Abstract
Herpes simplex virus 1 (HSV-1) rapidly eliminates mitochondrial DNA (mtDNA) from infected cells, an effect that is mediated by UL12.5, a mitochondrial isoform of the viral alkaline nuclease UL12. Our initial hypothesis was that UL12.5 directly degrades mtDNA via its nuclease activity. However, we show here that the nuclease activities of UL12.5 are not required for mtDNA loss. This observation led us to examine whether cellular nucleases mediate the mtDNA loss provoked by UL12.5. We provide evidence that the mitochondrial nucleases endonuclease G (ENDOG) and endonuclease G-like 1 (EXOG) play key redundant roles in UL12.5-mediated mtDNA depletion. Overall, our data indicate that UL12.5 deploys cellular proteins, including ENDOG and EXOG, to destroy mtDNA and contribute to a growing body of literature highlighting roles for ENDOG and EXOG in mtDNA maintenance.
INTRODUCTION
Many viruses, including herpesviruses, encode proteins that disrupt mitochondrial functions (reviewed in reference 1). Mitochondria are important targets for viral proteins due to their roles as key regulators of cellular energy production, apoptosis, antiviral signaling, intermediary metabolism, and calcium homeostasis (reviewed in reference 2). To perform these vital functions, mitochondria rely on imported proteins encoded by the nuclear genome as well as on a smaller set of proteins encoded by the mitochondrial genome. The 16.6-kb mitochondrial DNA (mtDNA) genome is present in hundreds to thousands of copies per cell (3, 4) and encodes 13 mitochondrial proteins involved in oxidative phosphorylation as well as 22 mitochondrial transfer RNAs and two mitochondrial ribosomal RNAs which are required for mitochondrial protein synthesis (5). Mutations of proteins encoded by the nuclear genome involved in mtDNA replication or nucleotide metabolism can cause mutations, deletions, or depletion of mtDNA, resulting in the collapse of oxidative phosphorylation and numerous debilitating or lethal diseases (reviewed in references 6 and 7). Therefore, it is important to understand what impact viral infection has on mtDNA and mitochondrial functions.
The only viruses known to directly cause a reduction in mtDNA levels during infection are Epstein-Barr virus (EBV) and herpes simplex virus 1 (HSV-1) and HSV-2 (8, 9). During EBV lytic infection, Zta associates with the mitochondrial single-stranded DNA binding protein (mtSSB), preventing mtSSB localization to mitochondria and leading to an inhibition of mtDNA replication (9). In the case of HSV-1, mtDNA is rapidly depleted from infected cells through the action of a mitochondrially targeted viral nuclease, UL12.5 (8, 10). UL12.5 is an amino-terminally truncated isoform of the HSV-1 UL12 alkaline nuclease. Both proteins are encoded by the UL12 gene but arise from distinct 3′ coterminal mRNAs (11, 12). The larger mRNA yields the full-length UL12 protein (11), which possesses strong exonuclease and much weaker endonuclease activity (13–16). Using an N-terminal nuclear localization signal, UL12 translocates to the nucleus (17), where it is proposed to process complex viral DNA structures into linear genomes suitable for packaging into mature viral particles (18, 19). Although alkaline nuclease is not essential for viral replication in cell culture, alkaline nuclease-null viruses are severely impaired (20, 21). The smaller mRNA encodes an in-frame, N-terminally truncated version of UL12 called UL12.5 which initiates at UL12 codon M127 (22). While UL12.5 displays enzymatic properties similar to those of UL12, it cannot functionally replace UL12 during viral replication (17, 22, 23). This lack of redundancy has been attributed to two factors: UL12 is much more abundant than UL12.5 (22), and UL12 and UL12.5 have distinct subcellular locations (17). Although a fraction of total UL12.5 is found in the nucleus, much of the protein is targeted to mitochondria using an N-proximal mitochondrial localization signal (MLS) (8, 10).
We recently mapped the UL12.5 MLS to residues M185 to R245 (of full-length UL12) and showed that an N-terminally truncated form initiating at residue M185 (UL12M185) localizes to mitochondria and depletes mtDNA (10). Intriguingly, a previous study had shown that two mutations in the N-terminal regions of UL12 and UL12.5 that are absent from UL12M185 inactivate detectable nuclease activity in vitro (23). Taken together, these data raised the possibility that UL12M185 lacks enzymatic activity and, by extension, that the nuclease activity of UL12.5 is not required for mtDNA depletion. Here we present data which support this hypothesis. We demonstrate that several nuclease-deficient mutants of UL12.5 retain the ability to deplete cells of mtDNA. Furthermore, we provide evidence that the mitochondrial proteins endonuclease G (ENDOG) and endonuclease G-like 1 (EXOG) contribute to the degradation of mtDNA following UL12.5 expression.
MATERIALS AND METHODS
Cells and transfections.
HeLa cells were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (complete medium). All plasmid transfections were performed using Lipofectamine 2000 (Invitrogen) unless otherwise stated and were followed by incubation in complete medium supplemented with 50 μg/ml uridine and 1 mM sodium pyruvate.
Plasmids.
HSV-1 UL12-SPA, UL12.5-SPA, and UL12.5-SPA mutants were generated in pMZS3F vector (24). Expression from this vector generates C-terminal sequential peptide affinity (SPA)-tagged proteins containing three FLAG epitopes. The UL12-SPA, UL12.5-SPA, and UL12M185-SPA open reading frames (ORFs) were amplified by PCR using DNA isolated from Vero cells infected with KOS37-SPA, which is a mutant virus that expresses C-terminal SPA-tagged UL12 and UL12.5 proteins (B. A. Duguay, H. A. Saffran, A. Ponomarev, H. E. Eaton, and J. R. Smiley, unpublished data). The PCRs introduced a 5′ EcoRI restriction site and either a 3′ XbaI or 3′ Bsu36I site to the ORFs using primers F1 and R1 (UL12-SPA), F2 and R2 (UL12.5-SPA), or F3 and R1 (UL12M185-SPA). Restriction enzyme-digested PCR products were ligated into pMZS3F to create pMZS3F UL12-SPA, pMZS3F UL12.5-SPA, and pMZS3F UL12M185-SPA.
Plasmids expressing untagged versions of UL12.5-L150K, UL12.5-ΔN, UL12.5-ΔMLS, and UL12.5-ΔC were first created in a pcDNA3.1 vector. Additional details regarding the construction of these plasmids are available upon request. The L150K mutation was introduced using a modified site-directed mutagenesis protocol (25) with primers F4 and R3. The ΔN mutation (deletes residues W128 to R148 [23]) and the ΔMLS mutation (deletes residues R188 to R212 [10]) were both introduced using a QuikChange XL site-directed mutagenesis kit (Stratagene) with primers F5 and R4 and primers F6 and R5, respectively. The ΔC mutation (deletes residues P578 to R626 [23]) was created by PCR using primers F2 and R6. The pcDNA3.1(-) vectors encoding UL12.5-L150K, UL12.5-ΔN, UL12.5-ΔMLS, and UL12.5-ΔC or pSAK vectors encoding UL12.5-D340E-mOrange (10) and UL12.5-G336A/S338A-mOrange (10) were used as the templates for PCR to add 5′ EcoRI and 3′ KpnI sites using the following primers: L150K, ΔMLS, UL12.5-D340E, UL12.5-G336A/S338A (F2 and R7), ΔN (F7 and R7), ΔC (F2 and R8), and UL12M185-D340E or UL12M185-G336A/S338A (F8 and R7). All PCR products were ligated into an EcoRI/KpnI-digested intermediate vector (pcDNA3.1 UL12.5-SPA) containing a KpnI site immediately upstream of the SPA tag coding sequence. This intermediate vector was created by digestion of pMZS3F UL12.5-SPA with Bsu36I, treatment with T4 DNA polymerase (Invitrogen), and then digestion with EcoRI. The UL12.5-SPA ORF was ligated into EcoRI/SmaI-digested pcDNA3.1/myc-His(-), creating pcDNA3.1 UL12.5-SPA. All intermediate vectors encoding SPA-tagged proteins were digested with EcoRI and Bsu36I and ligated into pMZS3F to generate pMZS3F UL12.5-L150K-SPA, pMZS3F UL12.5-D340E-SPA, pMZS3F UL12.5-G336A/S338A-SPA, pMZS3F UL12.5-ΔN-SPA, pMZS3F UL12.5-ΔMLS-SPA, pMZS3F UL12.5-ΔC-SPA, pMZS3F UL12M185-D340E-SPA, and pMZS3F UL12M185-G336A/S338A-SPA.
Plasmids encoding C-terminally myc-tagged ENDOG and EXOG were generated by PCR amplification of ENDOG cDNA (RG205089; OriGene) or EXOG cDNA (RG224222; OriGene). These PCRs introduced a 5′ EcoRI site (ENDOG) or 5′ BamHI site (EXOG) and 3′ HindIII sites to the ORFs. Amplification of the ENDOG ORF used primers F9 and R9. Amplification of the EXOG ORF used primers F10 and R10. Plasmids encoding nuclease-deficient versions of ENDOG (ENDOG-H141A [26]) and EXOG (EXOG-H140A [27]) were generated using overlap PCR. Briefly, the ENDOG ORF was amplified by PCR using primers F9 and R11 or primers F11 and R9. The resulting overlapping PCR products were combined in equimolar ratios and reamplified using primers F9 and R9 to generate the ENDOG-H141A ORF. Similarly, the EXOG-H140A ORF was amplified by PCR using primers F10 and R12 or F12 and R10 and the overlapping PCR products were reamplified using primers F10 and R10. All ENDOG and EXOG ORFs were ligated into pcDNA3.1/myc-His(-) to create pcDNA3.1-ENDOG-myc-His, pcDNA3.1-ENDOG-H141A-myc-His, pcDNA3.1-EXOG-myc-His, and pcDNA3.1-EXOG-H140A-myc-His. For simplicity, no subsequent references to the His epitope are made in describing these plasmids or proteins.
