rVSV RABV G particles by coated pits was indistinguishable from that of VSV (Fig. 6F). Collectively, the fixed and live-cell-imaging data support that RABV particles internalize into incompletely clathrin-coated pits with structures and a lifetime similar to those that internalize VSV.
Actin dependence of RABV entry.
We found previously that VSV also internalizes through partially coated clathrin pits which depend on actin to complete uptake (11). We therefore examined whether RABV entry is also actin dependent. Using the actin-depolymerizing drug latrunculin B (LatB), we disrupted formation of actin structures and monitored the effect on rVSV RABV G infection by cytofluorimetry (Fig. 7A). Inhibition of actin polymerization prior to the addition of virus resulted in a 62% reduction in rVSV RABV G infection, similar to that observed for VSV (Fig. 7A). Treatment of cells with LatB at 2 h postinoculation had a marginal effect on infection of both viruses, confirming that the requirement for actin is specific to entry (Fig. 7A). Inhibition of uptake was also observed for the single-cycle rabies virus, rRABV ΔG (Fig. 7B and C). Consistent with an entry block, viral particles fail to internalize and remain surface bound on cells pretreated with LatB (Fig. 7D). Previous work demonstrated that disruption of actin polymerization does not block coated-pit formation or alter the kinetics of internalization in BS-C-1 cells (11, 33). Accordingly, depolymerization of actin had a negligible effect on transferrin accumulation as detected by cytofluorimetry (Fig. 7A) and confocal microscopy (data not shown). Live-cell imaging of clathrin pits also confirmed that the formation and kinetics of coated pits are largely unchanged in the presence of LatB (see Movie S3 in the supplemental material). These data demonstrate a requirement for actin polymerization during the internalization of VSV and RABV that is not shared by pits that lack virus particles.
We next visualized the association of actin with clathrin-coated pits during rVSV RABV G entry by live confocal microscopy in BS-C-1 cells transiently expressing fluorescently labeled clathrin light chain and actin (see Movie S4 in the supplemental material). No detectable recruitment of actin was observed for pits lacking viral particles. In contrast, when the clathrin machinery was engaged in uptake of rVSV RABV G particles, we observed an accumulation of actin shortly following assembly of the clathrin coat. These results demonstrate that actin recruitment is integral to clathrin-mediated internalization of RABV particles. These data extend our findings on the clathrin-dependent uptake of VSV to another member of the family Rhabdoviridae, RABV, providing further support for the model that the particles are internalized through partially coated pits that require actin for internalization.
DISCUSSION
In this study, we used a combination of infectivity-based and single-particle-tracking approaches to examine how RABV productively enters epithelial cells. We employed a single-cycle rabies virus, rRABV ΔG, and a recombinant VSV in which the endogenous glycoprotein was replaced with that of RABV. We showed that uptake of rVSV RABV G mimics uptake of rRABV ΔG, demonstrating its usefulness as a surrogate for studying RABV entry. We present evidence that RABV internalization into BS-C-1 fibroblasts is primarily clathrin mediated. Furthermore, RABV uptake proceeds through partially coated clathrin pits which require actin for completion of envelopment, a mechanism shared for internalization of the related rhabdovirus VSV. Live-microscopy approaches revealed that the kinetics of internalization of RABV and VSV are indistinguishable. The presence of RABV G on the surfaces of the particles does not impact the kinetics of internalization of particles by clathrin-coated pits or their time to association with clathrin. This work therefore characterizes the clathrin-dependent process of RABV uptake at the plasma membrane and reveals commonalities with the internalization of VSV into epithelial cells. These results also further support our prior finding that the length of a rhabdovirus particle correlates with the lifetime of the coated pit and the involvement of the actin machinery for coated-pit internalization.
rVSV RABV G as a surrogate to study RABV entry.
Particle morphology as well as the identity, number, and abundance of viral surface proteins that bind cell surface attachment factors at the plasma membrane influences viral entry. The rVSV RABV G particle faithfully replicates the surface characteristics of authentic RABV. It incorporates the single glycoprotein efficiently and retains the characteristic bullet shape. Furthermore, its biological behavior during the entry stages of infection is identical to that of a “single-cycle” RABV. Therefore, rVSV RABV G is functionally indistinguishable from an authentic RABV particle and can be used as a surrogate to study viral uptake.
Viral factors that impact the mechanism and actin dependence of envelopment in a clathrin-coated pit.
