Abstract
The enrichment of HIV-1 macromolecules at the uropod of polarized T cells can significantly promote virus assembly and cell-mediated infection. Using live-cell fluorescence microscopy, we demonstrate that full-length HIV-1 RNA is enriched at the uropod membrane; furthermore, the presence of HIV-1 Gag containing a functional nucleocapsid domain is necessary for this HIV-1 RNA enrichment. The results from these studies provide novel insights into the mechanism of HIV-1 replication in polarized T cells.
TEXT
Most in vivo HIV-1 infection events occur in lymphoid tissues that contain densely packed CD4+ T cells. The majority of T cells within lymph nodes are polarized, with the leading edge enriched in β-actin and cytokine-detecting membrane proteins and with the uropod enriched in adhesion receptor molecules such as ICAM-1, -2, and -3, CD44, CD43, and PSGL-1 (1–4). In addition to cell-free infection, HIV-1 can also be transmitted from one infected cell to a target cell via a structure termed the virological synapse (5–9). Not only is the uropod more likely to form virological synapses than the leading edge, but the presence of the uropod can promote synapse formation (10). Interestingly, HIV-1 Gag and Env have been observed to be enriched in the membrane region of the uropod; furthermore, recent studies have concluded that HIV-1 assembly preferentially occurs at the uropod (10, 11). As the uropod promotes cell-cell transmission, the preferential localization of Gag at the uropod can facilitate viral spread in vivo. Currently, there are many unanswered questions regarding HIV-1 biology in polarized T cells. For example, does HIV-1 RNA target to the uropod region? Certain cellular mRNAs have been shown to distribute unevenly in polarized cells and are enriched in regions in which the encoded proteins are needed (12–18). For example, β-actin mRNA has been demonstrated to be enriched at the leading edge of fibroblast cells, where actin filaments are needed for forward movement of the cell (12–14). Similarly, the seven mRNAs that encode the actin-polymerization nucleator Arp2/3 complex, which is required for actin assembly, are also enriched at the leading protrusions of fibroblasts (18). Because HIV-1 Gag has been observed to be enriched at the uropod membrane, we sought to determine whether HIV-1 full-length RNA is unevenly distributed in polarized T cells and, if so, whether it is enriched at the uropod membrane.
Subcellular distribution of HIV-1 RNA in polarized T cells.
To determine the distribution of full-length HIV-1 RNA in polarized T cells, we visualized the RNA using a fluorescent protein tag fused to an RNA-binding protein that specifically recognizes sequences engineered into the HIV-1 genome (19). The general structure of the modified HIV-1 genome is shown in Fig. 1A; briefly, GagBSL-BCherry contains stem-loop sequences (BSL) recognized by BglG, an Escherichia coli RNA-binding protein (20). Additionally, this construct expresses a Gag fused with cerulean fluorescence protein (CeFP) and a Bgl-mCherry fusion protein; translation of the latter is directed by an internal ribosomal entry site (IRES) from encephalomyocarditis virus (EMCV) (21). We have previously shown that Bgl-mCherry expressed in trans can specifically and efficiently label HIV-1 RNA containing BSL sequences (19). In the current study, GagBSL-BCherry was introduced into the P2 T cell line (a kind gift from Akira Ono, University of Michigan) (10) by nucleofection; after 17 h, the cells were stained with a mouse antibody (BD Biosciences) against human CD43, a uropod marker, and then stained with an Alexa Fluor 514-labeled goat anti-mouse antibody (Invitrogen). Images were then captured using a Nikon fluorescence microscope. As shown in a set of representative images (Fig. 2A), HIV-1 Gag proteins, detected by CeFP signals, are enriched near the uropod membrane, which is defined by the CD43 staining. HIV-1 full-length RNAs, detected by the mCherry signals, are also enriched near the uropod membrane; because the Bgl-mCherry harbors a nuclear localization signal (NLS), proteins not bound to viral RNA are enriched in the nucleus, generating an intense red signal. To verify that the Bgl-mCherry signals enriched near the uropod membrane are from labeled HIV-1 RNA, we examined the distribution of the signals in P2 T cells from an HIV-1 genome, GagNoSL-BCherry (Fig. 1B), that lacks BSL sequences recognized by Bgl-mCherry but otherwise has a structure similar to that of GagBSL-BCherry. As shown in Fig. 2B, the Gag signals are enriched in the uropod membrane, whereas the Bgl-mCherry signals localize to the nuclear region. These results demonstrate that the mCherry signals detected near the uropod membrane in GagBSL-BCherry (Fig. 2A) are mediated by Bgl-mCherry binding to the BSL sequences in the viral RNA.
