Abstract
HLA-B*57 is strongly associated with immune control of HIV and delayed AIDS progression. The closely related, but less protective, HLA-B*58:01 presents similar epitopes, but HLA-B*58:01+ individuals do not generate CD8+ T cells targeting the KF11-Gag epitope, which has been linked to low viremia. Here we show that HLA-B*58:01 binds and presents KF11 peptide, but HIV-infected HLA-B*58:01+ cells fail to process KF11. This unexpected finding demonstrates that immunodominance patterns can be influenced by intracellular events independent of HLA binding motifs.
TEXT
Intracellular processing events critically influence the epitopes presented by HLA class I molecules on the surfaces of virus-infected cells to specific CD8+ T cells (21). HIV-specific CD8+ T-cell responses and their HLA restriction element together have a substantial impact on HIV disease progression (4, 5, 7, 18, 22). Recent analyses of three closely related HLA molecules, HLA-B*57:02, HLA-B*57:03, and HLA-B*58:01, suggested that the nature of responses to four immunodominant p24-Gag epitopes has a significant impact on viral load set point (13). Response to the KF11 (KAFSPEVIPMF) Gag epitope (6) is associated with improved HIV immune control (10) and was observed in HLA-B*57:03- but not in HLA-B*58:01-positive individuals, despite these HLA-B alleles' having apparently similar peptide binding motifs (13, 17, 19). Virus sequencing data show that this is not due to selection of viral escape in HLA-B*58:01-positive individuals (1, 13). To better understand the basis for this difference, we examined the presentation of KF11 by HLA-B*57:03 and the closely related HLA-B*57:01 and HLA-B*58:01 molecules. In this study, we identified an unexpected effect of the HLA molecule on intracellular processing of viral epitopes, overriding the capacity of that HLA molecule to bind and present particular peptides that is ultimately important for HIV immune control.
KF11 Gag is presented by HLA-B*57:01 and B*57:03, not by HLA-B*58:01.
We showed previously that KF11 Gag is dominantly targeted by HLA-B*57:03 in a large C-clade-infected cohort, with strong selection of A163X and S165X escape mutations (1, 3, 13). To further examine presentation of this epitope by closely related HLA-B molecules, we extended this analysis to include HLA-B*57:01, the HLA-B*57 allele most frequently expressed in Caucasians with B-clade infection together with HLA-B*58:01, expressed in 12% of C-clade-infected individuals (13, 16). HLA-B*57:01 differs from HLA-B*57:03 at only two HLA residues (residues 114 and 116) and from HLA-B*58:01 at three residues (HLA residues 45, 46, and 97) involved in forming the B and F pockets, respectively (Table 1).
Table 1.
HLA-B*57:01, -B*57:03, and -B*58:01 amino acid sequence differences
| Residue typea | Antigen | Residue at positionb |
Preferred residue atc: | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 9 | 24 | 45 | 46 | 63 | 66 | 67 | 99 | P2 | ||
| Residues determing B pocket | HLA-B*57:01 | Y | A | M | A | E | N | M | Y | S/T/A |
| HLA-B*57:03 | — | — | — | — | — | — | — | — | S/T/A | |
| HLA-B*58:01 | — | — | T | E | — | — | — | — | S/T/A | |
| 77 | 80 | 81 | 94 | 95 | 97 | 114 | 116 | PC | ||
| Residues determining F pocket | HLA-B*57:01 | N | I | A | I | I | V | D | S | F/W |
| HLA-B*57:03 | — | — | — | — | — | — | N | Y | F/W | |
| HLA-B*58:01 | — | — | — | — | — | R | — | — | F/W | |
| 103 | 156 | 194 | 282 | 305 | 325 | |||||
| Residues not in direct contact with B or F pockets | HLA-B*57:01 | V | L | I | V | A | C | |||
| HLA-B*57:03 | — | — | — | — | — | — | ||||
| HLA-B*58:01 | L | — | V | I | T | S | ||||
Strong cytotoxic-T-lymphocyte (CTL) responses by HLA-B*58:01-positive individuals are made only to the HIV p24-Gag epitopes TW10 and QW9, whereas HLA-B*57:03- and HLA-B*57:01-positive individuals also recognize the ISW9 and KF11 epitopes (13). A response to KF11 p24-Gag is specifically detected in 73% and 59% of HIV-infected subjects expressing HLA-B*57:01 and -B*57:03, respectively (P = 2 × 10−10 and 2 × 10−15) (Fig. 1) and not in subjects expressing HLA-B*58:01. In a previous study, 9 HIV-specific epitopes, out of 16 that were seen to be recognized in subjects expressing HLA-B*57:03, were also recognized in subjects expressing HLA-B*58:01, and two additional epitopes were recognized by subjects expressing HLA-B*58:01 that were not also recognized by subjects expressing HLA-B*57:03 (13). Thus, residues at positions 45, 46, and 97 of the HLA-B molecule affect immunogenicity of a number HIV-specific epitopes, including the KF11 Gag epitope, which is believed to contribute to immune control of HIV mediated by HLA-B*57:03 (10).
