Abstract
Emergence of viruses into the human population by transmission from nonhuman primates (NHPs) represents a serious potential threat to human health that is primarily associated with the increased bushmeat trade. Transmission of RNA viruses across primate species appears to be relatively frequent. In contrast, DNA viruses appear to be largely host specific, suggesting low transmission potential. Herein, we use a primate predator-prey system to study the risk of herpesvirus transmission between different primate species in the wild. The system was comprised of western chimpanzees (Pan troglodytes verus) and their primary (western red colobus, Piliocolobus badius badius) and secondary (black-and-white colobus, Colobus polykomos) prey monkey species. NHP species were frequently observed to be coinfected with multiple beta- and gammaherpesviruses (including new cytomegalo- and rhadinoviruses). However, despite frequent exposure of chimpanzees to blood, organs, and bones of their herpesvirus-infected monkey prey, there was no evidence for cross-species herpesvirus transmission. These findings suggest that interspecies transmission of NHP beta- and gammaherpesviruses is, at most, a rare event in the wild.
INTRODUCTION
Zoonotic transmission of animal pathogens into the human population is regarded as the major source of new human infectious disease (1–3). Such zoonoses have profoundly altered the course of human history, as reflected by the impact of the bubonic plague, Spanish flu, and HIV/AIDS on human society (4–6). Zoonoses are frequently transmitted to humans following an initial cross-species transmission into an intermediate animal host. Mechanisms underlying cross-species transmission and adaptation to new host species are far from clear but appear to be influenced by multiple factors, including the level and mode of interaction between animal reservoir/transmission source and humans, the phylogenetic relationship of these species, and the nature of the zoonotic pathogen (2, 7, 8). Zoonotic/enzootic cross-species transmission appears to be a relatively common characteristic of RNA viruses (8). In contrast, the efficiency of cross-species transmission for DNA viruses is unclear. For the Herpesviridae family, transmission appears to be a relatively rare event. In the few instances where virus transmission has been observed, the lack of onward transmission and the uncharacteristically highly pathogenic presentation of overt disease in the new species (e.g., ovine/caprine herpesvirus infection in free-ranging cervids and herpesvirus B in humans) suggest that herpesviruses poorly adapt to their new host environment (9–11).
To date, most studies examining cross-species transmission of herpesviruses have been based on phylogenetic analysis of genomic sequences. These studies reveal well-defined genotypic groupings of alpha-, beta-, and gammaherpesviruses within the respective herpesvirus subfamilies Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae (12). This phylogenetic distribution has been interpreted as coevolution (codivergence) of the major herpesvirus lineages with those of the mammalian host, with the absence of frequent cross-species transmission. More recent, sensitive methods of analysis using degenerate PCRs targeting common conserved regions of the herpesvirus genome support codivergence as the prominent mode of evolution of this virus family (13–15). However, these studies also reveal the presence of repeated cross-species beta- and gammaherpesvirus transmission over evolutionary time. Epstein-Barr virus (EBV) and a group of closely related African hominid gammaherpesviruses (genus Lymphocryptovirus [LCV]) were shown to be derived from at least two independent introductions from Old World monkey (OWM) LCVs within the past 12 million years (14). Similarly, transmission of betaherpesviruses (cytomegalovirus [CMV]) was observed between chimpanzees and gorillas, but the frequency of transmission and whether transmission had occurred within recent or historic time (within the last million years) could not be determined (15).
In the present study, we use several sensitive, degenerate-primer-based PCR assays for the detection of herpesviruses of different genera, in combination with phylogenetic analysis and specific PCR, to study cross-species beta- and gammaherpesvirus transmission in a large natural primate ecosystem in the Taï National Park, Côte d'Ivoire (western Africa). The study population is comprised of a great ape predator species (western chimpanzee, Pan troglodytes verus) and its primary (western red colobus, [WRC], Piliocolobus badius) and secondary (black-and-white colobus [BWC], Colobus polykomos) monkey prey, for which interspecies transmission of various retroviruses has been shown (16, 17). Our study shows that each primate species is infected with multiple species-specific beta- and gammaherpesviruses, but we find no evidence for cross-species transmission and persistence of these viruses between the interacting ape and monkey populations.
MATERIALS AND METHODS
Sample collection and DNA isolation. Necropsy samples were collected from 20 chimpanzees (Pan troglodytes verus) (bladder, bone, brain, buffy coat, heart, heart blood, intestine, kidney, liver, lung, lymph node, nasal swab, oral swab, pancreas, plasma, serum, spleen, thymus, tonsil, trachea, and whole blood; n = 78 samples). Necropsy samples were collected from 10 WRC animals (heart, kidney, liver, lung, lymph node, and spleen; n = 30 samples) and from 1 BWC animal (liver, spleen, and trachea; n = 3 samples). Buffy coat was sampled from live animals, with samples obtained from 8 WRC (n = 8 samples) and 8 BWC (n = 9 samples) monkeys. All animals originated from the same area of the Taï National Park in Côte d'Ivoire. Causes of death for chimpanzees were anthrax (18) and respiratory diseases (19) (two and three individuals, respectively) or undetermined (15 animals) (20), and death occurred between 2001 and 2009. The WRC and BWC samples were collected in the same time period. For all samples originating from Côte d'Ivoire, sample collection was performed using full-body protection suits and masks due to a history of Ebola virus and anthrax infections in these populations and to avoid any contamination of samples with human pathogens. Permission for sample collection from live and deceased wild primates was obtained from the Ministry of Research of Côte d'Ivoire and the Ivorian National Parks Office (Office Ivoirien des Parcs et Réserves), and tissue samples were exported with the appropriate CITES authorization from Côte d'Ivoire to Germany.
Sample importation adhered to German veterinary regulations for importation of organic materials. All samples were preserved in liquid nitrogen upon arrival at the research camps and were later transferred to −80°C at the Robert Koch Institute. DNA was isolated using a DNeasy Tissue Kit (Qiagen, Hilden, Germany).
Herpesvirus PCRs.
Details of the PCR are given below. Following PCR, all PCR products were purified by using a PCR purification kit (Qiagen) and directly sequenced with a BigDye Terminator cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom) in a 377 DNA automated sequencer (Applied Biosystems).
(i) Generic CMV PCR (PCR 1 and PCR 2).
For generic amplification of glycoprotein B (gB; UL55) and UL56 gene sequences of members of the genus Cytomegalovirus, nested sets of degenerate primers (Table 1) were derived from the gB gene (PCR 1) and the UL56 gene (PCR 2) of human cytomegalovirus (HCMV) (strain AD169; accession number X17403). The primer sites were located in regions conserved among the betaherpesviruses. The primers were only moderately degenerate in order to avoid amplification of roseoloviruses and alpha- and gammaherpesviruses. PCR was performed at an annealing temperature of 45°C under conditions used in PCR 5 (generic DNA polymerase [DPOL] PCR) as previously described (21) (see also below).
