Abstract
APOBEC3G (A3G) is a cytidine deaminase that restricts human immunodeficiency virus type 1 (HIV-1) and other lentiviruses. Most of these viruses encode a Vif protein that directly binds A3G and leads to its proteasomal degradation. Both Vif proteins of HIV-1 and African green monkey simian immunodeficiency virus (SIVagm) bind residue 128 of A3G. However, this position does not control the A3G degradation by Vif variants derived from HIV-2 and SIVmac, which both originated from SIV of sooty mangabey monkeys (SIVsmm), suggesting that the A3G binding site for Vif proteins of the SIVsmm/HIV-2 lineage differs from that of HIV-1. To map the SIVsmm Vif binding site of A3G, we performed immunoprecipitations of individual A3G domains, Vif/A3G degradation assays and a detailed mutational analysis of human A3G. We show that A3G residue 129, but not the adjacent position 128, confers susceptibility to degradation by SIVsmm Vif. An artificial A3G mutant, the P129D mutant, was resistant to degradation by diverse Vifs from HIV-1, HIV-2, SIVagm, and chimpanzee SIV (SIVcpz), suggesting a conserved lentiviral Vif binding site. Gorilla A3G naturally contains a glutamine (Q) at position 129, which makes its A3G resistant to Vifs from diverse lineages. We speculate that gorilla A3G serves as a barrier against SIVcpz strains. In summary, we show that Vif proteins from distinct lineages bind to the same A3G loop, which includes positions 128 and 129. The multiple adaptations within this loop among diverse primates underscore the importance of counteracting A3G in lentiviral evolution.
INTRODUCTION
Many Old World primate species among African primates are naturally infected with their own version of simian immunodeficiency virus (SIV) (1). The pandemic HIV-1 group M is believed to have originated from a single successful cross-species transmission event from SIV-infected chimpanzees to humans (2). Three additional transmission events of SIV from chimpanzees and gorillas resulted in nonpandemic HIV-1 groups N, O, and P (Fig. 1A) (3–5). In addition, SIV from naturally infected sooty mangabey monkeys (SIVsmm) was transmitted to humans on at least nine occasions, resulting in HIV-2 groups A through I (Fig. 1A) (6–8). SIVsmm has also been transmitted to Asian macaques in captivity, resulting in SIVmac (1).
Fig 1.
SIVsmm can overcome human APOBEC3G. (A) Many primates are naturally infected with SIV. SIVgor was transmitted to humans, resulting in HIV-1 groups O and P, and transmission of SIVcpz resulted in HIV-1 group N and the pandemic group M (subtypes A through J). Transmission of SIVsmm gave rise to nine HIV-2 groups (A to I). (B) SIVagm and HIV-1 Vif neutralize only their own species' A3G. Amino acid 128 in A3G controls this species specificity: humans contain an aspartate (D) at position 128, while A3Gagm variants carry a lysine (K) at this position. (C) C-terminally FLAG-tagged Vif expression plasmids (50 ng) were transfected with HA-tagged human A3G (hA3G) WT (100 ng) in 293T cells. Two days later, cells were lysed and analyzed by Western blotting for A3G (HA) and Vif (FLAG) expression. Tubulin served as a loading control. The asterisk denotes a nonspecific band. The Vif C133S mutant is unable to bind cullin 5 and serves as a negative control.
APOBEC3G (A3G) potently restricts HIV-1 and other lentiviruses by deaminating the viral DNA during reverse transcription, which subsequently becomes degraded or severely mutated (9–11). However, most lentiviruses encode the accessory protein Vif that mediates the proteasomal degradation of A3G (12–14). As a result of genetic conflicts between Vif and A3G, positive selection on both proteins has led to host-specific A3G/Vif adaptations (15–18). For example, Vifs from HIV-1 and SIV of African green monkeys (SIVagm) can efficiently degrade their cognate A3G but are unable to counteract A3G from the other species (Fig. 1B) (19–23). The determinant of this host specificity is a particular residue at position 128 (aspartic acid [D] in human A3G and lysine [K] in African green monkey A3G [AGM A3G]) of the A3G protein. Mutating human A3G 128D to K (as in AGM A3G) and vice versa fully reversed this specificity (Fig. 1B) (19, 20, 22, 23). The resistance of human A3G to SIVagm Vif could serve as an effective barrier and may explain why SIVagm has not colonized humans. Several studies have shown that A3G residue 128 directly affects the binding of HIV-1 and SIVagm Vif to their respective A3G proteins, suggesting that this residue is part of the Vif binding site (19, 20, 22, 24, 25). However, adjacent amino acids, such as those at positions 129 and 130, appear to also be required for HIV-1 Vif/A3G binding (25–27), and A3G position 130 is also involved in African green monkey subspecies-specific adaptions to Vif degradation (15). Current data suggest that A3G residues 128 to 130 are part of an exposed loop between the beta strand 4 and the alpha helix 3, but to date, no molecular Vif structure or information about the A3G-Vif protein interface is available (22).
In contrast to the well-established requirement of A3G position 128 for HIV-1 Vif binding (19, 22, 23), HIV-2 and SIVmac Vif are capable of recognizing A3G independently of the residue at position 128 (21, 23). Little is known about how the SIVsmm Vif protein counteracts human A3G. We thus considered the possibility that the Vif proteins of SIVsmm, HIV-2, and SIVmac strains use an A3G binding site that does not include position 128.
In this study, we show that residues at position 129 in A3G (adjacent to position 128) control Vif binding and mediate resistance to degradation by diverse Vifs from SIVsmm, HIV-2, HIV-1, and SIVagm lineages. A3G 129P is conserved among humans and most primates except gorillas. The gorilla A3G contains 129Q, which yields an A3G protein that is resistant to HIV-1 and SIV Vif-mediated degradation. Thus, our data indicate that Vif proteins from diverse HIV/SIV lineages use the same binding site in A3G to mediate its degradation.
MATERIALS AND METHODS
Plasmids.
The replication-competent molecular clones NL4-3 and NL4-3 ΔVif were provided by the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health NIH Reagent Program (28, 29). The SIVsmm PG molecular clone was a generous gift from Frank Kirchhoff, University of UIm, Ulm, Germany. SIVsmm ΔVif was produced by digesting the full-length molecular clone with XcmI, gel purifying the digested plasmid, and religating the gel-purified product to introduce a large deletion into the Vif sequence.