Fig 1.

KF11 Gag is targeted significantly more by individuals expressing HLA-B*57:01 or HLA-B*57:03 than by those expressing HLA-B*58:01. Data were extracted from a large C-clade cohort of 1,010 chronically HIV-infected individuals from Durban, South Africa, and a B-clade cohort from Oxford, United Kingdom. HLA-B*57:01-, HLA-B*57:03-, and HLA-B*58:01-expressing individuals were mutually excluded from groups expressing the other two HLA-B alleles. The 18-mer overlapping peptide containing KF11 (underlined), WVKVIEEKAFSPEVIPMF, was used to identify ex vivo IFN-γ ELISpot responses. P values were determined by Fisher's exact test.
The KF11 peptide can be presented by HLA-B*57:01, -B*57:03, and -B*58:01 to KF11-specific CD8+ T cells.
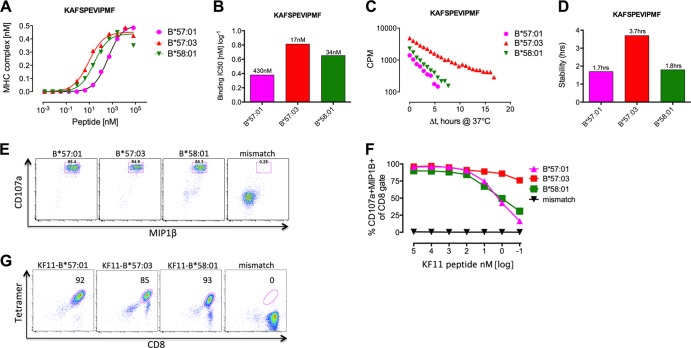
A potential explanation for this failure of KF11 to be recognized by subjects expressing HLA-B*58:01 would be reduced binding affinity of KF11 for HLA-B*58:01 compared to HLA-B*57:01 and HLA-B*57:03. Similar HLA-B molecules with apparently identical peptide binding motifs can exhibit very different peptide binding affinities and stability off-rates to several HIV epitopes (11, 13). To examine whether this could explain the differential KF11 epitope immunogenicity observed between HLA-B*57:01/03 and HLA-B*58:01, we tested the KF11 peptide binding affinity and binding half-life (stability) using previously published approaches based on refolding (8) or dissociation (9) of peptide, HLA, and β2 microglobulin (β2m) complexes, respectively, in a recombinant in vitro system. Unexpectedly KF11 has a very similar or even somewhat higher affinity, and a longer binding half-life, for HLA-B*58:01 than for HLA-B*57:01 (Fig. 2A to D). For comparison, when we assessed the TW10 peptide that is presented by HLA-B*57:01, HLA-B*57:03, and HLA-B*58:01, we also observed strong binding to all three HLA molecules (half-maximal inhibitory concentrations [IC50] [equilibrium dissociation {KD}], 185 nM, 6 nM, and 19 nM, respectively).
