Abstract
Primary infection with human cytomegalovirus (HCMV) is generally asymptomatic in healthy individuals and results in a lifelong infection of the host. In contrast, in immunosuppressed transplant recipients and late-stage AIDS patients, HCMV infection and reactivation can result in severe disease or death. In vivo, latency is established in bone marrow CD34+ progenitor cells with reactivation linked with their differentiation to macrophages and dendritic cells (DCs). However, previous analyses have relied on ex vivo differentiation of myeloid progenitor cells to DCs in culture. Here, we now report on the isolation and analysis of circulating blood myeloid DCs, resulting from natural differentiation in vivo, from healthy HCMV-seropositive carriers. We show that these in vivo-differentiated circulating DCs are fully permissive for HCMV and exhibit a phenotype similar to that of monocyte-derived DCs routinely used for in vitro studies of HCMV. Importantly, we also show that these DCs from healthy HCMV-seropositive donors carry HCMV genomes and, significantly, are typically positive for viral immediate-early (IE) gene expression, in contrast to circulating monocytes, which carry genomes with an absence of IE expression. Finally, we show that HCMV reactivation from these circulating DCs is enhanced by inflammatory stimuli. Overall, these data argue that the differentiation in vivo of myeloid progenitors to circulating DCs promotes the reactivation of HCMV lytic gene expression in healthy individuals, thereby providing valuable confirmation of studies performed using in vitro generation of DCs from myeloid precursors to study HCMV reactivation.
INTRODUCTION
Primary infection of healthy individuals with human cytomegalovirus (HCMV) normally results in an asymptomatic infection of the host. However, primary infection of neonates or infection with or reactivation of latent virus in immunosuppressed transplant patients and immunocompromised late-stage HIV patients, who have developed AIDS, can result in significant morbidity and mortality (1–3). Consequently, serious health risks posed by reactivation of latent HCMV have resulted in a concerted effort by a number of laboratories to further define the cell types and mechanisms involved in HCMV latency and reactivation.
The current consensus is that HCMV can establish a latent infection of pluripotent CD34+ mononuclear cells (4–8). However, carriage of the virus appears to be restricted only to certain cell types within the hematopoietic system, in particular, cells of the myeloid lineage such as monocytes, their circulating progenitors, and subsequent derivatives (9–14). This carriage of viral genomes occurs in the absence of any significant viral lytic gene expression (reviewed in reference 15); hence, cells of the myeloid lineage represent an important site of latency and persistence in the host. This is no more exemplified than by clinical observations that leukocyte depletion of peripheral blood prior to transfusion significantly diminished the incidence of HCMV transmission to recipients (16–18).
Pertinent to this report, a number of studies of both experimental and natural latency have illustrated that the in vitro differentiation of myeloid progenitor cells to terminally differentiated myeloid dendritic cell (DC) phenotypes results in the induction of HCMV reactivation from these latently infected myeloid cell types (5, 12, 19–23). These models have relied on in vitro differentiation to cell types that are defined as dermal (interstitial) or epidermal (Langerhans)-like cell types based on the expression of a panel of cell surface markers identified on corresponding cells directly isolated ex vivo (20, 24–29). Therefore, these data would predict that circulating DCs were sites of HCMV carriage in vivo and, furthermore, that these cells might be sites of reactivation in vivo. However, crucially, while a wealth of in vitro data supports this prevailing hypothesis, it has never been definitively shown that naturally occurring DCs derived from healthy seropositive individuals are sites of genome carriage and, importantly, sites of HCMV reactivation. Indeed, a previous study of CD11c+ dendritic cells isolated from buffy coats of healthy volunteers has suggested that the well-documented immunoparalysis observed following infection of in vitro-differentiated DCs (20, 30–37) is not observed when circulating DCs are similarly infected in vitro with HCMV (38). Although these data concern lytic infection, they exemplify the need for a direct analysis of HCMV latency and reactivation in DCs.
In this study, we have sought to formally address whether DCs directly isolated ex vivo from healthy seropositive donors are indeed sites of HCMV latency and reactivation. Here, we show that the purification of circulating blood DCs, which express a cell surface phenotype comparable to that of monocyte-derived DCs (MoDCs) ex vivo, results in the detection of HCMV genome-positive cells in healthy aviremic seropositive donors. Furthermore, the carriage of viral genomes is concomitant with the detection of HCMV lytic gene transcription in these cells and, ultimately, the recovery of infectious virus from them. Importantly, we show that monocytes isolated concomitantly from the same donors are immediate-early (IE) transcript negative, consistent with in vivo differentiation to a myeloid DC as a trigger for HCMV reactivation. Interestingly, we also observed that a number of inflammatory stimuli could significantly enhance the level of reactivation observed in these purified circulating DC populations, consistent with the concept that inflammation may play an important role in efficient reactivation, particularly in clinical scenarios (1, 23, 39).
MATERIALS AND METHODS
Ethics statement.
All research describing studies on primary human material with HCMV were assessed and approved by the Cambridge Local Research Ethics Committee. Informed consent was given for the collection of venous blood samples from healthy donors, and the collection was performed in accordance with established guidelines for the handling and processing of said tissue by the Cambridge Local Research Ethics Committee.
Cells and tissue culture.
Human foreskin fibroblasts (HFFs) were maintained in Eagle's minimal essential medium containing 10% fetal calf serum (EMEM-10) (Sigma-Aldrich, Poole, United Kingdom) and incubated at 37°C and in 5% CO2 by following standard procedure for tissue culture. Briefly, total peripheral blood mononuclear cells (PBMC) were isolated by Ficoll density-dependent centrifugation and then incubated with an antibody cocktail containing anti-CD3, -CD7, -CD16, -CD56, and -CD123 microbeads and separated on a magnetically activated cell sorting (MACS) column. The resultant flowthrough was confirmed to be T-cell receptor α/β (TCRα/β) negative and then incubated with an anti-CD4 microbead-conjugated antibody. The enriched fraction bound to the column was rescued and represented the DC fraction. Typically, the yield of DCs was around 0.5 to 1% of the total PBMC count. Where appropriate, DCs were stimulated with lipopolysaccharide (LPS; 500 ng/ml; Sigma-Aldrich, Poole, United Kingdom) or interleukin 6 (IL-6; 50 ng/ml; Peprotech EC, United Kingdom) to promote maturation or activation, respectively.
Virus infection and indirect immunofluorescence.
Following isolation, blood DCs were cultured on 8-well chamber slides in X-vivo 15 medium for 3 h and then infected overnight with HCMV strain TB40/e. To detect IE gene expression, infected cells were washed in phosphate-buffered saline (PBS) and fixed for 10 min in 4% paraformaldehyde at room temperature. After being permeabilized with 0.1% Triton X-100 in PBS, cells were incubated with monoclonal mouse anti-IE antibody (1:1,000 dilution in PBS; Millipore) for 1 h at room temperature. After a washing with PBS, the bound antibodies were detected using Alexa Fluor 594 (Millipore, Billerica, MA)-conjugated goat anti-mouse immunoglobulins (1:1,000 dilution in PBS) together with nuclear stain Hoechst (1:1,000 dilution in PBS) in the dark for 1 h at room temperature. To detect pp28 expression, cells 5 days postinfection were fixed and stained with rabbit anti-pp28 antibody (1:500 in PBS; Abcam, Cambridge, United Kingdom) and then detected using Alexa Fluor 594-conjugated goat anti-mouse immunoglobulins (1:1,000 dilution in PBS; Millipore) together with nuclear stain Hoechst (1:1,000 dilution in PBS; Sigma, Poole, United Kingdom) in the dark for 1 h at room temperature. After a washing with PBS, infected cells were visualized using a Nikon immunofluorescence microscope.
