Abstract
During placentation, mammals employ different strategies for nourishing and supporting fetuses. Members of the Bovidae family, consisting of cloven-hoofed ruminants, utilize multiple maternal attachment points on the placenta, known as cotyledons, and hybrid cells, named trinucleate cells or syncytial plaques, made up of a fusion of fetal trophoblasts and maternal endometrial cells to provide essential hormones and maintain long gestation periods. These hybrid cells are unique to the Bovidae, as fetomaternal borders are clearly separated by syncytiotrophoblasts or epithelial cells in the placenta of other mammals. Recently, it was reported that Syncytin-Rum1 was inserted into ruminant genomes, including cattle and sheep, and was possibly involved in fetomaternal cell-to-cell fusion in both species. However, Syncytin-Rum1 alone is insufficient to explain the morphological diversity of the fetomaternal hybrids between Bovinae and Caprinae (i.e., trinucleate cells in Bovinae and syncytial plaques in Caprinae). Here we report that the bovine endogenous retrovirus K1 (BERV-K1) envelope, which we term Fematrin-1, was specifically expressed in binucleated trophoblasts throughout gestation in cattle and induced fusion with bovine endometrial cells in vitro at a significantly higher level than Syncytin-Rum1 under physiological conditions. Fematrin-1 was found to be integrated into intron 18 of FAT tumor suppressor homolog 2 (FAT2) about 18.3 to 25.4 million years ago and has been subject to purifying selection through the evolution of Bovinae. Phylogenetically, Fematrin-1 is distinct from Syncytin genes found in other mammalian species that form syncytiotrophoblasts. Our results suggest that the newly acquired endogenous retroelement has contributed to generating placentation diversity through ruminant evolution.
INTRODUCTION
Analysis of the evolutionary development of the cetartiodactyl placenta suggests that the basic placenta in this taxon is a diffuse epitheliochorial type in which cell fusion does not take place. However, with the evolution of ruminants, a new type of placentation emerged, where cell fusions have become an integral element. There are three types of trophoblasts known in the bovine placenta: mononucleate trophoblast cells (MTCs), binucleate cells (BNCs), and trinucleate cells (TNCs) (1). While MTCs and BNCs have consistently been identified in the placental trophectoderm of many ruminants, TNCs have been discovered in bovine species, including cattle (1–3). Instead of TNCs, sheep and goats belonging to the subfamily Caprinae develop multinucleated syncytial plaques (SyPs) (4). Whereas BNCs are formed by differentiation of MTCs by endoreduplication, TNCs and SyPs are thought to be the result of cell-to-cell fusions between BNCs and maternal endometrial cells (1, 4, 5). These hybrid cells are unique to the Bovidae, as fetomaternal borders are clearly separated by syncytiotrophoblasts or epithelial cells in the placenta of other mammals (6, 7). These fused cells are essential for successful implantation in early gestation and for the transfer of various BNC-specific pregnancy-associated molecules to the maternal body (1, 4). However, the precise molecular mechanisms involved in the morphogenesis of these cells are unclear. Endogenous retroviruses (ERVs) are known to play a role in the developmental stages of many mammals (8–13). A hypothesis has been put forward that multinucleated sheep SyPs critical for the formation of placenta are formed by endogenous Jaagsiekte sheep retrovirus (enJSRV) envelope proteins (Envs) produced in BNCs; however, there is inadequate evidence to support the fusogenic potency of these viruses (4). We recently identified and described two novel Env-coding sequences (env genes) of bovine endogenous retroviruses (BERV-K1 and BERV-K2), which possess fusogenic motifs, or fusion peptides (FPs), in their N-terminal Env transmembrane (TM) domains (14). Because they were expressed in the bovine placenta and a cultured bovine trophoblast cell line (14, 15), we hypothesized that BERV-K Env proteins might be involved in the bovine fetomaternal fusogenic process. Here, we report that BERV-K1 Env is specifically expressed in binucleated trophoblasts throughout gestation and exhibits significant fusogenic activity with bovine endometrial cells in vitro. Recently, Cornelis et al. reported that a distinct ERV Env, termed Syncytin-Rum1, is possibly involved in ruminant placentation (16). We discuss the implications and relationship between Syncytin-Rum1 and fematrin-1 in Discussion.
MATERIALS AND METHODS
Cell cultures.
Cos-7, HEK293T, HeLa, TE671, MDTF, and D17 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2 in air. Bovine endometrial cells were cultured in DMEM-F12 Ham medium (Sigma) supplemented with 10% FBS, 100 IU/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2 in air.
Tissue collection.
Bovine tissues were collected from Japanese Black cattle as described previously (14, 15). Bovine conceptuses and placentas were obtained during early gestation (days 24, 30, 60, 78, 80, 90, and 92), midgestation (days 120, 120, 133, 156, 160, and 160 [i.e., 2 samples were used on day 120 and day 160]), and late gestation (days 174, 224, 250, 259, 268, and 270). Ovine tissues (liver, spleen, heart, muscle, kidney, brain, and peripheral blood mononuclear cells [PBMCs]) were harvested from three young Suffolk sheep. Ovine placentomes were collected on days 50, 50, and 100 of pregnancy and were kindly provided by Toru Takahashi (National Institute of Agrobiological Sciences). Caprine placentomes were collected from three goats, while other bovine-derived samples (PBMCs) were kindly provided by collaborators. All procedures for these animal experiments were carried out in accordance with guidelines approved by the ARC-Kyoto University, the Animal Committee of Iwate University, and the National Institute of Agrobiological Sciences for the use of experimental animals.
Generation of anti-BERV-K1 SU antibodies.
Antisera to the BERV-K1 surface (SU) domain (BK1SU) were raised by six immunizations of a rabbit with a commercially synthesized peptide cocktail (NH2-CQDWSNPDPGQDPMI-COOH and NH2-YKSLHFKSPPKYPNC-COOH). Purification of anti-BK1SU antibodies from rabbit total serum was achieved by using a column conjugated with the peptides used for immunization.
Immunoblotting.
Whole-cell lysates were prepared with radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], and 1% Nonidet P-40) and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on a 12% or commercial gradient gel. Electrophoresed samples were electrically blotted onto a polyvinylidene difluoride (PVDF) membrane, and the blots were reacted with appropriate antibodies. Signals were detected by using a Super Signal West Femto system (Thermo Fisher Scientific, Waltham, MA), and images were obtained and processed by using a luminescent image analyzer (LAS4000 Mini; Fujifilm, Tokyo, Japan).
Fusion assays.
A fusion assay to visualize fused foci was conducted as follows. A set of Cos-7 cells was cotransfected with an Env-coding vector and pCONT1 of the CoralHue Fluo-chase kit (MBL, Nagoya, Japan) by Lipofectamine 2000 (Invitrogen, Carlsbad, CA) in a 6-well plate. The other set of Cos-7 cells was independently transfected with pCONT2. After 24 h posttransfection, these two sets of Cos-7 cells were cocultured in a 6-well plate containing coverglasses at 37°C for 24 h. Cocultured cells were then fixed with 4% paraformaldehyde, permeabilized with 0.15% Triton X-100, stained with 4′,6-diamidino-2-phenylindole (DAPI), and analyzed by fluorescence microscopy.