Fig 2.
The KF11 Gag peptide is equally presented by HLA-B*57:01, -B*57:03, and -B*58:01. The affinity (A and B) and half-life (stability) (C and D) of binding of KF11 (KAFSPEVIPMF) to all three indicated HLA-B molecules was measured using a novel binding assay (8) and a novel peptide HLA dissociation assay (9); the numbers above the bars represent binding affinities (KD) (B) and peptide binding half-lives (D). (E and F) KF11 Gag peptide pulsing on four different B cell lines derived from 4 individuals (B*57:01 subject R039, B*57:03 subject R059, B*58:01 subject R007, and mismatch HLA subject N056) (data not shown) at a final concentration of 3 nM for 1 h (E) and titrated across 7 log10 concentrations of KF11 peptide (F) and cocultured with a KF11 Gag-specific CD8+ T-cell line (data not shown) for 6 h after thorough washing of the B cell lines. (G) HLA-B*57:01, -B*57:03, and -B*5801 tetramers loaded with KF11 was used to stain the KF11-specific CD8+ T-cell line and controlled using the HLA-B*44:02 mismatch tetramer.
To explore this further, we pulsed B cells expressing HLA-B*57:01, -B*57:03, and -B*58:01 (Table 2) with KF11 peptide and found equally strong recognition by HLA-B*57:01-restricted KF11-specific CD8+ T cells derived from a chronically HIV-infected HLA-B*57:01-positive individual (viremic controller) (Table 3 and Fig. 2). Peptide titration studies revealed equal peptide affinities to the T-cell receptor irrespective of the presenting HLA molecule (Fig. 2E and F), which was confirmed by HLA-B*57:01, -B*57:03, and -B*58:01 KF11 tetramer staining (15) of specific CD8+ T cells (Fig. 2G). Thus, a difference in peptide binding does not explain the differential targeting of KF11 by HLA-B*57:01, -B*57:03, and -B*58:01.
Table 2.
HLA-A, -B, and -C genotypes of cells used for KF11 peptide presentation
| Patient and cell typea | HLA genotypesb |
|---|---|
| R039, BCL | A*01:01/31:01, B*27:05/57:01, C*02:02/06:02 |
| R059, BCL | A*01:03/68:02, B*35:01/57:03, C*05:01/07:01 |
| R014, BCL | A*30:01/33:01, B*42:01/57:03, C*17:01/18:01 |
| R007, BCL | A*24:02/80:01, B*07:02/58:01, C*02:02/03:02 |
| R036, BCL | A*02:01/68:02, B*14:01/58:01, C*03:02/08:02 |
| N056, BCL (mismatch) | A*02:01/02:14, B*15:03/18:01, C*02:10/04:01 |
| Donor A, primary CD4+ T cells | ND/ND, B*15:03/57:01, ND/ND |
| Donor B, primary CD4+ T cells | A*02:01/02:01, B*39:10/57:03, C*07:02/12:03 |
| Donor C, primary CD4+ T cells | A*02:01/32:01, B*18:01/58:01, C*05:01/07:01 |
| Donor D, primary CD4+ T cells (mismatch) | A*01/24, B*07/44, C*05/07 |
BCL, B-cell line.
Bold type indicates relevant HLA-B*57/58:01 expression. ND, not done.
Table 3.
Clinical data and ex vivo enzyme-linked immunosorbent spot assay (ELISpot) responses for study subject R039, used to generate the KF11-specific CD8+ T cell line
| OLPb | SFU/106 PBMCsc | Optimal epitope | SFU/106 PBMCsc |
|---|---|---|---|
| 22 | 535 | KAFSPEVIPMF | 520 |
| 36 | 615 | KRWIILGLNK | 320 |
| 82 | 235 | KRKGGIGGY | 1,180 |
| 265 | 615 |
Subject R039 was antiretroviral treatment naive at the time of sampling and classified as a viremic controller. The HLA genotype was A*01:01/31:01, B*27:05/57:01, C*02:02/06:02. Infection occurred in October 2006. On 5 July 2007, the viral load was 204 RNA copies/ml of plasma, and the CD4 T cell count was 650 on 5 October 2007.
