Abstract
Reverse genetic analyses of negative-strand RNA (NSR) viruses have provided enormous advances in our understanding of animal viruses over the past 20 years, but technical difficulties have hampered application to plant NSR viruses. To develop a reverse genetic approach for analysis of plant NSR viruses, we have engineered Sonchus yellow net nucleorhabdovirus (SYNV) minireplicon (MR) reporter cassettes for Agrobacterium tumefaciens expression in Nicotiana benthamiana leaves. Fluorescent reporter genes substituted for the SYNV N and P protein open reading frames (ORFs) exhibited intense single-cell foci throughout regions of infiltrated leaves expressing the SYNV MR derivatives and the SYNV nucleocapsid (N), phosphoprotein (P), and polymerase (L) proteins. Genomic RNA and mRNA transcription was detected for reporter genes substituted for both the SYNV N and P ORFs. These activities required expression of the N, P, and L core proteins in trans and were enhanced by codelivery of viral suppressor proteins that interfere with host RNA silencing. As is the case with other members of the Mononegavirales, we detected polar expression of fluorescent proteins and chloramphenicol acetyltransferase substitutions for the N and P protein ORFs. We also demonstrated the utility of the SYNV MR system for functional analysis of SYNV core proteins in trans and the cis-acting leader and trailer sequence requirements for transcription and replication. This work provides a platform for construction of more complex SYNV reverse genetic derivatives and presents a general strategy for reverse genetic applications with other plant NSR viruses.
INTRODUCTION
A revolution in understanding of RNA viruses was initiated more than 25 years ago by the discovery that plus-strand viral RNA genomes could be reverse transcribed into cDNAs, and that purified plasmids containing these cDNAs could be faithfully transcribed in vitro to yield infectious “genomic” RNAs (gRNAs) (1). This finding permitted genetic manipulation of cDNAs and recovery of mutant RNAs whose effects on virus biology and pathology could be assessed. However, direct application of this strategy to negative-strand RNA (NSR) viruses was not possible because the minimal infectious unit of NSR viruses is the viral nucleocapsid (vNC), rather than naked viral gRNA (2, 3). Moreover, the development of methods for reconstitution of nucleocapsids (NCs) from cloned NSR cDNAs proved to be an enormous challenge that required nearly a decade to resolve (4, 5).
Generation of biologically active NCs was eventually solved with animal NSR viruses by cotransfecting permissive cell lines with plasmids designed to transcribe positive-strand or “antigenomic” RNA (agRNA) derivatives, along with plasmids able to express the three NC core proteins (6, 7, 8, 9). These approaches resulted in in vivo assembly of NCs that were able to replicate and jump-start the replication process. In seminal experiments to recover full-length recombinant Rabies virus and Vesicular stomatitis virus (VSV), ectopic expression of viral core proteins was facilitated by use of cell lines expressing bacteriophage T7 polymerase for transcription of the nucleocapsid protein (N), phosphoprotein (P), and polymerase protein (L) mRNAs and agRNAs from transfected plasmids (6, 8, 9). In practice, T7 promoters positioned for precise initiation at the 5′ leader ( ) sequence and generation of the exact 3′ terminus of the trailer (
) sequence and generation of the exact 3′ terminus of the trailer ( ) sequence by hepatitis delta virus ribozyme (ΔRz) cleavage were critical for recovery of recombinant virus, and a number of studies showed that agRNA transcripts yielded substantially more efficient recovery of recombinant virus than gRNA transcripts (7, 10). The three core proteins and processed agRNAs were able to assemble into antigenomic nucleocapsids (agNCs) that could function in replication of gRNAs and form genomic-sense nucleocapsids (gNCs) with the ectopic core proteins. The gNCs were biologically active and were able to initiate replication cycles due to their abilities to function in transcription of mRNAs and replication of agRNA intermediates. As replication of the biologically active recombinants proceeded, infected focal cells lysed and plaques formed as adjacent cells were infected with recombinant virus.
) sequence by hepatitis delta virus ribozyme (ΔRz) cleavage were critical for recovery of recombinant virus, and a number of studies showed that agRNA transcripts yielded substantially more efficient recovery of recombinant virus than gRNA transcripts (7, 10). The three core proteins and processed agRNAs were able to assemble into antigenomic nucleocapsids (agNCs) that could function in replication of gRNAs and form genomic-sense nucleocapsids (gNCs) with the ectopic core proteins. The gNCs were biologically active and were able to initiate replication cycles due to their abilities to function in transcription of mRNAs and replication of agRNA intermediates. As replication of the biologically active recombinants proceeded, infected focal cells lysed and plaques formed as adjacent cells were infected with recombinant virus.
Over the past decade, related strategies have been developed for reverse genetic analyses of members of all of the major animal NSR virus families (7, 11–16). These procedures are quite inefficient, but visible plaques that appear on cell lawns provide a valuable resource for phenotypic and biochemical studies (17). Moreover, these innovative strategies and subsequent second-generation adaptations not only have led to an increased understanding of the replication and disease potential of animal NSR viruses but also have permitted many practical applications such as vaccine production using recombinant viruses with attenuated virulence (18, 19) and the use of NSR virus vectors for foreign gene expression (20–22). Unfortunately, several technical problems associated with plants have hampered recovery of recombinant NSR plant viruses, even though these viruses have a number of physicochemical properties in common with their animal virus counterparts (23–28).