Fig 1.
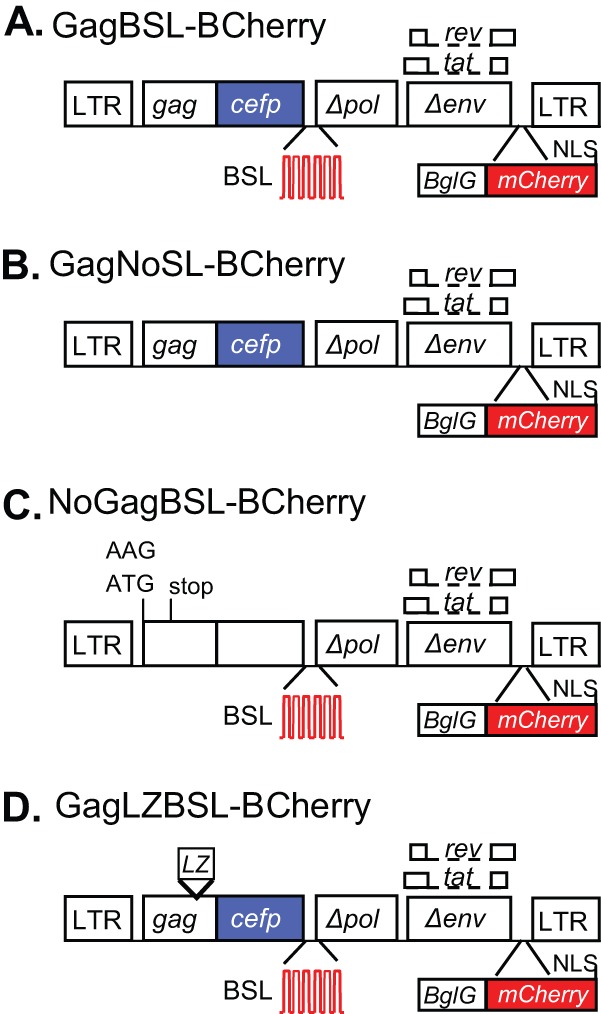
General structures of modified HIV-1 genomes. (A) GagBSL-BCherry is similar to the previously described construct GagCeFP-BglSL (19) but contains a Bgl-mCherry fusion protein in nef; the translation of Bgl-mCherry is directed by an IRES from EMCV. (B) GagNoSL-BCherry is similar to GagCeFP-BCherry but lacks the recognition sites (BSL) of the BglG protein. (C) NoGagBSL-BCherry is similar to GagBSL-BCherry but contains two mutations in gag to prevent Gag translation, an AAG substitution in place of the Gag translational start codon AUG, and a 4-bp insertion in CA that results in a premature stop codon in CA. (D) The NC domain of GagLZBSL-BCherry was replaced with a leucine zipper (LZ) motif. Open boxes, HIV-1 sequences; blue boxes, CeFP coding sequences; red stem-loops, recognition sites of the BglG protein (BSL); red boxes, mCherry coding sequences.
Fig 2.
