Abstract
Human bocavirus is a newly identified, globally prevalent, parvovirus that is associated with respiratory infection in infants and young children. Parvoviruses encode a large nonstructural protein 1 (NS1) that is essential for replication of the viral single-stranded DNA genome and DNA packaging and may play versatile roles in virus-host interactions. Here, we report the structure of the human bocavirus NS1 N-terminal domain, the first for any autonomous parvovirus. The structure shows an overall fold that is canonical to the histidine-hydrophobic-histidine superfamily of nucleases, which integrates two distinct DNA-binding sites: (i) a positively charged region mediated by a surface hairpin (residues 190 to 198) that is responsible for recognition of the viral origin of replication of the double-stranded DNA nature and (ii) the nickase active site that binds to the single-stranded DNA substrate for site-specific cleavage. The structure reveals an acidic-residue-rich subdomain that is present in bocavirus NS1 proteins but not in the NS1 orthologs in erythrovirus or dependovirus, which may mediate bocavirus-specific interaction with DNA or potential host factors. These results provide insights into recognition of the origin of replication and nicking of DNA during bocavirus genome replication. Mapping of variable amino acid residues of NS1s from four human bocavirus species onto the structure shows a scattered pattern, but the origin recognition site and the nuclease active site are invariable, suggesting potential targets for antivirals against this clade of highly diverse human viruses.
INTRODUCTION
Human bocavirus 1 (HBoV1) was originally identified in 2005 in nasopharyngeal aspirates of patients with acute respiratory tract infections (1), followed by the identification of three other HBoVs, namely, HBoV2 to HBoV4, in human feces (2–4). HBoV1 infection is mainly associated with pediatric respiratory disease and, in rare cases, shows gastrointestinal symptoms (5–9). HBoV1 is frequently detected in infants and young children less than 2 years of age. HBoV1 has been found worldwide (4, 10–12) and seroepidemiologic studies have shown that most children have IgG antibodies against HBoV1 by school age (13, 14). HBoV1 has been classified as a member of the genus Bocavirus of the family of Parvoviridae. It represents the first reported human pathogen of this genus, the third to human parvovirus B19 and PARV4 in the Parvoviridae family.
Virions of HBoV1 are small (18 to 26 nm in diameter), icosahedral, and nonenveloped. The capsid exhibits T=1 symmetry with 60 copies of coat protein (15, 16). The genome of HBoV1 is a single-stranded DNA (ssDNA) of 5,543 nucleotides (nt) with a left-end hairpin (LEH) and a right-end hairpin (REH), which appear to be hybrid remnants of the animal bocaviruses bovine parvovirus 1 (BPV1) and minute virus of canine (MVC), respectively (15, 17–19). All parvoviruses encode a nonstructural protein NS1 (termed Rep in adeno-associated virus [AAV]) that is essential for viral DNA replication (20–24) and packaging of viral DNA into capsid (25–27). It may play other versatile roles, for example, in the transactivation of viral (28–31) and cellular (32) gene expression, DNA damage response (33–36), cell cycle arrest, apoptosis (37–40), and/or the modulation of innate immunity (41). Like its parvovirus orthologs, HBoV1 NS1 contains an N-terminal DNA-binding/endonuclease domain, a central helicase domain, and a C-terminal zinc-finger domain (42–46).
The only structurally characterized parvovirus NS1 N-terminal nuclease domain is from AAV Rep (47, 48), which binds to the consecutive tetranucleotide repeats in the origin of replication (Ori). However, such tetranucleotide repeats are specific to AAV and are not present in HBoV1. Indeed, the LEH of the HBoV1 genome forms a loop with a three-way junction, whereas the REH is a hairpin with perfect base pairing, which are conserved in bocaviruses but distinct from terminal regions of the AAV and parvovirus B19 (B19V) genomes (15, 47, 49). These findings suggest that the mode of NS1 recognition of the Ori in HBoV is distinct from that in AAV. Moreover, AAV is not known to cause human disease and is a dependovirus because virus replication requires a helper virus such as herpesvirus or adenovirus. The HBoV NS1 shares as little as 14% sequence identity with AAV Rep. Here, we report the crystal structure of human parvovirus HBoV1 NS1 N-terminal origin-recognition/nickase domain (HBoV-NS1N). This represents the first structure of NS1 from any autonomous parvovirus. The structure demonstrates that HBoV-NS1 is a member of the histidine-hydrophobic-histidine (HUH) superfamily of endonucleases and provides insights into multiple roles of NS1 in DNA replication in this emerging human virus. HBoV species display high diversity and frequent mutation (4). Mapping of variable amino acid residues of NS1N from four HBoV species onto the structure shows a scattered pattern, but the origin recognition site and the nuclease active site are invariable, suggesting a high potential for mutation for the majority of surface residues and the suitability of the two invariable regions as potential targets for broad-spectrum anti-HBoV drugs or vaccines.
MATERIALS AND METHODS
Protein expression, purification, and crystallization.
The DNA fragment encoding HBoV-NS1N (residues 1 to 271; GenBank protein_id = AFR53039.1) was cloned into pET28a (Novagen) between NdeI/XhoI and a stop codon introduced after Gly271, based on predictions of protein secondary structure elements and disordered regions. The residue G271 is at the end of a predicted alpha helix preceding a predicted disordered region. The N-terminally His-tagged protein was overexpressed by growth of Escherichia coli strain B834(DE3) at 37°C in Luria-Bertani (LB) broth until reaching an optical density at 600 nm of 0.6, followed by induction at 25°C by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM. Cells were harvested after 16 h of growth and lysed on a French press in resuspension buffer (50 mM Na2HPO4/NaH2PO4 [pH 7.2], 500 mM NaCl, 10% glycerol, and 5 mM β-mercaptoethanol). The protein was purified with a Ni-NTA column (Qiagen), followed by gel filtration chromatography on a Hi-Load 16/60 Superdex 75 column (GE Healthcare), which showed an elution volume of 69.3 ml corresponding to a monomer. The eluted fractions were collected in tubes containing the gel filtration buffer (50 mM Na2HPO4/NaH2PO4 [pH 6.5], 150 mM NaCl, 2 mM dithiothreitol, 10% glycerol, and 1 mM EDTA) supplemented with a final concentration of 100 mM l-(+)-arginine (ACROS Organics) to prevent rapid protein precipitation. The protein was concentrated to ∼15 mg/ml using a Millipore Centricon (molecular mass cutoff, 10 kDa) prior to crystallization. Rod-shaped crystals were obtained by the hanging-drop method by mixing 1 μl of protein solution with 1 μl of the well solution containing 30% MPD (2-methyl-2,4-pentanediol). Crystals grew to the full size of approximately 0.4 mm by 0.04 mm by 0.04 mm within 1 to 2 days of incubation at 20°C. Crystals were cryoprotected by transferring into a solution with 30% MPD for 20 to 30 s prior to flash-freezing in liquid nitrogen. Heavy atom derivative crystals were obtained by soaking the native crystals in a 10 mM HgCl2 in 30% MPD for 5 to 15 s prior to freezing.