We show that clathrin-coated pits internalizing rVSV RABV G particles have an altered morphology compared to those lacking virus. The amounts of clathrin and adaptors recruited and the duration of the coating process observed by live confocal imaging as well as the appearance of the nascent coated pit by EM support rVSV RABV G uptake through partially coated pits. In addition, uptake of, and infection with, rVSV RABV G or rRABV ΔG are inhibited by chemicals that block actin polymerization. The related rhabdovirus VSV also internalizes via partially coated clathrin pits and requires actin polymerization (11, 12). The pits are indistinct in their appearance by EM and live-cell imaging, and the uptakes of the viruses follow similar kinetics. These studies also showed that the partially coated pit and the requirement for actin during viral internalization depend upon the size of the particle, as truncated VSV particles enter through fully coated pits and do not require actin polymerization (12). While we did not examine whether truncated RABV particles are taken up through fully clathrin-coated pits and bypass the need for actin polymerization, we predict this to be the case. The fact that both VSV and RABV particles of similar dimensions internalize through similar coated pits lends further support to this prediction.
Recent studies show that the structural adaptability of clathrin-coated pits is an intrinsic property of the host machinery. Supporting this view, the actin dependence of clathrin-mediated endocytosis is induced by artificially tensing the plasma membrane either by exposing cells to hypoosmotic media or by mechanical stretching of the plasma membrane (34). Under conditions of elevated membrane tension, actin is required to provide sufficient force to counteract membrane resistance and constrict the neck of nascent coated pits prior to the final recruitment of dynamin. The incoming viral particle, which represents a physical obstacle to pit constriction, may lead to the recruitment of actin by inducing membrane tension (11, 12, 34). Therefore, the presence of VSV or RABV G on the surface of the viral particle does not impact the formation of the clathrin-coated pit.
Commonalities in the uptake of two distinct rhabdoviruses.
Despite employing the same endocytic pathway, viruses internalized by clathrin-mediated endocytosis display different behaviors when associating with coated pits. Lateral diffusion into preformed pits has been reported for parvovirus and dengue virus entry (10, 15). In contrast, VSV and influenza A virus do not exhibit such properties and are internalized by pits that form at or in close proximity to viral particles (11, 14). The kinetics of uptake also vary, as parvovirus capture is very rapid (<20 s) (10) whereas dengue virus remains surface bound for several minutes prior to capture (15). For VSV and influenza A virus, the interval between binding and detection of a virus-associated clathrin signal is in the range of minutes (11, 14).
Like those of VSV, RABV particles rarely associate with preexisting pits and show no detectable lateral movement prior to the appearance of the AP2 signal. The interval between rVSV RABV G particle docking and clathrin appearance was indistinguishable from that observed with VSV. These similarities suggest that the clathrin uptake mechanisms for VSV and rVSV RABV G converge upon a common pathway. The rVSV RABV G glycoprotein is derived from the SAD B19 vaccine strain of RABV. Vaccine strains which have been extensively passaged in cell culture are believed to use ubiquitous molecules for cell attachment and entry. Although a recent report suggests that the low density lipoprotein receptor (LDLR) family serves as a VSV receptor, direct interaction of VSV G and LDLR has not been demonstrated and cells lacking LDLR are susceptible to VSV infection (35). Additional evidence supports a relatively nonspecific electrostatic interaction of VSV with the host cell surface (36). Although RABV and VSV engage the clathrin machinery with similar kinetics, we do not think it likely that this reflects the use of a shared cell surface attachment molecule. Whether the engagement of distinct receptor molecules can impact the kinetics with which a given particle associates with the clathrin machinery remains to be determined.
Implications for RABV internalization into different cell types.
RABV infects epithelial cells in the final stages of disease, when virus spreads from the CNS to a variety of extraneural tissues through innervation routes. Epithelial-cell infection is also a primary and necessary step in intranasal or oral infections in animals and in accidental exposures of laboratory workers to RABV (37–40). The BS-C-1 cells used in this study do not express the putative RABV receptors p75 neurotrophin receptor (p75NTR) and neural cell adhesion molecule (NCAM), as both were undetectable by reverse transcription (RT)-PCR (data not shown). Cells were not tested for the presence of the nicotinic acetylcholine receptor (nAChR) because it is known to be expressed only on the postsynaptic membrane of neuromuscular junctions and in the CNS (41). Infection was also unaffected by ganglioside depletion by neuraminidase treatment (data not shown). Consequently, the means by which RABV attaches to these cells is uncertain. Nevertheless, our observation of a primarily clathrin-mediated entry mechanism for RABV is consistent with previous EM studies with nonneuronal fibroblasts (9). We also observed, however, the AP2-independent uptake of a minor proportion (24%) of RABV particles. We do not know the entry route for these internalization event(s), but our pharmacological studies show that such events are unlikely to reflect a macropinocytic uptake.