In the current study, we have applied agroinfiltration to circumvent the most serious problems plaguing development of reverse genetic systems of plant NSR viruses. Our approach relies on our previous studies with Agrobacterium tumefaciens-mediated expression of Sonchus yellow net nucleorhabdovirus (SYNV) core N, P, and L proteins in Nicotiana benthamiana (29, 30). We have applied this methodology for delivery of the viral core protein genes into agroinfiltrated leaf cells in conjunction with plasmids containing a Cauliflower mosaic virus (CaMV) double 35S (35S2) promoter for transcription of SYNV agRNA derivatives. The transcribed agRNAs have ribozymes (Rzs) positioned for cleavage of the 5′ and 3′ termini corresponding to the SYNV  (31) and
(31) and  (32) sequences to produce a processed agRNA competent for encapsidation by the coexpressed N, P, and L core proteins. We also have developed applications for functional analyses of the core proteins and the virus-specific cis elements of the SYNV minireplicon (MR). Hence, our reverse genetic approach provides the basis for a more detailed understanding of replication of SYNV, a plant-adapted rhabdovirus, and for future construction of reverse genetic derivatives capable of autonomous virus replication.
(32) sequences to produce a processed agRNA competent for encapsidation by the coexpressed N, P, and L core proteins. We also have developed applications for functional analyses of the core proteins and the virus-specific cis elements of the SYNV minireplicon (MR). Hence, our reverse genetic approach provides the basis for a more detailed understanding of replication of SYNV, a plant-adapted rhabdovirus, and for future construction of reverse genetic derivatives capable of autonomous virus replication.
MATERIALS AND METHODS
General.
Plasmid DNA was extracted using an alkaline lysis procedure (33) and used for cloning and sequencing. DNAs for subcloning were recovered from gel slices using a Qiaex II gel extraction kit (Qiagen, Valencia, CA). Restriction enzymes were obtained from New England BioLabs (Beverly, MA), and chemicals were purchased from Sigma Chemical (St. Louis, MO) or Fisher Scientific (Springfield, NJ). Oligonucleotides used for PCR amplifications were purchased from Operon Technologies (Alameda, CA), and amplifications were performed using Pfu DNA polymerase (Stratagene, La Jolla, CA). PCR products were purified by gel electrophoresis, cloned into TOPO plasmids according to the recommendations of the manufacturer (Invitrogen, Carlsbad, CA), and subsequently transferred into A. tumefaciens pGD expression vectors (29). All clones were sequenced at the University of California—Berkeley DNA Sequencing Facility (Berkeley, CA) or Invitrogen (Shanghai, China).
Construction of an Rz processing cassette for reporter gene cloning.
Several steps were carried out to construct a bacteriophage T7 transcription plasmid (pSYNVHRz,ΔRz) for analysis of ribozyme (Rz) processing activities and use in cloning reporter genes (see Section SI in the supplemental material). The pSYNVHRz,ΔRz plasmid contains a bacteriophage T7 promoter (T7-Pro) inserted upstream of a hammerhead ribozyme (HRz), described by Herold and Andino (34), followed by the SYNV  sequence, the 5′ untranslated region (UTR) of the N gene, a multiple-cloning site, the 3′ UTR of the SYNV L gene, the
sequence, the 5′ untranslated region (UTR) of the N gene, a multiple-cloning site, the 3′ UTR of the SYNV L gene, the  sequence, and the hepatitis delta virus ribozyme (ΔRz). The HRz was designed to process transcribed RNAs at a cytosine residue immediately preceding the 5′ terminus of the
sequence, and the hepatitis delta virus ribozyme (ΔRz). The HRz was designed to process transcribed RNAs at a cytosine residue immediately preceding the 5′ terminus of the  sequence, and the ΔRz was positioned to cleave at the exact 3′ terminus of the
sequence, and the ΔRz was positioned to cleave at the exact 3′ terminus of the  sequence (35, 36). Details of the construction of pSYNVHRz,ΔRz and experiments to verify HRz and ΔRz cleavage of agRNA transcripts to produce termini complementary to the 3′ and 5′ ends of SYNV gRNA are described in Section SI in the supplemental material.
sequence (35, 36). Details of the construction of pSYNVHRz,ΔRz and experiments to verify HRz and ΔRz cleavage of agRNA transcripts to produce termini complementary to the 3′ and 5′ ends of SYNV gRNA are described in Section SI in the supplemental material.