X-ray data collection and structure determination.
HBoV-NS1N native and heavy atom derivative X-ray data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) and the Advanced Photon Source (APS), respectively. The data were processed with the program HKL2000 (50). Structure determination by molecular replacement using the AAV Rep structure (PDB code 1m55) as a search model failed to give any correct solution. The structure was determined by the single isomorphous replacement method using the mercury derivative data. The experimental electron density map calculated with SOLVE in PHENIX was of excellent quality, which allowed automated building of most of the model using PHENIX (51). Structure refinement coupled with manual model building was performed with the programs PHENIX (51) and COOT (52), respectively. The crystallographic asymmetric unit contains two molecules, corresponding to a Matthews coefficient of 4.6 Å3/Da and a solvent content as high as 73%. The refined model includes residues 6 to 271 for each molecule, with excellent refinement statistics and stereochemistry (Tables 1 and 2).
Table 1.
X-ray data collection
| Data collection | Valuesa |
|
|---|---|---|
| Native NS1N | Hg derivative | |
| Beamline | SSRL 11-1 | APS 23ID-B |
| Wavelength (Å) | 0.97945 | 0.99712 |
| Resolution range (Å) | 50–2.70 | 50–3.00 |
| No. of measurements | 283,360 | 153,760 |
| No. of unique reflections | 32,126 (2,214) | 23,495 (1,790) |
| Completeness (%) | 99.8 (98.9) | 99.5 (99.8) |
| I/σ | 27.4 (3.30) | 21.2 (2.53) |
| Rmergeb | 7.0 (51.0) | 12.0 (64.0) |
| Space group | P41212 | P41212 |
| Unit cell (Å) | a = 199.8, c = 56.5 | a = 199.8, c = 56.6 |
Values in the parentheses are for the outmost resolution shells.
Rmerge = ΣhklΣi|Ii(hkl) − <I(hkl)>/ΣhklΣiIi(hkl), where Ii(hkl) is the observed intensity of reflection hkl and <I(hkl)> is the averaged intensity of symmetry-equivalent measurements.
Table 2.
Refinement statistics for native NS1N structure
| Statistics | Value(s) |
|---|---|
| Resolution (Å) | 37.4–2.7 |
| Rwork/Rfreea | 0.18/0.22 |
| Avg B-factor | 58.07 |
| RMSD | |
| Bond length (Å) | 0.009 |
| Bond angle (°) | 1.155 |
| Ramachandran plot statistics (%) | |
| Most favored regions | 99.0 |
| Additionally allowed regions | 1.0 |
| Disallowed regions | 0.0 |
Rwork = Σhkl∥Fobs| − |Fcalc∥/Σhkl|Fobs|, where Fobs and Fcalc are structure factors of the observed reflections and those calculated from the refined model, respectively. Rfree has the same formula as Rwork, except that it was calculated against a test set of data that were not included in the refinement.
Accession codes.
The coordinates and reflection data have been deposited with the RCSB Protein Data Bank under accession code 4KW3.
RESULTS AND DISCUSSION
Overall structure.
The HBoV1 NS1 N-terminal domain encompassing residues 1 to 271 was overexpressed in E. coli, and purified. The protein existed as a monomer on the size exclusion column. Freshly purified protein must be immediately used for crystallization. Molecular replacement using the AAV Rep N-terminal domain structure (sequence identity ∼14%) as the search model failed, indicating major structural difference between the two proteins. The structure of HBoV-NS1N was determined at 2.7-Å resolution using the single isomorphous replacement method (Tables 1 and 2). The structure contains two molecules in the asymmetric unit (see below). Each HBoV-NS1N monomer consists of a central five-stranded antiparallel β-sheet surrounded by clusters of α-helices (Fig. 1a). The central β-sheet forms a cleft whose floor is formed by strands β1/β3/β2/β9, where the nuclease active site is located (see below), while its walls are formed by α8, β8, and a loop between β2 and β3 on one side and a β-hairpin formed by loop L14, β5, and β6 on the other side.
Fig 1.
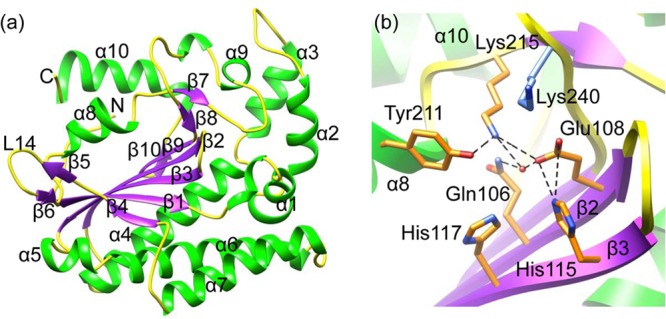
Overall structure and the nuclease active site. (a) Ribbon diagram of the structure. The α-helices are green, whereas loops and β-stands are yellow and purple, respectively. Helices, β-stands, and loop L14, as well as the N and C termini, are labeled. (b) Nuclease active site showing a network of H bonds (dashed lines in black). The color scheme is the same as in panel a. Side chains of the conserved residues are shown as sticks. The water molecule at the active site is shown as a sphere in red.
