Abstract
Varicella zoster virus (VZV) is the etiological agent of varicella (chickenpox) and herpes zoster (HZ [shingles]). Clinical observations suggest that VZV-specific T cell immunity plays a more critical role than humoral immunity in the prevention of VZV reactivation and development of herpes zoster. Although numerous studies have characterized T cell responses directed against select VZV open reading frames (ORFs), a comprehensive analysis of the T cell response to the entire VZV genome has not yet been conducted. We have recently shown that intrabronchial inoculation of young rhesus macaques with simian varicella virus (SVV), a homolog of VZV, recapitulates the hallmarks of acute and latent VZV infection in humans. In this study, we characterized the specificity of T cell responses during acute and latent SVV infection. Animals generated a robust and broad T cell response directed against both structural and nonstructural viral proteins during acute infection in bronchoalveolar lavage (BAL) fluid and peripheral blood. During latency, T cell responses were detected only in the BAL fluid and were lower and more restricted than those observed during acute infection. Interestingly, we identified a small set of ORFs that were immunogenic during both acute and latent infection in the BAL fluid. Given the close genome relatedness of SVV and VZV, our studies highlight immunogenic ORFs that may be further investigated as potential components of novel VZV vaccines that specifically boost T cell immunity.
INTRODUCTION
Varicella zoster virus (VZV) is a neurotropic alphaherpesvirus that causes varicella and establishes latency in the sensory ganglia (trigeminal and dorsal root) (2). Reactivation of VZV results in herpes zoster (HZ), a painful and debilitating disease that affects 1 million individuals in the United States alone (2), with 60% of these cases occurring in persons 50 years of age or older (3). Given that 22% of the U.S. population will be over the age of 60 by 2020 (www.census.gov/compendia/statab/2012/tables/12s0009.pdf), the incidence of HZ is likely to increase.
Successful resolution of primary VZV infection relies on both cell-mediated and humoral immunity. Specifically, the administration of immunoglobulins with high titers of IgG antibodies (Abs) to VZV can mitigate disease severity for up to 10 days after exposure (4, 5). Some clinical observations suggest that T cells may contribute more than antibodies in unvaccinated subjects. For instance, progressive varicella is most often reported in children with defects in cellular immunity or who are undergoing immunosuppressive treatment (7). In addition, children infected with HIV are at risk of prolonged viremia, continued formation of skin lesions, and dissemination of the virus to the lungs and other organs (7, 8) and early production of VZV antibodies by HIV-positive (HIV+) children does not prevent progressive varicella (9). On the other hand, children with agammaglobulinemia have uncomplicated varicella episodes (10–11). Similarly, it is believed that the increased risk of VZV reactivation among older individuals is due to an age-associated decrease in T cell immunity (13) as VZV-specific antibody titers do not significantly decline with age (14). However, despite its importance in the prevention of reactivation, the T cell response to VZV remains poorly defined.
Previous studies using cultured T cell lines from peripheral blood mononuclear cells (PBMCs) of healthy adults have demonstrated the presence of cytolytic CD4 and CD8 T cell responses to VZV open reading frames (ORFs) ORF4, ORF10, ORF29, ORF62, ORF63, and ORF67 (16–18). More recent studies also using T cell lines suggest that the anti-ORF4, -63, -67, and -68 responses are predominated by CD4 T cells (1, 19–21). However, to date, a comprehensive ex vivo analysis of the T cell response to the entire VZV genome has not been conducted. Consequently, our understanding of the global anti-VZV T cell response remains incomplete. This is illustrated by the fact that one study concluded that the CD4 T cell response to VZV is directed primarily against ORF4 (19), while a follow-up study suggested that the anti-VZV response is primarily mediated by CD4 T cells directed against ORF67 (20).
We have recently shown that intrabronchial infection of young rhesus macaques (RM) with simian varicella virus (SVV) results in disease characteristic of VZV infection in humans, with the appearance of a generalized varicella rash at 7 days postinfection (dpi), generation of T and B cell responses that peak at 7 to 14 days postinfection, resolution of viremia coincident with abatement of varicella at 21 to 35 days postinfection, and establishment of latency with limited transcriptional activity in the sensory ganglia (22, 23). SVV and VZV are evolutionarily related and are colinear with respect to genome organization (24–27). With a single exception (described below), each of the SVV ORFs has a corresponding VZV homolog which shares amino acid identities ranging from 75.4% (ORF31, gB) to 27.3% (ORF1), with an average 55.5% amino acid identity across the genome (26). The only significant difference between the SVV and VZV genomes occurs in the left terminus, where SVV lacks a VZV ORF2 homolog and encodes ORF A, which is absent from the VZV genome (24, 26, 28, 29). Indeed, immunization of Erythrocebus patas with VZV can protect the animals from SVV challenge, demonstrating the antigenic relatedness of the two viruses (30). SVV can also reactivate in animals exposed to social/environmental stress such as transportation or introduction of new animals into an existing group (31–34) as well as in animals subjected to immune system-suppressive treatments (35–38). Thus, infection of nonhuman primates (NHP) with SVV provides a translational model with which to gain insight into the anti-VZV T cell response.
In this study, we characterized the specificity of the T cell response generated during both acute and latent SVV infections by measuring the frequency of gamma interferon (IFN-γ)-producing T cells from PBMC or bronchial alveolar lavage (BAL) fluid samples following stimulation with overlapping peptide libraries for each SVV ORF. Despite the intersubject variability, we detected robust T cell responses to SVV ORF4, ORF9, ORF11, ORF37, and ORF47 during both acute and latent infection in BAL fluid and during acute infection in PBMC. Interestingly, no T cell responses were detected in PBMC during latency. SVV ORF4 encodes immediate early protein 2, functions as a transcriptional activator, and shares 43.2% amino acid similarity with VZV ORF4 (26). SVV ORF9, ORF11, and ORF37 encode tegument proteins (ORF9 and ORF11) and glycoprotein H (ORF37). Respectively, they share 59.4%, 50.8%, and 55.5% amino acid identity with VZV ORF9, ORF11, and ORF37 (26). SVV ORF47 is a putative protein kinase that shares 65% amino acid identity with VZV ORF47. Given the high homology between these SVV ORFs and their respective VZV counterparts, these results provide preclinical data that can help shape the development of second-generation vaccines to specifically boost key T cell responses to VZV.
METHODS AND MATERIALS
Animals and sample collection.
This study was carried out in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, the Office of Animal Welfare, and the United States Department of Agriculture. All animal work was approved by the Oregon National Primate Research Center Institutional Animal Care and Use Committee. All procedures were carried out under conditions of ketamine anesthesia in the presence of veterinary staff, and all efforts were made to minimize animal suffering. A total of 20 colony-bred rhesus macaques (RM [Macaca mulatta) (ages 2 to 12 years) of Indian origin were used in these studies. All animals were inoculated intrabronchially with 4 × 105 PFU SVV as previously described (22). For those animals in the acute group (n = 8), terminal blood and bronchial alveolar lavage (BAL) fluid samples were collected at 7, 10, and 14 days postinfection (dpi) (n = 2 to 3/time point). For those animals in the latent group (n = 12), terminal blood and BAL fluid samples were collected >72 dpi.
BAL fluid samples were pelleted and resuspended in RPMI medium supplemented with 10% fetal bovine serum (FBS), streptomycin/penicillin, and l-glutamine (RPMI complete). Peripheral blood mononuclear cells (PBMCs) were isolated by centrifugation over a Histopaque gradient (Sigma) per the manufacturer's recommendation. Cells were stored in FetalPlex (Gemini Bio Products, West Sacramento, CA) supplemented with 10% dimethyl sulfoxide (DMSO) and cryopreserved in liquid nitrogen. Cells were then thawed in 10 ml RPMI complete and 120 Kunitz units DNase for use in subsequent assays.