Table 1.
Generic and clade-specific primers
| PCR no. | Round no. | Primer sequence |
Product size (kb) | Target gene | Targeted herpesvirus(es) | Annealing temp (°C) | |
|---|---|---|---|---|---|---|---|
| Sense | Antisense | ||||||
| 1 | I | CGCAAATCGCAGA(N/I)KC(N/I)TGGTG | TGGTTGCCCAACAG(N/I)ATYTCRTT | 0.32 | gB | Members of the genus Cytomegalovirus | 45 |
| II | TTCAAGGAACTCAGYAARAT(N/I)AAYCC | CGTTGTCCTC(N/I)CC(N/I)ARYTG(N/I)CC | 0.25 | 45 | |||
| 2 | I | CCTGTCGCACAATGTGGACATG | CAGCTGTTTTCCGAA(N/I)GTTTCRTTAT | 0.25 | UL56 | Members of the genus Cytomegalovirus | 45 |
| II | TGGCCTACGCYTGYGAYAACG | GCGAACGTGC(N/I)TCCACATCTCC | 0.18 | 45 | |||
| 3 | I | CGTGAAATGTGCCGAGGGAACG | TCCGACATAACGGGCCGCGA | 2.2 | UL56-gB | WRC cytomegalovirusesa | 68 |
| II | GGATGGTCGTCAAGTATCAAGGCTTTT | GGCCGCGATCGGTTTGTCGTA | 2.2 | 68 | |||
| 4 | I | GCTCATCCGTTGGTGGCCTTCC | GCAACTGGGCATAAACCACACTTTGA | 0.58 | gB | WRC cytomegalovirusesa | 58 |
| II | ACTACAGCATGGTGGATTCCTACGG | CATCAGCCAGTTGCTTCATCTCCC | 0.39 | 58 | |||
| 5 | I | GAYTTYGC(N/I)AGYYT(N/I)TAYCC | 5′-GTCTTGCTCACCAG(N/I)TC(N/I)AC(N/I)CCYTT | 0.48–0.71 | DPOL | Members of the family Herpesviridae | 46 |
| TCCTGGACAAGCAGCAR(N/I)YSGC(N/I)MT(N/I)AA | |||||||
| II | TGTAACTCGGTGTAYGG(N/I)TTYAC(N/I)GG(N/I)GT | 5′-CACAGAGTCCGTRTC(N/I)CCRTA(N/I)AT | 0.21–0.235 | 46 | |||
| 6 | I | ACGGCCTCTTCCCCTGCCTA | GCAGCTGGCCTTCGGGGTTG | 0.165 | DPOL | WRC lymphocryptovirusesa | 62 |
| 7 | I | CCTCCCAGGTTCARTWYGCMTAYGA | CCGTTGAGGTTCTGAGTGTARTARTTRTAYTC | 0.55 | gB | Members of the genus Rhadinovirus | 45 |
| II | AAGATCAACCCCAC(N/I)AG(N/I)GT(N/I)ATG | GTGTAGTAGTTGTACTCCCTRAACAT(N/I)GTYTC | 0.5 | 45 | |||
| 8 | I | GCCAAGTTCGTGGGAGACGCC | CGCACGTCTCCTTGCACGTCT | 0.23 | gB | WRC and BWC rhadinovirusesa | 62 |
Identified in this study.
(ii) LD PCR for amplification of WRC CMV gB sequences (PCR 3).
Nested nondegenerate primers (Table 1) were designed using the sequences identified with PCR 1 and 2. Nested long-distance (LD) PCR of the nearly complete gB gene (approximately 2.2 kb) of the novel WRC CMVs was then performed using a TaKaRa-Ex PCR system according to the manufacturer's instructions (TaKaRa Bio, Inc., Japan).
(iii) Diagnostic PCR for amplification of WRC CMV gB sequences (PCR 4).
For differential amplification of all WRC CMVs, two specific nondegenerate primer pairs (Table 1) were designed following alignment of the 2.2-kb gB sequences obtained from the WRC CMVs. These primers were used in a nested format under the same PCR conditions as in PCR 5, except that AmpliTaq Gold was used at 0.2 μl/25 μl of reaction volume. Cycling conditions were as follows: 95°C for 12 min and 45 cycles of 95°C for 30 s, 58°C for 30 s, and 72°C for 1 min, followed by a 10-min final extension step at 72°C.
(iv) Generic herpesvirus PCR (PCR 5).
For generic detection of members of the Lymphocryptovirus and Rhadinovirus (RHV) genera, sequences of the herpesvirus DNA polymerase (DPOL) gene (UL30 in herpes simplex virus 1 [HSV-1], UL54 in HCMV, BALF5 in EBV, and ORF9 in human herpesvirus 8 [HHV-8]) were amplified with a nested set of degenerate primers (Table 1) as described previously (21).
(v) Diagnostic PCR for amplification of WRC LCV DPOL sequences (PCR 6).
For the differential detection of all WRC LCVs, specific primers (Table 1) were designed following alignment of the WRC LCV DPOL sequences identified with PCR 5 or available in GenBank. Amplification was performed under the PCR conditions of PCR 4, except that annealing was at 62°C.
(vi) Generic RHV PCR (PCR 7).
For generic detection of members of the genus Rhadinovirus, gB nucleic acid sequences were amplified with a nested set of degenerate primers (Table 1) as described previously (13).
(vii) Diagnostic PCR for amplification of WRC and BWC gB sequences (PCR 8).
For the differential detection of all WRC and BWC RHVs, specific primers (Table 1) were deduced from an alignment of the gB sequences of WRC and BWC RHVs identified with PCR 7. They were used under the PCR conditions of PCR 4, except that annealing was at 62°C.
Phylogenetic analysis.
Sets of nucleic acid sequences were aligned using the MAFFT (22) plug-in of the software Geneious Pro, version 5.5.7. Alignments were trimmed before being used for phylogenetic analysis by removal of regions that were considered not to be justifiably alignable and of loci with a gapping character in any sequence. Phylogenetic analysis was performed with the neighbor-joining module of Geneious Pro.
Nucleotide sequence accession numbers.
Names and abbreviations for newly detected herpesviruses were based on the host species name and the genus to which the virus was tentatively assigned (for example, Piliocolobus badius cytomegalovirus, PbadCMV). The sequences of the novel viruses were submitted to GenBank under the accession numbers given in Table 6.
Table 6.