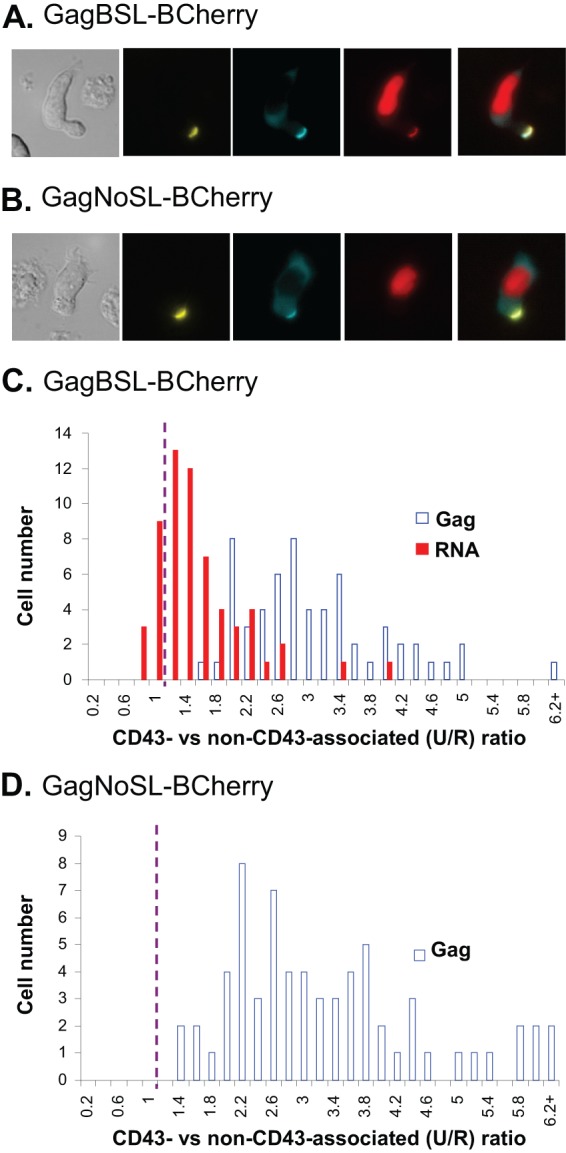
Subcellular distribution of the HIV-1 Gag and full-length RNA in polarized T cells. (A and B) Representative images of polarized T cells expressing GagBSL-BCherry (A) or GagNoSL-BCherry (B). Images were analyzed using Nikon Elements software, in which the sum intensities were measured using three regions of interests (ROIs): one around the CD43-stained region of the cell denoting the uropod; a second around the CD43-negative part of the cell; and a third around the nucleus. (C and D) The sum intensities were normalized to the area and then used to calculate the signal intensity per pixel near the uropod membrane (CD43-associated or U) and the signal intensity per pixel for the rest of the cytoplasm and plasma membrane (non-CD43-associated or R). When the signals are enriched near the uropod membrane, the U/R ratios are greater than 1. The distributions of Gag and RNA signals are summarized from cells expressing GagBSL-BCherry (C) and GagNoSL-BCherry (D). As a significant cytoplasmic mCherry signal was not detected in the GagNoSL-BCherry-expressing cells, only the distribution of the Gag U/R ratio is summarized in panel D. Data shown represent 60 cells collected from 7 experiments (C) and 66 cells collected from 8 experiments (D).
To quantify the enrichment of Gag and viral RNA at the uropod membrane, we measured the average signal intensity per pixel around the uropod membrane (U) as defined by CD43 staining and the average signal intensity per pixel for the rest of the cytoplasm and plasma membrane (R) using Element software (Nikon). We then calculated the level of enhancement of the signal around the uropod membrane by dividing the U value by the R value (U/R ratio). If the Gag or RNA signals are distributed evenly throughout the cytoplasm and plasma membrane, including the uropod, the U/R ratio should be close to 1. In contrast, if the signals are enriched near the uropod membrane, then the ratio should be much greater than 1. In Fig. 2C, we have summarized results from 60 cells expressing GagBSL-BCherry collected from seven independent experiments; the y axis value represents the number of cells whereas the x axis value represents the U/R ratio, which indicates the signals localized around the uropod membrane (CD43-associated) versus signals localized elsewhere in the cytoplasm and plasma membrane (non-CD43-associated). The GagCeFP signals were enriched near the uropod membrane in all 60 cells, with the U/R ratio ranging from 1.4 to 8. RNA signals in the majority of the cells were also enriched near the uropod membrane; 48 of 60 cells had U/R ratios of more than 1, whereas 12 of 60 cells had U/R ratios of less than 1 (P = 0.0001; Chi-square test), although the U/R ratios of the RNA signals were lower than those of the Gag signals. As a significant cytoplasmic mCherry signal was not detected in the GagNoSL-BCherry-expressing cells, only the distribution of the Gag U/R ratio is summarized in Fig. 2D. Our analyses showed that the CeFP signal distributions are similar for GagBSL-BCherry and GagNoSL-BCherry (Fig. 2C and D).