OLP, overlapping peptide (includes the corresponding optimal epitope).
SFU, spot-forming units; PBMCs, peripheral blood mononuclear cells.
The KF11 epitope is not intracellularly processed by HLA-B*58:01-expressing target cells.
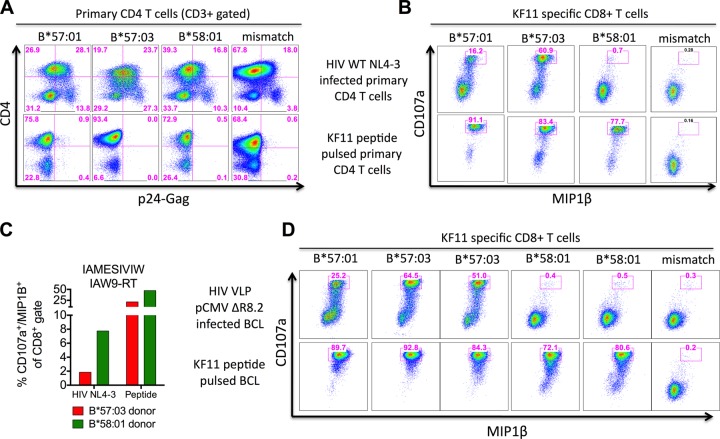
To determine whether KF11 Gag presentation is influenced upstream of peptide HLA loading, we synchronously infected primary CD4+ T cells derived from healthy individuals expressing either HLA-B*57:01, -B*57:03, -B*58:01, or irrelevant alleles (Table 2) with wild-type HIVNL4-3 to similar levels (Fig. 3A) (12). Endogenous processing and presentation of KF11 were then assessed by measuring activation (CD107a+/MIP1β+) of cocultured HLA-B*57:01-restricted KF11-specific CD8+ T cells, shown to recognize KF11 in the context of all three alleles (Fig. 2E), for the duration of 6 h at 48 h postinfection (Fig. 3B). From these data, it is clear that KF11 is well processed intracellularly and presented on the surfaces of HLA-B*57:01- and -B*57:03-positive but not HLA-B*58:01-positive cells. However, this was not a result of a general epitope processing failure by the HLA-B*58:01-positive target cells used here, illustrated by recognition of the IAW9-RT epitope following wild-type HIVNL4-3 infection (Fig. 3C).
Fig 3.
Lack of intracellular KF11 epitope processing within HLA-B*58:01-expressing target cells. (A) Primary CD4 T cells derived from healthy uninfected individuals expressing the indicated HLA-B molecule (B*57:01, donor A; B*57:03, donor B; B*58:01, donor C; HLA mismatch, donor D) (data not shown) were activated for 7 days and synchronously infected with wild-type HIVNL4-3 (data not shown). (B) After 48 h of infection, the cells were cocultured with the KF11-specific CD8+ T-cell line for 6 h in the presence of brefeldin A, stained for CD107a/MIP1β expression to measure KF11 epitope presentation on the surfaces of infected cells, and controlled by KF11 peptide pulsing of uninfected target cells at 3 nM. Representative results from one of two experiments are shown. (C) As described for panel B but using CD8+ T cells recognizing the Pol-RT epitope IAMESIVIW (AW9-RT) after HIVNL4-3 infection and shown with HLA mismatch background subtraction (8%). (D) B cell lines derived from 6 individuals expressing the relevant HLA-B molecule shown above each dot plot were infected with the HIV virus-like particle (HIV VLP pCMVΔR8.2) (data not shown) for 24 h, cocultured with the KF11-specific CD8+ T-cell line for 6 h, and controlled by KF11 peptide pulsing of uninfected B cell lines as described for panel B. Results from one of two independent experiments are shown.