A fusion-dependent luciferase assay was performed as described previously (17), with slight modifications. Briefly, Cos-7 cells were cotransfected with the env-encoded vector, pT7EMCVluc (18), and pRL-TK (Promega, Madison, WI). Cotransfections were carried out in 6-well plates by using Lipofectamine 2000 (Invitrogen). Plasmid-transfected Cos-7 cells were cocultured with target cells transfected with a T7 polymerase-encoded expression vector (pCAGT7pol) (18) in 96-well plates at 37°C for 24 h. Luciferase activities were measured for each sample by using a dual-luciferase reporter assay system (Promega).
BERV-K1 and -K2 SU binding assay.
HEK293T cells were transfected with the phCMV3 vector, phCMV3BERV-K1SUFLAG, and phCMV3BERV-K2SUFLAG. Cells were lysed with RIPA lysis buffer and immunoprecipitated by using anti-FLAG M2-conjugated agarose beads (Sigma). Precipitated proteins were competitively eluted with Tris-buffered saline containing FLAG peptide (Sigma) before being subjected to immunoblotting and binding assays as substrates. Cos-7 and bovine endometrial cells were incubated with these substrates at 4°C for 60 min before washing three times with phosphate-buffered saline (PBS). Cells were then reacted with anti-FLAG M2 on ice for 60 min and washed twice more with PBS before reacting them with anti-mouse IgG conjugated with Alexa Fluor 488 on ice for 60 min (Invitrogen). After two further PBS washes, cells were analyzed on a FACSCalibur instrument (Becton, Dickinson and Company, Franklin Lakes, NJ).
Measurement of Env expression levels.
Cos-7 cells were transfected with a pSG5 expression vector containing C-terminally FLAG-tagged BERV-K1 Env, BERV-K2 Env, or only the FLAG tag. Forty-eight hours after transfection, the cells were harvested and fixed with 4% paraformaldehyde (PFA) on ice for 30 min. The cells were then sequentially incubated with blocking buffer (PBS [pH 7.4] containing 3% bovine serum albumin [BSA]) on ice for 1 h, 90% methanol on ice for 30 min, anti-FLAG M2 diluted in blocking buffer on ice for 1 h, and anti-mouse IgG conjugated with Alexa Fluor 488 (Invitrogen) in blocking buffer on ice for 30 min. Finally, samples were analyzed by using a FACSCalibur instrument (BD Biosciences).
In situ hybridization.
BERV-K1 env and -K2 env mRNAs were localized with digoxigenin (DIG)-labeled single-strand cRNA probes, prepared by using a DIG RNA labeling kit (Roche Diagnostic GmbH, Mannheim, Germany) according to the manufacturer's instructions. In situ hybridization (ISH) was conducted as previously described (19). Briefly, formalin-fixed tissue was prepared for paraffin embedding. The tissues were then cut into sections 7 μm thick, and ISH was performed by using an automated Ventana HX Discovery system with a RiboMapKit and a BlueMapKit (Roche Diagnostic GmbH). The sections were hybridized with DIG-labeled probes in RiboHybe (Roche Diagnostic GmbH) hybridization solution at 61°C for 6 h, and the hybridized signal for each gene was detected by using rabbit monoclonal antidigoxin biotin conjugates (Sigma) and an AmpMapKit (Roche Diagnostic GmbH). Counterstaining was performed with Nuclear Fast Red (Roche Diagnostic GmbH). After the preparatory stages, hybridized slides were visualized by using a Leica DMRE HC microscope (Leica Microsystems, Wetzlar, Germany) equipped with a DSFi1 camera and a DS-L2 control unit (Nikon, Tokyo, Japan).
Immunohistochemical (IHC) analyses.
Paraffin-embedded tissues were cut into sections 7 μm thick, and the sections were incubated in 10 mM Tris-HCl (pH 8.0) for 30 min at 121°C after deparaffinization. Sections were then incubated in 0.3% (vol/vol) H2O2 in methanol for 20 min. Nonspecific antibody binding was minimized by treatment with 10% normal goat serum for 30 min, and sections were then incubated overnight at 4°C with a 1:1,000 dilution of anti-BK1SU polyclonal antibody. After washing, the tissue sections were incubated with peroxidase-conjugated goat anti-rabbit IgG (Sigma) as a secondary antibody for 60 min. Each section was rinsed and then treated with substrate solution (0.02% [wt/vol] 3,3′-diaminobenzidine tetrahydrochloride in 50 mM Tris-HCl with 0.01% [vol/vol] H2O2 [pH 7.5]) for 5 to 10 min at room temperature. This was followed by staining with Meyer's hematoxylin for 5 min. Control sections were treated with normal rabbit IgG instead of the antibody.
Genome DNA preparation and genomic PCR.
Tissues or cells were lysed in cell lysis buffer (50 mM Tris-HCl [pH 7.5], 20 mM EDTA [pH 8.0], 100 mM NaCl, and 1% SDS), and genomic DNA was purified from each lysate by phenol-chloroform-isoamyl alcohol (25:24:1) extraction and isopropanol precipitation. Genomic DNA pellets were resuspended in Tris-EDTA (TE) buffer containing 20 μg/ml RNase A and stored at −30°C prior to use. Genomic PCR was performed by using PrimeStar GXL polymerase (TaKaRa, Shiga, Japan) or LA Taq polymerase (TaKaRa), and amplicons were analyzed by electrophoresis in 0.8% agarose gels. The primers employed are listed in Table S1A in the supplemental material.
RT-PCR and quantitative real-time RT-PCR.
Total RNA was isolated from frozen tissues and cultured cells by using TRIzol reagent (Invitrogen). cDNA was synthesized from 1 μg of total RNA by using SuperScriptIII (Invitrogen) and stored at −80°C prior to experiments. Reverse transcription-PCR (RT-PCR) of bovine FAT2 (bFAT2) was performed by using PrimeStar GXL polymerase. The primers employed are listed in Table S1B in the supplemental material. Quantitative real-time RT-PCR was conducted by using TaqMan universal PCR master mix (Applied Biosystems, Foster City, CA) and an ABI Prism 7000 real-time PCR system (Applied Biosystems) according to the manufacturer's instructions. Primer pairs and probes used in this experiment were designed by the Primer Express program (Applied Biosystems) and are listed in Table S1C in the supplemental material. Assays were conducted in duplicate for each individual sample.
Genetic analyses.
To infer the phylogenetic relationship of Fematrin-1, an alignment was prepared, comprising Env amino acid sequences from a diverse range of retroviruses (14 exogenous and 19 endogenous retroviruses [see Table S1D in the supplemental material]). As the Env TM domain is known to be more conserved than the surface domain in many retroviruses (20), we used amino acid sequences from the former, generating our alignment using the multiple-sequence alignment program L-INS-i from the MAFFT suite (v.5) (21). Phylogenetic trees were estimated by using the maximum likelihood method (22) implemented in RAxML v7.2.6 (23). ProtTest 3 (24) specified the LG amino acid replacement model (25) with a discrete gamma distribution that accounted for heterogeneity in evolutionary rates among sites (α = 2.948), as identified by the Akaike information criterion. The robustness of the phylogenetic tree was evaluated by fast bootstrapping (26) with 1,000 pseudoreplicate data sets.