Nucleic acid isolation and analysis.
DNA isolation was performed using a sodium perchlorate method described previously which has been optimized for the isolation and detection of viral genomes from naturally latent mononuclear cells (40). Briefly, 106 cells were resuspended in 600 μl of buffer A (100 mM NaCl, 5 mM; pH 8.0), lysed with 10% SDS (125 μl), and then incubated with 5 M sodium perchlorate (150 μl). DNA was isolated by phenol-chloroform extraction and isopropanol precipitation. To eliminate salt contamination, total DNA was then dialyzed overnight at 4°C in circulating Tris-EDTA buffer. To analyze gene expression, total RNA was extracted from 106 cells using the RNeasy kit as described by the manufacturer (Qiagen, Sussex, United Kingdom) Contaminating genomic DNA was removed by a DNase I digestion (Promega, Madison, WI), followed by production of first-strand cDNA using the Promega RT system. Standard PCR was carried out using 2× PCR MasterMix (Promega) containing DNA polymerase, MgCl2, and deoxynucleoside triphosphates (dNTPs). To detect endogenous viral nucleic acids, two approaches were used; they have been described previously. Briefly, a single round (65 cycles) of IE-specific PCR (94°C for 40 s, 55°C for 40 s, and 72°C for 90 s) was used to amplify DNA (310 bp) or cDNA (196 bp) products using sense primer 5′-CGT CCT TGA CAC GAT GGA GT-3′ and antisense primer 5′-ATT CTT CGG CCA ACT CTG GA-3′. Following gel electrophoresis and Southern blotting, IE-specific sequences were detected using a 32P-radiolabeled probe generate using sense primer 5′-CCC TGA TAA TCC TGA CGA GG-3′ and antisense primer 5′-CAT AGT CTG CAG GAA CGT CGT-3′. Alternatively, nested IE primers were used to amplify target sequences by PCR with the following cycling conditions: 95°C for 5 min, then 20 to 35 cycles of 94°C for 1 min, 55°C for 40 s, and 72°C for 40 s, and then a final extension at 72°C for 10 min. IE72 was amplified with sense primer 5′-CAT CCA CAT CTC CCG CTT AT-3′ and antisense primer 5′-CAC GAC GTT CCT GCA GAC TAT G-3′ followed by nested PCR with sense primer 5′-GCG CCA GTG AAT TTC TCT TC and antisense primer 5′-ACG AGA ACC CCG AGA AAG ATG, yielding a final nested product of 302 bp (DNA) or 131 bp (cDNA). A 548-bp actin product was amplified using sense primer 5′-GCT CCG GCA TGT GCA-3′ and antisense primer 5′-AGG ATC TTC ATG AGG TAG T-3′ under the same PCR conditions.
ChIP and analysis.
All procedures were performed essentially as described previously (40). Briefly, DCs were fixed with 1% formaldehyde (10 min) and then lysed and sonicated to fragment DNA. DNA associated with histones was immunoprecipitated with control serum (Sigma, Poole, United Kingdom), anti-acetyl histone H4 antiserum (chromatin immunoprecipitation [ChIP] grade, 1:200 dilution; Upstate Biotechnology, Charlottesville, VA), anti-dimethyl lysine 4 histone H3 antiserum (ChIP grade, 1:200 dilution; Upstate Biotechnology), or anti-heterochromatin protein 1 (anti-HP1) antiserum (1:200 dilution; Serotec, Oxford, United Kingdom). For detection of the major immediate early promoter (MIEP) of HCMV, DNA from disrupted nucleosomes was precipitated and amplified by PCR with sense primer 5′-TGG GAC TTT CCT ACT TGG-3′ and antisense primer 5′-CCA GGC GAT CTG ACG GTT-3′, complementary to positions −272 and +13 relative to the MIEP start site. Amplified products were detected using a PCR product generated using sense primer 5′-ATT ACC ATG GTG ATG CGG TT-3′ and antisense primer 5′-GGC GGA GTT GTT ACG ACA T-3′, which was labeled with [32P]dCTP to allow detection by Southern blotting. All amplifications by PCR were performed with 2× Mastermix (Promega, Madison, WI). The cycle parameters for amplification by PCR were 95°C for 5 min and then 20 to 50 cycles at 94°C for 40 s, 50°C for 40 s, and 72°C for 90 s.
Amplification of the HS4 locus of the gamma globin gene by PCR was performed using sense primer 5′-TGG CAT CTA GCG CAA TGA CTT-3′ and antisense primer 5′-GGG CAA GCC ATC TCA TAG CTG-3′, which have been used in previous analyses of this region.
Cell surface phenotype flow cytometry analysis.
A total of 105 cells were pelleted at 400 × g for 5 min and were then resuspended in the residual volume. The cells were incubated with 3 μl of fluorescein isothiocyanate (FITC)-conjugated mouse anti-human CD1c, 5 μl of Alexa Fluor 488-conjugated mouse anti-human E-cadherin (R&D Systems, Abingdon, United Kingdom), 3 μl of phycoerythrin (PE)-conjugated mouse anti-human major histocompatibility complex class I, or 3 μl of allophycocyanin (APC)-conjugated mouse anti-human CD83, HLA-DR, or CD86 or the appropriate fluorochrome-conjugated mouse isotype control for 20 min in the dark. Following a washing in 10× volumes of PBS, the cells were pelleted at 400 × g for 5 min and were resuspended in 500 μl of PBS before analysis by flow cytometry (BD FACScalibur or BD FACSsort). Data handling was performed using WinMDI2.9 software. All antibodies were from BD Life Sciences (Franklin Lakes, NJ) unless otherwise stated. No differences in cell surface phenotype were observed whether DCs from seropositive or seronegative donors were analyzed.
RESULTS
Dendritic cells directly isolated from peripheral blood are permissive for HCMV infection.