To confirm these findings, we repeated the experiment setup by infecting 6 different B-cell lines (Table 2) with vesicular stomatitis virus G (VSV-G)-pseudotype-like particles (12) (Table 4) encoding all of the HIV proteins except envelope (VLP pCMVΔR8.2) (24). Again, only target cells expressing HLA-B*57:01 and B*57:03 were able to endogenously process and present KF11 on the cell surface (Fig. 3D). Thus, KF11 Gag presentation is abrogated by target cells expressing HLA-B*58:01, suggesting an upstream restriction prior to HLA loading and extracellular export. These data are consistent with the pattern of KF11 recognition summarized in Fig. 1.
Table 4.
Characteristics of HIV-specific CD8+ T cells obtained from subject R039 and viral vectors used for HIV infection of target cells
| Characteristic | Result |
|---|---|
| HLA restriction type | HLA-B*57:01 |
| Epitope | KF11 |
| Protein | Gag |
| HXB2 position | 162–172 |
| Viral vectors in NL4-3 and VLP | p83-2 and pCMVR8.2a |
| HIV clade B and C sequence | KAFSPEVIPMF |
| % CD8+ T-cell specificityb | 98.4 |
Both vectors contain the KF11 Gag epitope as listed in the Los Alamos database (www.hiv.lanl.gov).
Tetramer-positive CD3+ CD8+ gated T cells.
HLA-B*58:01 does not exclude endogenous KF11 processing in the presence of HLA-B*57.
Having established that KF11 is satisfactorily presented by HLA-B*58:01 after peptide pulsing, but not following intracellular processing of HIV Gag after infection of target cells, we addressed the question of whether HLA-B*58:01 abrogates KF11 processing in vivo. To test this, we identified 4 chronic HIV infected individuals expressing both HLA-B*57:03 and -B*58:01, each of whom had been tested for recognition of the 18-mer WVWVIEEKAFSPEVIPMF or the 15-mer EEKAFSPEVIPMFTA, both containing the optimal KF11 epitope, and/or of the optimal KF11 itself. We observed strong KF11 Gag-specific CD8+ T-cell responses ex vivo in all 4 individuals (Table 5). Thus, the presence of HLA-B*58:01 does not abrogate the processing of the optimal KF11 epitope in infected cells if HLA-B*57:03 is also present.
Table 5.
HLA-B*57:03/B*58:01 heterozygous HIV-infected, treatment-naive individuals with an IFN-γ enzyme-linked immunosorbent spot assay (ELISpot) response to the KF11 peptide
| Subject | HLA genotypea | 15- or 18-mer peptide sequence testedb | SFU/106 PBMCsc | Optimal KF11 epitope tested | SFU/106 PBMCsc |
|---|---|---|---|---|---|
| N087 (adult) | A*03:01/30:01, B*57:03/58:01, C*06:02/18:01 | WVKVIEEKAFSPEVIPMF | 820 | KAFSPEVIPMF | 820 |
| K140 (adult) | A*03:01/68:01, B*57:03/58:01, C*03:02/07:01 | EEKAFSPEVIPMFTA | 2,020 | ||
| K061 (child) | A*03:01/66:01, B*57:03/58:01, C*03:02/18:01 | EEKAFSPEVIPMFTA | 280 | ||
| 468 (child) | A*02:02/68:01, B*57:03/58:01, C*06:02/07:01 | KAFSPEVIPMF | 1,200 |
Boldface type indicates relevant HLA-B*57:03/58:01 expression.
The KF11 optimal epitope is underlined.
SFU, spot-forming units; PBMCs, peripheral blood mononuclear cells.