Pathogenic strains of RABV which have not been passaged in culture show a narrow and specific neurotropism. Although our studies were conducted with epithelial cells, RABV infection of neurons and RABV receptor biology support internalization by clathrin-mediated processes. For example, RABV particles are visible in clathrin-coated pits by electron microscopy of cultured hippocampal neurons (8). Furthermore, p75NTR and NCAM both internalize via clathrin-mediated endocytosis when bound to their endogenous ligands or when cross-linked with antibody (42–44). These observations suggest that clathrin-coated pits likely play a central role in RABV uptake in neuronal cells as well as in epithelial cells. Our ongoing studies are aimed at elucidating the internalization mechanism of RABV into primary neuronal cells and evaluating how engagement of specific receptors by RABV G can influence the mechanism and kinetics of particle internalization.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Cureton and S. Boulant for discussions and R. Massol for support using the IMAB analysis software.
This work was funded by NIH grant AI081842 and NERCE grant U54AI057159. S.P.J.W. is a recipient of the Burroughs Wellcome Investigators in the Pathogenesis of Infectious Disease Award, and S.P. is a recipient of the Giovanni Armenise-Harvard Ph.D. Award.
Footnotes
Published ahead of print 21 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01648-13.
REFERENCES
- 1.Dietzschold B, Schnell M, Koprowski H. 2005. Pathogenesis of rabies. Curr. Top. Microbiol. Immunol. 292:45–56 [DOI] [PubMed] [Google Scholar]
- 2.Kato S, Kobayashi K, Inoue K, Kuramochi M, Okada T, Yaginuma H, Morimoto K, Shimada T, Takada M, Kobayashi K. 2011. A lentiviral strategy for highly efficient retrograde gene transfer by pseudotyping with fusion envelope glycoprotein. Hum. Gene Ther. 22:197–206 [DOI] [PubMed] [Google Scholar]
- 3.Kato S, Kuramochi M, Takasumi K, Kobayashi K, Inoue K, Takahara D, Hitoshi S, Ikenaka K, Shimada T, Takada M, Kobayashi K. 2011. Neuron-specific gene transfer through retrograde transport of lentiviral vector pseudotyped with a novel type of fusion envelope glycoprotein. Hum. Gene Ther. 22:1511–1523 [DOI] [PubMed] [Google Scholar]
- 4.Morimoto K, Foley HD, McGettigan JP, Schnell MJ, Dietzschold B. 2000. Reinvestigation of the role of the rabies virus glycoprotein in viral pathogenesis using a reverse genetics approach. J. Neurovirol. 6:373–381 [DOI] [PubMed] [Google Scholar]
- 5.Gaudin Y. 2000. Rabies virus-induced membrane fusion pathway. J. Cell Biol. 150:601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaudin Y, Tuffereau C, Durrer P, Brunner J, Flamand A, Ruigrok R. 1999. Rabies virus-induced membrane fusion. Mol. Membr. Biol. 16:21–31 [DOI] [PubMed] [Google Scholar]
- 7.Roche S, Gaudin Y. 2004. Evidence that rabies virus forms different kinds of fusion machines with different pH thresholds for fusion. J. Virol. 78:8746–8752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis P, Lentz TL. 1998. Rabies virus entry into cultured rat hippocampal neurons. J. Neurocytol. 27:559–573 [DOI] [PubMed] [Google Scholar]
- 9.Superti F, Derer M, Tsiang H. 1984. Mechanism of rabies virus entry into CER cells. J. Gen. Virol. 65:781–789 [DOI] [PubMed] [Google Scholar]
- 10.Cureton DK, Harbison CE, Cocucci E, Parrish CR, Kirchhausen T. 2012. Limited transferrin receptor clustering allows rapid diffusion of canine parvovirus into clathrin endocytic structures. J. Virol. 86:5330–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. 2009. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 5:e1000394. 10.1371/journal.ppat.1000394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cureton DK, Massol RH, Whelan SP, Kirchhausen T. 2010. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 6:e1001127. 10.1371/journal.ppat.1001127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johannsdottir HK, Mancini R, Kartenbeck J, Amato L, Helenius A. 2009. Host cell factors and functions involved in vesicular stomatitis virus entry. J. Virol. 83:440–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rust MJ, Lakadamyali M, Zhang F, Zhuang X. 2004. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat. Struct. Mol. Biol. 11:567–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Schaar HM, Rust MJ, Chen C, van der Ende-Metselaar H, Wilschut J, Zhuang X, Smit JM. 2008. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 4:e1000244. 10.1371/journal.ppat.1000244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. 2004. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell 118:591–605 [DOI] [PubMed] [Google Scholar]