Plasmids with Vif sequences (Table 1) (28, 30–38) were used as templates for PCR amplification with Vif primers, containing NotI and EcoRI restriction sites, specific for the 5′ and 3′ regions of each variant, respectively. A carboxy-terminal FLAG tag was added to all cloned Vif sequences by overlapping PCR. Amplicons were digested with NotI/EcoRI, and the coding regions of the subcloned Vifs were inserted into pCRV1 as previously described (39, 40) Site-directed mutagenesis of Vif was performed using overlapping PCR as described previously (39). The mutated constructs were cloned into pCRV1 vector and confirmed by sequencing. Primer sequences are available upon request. NCBI reference sequence numbers for the Vifs used are U26942.1 (NL4-3 Vif), AF077017.1 (SIVsmm), U04005.1 (SIVagmSAB), U58991.1 (SIVagmTAN), EF394356.1 (SIVcpzTAN1), EF394357.1 (SIVcpzTAN2), DQ373065.1 (SIVcpzEK505), DQ373064.1 (SIVcpzLB715), EF535994.1 (SIVcpzMB897), M76764.1 (SIVmac239).
Table 1.
Summary of the Vif variants used in this study
| Vif variant | HIV or SIV origin | Plasmid sourcea | Reference |
|---|---|---|---|
| HIV-1 NL4-3 | Human | NIH 114 | 28 |
| SIVsmm (PBj) | Sooty mangabey monkey | NIH 2998 | 30 |
| SIVsmm (PGm) | Sooty mangabey monkey | F. Kirchhoff | 31 |
| HIV-2 Rod | Human | NIH 207 | 32 |
| SIVagm Sab | African green monkey | NIH 2614 | 33 |
| SIVagm Tan | African green monkey | NIH 3444 | 34 |
| SIVmac 239 | Rhesus macaque | NIH 210 | 35 |
| SIVgor | Gorilla | NIH 11722 | 36 |
| SIVpts1 (Tan1) | Chimpanzee (Pan troglodytes schweinfurthii) | B. H. Hahn | 37 |
| SIVpts2 (Tan2) | Chimpanzee (Pan troglodytes schweinfurthii) | B. H. Hahn | 37 |
| SIVptt1 (EK505) | Chimpanzee (Pan troglodytes troglodytes) | B. H. Hahn | 38 |
| SIVptt2 (LB715) | Chimpanzee (Pan troglodytes troglodytes) | B. H. Hahn | 38 |
| SIVptt3 (MB897) | Chimpanzee (Pan troglodytes troglodytes) | B. H. Hahn | 38 |
NIH strains were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health NIH Reagent Program.
Lowland gorilla untransformed fibroblasts were obtained from the Coriell Institute (catalog no. PR00950), and A3G was amplified from cDNA (SuperScript III first-strand synthesis kit; Invitrogen) using the primers 5′-TACAAGCTTATGACGYCTCAGTTCAGAAACACA (forward) and 5′-AACATCGTATGGGTAGTTTCCCTGATTCTGGAGAATGG (reverse). All A3G genes were C-terminally triple-hemagglutinin (3×HA) tagged and cloned as described previously (41). The African green monkey A3G expression plasmid was obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health (NIH) Reagent Program, and was used as a template for PCR amplification (33, 34). Sooty mangabey A3G was amplified from cDNA derived from peripheral blood mononuclear cells (PBMC) of two sooty mangabeys (provided by Guido Silvestri) using the primers 5′-GCCCTGGGAGGTCACTTTAAGGA and 5′-TGGCTCAACCCAGGTCTCTGCCT for the first round of amplification and 5′-CTTTAAGGAGGGCTGTCCTAAAA and 5′-CTTCCTTAGAGACTGAGGCCCAT for the final round of amplification. PCR products were cloned using a StrataClone Blunt kit (Stratagene). Sooty mangabey A3G haplotypes were amplified from StrataClone plasmids with PfuUltra II polymerase (Stratagene). A carboxyl-terminal 3×HA tag was added to all cloned A3G sequences by overlapping PCR, followed by cloning into the PTR600 expression plasmid (41). All DNA preparations were sequenced to confirm the integrity of the APOBEC3 sequences. NCBI reference sequence numbers for the A3G used are JN662548.1 (African green monkey), NP_068594.1 (human), and AH013828.1 (gorilla).
Culture of cell lines.
HEK 293T and TZM-bl reporter cells were maintained in Dulbecco's modified Eagle medium (Corning Cellgro) supplemented with 10% fetal bovine serum (FBS) and 100 U/ml penicillin-streptomycin. TZM-bl cells were provided by the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health NIH Reagent Program (42).
Vif-mediated A3G degradation.
HEK 293T cells were cotransfected with FLAG-tagged A3G expression vectors (100 ng) and increasing amounts of Vif pCRV1 expression plasmid (0, 2.5, 5, 10, 25, and 50 ng) and pCRV1 empty plasmid (total amount of pCRV1, 50 ng). The transfections were performed in a 24-well format using 3 mg/ml polyethylenimine (PEI; Polysciences, Inc.). Transfected cells were lysed 2 days posttransfection and analyzed by Western blotting.
Western blot analysis.
Transfected 293T cells were lysed (1% sodium dodecyl sulfate [SDS], 50 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 5 mM EDTA). Proteins were separated on 10% SDS-polyacrylamide gel (Invitrogen), transferred to a polyvinylidene difluoride (PVDF) membrane (Pierce), and probed with anti-HA polyclonal antiserum from rabbits (Sigma), anti-FLAG monoclonal from rabbits (Sigma), and anti-tubulin monoclonal antibody from mice (Sigma). Membranes were subsequently incubated with horseradish peroxidase-conjugated secondary antibodies (Sigma), developed with SuperSignal West Femto (Pierce), and detected by using the ProteinSimple FluorChem E imaging system.
Renilla luciferase-based A3G degradation assay.
Human A3G was C-terminally tagged with Renilla luciferase using standard overlapping PCR. HEK 293T cells in 24-well plates were transfected with 5 ng of the A3G-Renilla constructs and 60 ng of Vif expression plasmid. Twenty-four hours later, cells were lysed in 150 μl 1× Renilla-specific lysis buffer (Promega) for 10 min at room temperature. Lysates (20 μl) were transferred to black 96-well plates (Greiner), 75 μl diluted substrate (Promega) was added to each well, and luciferase activity was assayed on a Victor-3 1420 multilabel counter (PerkinElmer). Average values and standard deviations were calculated from data from three independent transfections.
Assessment of viral infectivity using single-cycle infectivity assays.
HA-tagged A3G expression vectors (50 ng) and FLAG-tagged Vif vectors (10 ng) were cotransfected with the HIV molecular clones NL4-3 WT and NL4-3 ΔVif (500 ng) in 293T cells. The transfections were performed in a 24-well format using 3 mg/ml polyethylenimine (PEI; Polysciences, Inc.), and culture medium was replenished after 24 h. The supernatants were harvested 48 h after transfection and used to infect 104 TZM-bl cells/well in black 96-well plates. TZM-bl cells were infected in triplicate with 20 μl of cell-free viral supernatants. β-Galactosidase activity was measured at 48 h postinfection using a Galacto-Star system (Applied Biosystems), as described previously (39). The data from three independent transfections were used to calculate average values and standard deviations.
Coimmunoprecipitation.