Cells and virus.
Wild-type SVV was propagated in primary rhesus fibroblasts (1oRF) at 37°C in 175-cm2 flasks with Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). SVV-infected 1oRF were frozen in FetalPlex with 10% DMSO, stored in liquid nitrogen (LN2), and assayed by plaque assay.
DNA extraction and quantitative real-time PCR (qPCR).
DNA was extracted from heparinized whole-blood (WB) and BAL fluid cells using an ArchivePure DNA cell/tissue kit (5 Prime, Gaithersburg, MD). SVV DNA loads were determined by real-time PCR using primers and probes specific for SVV ORF21 using an ABI StepOne instrument (Applied Biosystems, Foster City, CA) as previously described (22). The limit of detection for this assay is 10 copies per 100 ng DNA.
Peptide pool libraries.
Overlapping peptide pool libraries for each of the 70 SVV open reading frames (ORFs) were generated. Peptides were synthesized by Sigma-Aldrich (St. Louis, MO) as sequences 18 amino acids long and overlapping by 10 amino acids. All peptides were subjected to mass spectrometry to ensure the correct sequence and were provided as crude, albeit with a guaranteed purity range of 65% to 75%. Previous studies indicate that peptides ranging from 15 to 20 amino acids in length yield similar results in an IFN-γ–enzyme-linked immunosorbent spot (ELISpot) assay (39, 40), but there is a potential that CD8 T cell responses might be underestimated when using long peptides. Lyophilized peptides were reconstituted at a concentration of 10 mg/ml in sterile endotoxin-free DMSO and stored at −80°C. Peptide pools were prepared for each ORF by combining all peptides across an ORF. Peptide pools were composed of an average of 63 peptides per pool, with a range of from 9 (ORF49 and ORF65) to 331 (ORF22) peptides per pool. The concentration of each peptide within a given pool ranged from 1.1 mg/ml (ORF49 and ORF65) to 0.03 mg/ml (ORF22), with an average concentration of 0.29 mg/ml. PBMC or BAL fluid cells were stimulated using 1 μl (1 μg/ml final concentration) of each peptide pool.
ELISpot assay.
The ELISpot assay was carried out as previously described (40). Briefly, we used 96-well polyvinylidene fluoride-bottomed plates precoated with recombinant anti-rhesus IFN-γ monoclonal antibody (MAb; Mabtech AB, Sweden). The plates were blocked with RPMI medium supplemented with 10% FBS for 30 min at room temperature. PBMCs or BAL fluid cells (2 × 105/well) were stimulated in triplicate with overlapping peptide libraries for all 70 of the SVV ORFs (1 μg/well final concentration of the peptide pool), phorbol myristate acetate (PMA)/ionomycin (100 and 250 ng/ml; positive control), and DMSO (0.5% final concentration; negative control). After incubation at 37°C for 18 h, the plates were washed with phosphate-buffered saline (PBS). Biotinylated anti-human IFN-γ MAb (1 μg/ml) was added, and plates were incubated for 2 h at room temperature. After washing with PBS, streptavidin-alkaline phosphate (streptavidin-ALP) was added to all wells and the plates were incubated for 1 h at room temperature. Following washing with PBS, BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium (NBT)-plus substrate was added and the plates were allowed to develop in the dark for 5 to 15 min until spots appeared. Color development was stopped by washing in tap water, and the plates were allowed to dry. After drying, the numbers of SVV ORF-specific IFN-γ-secreting spot-forming cells (SFC) were counted by the use of an EliSpot reader (ELR06; AID GmbH, Germany) using AID EliSpot 6.0 software, LITE version. Data were normalized by first subtracting the number of SFC in negative-control wells (DMSO) from the number in antigen-stimulated wells and then subtracting the average number of SFC detected in stimulated wells from SVV-naive animals.
Intracellular cytokine staining.
PBMCs and BAL fluid cells (1 to 2 × 106/well) were stimulated with SVV overlapping peptide libraries (1 μg/well), PMA/ionomycin (1 μg/ml), or DMSO (1 μg/ml) for 1 h followed by addition of brefeldin A (Sigma) for an additional 14 h. After incubation, cells were stained with antibodies directed against CD4 (PerCp Cy5.5, clone OKT4, eBioscience, San Diego, CA) and CD8β (Pe Texas Red, clone 2ST8.5H7; Beckman Coulter, Brea, CA). Samples were then fixed and permeabilized with fixation buffer (BioLegend) before the addition of antibodies directed against IFN-γ (PeCy7, clone 4S.B3; eBioscience). Samples were analyzed using an LSRII instrument (Becton Dickinson and Company, San Jose, CA) and FlowJo software (TreeStar, Ashland, OR).
Statistical analysis.
Bootstrapping was used to establish the empirical 95% confidence intervals for the median, 2.5 percentile, and 97.5 percentile for responses generated by naive animals in BAL fluid and PBMC. Bootstrapping is a computer-based method for assigning measures of accuracy to sample estimates when the distribution of the data is not known (42). Reported point and interval estimates of median, 2.5 percentile, and 97.5 percentile were based on 10,000 bootstrap replications with unrestricted random sampling. These confidence intervals were then used to identify “positive” responses in the ELISpot assays. Pair-matched comparisons were performed using a two-tailed Student t test to analyze differences between (i) total SFC numbers between acute BAL fluid and PBMC samples within the same animal and (ii) CD4 and CD8 T cell responses to immediate early (IE)/early (E) and late (L) ORFs. An unpaired two-tailed Student t test was used to compare total SFC numbers between acute BAL fluid and latent BAL fluid samples as well as T cell responses to specific ORFs between acute and latent infections. Statistical significance was determined at the level of 0.05.
RESULTS
SVV loads.
All animals were infected intrabronchially with 4 × 105 PFU SVV. SVV loads were determined in whole-blood (WB) and bronchial alveolar lavage (BAL) fluid samples from all animals by quantitative real-time PCR (qPCR). We detected SVV DNA in BAL fluid at 7 (n = 3), 10 (n = 2), and 14 (n = 3) days postinfection (dpi) for all 8 animals (Table 1). SVV DNA was detected in WB in three animals at 7 dpi but in only one animal (27916) 10 dpi and in two animals (27180 and 27244) 14 dpi (Table 1). Collectively, SVV loads detected in BAL fluid were higher than those detected in WB during acute infection (P < 0.01), which is consistent with previous observations and reflects the route of infection (22, 23). SVV loads in WB and BAL fluid of all latently infected animals (n = 12) were below the level of detection (data not shown), although viral DNA was detected in at least one sensory ganglion in all latently infected animals (Table 2), which is a characteristic of viral latency (22).
Table 1.
SVV loads in BAL fluid and WB samples during acute infection
| Animal no. | dpi | Avg no. of SVV copies/100 ng DNA in: |
|
|---|---|---|---|
| BAL fluid | WB | ||
| 28138 | 7 | 109,107 | 473 |
| 27984 | 7 | 322,319 | 861 |
| 28552 | 7 | 1,144 | 32 |
| 26704 | 10 | 227 | NDa |
| 27916 | 10 | 268 | 80 |
| 27180 | 14 | 521 | 24 |
| 27244 | 14 | 51 | 36 |
| 28547 | 14 | 549 | ND |
ND, not detectable.
Table 2.