Nickase active site.
The HBoV-NS1N structure reveals an overall fold that is canonical to the HUH superfamily of endonucleases that are involved in rolling-circle replication, which contain signature motif HUH, also known as motif 2, and motif YUxxYx2-3K, also known as motif 3 (where Y is tyrosine, U is hydrophobic residue, x is any residue, and K is lysine) (53, 54). The motif 2 in the active site of HBoV-NS1N contains two His residues (H115/H117) separated by residue Cys116, followed by the three hydrophobic residues Ile118, Leu119, and Val120 (Fig. 1b). A network of hydrogen-bonds connects His115, His117, and Glu108 from motif 2, whereas Lys215 from motif 3 forms a salt bridge with the Glu108 (Fig. 1b). A zinc ion was observed in the active site of the AAV Rep nuclease domain and was engaged in the nuclease activity (47). However, no metal ion is observed in the HBoV-NS1N electron density map. A water molecule in proximity to His115 and His117 makes H bonds with side chains of Gln106, Glu108 and Lys 215 (Fig. 1b). The HBoV1-NS1N motif 3 contains only the second Tyr at position 211, and the position for the first Tyr (residue 207) is Phe. The two Tyr residues in motif 3 were shown to be catalytically essential in many HUH nucleases, where they were believed to alternate in the cleavage and joining reactions (55, 56). The presence of only one Tyr residue in HBoV-NS1N motif 3 is consistent with its function as a nickase with no need for the joining reaction. AAV Rep retains both conserved Tyr at positions 149 and 153; however, only the second Tyr (Tyr153) has been shown to be involved in the nuclease activity (47). It is tempting to speculate that the other Tyr is likely related to integration of viral DNA into the host chromosome in AAV, a function that is not required and may not exist in HBoV. In HBoV-NS1N structure, the Tyr211 side chain of molecule A points toward the two His residues and Glu108, making an H bond with the side chain amino group of Lys215 (Fig. 1b), whereas Tyr211 side chain of molecule B orientates away from the two His residues, potentially reflecting two different states. The Lys240 side chain is located on the top of the nickase active site, forming a lid above the active side (Fig. 1b). This Lys is absent in AAV Rep and may provide an additional, bocavirus-specific, regulatory mechanism for DNA substrate binding and cleavage.
Putative binding site for the origin of replication.
The parvovirus NS1 proteins triggers strand nicking and initiation of DNA replication by recognition of the Ori, which is achieved by specific binding of the NS1 N-terminal domain to DNA sequence motifs in the Ori as shown in AAV Rep (48). Analysis of the electrostatic potential surface shows a positively charged region formed by the β-hairpin (residues 190 to 198) and helix α5 (Fig. 2). This region superimposes well with that of AAV Rep (Fig. 3) (48). The β-hairpin contains the three positively charged residues Arg191, Arg193, and Arg194, which correspond to conserved residues Lys135, Lys137, and Lys138 in AAV Rep (Fig. 2b). The structure of AAV Rep complexed with the tetranucleotide repeats of the viral Ori shows that the corresponding hairpin and basic residues therein insert into the major groove of the bound DNA (48). In addition, helix α5 (residues 127 to 132) in HBoV-NS1N contains the three basic residues Lys127, Arg128, and Lys131, corresponding to a similar region in AAV Rep which interacts with the DNA minor groove. We generated a model for HBoV-NS1N:DNA binding by superimposing the AAV Rep-DNA complex structure onto HBoV-NS1N, showing a consistent mode of protein-DNA interaction (Fig. 2b). These results suggest that this positively charged region in HBoV-NS1N is the putative binding site for the Ori. AAV Rep contains an additional positively charged site for binding of the RBE stem-loop element, which strongly stimulates cleavage at the terminal resolution site (48). However, the corresponding region on HBoV-NS1N is negatively charged, and it remains to be determined whether it is engaged in binding of additional DNA elements.
Fig 2.
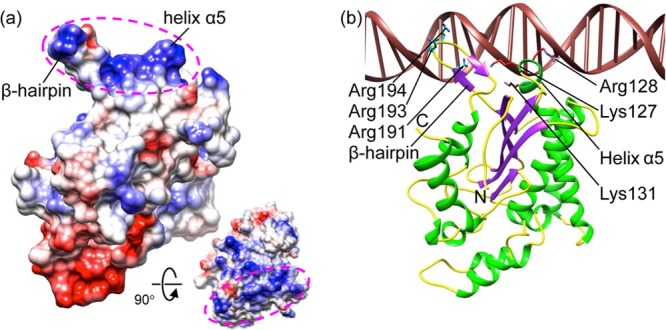
Putative Ori-recognition site. (a) Electrostatic potential rendition of HBoV-NS1N showing a positively charged surface region mediated by the β-hairpin and helix α5 (magenta ellipsoids). The inset shows a view 90° about the horizontal axis in which the positively charged region faces the reader. (b) Model of HBoV-NS1N (ribbon diagram) binding to dsDNA based on superposition of the AAV-5 Rep nuclease domain structure (PDB code 1RZ9) in the same view as in panel a. Side chains of conserved basic residues are shown as sticks. The N and C termini are labeled. The color scheme is the same as in Fig. 1a. The DNA is in brown.
Fig 3.