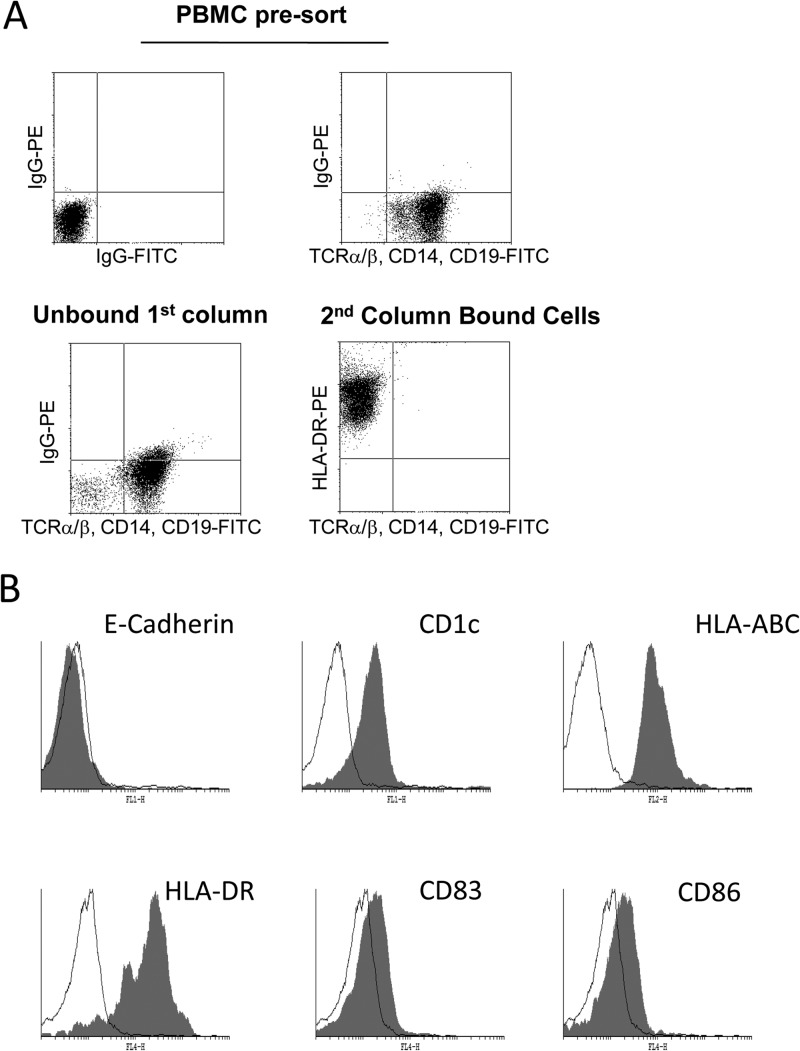
To isolate circulating DCs, we used a two-stage procedure that involved depletion of T lymphocytes, NK cells, and monocytes followed by a positive selection for CD4+ cells. A routine analysis of the isolated cells (which represented about 0.5% of the total PBMC) identified them as HLA-DR positive and TCRα/β, CD14, and CD19 negative (Fig. 1A), suggestive of enrichment of myeloid DCs (routinely >95%). Further characterization showed that these blood DCs were HLA-ABC, HLA-DR, and CD1c positive, CD83dim and CD86dim, and E-cadherin negative (Fig. 1B). This phenotype was consistent with a more immature dermal, and not Langerhans, DC phenotype (25), similar to that observed for classical MoDCs (20, 25), which have been used routinely to study HCMV latency and reactivation. Taken together, these data suggested that the circulating blood DCs isolated were predominantly of a dermal/interstitial-like phenotype, consistent with previous reports for the isolation of myeloid blood DCs from healthy donors (40, 41).
Fig 1.
Circulating myeloid dendritic cells can be isolated from peripheral blood. (A) Flow cytometric analysis was performed on peripheral blood mononuclear cells stained with FITC-conjugated anti-TCRα/β, -CD14, and -CD19 antibodies (top right) or isotype-matched antibody controls (top left). Following magnetic column depletion with anti-CD3, -CD7, -CD16, -CD56, and -CD123 antibodies, the unbound fraction was stained with FITC-conjugated anti-TCRα/β, -CD14, and -CD19 antibodies (bottom). CD4-positive cells were depleted from the unbound fraction and stained with FITC-conjugated anti-TCRα/β, -CD14, and -CD19 antibodies and a PE-conjugated anti-HLA-DR antibody. (B) Isolated DCs were stained with a panel of cell surface markers (filled histogram) or an isotype-matched control (open histogram) immediately postisolation.
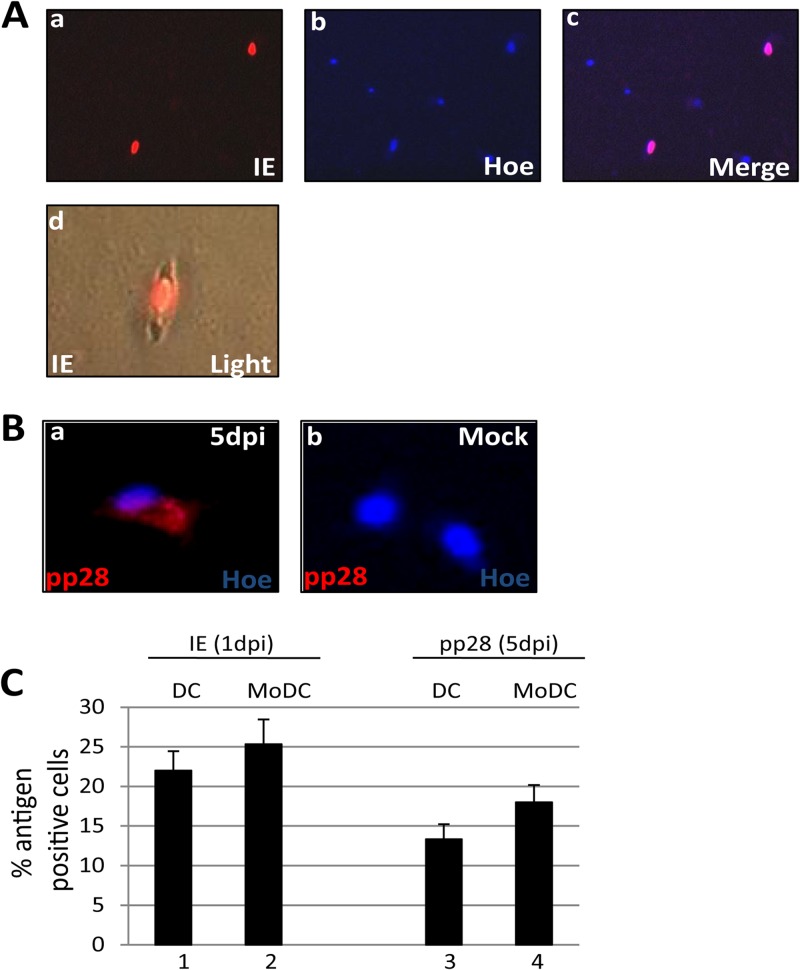
Given the phenotypic similarity between circulating blood DCs and MoDCs, we next asked whether these immature blood DCs were permissive for HCMV infection. Directly isolated blood DCs were infected with HCMV and stained for IE and pp28 gene expression at 1 and 5 days postinfection, respectively (Fig. 2). Indirect immunofluorescence for IE72/86 expression, 24 h postinfection (hpi), showed that directly isolated DCs supported HCMV lytic gene expression (Fig. 2A), consistent with a previous analysis of circulating DCs (38). Furthermore, we observed that the infection was not limited to IE protein expression, since the expression of the structural tegument protein, pp28, was clearly evident by 5 days postinfection (Fig. 2B). Finally, the level of infection of the purified DCs was similar to that observed following the infection of MoDCs generated from the same donor cells (Fig. 2C).
Fig 2.
Directly isolated DCs are permissive for HCMV gene expression. (A) Three hours postisolation, DCs were infected with a myelotropic stock of TB40/e and stained for IE gene (a) and nuclear DNA (b) 24 h postinfection. The merged image (c) shows nuclear IE gene expression. DC morphology after isolation and infection is shown in an IE gene-positive cell shown merged with bright-field image (d). (B) TB40/e-infected (a) or mock-infected (b) DCs were costained for pp28 expression and nuclear DNA 5 days postinfection. (C) DCs and monocyte-derived DCs infected with TB40/e were stained for IE (bars 1 and 2) or pp28 (bars 3 and 4) expression, and the number of antigen-positive cells was determined. Data show enumeration from 10 fields of view performed in triplicate. dpi, days postinfection.