SVV load in sensory ganglia
| Animal no. | Sample | No. of SVV copies/μg DNA |
|---|---|---|
| 23942 | TG | 108 |
| DRG-C | 676 | |
| DRG-T | NDa | |
| DRG-L/S | ND | |
| 23986 | TG | ND |
| DRG-C | 500 | |
| DRG-T | 487 | |
| DRG-L/S | 91 | |
| 26509 | TG | 116 |
| DRG-C | 116 | |
| DRG-T | 63 | |
| DRG-L/S | 75 | |
| 26100 | TG | 212 |
| DRG-C | 134 | |
| DRG-T | ND | |
| DRG-L/S | ND | |
| 27556 | TG | ND |
| DRG-C | 61 | |
| DRG-T | ND | |
| DRG-L/S | ND | |
| 24943 | TG | 90 |
| DRG-C | 89 | |
| DRG-T | 79 | |
| DRG-L/S | 67 | |
| 24953 | TG | 136 |
| DRG-C | 143 | |
| DRG-T | 174 | |
| DRG-L/S | 60 | |
| 25043 | TG | 27 |
| DRG-C | 12 | |
| DRG-T | ND | |
| DRG-L/S | ND | |
| 28428 | TG | 10 |
| DRG-C | 218 | |
| DRG-T | ND | |
| DRG-L/S | 162 | |
| 21211 | TG | ND |
| DRG-C | 93 | |
| DRG-T | 67 | |
| DRG-L/S | 83 | |
| 19559 | TG | ND |
| DRG-C | 87 | |
| DRG-T | 41 | |
| DRG-L/S | 60 | |
| 25339 | TG | 47 |
| DRG-C | 56 | |
| DRG-T | 51 | |
| DRG-L/S | 50 |
ND, not detected.
T cell responses to SVV ORFs during acute infection in BAL fluid.
Cryopreserved BAL fluid and PBMC samples from uninfected animals (n = 3) and from animals acutely infected with SVV (7, 10, and 14 dpi) were stimulated ex vivo with overlapping peptide pools corresponding to all 70 SVV ORFs. The number of responding T cells was determined using IFN-γ–ELISpot assays and is reported as the number of spot-forming cells (SFC). As expected, BAL fluid cells from uninfected animals show minimal response to stimulation with overlapping SVV peptide pools (median, 2.8 SFC/200,000 BAL fluid cells/ORF) (Fig. 1A). We used bootstrapping statistical analysis to establish the lower and upper 95% confidence intervals for the responses generated by naive animals. This analysis revealed an upper limit of 17 SFC for T cell responses in BAL fluid samples of naive animals, which was the value used as the cutoff to identify positive responses in infected animals.
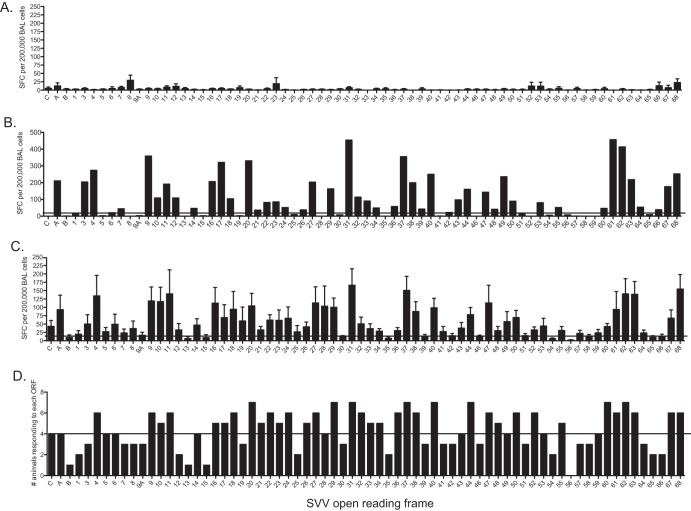
Fig 1.
Lung-resident T cells generate robust and broad responses following acute infection. T cell responses in bronchoalveolar lavage (BAL) fluid samples were determined using IFN-γ–ELISpot technology following stimulation with overlapping peptide libraries covering all 70 SVV ORFs. Each peptide was tested in triplicate, and average numbers of spot-forming cells (SFC) per ORF per 200,000 cells with the background subtracted are shown. (A) Average SFC responses ± standard errors of the means (SEM) of BAL fluid cells from three naive animals. (B) T cell responses from a representative SVV-infected 4-year-old animal. (C) Average SFC response ± SEM from all 8 acutely infected animals. (D) Numbers of animals that generated a positive response to each ORF. Horizontal lines indicate the threshold above which a response is defined as positive.
In SVV-infected animals, the numbers of ORFs targeted by the T cell response and the magnitudes of the response to each ORF differed between animals, resulting in a unique immunogenicity hierarchy for each animal. For instance, animal 28547 generated a T cell response to 55 ORFs, with roughly 30% of the overall responses being directed against ORF9, ORF31, ORF37, ORF61, and ORF62 (Fig. 1B). Overall, SVV-infected animals generate a robust (average, 4,555 total SFC/200,000 BAL fluid cells) and broad T cell response, with average positive responses detected against 55 SVV ORFs (Fig. 1C). No average positive responses were detected against ORF B, ORF9A, ORF13, ORF15, ORF30, ORF35, ORF39, ORF42, ORF46, ORF51, ORF54, ORF56, ORF58, ORF65, and ORF66 (Fig. 1C).
We also calculated the number of animals that generated a positive IFN-γ ELISpot response to each ORF. This analysis revealed that positive T cell responses to SVV ORF4, ORF9 to ORF11, ORF16 to ORF18, ORF20 to ORF24, ORF26, ORF27, ORF29, ORF31 to ORF34, ORF36 to ORF38, ORF40, ORF44, ORF47, ORF48, ORF50, ORF52, ORF55, ORF60 to ORF63, and ORF67 to ORF68 were detected in at least 5 of the 8 animals studied (Fig. 1D). T cell responses directed against these 35 immunogenic ORFs accounted for approximately 76% of the overall T cell response to SVV in each animal examined.
T cell responses to SVV ORFs during acute infection in PBMC.
We next characterized the anti-SVV T cell response in peripheral blood. PBMCs collected from naive animals generated minimal responses following stimulation with SVV ORFs (median, 1.78 SFC/200,000 PBMC/ORF) (Fig. 2A). We again used bootstrapping analysis to determine the upper limits of the T cell response generated by PBMC in naive animals. This approach defined a threshold of 4.78 SFC/200,000 PBMC to define a positive response. Similar to our analysis of BAL fluid cells, each animal generated a unique T cell response in terms of ORFs recognized and the magnitude of the response to each ORF. For example, animal 28547 generated a T cell response to 24 ORFs, with roughly 42% of the overall responses being directed against ORF A, ORF3, ORF9, ORF20, ORF27, and ORF37 (Fig. 2B). On average, T cell responses in peripheral blood were significantly lower than those observed in BAL fluid (average, 613 SFC/animal, P < 0.01) and slightly narrower, with average positive responses detected against 43 of the 70 SVV ORFs (Fig. 2C). The highest responses were directed against ORF3, ORF4, ORF9, ORF10, ORF11, ORF20, ORF28, ORF37, ORF47, ORF63, and ORF68 and together accounted for 42% of the total anti-SVV T cell response in PBMC.