HEK 293T cells were cotransfected with FLAG-Vif expression plasmids (100 ng), HA-A3G expression plasmid (900 ng) and NL4-3ΔVif molecular clone (1,000 ng) in a 6-well format (2 μg DNA total). Different amounts of Vif and A3G were tested in order to determine the optimal Vif:A3G transfection ratio (1:1, 1:4, and 1:9). Cells were lysed 2 days posttransfection in a mild lysis buffer (0.5% Triton X-100 in 1× phosphate-buffered saline [PBS] supplemented with EDTA-free protease inhibitor cocktail; Roche), and the cell lysates were cleared at 14,000 × g for 10 min at 4°C. Cleared lysates were incubated with EZ-View anti-HA beads (Sigma) at 4°C for 2 h. Beads were washed 4 times in mild lysis buffer, followed by 4 additional washes in stringent lysis buffer (1% Triton X, 0.1% SDS, 500 mM NaCl in PBS supplemented with EDTA-free protease inhibitor cocktail; Roche). Proteins were eluted from the beads by boiling in lithium dodecyl sulfate (LDS) loading buffer (Sigma). Proteins were analyzed by Western blotting for Vif (FLAG), A3G (HA), and tubulin.
Nucleotide sequence accession numbers.
Four A3G sequences obtained from representative RNA transcripts of the two sooty mangabeys have been submitted to GenBank (accession numbers KF591583, KF591584, KF591585, and KF591586.)
RESULTS
SIVsmm Vif efficiently degrades human A3G.
Although the importance of A3G position 128 is well established for HIV-1 and SIVagm Vif binding and A3G degradation (19, 20, 22, 23), little is known about the SIVsmm and HIV-2 Vif binding site. We first performed an A3G degradation assay with a panel of diverse SIV, HIV-1, and HIV-2 Vifs. Most of the Vif variants tested, including SIVsmm, could efficiently degrade human A3G compared to the no-Vif control or the inactive Vif mutant C133S, which cannot bind cullin 5 (43–45) (Fig. 1C). In agreement with previous studies (16, 19–23), SIVagm Vif failed to degrade human A3G. Taken together, our experimental system is suitable for efficiently discriminating between active and inactive Vif proteins and shows that human A3G is efficiently degraded by SIVsmm Vif.
A3Gsmm variants all contain 128K and restrict SIVsmm ΔVif.
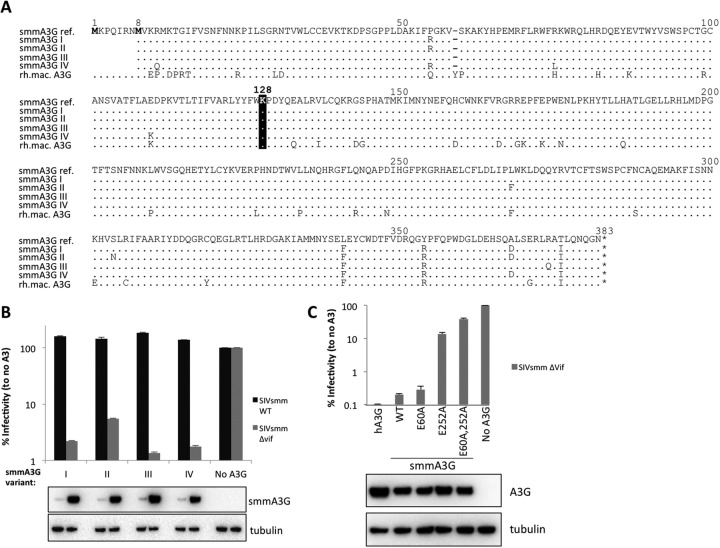
Given that human A3G was efficiently degraded by SIVsmm Vif (Fig. 1C), we speculated that the residues at position 128 of A3Gsmm and human A3G must be similar. However, at the start of this study, no information on A3Gsmm sequences was available. We therefore cloned and sequenced A3G transcripts from peripheral blood mononuclear cells (PBMC) of two sooty mangabeys. We identified four different A3Gsmm transcript variants, all of which encoded a lysine at position 128, like A3Gagm (Fig. 2A). Interestingly, all A3Gsmm variants started at a downstream start codon at position eight compared to the recently published A3Gsmm sequence (46) and the reference A3Gmac sequence (21).
Fig 2.
Sooty mangabey A3G contains a lysine at position 128. (A) A3G transcripts were amplified from PBMCs of two different sooty mangabey, and cDNAs were cloned and sequenced. Four different variants were observed, each containing a lysine at position 128 (A3Gsmm reference sequence, KC176186.1 [46]; A3Gmac sequence, NM_001198693.1 [21]). (B) SIVsmm WT and a ΔVif molecular clone were transfected with 100 ng of the different smmA3G variants in 293T cells. Two days later, supernatants were used to infect TZM-bl reporter cells, and β-galactosidase activity was measured 2 days postinfection. Error bars represent the standard deviations for triplicate transfections in a representative experiment. (C) 293T cells were cotransfected with 500 ng SIVsmm ΔVif molecular clone and 100 ng WT or mutant A3G. Two days later the supernatants were used to infect TZM-bl cells, and β-galactosidase activity was measured 2 days after infection. The same transfected 293T cells were lysed and analyzed by Western blotting. Tubulin served as a loading control.
We next performed single-cycle infectivity assays with SIVsmm and SIVsmm ΔVif to determine the antiviral potency of the A3Gsmm variants. Figure 2B shows that all four variants potently restrict SIVsmm ΔVif. Moreover, all four A3Gsmm variants were degraded with similar efficiency by SIVsmm Vif, resulting in complete rescue of viral infectivity (Fig. 2B).
We next explored the mode of A3Gsmm restriction by mutating the N-terminal, C-terminal, or both deaminase domains (Fig. 2C). Restriction of SIVsmm ΔVif was maintained upon mutation of the N-terminal A3G region but was largely lost when the C-terminal A3G domain or both deaminase domains were inactivated. These findings indicate that the C-terminal deaminase domain of A3Gsmm, much like in the human A3G, is essential for efficient lentiviral restriction (47–50).
SIVsmm Vif-mediated A3G degradation is unaffected by A3G residue 128.
Our results show that SIVsmm efficiently degrades human A3G, despite the residue difference at position 128 between A3Gsmm and human A3G (Fig. 1C). This indicates that SIVsmm, similarly to HIV-2 and SIVmac, can degrade A3G independently of the residue at position 128 (21, 23).