The role of Gag in the enrichment of HIV-1 full-length RNA at the uropod membrane.
We envision two possible scenarios regarding the relationship between HIV-1 Gag and RNA enrichment near the uropod membrane. Gag and HIV-1 RNA may be independently targeted to the uropod; alternatively, HIV-1 RNA may be brought to the uropod via Gag-RNA interactions. To distinguish between these two possibilities, we examined the distribution of HIV-1 RNA signals in the absence of Gag proteins. For this purpose, we generated a modified HIV-1 genome (NoGagBSL-BCherry, Fig. 1C) that contains two mutations in gag to abolish the start codon (AUG to AAG) and to introduce a stop codon in the capsid region via a frameshift mutation. These mutations have been shown to abolish Gag expression, but the resulting RNA genome can be efficiently packaged and undergo one round of replication when Gag and Gag-Pol are supplied in trans (22). As shown in a set of representative images (Fig. 3A), in the absence of Gag proteins, HIV-1 RNA signals were not enriched near the uropod membrane but displayed a diffusive pattern throughout the cytoplasm. We have analyzed the RNA signals of 52 cells expressing NoGagBSL-BCherry collected from five experiments; these results are summarized in Fig. 3B. In all 52 cells, the HIV-1 full-length RNAs were not enriched near uropod membranes and the U/R ratios were less than 1; these results are significantly different from the distribution shown in Fig. 2C (P < 0.00000001, Chi-square test). These results suggest that Gag is required for the enrichment of HIV-1 RNA at the uropod membrane and that the specific interactions between the Gag and the RNA genome are most likely required for the enrichment. To confirm this possibility, we generated an HIV-1 variant, GagLZBSL-BCherry, that expressed Gag proteins that do not specifically interact with the RNA genome by replacing the nucleocapsid (NC) domain of Gag with a leucine zipper (LZ) motif (23, 24). NC plays a key role in the specific interactions between HIV-1 Gag and the RNA genome. Gag proteins in which NC is replaced by a LZ motif maintain their ability to generate viral particles but are unable to package viral genomes (24, 25); as a result, the particles produced by these Gag proteins contain few, if any, viral RNA genomes (25). However, such mutant Gag proteins still enrich near the uropod membrane (10, 11). We examined the distribution of Gag and RNA signals from GagLZBSL-BCherry-expressing cells and found that the Gag signals, but not the RNA signals, were enriched near the uropod membrane (Fig. 3C). Quantitative results summarized from 57 cells collected from eight experiments are shown in Fig. 3D, indicating that in all 57 cells the HIV-1 RNA signals were not enriched near the uropod membrane. These results are significantly different from the results from the RNA signals shown in Fig. 2C (P < 0.00000001, Chi-square test). Therefore, the enrichment of HIV-1 RNA at the uropod membrane is dependent upon the presence of Gag proteins that can recognize and package the RNA genome.
Fig 3.