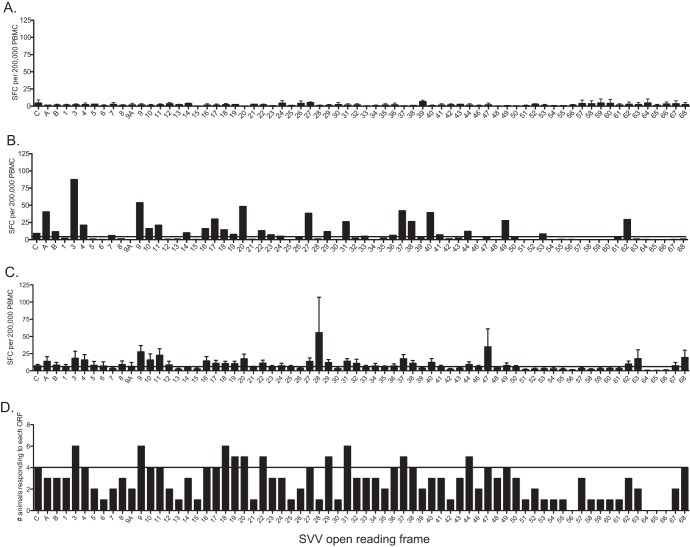
Fig 2.
T cell responses in peripheral blood are lower and narrower than those in BAL fluid. T cell responses in peripheral blood mononuclear cells (PBMC) were determined using IFN-γ–ELISpot technology following stimulation with overlapping peptide libraries covering all 70 SVV ORFs. Each peptide was tested in triplicate, and average numbers of spot-forming cells (SFC) per ORF per 200,000 cells with the background subtracted are shown. (A) Average SFC responses ± SEM of PBMCs from three naive animals. (B) T cell responses from a representative SVV-infected 4-year-old animal. (C) Average SFC response ± SEM of PBMC from all 8 acutely infected animals. (D) Number of animals that generated a positive response to each ORF. Horizontal lines indicate the threshold above which a response is defined as positive.
We next identified SVV ORFs that elicited a positive IFN-γ–ELISpot response in greater than 50% of the animals (Fig. 2C). SVV ORF3, ORF9, ORF18 to ORF20, ORF22, ORF29, ORF31, ORF37, and ORF44 met this criterion (Fig. 2C), and T cell responses directed against these 10 ORFs accounted for an average 26% of the overall response to SVV in PBMC. Since we measured T cell responses in two different anatomical sites (peripheral blood and lung) within each animal, we determined the correlation between the two responses. This analysis revealed an average r2 value of 0.56 (ranging from 0.41 to 0.66) and an average P value < 0.01 indicative of related responses in the periphery and the site of infection (lungs) during acute infection within each animal.
Contribution of CD4 versus CD8 T cells to the anti-SVV response during acute infection.
Both CD4 and CD8 T cells contribute to the anti-VZV/SVV response (22, 44–46). Therefore, we next assessed whether T cell responses to immunodominant ORFs in BAL fluid cells and PBMCs were primarily mediated by CD4 or CD8 T cells by measuring the frequency of IFN-γ+ CD4 and CD8 T cells by intracellular cytokine staining (ICS) following stimulation with overlapping peptide pools (Fig. 3A). We focused our analysis on the ORFs against which positive T cell responses were detected in BAL fluid cells and PBMCs in at least 4 of 8 animals: ORF4, ORF9, ORF10, ORF11, ORF16, ORF20, ORF22, ORF27, ORF31, ORF37, ORF38, ORF40, ORF44, ORF47, ORF62, ORF63, and ORF68 (Fig. 3). We were able to perform this analysis using BAL fluid and PBMC samples only from animals 6 and 7 of 8 animals, respectively, due to the paucity of available cells.
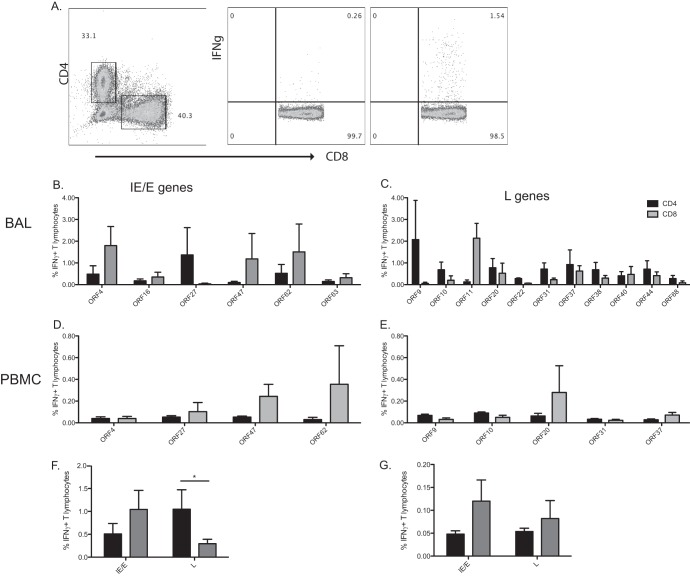
Fig 3.
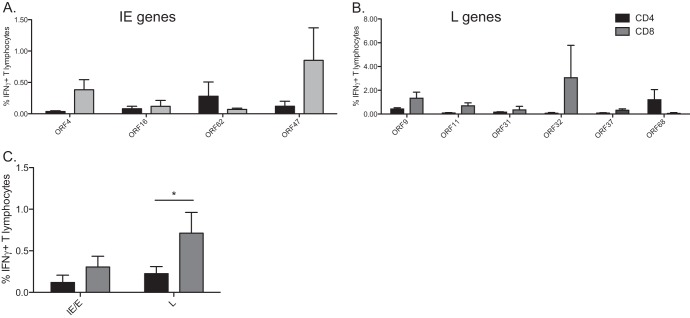
T cell responses to late SVV genes in BAL fluid are primarily mediated by CD4 T cells during acute infection. The frequency of IFN-γ-producing CD4 (black) and CD8 (gray) T cells in BAL fluid and PBMC in response to immunogenic SVV ORFs was measured by intracellular cytokine staining (ICS) following stimulation with the indicated overlapping peptide pools. (A) Representative example of the CD8 T cell response directed against ORF4 in BAL fluid. (Left panel) Delineation of CD4 and CD8 T cells. (Middle panel) CD8 T cell response following stimulation with 0.05% DMSO. (Right panel) CD8 T cell response following stimulation with overlapping peptide pool of ORF4. (B and C) Average percentages ± SEM of responding IFN-γ+ T cells against listed ORFs in BAL fluid. (D and E) Average percentages ± SEM of responding IFN-γ+ T cells against listed ORFs in PBMC. (F and G) Average percentages of responding IFN-γ+ T cells against putative immediate early/early (IE/E) or late (L) ORFs in BAL fluid (F) and PBMC (G). *, P < 0.05.
As observed with the IFN-γ ELISpot data, the average frequencies of responding T cells differed between ORFs and between animals responding to the same ORF in BAL fluid cells (Fig. 3B and C) and PBMCs (Fig. 3D and E). T cell responses to putative immediate early (IE) and early (E) gene products (ORF4, ORF16, ORF27, ORF47, ORF62, and ORF63) seemed to be primarily mediated by CD8 T cells in BAL fluid cells and/or PBMCs (Fig. 3B and D). On the other hand, T cell responses to putative late (L) ORFs (ORF9, ORF10, ORF11, ORF20, ORF22, ORF31, ORF37, ORF38, ORF40, ORF44, and ORF68) appeared to be primarily carried out by CD4 T cells in BAL fluid cells (Fig. 3C) and both CD4 and CD8 T cells in PBMCs (Fig. 3E). To test this observation, we compared the overall CD8 and CD4 T cell responses (Fig. 3F and G). This analysis showed that CD4 and CD8 T cell responses to putative IE/E genes in BAL fluid and PBMC were similar (P = 0.40 and 0.15, respectively) (Fig. 3F and G). However, overall CD4 T cell responses to late (structural) genes are significantly higher than those elicited by CD8 T cells in BAL fluid cells (P < 0.01) (Fig. 3F). Finally, there was no difference in the frequencies of CD4 and CD8 T cells responding to products of late genes in PBMCs (P = 0.48) (Fig. 3G).