Directly isolated DCs are HCMV genome positive and support the reactivation of HCMV ex vivo.
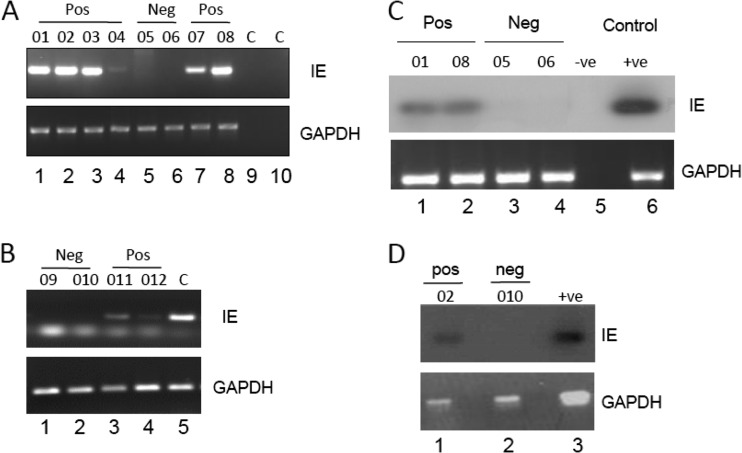
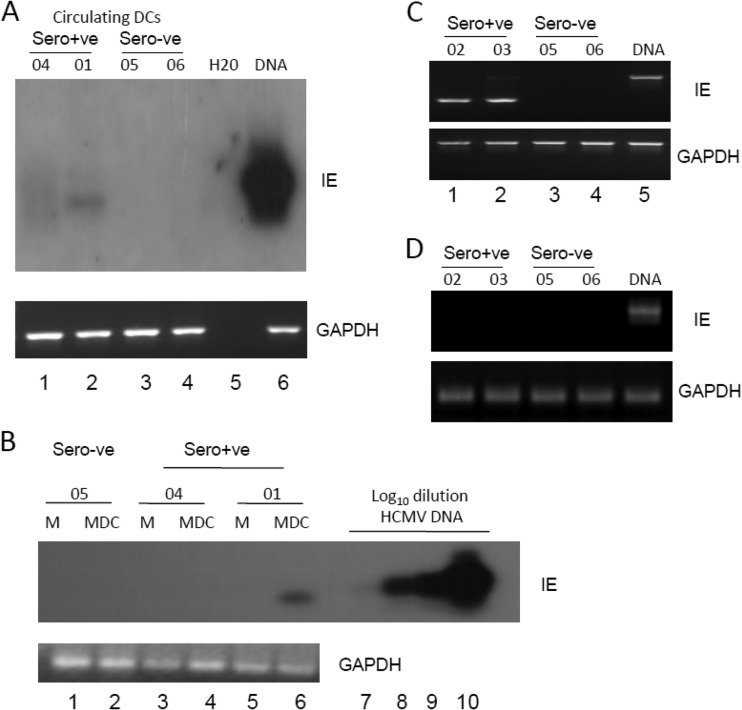
Having established that we could isolate purified populations of circulating DCs that supported lytic gene expression, we next asked whether these cells were sites of HCMV genome carriage in vivo. In order to test this, DCs were isolated directly from the blood concomitantly from seropositive and seronegative donors and analyzed using an IE-specific nested PCR in a number of independent analyses. The collated data clearly show that viral genomes were detectable in the DCs of 6 seropositive, but not seronegative, donors tested (Fig. 3A and B). It is interesting that although all seropositive donors were uniformly viral genome positive, the ability to detect viral genomes in the DCs of some donors compared to others differed. This was particularly evident with donor 04 (Fig. 3A, lane 4). It is interesting that this donor had recently seroconverted (within 1 year) around the time of our analyses, and the relative difficulty of detection of viral genomes in the donor's DCs was mirrored upon analyses of that individual's mobilized CD34+ cells or CD14+ monocyte populations also (12), suggesting that the latent load may accumulate with time. We then performed a second analysis using a single-round PCR followed by a radiolabeled probe detection step to provide further confirmation that we could detect HCMV genome sequences by PCR in circulating DCs directly isolated from selected healthy seropositive donors (Fig. 3C and D).
Fig 3.
Detection of HCMV DNA sequences in circulating blood DCs of seropositive donors. (A and B) Nested PCR analysis of DCs from multiple seropositive (donors 01 to 04, 07, 08, 011, and 012), seronegative (donors 05, 06, 09, and 010), or water (panel A, lanes 9 and 10) and DNA (panel B, lane 5) controls was performed using IE gene-specific (a) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) DNA-specific (b) PCRs. Nested PCR gave rise to a 312-bp product from HCMV DNA. (C and D) A single-round IE gene PCR analysis on three seropositive (donors 01, 02, and 08), three seronegative donors (05, 06, and 010), and a water (panel C, lane 5) or DNA (panel C, lane 6, and panel D, lane 3) control was analyzed by Southern blotting and probing with an IE gene-specific probe that detected a 310-bp product. A GAPDH PCR was used to confirm the isolation of DNA.
Confident that we could detect the carriage of HCMV genome in purified populations of circulating DCs isolated from healthy seropositive persons, we next addressed whether we could detect viral transcription. The carriage of viral genomes in the circulating monocytes of healthy seropositive donors has been shown to occur in an absence of detectable viral lytic gene expression (42). However, it is well established that differentiation of monocytes to MoDCs in ex vivo culture results in reactivation of latent virus, which would suggest that circulating DCs could be potential sites of reactivation in vivo. Thus, we next analyzed the transcriptional state of the HCMV genomes in the circulating DCs (Fig. 4). To do this, we performed a reverse transcription-PCR (RT-PCR) analysis of circulating DCs immediately after isolation from peripheral blood. The data show that IE gene expression is detectable in DCs isolated directly from seropositive donors (3/4 donors) (Fig. 4). Again, the donor in whom we failed to detect IE gene expression was donor 04, the donor of the DCs in which it was more difficult to detect viral genomes (Fig. 3). Furthermore, the in vitro differentiation of donor 04's monocytes to MoDCs also resulted in a failure to detect the expression of IE gene expression, unlike with donor 01 (Fig. 4B). Additionally, the failure to detect IE gene transcription in concomitantly isolated monocytes from the remaining donors (Fig, 4B and D) is indicative that these healthy volunteers were not viremic at the time of blood donation. In total, we were able to detect the expression of IE RNA in the circulating DCs of 6/7 donors tested for reactivation (Fig. 4; see also Fig. 6).
Fig 4.
Detection of IE RNA transcripts in circulating blood DCs of seropositive donors. (A) RNA prepared from DCs directly isolated from the peripheral blood of seropositive (donors 04 and 01) and seronegative (donors 05 and 06) donors was amplified in an IE gene RT-PCR. Water or HCMV DNA was amplified as a control for the viral PCRs (lane 5 and 6). Amplified products were then analyzed by Southern blotting with an IE gene-specific probe. (B) Monocytes isolated from donors 04, 01, and 05 were analyzed either directly postisolation (lanes 1, 3, and 5) or after differentiation to MoDCs (lanes 2, 4, and 6) for IE RNA or GAPDH expression by Southern blotting. A log dilution of viral DNA was used as a positive control (lanes 7 to 10). (C and D) RNA extracted from DCs (C) or monocytes (D) isolated from healthy seropositive (donors 02 and 03) or seronegative (donors 05 and 06) donors was analyzed using a nested intron-spanning RT-PCR for IE72 gene expression. Viral DNA was amplified as a control (lane 5). GAPDH controls are shown (lanes 1 to 5).