Engineering of SYNV minireplicon reporter cassettes.
pSYNV-MRrsGFP-CAT was engineered by substituting reporter genes between the NheI and BsrGI sites of pSYNVHRzΔRz (see Section SII in the supplemental material for cloning details). These intermediates were then inserted into pCAMBIA1300 to create the pSYNV-MR reporter cassettes used for cloning into pGD expression vectors (29). A red-shifted green fluorescent protein (rsGFP) gene from pGD-GFP (29) was substituted for the N protein open reading frame (ORF), and this was followed by gene junction (GJ) sequences separating the SYNV N and P genes (37). The chloramphenicol acetyltransferase (CAT) gene was then amplified from a Barley stripe mosaic virus (BSMV) gRNA-βbCAT construct described earlier (38) and inserted at a position corresponding to the P protein ORF to yield the pSYNV-MRrsGFP-CAT reporter intermediate (see Section SII in the supplemental material for details of the constructions).
For construction of pSYNV-MReGFP-CAT, MRCAT-eGFP, MReGFP-DsRed, and MRDsRed-eGFP, the pSYNV-MRrsGFP-CAT plasmid was used as a scaffold sequence and template. The enhanced GFP (eGFP) and DsRed reporter genes were amplified from plasmid pEGFP-N1 (Clontech, Palo Alto, CA) and pGD-DsRed (29), respectively. The reporter genes were fused to flanking SYNV regulatory sequences by ligation and PCR amplification. After four consecutive PCR amplifications, the resulting full-length expression cassettes were substituted for the rsGFP-CAT reporter cassette in pSYNV-MRrsGFP-CAT. Note that these manipulations resulted in substitution of new reporter genes in the N and P positions with the same flanking SYNV sequences as those in pSYNV-MRrsGFP-CAT (see Section SIII in the supplemental material).
SYNV N, P, and L genes used for agroinfiltration.
Plasmids harboring the SYNV N, P, and L genes used in these experiments were inserted into pGD vectors as described previously (29). Mutant N proteins with various nuclear localization and interaction phenotypes were described by Deng et al. (30). During the course of evaluating the P protein in pGD (29), we discovered a single nucleotide error in the initial sequence (39). This error resulted in the failure to identify a termination codon that reduces the predicted size of the P protein to 308 amino acids from the 346 residues previously deduced (39, 40). Sequencing of 10 independent P protein clones amplified from purified virus or SYNV-infected leaf RNA revealed several additional sequence variants that affected individual amino acids, but all of these had a predicted length of 308 residues. The L protein mRNA (41) was amplified from RNA of SYNV-infected N. benthamiana by reverse transcription-PCR (RT-PCR) with the primers “L-sense” and “L-anti” (see Table S1 in the supplemental material) and cloned into the SalI site of the TOPO-TA vector (Invitrogen, Carlsbad, CA). After verification by restriction analysis and sequencing, the L protein gene was subcloned into pGD.
The N protein mutant derivatives (F42A, Y51A, and YIY) that affect N-N and N-P heterologous interactions and subnuclear localization (30), as well as the nuclear localization signal (NLS) mutant (KKRR) constructed by Goodin et al. (40), were also analyzed in planta to assess their ability to function in MR amplification and reporter gene expression.
Mutant minireplicon derivatives.
To destroy HRz activity, a 16-nucleotide (nt) deletion preceding the normal HRz  sequence cleavage site was constructed in the 5′ HRz of the pSYNV-MReGFP-DsRed cassette. To assess the biological importance of the
sequence cleavage site was constructed in the 5′ HRz of the pSYNV-MReGFP-DsRed cassette. To assess the biological importance of the  and
and  regions, 18-nt deletions were incorporated into the
regions, 18-nt deletions were incorporated into the  and
and  termini of pSYNV-MReGFP-DsRed (see Section SIII in the supplemental material for construction of mutant minireplicon derivatives).
termini of pSYNV-MReGFP-DsRed (see Section SIII in the supplemental material for construction of mutant minireplicon derivatives).
Agroinfiltration-induced expression and confocal microscopy analyses of SYNV MR reporters in planta.
N. benthamiana plants used for routine SYNV maintenance were grown in a greenhouse at ambient temperature, and plants used for infiltration were maintained in a growth room at 23°C at 1,000 lumens with a 16-hour day/night regimen. The pSYNV-MR cassettes were cloned into pGD expression vectors (29) and transformed into A. tumefaciens strain EHA105 by a freeze-thaw method (42), and bacteria containing the cassettes were selected on LB agar plates containing rifampin (50 μg/ml) and kanamycin (50 μg/ml). To prepare for agroinfiltration, 1- to 2-day-old EHA105 cells were scraped from the plates and resuspended in buffer containing 10 mM MgCl2 and 10 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.6. Cell suspensions were adjusted to an A600 of 0.8 and incubated for 2 to 4 h at room temperature in the presence of 100 μM acetosyringone. Immediately before infiltration, the pGD:N, pGD:P, pGD:L, and pGD:MR cultures were mixed in the desired combinations, and bacteria containing the BSMV pGD:γb (43), Tomato bushy stunt virus (TBSV) pGD:p19 (44), and/or Tobacco etch virus (TEV) P1/HC-Pro pGD:HC-Pro (45) plasmid were included in the mix to minimize gene silencing. Bacterial mixtures were then infiltrated into the lower sides of N. benthamiana leaves with a 1-ml syringe (29).