T cell responses to SVV ORFs during latent infection.
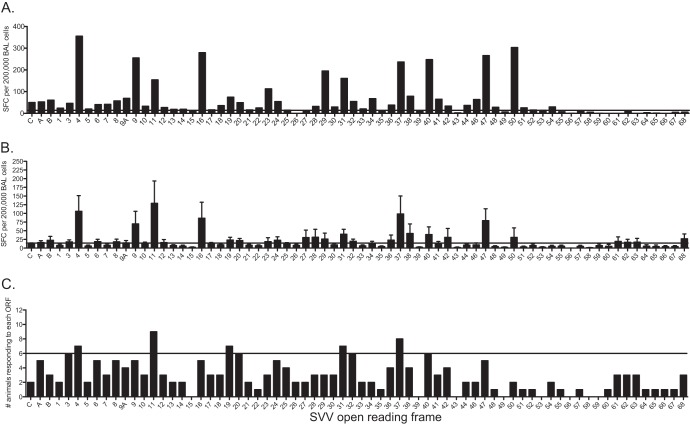
We next investigated the specificity of the anti-SVV T cell response during latency. We were able to detect SVV-specific T cells only in BAL fluid samples, as the frequency of responding T cells in PBMCs ex vivo was below the threshold of detection by IFN-γ–ELISpot assays. As was observed with acute infection, the numbers of ORFs targeted and the sizes of the T cell response to each ORF from BAL fluid samples of latently infected animals differed between animals. To illustrate, animal 25339 generated a response to 44 ORFs, with roughly 35% of the overall response being directed against SVV ORF4, ORF9, ORF16, ORF40, ORF47, and ORF50 (Fig. 4A). Overall, the T cell response in BAL fluid cells during latency is lower in magnitude than that observed during acute infection (average, 1,453 compared to 4,555 total SFC/200,000 BAL fluid cells, P < 0.01) (Fig. 4B and 1C, respectively). The breadth of the T cell response during latency was also reduced compared to that seen during acute infection, with average positive responses detected against 26 ORFs compared to 55 ORFs, respectively (Fig. 1C and 4B). Positive T cell responses to SVV ORF4, ORF11, ORF19, ORF31, and ORF37 were detected in >50% of the animals (Fig. 4C), and responses directed against these 5 ORFs accounted for 28% of the overall anti-SVV response during latency (Fig. 4C).
Fig 4.
Memory T cell responses are lower and more restricted than those observed during acute infection in BAL fluid samples. BAL fluid cells from latently infected young RM (2 to 4 years of age) were stimulated with overlapping peptide libraries covering all 70 SVV ORFs and T cell responses assessed using IFN-γ–ELISpot technology. Each peptide was tested in triplicate, and average numbers of spot-forming cells (SFC) per ORF per 200,000 cells with the background subtracted are shown. (A) Representative profile of one latently infected animal. (B) Average SFC responses ± SEM from all 12 latently infected animals. (C) Numbers of animals that generated a positive response to each ORF. Horizontal lines indicate the threshold above which a response is defined as positive.
Contribution of BAL fluid CD4 and CD8 T cells to the anti-SVV response during latent infection.
Using the ICS assay, we determined whether the BAL fluid T cell responses observed during latency were mediated by CD4 or by CD8 T cells. We were able to perform ICS analysis on only 8 of 12 animals and were able to test responses only to SVV ORF4, ORF9, ORF11, ORF16, ORF31, ORF32, ORF37, ORF47, ORF62, and ORF68 due to the scarcity of available cells. As noted above for the T cell response during acute infection, frequencies of responding T cells in the BAL fluid differed between ORFs and between animals responding to the same ORF (Fig. 5). CD8 T cell responses to ORF4 were slightly higher than the CD4 T cell responses (P = 0.07) (Fig. 5A). CD8 T cell responses to ORF9, ORF11, and ORF37 were also slightly higher than CD4 T cell responses (P = 0.07, 0.06, and 0.09, respectively) (Fig. 5B). Since we detected a trend toward higher CD8 T cell responses for some of the ORFs encoding putative L genes, we compared the overall CD8 and CD4 T cell responses to IE/E and L genes. This analysis showed that the overall CD8 T cell response to late proteins was significantly greater than the CD4 response (P = 0.04) (Fig. 5C).
Fig 5.
T cell responses to late proteins are primarily mediated by CD8 T cells during latent infection in BAL fluid samples. The frequency of IFN-γ-producing CD4 (black) and CD8 (gray) T cells in BAL fluid in response to immunogenic SVV ORFs was measured by intracellular cytokine staining (ICS) following stimulation with the indicated overlapping peptide pools. (A and B) The average percentages of responding IFN-γ+ T cells ± SEM in BAL fluid are shown. (C) Average overall CD4 and CD8 responses to all IE/E and L genes are shown.*, P < 0.05.
DISCUSSION
Varicella zoster virus (VZV), the causative agent of varicella, establishes latency in the sensory ganglia and cranial nerves, where it can reactivate and cause herpes zoster (HZ [shingles]) (2). Clinical observations strongly suggest that T cell immunity is critical in the successful resolution and control of VZV infection (47). However, our knowledge of the specificity of the anti-VZV T cell response during acute and latent infection remains incomplete. Simian varicella virus (SVV) is a neurotropic alphaherpesvirus that naturally infects nonhuman primates (NHP) and shares significant amino acid identity and genome colinearity with VZV (24–27). Intrabronchial inoculation of rhesus macaques with SVV recapitulates the following hallmarks of VZV infection: (i) rash; (ii) development of cellular and humoral immune responses; (iii) resolution of acute viremia; and (iv) establishment of latency in sensory ganglia (22). Similar to VZV, SVV transmission is believed to occur through contact with skin lesions or via exposure to virus-laden aerosolized droplets from infected animals (6, 48–50). Furthermore, reactivation of SVV has been observed in immune system-suppressed animals or those subjected to social and environmental stress (31–34). Therefore, SVV infection of nonhuman primates provides a robust animal model to gain insight into the anti-VZV T cell response.
In this study, we characterized the T cell response to the entire SVV proteome during both acute and latent infections. Our results show that each animal generates a unique T cell response with a distinctive immunogenicity profile. During acute infection, the T cell responses in peripheral blood exhibited reduced magnitude and breadth compared to the levels seen in BAL fluid cells, most likely a reflection of the increased viral replication in the lungs compared to peripheral blood. However, within the same animal, peripheral blood- and BAL fluid-resident T cells targeted similar ORFs. Despite the animal-to-animal variation, we were able to identify 35 immunogenic SVV ORFs to which a majority of animals generated a positive T cell response in BAL fluid samples. These ORFs represent all three kinetic classes of viral genes, ranging from transactivators to glycoproteins. T cell responses directed against these ORFs account on average for 75% of the total SVV-specific BAL fluid-resident T cell response. Although only 10 ORFs engendered positive T cell responses in the majority of the animals in PBMC, these ORFs also represented all three major kinetic classes and together accounted for 24% of the overall anti-SVV T cell response. Moreover, with the exception of ORF3, positive T cell responses to these ORFs were also detected in BAL fluid cells in the majority of animals studied. The higher and broader response observed in BAL fluid is in agreement with robust SVV replication and gene expression occurring primarily in the lung rather than peripheral blood during acute infection (22, 23).