Known and novel herpesviruses included in phylogenetic analysis
| Virus subfamily and name and/or strain | Virus abbreviation | Host species | Novel virus | Virus detected in this study | GenBank accession no. |
|---|---|---|---|---|---|
| Subfamily Betaherpesvirinae | |||||
| Human cytomegalovirus | Human | ||||
| strain Toledo | HCMV Toledo | GU937742 | |||
| strain AD169 | HCMV AD169 | X17403 | |||
| Chimpanzee cytomegalovirus | CCMV | Chimpanzee | + | NC_003521 | |
| Pan troglodytes cytomegalovirus 1 | PtroCMV1 | Western chimpanzee | + | FJ538485 | |
| Pan troglodytes cytomegalovirus 2 | PtroCMV2 | Western chimpanzee | + | FJ538487 | |
| Gorilla gorilla cytomegalovirus 1 | GgorCMV1 | Gorilla | FJ538492 | ||
| Gorilla gorilla cytomegalovirus 2 | GgorCMV2 | Gorilla | FJ538490 | ||
| Macaca fascicularis cytomegalovirus 1 | MfasCMV1 | Cynomolgus monkey | AY728171 | ||
| Mandrillus sphinx cytomegalovirus 1 | MsphCMV1 | Mandrill | AY129399 | ||
| Rhesus cytomegalovirus | RhCMV | Rhesus macaque | NC_006150 | ||
| Simian cytomegalovirus, strain Colburn | AGM-CMV | African green monkey | FJ483969 | ||
| Piliocolobus badius cytomegalovirus 1 | PbadCMV1 | Western red colobus | + | + | KF254800 |
| Piliocolobus badius cytomegalovirus 1b | PbadCMV1b | Western red colobus | + | + | KF254801 |
| Piliocolobus badius cytomegalovirus 2 | PbadCMV2 | Western red colobus | + | + | KF254799 |
| Piliocolobus badius cytomegalovirus 3 | PbadCMV3 | Western red colobus | + | KF318790 | |
| Colobus polykomos cytomegalovirus 1 | CpolCMV1 | Western black-and-white colobus | + | + | KF254796 |
| Colobus guereza cytomegalovirus 1.1 | CgueCMV1.1 | Guereza | AY129397 | ||
| Colobus guereza cytomegalovirus 1.2 | CgueCMV1.2 | Guereza | EU118147 | ||
| Subfamily Gammaherpesvirinae | |||||
| Epstein-Barr virus | EBV | Human | NC_007605 | ||
| Pan paniscus lymphocryptovirus 1 | PpanLCV1 | Bonobo | AF534220 | ||
| Pan troglodytes lymphocryptovirus 1 | PtroLCV1 | Western chimpanzee | + | AF534226 | |
| Gorilla gorilla lymphocryptovirus 1 | GgorLCV1 | Gorilla | AF534225 | ||
| Rhesus lymphocryptovirus | RhLCV | Rhesus macaque | NC_006146 | ||
| Piliocolobus badius lymphocryptovirus 1 | PbadLCV1 | Western red colobus | + | AF534228 | |
| Piliocolobus badius lymphocryptovirus 2 | PbadLCV2 | Western red colobus | GQ921927 | ||
| Colobus polykomos lymphocryptovirus 1 | CpolLCV1 | Western black-and-white colobus | + | GQ921923 | |
| Colobus guereza lymphocryptovirus 1 | CgueLCV1 | Guereza | AF534219 | ||
| Human herpesvirus 8 | HHV-8 | Human | NC_009333 | ||
| Pan troglodytes rhadinovirus 1 | PtroRHV1 | Western chimpanzee | + | AY138585 | |
| Pan troglodytes rhadinovirus 2 | PtroRHV2 | Western chimpanzee | + | EU085378 | |
| Pan troglodytes rhadinovirus 3 | PtroRHV3 | Western chimpanzee | GQ995451 | ||
| Gorilla gorilla rhadinovirus 1 | GgorRHV1 | Gorilla | AY177144 | ||
| Rhesus rhadinovirus | RRV | Rhesus monkey | NC_003401 | ||
| Retroperitoneal fibromatosis-associated herpesvirus | RFHV | Pig-tailed macaque | AF204166 | ||
| Piliocolobus badius rhadinovirus 1 | PbadRHV1 | Western red colobus | + | + | KF254798 |
| Colobus polykomos rhadinovirus 1 | CpolRHV1 | Western black-and-white colobus | + | + | KF254797 |
RESULTS
Detection of cytomegaloviruses in chimpanzees and colobus monkeys.
A core set of 128 nonhuman primate (NHP) samples, in total, were available for PCR-based analysis. The samples consisted of tissue and blood and of nasal/oral swabs from live or deceased members of the three primate species of the study. Samples were tested for the presence of CMVs by using generic primers that detect CMVs (but not roseoloviruses and alpha- and gammaherpesviruses) (Table 1, PCR 1). Bands of the expected product size were purified and sequenced. Fifteen of 78 (19%) chimpanzee samples, corresponding to 6 of 20 individuals (30%) (Table 2), were positive for CMV (Pan troglodyte CMV [PtroCMV]) (Table 3). The highest percentage (31%) was found in the lungs of deceased individuals (Table 3). The identified sequences originated from the known chimpanzee CMVs (CCMVs), PtroCMV1, PtroCMV2, and CCMV (Tables 4 and 5) (15). A number of animals were shown to be infected with multiple CMVs (Table 5).
Table 2.
NHP individuals positive in generic PCR
| Virus | No. of animals tested (no. of animals positive) in generic PCR by species |
||
|---|---|---|---|
| Chimpanzee | WRC | BWC | |
| Cytomegalovirus | 20 (6) | 18 (4) | 9 (1) |
| Lymphocryptovirus | 18 (10) | 9 (4) | 7 (1) |
| Rhadinovirus | 18 (5) | 9 (2) | 7 (5) |
Table 3.
Organs of NHP positive in generic PCR
| Virus group and sample type | No. of samples tested (no. of samples positive) in generic PCR by speciesa |
||
|---|---|---|---|
| Chimpanzee | WRC | BWC | |
| Cytomegaloviruses | |||
| Lung | 16 (5) | 8 (3) | |
| Spleen | 7 (1) | 7 (3) | 1 (1) |
| Liver | 10 (2) | 7 (2) | 1 (1) |
| Heart | 5 (1) | 2 (1) | |
| Lymph node | 10 (1) | 2 (1) | |
| Intestine | 12 | ||
| Tonsil | 6 (2) | ||
| Kidney | 3 | 2 (1) | |
| Whole blood | 4 (1) | ||
| Thymus | 1 (1) | ||
| Pancreas | 1 (1) | ||
| Brain | 2 | ||
| Bladder | 1 | 2 | |
| Trachea | 1 | ||
| Blood (buffy coat) | 8 | 9 | |
| Heart blood | 1 | ||
| Total | 78 (15) | 38 (11) | 12 (2) |
| Lymphocryptoviruses | |||
| Lung | 15 (5) | 7 (2) | |
| Spleen | 7 (4) | 7 (3) | 1 |
| Lymph node | 8 (4) | 2 | |
| Liver | ND | ND | 1 |
| Trachea | 1 | ||
| Blood (buffy coat) | ND | 7 (1) | |
| Total | 30 (13) | 16 (5) | 10 (1) |
| Rhadinoviruses | |||
| Lung | 15 (5) | 7 (1) | |
| Spleen | 7 (1) | 7 (1) | 1 (1) |
| Lymph node | 8 (2) | 2 (1) | |
| Liver | ND | ND | 1 (1) |
| Trachea | 1 | ||
| Blood (buffy coat) | ND | 7 (4) | |
| Total | 30 (8) | 16 (3) | 10 (6) |
ND, not determined.