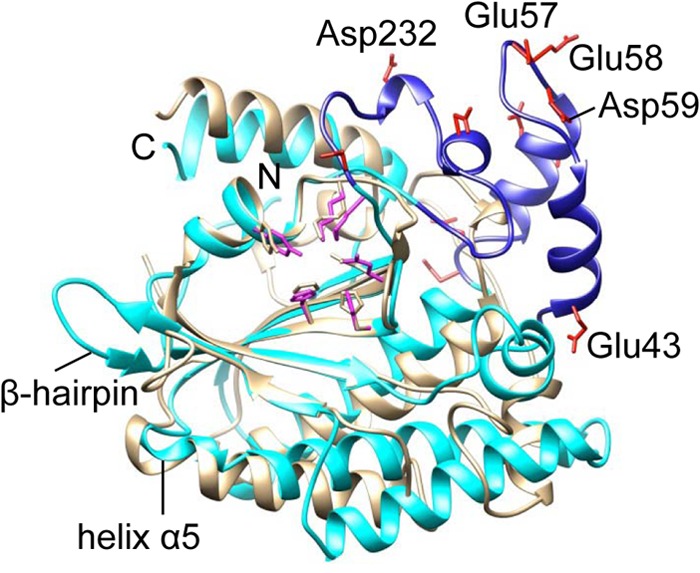
Subdomain of HBoV-NS1N. HBoV-NS1N (cyan) is superimposed with AAV-5 Rep (gray). The subdomain of HBoV-NS1N is blue, with side chains of characteristic acidic residues shown as red sticks. Some of the acidic residues in the subdomain are labeled. Side chains of the nuclease active site residues are shown as sticks for HBoV-NS1N (magenta) and AAV-5 Rep (gray). The β-hairpin and helix α5 of HBoV-NS1N are also indicated to show their positions with respect to the subdomain and the nuclease active site.
Bocavirus-specific subdomain.
HBoV-NS1N contains 271 residues in comparison to 197 residues in AAV Rep nuclease domain (47). The two structures are generally superimposable with a root mean square deviation (RMSD) of 2.49 Å for 144 Cα atoms, particularly for the central β-sheets and the nuclease active site with an RMSD of 0.80 Å for 49 Cα atoms. However, a subdomain is observed in HBoV that is not present in AAV Rep (Fig. 3). This subdomain contains residues 39 to 73 and residues 217 to 239, which are elongated insertions between α1 and α4 and between β7 and β8, respectively. This subdomain is highly negatively charged, studded by a characteristic array of a triplet and two duplets of consecutive acidic residue, including Asp57, Glu58, Asp59, Asp63, Asp64, Glu70, and Asp71, as well as residues Asp226, Asp232, and Glu239. Such a strongly acidic subdomain is located distal to the putative origin recognition region, thus may facilitate capturing of the Ori DNA by properly orienting the NS1 molecule with respect to the DNA. Sequence alignment shows that this subdomain is conserved among HBoV species, as well as in BPV and MVC, suggesting that it may be a characteristic feature of bocavirus NS1 proteins and is likely to be involved in bocavirus-specific NS1-DNA or NS1-host interaction.
Crystallographic dimer.
The asymmetric unit of the crystal contains two molecules related by a noncrystallographic 2-fold rotational symmetry (Fig. 4a). The two molecules are nearly identical, with an RMSD of 0.48 Å for superimposition of main chain backbones. The total solvent-accessible area for molecules A and B are 13,152 and 12,855 Å2, respectively. The buried surface area is ∼584 Å2 for each monomer, as calculated with the program PISA (57). The two nuclease active sites are facing the same side, forming a continuous, extended, highly positively charged area along the dimer (Fig. 4b). The two β-hairpins appear to embrace that extended positively charged area (Fig. 4b), forming an architecture that can snugly accommodate a double-stranded DNA (dsDNA), with the two β-hairpins inserted into the DNA major groove (data not shown). The Tyr211 residues in the two active sites are located within close proximity of the DNA and thus can approach the DNA and form a 5′-phosphotyrosyl linkage with it. Such an architecture is not incompatible with the NS1-DNA interaction for recognition of the Ori, which occurs along the other side of the β-hairpin. The biological implication of this dimeric architecture is not known. The substrate for NS1 nucleolytic cleavage may be ssDNA, as for many HUH superfamily nucleases (47, 48, 58–60), but NS1-DNA binding could involve structured DNA components such as stem-loops, as shown in structures of AAV Rep, relaxase TrwC/TraI, and DNA transposase TnpA (48, 59–61). DNA transposase TnpA, which also belongs to the HUH superfamily, forms a dimer with both nuclease active sites facing to the same side (60). Moreover, parvovirus NS1 proteins must assemble into ring oligomers in accordance with the helicase function. For example, AAV Rep was reported to form ring oligomers with variable stoichiometry (62). These data imply that NS1 can form oligomers and may partially be involved in interaction with dsDNA elements during nucleolytic catalysis. Nevertheless, further studies are required to elucidate the underlying mechanistic details and address whether the observed crystallographic dimer plays any role in HBoV-NS1N functions.
Fig 4.