Fluorescence in infiltrated leaves was visualized with a Zeiss Lumar epifluorescence dissecting microscope, and higher-resolution imaging of individual mesophyll cells was carried out with a Zeiss LSM 510 or LSM 780 confocal microscope (Thornwood, New York, NY). The excitation wavelength used for GFP was 470/40 nm, and the emission wavelength was 525/50 nm. DsRed was excited at 545/25 nm, and the emission between 605/70 nm was selected for visualization (46).
Analysis of proteins.
Proteins were extracted from agroinfiltrated N. benthamiana leaves harvested at 5 to 10 days postinfiltration (dpi) and evaluated by Western blotting (30). Proteins separated by SDS-PAGE were either stained with Coomassie blue or transferred to nitrocellulose membranes and detected with polyclonal antiserum specific to the SYNV N, P, or L protein (47) or monoclonal antibodies elicited against GFP (Clontech Laboratories, Mountain View, CA) and red fluorescent protein (RFP) (Huada, Beijing, China).
CAT assays.
CAT activity was assayed as described by Ausubel et al. (48) with modifications. Briefly, 1 g of infiltrated leaves was ground with liquid nitrogen in a mortar and pestle and then extracted with 3 ml of extraction buffer (25 mM Tris-HCl [pH 7.5], 2 mM MgCl2). The crude extract was centrifuged at 14,000 rpm in an Eppendorf microcentrifuge (5417C) for 5 min at room temperature, and the supernatant was transferred to a 1.5-ml Eppendorf tube and heated to 65°C for 10 min. The extract was then incubated with the fluorescent Bodipy FL chloramphenicol (BCAM) substrate (Molecular Probes, NY) and acetyl coenzyme A (acetyl-CoA) at 37°C for 2 h. Subsequently, the fluorescent substrate and the acetylated derivatives were extracted by addition of 0.5 ml ice-cold ethyl acetate and the ethyl acetate solvent recovered and evaporated in a speed vacuum apparatus. The nonvolatile reaction products were dissolved in a small volume of ethyl acetate and resolved by thin-layer chromatography (TLC) on silica gel plates using a solvent mixture of chloroform and methanol (∼87:13, vol/vol). The chromatogram was dried, and the reaction products were visualized by UV illumination and photographed.
RNA analyses.
For Northern hybridization, total RNA was isolated from infiltrated regions of leaves at 10 dpi using TRIzol reagent (Sigma), resolved in 1.2% agarose-formaldehyde gels, and probed with 32P-labeled GFP RNA probes. The probe designations and sequences used to amplify each probe are shown in Table 1. To transcribe probes specific for gRNA or agRNA detection, the pSYNV-MRrsGFP-CAT cassette was used as a template. Primers used for probe transcription were designed with T7 promoters in the desired genomic or antigenomic orientations, and the templates were transcribed in vitro with T7 RNA polymerase in the presence of [32P]UTP. Probes used to detect GFP gRNAs or agRNAs were amplified with the T7GFPF and 3′GFP primer pair or the 5′GFP and 3′T7rsGFPR primer pair, respectively. A full-length agRNA transcript probe to detect gRNAs was amplified with the T7 ldrF and  rev primers, and the full-length agRNA probe was amplified using the
rev primers, and the full-length agRNA probe was amplified using the  senseF and T7 trlR primers. To construct probes to detect the
senseF and T7 trlR primers. To construct probes to detect the  sequence, pSYNV-MRrsGFP-CAT was first amplified using the rz
sequence, pSYNV-MRrsGFP-CAT was first amplified using the rz sense and N5′UTR R primer pair to generate a 250-nt template for in vitro synthesis of the
sense and N5′UTR R primer pair to generate a 250-nt template for in vitro synthesis of the  genomic and antigenomic probes. The 250-nt product was then used as a template with either the
genomic and antigenomic probes. The 250-nt product was then used as a template with either the  sense or the
sense or the  rev primer in the presence of [32P]dCTP for synthesis of a 208-nt probe to target ag
rev primer in the presence of [32P]dCTP for synthesis of a 208-nt probe to target ag and agMR RNA sequences and a 193-nt probe to hybridize to gRNA products, respectively. To detect the genomic trailer (
and agMR RNA sequences and a 193-nt probe to hybridize to gRNA products, respectively. To detect the genomic trailer ( ) RNA, pSYNV-MRrsGFP-CAT was amplified using the
) RNA, pSYNV-MRrsGFP-CAT was amplified using the  sense and SYNV R primer pair to generate a 162-nt product. Next, the 162-nt product was used as a transcription template with the
sense and SYNV R primer pair to generate a 162-nt product. Next, the 162-nt product was used as a transcription template with the  sense primer in the presence of [32P]dCTP to generate a DNA probe. To detect the positive (mRNA)-sense CAT mRNA, pSYNV-MRrsGFP-CAT was amplified using the NPJF and L3′mRNAR primer pair to generate an 800-nt product. This PCR product was used as a template with the L3′mRNAR primer and [32P]dCTP to generate a probe to target the CAT agRNA sequence.
sense primer in the presence of [32P]dCTP to generate a DNA probe. To detect the positive (mRNA)-sense CAT mRNA, pSYNV-MRrsGFP-CAT was amplified using the NPJF and L3′mRNAR primer pair to generate an 800-nt product. This PCR product was used as a template with the L3′mRNAR primer and [32P]dCTP to generate a probe to target the CAT agRNA sequence.