Table 4.
Herpesviruses detected in chimpanzees and WRC and BWC monkeys by generic PCR
| Catarrhine host family and species | Virus | Abbreviation | Virus statusa | Target gene in generic PCRb | No. of PCR-positive animals | No. of PCR-positive samples |
|---|---|---|---|---|---|---|
| Hominidae | ||||||
| Western chimpanzee | Chimpanzee CMV | CCMV | K | gB | 2 | 3 |
| Pan troglodytes verus cytomegalovirus 1 | PtroCMV1 | K | gB | 2 | 5 | |
| Pan troglodytes verus cytomegalovirus 2 | PtroCMV2 | K | gB | 4 | 7 | |
| Pan troglodytes verus lymphocryptovirus 1 | PtroLCV1 | K | DPOL | 10 | 13 | |
| Pan troglodytes verus rhadinovirus 1 | PtroRHV1 | K | DPOL | 5 | 8 | |
| Pan troglodytes verus rhadinovirus 2 | PtroRHV2 | K | gB | 1 | 1 | |
| Cercopithecidae | ||||||
| Western red colobus | Piliocolobus badius cytomegalovirus 1 | PbadCMV1 | N | gB | 1 | 4 |
| Piliocolobus badius cytomegalovirus 1b | PbadCMV1b | N | gB | 2 | 2 | |
| Piliocolobus badius cytomegalovirus 2 | PbadCMV2 | N | gB | 2 | 5 | |
| Piliocolobus badius lymphocryptovirus 1 | PbadLCV1 | K | DPOL | 4 | 5 | |
| Piliocolobus badius rhadinovirus 1 | PbadRHV1 | N | gB | 2 | 3 | |
| Black-and-white colobus | Colobus polykomos cytomegalovirus 1 | CpolCMV1 | N | gB | 1 | 2 |
| Colobus polykomos lymphocryptovirus 1 | CpolLCV1 | K | DPOL | 1 | 1 | |
| Colobus polykomos rhadinovirus 1 | CpolRHV1 | N | gB | 5 | 6 |
K, known; N, novel.
gB, glycoprotein B; DPOL, DNA polymerase.
Table 5.
Coinfection status
| Host and animal number | Cause of death | Generic PCR-positive tissue(s) | Virus(es) detecteda |
||
|---|---|---|---|---|---|
| Cytomegalovirus | Lymphocryptovirus | Rhadinovirus | |||
| Chimpanzees (name) | |||||
| 2 (Leo) | Anthrax | Lung, lymph node, spleen | PtroCMV2 | − | PtroRHV1, PtroRHV2 |
| 10 (Noah) | Anthrax | Heart, liver, lung, pancreas, thymus | PtroCMV2, PtroCMV1, CCMV | PtroLCV1 | − |
| 21 (Ophelia) | Respiratory disease | Liver, lung, spleen, tonsils | PtroCMV1 | PtroLCV1 | − |
| 76 (Candy) | Respiratory disease | Lung, spleen | − | PtroLCV1 | − |
| 560 (Akwaba) | Respiratory disease | Lung, tonsils | PtroCMV1, PtroCMV2 | PtroLCV1 | − |
| Colobus monkeys (species) | |||||
| 66 (WRC) | Undetermined | Lung | PbadCMV1b | − | PbadRHV1 |
| 71 (WRC) | Undetermined | Lung | PbadCMV1b, PbadCMV2 | PbadLCV1 | − |
| 72 (WRC) | Undetermined | Spleen | PbadCMV2 | − | PbadRHV1 |
| 213 (WRC) | Undetermined | Spleen | PbadCMV2 | PbadLCV1 | − |
| 740 (BWC) | Undetermined | Spleen, trachea | CpolCMV1 | − | CpolRHV1 |
−, PCR negative.
Eleven of 38 (29%) WRC samples tested positive for CMV with the generic CMV PCR (Table 1, PCR 1), corresponding to 4 of 18 (22%) animals (Table 2). The organs with the highest positivity were lung and spleen, with six CMV-positive organs out of a total number of 15 lung and spleens tested (40%) (Table 3). Two of 12 (17%) BWC samples, both derived from a single animal (Table 2), were PCR positive for CMV (Table 3). Sequences of the 13 colobus-derived CMV PCR products were subjected to BLAST analysis and determined to originate from four formerly unknown CMVs (three from WRC samples and one from a BWC sample). These novel CMVs were named PbadCMV1, PbadCMV1b, PbadCMV2, and Colobus polykomos CMV1 (CpolCMV1) (Table 4). One animal showed the presence of multiple CMVs (Table 5). The analysis also confirmed the absence of chimpanzee CMV in any of the monkey samples.
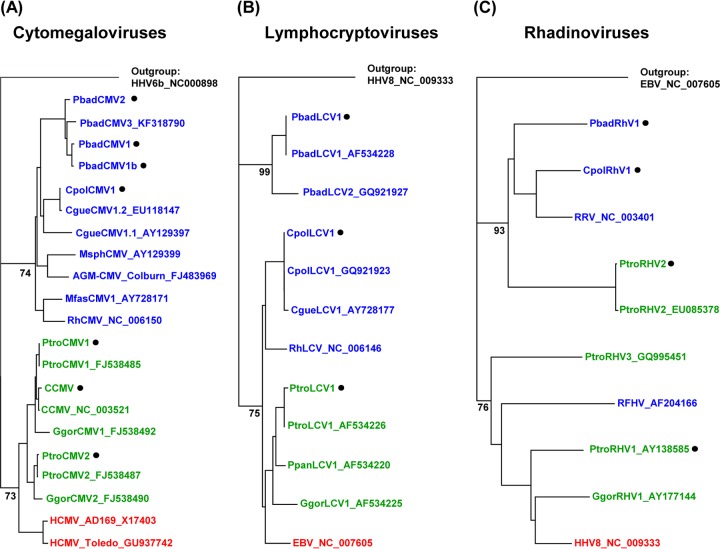
A phylogenetic tree was constructed using a MAFFT alignment of sequences from human CMV (HCMV) (strains Toledo and AD169), great ape CMVs (chimpanzee and gorilla), and OWM CMVs (African green monkey, mandrill, rhesus macaque, and multiple colobus monkey species). Inspection of the tree revealed the presence of two distinct clades: one clade was comprised of human and great ape CMVs, and the other clade was comprised of OWM CMVs. In the OWM clade, the novel WRC CMVs (PbadCMV1, PbadCMV1b, and PbadCMV2) formed a distinct subclade together with a WRC CMV (designated PbadCMV3) that we had detected previously in spleens of two WRC animals (B. Ehlers, unpublished data). The novel BWC CMV (CpolCMV1) was closely related to the Colobus guereza virus, CgueCMV1.2 (Fig. 1a).