To examine evolutionary changes among four Fematrin-1 genes (derived from Bos taurus, Bos javanicus, Bubalus bubalis, and Tragelaphus spekii), the amino acid sequences of products of these genes were utilized to estimate their phylogenies, as described above. Pairwise amino acid identities and the ratios of substitution rates at nonsynonymous and synonymous sites (dN/dS ratios) were calculated by using MEGA5 (27). The probability of rejecting the null hypothesis of strict neutrality (dN = dS) was calculated by using the Nei-Gojobori method (28). To examine branch-specific positive selection, the dN/dS ratio for each tree branch was examined by maximum likelihood using the codeml program from PAMLv4.5 (29), with codon frequencies as free parameters (CodonFreq = 3).
RESULTS
Fusion activities of BERV-K Env proteins.
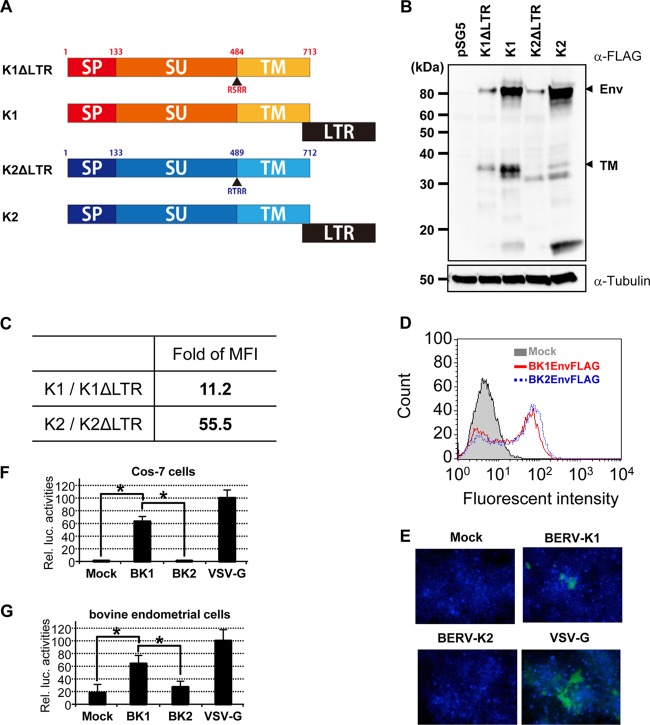
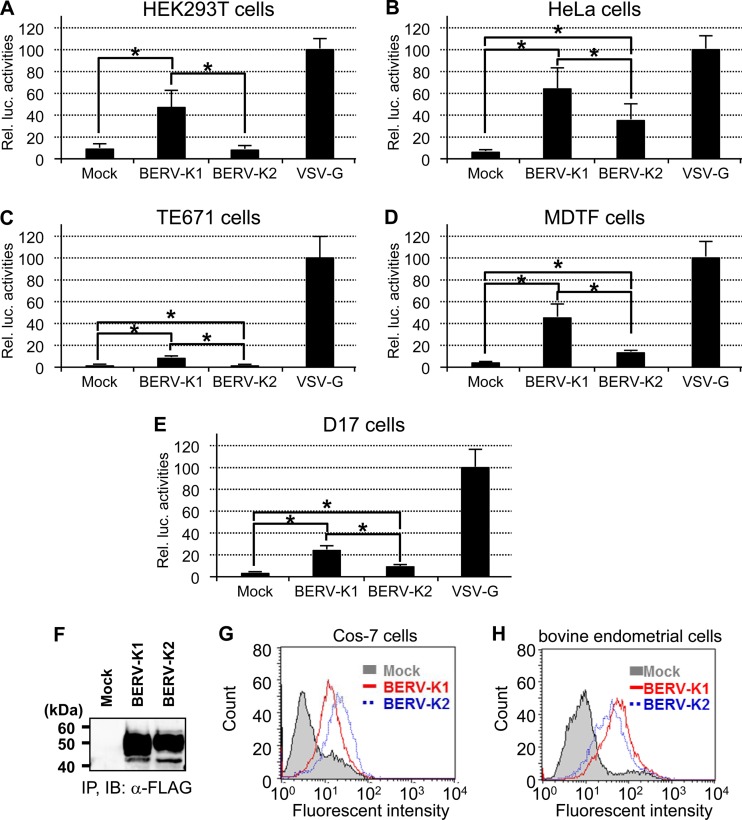
Having previously identified that 3′ long terminal repeats (LTRs) are necessary for nuclear export of BERV-K1 and -K2 env mRNAs (14), we constructed C-terminally FLAG-tagged BERV-K1 and -K2 Env expression vectors either flanked or unflanked by their respective 3′ LTRs (Fig. 1A). A FLAG tag was inserted just downstream of the C-terminal amino acids of both Envs without any additional sequences. These plasmids were transfected into Cos-7 cells, and subsequently, cell lysates were subjected to immunoblotting analysis. Consistent with our previous findings, BERV-K Env precursors were expressed more efficiently when the genes were flanked by their respective 3′ LTRs (Fig. 1B and C). To investigate the fusogenicity of these BERV-K Envs, Cos-7 cells expressing both KUSABIRA-Green (Gr) positive control 1 (pCONT1) and the indicated Env were cocultured with Cos-7 cells expressing KUSABIRA-Gr positive control 2 (pCONT2). If cell-to-cell fusion is induced, fused cells should be observed as green foci by fluorescence microscopy. Despite similar expression levels of BERV-K1 and -K2 Envs (Fig. 1D), only BERV-K1 Env formed fusion foci similar to those observed for the highly fusogenic glycoprotein of vesicular stomatitis virus (VSV-G) (Fig. 1E). To further investigate the fusion activities of these viral glycoproteins, we conducted a cell-to-cell fusion-dependent luciferase assay, in which luciferase activity reflects fusogenicity. Results revealed a significantly higher fusogenic activity for BERV-K1 Env than that observed for BERV-K2, not only in Cos-7 cells but also in several other cell lines, including bovine endometrial cells (Fig. 1F and G and 2A to E) (30). Similar properties of binding of the BERV-K1 and -K2 Env surface (SU) domains to the cell types examined (Fig. 2F to H) indicate that the specific fusion activity of BERV-K1 Env is not due to different expression levels of unidentified receptors for these Envs. Retroviral envelope glycoproteins are generally divided into three subunits: the signal peptide (SP), SU, and TM components. Given that fusogenicity requires the cleavage of SU and TM domains by cellular proteases (31), the different fusion activities of the two BERV proteins described here may be explained by an insufficient cleavage of the BERV-K2 Env domains (Fig. 1B).
Fig 1.