A pcDNA3.1 plasmid expressing monomeric orange (mOrange) fluorescent protein has been previously described (8). A pcDNA4 plasmid expressing a catalytically inactive, myc-tagged version of sirtuin 3 (Sirt3-H248Y-myc) (plasmid 24917) was obtained from Addgene (28). All oligonucleotides (see Table 1) were synthesized by Integrated DNA Technologies. All PCRs were performed using Platinum Pfx DNA polymerase (Invitrogen) unless otherwise stated. Protein residue numbering of all UL12.5 mutants is relative to that of the full-length HSV-1 UL12 protein.
Table 1.
Oligonucleotides used to generate expression plasmids for this study
| Primer name | Sequence (5′→3′)a |
|---|---|
| F1 | GGAATTCCGCCACCATGGAGTCCACGGGAGGCC |
| F2 | GGAATTCCGCCACCATGTGGTCGGCGTCGGTGAT |
| F3 | GGAATTCCGCCACCATGGTGGACCGCGGACTCGG |
| F4 | CGAGACGTTCGAGCGCCACAAACGCGGGTTGCTGC |
| F5 | CGAATTCCGCCACCATGCACCTGCGCGGG |
| F6 | GGTATGGTGGACGCATATGCACCCCTCATGGGG |
| F7 | AGAATTCCGCCACCATGCACCTGCGCGG |
| F8 | GGAATTCCGCCACCATGGTGGACCGCGGACTCGG |
| F9 | TAGTCGAATTCCGCCACCATGCGGGCGCTGCG |
| F10 | TACGGATCCCGCCACCATGGCTATCAAGAGTATCGCTTCCCG |
| F11 | CCGCGGTGCCCTGGCC |
| F12 | GTCACGAGGAGCCATGGCTCCAG |
| R1 | CCTCTAGAGCCTACTTGTCATCGTCATCCTTGTAGTCG |
| R2 | GGCACAGCCTCAGGTCACTACTTGTCATCGTCATCC |
| R3 | GCAGCAACCCGCGTTTGTGGCGCTCGAACGTCTCG |
| R4 | CCCGCGCAGGTGCATGGTGGCGGAATTCG |
| R5 | CCCCATGAGGGGTGCATATGCGTCCACCATACC |
| R6 | CCGATATCTCAATCGATGCGGACGGGGGTAATG |
| R7 | GGGGTACCGCGAGACGACCTCCCCGTCGTCGGTG |
| R8 | GGGGTACCATCGATGCGGACGGGGGTAATGATCAGGGCGATCG |
| R9 | AGCAAGCTTCTTACTGCCCGCCGTGATGG |
| R10 | TGCAAGCTTGGATGGCTTTCTTATCTGGGTTCCTGA |
| R11 | GGCCAGGGCACCGCGG |
| R12 | CTGGAGCCATGGCTCCTCGTGAC |
Start/stop codons are indicated in bold italics, point mutations/insertions are indicated in bold, and restriction sites are underlined.
Cell lysis and Western blotting.
Transfected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8.0], 1% Igepal CA-630, 150 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate, Roche Complete [EDTA-free] protease inhibitor cocktail) for 20 to 30 min on ice. Lysates were cleared by centrifugation at 20,800 × g for 5 min at 4°C. Lysate protein concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (Pierce). Protein sample buffer (5×) (200 mM Tris-HCl [pH 6.8], 5% SDS, 50% glycerol, 1.43 M β-mercaptoethanol) was added to each lysate. For immunoprecipitations (IP), transfected cells were harvested at 48 h posttransfection (hpt), lysed using IP lysis buffer (50 mM Tris-HCl [pH 7.5], 1% Igepal CA-630, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EGTA, plus protease inhibitors), and cleared by centrifugation at 20,800 × g for 15 min.
All lysates were subjected to SDS-PAGE and transferred to nitrocellulose membranes (Hybond ECL; GE Healthcare). Membranes for chemiluminescence detection were blocked in phosphate-buffered saline (PBS) containing 5% skim milk for 1 h at room temperature. The primary antibodies mouse anti-FLAG M2 (Sigma) and rabbit anti-c-myc (Sigma) were incubated with membranes overnight at 4°C, and horseradish peroxidase (HRP)-conjugated goat anti-mouse or goat anti-rabbit secondary antibodies (Bio-Rad) were incubated with membranes for 1 h at room temperature. All antibodies were diluted in PBS containing 1% skim milk. Proteins were visualized using ECL Plus (GE Healthcare) or ECL2 (Pierce). Membranes for infrared imaging were blocked in Odyssey blocking buffer (Li-COR)/Tris-buffered saline (1:1) containing 0.1% Tween 20 (OBBT) for 1 h at room temperature. Primary antibodies were incubated overnight at 4°C, and secondary antibodies (Alexa Fluor 680 goat anti-rabbit [Invitrogen] and IRDye 800 donkey anti-mouse [Rockland]) were incubated for 1 h at room temperature. All antibodies were diluted in OBBT. Proteins were detected using an Odyssey infrared imaging system (Li-COR).
Immunofluorescence microscopy.
HeLa cells were grown on coverslips, transfected for 48 h, and processed for immunofluorescence as previously described (17). Cells were costained with rabbit anti-FLAG (Sigma) and mouse anti-cytochrome c (556432; BD Biosciences) followed by incubation with Alexa Fluor 555-conjugated goat anti-rabbit and Alexa Fluor 488-conjugated goat anti-mouse secondary antibodies (Invitrogen). Stained cells were mounted onto slides using Vectashield (Vector Laboratories) containing 1 mg/ml 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes). Images were obtained using a fluorescence microscope with a 63× oil immersion objective and an ApoTome optical sectioning device (Zeiss).
Live-cell imaging of mtDNA depletion.
Chambered coverglass dishes (155383; Nunc) were used for all live-cell imaging. To quantitate mtDNA depletion by UL12.5 mutants, 200 ng of plasmid DNA was cotransfected with 100 ng of pcDNA-Orange. To assess the effect of ENDOG and/or EXOG overexpression on mtDNA depletion by UL12M185-G336A/S338A-SPA, 100 ng of pMZS3F plasmid DNA (empty vector or UL12M185-G336A/S338A-SPA) was cotransfected with 200 ng of pcDNA3.1 plasmid DNA (empty vector alone, empty vector and ENDOG/EXOG/Sirt3 plasmid DNA, or ENDOG and EXOG plasmid DNA) and 100 ng of pcDNA-Orange. To determine if ENDOG and/or EXOG knockdown had an effect on mtDNA depletion, HeLa cells were first seeded into 24-well plates and transfected with 10 nM total small interfering RNA (siRNA) using DharmaFECT 1 (Thermo Scientific) for 48 h. These cells were trypsinized and reverse transfected into chambered coverglass dishes for an additional 24 h with the indicated siRNAs, 100 ng of pcDNA-Orange, and 100 ng of empty vector or a plasmid expressing UL12.5-SPA or UL12M185-G336A/S338A-SPA using DharmaFECT Duo (Thermo Scientific). For all live-cell imaging, mtDNA was visualized in mOrange-positive cells using PicoGreen (Invitrogen) staining as previously described (10). Images were obtained using a fluorescence microscope and a 40× oil immersion objective (Zeiss).
Immunoprecipitations.
All incubations were carried out at 4°C. Mouse anti-FLAG M2 (Sigma)-protein G-agarose conjugates were formed overnight in PBS and washed three times with IP lysis buffer before use. Transfected cell lysates were incubated for 30 min with protein G-agarose (Roche) prior to incubating with the antibody-agarose conjugates for 2 h. The immunoprecipitates were washed three times in IP lysis buffer followed by three washes with 50 mM Tris (pH 8.8). The agarose-conjugated protein complexes were resuspended in 50 mM Tris (pH 8.8) containing protease inhibitors. Protein sample buffer was added to an aliquot of each immunoprecipitate and subjected to infrared Western blotting using rabbit anti-FLAG (Sigma).
In vitro IP nuclease assays.