Fig 1.
Phylogenetic analysis. Partial gene sequences from the herpesviruses detected in this study and from published herpesviruses were aligned using MAFFT and subjected to phylogenetic construction of trees using the Geneious, version 5.5.7, tree builder (neighbor-joining module). Human, great ape, and OWM herpesviruses are shown in red, green, and blue, respectively. Viruses detected in this study are marked (black dot). (A) Tree based on glycoprotein B sequences of CMVs. (B) Tree based on DNA polymerase sequences of LCVs. (C) Tree based on gB sequences of RHVs. Bootstrap values are indicated at the basis of major clades and are suppressed at the tips of the clades.
WRC is the major monkey prey species of chimpanzees in the Taï National Park. To assess whether CMVs of WRC monkeys were present in chimpanzees, we used nested PCR primers that were designed to specifically target WRC CMVs without amplifying chimpanzee CMVs. Since the gB sequences of the novel WRC CMVs were too short for design of the necessary primers, the UL56 3′ region of three of the WRC CMVs (PbadCMV1, PbadCMV1b, and PbadCMV2) was amplified by generic UL56 PCR (Table 1, PCR 2), followed by amplification of a 2.2-kbp region between gB and UL56 by using long-distance PCR (Table 1, PCR 3). This 2.2-kbp section of PbadCMV sequence was then used for primer design (Table 1, PCR 4). Analysis using PCR 4 did not detect colobus CMVs in any of the chimpanzee samples (data not shown).
Detection of lymphocryptoviruses in chimpanzees and colobus monkeys.
A generic herpesvirus PCR targeting the highly conserved DNA polymerase gene (DPOL) (Table 1, PCR 5) was used to analyze 30 chimpanzee samples for the presence of lymphocryptoviruses (LCV; subfamily Gammaherpesvirinae). Lung, spleen, and lymph node samples were selected for analysis since they had been shown to be prominent sources of gammaherpesviruses in previous studies (13, 14, 23). Sequencing of the amplified products showed 13 of 30 samples (43%) to be positive for LCV (Table 3), corresponding to 10 of 18 (56%) animals. Spleens and lymph nodes of deceased chimpanzees showed the highest levels of LCV positivity (57% and 50%, respectively). All sequences originated from an LCV that had 99% identity with PtroLCV1, an LCV previously identified in chimpanzees (Tables 4 and 6) (23). WRC (16 samples) and BWC (10 samples; including buffy coat) samples were analyzed for the presence of LCV (Table 1, PCR 5). Five of 16 WRC samples (31%) (all from the lung and spleen) and 1 of 10 BWC samples (from buffy coat) tested positive for OWM LCV, corresponding to 5 of 16 monkeys (31%) (four WRC monkeys and one BWC monkey) (Tables 2 and 3). BLAST analysis identified a previously reported WRC LCV (PbadLCV1) (14) and a BWC LCV (CpolLCV1). This PCR analysis also confirmed the absence of chimpanzee LCV in the animals.
Phylogenetic analysis was performed using the corresponding DPOL region of LCV sequences from human (Epstein-Barr virus [EBV]), great ape (chimpanzee and gorilla), and OWM (Colobus guereza and rhesus macaque) viruses. In the tree (Fig. 1B), WRC LCVs (PbadLCV1 and the previously identified PbadLCV2) formed a clade distinct from a mixed clade comprising human, great ape, rhesus, and BWC LCVs. Primers targeting WRC LCVs (and excluding chimpanzee LCVs) were selected to test for the presence of WRC LCV in chimpanzees (Table 1, PCR 6). This PCR did not detect WRC LCVs in any chimpanzee sample (data not shown).
Detection of rhadinoviruses in chimpanzees and colobus monkeys.
Generic DPOL PCR (Table 1, PCR 5) was used to analyze the 30 lung, spleen, and lymph node chimpanzee samples for the presence of rhadinoviruses (RHV; subfamily Gammaherpesvirinae). Eight of 30 samples (27%) were positive for RHV (Table 3), corresponding to 5 of 18 (28%) animals. Similar to the distribution of CMV, lungs of deceased chimpanzees showed a high level of RHV positivity (33%). The detected virus was in all cases identical to the previously identified PtroRHV1 (Tables 4 and 6) (24). All chimpanzee samples were also tested by using a generic gB PCR (Table 1, PCR 7), which resulted in detection of the known PtroRHV2 (Tables 4 and 6) (24). Similar to findings for CMV, one animal showed the presence of multiple RHVs (Table 5).
In the final analysis, the 16 WRC and 10 BWC samples were tested for the presence of RHV with PCR 7. Three of 16 WRC samples (19%) and 6 of 10 BWC samples (60%) were positive for OWM RHV, corresponding to 7 of 16 (44%) monkeys (2 WRC and 5 BWC animals) (Table 2). Viruses were distributed between lymphoid organs (spleen and lymph nodes), lung, liver, and blood (buffy coat) (Table 3). BLAST analysis identified the presence of two novel viruses (designated PbadRHV1 and CpolRHV1) and confirmed the absence of any chimpanzee RHV in the monkeys. With PCR 5, the same RHV (CpolRHV1) was detected in the BWC samples. The WRC samples were negative (data not shown).
Phylogenetic analysis was performed using published gB sequences from human herpesvirus 8 (HHV-8), great ape RHVs (chimpanzees and gorilla), and OWM RHVs (WRC, BWC, rhesus, and pig-tailed macaque). Distinct clades of great ape RHVs and OWM RHVs were apparent (Fig. 1C). The presence of a distinct clade containing WRC, BWC, and rhesus RHVs enabled the design of clade-specific primers that detected colobus RHVs but excluded great ape RHVs (Table 1, PCR 8). Use of these primers confirmed the absence of colobus RHVs in all chimpanzee samples (data not shown).
Summary of DNA viruses detected and novel viruses discovered.