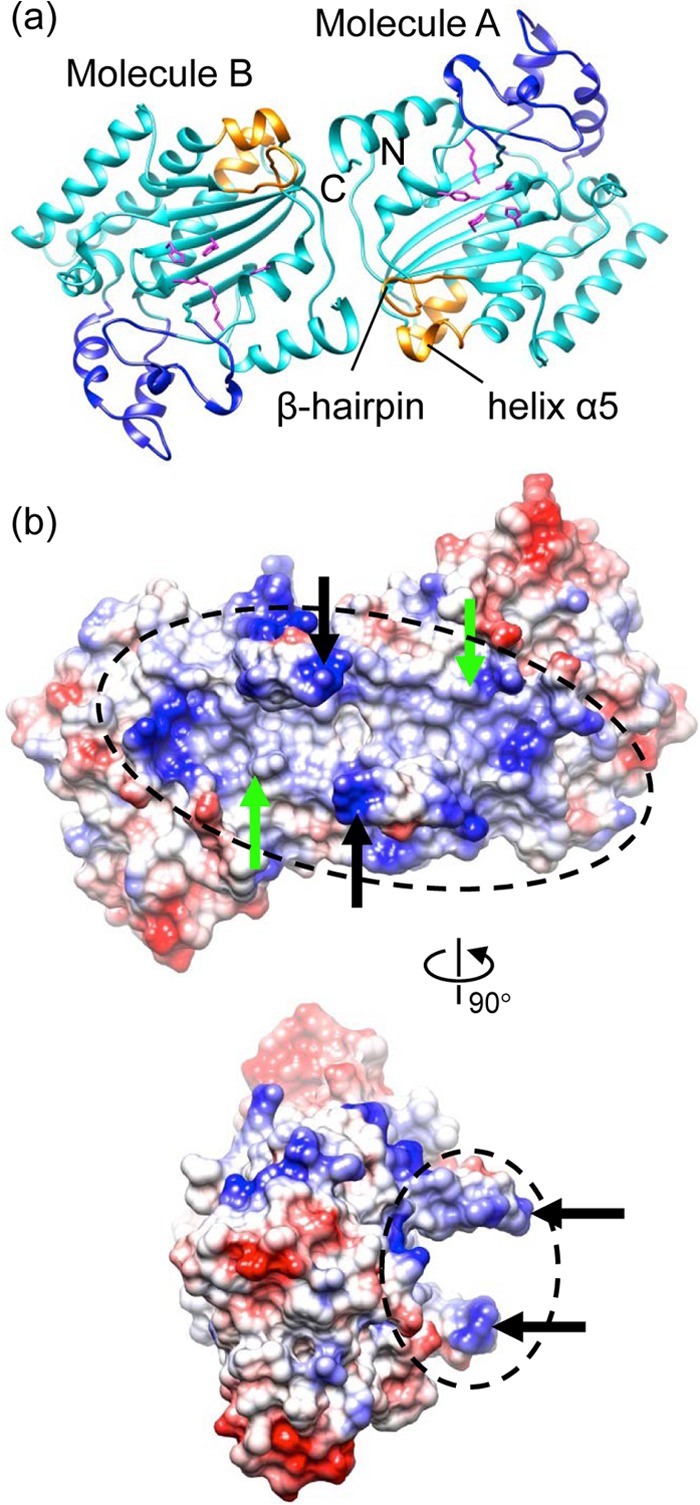
Crystallographic dimer. (a) Ribbon diagram of the dimer (cyan) with the subdomain, the putative Ori-binding site, and the nuclease active site residues shown in blue, gold, and magenta, respectively. (b) Electrostatic potential molecular surface of the dimer showing an extended positively charged area (dashed ellipsoids) spanning two nuclease active sites (active Tyr indicated with green arrows) and the putative Ori recognition sites (β-hairpin indicated with black arrows). The bottom view is 90° about the vertical axis from the top view.
Structural mapping of the diversity of HBoV NS1 proteins.
Parvoviruses have been shown to evolve rapidly, having a mutation or substitution rate approaching that of RNA viruses (63), as was demonstrated by the emergence of canine parvovirus from feline panleukopenia virus in the last ∼40 years (64) and by studies of the B19V evolution (65, 66). Consistent with this, genetic characterization has shown that HBoV is a group of highly diverse, recombination-prone, dynamic viruses (4). We have mapped the amino acid residues that are variable in HBoV NS1 nuclease domains onto the present structure (Fig. 5). A highly scattered pattern is observed, with changed residues distributing quite uniformly all over the molecule, implying that these residues are less likely associated with NS1 functions critical for virus replication thus may be more prone to mutation. However, two extended invariable regions are observed: (i) the putative origin recognition site encompassing the β-hairpin and the helix α5 and (ii) the region encompassing the nuclease active site (Fig. 5). The β-hairpin and the helix α5 are conserved among the four HBoV species (data not shown), suggesting that these viruses use identical molecular elements for the Ori recognition by the NS1 proteins. These data indicate that the recognition of Ori and the nickase activity are two highly conserved features of HBoV and that the underlying structural elements are less prone to mutation and so can serve as valuable targets for the development of broad-spectrum antivirals that are compatible with the high diversity of HBoV. Residue Lys112, which is located upstream of the two His residues in motif 2 next to the nuclease active site, is among a few residues that show the highest variability, changing to acidic residues in HBoV2 and HBoV3 (Fig. 5). Nevertheless, the potential impact of such variability of Lys112 on the NS1 function is yet to be understood.
Fig 5.
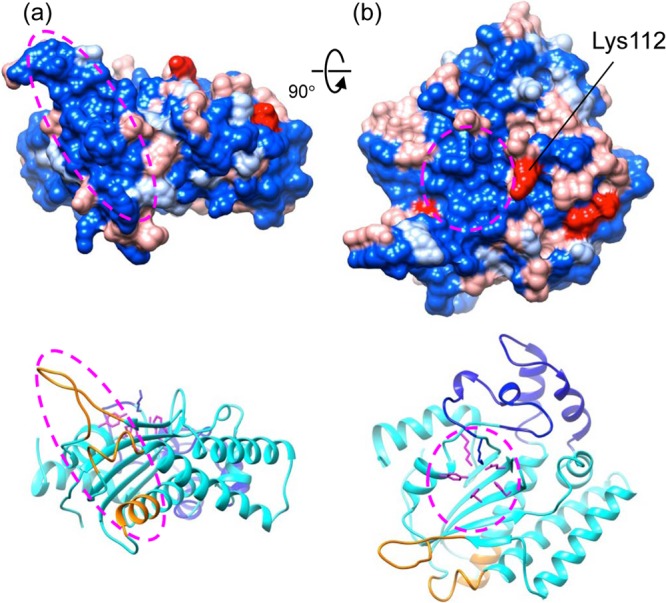
Mapping the variable amino acid residues onto the HBoV-NS1N structure. The amino acid sequences of NS1 from the four HBoV species were aligned, and the residues that are constant among four, three, and two sequences and different for all four sequences are colored blue, light blue, light red, and red, respectively. The putative Ori recognition site and the nuclease active site are indicated with an ellipsoid in panels a and b, respectively. One of the highly variable residues, Lys112, which is next to the active site, is indicated. The view in panel a is 90° about the horizontal axis from that in panel a. In panels a and b, ribbon diagrams of the structure in the same view colored in the same scheme as in Fig. 4a are shown at the bottom.
Model for recognition of HBoV1 Ori by NS1.