Requirement of BERV-K1 and -K2 3′ LTRs for efficient translation of the respective retroviral Env proteins and fusion activities of BERV-K Env proteins. (A) Schematic representation of each construct transfected into Cos-7 cells. SP, SU, and TM indicate the signal peptide, surface domain, and transmembrane domain, respectively. All constructs harbor FLAG epitope tags adjacent to the C termini of the TM domains. (B) Cos-7 cell lysates transfected with each construct were subjected to SDS-PAGE followed by blotting onto PVDF membranes. FLAG-tagged proteins were detected by using anti-FLAG M2 and horseradish peroxidase-conjugated anti-mouse IgG antibodies. pSG5, which is a backbone plasmid, was used as a negative control. (C) Expression ratio of BERV Env precursors produced where plasmids were flanked by their respective 3′ LTRs (K1 and K2) to those not flanked by 3′ LTRs (K1ΔLTR and K2ΔLTR) determined by flow cytometric analysis using anti-FLAG M2 and Alexa Fluor 488-conjugated anti-mouse IgG antibodies. Values represent the ratios of mean fluorescence intensities (MFI). (D) Expression of BERV-K1 and -K2 Env in Cos-7 cells transfected with expression plasmids. Cos-7 cells were transfected and analyzed by flow cytometry using anti-FLAG M2 and Alexa Fluor 488-conjugated anti-mouse IgG antibodies. (E) Fluorescent focus of fused cells. Cos-7 cells were cotransfected with the indicated Env expression vectors and pCONT-1 and cocultured with Cos-7 cells transfected with pCONT-2. The fusion focus (green) was analyzed by fluorescence microscopy. Nuclei were stained with DAPI (blue). (F and G) Fusion-dependent luciferase assay of FLAG-tagged BERV-K1 and -K2 Envs. Cos-7 cells cotransfected with the pT7EMCVluc, pRL-TK, and pSG5 expression vectors containing the indicated FLAG-tagged Env or only the FLAG tag were cocultured with Cos-7 (F) or bovine endometrial (G) cells transfected with pCAGT7pol. The resultant cell lysates were subjected to dual-luciferase reporter assays, which were performed in triplicate and repeated as 3 independent experiments (n = 9). Each value for relative luciferase (Rel. luc.) activity is shown as the mean ± standard error. Statistical analysis was performed by a Student t test, and differences were considered significant at a P value of <0.05 (indicated by an asterisk).
Fig 2.
Relative fusion activities of BERV-K1 and -K2 Envs in several cell lines, and properties of binding of BERV-K1 and -K2 SU Envs to Cos-7 and bovine endometrial cells. (A to E) Cos-7 cells cotransfected with pT7EMCVluc, pRL-TK, and the indicated Env expression vectors were cocultured with HEK293T (human) (A) HeLa (human) (B), TE671 (human) (C), MDTF (Mus dunni) (D), and D17 (dog) (E) cells transfected with pCAGT7pol. Cell lysates were then subjected to dual-luciferase reporter assays. Assays were performed in triplicate and repeated as 3 independent experiments (n = 9), and each value of luciferase activity is shown as the mean ± standard error. Statistical analysis was performed by the Student t test, and differences were considered significant at a P value of <0.05 (indicated by an asterisk). (F) Preparation of C-terminally FLAG-tagged BERV-K1 and -K2 SU Envs. Cell lysates from HEK293T cells transfected with the phCMV3 (mock), phCMV3BERV-K1SUFLAG (BERV-K1), or phCMV3BERV-K2SUFLAG (BERV-K2) expression vector were immunoprecipitated (IP) by using ant-FLAG M2 antibody conjugated to agarose beads. Elution of precipitated proteins was achieved by using an excess of FLAG peptides and was monitored by SDS-PAGE and immunoblotting (IB) using anti-FLAG M2 and horseradish peroxidase-conjugated anti-mouse IgG antibodies. (G and H) Binding assays of C-terminally FLAG-tagged BERV-K1 and -K2 SU Envs. Cos-7 (G) or bovine endometrial (H) cells were incubated with each of the purified products and anti-FLAG M2 and Alexa Fluor 488-conjugated anti-mouse IgG antibodies.
Expression profiles of BERV-K1 and -K2 Envs.
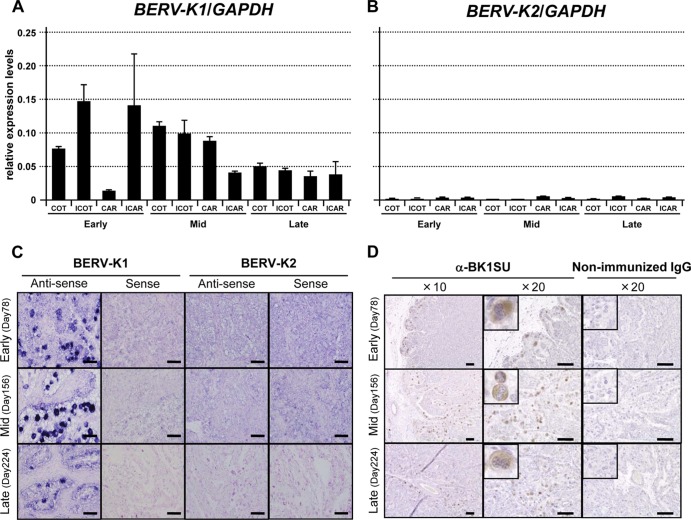
Expression levels of BERV-K1 and -K2 env mRNAs were monitored in the bovine placenta at different stages of gestation (early, mid-, and late). At all points during gestation, the expression level of BERV-K1 env mRNA far exceeded that of BERV-K2 (Fig. 3A and B). ISH was utilized to establish which cells were expressing BERV-K env mRNAs in the placenta. The results revealed that BERV-K1 env mRNA was expressed specifically in BNCs throughout gestation, in contrast to BERV-K2 env mRNA, which was not detected at any stage (Fig. 3C). Anti-BERV-K1 SU polyclonal antibodies were generated for use in IHC analyses, and the results confirmed the ISH findings that BERV-K1 Env was predominantly expressed in BNCs throughout pregnancy (Fig. 3D). Three novel BERV-K env-related sequences, also bearing FPs, were identified by data mining of the NCBI genomic database and were named BERV-K env rel-1, rel-2, and rel-3 (Fig. 4G). BERV-K env rel-1 to rel-3 were not identical to bos–env-1 to -5, identified previously (16). Expression levels of these sequences were low in all bovine tissues (similar to those seen for BERV-K2 env) and not specific to the placenta (data not shown). We therefore concluded that staining of molecules during ISH and IHC analysis is likely to demonstrate the presence of BERV-K1 env mRNA and Env in the bovine placenta.
Fig 3.