Immunoprecipitates were incubated with EcoRI-linearized pUC19 DNA or uncut pEGFP-C1 DNA (Clontech) in nuclease assay buffer (50 mM Tris-Cl [pH 8.8], 10 mM MgCl2, 5 mM β-mercaptoethanol) for 2 h at 37°C. The remaining pUC19 DNA was resolved by agarose gel electrophoresis, stained with SYBR gold (Invitrogen), and visualized with a FLA-5100 imaging system (Fujifilm). Immunoprecipitates incubated with circular pEGFP-C1 were processed by SYBR gold staining to visualize DNA quantity and migration or by nick translation to visualize endonuclease activity. For nick translation, the immunoprecipitates were removed by centrifugation and the supernatants were incubated at room temperature for 1 h in 33 mM Tris-Cl (pH 7.9)–10 mM MgCl2–50 mM NaCl–33 μM dATP/dGTP/dTTP with 5 U of Escherichia coli DNA polymerase I (NEB) and 20 μCi of [32P]dCTP. DNA was isolated by phenol-chloroform extraction and resolved by agarose gel electrophoresis. The agarose gel was dried overnight and exposed to a phosphor screen, and radiolabeled DNA was visualized using a FLA-5100 imaging system.
siRNA knockdown of EXOG and ENDOG.
Small interfering RNAs (siRNAs) targeting ENDOG (s707), EXOG (s19298), or a nontargeting control (4390847) were obtained from Ambion. Endogenous EXOG knockdown was evaluated using HeLa cells transfected for 48 h with 10 nM total siRNA (10 nM negative-control [N.C.] siRNA, 5 nM EXOG plus 5 nM N.C. siRNAs, or 5 nM ENDOG plus 5 nM N.C. siRNAs) using DharmaFECT 1. Total cell lysates were prepared and processed for infrared Western blotting using rabbit anti-EXOG (Invitrogen) and mouse anti-actin (Sigma) primary antibodies. Alternatively, siRNA-transfected cells were lysed for 10 min on ice with digitonin lysis buffer (1 mM NaH2PO4, 8 mM Na2HPO4, 75 mM NaCl, 250 mM sucrose, 190 μg/ml digitonin, plus protease inhibitors) and centrifuged at 20,800 × g for 5 min at 4°C. To the supernatants (cytoplasmic fractions), 5× protein sample buffer was added. The pellets (mitochondrial fractions) were resuspended in 25 mM Tris (pH 8.0)–0.1% Triton X-100, to which 5× protein sample buffer was added. Lysates were processed for infrared Western blotting using rabbit anti-EXOG (Invitrogen), mouse anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Biodesign International), mouse anti-manganese superoxide dismutase (MnSOD) (Stressgen), and mouse anti-cytochrome c (556433; BD Biosciences).
SiRNAs were also evaluated for their ability to suppress overexpression of the desired protein. HeLa cells were cotransfected with 10 nM total siRNA (10 nM N.C. siRNA, 5 nM EXOG plus 5 nM N.C. siRNAs, or 5 nM ENDOG plus 5 nM N.C. siRNAs) and 1 μg plasmid DNA (pcDNA3.1-EXOG-myc or pcDNA3.1-ENDOG-myc) using DharmaFECT Duo. Lysates were prepared at 48 hpt and processed for infrared Western blotting using mouse anti-c-myc (Sigma) and rabbit anti-actin (Sigma).
Image processing.
All microscopy images were acquired with an Axiovert 200 M fluorescence microscope using the AxioVision 4.5 program (Zeiss). Image processing was performed using Illustrator CS2 and Photoshop CS2 (Adobe).
RESULTS AND DISCUSSION
Construction, expression, and mitochondrial localization of nuclease-deficient UL12.5-SPA mutants.
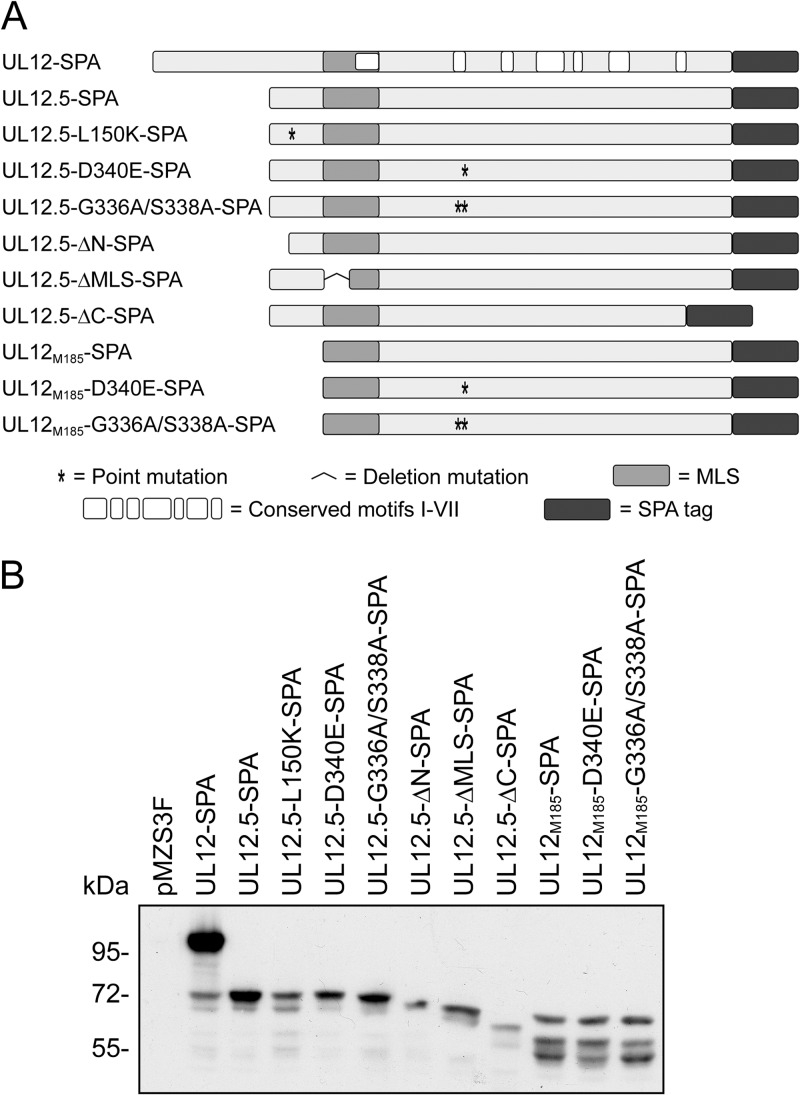
We previously demonstrated that HSV-1 UL12.5 is responsible for the rapid loss of mtDNA following infection (8). To better understand the relationship between UL12.5 nuclease activity and UL12.5-mediated mtDNA depletion, we constructed a series of plasmids containing known nuclease-inactivating mutations (Fig. 1A) (23, 29). These included two well-characterized mutations in invariant residues of conserved motif II (29), which, based on the structures of the Kaposi's sarcoma-associated herpesvirus (KSHV) and EBV orthologs, lie in the active site of the enzyme (30, 31). Mutational analysis has demonstrated that the G336A/S338A double-substitution mutation eliminates detectable exo- and endonuclease activities of UL12 whereas the D340E substitution mutation disrupts only exonuclease activity (29). Both mutations abolish the ability of UL12 to complement the growth defect of a UL12-null mutant virus (29). Mutagenesis of EBV, HCMV, and KSHV UL12 orthologs has also demonstrated that residues analogous to UL12 G336, S338, and D340 are critical for nuclease activity (30, 32–34). We included three additional mutations that have also been shown to eliminate detectable nuclease activity in vitro and abolish complementing activity in vivo; ΔN, L150K, and ΔC (23). A partial deletion of the UL12.5 MLS (ΔMLS) adjacent to conserved motif I was also included to assess the importance of mitochondrial localization during mtDNA depletion.
Fig 1.
Carboxy-terminally tagged UL12- and UL12.5-derived expression constructs. (A) Schematic of sequential peptide affinity (SPA)-tagged proteins used in this study. The locations of point and deletion mutations, the MLS, conserved alkaline nuclease motifs I to VII (29), and the carboxy-terminal SPA tag are indicated. (B) Expression of SPA-tagged constructs in transfected HeLa cells. Cells were transfected with the indicated plasmids and harvested 24 hpt. Lysates were assessed for protein expression by immunoblotting with the mouse anti-FLAG M2 antibody and chemiluminescence detection.
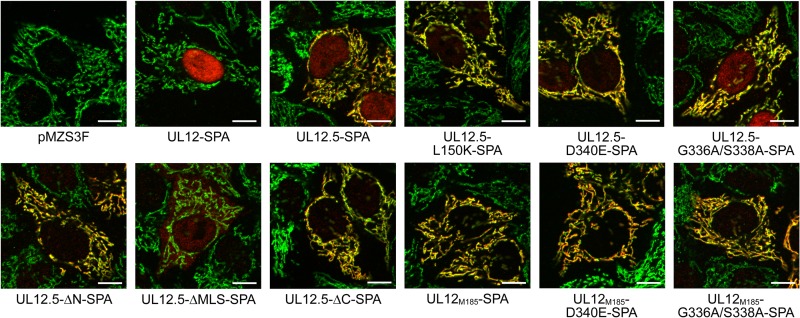
The mutant proteins were expressed in transfected HeLa cells, and their steady-state levels were observed via Western blotting (Fig. 1B). The largest protein product of each expression vector migrated at the predicted mobility. The UL12-SPA expression plasmid also expressed a low level of UL12.5-SPA from the native UL12.5 promoter contained within the UL12 open reading frame (Fig. 1B) (35). To determine whether the nuclease-inactivating mutations had an effect on mitochondrial targeting, we compared the localization of SPA-tagged proteins (shown in red) with that of the mitochondrial protein cytochrome c (shown in green) using immunofluorescence microscopy. UL12-SPA demonstrated strong nuclear localization in most cells (Fig. 2); however, a minority (ca. 20%) of cells also displayed a clear mitochondrial signal (data not shown), presumably due to the UL12.5 that is coexpressed from this plasmid using the native UL12.5 promoter. In contrast, UL12.5-SPA was predominantly mitochondrial in all cells, displaying a weaker nuclear signal than UL12 (Fig. 2) consistent with our previous observations (8, 10). All but one of the UL12.5 mutants displayed nuclear/mitochondrial localization similar to that seen with the wild-type UL12.5-SPA protein (Fig. 2). The exception was UL12.5-ΔMLS-SPA, which lacks a portion (R188 to R212) of the MLS and was excluded from mitochondria. This observation supports our previous data highlighting the importance of N-proximal residues M185 to R245 in targeting UL12.5 to mitochondria (10). UL12M185-SPA and its mutant derivatives localized exclusively to mitochondria, as did UL12.5-ΔN-SPA (Fig. 2). The failure of these N-terminally truncated mutants of UL12.5 to target the nucleus suggests that residues in the N terminus of UL12.5 are needed for the partial nuclear localization of wild-type UL12.5, consistent with our previous data (10).