Our previous characterization of acute SVV gene expression in BAL fluid cells showed that the transcripts associated with ORF63 (transcriptional activator), ORF49 (putative myristylated virion protein), ORF41 (capsid protein), ORF23 (putative capsid protein), ORF53 (putative gamma-1 protein), and ORF65 (putative tegument phosphoprotein) were the most abundant (23). However, with the exception of ORF63, which engendered the second-highest T cell response in BAL fluid samples, responses to the remaining ORFs were rather modest (ORF23, ORF41, ORF49, and ORF53) or below the detection limit (ORF65). One potential explanation for the discrepancy between transcript level and immunogenicity is that the gene expression and immunogenicity data were not generated in the same animals. Therefore, it is possible that the animals in this study had a transcriptional profile different from that of the animals in the previous study. Another explanation is that transcript levels do not always correlate directly to protein levels. Moreover, immunogenicity is influenced by antigen abundance (52), antigen processing and presentation (12), antigenic competition for major histocompatibility (MH) molecules (15), and the diversity of the T cell repertoire available at the time of infection (54).
Competition for MH complex (MHC) binding can be influenced by the number of peptides included in each pool. Previous studies have suggested that peptide pools consisting of fewer than 50 peptides are ideal for the stimulation of T cell responses (55). Given that 33 of the peptide pools used in this current study were composed of more than 50 peptides/pool, it is possible that the larger mixtures of peptides could have resulted in lower responses. Specifically, the ORF22 pool had 331 peptides; the ORF40 and ORF62 pools had 160 peptides; and the ORF21, ORF28, ORF29, ORF31, ORF37, and ORF55 pools had on average 100 peptides. Consequently, T cell responses to these ORFs could be underestimated in our study. However, it is unlikely that large peptide pool complexity was a factor in the low responses to ORF B, ORF9A, ORF13, ORF15, ORF30, ORF35, ORF39, ORF42, ORF46, ORF51, ORF54, ORF56, ORF58, ORF65, and ORF66 detected during acute infection in the BAL fluid since the average pool size for members of this group is 45 peptides. Similarly, the peptide pools of ORF1, ORF B, ORF9A, ORF25, ORF32, ORF46, ORF49, ORF57, and ORF58 had <25 peptides/pool, and yet T cell responses to these small pools were also on average low and often fell below the threshold established for positive responses. Finally, we observed no correlation between the number of peptides/ORF and the average number of SFC/ORF (data not shown). Thus, for the purposes of this study, a high number of peptides present in a peptide pool did not automatically result in lower T cell responses.
As expected, we detected fewer SVV-specific T cells in BAL fluid samples during latent infection. The number of SVV ORFs that elicited positive T cell responses in >50% of the animals was also reduced, with only 5 ORFs meeting this criterion during latent infection compared to 35 ORFs during acute infection. The T cell response to these 5 ORFs accounted for about 28% of the total SVV-specific T cell response during latency. This narrowing in the T cell response between acute infection and latent infection is most likely due to the reduced magnitude of the memory T cell response. We do, however, acknowledge that the cross-sectional nature of our study design (different cohorts of animals were examined during acute infection and latent infection) could be a confounding factor. Interestingly, we were not able to detect any SVV-specific T cells in peripheral blood during latent infection. Several key studies have shown that memory T cells can primarily reside in nonlymphoid tissues such as the lung (56). Moreover, tissue-specific programming during priming leads to directed migration of memory T cells to that tissue (57–59). Therefore, it is likely that the intrabronchial route of infection used in our model favored the development of memory T cell population in the BAL fluid rather than the blood.
The immunogenic SVV ORFs identified in this study share significant similarities with previous data on human T cell responses. T cell clones derived from ocular fluid from patients with VZV uveitis recognize VZV ORF4, ORF10, ORF14, ORF18, ORF29, ORF31, ORF61, ORF62, ORF63, ORF67, and ORF68 (60), the homologues of which were all targeted by T cells in our analysis of acute infection in BAL fluid. Moreover, we also measured robust responses to SVV ORF4, ORF62, and ORF63 during acute infection that were in line with earlier studies using human T cell lines (18, 19).
The data presented in the paper indicate that during acute infection, both CD4 and CD8 T cells respond to putative immediate early and early (IE and E) proteins, with a trend toward higher CD8 T cell responses. On the other hand, CD4 T cells are predominant in the acute T cell response to putative late structural proteins in the BAL fluid. We also detected a higher CD8 T cell response to late proteins during latency. Given that CD4 T cells are predominant in the response to these structural proteins during acute infection, this suggests that the anti-SVV CD4 T cell response may be preferentially lost over time. This observation is in line with those of previous studies showing that VZV-seropositive young adults (18 to 40 years of age) have higher frequencies of VZV-specific CD4 T cells than older individuals (>55 years) (61) but a comparable frequency of VZV-specific CD8 T cells (43).
One possible approach to boost T cell immunity against VZV and prevent herpes zoster would be to use subunit vaccines expressing highly immunogenic viral antigens such as the ones identified in this study. Previous studies that investigated the ability of subunit vaccines to boost VZV-specific T cell immunity have almost exclusively focused on VZV glycoprotein E (gE, ORF68) (63). The administration of recombinant gE to naive or VZV-vaccinated rodents results in the development or enhancement of gE-specific humoral responses (41, 64–66) and CD4 T cell responses (41). Interestingly, responses to SVV gE are significantly decreased during latent SVV infection compared to acute infection (mean SFC, 155 versus 27, P = 0.005).
Given that VZV reactivation is believed to be largely due to a loss in T cell immunity, boosting T cell responses that are robust during acute infection but are reduced or lost during latency may be a more promising direction for vaccine development. Our study identified several additional ORFs against which T cell responses in the BAL fluid were significantly reduced during latency. Specifically, T cell responses to ORF10, ORF20, ORF29, ORF31, ORF62, and ORF63 showed an average 83% decrease between acute infection and latent infection (P < 0.03 for each of these ORFs). Our report paves the way for a similar analysis to be carried out using human samples. The genetic homology between SVV and VZV and the close relatedness between rhesus macaques and humans provide a robust model in which to evaluate novel vaccines to boost specific anti-VZV T cell responses, thereby preventing herpes zoster in a preclinical setting.
ACKNOWLEDGMENTS
This work was supported by American Heart Association career development grant 0930234N, NIH R01AG037042, 2T32AI007472-16, and NIH 8P51 OD011092-53.
We thank the veterinarians and the husbandry staff at the Oregon National Primate Research Center for their expert care of the animals. We also thank Miranda Fischer, Reed Norris, and Flora Engelmann for their assistance in sample collection and tissue processing. We thank D. N. Streblow and J. B. Sacha for helpful discussion and critical reading of the manuscript.