Expression profiles of BERV-K1 and -K2 env mRNAs and BERV-K1 Env in the bovine placenta. (A and B) Quantitative real-time RT-PCR analysis of BERV-K1 and -K2 env mRNA levels through gestation in the bovine placenta. COT, ICOT, CAR, and ICAR denote cotyledon, intercotyledon, caruncular, and intercaruncular tissues, respectively. Early, Mid, and Late on the x axis relate to stages of gestation. Each value represents the mean ± standard error of the ratio of BERV-K1 and -K2 env expression levels to that of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) internal control gene. Assays were conducted in duplicate for 2 or 3 individual samples (n = 4 or 6). (C) ISH analysis of BERV-K1 and -K2 env mRNAs in the bovine placenta. Analyses were performed by using sense and antisense probes for both BERV-K1 and -K2 env genes. The y axis shows stages of gestation during which ISH was performed. Scale bars represent 100 μm. (D) IHC of BERV-K1 Env in the bovine placenta. Analyses were performed by using an anti-BK1SU antibody. The y axis shows stages of gestation during which ISH was performed. Scale bars represent 100 μm. BNCs are enlarged 5 times in small panels.
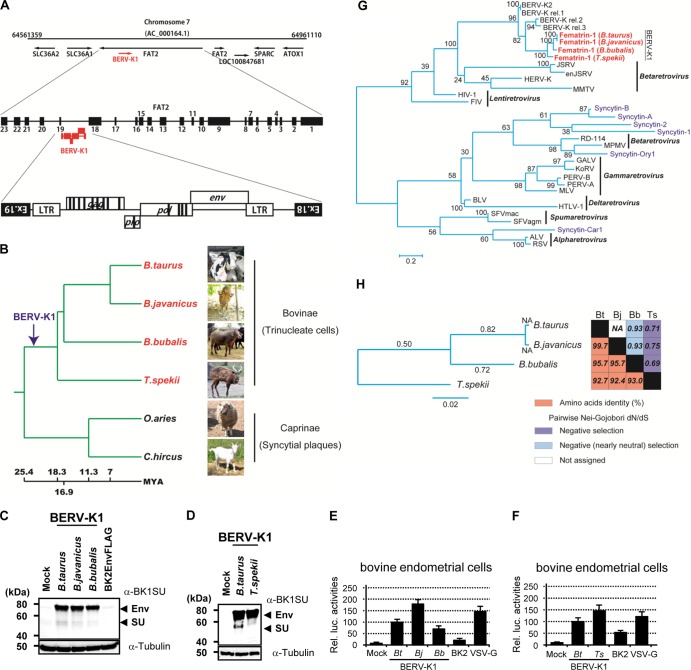
Fig 4.
Detection of BERV-K1 and Fematrin-1 in the genomes of Bovinae. (A) Schematic representation of the BERV-K1 integrated locus in the bovine genome. (B) Correlation of BERV-K1 insertion in the FAT2 locus and ruminant evolution. The phylogenic tree was inferred from data reported previously by Hernández Fernández and Vrba (33) and modified. The arrow indicates the proposed point of BERV-K1 integration into bovine genomes. The horizontal branch length is proportional to the length of time from the present, and the time scale is millions of years ago (MYA). (C and D) Immunoblotting of Fematrin-1 proteins from B. taurus, B. javanicus, B. bubalis, and T. spekii. Cos-7 cell lysates transfected with expression vectors carrying Fematrin-1 from each species were electrophoresed and then immunoblotted by using an anti-BK1SU antibody. (E and F) Fusion-dependent luciferase assays of fusion of B. taurus, B. javanicus, B. bubalis, and T. spekii Fematrin-1 proteins to bovine endometrial cells. Assays were performed in triplicate and were repeated in 6 independent experiments (n = 18). Each value of luciferase activity is shown as the mean ± standard error. Statistical analysis was performed by a Student t test, and differences were considered significant at a P value of <0.05 (indicated by an asterisk). B. taurus, B. javanicus, B. bubalis, and T. spekii are abbreviated Bt, Bj, Bb, and Ts, respectively. (G) Maximum likelihood phylogenetic tree of Fematrin-1 and other retroviral Env sequences. The tree was built by comparing each retroviral Env TM domain by using RAxML. Branch lengths are proportional to the percentage of amino acid substitutions from the node. Fast bootstrapping was repeated 1,000 times, and percent bootstrap support values are displayed at the nodes. HERV-K, human endogenous retrovirus K; MMTV, mouse mammary tumor virus; FIV, feline immunodeficiency virus; MPMV, Mason-Pfizer monkey virus; GALV, gibbon ape leukemia virus; KoRV, koala retrovirus; PERV-A, porcine endogenous retrovirus subgroup A; MLV, murine leukemia virus; HTLV-1, human T cell leukemia virus type 1; SFVmac, macaque simian foamy virus; ALV, avian leukosis virus; RSV, Rous sarcoma virus. (H) Evolutionary conservation of Fematrin-1 amino acid sequences. (Left) The maximum likelihood phylogenetic tree was built by comparing the entire amino acid sequences of Fematrin-1 from each species. Estimated dN/dS ratios for each branch are shown. (Right) Percent amino acid identities and dN/dS ratios are presented in the accompanying grid. NA, not assigned.
Detection of BERV-K1 in the genomes of Bovinae.
The variability of fetomaternal hybrid cells like TNCs and SyPs in ruminants could depend on the different ERVs involved in their formation processes. To this end, we surveyed which mammals possessed intact BERV-K1 env, both in silico and by genomic PCR. The bovine genomic database provided evidence that BERV-K1 was integrated in a reverse orientation into intron 18 of bovine FAT tumor suppressor homolog 2 (bFAT2) (Fig. 4A), a homolog of the Drosophila melanogaster fat gene that is conserved through mammalian evolution. The other genomes examined, including human, lacked a BERV-K1 insertion in this gene. Although the primer pair used in our genomic PCR experiment was designed to amplify a region spanning exons 18 and 19 of Bos taurus FAT2, we could successfully amplify the same locus in domestic Bali cattle (Bos javanicus) (32), Asian water buffalo (Bubalus bubalis), marshbuck (Tragelaphus spekii), domestic sheep (Ovis aries), and domestic goat (Capra hircus) (data not shown). Sequence analysis of the resultant amplicons indicated that BERV-K1 was integrated at this locus in B. taurus, B. javanicus, B. bubalis, and T. spekii but not in O. aries and C. hircus. These results support the assertion that BERV-K1 integrated into the bovine genome after the appearance of the subfamily Bovinae, which is estimated at 18.3 to 25.4 million years ago (Fig. 4B) (33).
Phylogenetic analyses of Fematrin-1.
The BERV-K1 env gene was conserved as an intact coding sequence in all four species examined, and the amino acid sequence of BERV-K1 Env in these species was highly conserved (Fig. 4H). Furthermore, each BERV-K1 Env exhibited fusogenicity with bovine endometrial cells (Fig. 4C to F). Here, we designated the BERV-K1 Env protein Fematrin-1 (fetomaternal trinucleate cell inducer 1). Phylogenetically, Fematrin-1 is distinct from all Syncytin genes found in previous studies (Fig. 4G). The pairwise ratio of nonsynonymous to synonymous sites (dN/dS ratio), based on the Nei-Gojobori method, was <1 for each Fematrin-1 protein (Fig. 4H, right), indicating that this protein has been subject to negative selection and functionally conserved throughout the evolution of the subfamily Bovinae. Such negative selection pressures were also observed in the evolutionary lineage of bovine species (Fig. 4H, left). These data support our earlier hypothesis since TNCs exist in the placenta of B. bubalis (3) and Tragelaphus placental morphology is very similar to that of the genus Bos (7).