- 17.Wickersham IR, Finke S, Conzelmann KK, Callaway EM. 2007. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 4:47–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong AC, Sandesara RG, Mulherkar N, Whelan SP, Chandran K. 2010. A forward genetic strategy reveals destabilizing mutations in the Ebolavirus glycoprotein that alter its protease dependence during cell entry. J. Virol. 84:163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whelan SP, Barr JN, Wertz GW. 2000. Identification of a minimal size requirement for termination of vesicular stomatitis virus mRNA: implications for the mechanism of transcription. J. Virol. 74:8268–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whelan SP, Ball LA, Barr JN, Wertz GT. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. U. S. A. 92:8388–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Massol RH, Boll W, Griffin AM, Kirchhausen T. 2006. A burst of auxilin recruitment determines the onset of clathrin-coated vesicle uncoating. Proc. Natl. Acad. Sci. U. S. A. 103:10265–10270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maupin P, Pollard TD. 1983. Improved preservation and staining of HeLa cell actin filaments, clathrin-coated membranes, and other cytoplasmic structures by tannic acid-glutaraldehyde-saponin fixation. J. Cell Biol. 96:51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefrancios L, Lyles DS. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology 121:157–167 [PubMed] [Google Scholar]
- 25.Lefrancois L, Lyles DS. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology 121:168–174 [DOI] [PubMed] [Google Scholar]
- 26.Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Moller P, Wagner R, Volchkov V, Klenk HD, Feldmann H, Stroher U. 2004. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J. Virol. 78:5458–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guichard P, Krell T, Chevalier M, Vaysse C, Adam O, Ronzon F, Marco S. 2011. Three dimensional morphology of rabies virus studied by cryo-electron tomography. J. Struct. Biol. 176:32–40 [DOI] [PubMed] [Google Scholar]
- 28.Mebatsion T, Konig M, Conzelmann KK. 1996. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84:941–951 [DOI] [PubMed] [Google Scholar]
- 29.Koivusalo M, Welch C, Hayashi H, Scott CC, Kim M, Alexander T, Touret N, Hahn KM, Grinstein S. 2010. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J. Cell Biol. 188:547–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. 2006. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell 10:839–850 [DOI] [PubMed] [Google Scholar]
- 31.Cureton DK, Burdeinick-Kerr R, Whelan SP. 2012. Genetic inactivation of COPI coatomer separately inhibits vesicular stomatitis virus entry and gene expression. J. Virol. 86:655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirchhausen T, Boll W, van Oijen A, Ehrlich M. 2005. Single-molecule live-cell imaging of clathrin-based endocytosis. Biochem. Soc. Symp. 71–76 [DOI] [PubMed] [Google Scholar]
- 33.Boucrot E, Saffarian S, Massol R, Kirchhausen T, Ehrlich M. 2006. Role of lipids and actin in the formation of clathrin-coated pits. Exp. Cell Res. 312:4036–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boulant S, Kural C, Zeeh JC, Ubelmann F, Kirchhausen T. 2011. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat. Cell Biol. 13:1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finkelshtein D, Werman A, Novick D, Barak S, Rubinstein M. 2013. LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 110:7306–7311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coil DA, Miller AD. 2004. Phosphatidylserine is not the cell surface receptor for vesicular stomatitis virus. J. Virol. 78:10920–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler WG, Baker EF, Jr, Hopkins CC. 1972. An outbreak of non-bite transmitted rabies in a laboratory animal colony. Am. J. Epidemiol. 95:267–277 [DOI] [PubMed] [Google Scholar]
- 38.Winkler WG, Fashinell TR, Leffingwell L, Howard P, Conomy P. 1973. Airborne rabies transmission in a laboratory worker. JAMA 226:1219–1221 [PubMed] [Google Scholar]
- 39.Conomy JP, Leibovitz A, McCombs W, Stinson J. 1977. Airborne rabies encephalitis: demonstration of rabies virus in the human central nervous system. Neurology 27:67–69 [DOI] [PubMed] [Google Scholar]
- 40.Johnson N, Phillpotts R, Fooks AR. 2006. Airborne transmission of lyssaviruses. J. Med. Microbiol. 55:785–790 [DOI] [PubMed] [Google Scholar]
- 41.Gotti C, Fornasari D, Clementi F. 1997. Human neuronal nicotinic receptors. Prog. Neurobiol. 53:199–237 [DOI] [PubMed] [Google Scholar]
- 42.Deinhardt K, Reversi A, Berninghausen O, Hopkins CR, Schiavo G. 2007. Neurotrophins redirect p75NTR from a clathrin-independent to a clathrin-dependent endocytic pathway coupled to axonal transport. Traffic 8:1736–1749 [DOI] [PubMed] [Google Scholar]
- 43.Diestel S, Schaefer D, Cremer H, Schmitz B. 2007. NCAM is ubiquitylated, endocytosed and recycled in neurons. J. Cell Sci. 120:4035–4049 [DOI] [PubMed] [Google Scholar]
- 44.Minana R, Duran JM, Tomas M, Renau-Piqueras J, Guerri C. 2001. Neural cell adhesion molecule is endocytosed via a clathrin-dependent pathway. Eur. J. Neurosci. 13:749–756 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.