Fig 2.
Mutations that disrupt nuclease activity do not affect mitochondrial localization. Transfected HeLa cells were costained with rabbit anti-FLAG (red) and mouse anti-cytochrome c (green) and visualized using fluorescence microscopy. Colocalizations of SPA-tagged proteins (containing three FLAG epitopes) with the mitochondrial protein cytochrome c are indicated in yellow. For clarity, the DAPI channel has been omitted. Scale bars, 10 μm.
Some nuclease-inactivating mutations do not prevent mtDNA depletion.
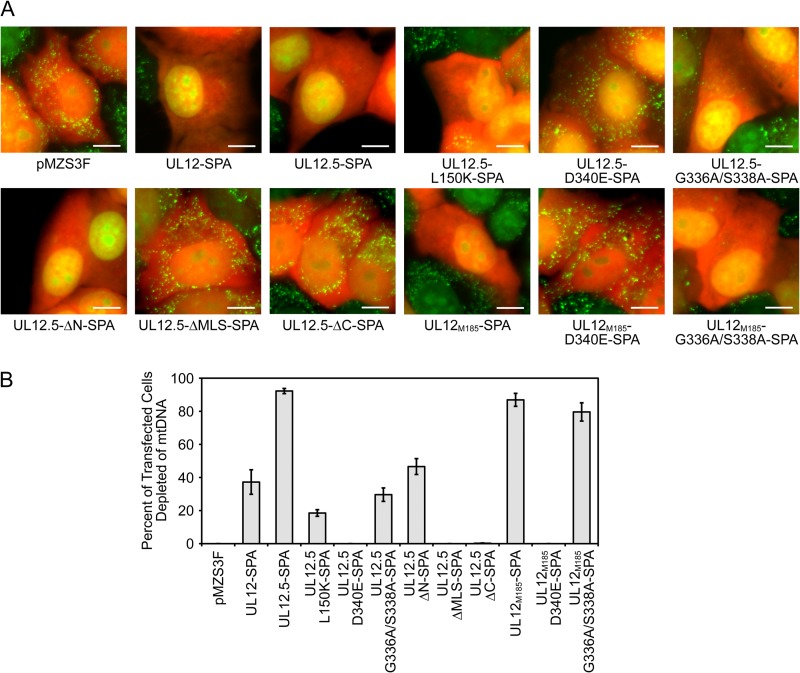
We next determined if the nuclease-inactivating mutations eliminated mtDNA depletion using a live-cell-imaging assay (10). HeLa cells were cotransfected with plasmids expressing monomeric orange fluorescent protein (mOrange, to identify transfected cells) and UL12-SPA, UL12.5-SPA, or UL12.5-SPA mutants and stained with PicoGreen to determine the presence or absence of mtDNA. Consistent with our previous observations (10), UL12.5-SPA and UL12M185-SPA depleted mtDNA from the majority of transfected cells (Fig. 3). The UL12-SPA expression plasmid also caused mtDNA depletion in a minority of cells, likely due to expression of UL12.5-SPA from the UL12.5 promoter in a subset of transfected cells (see above). Surprisingly, although two of the nuclease-inactivating mutations (D340E and ΔC) abrogated mtDNA depletion, the L150K, G336A/S338A, and ΔN mutants retained significant activity (Fig. 3).
Fig 3.
Some UL12.5 nuclease-deficient mutants retain the ability to cause mtDNA depletion. (A) HeLa cells were cotransfected with empty vector (pMZS3F), UL12-SPA, UL12.5-SPA, or UL12.5-SPA mutants and a plasmid expressing mOrange to identify transfected cells. PicoGreen staining was used to identify mtDNA foci in live cells using fluorescence microscopy at 48 hpt. Scale bars, 10 μm. (B) The extent of mtDNA depletion was determined by scoring for the presence (not depleted) or absence (depleted) of cytoplasmic PicoGreen staining in >100 randomly selected mOrange-positive cells. Data from three separate experiments were averaged, and standard errors are indicated.
Purified UL12 bearing the G336A/S338A double substitution has no detectable nuclease activity (29). It is therefore striking that mutating these residues did not prevent mtDNA depletion by UL12.5 or UL12M185 (Fig. 3): UL12.5-G336A/S338A-SPA caused mtDNA depletion in approximately one-third as many cells as UL12.5-SPA, while UL12M185-G336A/S338A-SPA was as active as UL12M185-SPA. The enhanced ability of UL12M185-G336A/S338A-SPA to cause mtDNA depletion relative to UL12.5-G336A/S338A-SPA may be due to its more efficient mitochondrial localization. Overall, these data indicate that neither the exonuclease nor endonuclease activities of UL12.5 are required for mtDNA depletion in transfected cells.
The observation that the G336A/S338A mutations did not prevent mtDNA depletion seemed to conflict with our previous finding that a UL12.5-G336A/S338A-mOrange mutant was inactive in this live-cell-imaging assay (10). We therefore reassessed the activity of this construct and found that it caused mtDNA depletion in a small percentage of transfected cells in some but not all experiments (data not shown). The reduced activity of this construct may stem from the use of the larger mOrange tag (236 residues versus 71 residues for the SPA tag used in this study).
Cellular endonuclease activity associates with some nuclease-deficient UL12.5 mutants.
The rapid depletion of mtDNA following infection (8) argues against a defect in mtDNA replication being responsible for mtDNA loss and instead suggests that mtDNA is actively degraded by one or more nucleases. The ability of several nuclease-deficient mutants of UL12.5 to deplete mtDNA implied that cellular nucleases are involved. Indeed, UL12 and UL12 orthologs are known to harness cellular nucleases to assist in their functions. HSV-1 UL12 directly interacts with components of the MRE11-RAD50-NBS1 (MRN) complex, including the exo/endonuclease Mre11, which may facilitate viral DNA recombination during infection (36). Additionally, KSHV SOX facilitates mRNA turnover in part through recruitment of the host 5′→3′ exoribonuclease 1 (XRN1) (37). Therefore, we examined whether cellular nucleases contribute to mtDNA loss following UL12.5 expression.
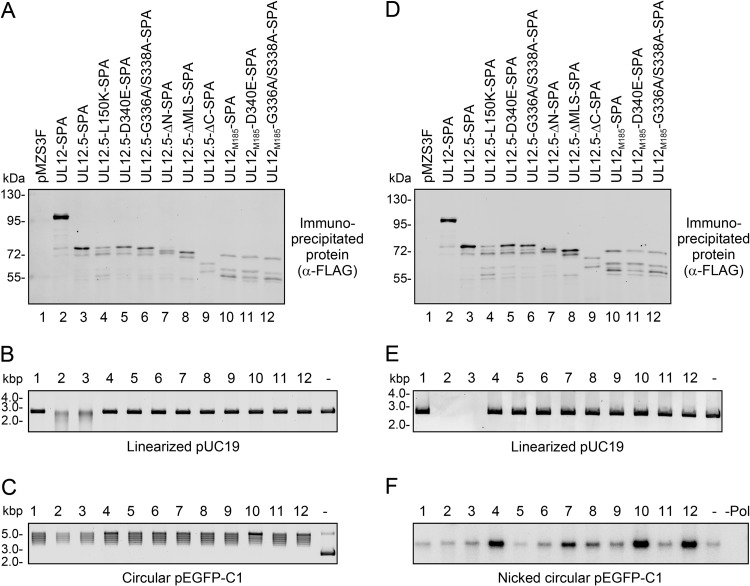
As one test of this hypothesis, we expressed SPA-tagged UL12, UL12.5, and UL12.5 mutants in HeLa cells and isolated the protein complexes containing these proteins by immunoprecipitation (Fig. 4A and D). The resulting immunoprecipitates were tested for their ability to degrade linearized plasmid DNA or to nick circular plasmid DNA in vitro. These nuclease assays were performed under alkaline pH conditions that were conducive to the detection of UL12/UL12.5 nuclease activity.
Fig 4.