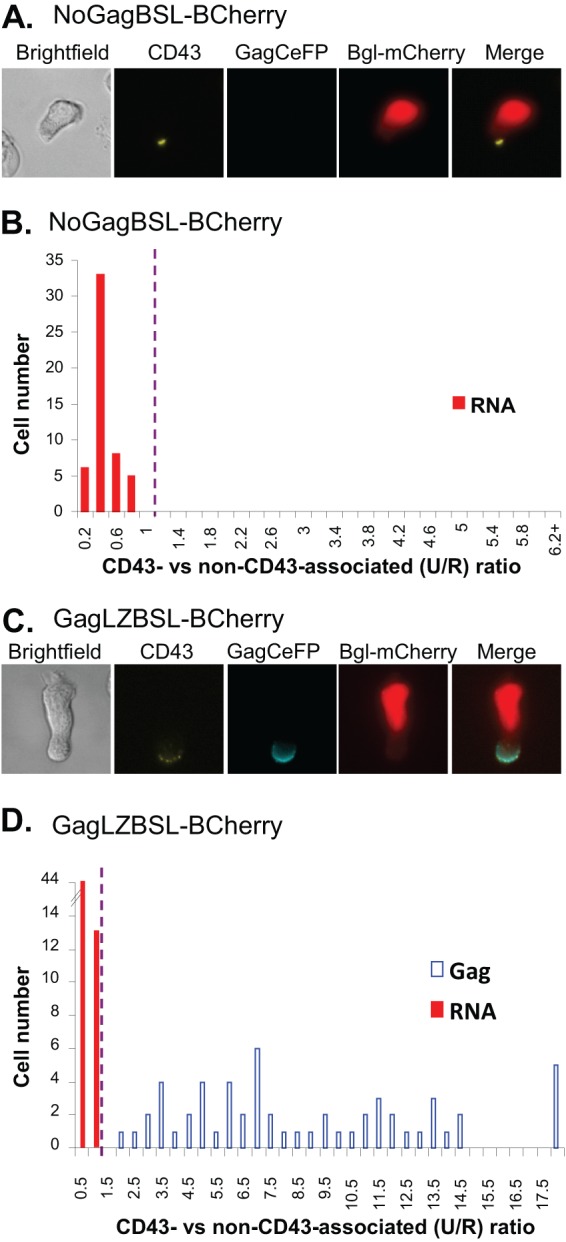
Subcellular distribution of the HIV-1 full-length RNAs of Gag mutants in polarized T cells. (A and C) Representative images of polarized T cells expressing NoGagBSL-BCherry (A) or GagLZBSL-BCherry (C). (B and D) The distributions of Gag and RNA signals summarized from 5 experiments with 52 cells expressing NoGagBSL-BCherry (B) and 8 experiments with 57 cells expressing GagLZBSL-BCherry (D) are shown. Calculations and abbreviations are the same as described for Fig. 2.
The enrichment of HIV-1 RNA at the uropod and its implications.
Polarized T cells not only constitute a majority of HIV-1 target cells in vivo but also play a critical role in the spread of HIV-1 via cell-to-cell infection. Therefore, determining the mechanisms of HIV-1 macromolecule trafficking in polarized T cells is crucial to understanding HIV-1 replication in these key target cells. In this report, we studied the subcellular distribution of the HIV-1 RNA genome in polarized T cells and found that full-length RNAs are enriched near the uropod plasma membrane. In contrast to the direct RNA targeting mechanism used by certain cellular mRNAs, HIV-1 RNA enrichment is dependent on Gag and its ability to interact with the full-length RNA. These results indicate that HIV-1 RNA is enriched during the process of virus assembly. We speculate that HIV-1 Gag interacts with the RNA genome in the cytoplasm prior to membrane targeting, as cytoplasmic Gag-RNA associations have been shown or suggested by other groups (26, 27). These studies provide additional molecular insights into the preferential assembly of HIV-1 virions at the uropod of polarized T cells. As the Gag-enriched uropod is more likely to form a virological synapse than regions at the leading edge, such targeting facilitates cell-mediated infection and virus spread in vivo.
ACKNOWLEDGMENTS
We thank Eric Freed and Steve Hughes for valuable discussions and Akira Ono (University of Michigan) for the P2 T cell line.
This work was funded in part by the Intramural AIDS Targeted Antiviral Program, NIH, and the Intramural Research Program of the Center for Cancer Research, NCI, and in part with federal funds from the NCI, NIH, under contract no. HHSN261200800001E with SAIC-Frederick, Inc.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government.