To further explore the role of A3G position 128 in SIVsmm Vif-mediated degradation, we tested the efficiencies of different lentiviral Vif variants (two different SIVsmm, HIV-2, SIVagm, and HIV-1 NL4-3 Vifs) to degrade human A3G, A3Gsmm, A3Gagm, and the corresponding variants in which position 128 was mutated to either a D or K. Figure 3 shows that SIVsmm and HIV-2 Vifs both efficiently degraded A3G proteins from humans, African green monkeys, and sooty mangabeys regardless of their 128 residue. Of note, SIVagm Vif degraded only A3G variants that contain 128K, whereas HIV-1 Vif specifically degraded A3G 128D variants. These data indicate that SIVsmm and HIV-2 Vif are able to counteract different A3G proteins irrespective of the identity of the residue at position 128.
Fig 3.

SIVsmm and HIV-2 Vif degrade A3G regardless of position 128. Vif expression plasmids (50 ng) were transfected with 100 ng A3G WT or position A3G 128 mutants in 293T cells. Two days later, cells were lysed and analyzed by Western blotting for A3G (HA) or Vif (FLAG). Tubulin served as a loading control.
SIVsmm Vif-mediated A3G degradation requires a region between A3G amino acid positions 122 and 148.
We next investigated whether SIVsmm and HIV-2 Vifs are more tolerant to variation at position 128 or whether they use another region within A3G for binding. Precedents for other APOBEC3 binding sites being used by Vif exist: HIV-1 Vif binds the N-terminal domain of human A3G around position 128 (19, 20, 22, 23) but associates with the C-terminal domains of human A3D and human A3F to induce their proteasomal degradation (51–54).
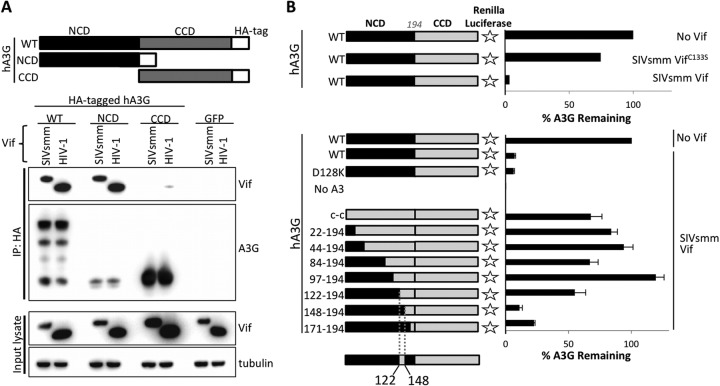
To determine which A3G domain is utilized for SIVsmm binding, we performed coimmunoprecipitations with SIVsmm Vif and the full-length human A3G or its N- and C-terminal domains. We found that SIVsmm Vif specifically coimmunoprecipitates with full-length A3G and its N-terminal domain, suggesting that SIVsmm Vif binds to the N-terminal domain of human A3G (Fig. 4A).
Fig 4.
SIVsmm Vif interacts between positions 122 and 148 in the N-terminal domain of human A3G. (A) 293T cells were transfected with SIVsmm Vif and HA-tagged human A3G WT and the N-terminal and C-terminal A3G domains. Cells were lysed in a gentle lysis buffer 2 days later, and the cleared lysates were incubated with anti-HA beads (Sigma) at 4°C for 2 h. Beads were thoroughly washed in lysis buffer, and proteins were eluted by boiling in sample loading buffer. Proteins were analyzed by Western blotting. (B) C-terminally Renilla luciferase-tagged chimeras were made by replacing portions of the N-terminal cytidine deaminase domain (NCD) with corresponding regions from the C-terminal cytidine deaminase domain (CCD). 293T cells were transfected with 5 ng of the A3G construct and 60 ng of Vif expression plasmid. The next day, cells were lysed and analyzed for Renilla luciferase activity as a measure of A3G expression.
In order to map the region of interaction more precisely, we established a rapid, quantitative, and sensitive degradation assay based on degradation of human A3G fused to Renilla luciferase at the C terminus. The A3G-luciferase fusion protein was stably expressed when transfected in the absence of Vif or cotransfected with the SIVsmm VifC133S mutant, which cannot bind cullin 5 (Fig. 4; high numbers of relative light units [RLU] indicate high levels of expression) (43–45). However, in the presence of SIVsmm Vif, A3G was degraded and luciferase expression was reduced (Fig. 4B). Since SIVsmm Vif fails to bind the C-terminal A3G domain (Fig. 4A), we constructed chimeras of the N-terminal and C-terminal domains, which were then tested for Renilla luciferase expression in the presence of SIVsmm Vif. The Renilla luciferase expression of most of these chimeras was not affected by SIVsmm Vif, indicating that they were resistant to Vif-mediated degradation (Fig. 4B). However, inclusion of N-terminal A3G residues 122 to 148 conferred sensitivity to SIVsmm Vif-mediated degradation, suggesting that this region contains a binding site for SIVsmm Vif (Fig. 4B).
Position 129 in human A3G affects SIVsmm Vif-mediated degradation.
Our data imply that residues between positions 122 and 148 of human A3G are necessary and required for SIVsmm Vif degradation. This region includes the A3G loop containing position 128, which is important for HIV-1 and SIVagm Vif binding and degradation (19, 20, 22–24, 26, 27, 55, 56). We thus performed a comprehensive mutational analysis of this A3G loop by alanine-scanning mutagenesis of the residues between positions 126 and 130. We subsequently tested five mutants in the Renilla luciferase degradation assay (Fig. 5A). Cotransfection of the A3G-luciferase constructs with HIV-1 Vif showed that A3G containing 129A was as resistant to HIV-1 degradation as A3G containing 128K (control; Fig. 5B), an observation which is consistent with results from previous studies (19, 20, 22, 23). A3G 127A and 129A mutants displayed some resistance to SIVsmm-mediated degradation, albeit to a lesser degree than HIV-1 Vif (Fig. 5C). Interestingly, position 129 in human A3G has been described as conferring resistance to HIV-1 Vif-mediated degradation (25, 27).
Fig 5.
A3G position 129 affects SIVsmm Vif-mediated degradation. (A) A3G amino acids 126 to 130 were individually mutated to alanine and tested in the luciferase degradation assay. (B) Luciferase-tagged hA3G alanine mutants (10 ng) were cotransfected with 20 ng NL4-3 Vif or the NL4-3 Vif mutant C133A. A3G expression is plotted as the ratio of WT RLU to RLU of the inactive C133S Vif mutant. A3G D128K served as a control. (C) Luciferase-tagged hA3G alanine mutants (10 ng) were cotransfected with 60 ng SIVsmm Vif in 293T cells, and Renilla luciferase activity was measured the next day. (D) The indicated amino acids were introduced into human A3G at position 129 and tested for Renilla luciferase activity the next day.
We chose to focus next on position 129 for in-depth analysis, since A3G position 127 is part of the RNA binding domain, which also mediates packaging into virions (25, 26). We tested nine different residues with various biochemical properties (e.g., hydrophobic, polar, acidic, or basic) at this position within the human A3G luciferase fusion construct. Several residues resulted in resistance against SIVsmm Vif-mediated degradation, with the A3G mutant containing an acidic aspartic acid (D) at position 129 displaying the highest level of resistance (Fig. 5D). In summary, our data identify A3G position 129 as an important determinant for degradation by SIVsmm Vif.