Some nuclease-deficient mutants associate with cellular endonuclease activity in transfected cells. (A and D) SPA-tagged proteins were immunoprecipitated from transfected HeLa cells and visualized by immunoblotting with the rabbit anti-FLAG antibody followed by infrared detection. (B and C) Immunoprecipitates prepared as described for panel A were incubated with 50 ng linearized pUC19 (B) or 50 ng pEGFP-C1 (C) to visualize nuclease activity. Samples are labeled as indicated in panel A. Plasmid DNA incubated in the absence of immunoprecipitate (DNA only) is indicated as “-.” DNA was visualized using SYBR gold staining. (E) Immunoprecipitates from the experiment whose results are shown in panel D were incubated with 20 ng linearized pUC19 to visualize nuclease activity. Samples are labeled as indicated in panel D. Linearized pUC19 DNA incubated in the absence of immunoprecipitate (DNA only) is indicated as “-.” DNA was visualized using SYBR gold staining. (F) Immunoprecipitates from the experiment whose results are shown in panel D were also incubated with 50 ng circular pEGFP-C1, and the resulting nicked plasmid DNAs were radiolabeled using nick translation, separated by agarose gel electrophoresis, and visualized using a phosphorimager. Samples are labeled as indicated in panel D. pEGFP-C1 DNA incubated in the absence of immunoprecipitate (DNA only) is indicated as “-.” Circular pEGFP-C1 DNA incubated in the absence of immunoprecipitate and processed for nick translation without E. coli DNA polymerase I is indicated as “-Pol.”
As expected, immunoprecipitates containing UL12-SPA and UL12.5-SPA were capable of degrading linearized plasmid DNA, consistent with their known nuclease activity (Fig. 4B and E), while immunoprecipitates from cells transfected with empty pMZS3F vector displayed no activity. The degradation of the linear DNA substrate could be due to exo- and/or endonuclease activity; however, it is plausible that the much stronger exonuclease activity (16) is the major contributor. In contrast, the immunoprecipitates containing UL12M185 or the substitution/truncation mutants did not detectably degrade the linearized DNA substrate (Fig. 4B and E). These data indicated that none of these mutant proteins possessed, or were associated with, detectable nuclease activity against linear DNA substrates as measured in this assay. However, note that this assay would not detect low levels of endonuclease activity, as a large number of nicks would be required to alter the mobility of the linear substrate in nondenaturing gels.
We also specifically tested the same immunoprecipitates for endonuclease activity using a covalently closed circular plasmid DNA substrate. The outcome was visualized by monitoring the overall loss of DNA and the generation of nicked circles using SYBR gold staining (Fig. 4C) or nick translation (Fig. 4F). With all immunoprecipitates, the circular plasmid DNA migrated as a cluster of closely spaced bands (Fig. 4C). These bands collapsed into a single linear species following digestion with a unique cutting restriction enzyme (data not shown), suggesting the presence of contaminating topoisomerase activity. Comparable topoisomerase activity was detected after incubating HeLa cell lysate with protein G agarose beads in the absence of antibody (data not shown). As expected, immunoprecipitates containing UL12-SPA and UL12.5-SPA caused an appreciable decrease in the total amount of DNA as revealed by SYBR gold staining (Fig. 4C), likely due to endonucleolytic nicking followed by exonuclease activity. However, when these immunoprecipitates were scored for endonuclease activity using nick translation, there was no increase compared to the levels seen with the controls (pMZS3F or DNA only; Fig. 4F), suggesting that the nicked DNA is rapidly degraded by the powerful exonuclease activity of UL12, as previously observed (16). Interestingly, immunoprecipitates containing UL12.5-L150K-SPA, UL12.5-ΔN-SPA, UL12M185-SPA, and UL12M185-G336A/S338A-SPA yielded consistently higher levels of nick translation indicative of the presence of endonuclease activity (Fig. 4F). Some of these immunoprecipitates also generated higher levels of relaxed circles when assessed by the less sensitive SYBR gold detection method (UL12.5-L150K-SPA, UL12M185-SPA, and, to a lesser extent, UL12M185-G336A/S338A-SPA; Fig. 4C). A low level of endonuclease activity was observed in immunoprecipitates containing the UL12.5-ΔMLS-SPA protein, and no appreciable endonuclease activity was observed by nick translation for immunoprecipitates containing UL12.5-D340E-SPA, UL12.5-G336A/S338A-SPA, UL12.5-ΔC-SPA, or UL12M185-D340E-SPA (Fig. 4F).
It might be argued that the enhanced nicking activity detected in the immunoprecipitates of some mutant proteins is due to residual UL12.5 endonuclease activity. However, the mutational sensitivity profile of the associated endonuclease activity is incompatible with this suggestion: UL12M185-G336A/S338A-SPA immunoprecipitates were among the most active in our assay, and yet purified UL12 protein bearing the G336A/S338A double substitution lacks detectable endo- and exonuclease activity (29); in contrast, UL12M185-D340E-SPA displayed only background levels of nicking in our assay, and yet the D340E substitution does not greatly impair the endonuclease activity of purified UL12 (29). These considerations indicate that our immunoprecipitation assay did not detect the weak endonuclease activity of UL12.5 and strongly argue that the nicking activity that we observed was due to the presence of one or more associated cellular endonucleases.
Interestingly, the nuclease-deficient UL12.5 mutants that associated with presumably cellular endonuclease activity tended to be those capable of causing mtDNA depletion (UL12.5-L150K-SPA, UL12.5-ΔN-SPA, UL12M185-SPA, and UL12M185-G336A/S338A-SPA; Fig. 3). The exceptions to this correlation were as follows: (i) UL12.5-G336A/S338A-SPA, which, although capable of causing mtDNA depletion, did not associate with appreciable endonuclease activity, and (ii) UL12.5-ΔMLS-SPA, which associated with some endonuclease activity but was unable to cause mtDNA loss (Fig. 3 and 4F). The observation that the UL12M185 double-substitution mutant exhibits stronger mitochondrial localization (Fig. 2), causes more mtDNA depletion (Fig. 3), and associates with more endonuclease activity (Fig. 4F) than the UL12.5 double-substitution mutant is consistent with the possibility that the associated nuclease is mitochondrial.
The mitochondrial nucleases ENDOG and EXOG participate in mtDNA depletion mediated by UL12.5.
Mammalian mitochondria contain a small number of endonucleases, including ENDOG (38, 39), EXOG (27), apurinic-apyrimidinic endonucleases 1 and 2 (APEX1/2) (40–42), flap endonuclease 1 (FEN1) (43, 44), DNA replication ATP-dependent helicase/nuclease (DNA2) (45), and the recently described endo/exonuclease Ddk1 (46). Of these, APEX1/2, FEN1, and DNA2 seemed unlikely to be involved in mtDNA loss provoked by UL12.5 since they act on specialized DNA substrates, while Ddk1 had not been discovered at the onset of our research. We therefore examined ENDOG and EXOG as potential candidates. Unlike in Neurospora crassa and Saccharomyces cerevisiae, where a single ENDOG homolog possesses both endonuclease and exonuclease activity (47–49), the majority of mammalian mitochondrial endonuclease and exonuclease activities are thought to be due to the combined action of ENDOG and its highly related paralog EXOG (27). For this reason, we considered the possibility that ENDOG and EXOG may play redundant roles in UL12.5-SPA-mediated mtDNA depletion.
ENDOG is a non-sugar-specific, magnesium-dependent endonuclease which preferentially introduces single-stranded nicks adjacent to guanine residues (50, 51). The biochemical properties of ENDOG are remarkably similar to those of UL12.5 in that ENDOG functions in the presence of magnesium, alkaline pH, and low ionic strength (22, 51). Aside from its role in apoptosis, ENDOG is responsible for the majority of mammalian mitochondrial nuclease activity and has been observed to associate with mtDNA via chromatin immunoprecipitation (38, 39, 52). Interestingly, ENDOG has already been shown to participate in HSV-1 biology from its proposed role in HSV-1 a sequence recombination (53, 54). EXOG differs from ENDOG in that it possesses both endonuclease and weak 5′→3′ exonuclease activity and exhibits a preference for single-stranded DNA (ssDNA) (27).
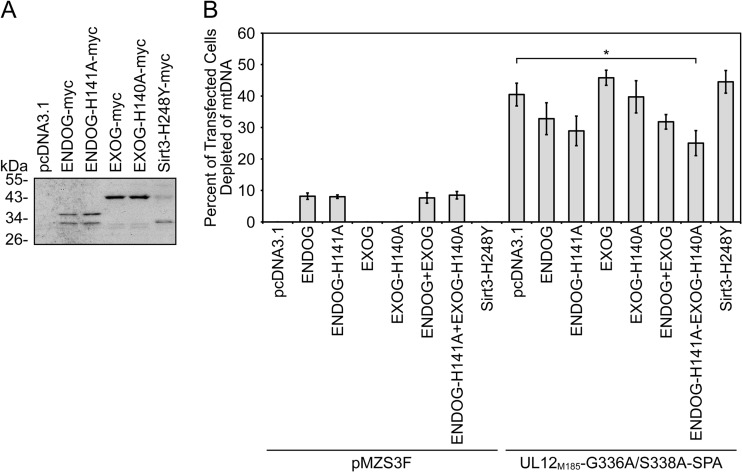
As one approach to assessing the potential roles of ENDOG and EXOG in UL12.5-mediated mtDNA depletion, we examined the effect of overexpressing mutant forms of ENDOG and EXOG along with the mtDNA depletion-competent and nuclease-deficient mutant UL12M185-G336A/S338A-SPA in our live-cell mtDNA depletion assay. We created plasmids which express C-terminally epitope-tagged wild-type and catalytically inactive forms (26, 27) of ENDOG and EXOG (Fig. 5A). The addition of a C-terminal epitope tag does not interfere with the enzymatic activity of ENDOG or EXOG (27, 51, 55), and we verified by coimmunoprecipitation that the catalytically inactive mutants (ENDOG-H141A and EXOG-H140A) retained the ability to form homomultimers with their wild-type counterparts (26, 27, 56) (data not shown). As a negative control, we used a plasmid expressing an epitope-tagged, catalytically inactive version of the mitochondrial protein sirtuin 3 (Sirt3-H248Y) (28). The expression and apparent molecular masses of all myc-tagged proteins were confirmed by Western blotting (Fig. 5A). The larger and smaller protein species observed following transfection of the ENDOG and Sirt3 plasmids likely correspond to previously described precursor and mature forms of the proteins, respectively (Fig. 5A) (57, 58). Wild-type and mutant EXOG-myc proteins also appeared to migrate as doublets during SDS-PAGE.