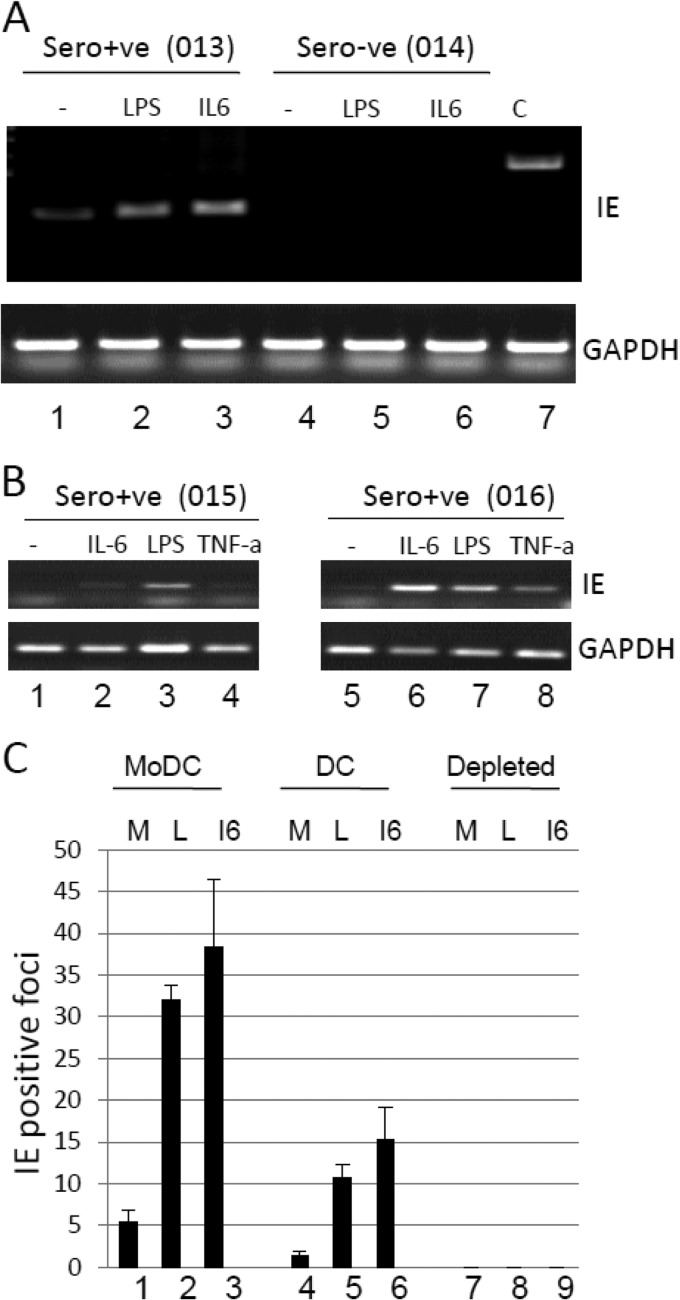
Fig 6.
Inflammatory stimulation of DCs enhances reactivation of HCMV. (A) Isolated DCs from a seropositive (donor 013) or seronegative (donor 014) donor were cultured alone (lane 1) or with LPS (lane 2) or IL-6 (lane 3) and analyzed for IE72 and GAPDH RNA expression 16 h poststimulation. (B) Isolated DCs from two further seropositive donors (donors 015 and 016) were left unstimulated (lanes 1 and 5) or stimulated with IL-6 (lanes 2 and 6), LPS (lanes 3 and 7), or TNF-α (lanes 4 and 8) and analyzed for IE72 or GAPDH RNA expression 16 h poststimulation. (C) To test for viral reactivation, 5 × 105 MoDCs (lanes 1 to 3), directly isolated DCs (lanes 4 to 6), or DC- and monocyte-depleted cells (lanes 7 to 9) were cultured alone (M) or with LPS (L) or IL-6 (I6) and then cocultured with fibroblasts for 21 days. The fibroblast monolayer was then stained for IE gene expression and the average number of IE gene-positive foci scored from 5 fields of view.
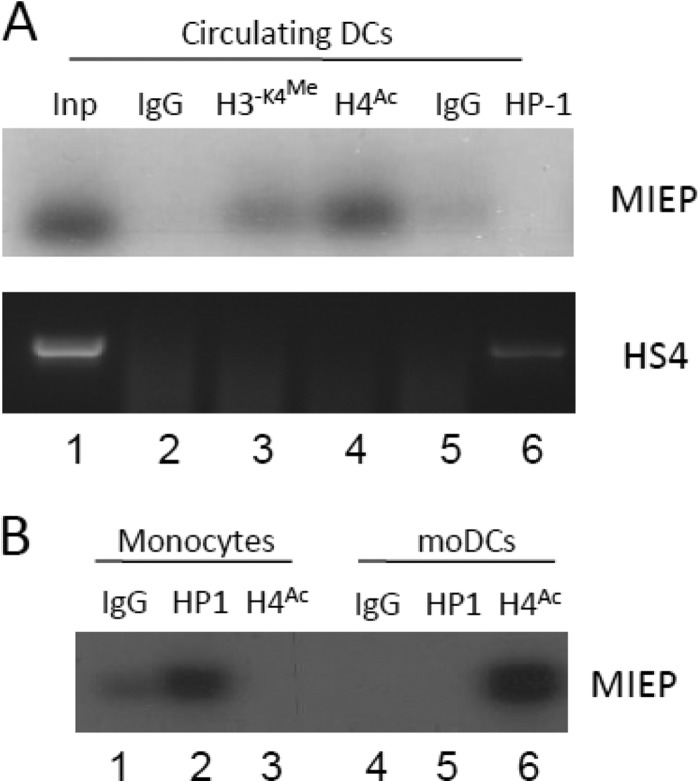
Consistent with the detection of viral lytic gene expression in the isolated DCs, chromatin immunoprecipitation analyses of the histone occupancy of the MIEP in the circulating DCs of a healthy seropositive donor was also consistent with IE transcription (Fig. 5). Specifically, the MIEP was predominantly immunoprecipitated with antibodies recognizing dimethylated H3-K4 and acetylated H4 histones (both markers of transcriptional activation) but not the marker of transcriptionally repressed chromatin HP1 (Fig. 5A). Again, an analysis of the same donor monocytes exhibited the opposite phenotype. The MIEP in these cells was associated predominantly with the marker of transcriptional repression, HP1 (Fig. 5B). Furthermore, the in vitro differentiation to a DC phenotype was concomitant with the MIEP being predominantly associated with acetylated histones, consistent with these cells supporting HCMV reactivation (12, 21) and our previous studies of CD34+ cells and the DCs derived from these (12).
Fig 5.