Footnotes
Published ahead of print 28 August 2013
REFERENCES
- 1.Hickling JK, Borysiewicz LK, Sissons JG. 1987. Varicella-zoster virus-specific cytotoxic T lymphocytes (Tc): detection and frequency analysis of HLA class I-restricted Tc in human peripheral blood. J. Virol. 61:3463–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver BA. 2009. Herpes zoster overview: natural history and incidence. J. Am. Osteopath. Assoc. 109:S2–S6 [PubMed] [Google Scholar]
- 3.Insinga RP, Itzler RF, Pellissier JM, Saddier P, Nikas AA. 2005. The incidence of herpes zoster in a United States administrative database. J. Gen. Intern. Med. 20:748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paryani SG, Arvin AM, Koropchak CM, Dobkin MB, Wittek AE, Amylon MD, Budinger MD. 1984. Comparison of varicella zoster antibody titers in patients given intravenous immune serum globulin or varicella zoster immune globulin. J. Pediatr. 105:200–205 [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention 2012. FDA approval of an extended period for administering VariZIG for postexposure prophylaxis of varicella. MMWR Morb. Mortal. Wkly. Rep. 61:212. [PubMed] [Google Scholar]
- 6.Dueland AN, Martin JR, Devlin ME, Wellish M, Mahalingam R, Cohrs R, Soike KF, Gilden DH. 1992. Acute simian varicella infection: clinical, laboratory, pathologic, and virologic features. Lab. Invest. 66:762–773 [PubMed] [Google Scholar]
- 7.Arvin AM, Koropchak CM, Williams BR, Grumet FC, Foung SK. 1986. Early immune response in healthy and immunocompromised subjects with primary varicella-zoster virus infection. J. Infect. Dis. 154:422–429 [DOI] [PubMed] [Google Scholar]
- 8.Jura E, Chadwick EG, Josephs SH, Steinberg SP, Yogev R, Gershon AA, Krasinski KM, Borkowsky W. 1989. Varicella-zoster virus infections in children infected with human immunodeficiency virus. Pediatr. Infect. Dis. J. 8:586–590 [DOI] [PubMed] [Google Scholar]
- 9.Weigle KA, Grose C. 1984. Molecular dissection of the humoral immune response to individual varicella-zoster viral proteins during chickenpox, quiescence, reinfection, and reactivation. J. Infect. Dis. 149:741–749 [DOI] [PubMed] [Google Scholar]
- 10.Myers MG. 1979. Viremia caused by varicella-zoster virus: association with malignant progressive varicella. J. Infect. Dis. 140:229–233 [DOI] [PubMed] [Google Scholar]
- 11.Zerboni L, Nader S, Aoki K, Arvin AM. 1998. Analysis of the persistence of humoral and cellular immunity in children and adults immunized with varicella vaccine. J. Infect. Dis. 177:1701–1704 [DOI] [PubMed] [Google Scholar]
- 12.Yewdell JW. 2006. Confronting complexity: real-world immunodominance in antiviral CD8+ T cell responses. Immunity 25:533–543 [DOI] [PubMed] [Google Scholar]
- 13.Thomas SL, Hall AJ. 2004. What does epidemiology tell us about risk factors for herpes zoster? Lancet Infect. Dis. 4:26–33 [DOI] [PubMed] [Google Scholar]
- 14.Amanna IJ, Carlson NE, Slifka MK. 2007. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 357:1903–1915 [DOI] [PubMed] [Google Scholar]
- 15.Pamer EG. 1999. Antigen presentation in the immune response to infectious diseases. Clin. Infect. Dis. 28:714–716 [DOI] [PubMed] [Google Scholar]
- 16.Arvin AM, Sharp M, Smith S, Koropchak CM, Diaz PS, Kinchington P, Ruyechan W, Hay J. 1991. Equivalent recognition of a varicella-zoster virus immediate early protein (IE62) and glycoprotein I by cytotoxic T lymphocytes of either CD4+ or CD8+ phenotype. J. Immunol. 146:257–264 [PubMed] [Google Scholar]
- 17.Sadzot-Delvaux C, Kinchington PR, Debrus S, Rentier B, Arvin AM. 1997. Recognition of the latency-associated immediate early protein IE63 of varicella-zoster virus by human memory T lymphocytes. J. Immunol. 159:2802–2806 [PubMed] [Google Scholar]
- 18.Arvin AM, Sharp M, Moir M, Kinchington PR, Sadeghi-Zadeh M, Ruyechan WT, Hay J. 2002. Memory cytotoxic T cell responses to viral tegument and regulatory proteins encoded by open reading frames 4, 10, 29, and 62 of varicella-zoster virus. Viral Immunol. 15:507–516 [DOI] [PubMed] [Google Scholar]
- 19.Jones L, Black AP, Malavige GN, Ogg GS. 2006. Persistent high frequencies of varicella-zoster virus ORF4 protein-specific CD4+ T cells after primary infection. J. Virol. 80:9772–9778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malavige GN, Jones L, Black AP, Ogg GS. 2007. Rapid effector function of varicella-zoster virus glycoprotein I-specific CD4 + T cells many decades after primary infection. J. Infect. Dis. 195:660–664 [DOI] [PubMed] [Google Scholar]
- 21.Malavige GN, Jones L, Black AP, Ogg GS. 2008. Varicella zoster virus glycoprotein E-specific CD4+ T cells show evidence of recent activation and effector differentiation, consistent with frequent exposure to replicative cycle antigens in healthy immune donors. Clin. Exp. Immunol. 152:522–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Messaoudi I, Barron A, Wellish M, Engelmann F, Legasse A, Planer S, Gilden D, Nikolich-Zugich J, Mahalingam R. 2009. Simian varicella virus infection of rhesus macaques recapitulates essential features of varicella zoster virus infection in humans. PLoS Pathog. 5:e1000657. 10.1371/journal.ppat.1000657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer C, Kerns A, Barron A, Kreklywich C, Streblow DN, Messaoudi I. 2011. Simian varicella virus gene expression during acute and latent infection of rhesus macaques. J. Neurovirol. 17:600–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray WL, Pumphrey CY, Ruyechan WT, Fletcher TM. 1992. The simian varicella virus and varicella zoster virus genomes are similar in size and structure. Virology 186:562–572 [DOI] [PubMed] [Google Scholar]
- 25.Pumphrey CY, Gray WL. 1992. The genomes of simian varicella virus and varicella zoster virus are colinear. Virus Res. 26:255–266 [DOI] [PubMed] [Google Scholar]
- 26.Gray WL, Starnes B, White MW, Mahalingam R. 2001. The DNA sequence of the simian varicella virus genome. Virology 284:123–130 [DOI] [PubMed] [Google Scholar]
- 27.Gray WL. 2004. Simian varicella: a model for human varicella-zoster virus infections. Rev. Med. Virol. 14:363–381 [DOI] [PubMed] [Google Scholar]
- 28.Gray WL, Oakes JE. 1984. Simian varicella virus DNA shares homology with human varicella-zoster virus DNA. Virology 136:241–246 [DOI] [PubMed] [Google Scholar]
- 29.Mahalingam R, Gray WL. 2007. The simian varicella virus genome contains an invertible 665 base pair terminal element that is absent in the varicella zoster virus genome. Virology 366:387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felsenfeld AD, Schmidt NJ. 1979. Varicella-zoster virus immunizes patas monkeys against simian varicella-like disease. J. Gen. Virol. 42:171–178 [DOI] [PubMed] [Google Scholar]
- 31.Soike KF, Rangan SR, Gerone PJ. 1984. Viral disease models in primates. Adv. Vet. Sci. Comp. Med. 28:151–199 [DOI] [PubMed] [Google Scholar]
- 32.Clarkson MJ, Thorpe E, Mccarthy K. 1967. A virus disease of captive vervet monkeys (Cercopithecus aethiops) caused by a new herpesvirus. Arch. Gesamte Virusforsch. 22:219–234 [DOI] [PubMed] [Google Scholar]