Fig 5.
Concurrent overexpression of nuclease-deficient ENDOG and EXOG inhibits mtDNA depletion by a mutant UL12.5-SPA protein. (A) Expression of myc-tagged constructs in transfected HeLa cells was visualized at 24 hpt. Lysates were assessed for protein expression by immunoblotting with the rabbit anti-c-myc antibody and chemiluminescence detection. (B) HeLa cells were cotransfected for 48 h with empty vector (pcDNA3.1) or plasmids expressing myc-tagged ENDOG, nuclease-deficient ENDOG (ENDOG-H141A), EXOG, nuclease-deficient EXOG (EXOG-H140A), or inactive sirtuin 3 (Sirt3-H248Y) along with an empty vector (pMZS3F) or a plasmid expressing UL12M185-G336A/S338A-SPA. MtDNA depletion was measured as described in the Fig. 3 legend. Data are from four separate experiments, except those data that include the Sirt3 mutant, which are from three separate experiments. All data were averaged, and standard errors are indicated. Data denoted with an asterisk (*) indicate statistical significance (P < 0.05) compared to the control data (UL12M185-G336A/S338A-SPA/pcDNA) as determined using a two-tailed t test with equal variances.
Expression of EXOG, EXOG-H140A, or Sirt3-H248Y alone had no observable effect on mtDNA levels in our live-cell mtDNA depletion assay. However, expression of ENDOG or ENDOG-H141A caused mtDNA depletion in approximately 8% of transfected cells (Fig. 5B). In the case of wild-type ENDOG, this effect was eliminated by cotransfecting ENDOG siRNA (data not shown). The observation that overexpression of wild-type or mutant ENDOG causes mtDNA depletion is novel and very intriguing. Although it is currently the subject of debate, several studies support the notion of a role of ENDOG in mtDNA maintenance (52, 59–62). These observations suggest that, although the nuclease activity of ENDOG is not responsible for mtDNA loss following overexpression, ENDOG is involved in regulating mtDNA metabolism.
Expression of wild-type or mutant ENDOG or EXOG had no significant effects on UL12M185-G336A/S338A-SPA-mediated mtDNA depletion (Fig. 5B). However, when cells simultaneously expressed ENDOG-H141A and EXOG-H140A, mtDNA depletion was reduced by 38% (P = 0.029; Fig. 5B). In contrast, overexpression of the unrelated mitochondrial protein Sirt3-H248Y had no effect on mtDNA depletion by UL12M185-G336A/S338A-SPA (Fig. 5B). Interestingly, overexpression of both mutant ENDOG and mutant EXOG was needed to significantly impair mtDNA depletion by UL12M185-G336A/S338A-SPA. Therefore, ENDOG and EXOG likely play at least partially redundant roles in the mtDNA degradation pathway that is stimulated following the expression of UL12M185-G336A/S338A-SPA. The inhibitory effect of the combination of ENDOG-H141A and EXOG-H140A appears to be dose dependent, as increasing the amount of ENDOG-H141A/EXOG-H140A plasmid DNA 2.5-fold reduced mtDNA depletion by ca. 68% (P = 0.001) in a separate series of experiments (data not shown).
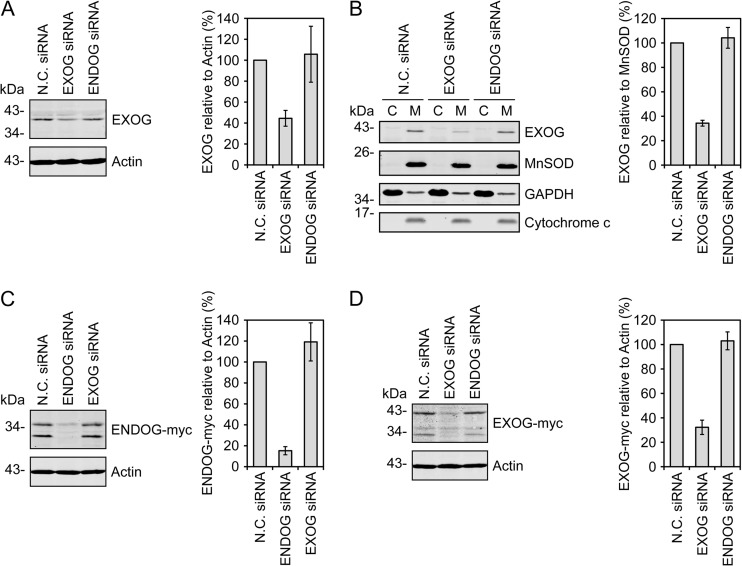
To complement the overexpression experiments described above, we employed RNA interference to reduce ENDOG and/or EXOG levels in HeLa cells prior to mtDNA depletion. The siRNA targeting EXOG reduced total protein expression by 56% (Fig. 6A) and reduced EXOG levels in mitochondria by 66% (Fig. 6B). We have not been able to identify an antibody that reliably detects endogenous ENDOG and therefore tested our ENDOG siRNA for its ability to prevent de novo expression of exogenous tagged ENDOG. Using siRNA/plasmid DNA cotransfections, we observed that the ENDOG siRNA reduced ENDOG-myc expression by 85% relative to treatment with a negative-control siRNA, indicating that the siRNA targets the ENDOG transcript (Fig. 6C). Similar suppression of EXOG-myc expression by the EXOG siRNA was also observed (68% reduction; Fig. 6D). Importantly, the EXOG and ENDOG siRNAs specifically target their respective transcripts (Fig. 6C and D). It therefore seems likely that the ENDOG siRNA depletes endogenous ENDOG in a fashion similar to the effect of EXOG siRNA on endogenous EXOG.
Fig 6.
Knockdown of ENDOG and EXOG using siRNA. (A) Endogenous EXOG knockdown was measured in HeLa cells transfected with the indicated siRNAs. (B) Lysates from HeLa cells transfected with the indicated siRNAs were separated into cytoplasmic (C) and mitochondrial (M) fractions to visualize knockdown of EXOG in the mitochondrial fraction. (C and D) Alternatively, HeLa cells were cotransfected with the indicated siRNAs and ENDOG-myc (C) or EXOG-myc (D) for 48 h to verify siRNA function. EXOG, ENDOG-myc, and EXOG-myc levels were quantitated following immunoblotting and infrared detection. Data from three separate experiments were averaged, plotted relative to actin or MnSOD, and normalized to the N.C. siRNA treatment. Standard errors are indicated.
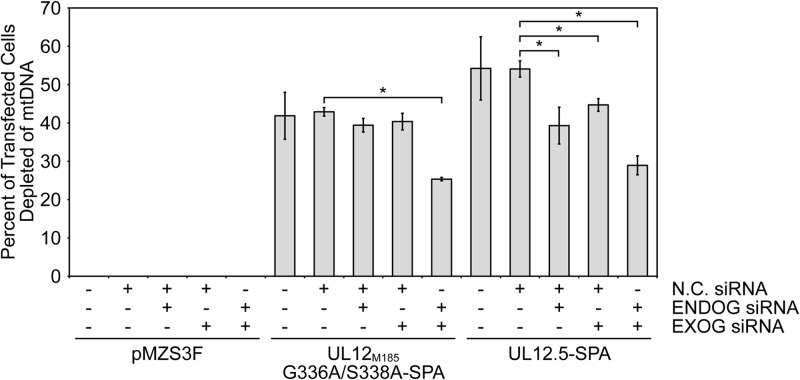
For the knockdown/mtDNA depletion experiments, we pretreated HeLa cells with negative control (N.C.) siRNA, ENDOG siRNA, EXOG siRNA, or a mixture of ENDOG and EXOG siRNAs for 48 h. Cells were then trypsinized and reverse cotransfected with additional siRNA and the indicated effector plasmids, and mtDNA depletion was evaluated 24 h later using the PicoGreen live-cell-imaging assay. When tested individually, the ENDOG and EXOG siRNAs did not significantly affect mtDNA depletion by UL12M185-G336A/S338A-SPA; however, concurrent knockdown of ENDOG and EXOG led to a 41% reduction of mtDNA depletion (P = 0.0001; Fig. 7). In the case of nuclease-competent UL12.5-SPA, mtDNA depletion was significantly inhibited by both ENDOG siRNA (27% reduction, P = 0.048) and EXOG siRNA (17% reduction, P = 0.025), an effect that was enhanced by simultaneous knockdown of both ENDOG and EXOG (47% reduction, P = 0.0015; Fig. 7). These data further implicate ENDOG and EXOG as playing key and at least partially overlapping roles in mediating mtDNA loss triggered by UL12.5. Note that the inhibition of mtDNA depletion following ENDOG and EXOG knockdown is not complete, likely due at least in part to incomplete loss of the target proteins. Additional studies using cells derived from ENDOG/EXOG double-knockout mice could address this possibility, if such mice prove to be viable.