Footnotes
Published ahead of print 21 August 2013
REFERENCES
- 1.Alonso-Lebrero JL, Serrador JM, Dominguez-Jimenez C, Barreiro O, Luque A, del Pozo MA, Snapp K, Kansas G, Schwartz-Albiez R, Furthmayr H, Lozano F, Sanchez-Madrid F. 2000. Polarization and interaction of adhesion molecules P-selectin glycoprotein ligand 1 and intercellular adhesion molecule 3 with moesin and ezrin in myeloid cells. Blood 95:2413–2419 [PubMed] [Google Scholar]
- 2.del Pozo MA, Nieto M, Serrador JM, Sancho D, Vicente-Manzanares M, Martinez C, Sanchez-Madrid F. 1998. The two poles of the lymphocyte: specialized cell compartments for migration and recruitment. Cell Adhes. Commun. 6:125–133 [DOI] [PubMed] [Google Scholar]
- 3.Miller MJ, Wei SH, Parker I, Cahalan MD. 2002. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science 296:1869–1873 [DOI] [PubMed] [Google Scholar]
- 4.Sánchez-Madrid F, Serrador JM. 2009. Bringing up the rear: defining the roles of the uropod. Nat. Rev. Mol. Cell Biol. 10:353–359 [DOI] [PubMed] [Google Scholar]
- 5.Chen BK. 2012. T cell virological synapses and HIV-1 pathogenesis. Immunol. Res. 54:133–139 [DOI] [PubMed] [Google Scholar]
- 6.Chen P, Hubner W, Spinelli MA, Chen BK. 2007. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 81:12582–12595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. 2004. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 199:283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sattentau QJ. 2010. Cell-to-cell spread of retroviruses. Viruses 2:1306–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waki K, Freed EO. 2010. Macrophages and cell-cell spread of HIV-1. Viruses 2:1603–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llewellyn GN, Hogue IB, Grover JR, Ono A. 2010. Nucleocapsid promotes localization of HIV-1 gag to uropods that participate in virological synapses between T cells. PLoS Pathog. 6:e1001167. 10.1371/journal.ppat.1001167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen B, Fang Y, Wu N, Gould SJ. 2011. Biogenesis of the posterior pole is mediated by the exosome/microvesicle protein-sorting pathway. J. Biol. Chem. 286:44162–44176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Condeelis J, Singer RH. 2005. How and why does beta-actin mRNA target? Biol. Cell 97:97–110 [DOI] [PubMed] [Google Scholar]
- 13.Kislauskis EH, Zhu X, Singer RH. 1997. beta-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 136:1263–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence JB, Singer RH. 1986. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45:407–415 [DOI] [PubMed] [Google Scholar]
- 15.Macara IG, Iioka H, Mili S. 2009. Axon growth-stimulus package includes local translation. Nat. Cell Biol. 11:919–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mili S, Macara IG. 2009. RNA localization and polarity: from A(PC) to Z(BP). Trends Cell Biol. 19:156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mili S, Moissoglu K, Macara IG. 2008. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature 453:115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mingle LA, Okuhama NN, Shi J, Singer RH, Condeelis J, Liu G. 2005. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 118:2425–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U. S. A. 106:13535–13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houman F, Diaz-Torres MR, Wright A. 1990. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 62:1153–1163 [DOI] [PubMed] [Google Scholar]
- 21.Jang SK, Wimmer E. 1990. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 4:1560–1572 [DOI] [PubMed] [Google Scholar]
- 22.Nikolaitchik O, Rhodes TD, Ott D, Hu WS. 2006. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J. Virol. 80:4691–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Accola MA, Strack B, Gottlinger HG. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Qian H, Love Z, Barklis E. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crist RM, Datta SA, Stephen AG, Soheilian F, Mirro J, Fisher RJ, Nagashima K, Rein A. 2009. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: implications for retrovirus assembly. J. Virol. 83:2216–2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemler I, Meehan A, Poeschla EM. 2010. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J. Virol. 84:6352–6366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kutluay SB, Bieniasz PD. 2010. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 6:e1001200. 10.1371/journal.ppat.1001200 [DOI] [PMC free article] [PubMed] [Google Scholar]