Expression of the FAT2 gene in the fetal placenta.
To investigate why Fematrin-1 came to be specifically expressed in bovine placenta, we conducted quantitative real-time RT-PCR and revealed that bFAT2 expression was specific to the placentome during gestation (Fig. 5A). O. aries, which lacked Fematrin-1, also predominantly expressed ovine FAT2 (oFAT2) in the placentome (Fig. 5B). This integrated locus might thus provide a great advantage for Fematrin-1 in being preferentially expressed in the placenta.
Fig 5.
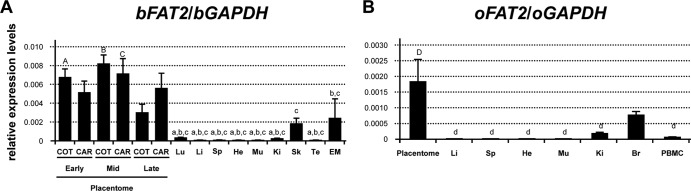
Expression of the FAT2 gene in the fetal placenta. (A and B) Quantitative real-time RT-PCR analysis of bFAT2 and oFAT2 mRNA levels in each indicated tissue. Values shown represent the means ± standard errors of bFAT2 and oFAT2 expression levels relative to that of GAPDH in each tissue. COT, ICOT, CAR, ICAR, Lu, Li, Sp, He, Mu, Ki, Sk, Te, EM, Br, and PBMC are abbreviations of cotyledon, intercotyledon, caruncular, intercaruncular, lung, liver, spleen, heart, muscle, kidney, skin, testis, endometrium, brain, and peripheral blood mononuclear cells, respectively. Assays were conducted in duplicate with 2, 3, or 6 individual samples (n = 4, 6, or 12). Results were statistically analyzed by means of one-way analysis of variance and Tukey-Kramer multiple-comparison tests. Differences were considered significant at a P value of <0.05 and are denoted by letters. These letters indicate statistical significances between A and a, B and b, C and c, and D and d, respectively.
Comparison of the fusogenic activities of Fematrin-1 and Syncytin-Rum1 under physiological conditions.
Quite recently, Syncytin-Rum1, which is phylogenetically distinct from Fematrin-1, was suggested to play a central role in fetomaternal cell-to-cell fusion in most ruminant placentas, including both Bovinae and Caprinae (16); however, that study did not confirm protein expression in placental tissues. Moreover, the authors of that study found that Syncytin-Rum1 failed to induce cell-to-cell fusion under a physiological pH of 7.0 but exhibited fusogenicity under acidic conditions (pH 5.0). We also conducted a quantitative fusion assay using bovine Syncytin-Rum1 and bovine endometrial cells. We expressed C-terminally FLAG-tagged Syncytin-Rum1 in Cos-7 cells (Fig. 6A) and performed a fusion-dependent luciferase assay using bovine endometrial cells as target cells. The results revealed that Syncytin-Rum1 fusogenicity was much lower than that of Fematrin-1 under physiological conditions (Fig. 6B). We also confirmed that the fusion activity of Syncytin-Rum1 increased when cells were subjected to an acidic shock (pH 5.0) (data not shown); however, evidence of such a low pH in bovine placenta has not been reported.
Fig 6.
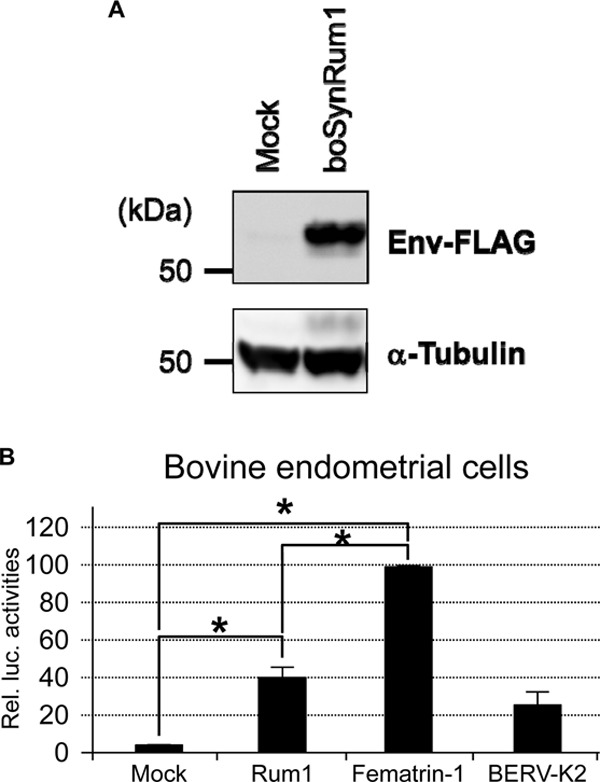
Expression and fusogenic activity of bovine Syncytin-Rum1 in vitro. (A) Immunoblotting of C-terminally FLAG-tagged bovine Syncytin-Rum1 (boSynRum1). Bovine Syncytin-Rum1 was cloned into the pSG5 vector and C-terminally FLAG tagged by using In-Fusion HD. Cos-7 cell lysates transfected with the pSG5 vector or pSG5boSynRum1FLAG were subjected to SDS-PAGE followed by blotting onto a PVDF membrane. FLAG-tagged proteins were detected by using anti-FLAG M2 and horseradish peroxidase-conjugated anti-mouse IgG antibodies. pSG5, which is a backbone plasmid, was used as a negative control. (B) Fusion assay of bovine Syncytin-Rum1 (Rum1). Cos-7 cells cotransfected with pT7EMCVluc, pRL-TK, and the indicated Env expression vectors were cocultured with bovine endometrial cells transfected with pCAGT7pol. Cell lysates were then subjected to dual-luciferase reporter assays. Assays were repeated as 3 independent experiments (n = 3), and each value for luciferase activity is shown as the mean ± standard error. Statistical analysis was performed by a Student t test, and differences were considered significant at a P value of <0.05 (indicated by an asterisk).