Taken together, these data show that, in HLA-B*58:01-positive individuals, the targeting of the protective KF11 Gag epitope is prevented by events upstream of peptide HLA loading during intracellular processing, which may ultimately impact HIV control (10, 13), resulting either from efficient killing of virus-infected targets (12) or through reduced viral replicative capacity from HLA-B*57-driven escape mutants within KF11-Gag (2, 3). This example also applies to HLA-B*57:02, which differs from HLA-B*57:03 by one amino acid only (Leu156 in HLA-B*57:03, Arg-156R in HLA-B*57:02), and binds the KF11 peptide with very acceptable affinity (IC50, 87 nM; also data not shown), yet no responses to KF11 restricted by HLA-B*57:02 are detectable and no KF11 escape mutations are evident in HLA-B*57:02-positive subjects (13). In addition, we have observed multiple examples of other peptides which exhibited strong binding affinities for the relevant HLA molecule yet which were nonimmunogenic when peptide recognition was tested in a cohort of >1,000 HIV-infected study subjects (data not shown). This strongly suggests that the observation described here of KF11 binding to HLA-B*58:01, but lack of immunogenicity, is an example of a phenomenon that exists more widely.
Potential explanations for this novel phenomenon include the following. First, it is possible that the HLA-B*58:01-KF11 complex has a lower binding affinity for the T-cell receptor (TCR) than do the HLA-B*57:01-KF11 and HLA-B*57:03-KF11 complexes. Second, we cannot rule out the possibility that the absence of KF11-specific CD8+ T cell responses in HLA-B*58:01-positive individuals may result from a “gap” in the TCR repertoire. Third, lack of intracellular processing of KF11 after HIV infection suggests that HLA-B*58:01 interferes with processing events prior to peptide transport into the endoplasmic reticulum (ER), although this seems unlikely, as similar degradation patterns for Gag peptides have been observed for multiple donors with different HLA expression (14). The most likely explanation may be abrogation of actual peptide loading within the peptide loading complex (PLC) in the ER, resulting in failure of the normal downstream egress from the ER, exit, and Golgi transport to the cell surface (21).
The strong KF11-specific CD8+ T-cell responses within individuals heterozygous for the presenting and nonpresenting alleles HLA-B*57:03 and -B*58:01, respectively, might indicate that the critical event occurs after proteosomal cleavage of the Gag-p24 protein, transporter associated with antigen processing (TAP) transport to the ER lumen, and ERAP-1 N-terminal trimming of the KF11 precursor peptide but prior to tapasin-ERp57 conjugate loading onto the HLA molecule. Peptide loading onto HLA class I molecules within the ER happens in the PLC, in which the exact regulation of HLA maturation and interaction with PLC proteins is still debated. The core of the PLC is composed of the TAP heterodimer in conjunction with the ERp57/tapasin conjugate, and multiple weak interactions within the PLC are necessary for optimal peptide loading, which is severely impaired for most HLA alleles after prevention of the interaction with tapasin (21). Tapasin interacts only with alpha domain 2 of the HLA heavy chain, and in particular, residues 114 and 116 play an important role (20, 23). Interestingly, while HLA-B*57:03 differs at these residues from HLA-B*57:01, HLA-B*57:01 and -B*58:01 do not, suggesting that tapasin dependence does not play a role here (21). Furthermore, in this study we used wild-type primary CD4+ T cells and wild-type B cell lines derived from multiple different human donors to study the intracellular processing of KF11 and therefore assume that these cells are equipped with fully functional tapasin along with a complete set of PLC proteins to ensure optimal regulation and processing of epitopes. Finally, another potential mechanism for the absence of KF11 epitope presentation on the surface of HLA-B*58:01 target cells may be that KF11 is outcompeted by other HLA-B*57/58:01-binding peptides, such as TW10, which has very strong binding affinity for both HLA-B*57:03 and -B*58:01 (IC50 [KD], 6 nM and 19 nM, respectively). However, while HLA-B*57:03-restricted KF11 responses dominate in chronic infection, HLA-B*58:01-restricted KF11 responses are not detectable, suggesting that outcompetition by high-avidity peptides such as TW10 is not the explanation.
This novel example shows how peptide presentation, immunogenicity, and ultimately clinical disease outcome can be governed at the level of intracellular processing, which is unrelated to optimal peptide binding affinity. Furthermore, it strongly implies the existence of yet-unknown events or mechanisms located within the PLC that influence peptide presentation by HLA class I alleles and raises the question of exactly how peptides are loaded onto HLA class I molecules by the PLC. Future studies are needed to determine how widespread this phenomenon is and to what extent it explains the observed difference in epitope repertoire of closely related HLA alleles.