In the present study, we detected four novel OWM CMVs (PbadCMV1, PbadCMV1b, PbadCMV2, and CpolCMV1) and two novel OWM RHVs (CpolRHV1 and PbadRHV1) in the colobus monkey study group. Together with previously identified herpesviruses detected in this study, the novel viruses are listed in Table 4 and are presented phylogenetically in Fig. 1. The detection frequencies of herpesviruses in chimpanzees were 30% (CMV), 56% (LCV), and 28% (RHV); in WRC monkeys the frequencies were 22% (CMV), 44% (LCV), and 22% (RHV); and in BWC monkeys the frequencies were 11% (CMV), 14% (LCV), and 71% (RHV) (Table 2). Individual chimpanzees and monkeys were shown to be infected by multiple herpesviruses but with no apparent bias toward coinfection with particular viruses (Table 5). Finally, although all primate species were infected to a substantial level with their own species-specific beta- and gammaherpesviruses, there was no evidence for cross-species transmission.
DISCUSSION
We have used highly sensitive degenerate PCRs in combination with specific PCRs and phylogenetic analysis to analyze primate betaherpesviruses (CMVs) and gammaherpesviruses (LCVs and RHVs) in a great ape predator (western chimpanzee) and its primary (WRC) and secondary (BWC) monkey prey species. Our results show that all three primate species are commonly infected (and frequently coinfected) with multiple species-specific CMV, LCV, and RHV herpesviruses. The lung and spleen of WRC and BWC monkeys were observed to be the most frequent sites of herpesvirus infection. Six of the herpesviruses detected in this study represent new viruses described for the first time.
Hunting frequently involves biting (both of monkeys by chimpanzees and, on occasion, of chimpanzees by monkeys). Most monkey tissues, organs, and bone marrow are consumed in their entirety by chimpanzees. Marrow is extracted by crushing of bones, which furthers the possibility for direct blood-to-blood contact by oral laceration. This predator-prey system, in which chimpanzees are exposed on a continual basis to monkey blood and tissues, therefore represents a unique natural primate ecosystem in which to assess microbe cross-species transmission in the wild. This intensive level of interaction has been shown to lead to transmission of retroviruses such as simian T-cell leukemia virus type 1 (STLV-1) and simian foamy virus (SFV) between chimpanzees and monkeys (16, 17, 25, 26). However, despite this extensive exposure, there was no evidence for cross-species transmission of herpesviruses between these species.
Following primary infection, herpesviruses establish life-long infection within their respective host species (27). Our PCR-based analysis is therefore both a measure of cross-species herpesvirus transmission and of the ability of transmitted viruses to establish themselves within the new host. Excluding the period of acute infection, this method of analysis will detect cross-species transmission only if the virus persists following transmission, thereby avoiding “background” from transient exposure to herpesviruses (such as would be detected using serologically based approaches). Beta- and gammaherpesviruses are phylogenetically closely grouped into distinct clades based on the specific primate species they infect. Our phylogenetic analysis is therefore able to detect persistence of transmitted herpesviruses not only in contemporary time but also at the population level extending over the past 20 million years (assuming an ability of transmitted viruses to be maintained within the new host species population) (see below). By both measures, cross-species transmission/persistence of beta- and gammaherpesviruses was not detected between the different primate study populations.
The considerable level of interaction between prey and predator species in the primate ecosystem studied here, combined with the high prevalence of herpesviruses within the two species, would be expected to promote the possibility for transmission such that exposure to herpesviruses would not be the limiting factor. The absence of transmission more likely reflects the inability of herpesviruses to genetically adapt to a level sufficient to infect and then persist within the new primate host. Following exposure, a microbe must be able to persist and spread within the new population, represented by the basic reproduction number R0 (new infections per unit time). R0 is a critical measure of the potential for success of the pathogen within its new host population, with only R0 values of >1 being generally consistent with maintenance of an enzootic/zoonotic cross-species transmission (8). Given the predator-prey nature of the relationship, the possibility for transmission of microbes from chimpanzees into the monkey population would be limited. However, the calculation that the average adult male chimpanzee in the Taï National Forest consumes nearly 250 kg of colobus meat during its 20-year lifetime suggests extensive exposure of chimpanzees to herpesvirus-infected monkey tissue (28). Thus, the inability to detect monkey-derived herpesviruses in chimpanzees suggests that primate herpesviruses maintain a high degree of species specificity, even between related primate species. It is possible that within the limits imposed by our animal group size of 20 chimpanzees, we were unable to observe transmission/persistence events that were occurring at a low frequency. Our results therefore do not rule out the possibility for herpesvirus transmission between these interacting primate populations but indicate that transmission is, at most, rare.
The level of genetic similarity between reservoir/transmission species and a new host has been suggested to play an important role in facilitating enzootic/zoonotic cross-species transmission by weakening the species barrier and thereby potentially increasing both the number of primary infections (I0) and R0. This effect of host phylogenetic similarity on transmission is reflected in the high incidence of tropical zoonotic diseases that have an NHP source (2, 29–31). Genetic similarity between these primate species is thought to facilitate preadaptation or rapid adaptation of the microbe, promoting its transmission and establishment within humans; this can be compared to the relative inefficiency of microbes moving to humans from more distantly related animal species (i.e., H5N1 avian flu virus from birds). In the system studied here, even the presumed weak species barrier resulting from the close phylogenetic relationship between the chimpanzees and their interacting monkey species appears to be sufficient to prevent herpesviruses from transmitting and persisting within a new primate host species.
RNA viruses appear to be particularly prone to cross-species epizootic/zoonotic transmission (8, 32). This propensity of RNA viruses for cross-species transmission is believed to correspond to the rapid replication and high mutation rate of these viruses, facilitating adaptation to the new host environment (8, 32). In contrast, replication of DNA viruses such as herpesviruses is characterized by low-level “smouldering” or latent infection with periodic reactivation (increased levels of herpesvirus replication and overt disease during the chronic phase of infection are generally seen only associated with immunosuppression). DNA viruses also have a far higher fidelity of replication than observed for RNA viruses (32). Both of these factors may result in reducing the potential for adaptation of herpesviruses to a new host, negatively impacting I0 and R0 and reducing the capacity for cross-species epizootic/zoonotic transmission.
Our current study would suggest that relatively strict species specificity exists for primate beta- and gammaherpesviruses. Previous results from in vitro studies are consistent with our findings, with species-specific CMVs replicating poorly in cells from other species (33–35). In these studies, the genetic similarity between host species appeared to influence the replicative capacity of the respective CMVs, with HCMV replication being reduced only 10-fold in chimpanzee cells, compared to being nondetectable in cells from mice. In vivo studies support this level of species specificity, with no cross-species transmission/persistence being observed for murine CMV (MCMV) from naturally infected Mus musculus domesticus (house mouse) to native Leggadina lakedownensis (short-tailed mice) following the release of MCMV-infected house mice into the Thevenard Island natural reserve (36). In the Thevenard Island study, MCMV did not replicate even following direct inoculation of virus into L. lakedownensis.