DISCUSSION
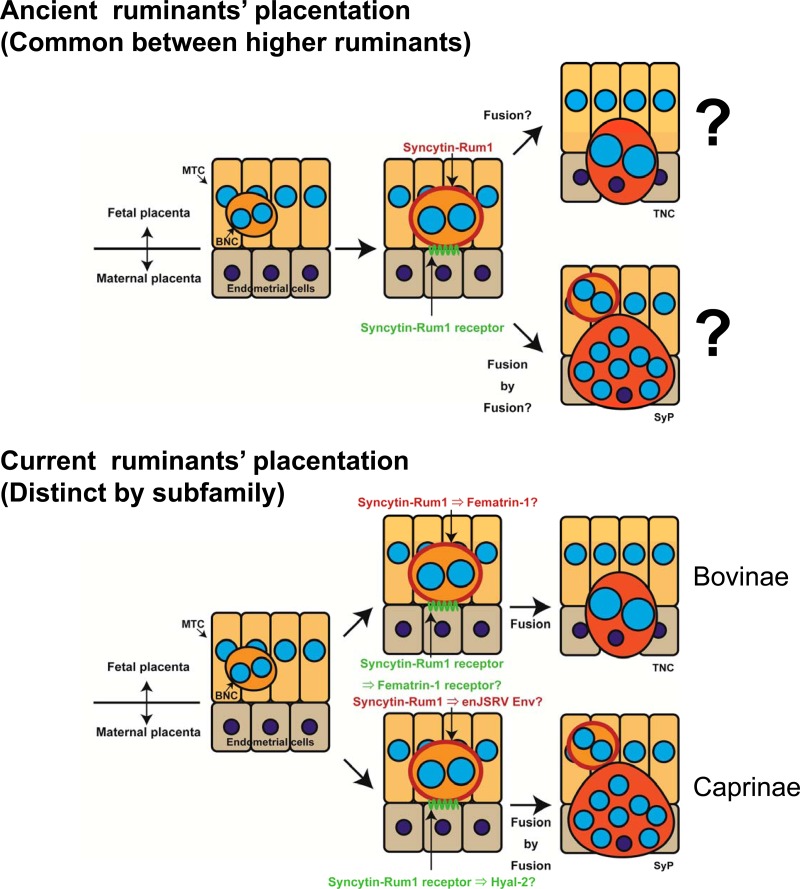
Based on our findings, we suggest a model for the mechanism of TNC generation in the bovine placenta (Fig. 7). The current dogma for the formation of TNCs in the bovine placenta is that cells from the fetal placenta develop into BNCs by acytokinetic mitoses and that these cells migrate through the surrounding fetal trophoblast until they come into contact with maternal uterine epithelial cells, with which they then fuse, forming a TNC. The TNCs do not survive long before degenerating (34). We propose that it is in fact Fermatrin-1 that is predominantly responsible for this fusion process. Our study provides convincing evidence for the fusogenic characteristics of this protein and its specific expression in appropriate tissues in the bovine placenta to support this proposal. The protein is also present in Bovinae that are evolutionarily and closely related to cattle and might express the same fusogenic phenotype in their placentas. Fematrin-1 is not present in more distantly related ruminants (sheep and goats) that phenotypically form large multinucleated SyPs rather than TNCs (Fig. 3B and data not shown), suggesting that Fematrin-1 is one of the crucial elements generating diversity of placentas in Bovidae.
Fig 7.
Schematic representation of the proposed model for functional replacement (or substitution) of Syncytin-Rum1 by Fematrin-1's and enJSRV's Env, which might be nascent ERV Envs.
It is plausible that the acquisition of Fematrin-1 in Bovinae is an example of the “baton pass hypothesis” that was previously proposed by Nakamura and Imakawa (35). In the baton pass hypothesis, a new gene, such as an ERV, gradually replaces the function of a preexisting gene during endogenization and placental evolution. In this study, we clearly showed that Fematrin-1 is more fusogenic than Syncytin-Rum1 under physiologically relevant conditions. This evidence indicates that Fematrin-1 rather than Syncytin-Rum1 might be the main player involved in the formation of TNCs in current Bovinae species. Previously, Cornelis et al. reported that Syncytin-Rum1 exhibited fusogenicity at pH 5.0 (16). However, to the best of our knowledge, such a low pH in bovine placenta has not been reported. Nonetheless, the fusogenicity of Syncytin-Rum1 might have a different role from that of Fematrin-1. Additional studies are needed in order to link Syncytin-Rum1 with fetomaternal cell-to-cell fusion. In addition, Cornelis et al. failed to detect fusogenicity of Fematrin-1 (termed bos–env-4 in their study) (16). This discrepancy may be due to several reasons: (i) our fusion assay may be more sensitive than that used in their study because we used luciferase as a reporter to measure cell-to-cell fusion activity; (ii) Fematrin-1 expression is complicated because BERV-K1 needs its 3′ LTR to produce the protein efficiently, as indicated in Fig. 1B; and (iii) efficient overexpression of Fematrin-1 protein is dependent on the cell line being used (data not shown). Although we do not know how Cornelis et al. constructed the expression vector of Fematrin-1 and which cell lines they used, they might have failed in efficient overexpression of Fematrin-1. It is unclear how Syncytin-Rum1 and enJSRV Env (4, 13) cooperated in the formation of SyPs in current Caprinae species, because the cell-to-cell fusogenic potency of enJSRV Env has not been shown.
We believe that BERV-K1 infected a common ancestral Bovinae species and integrated into an appropriate locus to function in placenta, such as in the FAT2 gene, which is preferentially expressed in cattle (Bovinae) and sheep (Caprinae) placentas. It is possible that Fematrin-1 generated “Bovinae-specific” hybrid TNCs in bovine placentas after replacing Syncytin-Rum1 during the fusion process (Fig. 7). Presumably, the formation of TNCs in Bovinae was advantageous over SyPs in Caprinae to maintain a longer gestation period in Bovinae (280 days in cattle and water buffalos and 210 days in marshbuck) than in Caprinae (150 days in sheep and goats), because the turnover of TNCs is more efficient. Syncytin-Rum1 could have played a central role in the fetomaternal cell-to-cell fusion of ancient Bovidae; however, the fusogenic activity of Syncytin-Rum1 might have been attenuated during the evolution of Bovinae after the acquisition of Fematrin-1, perhaps changing its role along the way. Indeed, Syncytins are considered to have immunosuppressive function to subdue maternal immunity for avoiding fetal rejection (36). Furthermore, Syncytin-Rum1 but not Fematrin-1 has a putative immunosuppressive domain within its TM region, which has been subjected to purifying selection, suggesting that it may be functional (16). A similar relationship between Syncytin-1 and Synctyin-2 has been observed (9, 36). Syncytin-2 was integrated into primate genomes much earlier than Syncytin-1, and its fusogenic activity was found to be much lower than that of Syncytin-1 (36). Furthermore, the immunosuppressive activity of Syncytin-2 was much higher than that of Syncytin-1 (36). Baton pass evolution might have continuously occurred throughout the evolution of mammalian placentas, and it may well explain why cell-to-cell fusion mechanisms of trophoblasts seem to have evolved in a convergent manner using distinct ERVs in mammals. Baton pass evolution by acquisition of different ERV Envs would occur faster than gene multiplication by retrotransposition or gene duplication because the animals can try a set of different Envs that utilize different cellular receptors to initiate cell fusion.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to Rachael Tarlinton (University of Nottingham, Loughborough, United Kingdom), and Joanne Martin (LGC, Twickenham, United Kingdom) for their helpful critical reading. We thank Y. Kitagawa (Shiga University of Medical Science, Shiga, Japan) for providing plasmids for fusion assays. We also thank E. Hondo, I. Nishizawa, N. Yokoyama, and S. Zakimi for helping us in collecting tissue samples. We are grateful to P. Gee and K. Imakawa for helpful discussion and preparation of the manuscript.