ACKNOWLEDGMENTS
We declare that no competing interests exist.
We are grateful to Sylvie Le Gall for critical reading of the manuscript.
This work was supported by the Wellcome Trust (P.G.) and the National Institutes of Health, grant R01 AI46995. H.N.K holds a grant from the Danish Agency for Science, Technology and Innovation, no. 12-132295.
Footnotes
Published ahead of print 17 July 2013
REFERENCES
- 1.Carlson JM, Listgarten J, Pfeifer N, Tan V, Kadie C, Walker BD, Ndung'u T, Shapiro R, Frater J, Brumme ZL, Goulder PJ, Heckerman D. 2012. Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J. Virol. 86:5230–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford H, Lumm W, Leslie A, Schaefer M, Boeras D, Prado JG, Tang J, Farmer P, Ndung'u T, Lakhi S, Gilmour J, Goepfert P, Walker BD, Kaslow R, Mulenga J, Allen S, Goulder PJ, Hunter E. 2009. Evolution of HLA-B*5703 HIV-1 escape mutations in HLA-B*5703-positive individuals and their transmission recipients. J. Exp. Med. 206:909–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford H, Prado JG, Leslie A, Hue S, Honeyborne I, Reddy S, van der Stok M, Mncube Z, Brander C, Rousseau C, Mullins JI, Kaslow R, Goepfert P, Allen S, Hunter E, Mulenga J, Kiepiela P, Walker BD, Goulder PJ. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 81:8346–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, Urban TJ, Zhang K, Gumbs CE, Smith JP, Castagna A, Cozzi-Lepri A, De Luca A, Easterbrook P, Gunthard HF, Mallal S, Mussini C, Dalmau J, Martinez-Picado J, Miro JM, Obel N, Wolinsky SM, Martinson JJ, Detels R, Margolick JB, Jacobson LP, Descombes P, Antonarakis SE, Beckmann JS, O'Brien SJ, Letvin NL, McMichael AJ, Haynes BF, Carrington M, Feng S, Telenti A, Goldstein DB. 2009. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 5:e1000791. 10.1371/journal.pgen.1000791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. 2007. A whole-genome association study of major determinants for host control of HIV-1. Science 317:944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goulder PJ, Bunce M, Krausa P, McIntyre K, Crowley S, Morgan B, Edwards A, Giangrande P, Phillips RE, McMichael AJ. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retroviruses 12:1691–1698 [DOI] [PubMed] [Google Scholar]
- 7.Goulder PJ, Walker BD. 2012. HIV and HLA class I: an evolving relationship. Immunity 37:426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harndahl M, Justesen S, Lamberth K, Roder G, Nielsen M, Buus S. 2009. Peptide binding to HLA class I molecules: homogenous, high-throughput screening, and affinity assays. J. Biomol. Screen. 14:173–180 [DOI] [PubMed] [Google Scholar]
- 9.Harndahl M, Rasmussen M, Roder G, Buus S. 2010. Real-time, high-throughput measurements of peptide-MHC-I dissociation using a scintillation proximity assay. J. Immunol. Methods 374:5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N, Bishop K, van der Stok M, Nair K, Khan N, Crawford H, Payne R, Leslie A, Prado J, Prendergast A, Frater J, McCarthy N, Brander C, Learn GH, Nickle D, Rousseau C, Coovadia H, Mullins JI, Heckerman D, Walker BD, Goulder P. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53 [DOI] [PubMed] [Google Scholar]