Human A3G 129D is resistant to HIV-1, HIV-2, and SIV Vif-mediated degradation.
We next analyzed the effect of the aspartic acid at A3G position 129 within full-length human A3G using degradation and infectivity assays. Increasing amounts of SIVsmm Vif (0, 10, 25, 50, or 100 ng) were cotransfected with A3G WT (100 ng), and Vif-mediated degradation was assessed by Western blotting 2 days posttransfection. SIVsmm Vif efficiently degraded human A3G, whereas A3G 129D was resistant to SIVsmm Vif degradation even at high Vif levels, which indicates that 129D confers resistance to degradation (Fig. 6A).
Fig 6.
Human A3G P129D mutant resists degradation of diverse Vifs. (A) 293T cells were transfected with 0, 10, 25, 50, or 100 ng SIVsmm Vif and 100 ng hA3G. Two days later, cells were lysed and analyzed by Western blotting. (B) HA-tagged hA3G plasmid (0, 10, 25, 50 or 100 ng) was transfected with 500 ng SIVsmm ΔVif or SIVsmm WT molecular clone in 293T cells; supernatants were used to infect TZM-bl cells 2 days later, and β-galactosidase activity was measured 2 days postinfection. The same transfected 293T cells were also lysed and analyzed by Western blotting. (C) A total of 10 ng of the indicated Vifs, 50 ng of human WT, 129D, and 128K A3G, and 500 ng NL4-3 ΔVif were used to transfect 293T cells. Supernatants were used to infect TZM-bl cells 2 days later and β-galactosidase activity was measured 2 days postinfection. The same transfected 293T cells were also lysed and analyzed by Western blotting.
Infectivity assays were performed using full-length infectious SIVsmm and SIVsmm ΔVif molecular clones transfected in the presence of increasing amounts of human wild-type A3G (A3G WT; 129 proline [P]) and A3G 129D. Assessment of viral infectivity showed that human A3G WT and A3G 129D both restrict SIVsmm ΔVif efficiently (Fig. 6B, left), indicating that 129D does not affect A3G activity in the absence of Vif. However, only the activity of A3G WT was counteracted by WT SIVsmm, while A3G 129D was fully resistant (Fig. 6B, right).
In order to determine the sensitivity of human A3G 129D to other Vif proteins, we performed infectivity assays with HIV-1 ΔVif complemented with a comprehensive panel of lentiviral Vifs in the presence of human A3G WT, A3G129D, and A3G128K. Five different Vifs (except SIVagm Vifs) efficiently counteracted and degraded A3G WT (Fig. 6C). However, human A3G 129D was found to be resistant to all Vifs except SIVmac Vif. Lastly, as previously shown, A3G 128K was resistant only to SIVcpz and HIV-1 Vifs (19–23). Thus, an aspartic acid at position 129 in human A3G confers resistance to Vif variants from most lineages, suggesting that these diverse viral proteins all recognize the same interface within A3G.
Gorilla A3G is naturally resistant to diverse lentiviral Vifs because it contains a glutamine at position 129.
The importance of residue 129 in resistance to Vif-mediated degradation prompted us to look at the conservation of position 129 in A3G across different nonhuman primates. We found that the P at position 129 was highly conserved among most primates with the exception of gorilla A3G, which contains a glutamine (Q) at position 129 (Fig. 7A) (17, 18, 46).
Fig 7.
A glutamine residue at position 129 in the gorilla A3G protects against degradation by noncognate SIV Vifs. (A) SIVcpzPtt has been transmitted to both humans and gorillas, giving rise to HIV-1 groups M and N as well as SIVgor. SIVgor has also been transmitted to humans, giving rise to HIV-1 groups O and P. A3G 129P is conserved among humans, chimps, sooty mangabeys, African green monkeys, and rhesus macaques, but gorillas contain A3G 129Q. (B) Human A3G WT and gorilla A3G were cotransfected with the indicated Vif plasmids and NL4-3Δvif molecular clone in 293T cells. Two days later, the supernatants were used to infect TZM-bl cells, and β-galactosidase activity was measured 2 days postinfection. Transfected 293T cells were lysed and analyzed by Western blotting. (C) Human A3G 129Q and gorilla A3G 129P were cotransfected with the indicated Vif plasmids and NL4-3Δvif molecular clone in 293T cells. Two days later the supernatants were used to infect TZM-bl cells, and β-galactosidase activity was measured 2 days postinfection. Transfected 293T cells were lysed and analyzed by Western blotting. (D) Human A3G 129D was cotransfected with the indicated Vif plasmid and NL4-3Δvif molecular clone in 293T cells. Transfected 293T cells were lysed and analyzed by Western blotting.
Gorillas are naturally infected with SIVgor, which was transmitted originally from SIVcpz-infected chimpanzees living in the same habitat (Fig. 7A) (36). In addition, SIVgor has been transmitted to humans at least once, resulting in HIV-1 group P (3, 57). SIVgor has also been proposed to be the source of HIV-1 group O in humans (36, 58).
In order to analyze the effect of residue 129Q on the antiviral activity of gorilla A3G, we cloned gorilla A3G and generated its Q129P mutant. We compared the sensitivity of the gorilla A3G to various lentiviral Vifs with the sensitivity of human A3G WT and its corresponding P129Q mutant. SIVgor Vif counteracted human A3G with an efficiency similar to that of most other Vifs (Fig. 7B, left). However, most Vifs tested failed to counteract gorilla A3G with the exception of SIVgor Vif itself (Fig. 7B, right). To exclude the possibility that other residue differences between human and gorilla A3G affected the resistance to Vif, we exchanged both 129 residues and tested their sensitivity to various Vif proteins. The gorilla Q129P mutant behaved like human A3G inasmuch as both were counteracted by all Vifs except SIVagm Vif (Fig. 7C, left). Conversely, introducing 129Q into human A3G mimicked the resistance pattern observed for gorilla A3G (Fig. 7C, right). A comparison of the same Vifs cotransfected with A3G 129D showed a resistance pattern similar to that observed with A3G 129Q (Fig. 7D). Taken together, these results indicate that residue 129Q in gorilla A3G confers resistance to a diverse panel of Vif proteins.
A3G 129Q confers resistance to divers SIVcpz Vifs.
SIVgor originated from SIVcpz (36), but the SIVcpz Vif tested did not efficiently counteract gorilla A3G (Fig. 7B). We therefore tested five additional SIVcpz Vifs derived from members of both the Pan troglodytes troglodytes (SIVcpzPtt) and Pan troglodytes schweinfurthii (SIVcpzPts) subspecies of chimpanzees for their efficiency to counteract gorilla A3G. All SIVcpz Vifs counteracted the gorilla A3G only poorly (Fig. 8A) but efficiently degraded the “humanized” A3G version Q129P (Fig. 8B). Of note, SIVgor Vif efficiently antagonized both gorilla A3G and its corresponding 129P mutant (compare Fig. 8A and B). In conclusion, all the SIVcpz Vifs tested failed to efficiently counteract gorilla A3G because of a protective glutamine residue at position 129.