Replication mechanisms of HBoV have not been well characterized, largely owing to lack of an animal model and difficulty in tissue culture (15). Our structural results indicate that HBoV-NS1 is a member of the HUH superfamily of nucleases and contains a positively charged surface that is the putative binding site for the Ori. This supports that the HBoV DNA replication is directed by NS1, as in the common rolling-hairpin mechanism proposed for parvoviruses (67). Nevertheless, details of initiation of HBoV DNA replication by NS1 display differences from that known for AAV (48). Like AAV Rep, binding of NS1 via its N-terminal domain on the Ori serves as the first step. This binding involves the highly basic region formed by the beta-hairpin and helix α5. While the mode of DNA binding for the monomeric NS1 would be similar to that of AAV Rep (48), HBoV-NS1 may not form the multimeric protein-DNA complex in the same manner as seen in AAV Rep crystal structure. This is because AAV Ori contains a series of tandem tetranucleotide motifs that serve as the Rep-binding sites, but there are no apparent repeated DNA sequence motifs in the HBoV LEH or REH regions where the potential Ori is located (15). Instead, the HBoV LEH and REH share conserved features with other two bocaviruses, BPV and MVC, in that the LEHs contain cruciform structures and the REHs are hairpins (15, 17, 68). This indicates that bocaviruses use a conserved mode of NS1-Ori recognition, which is distinct from that of dependoviruses. In AAV, it was proposed that binding of multiple Rep molecules at the Ori would trigger oligomerization of the helicase domain, leading to unwinding of dsDNA and the generation of ssDNA at the terminal-resolution site situated at a different location in the Ori, which is to be bound and cleaved by the nuclease domain (48). HBoV NS1 may undergo similar oligomerization for the helicase domain, enabling DNA nicking. Oligomerization of the helicase domain is a common feature in viral origin-binding proteins such as simian virus 40 LTag and papillomavirus E1, which are required for melting of dsDNA to enable DNA replication (69, 70). However, parvovirus NS1 proteins are unique in that they integrate an N-terminal domain that is not only responsible for origin binding but also nicks the DNA to generate a free 3′-hydroxyl group for DNA replication.
The HBoV-NS1N structure reported here raises numerous questions about the versatile functions of NS1. How, for example, do multiple copies of NS1 bind to the Ori in a manner likely different from AAV Rep? What is the architecture of the potential high-order nucleoprotein complex formed by the NS1 and the Ori? What is the mechanism that coordinates the steps of NS1-Ori binding, oligomerization of the NS1 helicase domain, DNA nicking, and subsequent DNA replication? What is the exact role of the subdomain that potentially distinguishes bocavirus from dependovirus? If and how does the crystallographic dimer of HBoV-NS1N take part in NS1 functions? The HBoV-NS1N structure provides a framework for further studies to understand biological roles of the NS1 proteins in parvoviruses in detail.
ACKNOWLEDGMENTS
We thank the staffs at the APS and the SSRL for assistance in X-ray data collection.
The research in the L.T. and J.Q. laboratories was supported by National Institutes of Health awards R01GM090010 (National Institute of General Medical Sciences) and R01AI070723 (National Institute of Allergy and Infectious Diseases), respectively.
Footnotes
Published ahead of print 21 August 2013
REFERENCES
- 1.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. 2005. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. U. S. A. 102:12891–12896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapoor A, Slikas E, Simmonds P, Chieochansin T, Naeem A, Shaukat S, Alam MM, Sharif S, Angez M, Zaidi S, Delwart E. 2009. A newly identified bocavirus species in human stool. J. Infect. Dis. 199:196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur JL, Higgins GD, Davidson GP, Givney RC, Ratcliff RM. 2009. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 5:e1000391. 10.1371/journal.ppat.1000391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapoor A, Simmonds P, Slikas E, Li L, Bodhidatta L, Sethabutr O, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser J, Bartkus J, Delwart E. 2010. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 201:1633–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allander T. 2008. Human bocavirus. J. Clin. Virol. 41:29–33 [DOI] [PubMed] [Google Scholar]
- 6.Schildgen O, Muller A, Allander T, Mackay IM, Volz S, Kupfer B, Simon A. 2008. Human bocavirus: passenger or pathogen in acute respiratory tract infections? Clin. Microbiol. Rev. 21:291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JI, Chung JY, Han TH, Song MO, Hwang ES. 2007. Detection of human bocavirus in children hospitalized because of acute gastroenteritis. J. Infect. Dis. 196:994–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau SK, Yip CC, Que TL, Lee RA, Au-Yeung RK, Zhou B, So LY, Lau YL, Chan KH, Woo PC, Yuen KY. 2007. Clinical and molecular epidemiology of human bocavirus in respiratory and fecal samples from children in Hong Kong. J. Infect. Dis. 196:986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albuquerque MC, Rocha LN, Benati FJ, Soares CC, Maranhao AG, Ramirez ML, Erdman D, Santos N. 2007. Human bocavirus infection in children with gastroenteritis, Brazil. Emerg. Infect. Dis. 13:1756–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kesebir D, Vazquez M, Weibel C, Shapiro ED, Ferguson D, Landry ML, Kahn JS. 2006. Human bocavirus infection in young children in the United States: molecular epidemiological profile and clinical characteristics of a newly emerging respiratory virus. J. Infect. Dis. 194:1276–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peltola V, Soderlund-Venermo M, Jartti T. 2013. Human bocavirus infections. Pediatr. Infect. Dis. J. 32:178–179 [DOI] [PubMed] [Google Scholar]
- 12.Jartti T, Hedman K, Jartti L, Ruuskanen O, Allander T, Soderlund-Venermo M. 2012. Human bocavirus: the first 5 years. Rev. Med. Virol. 22:46–64 [DOI] [PubMed] [Google Scholar]