The capacity for transmission of gammaherpesviruses has not been empirically examined. However, degenerate-PCR-based approaches, similar to those used in the present study, indicate that cross-species transmission/persistence has occurred for both beta- and gammaherpesviruses at least on an evolutionary time scale although the scarcity of these events would support that such transmission is rare. Specifically, phylogenetic analysis provides evidence for transmission of CMV between ape species (chimpanzees and gorillas) within the last million years and of at least two independent introductions of OWM LCV into ape populations around 12 million years ago (14, 15). An OWM LCV transmission into Asian apes (orangutans and gibbons) is also believed to have occurred more recently, approximately 1 million years ago (14). There is also evidence for transmission of nonprimate herpesviruses (specifically, RHVs) (13). Interestingly, the close phylogenetic relationship of RHVs from spotted hyena with those of zebra/horses and of lion RHV with RHVs of wild pig/rhino species suggests that a predator and prey interaction may be one scenario that favors cross-species herpesvirus transmission. Together with results from these earlier studies, the lack of evidence for transmission/persistence of primate beta- and gammaherpesviruses in our current study suggests that although viruses from these two herpesvirus families are capable of cross-species transmission/persistence, such events are rare on both a contemporary and evolutionary time scale.
Due to high immunogenicity and effector T cell memory bias of CMV-induced immune responses, a number of laboratories are developing CMV as a vaccine platform (37–41). CMVs have evolved to spread through their target host population and have a remarkable capacity to reinfect the host, regardless of prior CMV immunity (42). We along with others are therefore beginning to exploit this ability of CMV to spread for the development of “disseminating” vaccines to target animal populations that are geographically or economically inaccessible to standard vaccination strategies (for example, to prevent Ebola virus infection in great apes in Central Africa or for immunocontraception in mice to prevent mouse plagues) (38, 40). The present study furthers our understanding of the capacity for cross-species transmission of CMV between closely related species in a natural ecosystem, which will be critical as these vaccine strategies move toward potential application.
ACKNOWLEDGMENTS
We are grateful for the excellent technical assistance provided by Sonja Liebmann, Cornelia Walter, and Nezlisah Yasmum. For work in Côte d'Ivoire we thank the Ivorian authorities for their long-term support, especially the Ministry of the Environment and Forests, the Ministry of Research, the Directorship of the Taï National Park, and the Swiss Research Centre in Abidjan.
For funding, we thank the Deutsche Forschungsgemeinschaft (grant number LE1813/4-1) and the University of Plymouth, School of Biomedical and Biological Sciences (M.A.J.).
Footnotes
Published ahead of print 24 July 2013
REFERENCES
- 1.Calvignac-Spencer S, Leendertz SA, Gillespie TR, Leendertz FH. 2012. Wild great apes as sentinels and sources of infectious disease. Clin. Microbiol. Infect. 18:521–527 [DOI] [PubMed] [Google Scholar]
- 2.Wolfe ND, Dunavan CP, Diamond J. 2007. Origins of major human infectious diseases. Nature 447:279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451:990–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drancourt M, Raoult D. 2002. Molecular insights into the history of plague. Microbes Infect. 4:105–109 [DOI] [PubMed] [Google Scholar]
- 5.Taubenberger JK, Morens DM. 2009. Pandemic influenza–including a risk assessment of H5N1. Rev. Sci. Tech. 28:187–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piot P, Bartos M, Ghys PD, Walker N, Schwartlander B. 2001. The global impact of HIV/AIDS. Nature 410:968–973 [DOI] [PubMed] [Google Scholar]
- 7.Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, Laughlin CA, Saif LJ, Daszak P. 2008. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol. Mol. Biol. Rev. 72:457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woolhouse ME, Haydon DT, Antia R. 2005. Emerging pathogens: the epidemiology and evolution of species jumps. Trends Ecol. Evol. 20:238–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vikoren T, Li H, Lillehaug A, Jonassen CM, Bockerman I, Handeland K. 2006. Malignant catarrhal fever in free-ranging cervids associated with OvHV-2 and CpHV-2 DNA. J. Wildl. Dis. 42:797–807 [DOI] [PubMed] [Google Scholar]
- 10.Huff JL, Barry PA. 2003. B-virus (Cercopithecine herpesvirus 1) infection in humans and macaques: potential for zoonotic disease. Emerg. Infect. Dis. 9:246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wittmann G, Rziha RJ. 1989. Aujeszky's disease (pseudorabies) in pigs, p 230–325 In Wittmann G. (ed), Herpesvirus diseases of cattle, horses and pigs. Kluwer, Boston, MA [Google Scholar]
- 12.McGeoch DJ, Dolan A, Ralph AC. 2000. Toward a comprehensive phylogeny for mammalian and avian herpesviruses. J. Virol. 74:10401–10406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehlers B, Dural G, Yasmum N, Lembo T, de Thoisy B, Ryser-Degiorgis MP, Ulrich RG, McGeoch DJ. 2008. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: cospeciation and interspecies transfer. J. Virol. 82:3509–3516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehlers B, Spiess K, Leendertz F, Peeters M, Boesch C, Gatherer D, McGeoch DJ. 2010. Lymphocryptovirus phylogeny and the origins of Epstein-Barr virus. J. Gen. Virol. 91:630–642 [DOI] [PubMed] [Google Scholar]
- 15.Leendertz FH, Deckers M, Schempp W, Lankester F, Boesch C, Mugisha L, Dolan A, Gatherer D, McGeoch DJ, Ehlers B. 2009. Novel cytomegaloviruses in free-ranging and captive great apes: phylogenetic evidence for bidirectional horizontal transmission. J. Gen. Virol. 90:2386–2394 [DOI] [PubMed] [Google Scholar]