Fig 8.

A3G 129Q confers resistance to divers SIVcpz Vifs. A panel of SIVcpz Vif expression plasmids were cotransfected with gorilla A3G WT (A) and gorilla A3G 129P (B) and the NL4-3Δvif molecular clone in 293T cells. Two days later, the supernatants were used to infect TZM-bl cells, and β-galactosidase activity was measured 2 days postinfection. Transfected 293T cells were lysed and analyzed by Western blotting. Pts1, SIVcpzTAN1; Pts2, SIVcpzTAN2; Ptt1, SIVcpzEK505; Ptt2, SIVcpzLB715; Ptt3, SIVcpzMB897.
A3G 129D and 129Q reduce Vif binding.
We next examined whether sensitivity to degradation would correlate with association between Vif and APOBEC3. Toward this end, we titrated the DNA Vif/A3G ratios in immunoprecipitation experiments (1:1, 1:4, and 1:9). The 1:1 and 1:4 ratios showed no differences in HIV-1 Vif binding to human A3G WT and A3G 128K, but the lowest Vif:A3G ratio (1:9) showed a clear difference in binding (Fig. 9A). The latter result is in good agreement with other studies (19, 20, 22, 25, 43, 59).
Fig 9.

SIVsmm Vif binds human A3G position 129. (A) A total of 500 ng Vif and 500 ng A3G (1:1), 250 ng Vif and 750 ng A3G (1:4), and 100 ng Vif and 900 ng A3G (1:9) were used to cotransfected 293T cells. Cells were lysed in a gentle lysis buffer 2 days later, and the cleared lysates were incubated with anti-HA beads (Sigma) at 4°C for 2 h. Beads were thoroughly washed in lysis buffer and proteins were eluted by boiling in sample loading buffer. Proteins were analyzed by Western blotting for Vif (FLAG) and hA3G (HA). (B) A total of 100 ng Vif and 900 ng A3G plasmids (1:9) were transfected, and cell lysates were subjected to coimmunoprecipitation as described above. (C) The predicted structure for the NTD of hA3G was modeled with SwissModel using APOBEC3C as a reference (61). SIVgor Vif is sensitive only to variation at position 128, whereas HIV-1 and SIVagm Vif are sensitive to changes in both positions 128 and 129; HIV-2 and SIVsmm Vif interact only with position 129.
We performed immunoprecipitations using the optimal 1:9 Vif/A3G ratio, with HIV-1, SIVsmm, and HIV-2 Vifs cotransfected with human A3G WT, human A3G 128K, human A3G 129D, and human A3G 129Q. HIV-1 Vif association was decreased for A3G 128K, 129D, and 129Q mutants compared to A3G WT, despite the lower expression levels of A3G (which was due to Vif-mediated A3G degradation) (Fig. 9B). SIVsmm and HIV-2 Vif association was reduced only for A3G 129D and 129Q, which corresponded well with their resistance to Vif mediated degradation. Thus, the immunoprecipitation results show that human A3G 129D and human A3G 129Q escape Vif-mediated degradation because of reduced association with Vif.
DISCUSSION
HIV-1 and HIV-2 originated from SIV of chimpanzees and sooty mangabeys, respectively (2). Successful transmission of these simian viruses required adaptation to human restriction factors (2, 15, 17). The HIV-1 Vif binding site of A3G includes position 128 (19, 20, 22, 23), which constitutes a barrier between humans and African green monkeys. However, degradation by HIV-2 and SIVmac Vif proteins from the distinct SIVsmm lineage was unaffected by this position, raising the possibility that another binding site might be used (22, 23). Our data now show that this is not the case. We find that residue 129 in human A3G confers resistance to Vif-mediated degradation and leads to reduced association to a panel of diverse Vif proteins, including SIVsmm, HIV-2, HIV-1, SIVcpz, and SIVagm Vifs. Our results suggest that HIV and SIV Vifs adapted differently to the various primate A3G variants, resulting in HIV-1 and SIVagm Vifs requiring certain residues at positions 128 and 129, whereas HIV-2 and SIVsmm Vifs interact only with position 129. SIVgor Vif binding, however, depends only on the residue at A3G position 128 (Fig. 9C). However, all of these diverse Vif proteins bind the same exposed loop in A3G.
Although most reports showed that A3G position 128 affected Vif binding directly (19, 20, 22), some reports speculated that Vif-mediated A3G degradation was affected post-Vif binding (23, 52). Our data indicate that Vif/A3G DNA ratios used in the immunoprecipitation experiments greatly affect the results (Fig. 9A). Moreover, Vif binding differences between A3G WT and A3G 128K mutants could be observed only at low Vif/A3G ratios (Fig. 9A). These DNA ratios also better reflect the actual DNA concentrations used in the degradation and infectivity assays. Indeed, a study showing that A3G position 128 did not affect Vif binding used more Vif than A3G expression plasmids (23), which, according to our results, may lead to underestimation of the differences in Vif binding efficiencies. Of note, numerous other differences between the aforementioned immunoprecipitations exist, such as salt and detergent concentrations, which could also account for the discrepancy between studies.
In contrast to chimpanzees, who frequently hunt monkeys, gorillas are strict herbivores, suggesting that their risk of exposure to SIV from infected primates is limited (36, 37, 60). Our findings suggest that gorilla A3G resists degradation by most Vifs, effectively serving as a barrier to SIV transmission. Despite a low risk of exposure and Vif-resistant A3G, gorillas acquired SIVcpzPtt once (36), implying that SIVcpz Vif must have bypassed gorilla A3G. Most of the SIVcpz Vif alleles tested in this study failed to efficiently counteract gorilla A3G (Fig. 8). However, two of the SIVcpz Vifs (SIVcpzPtt1 and SIVcpzPtt2) exhibited some infectivity in the presence of gorilla A3G (Fig. 8A), suggesting that the restriction is not absolute. It is possible that some SIVcpz strains exist that can naturally counteract gorilla A3G. Indeed, SIVcpzPts, which encode Vifs with minimal activity against gorilla A3G (Fig. 8A), were never transmitted to gorillas, although their natural habitats overlap. Furthermore, we lack information to comprehensively assess natural gorilla A3G variation (A3G sequences from only two gorillas exist). Based on the extensive A3G diversity observed in rhesus and sooty mangabeys, it is conceivable that A3G position 129 is polymorphic in certain gorillas, making them more susceptible to transmissions of SIV from chimpanzees.