Fig 7.
Knockdown of ENDOG and/or EXOG inhibits mtDNA depletion by UL12.5-SPA. HeLa cells cotransfected with the indicated siRNAs and either empty vector (pMZS3F) or plasmids expressing UL12.5-SPA or UL12M185-G336A/S338A-SPA were assessed for mtDNA depletion as described for Fig. 3. Data from three separate experiments were averaged, and standard errors are indicated. Data denoted with an asterisk (*) indicate statistical significance (P < 0.05) compared to the respective control data (UL12M185-G336A/S338A-SPA/N.C. siRNA or UL12.5-SPA/N.C. siRNA) as determined using a two-tailed t test with equal variances.
Possible mechanisms and outstanding questions.
We have been unable to demonstrate a specific association between UL12.5-SPA and ENDOG or EXOG by coimmunoprecipitation (data not shown). Therefore, we cannot say with certainty whether the endonuclease activity observed in our IP nuclease assays is due to ENDOG or EXOG (Fig. 4C and F). It is possible that UL12.5 associates with these endogenous nucleases transiently or weakly, thus precluding their detection using Western blotting. Alternatively, UL12.5 may indirectly enhance the nuclease activity of ENDOG and EXOG against mtDNA. Recent studies on ENDOG and EXOG have proposed that these enzymes have an important role in mtDNA maintenance and repair (52, 61–63). In one particularly elegant study, ENDOG was demonstrated to be the enzyme responsible for depleting Drosophila melanogaster mtDNA from developing sperm, which in turn promotes the maternal inheritance of mtDNA (61). This work provides a clear example of the influence of ENDOG on mtDNA maintenance. Mitochondria also contain DNA repair pathways similar to those found in the nucleus which defend against genotoxic insults and oxidative damage (reviewed in reference 64). UL12 interacts with components of the host nuclear DNA repair MRN complex (36); therefore, it would be interesting to investigate whether UL12.5 directly interacts with one or more mtDNA repair proteins other than ENDOG and EXOG.
HSV infection also leads to rapid loss of mitochondrial mRNAs (mt-mRNAs) (8, 65), a process that is at least as rapid as mtDNA loss and depends on UL12 gene products (8). Although UL12 orthologs EBV BGLF5 and KSHV SOX have been shown to mediate cytoplasmic mRNA turnover (34, 66), HSV-1 UL12 does not appear to possess this activity (34). Note that ENDOG and EXOG both possess low levels of RNase activity (26, 27). Indeed, earlier work on ENDOG had suggested that its RNase activity generates the RNA primers for mtDNA replication (59). Thus, it is possible that ENDOG and or EXOG directly degrade mt-mRNAs during HSV infection. Further studies are required to test this possibility.
At this time, it is still unknown what roles mtDNA depletion serves during HSV-1 replication or pathogenesis. Unfortunately, this issue cannot be cleanly addressed using the mutant forms of UL12.5 that we have studied here because these mutations also inactivate the nuclease activity of full-length UL12 and hence severely reduce viral yields (23, 29). We are therefore currently exploring strategies for generating HSV-1 isolates that retain the nuclear functions of UL12 and yet fail to deplete mtDNA.
ACKNOWLEDGMENTS
We thank Holly Saffran for technical assistance and Lori Frappier for the gift of pMZS3F plasmid DNA.
This research was funded by an operating grant from the Canadian Institutes for Health Research to J.R.S. (funding reference number: MOP 37995). B.A.D. was supported by a full-time studentship from Alberta Innovates-Health Solutions and a QEII Graduate Scholarship from the government of the Province of Alberta. J.R.S. holds a Canada Research Chair in Molecular Virology (Tier I).
Footnotes
Published ahead of print 28 August 2013
REFERENCES
- 1.Ohta A, Nishiyama Y. 2011. Mitochondria and viruses. Mitochondrion 11:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. 2012. Mitochondrial control of cellular life, stress, and death. Circ. Res. 111:1198–1207 [DOI] [PubMed] [Google Scholar]
- 3.Bogenhagen D, Clayton DA. 1974. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J. Biol. Chem. 249:7991–7995 [PubMed] [Google Scholar]
- 4.Shmookler Reis RJ, Goldstein S. 1983. Mitochondrial DNA in mortal and immortal human cells. Genome number, integrity, and methylation. J. Biol. Chem. 258:9078–9085 [PubMed] [Google Scholar]
- 5.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. 1981. Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- 6.Copeland WC. 2008. Inherited mitochondrial diseases of DNA replication. Annu. Rev. Med. 59:131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Copeland WC. 2012. Defects in mitochondrial DNA replication and human disease. Crit. Rev. Biochem. Mol. Biol. 47:64–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saffran HA, Pare JM, Corcoran JA, Weller SK, Smiley JR. 2007. Herpes simplex virus eliminates host mitochondrial DNA. EMBO Rep. 8:188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiedmer A, Wang P, Zhou J, Rennekamp AJ, Tiranti V, Zeviani M, Lieberman PM. 2008. Epstein-Barr virus immediate-early protein Zta co-opts mitochondrial single-stranded DNA binding protein to promote viral and inhibit mitochondrial DNA replication. J. Virol. 82:4647–4655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corcoran JA, Saffran HA, Duguay BA, Smiley JR. 2009. Herpes simplex virus UL12.5 targets mitochondria through a mitochondrial localization sequence proximal to the N terminus. J. Virol. 83:2601–2610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa RH, Draper KG, Banks L, Powell KL, Cohen G, Eisenberg R, Wagner EK. 1983. High-resolution characterization of herpes simplex virus type 1 transcripts encoding alkaline exonuclease and a 50,000-dalton protein tentatively identified as a capsid protein. J. Virol. 48:591–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez R, Shao L, Bronstein JC, Weber PC, Weller SK. 1996. The product of a 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology 215:152–164 [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann PJ, Cheng YC. 1978. The deoxyribonuclease induced after infection of KB cells by herpes simplex virus type 1 or type 2. I. Purification and characterization of the enzyme. J. Biol. Chem. 253:3557–3562 [PubMed] [Google Scholar]
- 14.Hoffmann PJ, Cheng YC. 1979. DNase induced after infection of KB cells by herpes simplex virus type 1 or type 2. II. Characterization of an associated endonuclease activity. J. Virol. 32:449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banks L, Purifoy DJ, Hurst PF, Killington RA, Powell KL. 1983. Herpes simplex virus non-structural proteins. IV. Purification of the virus-induced deoxyribonuclease and characterization of the enzyme using monoclonal antibodies. J. Gen. Virol. 64(Pt 10):2249–2260 [DOI] [PubMed] [Google Scholar]
- 16.Bronstein JC, Weber PC. 1996. Purification and characterization of herpes simplex virus type 1 alkaline exonuclease expressed in Escherichia coli. J. Virol. 70:2008–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reuven NB, Antoku S, Weller SK. 2004. The UL12.5 gene product of herpes simplex virus type 1 exhibits nuclease and strand exchange activities but does not localize to the nucleus. J. Virol. 78:4599–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez R, Sarisky RT, Weber PC, Weller SK. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao L, Rapp LM, Weller SK. 1993. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196:146–162 [DOI] [PubMed] [Google Scholar]
- 20.Weller SK, Seghatoleslami MR, Shao L, Rowse D, Carmichael EP. 1990. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J. Gen. Virol. 71(Pt 12):2941–2952 [DOI] [PubMed] [Google Scholar]
- 21.Feederle R, Bannert H, Lips H, Muller-Lantzsch N, Delecluse HJ. 2009. The Epstein-Barr virus alkaline exonuclease BGLF5 serves pleiotropic functions in virus replication. J. Virol. 83:4952–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bronstein JC, Weller SK, Weber PC. 1997. The product of the UL12.5 gene of herpes simplex virus type 1 is a capsid-associated nuclease. J. Virol. 71:3039–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson JO, Ball-Goodrich LJ, Parris DS. 1998. Structure-function analysis of the herpes simplex virus type 1 UL12 gene: correlation of deoxyribonuclease activity in vitro with replication function. Virology 243:247–259 [DOI] [PubMed] [Google Scholar]
- 24.Zeghouf M, Li J, Butland G, Borkowska A, Canadien V, Richards D, Beattie B, Emili A, Greenblatt JF. 2004. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3:463–468 [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Malcolm BA. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. Biotechniques 26:680–682 [DOI] [PubMed] [Google Scholar]