- 33.Treuting PM, Johnson-Delaney C, Birkebak TA. 1998. Diagnostic exercise: vesicular epidermal rash, mucosal ulcerations, and hepatic necrosis in a cynomolgus monkey (Macaca fascicularis). Lab. Anim. Sci. 48:384–386 [PubMed] [Google Scholar]
- 34.Mccarthy K, Thorpe E, Laursen AC, Heymann CS, Beale AJ. 1968. Exanthematous disease in patas monkeys caused by a herpes virus. Lancet ii:856–857 [DOI] [PubMed] [Google Scholar]
- 35.Kolappaswamy K, Mahalingam R, Traina-Dorge V, Shipley ST, Gilden DH, Kleinschmidt-Demasters BK, McLeod CG, Jr, Hungerford LL, DeTolla LJ. 2007. Disseminated simian varicella virus infection in an irradiated rhesus macaque (Macaca mulatta). J. Virol. 81:411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahalingam R, Traina-Dorge V, Wellish M, Lorino R, Sanford R, Ribka EP, Alleman SJ, Brazeau E, Gilden DH. 2007. Simian varicella virus reactivation in cynomolgus monkeys. Virology 368:50–59 [DOI] [PubMed] [Google Scholar]
- 37.Mahalingam R, Traina-Dorge V, Wellish M, Deharo E, Singletary ML, Ribka EP, Sanford R, Gilden D. 2010. Latent simian varicella virus reactivates in monkeys treated with tacrolimus with or without exposure to irradiation. J. Neurovirol. 16:342–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schoeb TR, Eberle R, Black DH, Parker RF, Cartner SC. 2008. Diagnostic exercise: papulovesicular dermatitis in rhesus macaques (Macaca mulatta). Vet. Pathol. 45:592–594 [DOI] [PubMed] [Google Scholar]
- 39.Draenert R, Altfeld M, Brander C, Basgoz N, Corcoran C, Wurcel AG, Stone DR, Kalams SA, Trocha A, Addo MM, Goulder PJ, Walker BD. 2003. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods 275:19–29 [DOI] [PubMed] [Google Scholar]
- 40.Pahar B, Li J, Rourke T, Miller CJ, McChesney MB. 2003. Detection of antigen-specific T cell interferon γ expression by ELISPOT and cytokine flow cytometry assays in rhesus macaques. J. Immunol. Methods 282:103–115 [DOI] [PubMed] [Google Scholar]
- 41.Dendouga N, Fochesato M, Lockman L, Mossman S, Giannini SL. 2012. Cell-mediated immune responses to a varicella-zoster virus glycoprotein E vaccine using both a TLR agonist and QS21 in mice. Vaccine 30:3126–3135 [DOI] [PubMed] [Google Scholar]
- 42.Efron B, Tibshirani RJ. 1993. An introduction to the Bootstrap. Chapman & Hall, New York, NY [Google Scholar]
- 43.Weinberg A, Zhang JH, Oxman MN, Johnson GR, Hayward AR, Caulfield MJ, Irwin MR, Clair J, Smith JG, Stanley H, Marchese RD, Harbecke R, Williams HM, Chan IS, Arbeit RD, Gershon AA, Schödel F, Morrison VA, Kauffman CA, Straus SE, Schmader KE, Davis LE, Levin MJUS Department of Veterans Affairs (VA) Cooperative Studies Program Shingles Prevention Study Investigators 2009. Varicella-zoster virus-specific immune responses to herpes zoster in elderly participants in a trial of a clinically effective zoster vaccine. J. Infect. Dis. 200:1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arvin AM. 1992. Cell-mediated immunity to varicella-zoster virus. J. Infect. Dis. 166(Suppl 1):S35–S41 [DOI] [PubMed] [Google Scholar]
- 45.Arvin AM. 1996. Immune responses to varicella-zoster virus. Infect. Dis. Clin. North Am. 10:529–570 [DOI] [PubMed] [Google Scholar]
- 46.Abendroth A, Slobedman B, Lee E, Mellins E, Wallace M, Arvin AM. 2000. Modulation of major histocompatibility class II protein expression by varicella-zoster virus. J. Virol. 74:1900–1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arvin AM. 2008. Humoral and cellular immunity to varicella-zoster virus: an overview. J. Infect. Dis. 197:S58–S60 [DOI] [PubMed] [Google Scholar]
- 48. Reference deleted.
- 49.Sawyer MH, Chamberlin CJ, Wu YN, Aintablian N, Wallace MR. 1994. Detection of varicella-zoster virus DNA in air samples from hospital rooms. J. Infect. Dis. 169:91–94 [DOI] [PubMed] [Google Scholar]
- 50.Suzuki K, Yoshikawa T, Tomitaka A, Matsunaga K, Asano Y. 2004. Detection of aerosolized varicella-zoster virus DNA in patients with localized herpes zoster. J. Infect. Dis. 189:1009–1012 [DOI] [PubMed] [Google Scholar]
- 51.Mehta SK, Cohrs RJ, Forghani B, Zerbe G, Gilden DH, Pierson DL. 2004. Stress-induced subclinical reactivation of varicella zoster virus in astronauts. J. Med. Virol. 72:174–179 [DOI] [PubMed] [Google Scholar]
- 52.Jing L, Davies DH, Chong TM, Chun S, McClurkan CL, Huang J, Story BT, Molina DM, Hirst S, Felgner PL, Koelle DM. 2008. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J. Virol. 82:7120–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reference deleted.
- 54.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. 2007. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27:203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hosken N, McGowan P, Meier A, Koelle DM, Sleath P, Wagener F, Elliott M, Grabstein K, Posavad C, Corey L. 2006. Diversity of the CD8+ T-cell response to herpes simplex virus type 2 proteins among persons with genital herpes. J. Virol. 80:5509–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reinhardt RL, Khoruts A, Merica R, Zell T, Jenkins MK. 2001. Visualizing the generation of memory CD4 T cells in the whole body. Nature 410:101–105 [DOI] [PubMed] [Google Scholar]
- 57.Teijaro JR, Turner D, Pham Q, Wherry EJ, Lefrancois L, Farber DL. 2011. Cutting edge: tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187:5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sheridan BS, Lefrancois L. 2011. Regional and mucosal memory T cells. Nat. Immunol. 12:485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cauley LS, Lefrancois L. 2013. Guarding the perimeter: protection of the mucosa by tissue-resident memory T cells. Mucosal Immunol. 6:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Milikan JC, Kinchington PR, Baarsma GS, Kuijpers RW, Osterhaus AD, Verjans GM. 2007. Identification of viral antigens recognized by ocular infiltrating T cells from patients with varicella zoster virus-induced uveitis. Invest. Ophthalmol. Vis. Sci. 48:3689–3697 [DOI] [PubMed] [Google Scholar]
- 61.Asanuma H, Sharp M, Maecker HT, Maino VC, Arvin AM. 2000. Frequencies of memory T cells specific for varicella-zoster virus, herpes simplex virus, and cytomegalovirus by intracellular detection of cytokine expression. J. Infect. Dis. 181:859–866 [DOI] [PubMed] [Google Scholar]
- 62. Reference deleted.
- 63.Mo C, Lee J, Sommer M, Grose C, Arvin AM. 2002. The requirement of varicella zoster virus glycoprotein E (gE) for viral replication and effects of glycoprotein I on gE in melanoma cells. Virology 304:176–186 [DOI] [PubMed] [Google Scholar]
- 64.Vafai A. 1993. Antigenicity of a candidate varicella-zoster virus glycoprotein subunit vaccine. Vaccine 11:937–940 [DOI] [PubMed] [Google Scholar]
- 65.Ludvíková V, Kunke D, Hamsíková E, Kutinová L, Vonka V. 1991. Immunogenicity in mice of varicella-zoster virus glycoprotein I expressed by a vaccinia virus-varicella-zoster virus recombinant. J. Gen. Virol. 72(Pt 6):1445–1449 [DOI] [PubMed] [Google Scholar]
- 66.Leroux-Roels I, Leroux-Roels G, Clement F, Vandepapeliere P, Vassilev V, Ledent E, Heineman TC. 2012. A phase 1/2 clinical trial evaluating safety and immunogenicity of a varicella zoster glycoprotein e subunit vaccine candidate in young and older adults. J. Infect. Dis. 206:1280–1290 [DOI] [PubMed] [Google Scholar]