As a result of adaptations to overcoming gorilla A3G 129Q, SIVgor Vif appears to have lost its specificity regarding position 129. In contrast, SIVmac Vif is the only Vif that efficiently degrades all A3G variants independently of specific residues at 128 and 129 (Fig. 6C). Possibly, SIVmac Vif binds a distinct A3G region outside the A3G loop containing positions 128 and 129. A recent study showed that Vif from SIV in mantled colobus monkeys (Colobus guereza) (SIVcol) shifted its A3G binding site away from A3G positions 128 and 129 as a result of an A3G insertion near position 60 (46). Interestingly, rhesus macaque A3G also contains an insertion at a similar A3G position (Fig. 2A), which could also have resulted in a shifted binding site in A3G.
Overall, our analysis demonstrates that Vif from HIV-1, HIV-2, and several SIV strains bind a common, conserved region within A3G. The requirement of diverse Vifs to bind the same A3G region could potentially make A3G a better drug target than Vif itself, which is more variable and will quickly adapt leading to drug resistance. Structural information on the A3G-Vif interface will help define specific residues involved in this interaction leading to the identification of important A3G structural elements likely encompassing position 129 in A3G.
ACKNOWLEDGMENTS
We thank F. Kirchhoff for providing the SIVsmm PGm full-length molecular clone. We also kindly thank V. Pathak for productive discussions.
This work was funded in part by NIH grants R01 AI089246 (V.S.), R01 AI064001 (V.S.), R37 AI50529 (B.H.H.), R01 AI58715 (B.H.H.), and R37 AI66998 (G.S.) and Public Health Service Institutional Research Training Award T32A107647 (M.L.).
Footnotes
Published ahead of print 28 August 2013
REFERENCES
- 1.Apetrei C, Robertson DL, Marx PA. 2004. The history of SIVS and AIDS: epidemiology, phylogeny and biology of isolates from naturally SIV infected non-human primates (NHP) in Africa. Front. Biosci. 9:225–254 [DOI] [PubMed] [Google Scholar]
- 2.Sharp PM, Hahn BH. 2011. Origins of HIV and the AIDS pandemic. Cold Spring Harbor Perspect. Med. 1:a006841. 10.1101/cshperspect.a006841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plantier J-C, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemée V, Damond F, Robertson DL, Simon F. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871–872 [DOI] [PubMed] [Google Scholar]
- 4.Simon F, Mauclère P, Roques P, Loussert-Ajaka I, Müller-Trutwin MC, Saragosti S, Georges-Courbot MC, Barré-Sinoussi F, Brun-Vézinet F. 1998. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat. Med. 4:1032–1037 [DOI] [PubMed] [Google Scholar]
- 5.Vanden Haesevelde M, Decourt JL, De Leys RJ, Vanderborght B, van der Groen G, van Heuverswijn H, Saman E. 1994. Genomic cloning and complete sequence analysis of a highly divergent African human immunodeficiency virus isolate. J. Virol. 68:1586–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayouba A, Akoua-Koffi C, Calvignac-Spencer S, Esteban A, Locatelli S, Li H, Li Y, Hahn BH, Delaporte E, Leendertz FH, Peeters M. 2013. Evidence for continuing cross-species transmission of SIVsmm from sooty mangabeys to humans: full-length genome characterization of a new HIV-2 lineage in rural Côte d' Ivoire. AIDS [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z, Telfier P, Gettie A, Reed P, Zhang L, Ho DD, Marx P. 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J. Virol. 70:3617–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao F, Yue L, White A, Pappas P. 1992. Human infection by genetically diverse SIVsm-related HIV-2 in West Africa. Nature 358:495–499 [DOI] [PubMed] [Google Scholar]
- 9.Harris R, Bishop K, Sheehy A. 2003. DNA Deamination Mediates Innate Immunity to Retroviral Infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 10.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 11.Zhang H, Yang B, Pomerantz R. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marin M, Rose KM, Kozak SL, Kabat D. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398–1403 [DOI] [PubMed] [Google Scholar]
- 13.Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404–1407 [DOI] [PubMed] [Google Scholar]
- 14.Stopak K, de Noronha C, Yonemoto W, Greene WC. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12:591–601 [DOI] [PubMed] [Google Scholar]
- 15.Compton A, Hirsch VM, Emerman M. 2012. The host restriction factor APOBEC3G and retroviral Vif protein coevolve due to ongoing genetic conflict. Cell Host Microbe 11:91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaddis NC, Sheehy AM, Ahmad KM, Swanson CM, Bishop KN, Beer BE, Marx PA, Gao F, Bibollet-Ruche F, Hahn BH, Malim MH. 2004. Further investigation of simian immunodeficiency virus Vif function in human cells. J. Virol. 78:12041–12046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawyer SL, Emerman M, Malik HS. 2004. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2:e275. 10.1371/journal.pbio.0020275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang J, Webb DM. 2004. Rapid evolution of primate antiviral enzyme APOBEC3G. Hum. Mol. Genet. 13:1785–1791 [DOI] [PubMed] [Google Scholar]
- 19.Bogerd HP, Doehle BP, Wiegand HL, Cullen BR. 2004. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. U. S. A. 101:3770–3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangeat B, Turelli P, Liao S, Trono D. 2004. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 279:14481–14483 [DOI] [PubMed] [Google Scholar]
- 21.Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, Münk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31 [DOI] [PubMed] [Google Scholar]
- 22.Schröfelbauer B, Chen D, Landau NR. 2004. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. U. S. A. 101:3927–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu H, Svarovskaia ES, Barr R, Zhang Y, Khan MA, Strebel K, Pathak VK. 2004. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. U. S. A. 101:5652–5657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulliard Y, Turelli P, Röhrig UF, Zoete V, Mangeat B, Michielin O, Trono D. 2009. Functional analysis and structural modeling of human APOBEC3G reveal the role of evolutionarily conserved elements in the inhibition of human immunodeficiency virus type 1 infection and Alu transposition. J. Virol. 83:12611–12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huthoff H, Malim MH. 2007. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J. Virol. 81:3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. 2009. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 5:e1000330. 10.1371/journal.ppat.1000330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lavens D, Peelman F, Van der Heyden J, Uyttendaele I, Catteeuw D, Verhee A, Van Schoubroeck B, Kurth J, Hallenberger S, Clayton R, Tavernier J. 2010. Definition of the interacting interfaces of Apobec3G and HIV-1 Vif using MAPPIT mutagenesis analysis. Nucleic Acids Res. 38:1902–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbs JS, Regier DA, Desrosiers RC. 1994. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retroviruses 10:343–350 [DOI] [PubMed] [Google Scholar]