The MIEP in circulating DCs is predominantly associated with markers of transcriptional activation. (A) Chromatin immunoprecipitation assays were performed on seropositive DCs with rabbit IgG (lane 2), anti-dimethylated histone H3-K4 (lane 3), anti-acetylated H4 (lane 4), anti-rat IgG (lane 5), or anti-HP1β (lane 6) antibodies. DNA amplified in an MIEP-specific PCR was then analyzed by Southern blotting using an MIEP-specific probe. Alternatively, samples were amplified using primers against the HS4 region of the gamma globulin gene. (B) Monocytes isolated from the same donor were either subjected to ChIP (lanes 1 to 3) or differentiated to MoDCs and then subjected to ChIP analysis (lanes 4 to 6). Immunoprecipitation with IgG (lanes 1 and 3), anti-HP-1 (lanes 2 and 5) or anti-acetylated histone H4 (lanes 3 and 6) was performed, and samples were amplified in an MIEP PCR and analyzed by Southern blotting.
Reactivation of HCMV from DCs is elevated upon inflammatory stimulation and results in the formation of infectious centers.
These data suggested that during the natural differentiation of blood DCs in vivo, a switch from an HCMV latent to an HCMV reactivating phenotype could be occurring. However, in vivo, symptomatic HCMV reactivation and disease are often associated with immunosuppression and highly inflammatory environments, and thus, we next asked whether known inflammatory mediators impacted the level of HCMV reactivation from circulating DCs ex vivo. Circulating DCs, isolated from healthy donors, were stimulated with a number of inflammatory mediators and analyzed for IE RNA (24 h poststimulation) or IE gene-positive fibroblasts (21 days after coculture with 5 × 105 mononuclear cells) (Fig. 6). The latter approach, the direct staining of cocultures rather than 50% tissue culture infective dose (TCID50) analysis on supernatants for infectious virus, was employed because we could obtain only limited numbers of circulating DCs and only a small minority of these would likely be HCMV genome positive (13) and is an approach that has been used previously for studying reactivation events from natural latency with limited cell numbers (12, 20, 43). As observed before (Fig. 4A), circulating DCs showed detectable IE gene expression (Fig. 6A), and this was elevated following maturation with LPS or stimulation with IL-6 (Fig. 6A and B) or tumor necrosis factor alpha (TNF-α) (Fig. 6B), two cytokines that correlate with HCMV reactivation episodes in vivo (11, 44). We next analyzed infectious-virus production in the DC cocultures, and as an additional set of controls, we analyzed infectious virus production from MoDCs and the monocyte- and DC-depleted fraction (predominantly lymphocytes) from naturally latent donors. Consistent with previous analyses (12, 21), HCMV reactivation was detectable in immature MoDCs after ex vivo differentiation of monocytes to MoDCs in culture, and this was elevated following activation with LPS and IL-6 (Fig. 6C). Similarly, directly isolated DCs exhibited a similar response: virus reactivation was elevated by further stimulation with LPS or IL-6 (Fig. 6C). In contrast, no HCMV reactivation was detectable in the monocyte- and DC-depleted fraction (predominantly lymphocytes), consistent with these cells not being major sites of viral persistence in the peripheral blood (14). Taken together, these data argue that naturally circulating DCs can support the reactivation of HCMV ex vivo and that further inflammatory stimulation augments the reactivation of virus observed in DCs.
DISCUSSION
There exists a wealth of data from a number of laboratories that strongly supports the myeloid lineage as a major site of HCMV latency and reactivation in vivo (reviewed in references 19 and 45). These data are consistent with the view that HCMV latency is established in early progenitor cells in the bone marrow, the viral genome then persists in the myeloid/monocyte compartment in the peripheral blood without detectable viremia, and finally, terminal differentiation of latent monocytes to a macrophage or DC phenotype instigates a chain of events leading to reactivation of the virus in the periphery due to a more favorable transcriptional environment for MIEP activity and that this is likely augmented by inflammatory signaling in vivo (1, 46). The paucity of circulating myeloid DCs in the peripheral blood compartment has led many investigators to use in vitro differentiation of early myeloid progenitors to model HCMV reactivation (4, 5, 20–22, 47–49). However, we wished to determine whether the robust link between HCMV reactivation and experimental differentiation of monocytes to DCs ex vivo was recapitulated in differentiated circulating myeloid DCs in vivo: in that case, direct isolation of the DC compartment directly from peripheral blood of healthy seropositive donors should yield genome-positive cells that can support HCMV reactivation without further ex vivo differentiation.
Herein, we have shown that CD1c-positive myeloid DCs from peripheral blood are phenotypically similar to MoDCs based on cell surface marker expression (20, 40). Consistent with their cellular phenotype, the circulating DCs exhibited the same permissiveness for HCMV as MoDCs (50): infection of the cells directly postisolation resulted in the detection of IE protein-positive cells and, by day 5, pp28-positive cells. Thus, circulating DCs encountering HCMV in vivo are likely to be fully permissive for HCMV infection.
More pertinent to our ongoing studies of HCMV latency and reactivation, the detection of transcriptionally active genomes in the circulating DCs of healthy seropositive donors is highly supportive of the hypothesis that predicts (based on data derived from studies using in vitro differentiation) that terminally differentiated myeloid cells such as DCs support HCMV reactivation in vivo. We note that these analyses represent a global analysis of purified DC populations, and thus, there is the potential that during the purification we may isolate a minor contaminating fraction. Arguing against this is that the isolation of CD34+ hematopoietic cells and their derivatives, CD14+ monocytes, lymphocyte populations, or granulocytes using similar procedures does not result in the detection of IE RNA-positive cells (5–7, 12, 14, 21, 42, 51, 52). Isolation of a contaminating cell only in our DC isolations and not any of these other procedures appears highly unlikely but cannot be dismissed. These caveats aside, we were particularly intrigued by the identification of IE gene expression in purified DCs in the absence of any additional stimulation, which suggests that the normal differentiation of DCs in vivo is sufficient to trigger HCMV reactivation. While we cannot formally rule out the possibility that the protocol for isolating circulating DCs resulted in the activation of the cells and, pertinently, activation of the MIEP, we note that the isolation of circulating DCs was complete in less than 3 h and that activation of the MIEP does not occur in monocytes isolated in a similar way (data shown herein and references 12 and 14). This suggests that circulating DCs are capable of supporting reactivation of viral lytic gene expression in vivo. Furthermore, we fully appreciate that it is impossible to formally preclude the possibility that the isolated circulating DCs were undergoing a de novo infection of HCMV in vivo at the time of harvest. However, we have performed our analyses on healthy aviremic donors, and thus, we assume that there is minimal de novo infection of circulating DCs occurring at the times of analyses. Furthermore, concomitant analyses of donors' monocytes showed that these cells were not IE transcript positive, suggesting that these donors were not viremic at the time of analysis. Notwithstanding this, if circulating DCs do express IE RNA, then our data support the view that DCs could be an important site of viral reactivation in vivo. Indeed, it is very likely that this reactivation of viral IE gene expression in circulating DCs in vivo is driven by the differentiation of DC precursors to a DC phenotype and would argue that sporadic reactivation events, albeit at very low frequency, routinely occur upon DC differentiation in vivo as HCMV genome-positive myeloid progenitors differentiate into DCs. Whether this reactivation in vivo is abortive or fully permissive is still uncertain, but our data would suggest that circulating DCs are potential sites of reactivation of the initial events necessary for infectious-virus production. It is possible, however, that the reactivation of IE gene expression in the circulating DCs of healthy individuals is sufficient to trigger their recognition by the high numbers of IE-specific cytotoxic T cells (CTLs) known to be present in healthy seropositive donors, which results in their rapid elimination by a normal IE-specific CTL response. This model would be consistent with the well-established high frequency of memory CTLs directed against HCMV—the possible result of a constant repriming of the immune system (53). Indeed, in the absence of this normal CTL response, such reactivation events would become uncontrolled, resulting in the severe disease observed in immunocompromised patients (1, 2).