Table 1.
Primers used to create probes for detection of genomic and antigenomic RNA transcripts from the SYNV minireplicon
| Clone name | Directiona | Primer name | Primer sequence | Purpose |
|---|---|---|---|---|
| T7rsGFP | F | T7rsGFPF | TAATACGACTCACTATAGGCGGAGAAGAACTTTTCACTGG | To amplify a plus-sense rsGFP probe for detection of minus-sense transcripts harboring rsGFP |
| R | 3GFP | TTAGTATAGTTCATCCAT | ||
| rsGFPT7 | F | 5GFP | ATGGGTAAAGGAGAAGAAC | To amplify a minus-sense rsGFP probe to detect plus-sense rsGFP transcripts |
| R | T7rsGFPR | AATACGACTCACTATAGGCCCCAGCAGCTGTTACAAAC | ||
| T7MR | F | T7 F F |
TAATACGACTCACTATAGGCAGAGACAGAAACTCAGAAAATACAATCACCG | Template to create a size marker to assess mobility of minus-sense MR transcripts |
| R |
 rev rev |
TTATACTTCAAGAGATATAGATGTACACAATTTCCTGGAAG | ||
| MRT7 | F |
 sense sense |
AGAGACAGAAACTCAGAAAATACAATCACCG | Template to create a size marker standard for the plus-sense MR transcript |
| R | T7
|
TAATACGACTCACTATAGGCAGAGACAAAAGCTCAGAACAATCCC | ||
| F | rz sense sense |
GTCTCTCTGATGAGGCCGAAAGGCCGAAAACCCGGTATCCCGGGTTC | To amplify the leader probe transcription template | |
| R | N5′UTR R | ATGCTAGCGATTACCTGCAATTAAAATAC | ||
 for positive-sense for positive-sense |
R |
 rev rev |
TTTTTCTTTAATCTACTAATCAGTTAAAATCAC | To amplify genomic probes for detection of the leader RNA and full-length antigenomic RNAs |
 for negative-sense for negative-sense |
F |
 sense sense |
AGAGACAGAAACTCAGAAAATACAATCACCG | To detect full-length genomic transcript with the plus-sense leader probe |
| CAT | F | NPJF | ACTACAGCCACAACTCTACC | Template to prepare the CAT probe |
| R | L3′mRNAR | TTTTTCTTATACTTTCAAGAGATATAG | ||
| CAT for positive sense | R | L3′mRNAR | TTTTTCTTATACTTTCAAGAGATATAG | To detect antigenomic CAT transcript |
| F |
 sense sense |
ATCGCTAGCTGGCCATGTACATCTATATCTCTTGAA | Template to prepare the trailer probe | |
| R | SYNV R | AGAGACAAAAGCTCAGAACAATCCC | ||
| Trailer for negative sense | F |
 sense sense |
ATCGCTAGCTGGCCATGTACATCTATATCTCTTGAA | To synthesize a trailer probe for detection of genomic RNAs |
| RT-PCR | ||||
| Reverse transcription | F |
 sense sense |
AGAGACAGAAACTCAGAAAATACAATCACCG | To make a genomic sense cDNA template |
| PCR | F | 5eGFP-NheI | GCTAGCATGGTGAGCAAGGGCGAGGAG | For amplification of minus-sense eGFP genomic transcripts |
| R | 3eGFP-NPJ | GTAGAGTTGTGGCTGTAGTTTATTACTTGTACAGCTCGTCCAT |
F, forward; R, reverse.
Nucleotide sequence accession numbers.
The complete sequence from the transfer DNA (T-DNA) right border to the left border of pSYNV-MRrsGFP-CAT has been submitted to GenBank and can be found under accession number JNO38404. A revised P protein sequence has been submitted to GenBank under accession number AY971951. The L protein gene has GenBank accession number M87829.
RESULTS
Reporter gene expression from SYNV minireplicons in planta.
Reverse genetic strategies devised for NSR viruses have usually relied on transcription of viral agRNA derivatives with authentic 5′ and 3′ termini in cells coexpressing the N, P, and L core proteins (49). Our ribozyme processing results (see Fig. S2 in the supplemental material) show that HRz and ΔRz ribozyme processing is active, and the processed agRNA products are predicted to consist of a single nucleotide preceding the 5′ terminus of the  sequence and an exact
sequence and an exact  terminus. A central requirement for expression of the reporter genes is that the coexpressed N, P, and L core proteins assemble in vivo at the 5′ termini of the nascent viral agRNAs to produce biologically active agNCs. The agNCs then initiate replication of gRNAs, which assemble with the core proteins to generate gNCs able to transcribe reporter gene mRNAs.
terminus. A central requirement for expression of the reporter genes is that the coexpressed N, P, and L core proteins assemble in vivo at the 5′ termini of the nascent viral agRNAs to produce biologically active agNCs. The agNCs then initiate replication of gRNAs, which assemble with the core proteins to generate gNCs able to transcribe reporter gene mRNAs.