This study was supported by the Programme for Promotion of Basic and Applied Research for Innovations Bio-Oriented Industry, Ministry of Agriculture, Forestry, and Fisheries of Japan. Y.N. is a research fellow of the Japan Society for the Promotion of Science. Y.N. is currently and S.N. was supported by grants-in-aid from the Japanese Society for the Promotion of Science under fellow and research project numbers 25-5281 and 11J03243, respectively.
Footnotes
Published ahead of print 17 July 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01398-13.
REFERENCES
- 1.Klisch K, Pfarrer C, Schuler G, Hoffmann B, Leiser R. 1999. Tripolar acytokinetic mitosis and formation of feto-maternal syncytia in the bovine placentome: different modes of the generation of multinuclear cells. Anat. Embryol. 200:229–237 [DOI] [PubMed] [Google Scholar]
- 2.Wooding FB, Morgan G, Adam CL. 1997. Structure and function in the ruminant synepitheliochorial placenta: central role of the trophoblast binucleate cell in deer. Microsc. Res. Tech. 38:88–99 [DOI] [PubMed] [Google Scholar]
- 3.Carvalho AF, Klisch K, Miglino MA, Pereira FT, Bevilacqua E. 2006. Binucleate trophoblast giant cells in the water buffalo (Bubalus bubalis) placenta. J. Morphol. 267:50–56 [DOI] [PubMed] [Google Scholar]
- 4.Black SG, Arnaud F, Palmarini M, Spencer TE. 2010. Endogenous retroviruses in trophoblast differentiation and placental development. Am. J. Reprod. Immunol. 64:255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klisch K, Hecht W, Pfarrer C, Schuler G, Hoffmann B, Leiser R. 1999. DNA content and ploidy level of bovine placentomal trophoblast giant cells. Placenta 20:451–458 [DOI] [PubMed] [Google Scholar]
- 6.Wildman DE, Chen C, Erez O, Grossman LI, Goodman M, Romero R. 2006. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc. Natl. Acad. Sci. U. S. A. 103:3203–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klisch K, Mess A. 2007. Evolutionary differentiation of cetartiodactyl placentae in the light of the viviparity-driven conflict hypothesis. Placenta 28:353–360 [DOI] [PubMed] [Google Scholar]
- 8.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, Keith JC, Jr, McCoy JM. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785–789 [DOI] [PubMed] [Google Scholar]
- 9.Blaise S, de Parseval N, Bénit L, Heidmann T. 2003. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. U. S. A. 100:13013–13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupressoir A, Marceau G, Vernochet C, Bénit L, Kanellopoulos C, Sapin V, Heidmann T. 2005. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. U. S. A. 102:725–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. 2009. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology 6:107. 10.1186/1742-4690-6-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Véron G, Mulot B, Dupressoir A, Heidmann T. 2012. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl. Acad. Sci. U. S. A. 109:E432–E441. 10.1073/pnas.1115346109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, Spencer TE. 2006. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc. Natl. Acad. Sci. U. S. A. 103:14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baba K, Nakaya Y, Shojima T, Muroi Y, Kizaki K, Hashizume K, Imakawa K, Miyazawa T. 2011. Identification of novel endogenous betaretroviruses which are transcribed in the bovine placenta. J. Virol. 85:1237–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koshi K, Suzuki Y, Nakaya Y, Imai K, Hosoe M, Takahashi T, Kizaki K, Miyazawa T, Hashizume K. 2012. Bovine trophoblastic cell differentiation and binucleation involves enhanced endogenous retrovirus element expression. Reprod. Biol. Endocrinol. 10:41. 10.1186/1477-7827-10-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornelis G, Heidmann O, Degrelle SA, Vernochet C, Lavialle C, Letzelter C, Bernard-Stoecklin S, Hassanin A, Mulot B, Guillomot M, Hue I, Heidmann T, Dupressoir A. 2013. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc. Natl. Acad. Sci. U. S. A. 110:E828–E837. 10.1073/pnas.1215787110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arii J, Goto H, Suenaga T, Oyama M, Kozuka-Hata H, Imai T, Minowa A, Akashi H, Arase H, Kawaoka Y, Kawaguchi Y. 2010. Non-muscle myosin IIA is a functional entry receptor for herpes simplex virus-1. Nature 467:859–862 [DOI] [PubMed] [Google Scholar]
- 18.Aoki Y, Aizaki H, Shimoike T, Tani H, Ishii K, Saito I, Matsuura Y, Miyamura T. 1998. A human liver cell line exhibits efficient translation of HCV RNAs produced by a recombinant adenovirus expressing T7 RNA polymerase. Virology 250:140–150 [DOI] [PubMed] [Google Scholar]
- 19.Koshi K, Ushizawa K, Kizaki K, Takahashi T, Hashizume K. 2011. Expression of endogenous retrovirus-like transcripts in bovine trophoblastic cells. Placenta 32:493–499 [DOI] [PubMed] [Google Scholar]
- 20.Bénit L, Dessen P, Heidmann T. 2001. Identification, phylogeny, and evolution of retroviral elements based on their envelope genes. J. Virol. 75:11709–11719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33:511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felsenstein J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17:368–376 [DOI] [PubMed] [Google Scholar]
- 23.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 24.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25:1307–1320 [DOI] [PubMed] [Google Scholar]
- 26.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 57:758–771 [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 29.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591 [DOI] [PubMed] [Google Scholar]
- 30.Yamauchi N, Takezawa T, Kizaki K, Herath CB, Hashizume K. 2003. Proliferative potential of endometrial stromal cells, and endometrial and placental expression of cyclin in the bovine. J. Reprod. Dev. 49:553–560 [DOI] [PubMed] [Google Scholar]
- 31.Garten W, Hallenberger S, Ortmann D, Schäfer W, Vey M, Angliker H, Shaw E, Klenk HD. 1994. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie 76:217–225 [DOI] [PubMed] [Google Scholar]
- 32.Mohamad K, Olsson M, van Tol HT, Mikko S, Vlamings BH, Andersson G, Rodríguez-Martínez H, Purwantara B, Paling RW, Colenbrander B, Lenstra JA. 2009. On the origin of Indonesian cattle. PLoS One 4:e5490. 10.1371/journal.pone.0005490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernández Fernández M, Vrba ES. 2005. A complete estimate of the phylogenetic relationships in Ruminantia: a dated species-level supertree of the extant ruminants. Biol. Rev. Camb. Philos. Soc. 80:269–302 [DOI] [PubMed] [Google Scholar]
- 34.Wooding FB. 1992. Current topic. The synepitheliochorial placenta of ruminants: binucleate cell fusions and hormone production. Placenta 13:101–113 [DOI] [PubMed] [Google Scholar]
- 35.Nakamura Y, Imakawa K. 2011. Retroviral endogenization and its role in the genital tract during mammalian evolution. J. Mamm. Ova Res. 28:203–218 [Google Scholar]
- 36.Mangeney M, Renard M, Schlecht-Louf G, Bouallaga I, Heidmann O, Letzelter C, Richaud A, Ducos B, Heidmann T. 2007. Placental syncytins: genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl. Acad. Sci. U. S. A. 104:20534–20539 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.