- 13.Meriluoto M, Hedman L, Tanner L, Simell V, Makinen M, Simell S, Mykkanen J, Korpelainen J, Ruuskanen O, Ilonen J, Knip M, Simell O, Hedman K, Soderlund-Venermo M. 2012. Association of human bocavirus 1 infection with respiratory disease in childhood follow-up study, Finland. Emerg. Infect. Dis. 18:264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn JS, Kesebir D, Cotmore SF, D'Abramo A, Jr, Cosby C, Weibel C, Tattersall P. 2008. Seroepidemiology of human bocavirus defined using recombinant virus-like particles. J. Infect. Dis. 198:41–50 [DOI] [PubMed] [Google Scholar]
- 15.Huang Q, Deng X, Yan Z, Cheng F, Luo Y, Shen W, Lei-Butters DC, Chen AY, Li Y, Tang L, Soderlund-Venermo M, Engelhardt JF, Qiu J. 2012. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog. 8:e1002899. 10.1371/journal.ppat.1002899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gurda BL, Parent KN, Bladek H, Sinkovits RS, DiMattia MA, Rence C, Castro A, McKenna R, Olson N, Brown K, Baker TS, Agbandje-McKenna M. 2010. Human bocavirus capsid structure: insights into the structural repertoire of the Parvoviridae. J. Virol. 84:5880–5889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Chen AY, Cheng F, Guan W, Johnson FB, Qiu J. 2009. Molecular characterization of infectious clones of the minute virus of canines reveals unique features of bocaviruses. J. Virol. 83:3956–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen KC, Shull BC, Moses EA, Lederman M, Stout ER, Bates RC. 1986. Complete nucleotide sequence and genome organization of bovine parvovirus. J. Virol. 60:1085–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schildgen O, Qiu J, Soderlund-Venermo M. 2012. Genomic features of the human bocaviruses. Future Virol. 7:31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han Y, Wang Q, Qiu Y, Wu W, He H, Zhang J, Hu Y, Zhou X. 2013. Periplaneta fuliginosa densovirus nonstructural protein NS1 contains an endonuclease activity that is regulated by its phosphorylation. Virology 437:1–11 [DOI] [PubMed] [Google Scholar]
- 21.Cotmore SF, Gottlieb RL, Tattersall P. 2007. Replication initiator protein NS1 of the parvovirus minute virus of mice binds to modular divergent sites distributed throughout duplex viral DNA. J. Virol. 81:13015–13027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhi N, Mills IP, Lu J, Wong S, Filippone C, Brown KE. 2006. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J. Virol. 80:5941–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Rhode SL., III 1990. Mutation of lysine 405 to serine in the parvovirus H-1 NS1 abolishes its functions for viral DNA replication, late promoter trans activation, and cytotoxicity. J. Virol. 64:4654–4660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niskanen EA, Ihalainen TO, Kalliolinna O, Hakkinen MM, Vihinen-Ranta M. 2010. Effect of ATP binding and hydrolysis on dynamics of canine parvovirus NS1. J. Virol. 84:5391–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cotmore SF, Tattersall P. 2005. Encapsidation of minute virus of mice DNA: aspects of the translocation mechanism revealed by the structure of partially packaged genomes. Virology 336:100–112 [DOI] [PubMed] [Google Scholar]
- 26.King JA, Dubielzig R, Grimm D, Kleinschmidt JA. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bleker S, Pawlita M, Kleinschmidt JA. 2006. Impact of capsid conformation and Rep-capsid interactions on adeno-associated virus type 2 genome packaging. J. Virol. 80:810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsay GJ, Zouali M. 2006. Unscrambling the role of human parvovirus B19 signaling in systemic autoimmunity. Biochem. Pharmacol. 72:1453–1459 [DOI] [PubMed] [Google Scholar]
- 29.Ohshima T, Yoshida E, Nakajima T, Yagami KI, Fukamizu A. 2001. Effects of interaction between parvovirus minute virus of mice NS1 and coactivator CBP on NS1- and p53-transactivation. Int. J. Mol. Med. 7:49–54 [DOI] [PubMed] [Google Scholar]
- 30.Deleu L, Pujol A, Faisst S, Rommelaere J. 1999. Activation of promoter P4 of the autonomous parvovirus minute virus of mice at early S phase is required for productive infection. J. Virol. 73:3877–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gareus R, Gigler A, Hemauer A, Leruez-Ville M, Morinet F, Wolf H, Modrow S. 1998. Characterization of cis-acting and NS1 protein-responsive elements in the p6 promoter of parvovirus B19. J. Virol. 72:609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffatt S, Tanaka N, Tada K, Nose M, Nakamura M, Muraoka O, Hirano T, Sugamura K. 1996. A cytotoxic nonstructural protein, NS1, of human parvovirus B19 induces activation of interleukin-6 gene expression. J. Virol. 70:8485–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo Y, Chen AY, Qiu J. 2011. Bocavirus infection induces a DNA damage response that facilitates viral DNA replication and mediates cell death. J. Virol. 85:133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo Y, Deng X, Cheng F, Li Y, Qiu J. 2013. SMC1-mediated intra-S-phase arrest facilitates bocavirus DNA replication. J. Virol. 87:4017–4032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lou S, Luo Y, Cheng F, Huang Q, Shen W, Kleiboeker S, Tisdale JF, Liu Z, Qiu J. 2012. Human parvovirus B19 DNA replication induces a DNA damage response that is dispensable for cell cycle arrest at phase G2/M. J. Virol. 86:10748–10758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kivovich V, Gilbert L, Vuento M, Naides SJ. 2012. The putative metal coordination motif in the endonuclease domain of human parvovirus B19 NS1 is critical for NS1 induced S phase arrest and DNA damage. Int. J. Biol. Sci. 8:79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun B, Cai Y, Li Y, Li J, Liu K, Yang Y. 2013. The nonstructural protein NP1 of human bocavirus 1 induces cell cycle arrest and apoptosis in HeLa cells. Virology 440:75–83 [DOI] [PubMed] [Google Scholar]
- 38.Wizla P, Begue A, Loison I, Richard A, Caillet-Fauquet P, Stehelin D. 2010. Ectopic expression of H-1 parvovirus NS1 protein induces alterations in actin filaments and cell death in human normal MRC-5 and transformed MRC-5 SV2 cells. Arch. Virol. 155:771–775 [DOI] [PubMed] [Google Scholar]