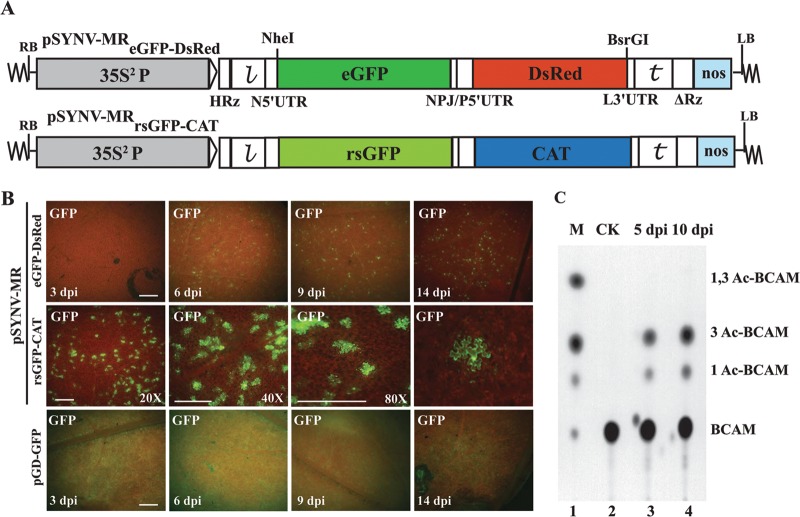
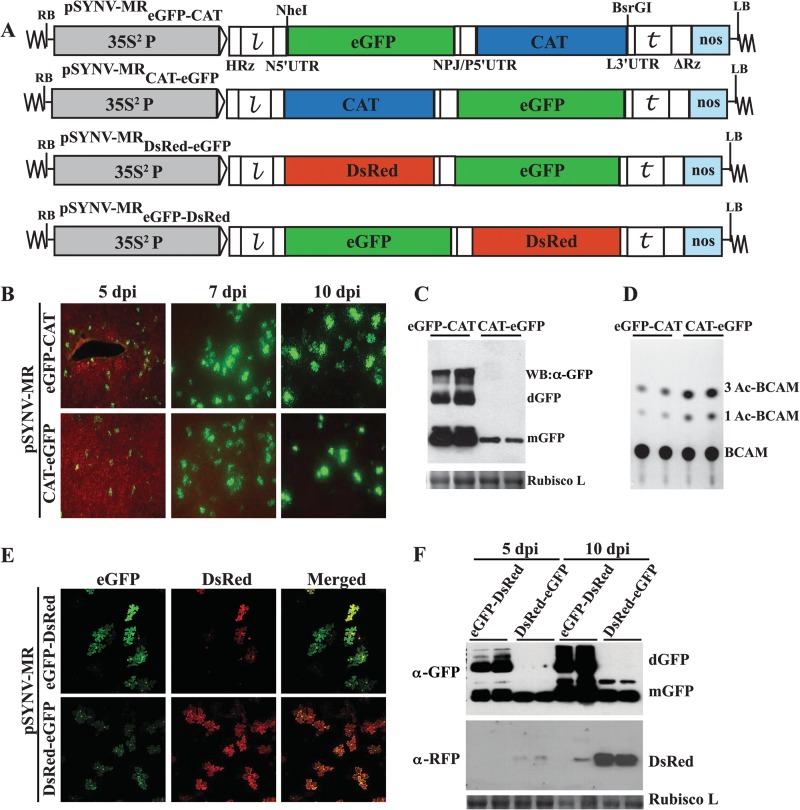
To assess GFP reporter gene expression, experiments were first carried out with pSYNV-MReGFP-DsRed, which contains eGFP and DsRed reporter genes substituted for the N and P protein ORFs, respectively, and pSYNV-MRrsGFP-CAT, which contains an rsGFP reporter gene substituted for the N protein ORF and a CAT reporter gene substituted for the P protein ORF (Fig. 1A). At about 4 to 5 days postinfiltration (dpi) of Agrobacterium harboring pSYNV-MRrsGFP-DsRed or pSYNV-MReGFP-CAT and plasmids for expression of the SYNV N, P, and L proteins and the γb and p19 suppressor proteins, GFP foci scattered throughout the infiltrated tissue began to appear (Fig. 1B). The number of fluorescent cells expressing both eGFP and rsGFP increased until about 9 to 10 dpi, when approximately 15% of the epidermal cells were estimated to exhibit fluorescence (data not shown, but based on counts of ∼900 fluorescing cells). Higher magnifications of the tissue by confocal microscopy also indicated that the foci were normally restricted to single cells (Fig. 1B). These experiments thus reveal that under conditions conducive for formation of NCs, GFP foci appear in a substantial proportion of leaf cells in regions infiltrated with A. tumefaciens containing the three SYNV MR plasmids.