- 16.Leendertz FH, Zirkel F, Couacy-Hymann E, Ellerbrok H, Morozov VA, Pauli G, Hedemann C, Formenty P, Jensen SA, Boesch C, Junglen S. 2008. Interspecies transmission of simian foamy virus in a natural predator-prey system. J. Virol. 82:7741–7744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvignac-Spencer S, Adjogoua EV, Akoua-Koffi C, Hedemann C, Schubert G, Ellerbrok H, Leendertz SA, Pauli G, Leendertz FH. 2012. Origin of human T-lymphotropic virus type 1 in rural Cote d'Ivoire. Emerg. Infect. Dis. 18:830–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leendertz FH, Ellerbrok H, Boesch C, Couacy-Hymann E, Matz-Rensing K, Hakenbeck R, Bergmann C, Abaza P, Junglen S, Moebius Y, Vigilant L, Formenty P, Pauli G. 2004. Anthrax kills wild chimpanzees in a tropical rainforest. Nature 430:451–452 [DOI] [PubMed] [Google Scholar]
- 19.Kondgen S, Kuhl H, N′Goran PK, Walsh PD, Schenk S, Ernst N, Biek R, Formenty P, Matz-Rensing K, Schweiger B, Junglen S, Ellerbrok H, Nitsche A, Briese T, Lipkin WI, Pauli G, Boesch C, Leendertz FH. 2008. Pandemic human viruses cause decline of endangered great apes. Curr. Biol. 18:260–264 [DOI] [PubMed] [Google Scholar]
- 20.Leendertz FH, Pauli G, Maetz-Rensing K, Boardman W, Nunn C, Ellerbrok H, Jensen SA, Junglen S, Boesch C. 2006. Pathogens as drivers of population declines: the importance of systematic monitoring in great apes and other threatened mammals. Biol. Conserv. 131:325–337 [Google Scholar]
- 21.Chmielewicz B, Goltz M, Lahrmann KH, Ehlers B. 2003. Approaching virus safety in xenotransplantation: a search for unrecognized herpesviruses in pigs. Xenotransplantation 10:349–356 [DOI] [PubMed] [Google Scholar]
- 22.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehlers B, Ochs A, Leendertz F, Goltz M, Boesch C, Matz-Rensing K. 2003. Novel simian homologues of Epstein-Barr virus. J. Virol. 77:10695–10699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prepens S, Kreuzer KA, Leendertz F, Nitsche A, Ehlers B. 2007. Discovery of herpesviruses in multi-infected primates using locked nucleic acids (LNA) and a bigenic PCR approach. Virol. J. 4:84. 10.1186/1743-422X-4-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Junglen S, Hedemann C, Ellerbrok H, Pauli G, Boesch C, Leendertz FH. 2010. Diversity of STLV-1 strains in wild chimpanzees (Pan troglodytes verus) from Cote d'Ivoire. Virus Res. 150:143–147 [DOI] [PubMed] [Google Scholar]
- 26.Leendertz FH, Junglen S, Boesch C, Formenty P, Couacy-Hymann E, Courgnaud V, Pauli G, Ellerbrok H. 2004. High variety of different simian T-cell leukemia virus type 1 strains in chimpanzees (Pan troglodytes verus) of the Tai National Park, Cote d'Ivoire. J. Virol. 78:4352–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roizman B. 1996. Herpesviridae, p 2221–2230 In Fields BN, Knipe DM, Howley PM, Channock RM, Melnick JL, Monath TP, Roizman BStrauss. (ed), Virology, 3rd ed. Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 28.Leendertz SA, Locatelli S, Boesch C, Kucherer C, Formenty P, Liegeois F, Ayouba A, Peeters M, Leendertz FH. 2011. No evidence for transmission of SIVwrc from western red colobus monkeys (Piliocolobus badius badius) to wild West African chimpanzees (Pan troglodytes verus) despite high exposure through hunting. BMC Microbiol. 11:24. 10.1186/1471-2180-11-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gnanadurai CW, Pandrea I, Parrish NF, Kraus MH, Learn GH, Salazar MG, Sauermann U, Topfer K, Gautam R, Munch J, Stahl-Hennig C, Apetrei C, Hahn BH, Kirchhoff F. 2010. Genetic identity and biological phenotype of a transmitted/founder virus representative of nonpathogenic simian immunodeficiency virus infection in African green monkeys. J. Virol. 84:12245–12254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouquet P, Froment JM, Bermejo M, Kilbourn A, Karesh W, Reed P, Kumulungui B, Yaba P, Delicat A, Rollin PE, Leroy EM. 2005. Wild animal mortality monitoring and human Ebola outbreaks, Gabon and Republic of Congo, 2001–2003. Emerg. Infect. Dis. 11:283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groseth A, Feldmann H, Strong JE. 2007. The ecology of Ebola virus. Trends Microbiol. 15:408–416 [DOI] [PubMed] [Google Scholar]
- 32.Holmes EC. 2008. Evolutionary history and phylogeography of human viruses. Annu. Rev. Microbiol. 62:307–328 [DOI] [PubMed] [Google Scholar]
- 33.Lafemina RL, Hayward GS. 1988. Differences in cell-type-specific blocks to immediate early gene expression and DNA replication of human, simian and murine cytomegalovirus. J. Gen. Virol. 69:355–374 [DOI] [PubMed] [Google Scholar]
- 34.Perot K, Walker CM, Spaete RR. 1992. Primary chimpanzee skin fibroblast cells are fully permissive for human cytomegalovirus replication. J. Gen. Virol. 73:3281–3284 [DOI] [PubMed] [Google Scholar]
- 35.Jurak I, Brune W. 2006. Induction of apoptosis limits cytomegalovirus cross-species infection. EMBO J. 25:2634–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moro D, Lloyd ML, Smith AL, Shellam GR, Lawson MA. 1999. Murine viruses in an island population of introduced house mice and endemic short-tailed mice in Western Australia. J. Wildl. Dis. 35:301–310 [DOI] [PubMed] [Google Scholar]
- 37.Tierney R, Nakai T, Parkins CJ, Caposio P, Fairweather NF, Sesardic D, Jarvis MA. 2012. A single-dose cytomegalovirus-based vaccine encoding tetanus toxin fragment C induces sustained levels of protective tetanus toxin antibodies in mice. Vaccine 30:3047–3052 [DOI] [PubMed] [Google Scholar]
- 38.Tsuda Y, Caposio P, Parkins CJ, Botto S, Messaoudi I, Cicin-Sain L, Feldmann H, Jarvis MA. 2011. A replicating cytomegalovirus-based vaccine encoding a single Ebola virus nucleoprotein CTL epitope confers protection against Ebola virus. PLoS Negl. Trop. Dis. 5:e1275. 10.1371/journal.pntd.0001275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M, Jr, Lifson JD, Picker LJ. 2011. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redwood AJ, Messerle M, Harvey NL, Hardy CM, Koszinowski UH, Lawson MA, Shellam GR. 2005. Use of a murine cytomegalovirus K181-derived bacterial artificial chromosome as a vaccine vector for immunocontraception. J. Virol. 79:2998–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizvanov AA, van Geelen AG, Morzunov S, Otteson EW, Bohlman C, Pari GS, St Jeor SC. 2003. Generation of a recombinant cytomegalovirus for expression of a hantavirus glycoprotein. J. Virol. 77:12203–12210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen SG, Powers CJ, Richards R, Ventura AB, Ford JC, Siess D, Axthelm MK, Nelson JA, Jarvis MA, Picker LJ, Fruh K. 2010. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 328:102–106 [DOI] [PMC free article] [PubMed] [Google Scholar]