- 11.Kloverpris HN, Harndahl M, Leslie AJ, Carlson JM, Ismail N, van der Stok M, Huang KH, Chen F, Riddell L, Steyn D, Goedhals D, van Vuuren C, Frater J, Walker BD, Carrington M, Ndung'u T, Buus S, Goulder P. 2012. HIV control through a single nucleotide on the HLA-B locus. J. Virol. 86:11493–11500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kloverpris HN, Payne RP, Sacha JB, Rasaiyaah JT, Chen F, Takiguchi M, Yang OO, Towers GJ, Goulder P, Prado JG. 2013. Early antigen presentation of protective HIV-1 KF11Gag and KK10Gag epitopes from incoming viral particles facilitates rapid recognition of infected cells by specific CD8+ T cells. J. Virol. 87:2628–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kloverpris HN, Stryhn A, Harndahl M, van der Stok M, Payne RP, Matthews PC, Chen F, Riddell L, Walker BD, Ndung'u T, Buus S, Goulder P. 2012. HLA-B*57 micropolymorphism shapes HLA allele-specific epitope immunogenicity, selection pressure, and HIV immune control. J. Virol. 86:919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazaro E, Kadie C, Stamegna P, Zhang SC, Gourdain P, Lai NY, Zhang M, Martinez SA, Heckerman D, Le Gall S. 2011. Variable HIV peptide stability in human cytosol is critical to epitope presentation and immune escape. J. Clin. Invest. 121:2480–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leisner C, Loeth N, Lamberth K, Justesen S, Sylvester-Hvid C, Schmidt EG, Claesson M, Buus S, Stryhn A. 2008. One-pot, mix-and-read peptide-MHC tetramers. PLoS One 3:e1678. 10.1371/journal.pone.0001678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leslie A, Matthews PC, Listgarten J, Carlson JM, Kadie C, Ndung'u T, Brander C, Coovadia H, Walker BD, Heckerman D, Goulder PJ. 2010. Additive contribution of HLA class I alleles in the immune control of HIV-1 infection. J. Virol. 84:9879–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marsh SGE, Parham P, Barber LD. 2000. The HLA facts book. Academic Press, London, United Kingdom [Google Scholar]
- 18.Mothe B, Llano A, Ibarrondo J, Daniels M, Miranda C, Zamarreno J, Bach V, Zuniga R, Perez-Alvarez S, Berger CT, Puertas MC, Martinez-Picado J, Rolland M, Farfan M, Szinger JJ, Hildebrand WH, Yang OO, Sanchez-Merino V, Brumme CJ, Brumme ZL, Heckerman D, Allen TM, Mullins JI, Gomez G, Goulder PJ, Walker BD, Gatell JM, Clotet B, Korber BT, Sanchez J, Brander C. 2011. Definition of the viral targets of protective HIV-1-specific T cell responses. J. Transl. Med. 9:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrov DA, Grant BJ, Pompeu YA, Sidney J, Harndahl M, Southwood S, Oseroff C, Lu S, Jakoncic J, de Oliveira CA, Yang L, Mei H, Shi L, Shabanowitz J, English AM, Wriston A, Lucas A, Phillips E, Mallal S, Grey HM, Sette A, Hunt DF, Buus S, Peters B. 2012. Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire. Proc. Natl. Acad. Sci. U. S. A. 109:9959–9964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park B, Lee S, Kim E, Ahn K. 2003. A single polymorphic residue within the peptide-binding cleft of MHC class I molecules determines spectrum of tapasin dependence. J. Immunol. 170:961–968 [DOI] [PubMed] [Google Scholar]
- 21.Peaper DR, Cresswell P. 2008. Regulation of MHC class I assembly and peptide binding. Annu. Rev. Cell Dev. Biol. 24:343–368 [DOI] [PubMed] [Google Scholar]
- 22.Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O'Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, et al. 2010. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330:1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zernich D, Purcell AW, Macdonald WA, Kjer-Nielsen L, Ely LK, Laham N, Crockford T, Mifsud NA, Bharadwaj M, Chang L, Tait BD, Holdsworth R, Brooks AG, Bottomley SP, Beddoe T, Peh CA, Rossjohn J, McCluskey J. 2004. Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J. Exp. Med. 200:13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871–875 [DOI] [PubMed] [Google Scholar]