Fig 1.
pSYNV-MR reporter gene expression in agroinfiltrated N. benthamiana leaves. (A) Illustration of pSYNV-MReGFP-DsRed and pSYNV-MRrsGFP-CAT with reporter genes substituted between the NheI and BsrGI sites of pSYNVHRz,ΔRz (see Sections SI to SIV and Fig. S1, S3, and S4 in the supplemental material for construction details). The first reporter in pSYNV-MReGFP-DsRed was derived from an enhanced green fluorescent protein (eGFP) gene substituted for the SYNV N gene, and the second reporter was a red fluorescent protein (DsRed) gene replacement of the SYNV P gene. The pSYNV-MRrsGFP-CAT reporter contained an N gene substitution with a red-shifted green fluorescent protein (rsGFP) gene and a chloramphenicol acetyltransferase (CAT) reporter substitution for the P protein gene. RB and LB, right and left border sequences of pGD, respectively; 35S2P, double CaMV 35S promoter; HRz, = hammerhead ribozyme;  , leader sequence; N5′UTR, 5′ untranslated region of N protein mRNA; NPJ/P5′UTR, gene junction separating the N and P protein ORFs and the 5′ untranslated region of the P protein mRNA; L3′UTR, 3′ untranslated region of the L protein mRNA;
, leader sequence; N5′UTR, 5′ untranslated region of N protein mRNA; NPJ/P5′UTR, gene junction separating the N and P protein ORFs and the 5′ untranslated region of the P protein mRNA; L3′UTR, 3′ untranslated region of the L protein mRNA;  , SYNV trailer sequence; ΔRz = hepatitis delta virus ribozyme; NOS, pGD plasmid synthetase terminator. (B) Comparisons of GFP expression from the pSYNV-MRrsGFP-CAT minireplicon and pGD-GFP. Top panels, appearance of punctate GFP foci in N. benthamiana leaf tissue monitored with a Zeiss Lumar epifluorescence microscope for 14 days postinfiltration (dpi) with bacteria harboring the pSYNV-MReGFP-DsRed reporter cassette and the SYNV N, P, and L plasmids. The infiltration mixtures also contained bacteria with plasmids encoding the p19 and γb host gene silencing suppressors. The magnification bar represents 100 μm. GFP fluorescence from pSYNV-MReGFP-DsRed was evident at 4 to 6 dpi, and the foci continued to express GFP for up to 3 weeks postinfiltration until the infiltrated tissue began to senesce (not shown). Middle panels, micrographs at 10 dpi, showing punctate GFP foci in single cells of N. benthamiana leaf tissue expressing the pSYNV-MRrsGFP-CAT reporter cassette. Aside from the pSYNV-MRrsGFP-CAT reporter cassette, the infiltration mixtures were as in the top panels. Different magnifications are shown to illustrate the distribution of the foci and to demonstrate that the foci are confined to single cells. The magnification bar represents 100 μm. Bottom panels, fluorescence from leaf tissue infiltrated with pGD-GFP. Fluorescence was evident by 3 dpi and was evenly distributed throughout infiltrated zones rather than in punctate foci. Fluorescence was most intense at 6 dpi and declined in the 9- and 14-dpi periods. (C) TLC analysis of CAT activity. CAT activities from leaf extracts were analyzed by thin-layer chromatography (TLC) as described in Materials and Methods. M indicates marker chloramphenicol derivatives separated by TLC. CK denotes control leaf tissue infiltrated with pSYNV-MRrsGFP-CAT alone; 5 dpi and 10 dpi show CAT activity in N. benthamiana leaves harvested at 5 and 10 dpi. Positions of BCAM (nonacetylated fluorescent chloramphenicol substrate) and the CAT products 1Ac-BCAM (fluorescent 1-monoacetylated chloramphenicol), 3Ac-BCAM (fluorescent 3-monoacetylated chloramphenicol), and 1,3Ac-BCAM (fluorescent 1,3-diacetylated chloramphenicol) are indicated.
, SYNV trailer sequence; ΔRz = hepatitis delta virus ribozyme; NOS, pGD plasmid synthetase terminator. (B) Comparisons of GFP expression from the pSYNV-MRrsGFP-CAT minireplicon and pGD-GFP. Top panels, appearance of punctate GFP foci in N. benthamiana leaf tissue monitored with a Zeiss Lumar epifluorescence microscope for 14 days postinfiltration (dpi) with bacteria harboring the pSYNV-MReGFP-DsRed reporter cassette and the SYNV N, P, and L plasmids. The infiltration mixtures also contained bacteria with plasmids encoding the p19 and γb host gene silencing suppressors. The magnification bar represents 100 μm. GFP fluorescence from pSYNV-MReGFP-DsRed was evident at 4 to 6 dpi, and the foci continued to express GFP for up to 3 weeks postinfiltration until the infiltrated tissue began to senesce (not shown). Middle panels, micrographs at 10 dpi, showing punctate GFP foci in single cells of N. benthamiana leaf tissue expressing the pSYNV-MRrsGFP-CAT reporter cassette. Aside from the pSYNV-MRrsGFP-CAT reporter cassette, the infiltration mixtures were as in the top panels. Different magnifications are shown to illustrate the distribution of the foci and to demonstrate that the foci are confined to single cells. The magnification bar represents 100 μm. Bottom panels, fluorescence from leaf tissue infiltrated with pGD-GFP. Fluorescence was evident by 3 dpi and was evenly distributed throughout infiltrated zones rather than in punctate foci. Fluorescence was most intense at 6 dpi and declined in the 9- and 14-dpi periods. (C) TLC analysis of CAT activity. CAT activities from leaf extracts were analyzed by thin-layer chromatography (TLC) as described in Materials and Methods. M indicates marker chloramphenicol derivatives separated by TLC. CK denotes control leaf tissue infiltrated with pSYNV-MRrsGFP-CAT alone; 5 dpi and 10 dpi show CAT activity in N. benthamiana leaves harvested at 5 and 10 dpi. Positions of BCAM (nonacetylated fluorescent chloramphenicol substrate) and the CAT products 1Ac-BCAM (fluorescent 1-monoacetylated chloramphenicol), 3Ac-BCAM (fluorescent 3-monoacetylated chloramphenicol), and 1,3Ac-BCAM (fluorescent 1,3-diacetylated chloramphenicol) are indicated.