- 39.Best SM, Shelton JF, Pompey JM, Wolfinbarger JB, Bloom ME. 2003. Caspase cleavage of the nonstructural protein NS1 mediates replication of Aleutian mink disease parvovirus. J. Virol. 77:5305–5312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen AY, Luo Y, Cheng F, Sun Y, Qiu J. 2010. Bocavirus infection induces mitochondrion-mediated apoptosis and cell cycle arrest at G2/M phase. J. Virol. 84:5615–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu GJ, Tzang BS, Tsai CC, Chiu CC, Huang CY, Hsu TC. 2011. Effects of human parvovirus B19 on expression of defensins and Toll-like receptors. Chin. J. Physiol. 54:367–376 [PubMed] [Google Scholar]
- 42.Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. 2009. Human bocavirus can be cultured in differentiated human airway epithelial cells. J. Virol. 83:7739–7748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen AY, Cheng F, Lou S, Luo Y, Liu Z, Delwart E, Pintel D, Qiu J. 2010. Characterization of the gene expression profile of human bocavirus. Virology 403:145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Astell CR, Brunstein J, St Amannd J. 1997. B19 parvovirus: biochemical and molecular features, p 16–41 In Anderson LJ, Young N. (ed), Human parvovirus B19, vol 20 Karger, Basel, Switzerland [Google Scholar]
- 45.Doerig C, Hirt B, Antonietti JP, Beard P. 1990. Nonstructural protein of parvoviruses B19 and minute virus of mice controls transcription. J. Virol. 64:387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raab U, Beckenlehner K, Lowin T, Niller HH, Doyle S, Modrow S. 2002. NS1 protein of parvovirus B19 interacts directly with DNA sequences of the p6 promoter and with the cellular transcription factors Sp1/Sp3. Virology 293:86–93 [DOI] [PubMed] [Google Scholar]
- 47.Hickman AB, Ronning DR, Kotin RM, Dyda F. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol. Cell 10:327–337 [DOI] [PubMed] [Google Scholar]
- 48.Hickman AB, Ronning DR, Perez ZN, Kotin RM, Dyda F. 2004. The nuclease domain of adeno-associated virus rep coordinates replication initiation using two distinct DNA recognition interfaces. Mol. Cell 13:403–414 [DOI] [PubMed] [Google Scholar]
- 49.Guan W, Wong S, Zhi N, Qiu J. 2009. The genome of human parvovirus b19 can replicate in nonpermissive cells with the help of adenovirus genes and produces infectious virus. J. Virol. 83:9541–9553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66:486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ilyina TV, Koonin EV. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes, and archaebacteria. Nucleic Acids Res. 20:3279–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koonin EV, Ilyina TV. 1993. Computer-assisted dissection of rolling-circle DNA replication. Biosystems 30:241–268 [DOI] [PubMed] [Google Scholar]
- 55.Hanai R, Wang JC. 1993. The mechanism of sequence-specific DNA cleavage and strand transfer by ϕX174 gene A* protein. J. Biol. Chem. 268:23830–23836 [PubMed] [Google Scholar]
- 56.Odegrip R, Haggard-Ljungquist E. 2001. The two active-site tyrosine residues of the a protein play non-equivalent roles during initiation of rolling circle replication of bacteriophage p2. J. Mol. Biol. 308:147–163 [DOI] [PubMed] [Google Scholar]
- 57.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 58.Boer R, Russi S, Guasch A, Lucas M, Blanco AG, Perez-Luque R, Coll M, de la Cruz F. 2006. Unveiling the molecular mechanism of a conjugative relaxase: the structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J. Mol. Biol. 358:857–869 [DOI] [PubMed] [Google Scholar]
- 59.Guasch A, Lucas M, Moncalian G, Cabezas M, Perez-Luque R, Gomis-Ruth FX, de la Cruz F, Coll M. 2003. Recognition and processing of the origin of transfer DNA by conjugative relaxase TrwC. Nat. Struct. Biol. 10:1002–1010 [DOI] [PubMed] [Google Scholar]
- 60.Barabas O, Ronning DR, Guynet C, Hickman AB, Ton-Hoang B, Chandler M, Dyda F. 2008. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell 132:208–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larkin C, Datta S, Harley MJ, Anderson BJ, Ebie A, Hargreaves V, Schildbach JF. 2005. Inter- and intramolecular determinants of the specificity of single-stranded DNA binding and cleavage by the F factor relaxase. Structure 13:1533–1544 [DOI] [PubMed] [Google Scholar]
- 62.Mansilla-Soto J, Yoon-Robarts M, Rice WJ, Arya S, Escalante CR, Linden RM. 2009. DNA structure modulates the oligomerization properties of the AAV initiator protein Rep68. PLoS Pathog. 5:e1000513. 10.1371/journal.ppat.1000513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat. Rev. Genet. 9:267–276 [DOI] [PubMed] [Google Scholar]
- 64.Shackelton LA, Parrish CR, Truyen U, Holmes EC. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. U. S. A. 102:379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shackelton LA, Holmes EC. 2006. Phylogenetic evidence for the rapid evolution of human B19 erythrovirus. J. Virol. 80:3666–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Norja P, Eis-Hubinger AM, Soderlund-Venermo M, Hedman K, Simmonds P. 2008. Rapid sequence change and geographical spread of human parvovirus B19: comparison of B19 virus evolution in acute and persistent infections. J. Virol. 82:6427–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cotmore SF, Tattersall P. 2005. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses, p 171–181 In Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Hodder Arnold, London, United Kingdom [Google Scholar]
- 68.Qiu J, Cheng F, Johnson FB, Pintel D. 2007. The transcription profile of the bocavirus bovine parvovirus is unlike those of previously characterized parvoviruses. J. Virol. 81:12080–12085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li D, Zhao R, Lilyestrom W, Gai D, Zhang R, DeCaprio JA, Fanning E, Jochimiak A, Szakonyi G, Chen XS. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512–518 [DOI] [PubMed] [Google Scholar]
- 70.Chen G, Stenlund A. 1998. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J. Virol. 72:2567–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]