In conclusion, to date it has been hypothesized that differentiation of myeloid progenitor cells to differentiated macrophages and DCs in vivo results in the reactivation of latent virus (5, 8, 12, 23, 42, 54). However, this hypothesis has been based on the ex vivo differentiation of myeloid progenitors. We now show that differentiated myeloid DCs of healthy seropositive HCMV carriers in vivo are also sites of HCMV reactivation. These data provide direct evidence for the hypothesis that myeloid cell differentiation in vivo is an important event for the initiation of HCMV reactivation. Furthermore, they provide strong support for the use of experimental latency models employed by a number of laboratories looking to understand the mechanisms essential for the maintenance of HCMV latency and subsequent control of reactivation in the cells of the myeloid lineage.
ACKNOWLEDGMENTS
J.H.S. is supported by an MRC programme grant (G:0701279) and M.B.R. by an MRC CDA Fellowship (G:0900466).
Footnotes
Published ahead of print 24 July 2013
REFERENCES
- 1.Limaye AP, Kirby KA, Rubenfeld GD, Leisenring WM, Bulger EM, Neff MJ, Gibran NS, Huang ML, Santo Hayes TK, Corey L, Boeckh M. 2008. Cytomegalovirus reactivation in critically ill immunocompetent patients. JAMA 300:413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peggs KS, Mackinnon S. 2004. Cytomegalovirus: the role of CMV post-haematopoietic stem cell transplantation. Int. J. Biochem. Cell Biol. 36:695–701 [DOI] [PubMed] [Google Scholar]
- 3.Revello MG, Gerna G. 2004. Pathogenesis and prenatal diagnosis of human cytomegalovirus infection. J. Clin. Virol. 29:71–83 [DOI] [PubMed] [Google Scholar]
- 4.Goodrum FD, Jordan CT, High K, Shenk T. 2002. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc. Natl. Acad. Sci. U. S. A. 99:16255–16260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn G, Jores R, Mocarski ES. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. U. S. A. 95:3937–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendelson M, Monard S, Sissons P, Sinclair J. 1996. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J. Gen. Virol. 77(Part 12):3099–3102 [DOI] [PubMed] [Google Scholar]
- 7.Sindre H, Tjoonnfjord GE, Rollag H, Ranneberg-Nilsen T, Veiby OP, Beck S, Degre M, Hestdal K. 1996. Human cytomegalovirus suppression of and latency in early hematopoietic progenitor cells. Blood 88:4526–4533 [PubMed] [Google Scholar]
- 8.Zhuravskaya T, Maciejewski JP, Netski DM, Bruening E, Mackintosh FR, St Jeor S. 1997. Spread of human cytomegalovirus (HCMV) after infection of human hematopoietic progenitor cells: model of HCMV latency. Blood 90:2482–2491 [PubMed] [Google Scholar]
- 9.Bevan IS, Daw RA, Day PJ, Ala FA, Walker MR. 1991. Polymerase chain reaction for detection of human cytomegalovirus infection in a blood donor population. Br. J. Haematol. 78:94–99 [DOI] [PubMed] [Google Scholar]
- 10.Kondo K, Kaneshima H, Mocarski ES. 1994. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc. Natl. Acad. Sci. U. S. A. 91:11879–11883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prosch S, Docke WD, Reinke P, Volk HD, Kruger DH. 1999. Human cytomegalovirus reactivation in bone-marrow-derived granulocyte/monocyte progenitor cells and mature monocytes. Intervirology 42:308–313 [DOI] [PubMed] [Google Scholar]
- 12.Reeves MB, MacAry PA, Lehner PJ, Sissons JG, Sinclair JH. 2005. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc. Natl. Acad. Sci. U. S. A. 102:4140–4145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slobedman B, Mocarski ES. 1999. Quantitative analysis of latent human cytomegalovirus. J. Virol. 73:4806–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor-Wiedeman J, Sissons JG, Borysiewicz LK, Sinclair JH. 1991. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 72(Part 9):2059–2064 [DOI] [PubMed] [Google Scholar]
- 15.Slobedman B, Cao JZ, Avdic S, Webster B, McAllery S, Cheung AK, Tan JC, Abendroth A. 2010. Human cytomegalovirus latent infection and associated viral gene expression. Future Microbiol. 5:883–900 [DOI] [PubMed] [Google Scholar]
- 16.De Witte T, Schattenberg A, Van Dijk BA, Galama J, Olthuis H, Van der Meer JW, Kunst VA. 1990. Prevention of primary cytomegalovirus infection after allogeneic bone marrow transplantation by using leukocyte-poor random blood products from cytomegalovirus-unscreened blood-bank donors. Transplantation 50:964–968 [DOI] [PubMed] [Google Scholar]
- 17.Gilbert GL, Hayes K, Hudson IL, James J. 1989. Prevention of transfusion-acquired cytomegalovirus infection in infants by blood filtration to remove leucocytes. Neonatal Cytomegalovirus Infection Study Group. Lancet i:1228–1231 [DOI] [PubMed] [Google Scholar]
- 18.Jordan MC. 1983. Latent infection and the elusive cytomegalovirus. Rev. Infect. Dis. 5:205–215 [DOI] [PubMed] [Google Scholar]
- 19.Bego MG, St Jeor S. 2006. Human cytomegalovirus infection of cells of hematopoietic origin: HCMV-induced immunosuppression, immune evasion, and latency. Exp. Hematol. 34:555–570 [DOI] [PubMed] [Google Scholar]
- 20.Huang MM, Kew VG, Jestice K, Wills MR, Reeves MB. 2012. Efficient human cytomegalovirus reactivation is maturation dependent in the Langerhans dendritic cell lineage and can be studied using a CD14+ experimental latency model. J. Virol. 86:8507–8515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reeves MB, Compton T. 2011. Inhibition of inflammatory interleukin-6 activity via extracellular signal-regulated kinase-mitogen-activated protein kinase signaling antagonizes human cytomegalovirus reactivation from dendritic cells. J. Virol. 85:12750–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeves MB, Lehner PJ, Sissons JG, Sinclair JH. 2005. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J. Gen. Virol. 86:2949–2954 [DOI] [PubMed] [Google Scholar]
- 23.Söderbérg-Naucler C, Fish KN, Nelson JA. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119–126 [DOI] [PubMed] [Google Scholar]
- 24.Geissmann F, Prost C, Monnet JP, Dy M, Brousse N, Hermine O. 1998. Transforming growth factor beta1, in the presence of granulocyte/macrophage colony-stimulating factor and interleukin 4, induces differentiation of human peripheral blood monocytes into dendritic Langerhans cells. J. Exp. Med. 187:961–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grassi F, Dezutter-Dambuyant C, McIlroy D, Jacquet C, Yoneda K, Imamura S, Boumsell L, Schmitt D, Autran B, Debre P, Hosmalin A. 1998. Monocyte-derived dendritic cells have a phenotype comparable to that of dermal dendritic cells and display ultrastructural granules distinct from Birbeck granules. J. Leukoc. Biol. 64:484–493 [DOI] [PubMed] [Google Scholar]