- 26.Schäfer P, Scholz SR, Gimadutdinow O, Cymerman IA, Bujnicki JM, Ruiz-Carrillo A, Pingoud A, Meiss G. 2004. Structural and functional characterization of mitochondrial EndoG, a sugar non-specific nuclease which plays an important role during apoptosis. J. Mol. Biol. 338:217–228 [DOI] [PubMed] [Google Scholar]
- 27.Cymerman IA, Chung I, Beckmann BM, Bujnicki JM, Meiss G. 2008. EXOG, a novel paralog of endonuclease G in higher eukaryotes. Nucleic Acids Res. 36:1369–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. 2008. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. U. S. A. 105:14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldstein JN, Weller SK. 1998. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology 244:442–457 [DOI] [PubMed] [Google Scholar]
- 30.Buisson M, Geoui T, Flot D, Tarbouriech N, Ressing ME, Wiertz EJ, Burmeister WP. 2009. A bridge crosses the active-site canyon of the Epstein-Barr virus nuclease with DNase and RNase activities. J. Mol. Biol. 391:717–728 [DOI] [PubMed] [Google Scholar]
- 31.Dahlroth SL, Gurmu D, Schmitzberger F, Engman H, Haas J, Erlandsen H, Nordlund P. 2009. Crystal structure of the shutoff and exonuclease protein from the oncogenic Kaposi's sarcoma-associated herpesvirus. FEBS J. 276:6636–6645 [DOI] [PubMed] [Google Scholar]
- 32.Kuchta AL, Parikh H, Zhu Y, Kellogg GE, Parris DS, McVoy MA. 2012. Structural modelling and mutagenesis of human cytomegalovirus alkaline nuclease UL98. J. Gen. Virol. 93:130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horst D, Burmeister WP, Boer IG, van Leeuwen D, Buisson M, Gorbalenya AE, Wiertz EJ, Ressing ME. 2012. The “bridge” in the Epstein-Barr virus alkaline exonuclease protein BGLF5 contributes to shutoff activity during productive infection. J. Virol. 86:9175–9187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glaunsinger B, Ganem D. 2004. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol. Cell 13:713–723 [DOI] [PubMed] [Google Scholar]
- 35.Draper KG, Devi-Rao G, Costa RH, Blair ED, Thompson RL, Wagner EK. 1986. Characterization of the genes encoding herpes simplex virus type 1 and type 2 alkaline exonucleases and overlapping proteins. J. Virol. 57:1023–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balasubramanian N, Bai P, Buchek G, Korza G, Weller SK. 2010. Physical interaction between the herpes simplex virus type 1 exonuclease, UL12, and the DNA double-strand break-sensing MRN complex. J. Virol. 84:12504–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Covarrubias S, Gaglia MM, Kumar GR, Wong W, Jackson AO, Glaunsinger BA. 2011. Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1. PLoS Pathog. 7:e1002339. 10.1371/journal.ppat.1002339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cummings OW, King TC, Holden JA, Low RL. 1987. Purification and characterization of the potent endonuclease in extracts of bovine heart mitochondria. J. Biol. Chem. 262:2005–2015 [PubMed] [Google Scholar]
- 39.Gerschenson M, Houmiel KL, Low RL. 1995. Endonuclease G from mammalian nuclei is identical to the major endonuclease of mitochondria. Nucleic Acids Res. 23:88–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chattopadhyay R, Wiederhold L, Szczesny B, Boldogh I, Hazra TK, Izumi T, Mitra S. 2006. Identification and characterization of mitochondrial abasic (AP)-endonuclease in mammalian cells. Nucleic Acids Res. 34:2067–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tell G, Crivellato E, Pines A, Paron I, Pucillo C, Manzini G, Bandiera A, Kelley MR, Di Loreto C, Damante G. 2001. Mitochondrial localization of APE/Ref-1 in thyroid cells. Mutat. Res. 485:143–152 [DOI] [PubMed] [Google Scholar]
- 42.Tsuchimoto D, Sakai Y, Sakumi K, Nishioka K, Sasaki M, Fujiwara T, Nakabeppu Y. 2001. Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 29:2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, III, Shen B, Demple B. 2008. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol. Cell. Biol. 28:4975–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S. 2008. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 283:26349–26356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, Shen B. 2008. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol. Cell 32:325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szczesny RJ, Hejnowicz MS, Steczkiewicz K, Muszewska A, Borowski LS, Ginalski K, Dziembowski A. 2013. Identification of a novel human mitochondrial endo-/exonuclease Ddk1/c20orf72 necessary for maintenance of proper 7S DNA levels. Nucleic Acids Res. 41:3144–3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dake E, Hofmann TJ, McIntire S, Hudson A, Zassenhaus HP. 1988. Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 263:7691–7702 [PubMed] [Google Scholar]
- 48.Zassenhaus HP, Hofmann TJ, Uthayashanker R, Vincent RD, Zona M. 1988. Construction of a yeast mutant lacking the mitochondrial nuclease. Nucleic Acids Res. 16:3283–3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chow TY, Fraser MJ. 1983. Purification and properties of single strand DNA-binding endo-exonuclease of Neurospora crassa. J. Biol. Chem. 258:12010–12018 [PubMed] [Google Scholar]
- 50.Ruiz-Carrillo A, Renaud J. 1987. Endonuclease G: a (dG)n X (dC)n-specific DNase from higher eukaryotes. EMBO J. 6:401–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Widlak P, Li LY, Wang X, Garrard WT. 2001. Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates: cooperation with exonuclease and DNase I. J. Biol. Chem. 276:48404–48409 [DOI] [PubMed] [Google Scholar]
- 52.McDermott-Roe C, Ye J, Ahmed R, Sun XM, Serafin A, Ware J, Bottolo L, Muckett P, Canas X, Zhang J, Rowe GC, Buchan R, Lu H, Braithwaite A, Mancini M, Hauton D, Marti R, Garcia-Arumi E, Hubner N, Jacob H, Serikawa T, Zidek V, Papousek F, Kolar F, Cardona M, Ruiz-Meana M, Garcia-Dorado D, Comella JX, Felkin LE, Barton PJ, Arany Z, Pravenec M, Petretto E, Sanchis D, Cook SA. 2011. Endonuclease G is a novel determinant of cardiac hypertrophy and mitochondrial function. Nature 478:114–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang KJ, Ku CC, Lehman IR. 2006. Endonuclease G: a role for the enzyme in recombination and cellular proliferation. Proc. Natl. Acad. Sci. U. S. A. 103:8995–9000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang KJ, Zemelman BV, Lehman IR. 2002. Endonuclease G, a candidate human enzyme for the initiation of genomic inversion in herpes simplex type 1 virus. J. Biol. Chem. 277:21071–21079 [DOI] [PubMed] [Google Scholar]
- 55.Parrish J, Li L, Klotz K, Ledwich D, Wang X, Xue D. 2001. Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature 412:90–94 [DOI] [PubMed] [Google Scholar]
- 56.Cote J, Renaud J, Ruiz-Carrillo A. 1989. Recognition of (dG)n.(dC)n sequences by endonuclease G. Characterization of the calf thymus nuclease. J. Biol. Chem. 264:3301–3310 [PubMed] [Google Scholar]
- 57.Li LY, Luo X, Wang X. 2001. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412:95–99 [DOI] [PubMed] [Google Scholar]
- 58.Schwer B, North BJ, Frye RA, Ott M, Verdin E. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158:647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cote J, Ruiz-Carrillo A. 1993. Primers for mitochondrial DNA replication generated by endonuclease G. Science 261:765–769 [DOI] [PubMed] [Google Scholar]
- 60.Tiranti V, Rossi E, Ruiz-Carrillo A, Rossi G, Rocchi M, DiDonato S, Zuffardi O, Zeviani M. 1995. Chromosomal localization of mitochondrial transcription factor A (TCF6), single-stranded DNA-binding protein (SSBP), and endonuclease G (ENDOG), three human housekeeping genes involved in mitochondrial biogenesis. Genomics 25:559–564 [DOI] [PubMed] [Google Scholar]
- 61.DeLuca SZ, O'Farrell PH. 2012. Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell 22:660–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dzierzbicki P, Kaniak-Golik A, Malc E, Mieczkowski P, Ciesla Z. 2012. The generation of oxidative stress-induced rearrangements in Saccharomyces cerevisiae mtDNA is dependent on the Nuc1 (EndoG/ExoG) nuclease and is enhanced by inactivation of the MRX complex. Mutat. Res. 740:21–33 [DOI] [PubMed] [Google Scholar]
- 63.Tann AW, Boldogh I, Meiss G, Qian W, Van Houten B, Mitra S, Szczesny B. 2011. Apoptosis induced by persistent single-strand breaks in mitochondrial genome: critical role of EXOG (5′-EXO/endonuclease) in their repair. J. Biol. Chem. 286:31975–31983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kazak L, Reyes A, Holt IJ. 2012. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat. Rev. Mol. Cell Biol. 13:659–671 [DOI] [PubMed] [Google Scholar]
- 65.Latchman DS. 1988. Effect of herpes simplex virus type 2 infection on mitochondrial gene expression. J. Gen. Virol. 69(Pt 6):1405–1410 [DOI] [PubMed] [Google Scholar]
- 66.Rowe M, Glaunsinger B, van Leeuwen D, Zuo J, Sweetman D, Ganem D, Middeldorp J, Wiertz EJ, Ressing ME. 2007. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc. Natl. Acad. Sci. U. S. A. 104:3366–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]