- 30.Dewhurst S, Embretson JE, Anderson DC, Mullins JI, Fultz PN. 1990. Sequence analysis and acute pathogenicity of molecularly cloned SIVSMM-PBj14. Nature 345:636–640 [DOI] [PubMed] [Google Scholar]
- 31.November FJ, De Rosayro J, O'Neil SP, Anderson DC, Klumpp SA, McClure HM. 1998. Isolation and characterization of a neuropathogenic simian immunodeficiency virus derived from a sooty mangabey. J. Virol. 72:8841–8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naidu YM, Kestler HW, III, Li Y, Butler CV, Silva DP, Schmidt DK, Troup CD, Sehgal PK, Sonigo P, Daniel MD, et al. 1988. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J. Virol. 62:4691–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin MJ, Hui H, Robertson DL, Muller MC, Barre-Sinoussi F, Hirsch VM, Allan JS, Shaw GM, Sharp PM, Hahn BH. 1994. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 13:2935–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soares MA, Robertson DL, Hui H, Allan JS, Shaw GM, Hahn BH. 1997. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology 228:394–399 [DOI] [PubMed] [Google Scholar]
- 35.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, et al. 1990. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 248:1109–1112 [DOI] [PubMed] [Google Scholar]
- 36.Takehisa J, Kraus MH, Ayouba A, Bailes E, Van Heuverswyn F, Decker JM, Li Y, Rudicell RS, Learn GH, Neel C, Ngole EM, Shaw GM, Peeters M, Sharp PM, Hahn BH. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takehisa J, Kraus MH, Decker JM, Li Y, Keele BF, Bibollet-Ruche F, Zammit KP, Weng Z, Santiago ML, Kamenya S, Wilson ML, Pusey AE, Bailes E, Sharp PM, Shaw GM, Hahn BH. 2007. Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. J. Virol. 81:7463–7475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Ndjango JB, Learn GH, Ramirez MA, Keele BF, Bibollet-Ruche F, Liu W, Easlick JL, Decker JM, Rudicell RS, Inogwabini BI, Ahuka-Mundeke S, Leendertz FH, Reynolds V, Muller MN, Chancellor RL, Rundus AS, Simmons N, Worobey M, Shaw GM, Peeters M, Sharp PM, Hahn BH. 2012. Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir. J. Virol. 86:10776–10791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon V, Zennou V, Murray D, Huang Y, Ho DD, Bieniasz PD. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1:e6. 10.1371/journal.ppat.0010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78:12058–12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harari A, Ooms M, Mulder LC, Simon V. 2009. Polymorphisms and splice variants influence the antiretroviral activity of human APOBEC3H. J. Virol. 83:295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi M, Takaori-Kondo A, Miyauchi Y, Iwai K, Uchiyama T. 2005. Ubiquitination of APOBEC3G by an HIV-1 Vif-cullin 5-elongin B-elongin C complex is essential for Vif function. J. Biol. Chem. 280:18573–18578 [DOI] [PubMed] [Google Scholar]
- 44.Luo K, Xiao Z, Ehrlich E, Yu Y, Liu B, Zheng S, Yu XF. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. U. S. A. 102:11444–11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu Y, Xiao Z, Ehrlich ES, Yu X, XF Yu. 2004. Selective assembly of HIV-1 Vif–Cul5–elongin B–elongin C E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 18:2867–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Compton AA, Emerman M. 2013. Convergence and divergence in the evolution of the APOBEC3G-Vif interaction reveal ancient origins of simian immunodeficiency viruses. PLoS Pathog. 9:e1003135. 10.1371/journal.ppat.1003135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haché G, Liddament MT, Harris RS. 2005. The retroviral hypermutation specificity of APOBEC3F and APOBEC3G is governed by the C-terminal DNA cytosine deaminase domain. J. Biol. Chem. 280:10920–10924 [DOI] [PubMed] [Google Scholar]
- 48.Iwatani Y, Chan D, Liu L. 2009. HIV-1 Vif-mediated ubiquitination/degradation of APOBEC3G involves four critical lysine residues in its C-terminal domain. Proc. Natl. Acad. Sci. U. S. A. 106:19539–19544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Navarro F, Bollman B, Chen H, König R, Yu Q, Chiles K, Landau NR. 2005. Complementary function of the two catalytic domains of APOBEC3G. Virology 333:374–386 [DOI] [PubMed] [Google Scholar]
- 50.Newman ENC, Holmes RK, Craig HM, Klein KC, Lingappa JR, Malim MH, Sheehy AM. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170 [DOI] [PubMed] [Google Scholar]
- 51.Albin JS, Laruen RS, Weaver JA, Brown WL, Shindo K, Harjes E, Matsuo H, Harris RS. 2010. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J. Biol. Chem. 285:40785–40792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Russell Ra., Pathak VK. 2007. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 81:8201–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russell Ra., Smith J, Barr R, Bhattacharyya D, Pathak VK. 2009. Distinct domains within APOBEC3G and APOBEC3F interact with separate regions of human immunodeficiency virus type 1 Vif. J. Virol. 83:1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith JL, Pathak VK. 2010. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J. Virol. 84:12599–12608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harjes E, Gross PJ, Chen KM, Lu Y, Shindo K, Nowarski R, Gross JD, Kotler M, Harris RS, Matsuo H. 2009. An extended structure of the APOBEC3G catalytic domain suggests a unique holoenzyme model. J. Mol. Biol. 389:819–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang K-L, Mangeat B, Ortiz M, Zoete V, Trono D, Telenti A, Michielin O. 2007. Model structure of human APOBEC3G. PLoS One 2:e378. 10.1371/journal.pone.0000378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vallari A, Holzmayer V, Harris B, Yamaguchi J, Ngansop C, Makamche F, Mbanya D, Kaptué L, Ndembi N, Gürtler L, Devare S, Brennan Ca. 2011. Confirmation of putative HIV-1 group P in Cameroon. J. Virol. 85:1403–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Loul S, Butel C, Liegeois F, Bienvenue Y, Ngolle EM, Sharp PM, Shaw GM, Delaporte E, Hahn BH, Peeters M. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. [DOI] [PubMed] [Google Scholar]
- 59.Mehle A, Wilson H, Zhang C, Brazier AJ, McPike M, Pery E, Gabuzda D. 2007. Identification of an APOBEC3G binding site in human immunodeficiency virus type 1 Vif and inhibitors of Vif-APOBEC3G binding. J. Virol. 81:13235–13241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neel C, Etienne L, Li Y, Takehisa J, Rudicell RS, Bass IN, Moudindo J, Mebenga A, Esteban A, Van Heuverswyn F, Liegeois F, Kranzusch PJ, Walsh PD, Sanz CM, Morgan DB, Ndjango JB, Plantier JC, Locatelli S, Gonder MK, Leendertz FH, Boesch C, Todd A, Delaporte E, Mpoudi-Ngole E, Hahn BH, Peeters M. 2010. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J. Virol. 84:1464–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, Sugiura W, Iwatani Y. 2012. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 19:1005–1010 [DOI] [PubMed] [Google Scholar]