- 26.MacAry PA, Lindsay M, Scott MA, Craig JI, Luzio JP, Lehner PJ. 2001. Mobilization of MHC class I molecules from late endosomes to the cell surface following activation of CD34-derived human Langerhans cells. Proc. Natl. Acad. Sci. U. S. A. 98:3982–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merad M, Ginhoux F, Collin M. 2008. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol. 8:935–947 [DOI] [PubMed] [Google Scholar]
- 28.Peiser M, Wanner R, Kolde G. 2004. Human epidermal Langerhans cells differ from monocyte-derived Langerhans cells in CD80 expression and in secretion of IL-12 after CD40 cross-linking. J. Leukoc. Biol. 76:616–622 [DOI] [PubMed] [Google Scholar]
- 29.Sallusto F, Lanzavecchia A. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 179:1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang WL, Baumgarth N, Yu D, Barry PA. 2004. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J. Virol. 78:8720–8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grigoleit U, Riegler S, Einsele H, Laib Sampaio K, Jahn G, Hebart H, Brossart P, Frank F, Sinzger C. 2002. Human cytomegalovirus induces a direct inhibitory effect on antigen presentation by monocyte-derived immature dendritic cells. Br. J. Haematol. 119:189–198 [DOI] [PubMed] [Google Scholar]
- 32.Hertel L, Lacaille VG, Strobl H, Mellins ED, Mocarski ES. 2003. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J. Virol. 77:7563–7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moutaftsi M, Brennan P, Spector SA, Tabi Z. 2004. Impaired lymphoid chemokine-mediated migration due to a block on the chemokine receptor switch in human cytomegalovirus-infected dendritic cells. J. Virol. 78:3046–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moutaftsi M, Mehl AM, Borysiewicz LK, Tabi Z. 2002. Human cytomegalovirus inhibits maturation and impairs function of monocyte-derived dendritic cells. Blood 99:2913–2921 [DOI] [PubMed] [Google Scholar]
- 35.Raftery MJ, Schwab M, Eibert SM, Samstag Y, Walczak H, Schonrich G. 2001. Targeting the function of mature dendritic cells by human cytomegalovirus: a multilayered viral defense strategy. Immunity 15:997–1009 [DOI] [PubMed] [Google Scholar]
- 36.Sénéchal B, Boruchov AM, Reagan JL, Hart DN, Young JW. 2004. Infection of mature monocyte-derived dendritic cells with human cytomegalovirus inhibits stimulation of T-cell proliferation via the release of soluble CD83. Blood 103:4207–4215 [DOI] [PubMed] [Google Scholar]
- 37.Varani S, Frascaroli G, Homman-Loudiyi M, Feld S, Landini MP, Soderberg-Naucler C. 2005. Human cytomegalovirus inhibits the migration of immature dendritic cells by down-regulating cell-surface CCR1 and CCR5. J. Leukoc. Biol. 77:219–228 [DOI] [PubMed] [Google Scholar]
- 38.Kvale EO, Dalgaard J, Lund-Johansen F, Rollag H, Farkas L, Midtvedt K, Jahnsen FL, Brinchmann JE, Olweus J. 2006. CD11c+ dendritic cells and plasmacytoid DCs are activated by human cytomegalovirus and retain efficient T cell-stimulatory capability upon infection. Blood 107:2022–2029 [DOI] [PubMed] [Google Scholar]
- 39.Prosch S, Wuttke R, Kruger DH, Volk HD. 2002. NF-kappaB—a potential therapeutic target for inhibition of human cytomegalovirus (re)activation? Biol. Chem. 383:1601–1609 [DOI] [PubMed] [Google Scholar]
- 40.MacDonald KP, Munster DJ, Clark GJ, Dzionek A, Schmitz J, Hart DN. 2002. Characterization of human blood dendritic cell subsets. Blood 100:4512–4520 [DOI] [PubMed] [Google Scholar]
- 41.Robinson SP, Patterson S, English N, Davies D, Knight SC, Reid CD. 1999. Human peripheral blood contains two distinct lineages of dendritic cells. Eur. J. Immunol. 29:2769–2778 [DOI] [PubMed] [Google Scholar]
- 42.Taylor-Wiedeman J, Sissons P, Sinclair J. 1994. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 68:1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weekes MP, Tan SY, Poole E, Talbot S, Antrobus R, Smith DL, Montag C, Gygi SP, Sinclair JH, Lehner PJ. 2013. Latency-associated degradation of the MRP1 drug transporter during latent human cytomegalovirus infection. Science 340:199–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Humar A, St Louis P, Mazzulli T, McGeer A, Lipton J, Messner H, MacDonald KS. 1999. Elevated serum cytokines are associated with cytomegalovirus infection and disease in bone marrow transplant recipients. J. Infect. Dis. 179:484–488 [DOI] [PubMed] [Google Scholar]
- 45.Sinclair J, Sissons P. 2006. Latency and reactivation of human cytomegalovirus. J. Gen. Virol. 87:1763–1779 [DOI] [PubMed] [Google Scholar]
- 46.Reinke P, Prosch S, Kern F, Volk HD. 1999. Mechanisms of human cytomegalovirus (HCMV) (re)activation and its impact on organ transplant patients. Transpl. Infect. Dis. 1:157–164 [DOI] [PubMed] [Google Scholar]
- 47.Hargett D, Shenk TE. 2010. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc. Natl. Acad. Sci. U. S. A. 107:20039–20044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khaiboullina SF, Maciejewski JP, Crapnell K, Spallone PA, Dean Stock A, Pari GS, Zanjani ED, Jeor SS. 2004. Human cytomegalovirus persists in myeloid progenitors and is passed to the myeloid progeny in a latent form. Br. J. Haematol. 126:410–417 [DOI] [PubMed] [Google Scholar]
- 49.Slobedman B, Mocarski ES, Arvin AM, Mellins ED, Abendroth A. 2002. Latent cytomegalovirus down-regulates major histocompatibility complex class II expression on myeloid progenitors. Blood 100:2867–2873 [DOI] [PubMed] [Google Scholar]
- 50.Riegler S, Hebart H, Einsele H, Brossart P, Jahn G, Sinzger C. 2000. Monocyte-derived dendritic cells are permissive to the complete replicative cycle of human cytomegalovirus. J. Gen. Virol. 81:393–399 [DOI] [PubMed] [Google Scholar]
- 51.Kondo K, Xu J, Mocarski ES. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. U. S. A. 93:11137–11142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taylor-Wiedeman J, Hayhurst GP, Sissons JG, Sinclair JH. 1993. Polymorphonuclear cells are not sites of persistence of human cytomegalovirus in healthy individuals. J. Gen. Virol. 74(Part 2):265–268 [DOI] [PubMed] [Google Scholar]
- 53.Jackson SE, Mason GM, Wills MR. 2011. Human cytomegalovirus immunity and immune evasion. Virus Res. 157:151–160 [DOI] [PubMed] [Google Scholar]
- 54.Minton EJ, Tysoe C, Sinclair JH, Sissons JG. 1994. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J. Virol. 68:4017–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]