During the course of the experiments, we noted that GFP expression from leaf tissue infiltrated with bacteria containing pGD:GFP along with bacteria harboring plasmids for expression of the γb and p19 suppressors of RNA silencing appeared about 2 days earlier than in tissue infiltrated with bacteria containing the two MR plasmids, the viral core protein plasmids, and the γb and p19 plasmids. The kinetics of GFP expression from pGD:GFP were compared with those from pSYNV-MReGFP-DsRed for 2 weeks (Fig. 1B). Initially, GFP expression with pGD:GFP and the γb and p19 suppressors appeared throughout the infiltrated regions by 2 to 3 dpi (Fig. 1B). In marked contrast, GFP fluorescence from pSYNV-MReGFP-DsRed was not evident until 4 to 5 dpi, and rather than being generally distributed throughout the infiltrated area, fluorescence appeared as intense single-cell foci. Reductions in intensity of the GFP foci also began to diminish after ∼6 dpi, whereas most of the fluorescent foci from the SYNV MR were still intense at 14 dpi (Fig. 1B), and some foci persisted for up to 21 dpi even as the infiltrated tissue began to senesce (data not shown). These experiments thus reveal that the kinetics and distribution of GFP expression from the pSYNV-MR-infiltrated cells differed substantially from fluorescence in tissue agroinfiltrated with pGD:GFP.
In separate assays to assess CAT expression from the second ORF of pSYNV-MRrsGFP-CAT, tissue extracts were incubated with the BCAM substrate and the 1Ac-BCAM, 3Ac-BCAM, and 1,3Ac-BCAM reaction products were resolved by thin-layer chromatography (TLC) on silica gel plates (Fig. 1C). As anticipated, CAT activity was not evident in infiltrated tissues at 5 dpi with pSYNV-MRrsGFP-CAT alone (Fig. 1C, lane 2), indicating that detectable levels of CAT were not expressed from the pSYNV-MRrsGFP-CAT primary transcripts. However, in the presence of the N, P, and L proteins, CAT was readily detectable at 5 dpi, and higher activities were present at 10 dpi (Fig. 1C, lanes 3 and 4). Taken together, these cumulative data provide convincing evidence for pSYNV-MR-driven reporter gene activity during transient expression from the binary agrobacterium vector and that reporter gene expression requires the presence of the SYNV core proteins.
N, P, and L protein requirements for reporter gene expression.
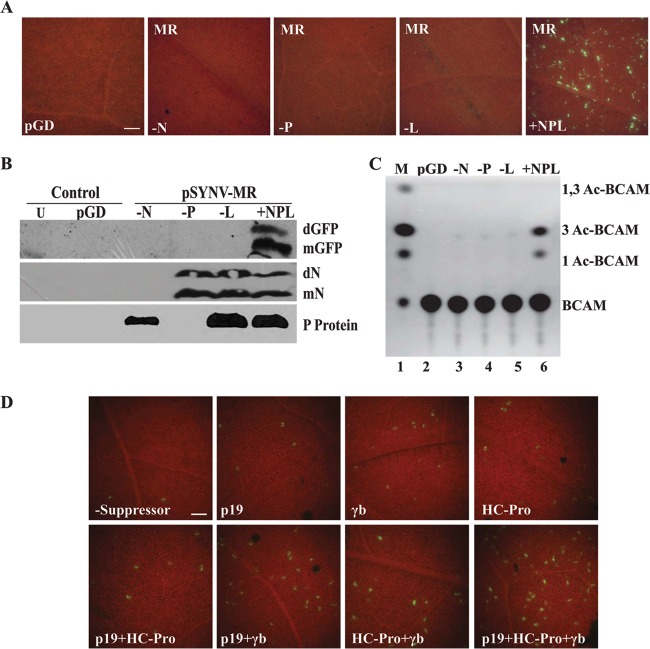
The results above suggest that reporter gene activities from the SYNV MR derivatives require ectopic expression of the core proteins for formation of NCs capable of transcribing the reporter mRNAs. To more precisely determine the individual N, P, and L requirements for reporter gene expression, Agrobacterium mixtures harboring pSYNV-MRrsGFP-CAT and plasmids for expression of the p19 and γb RNA silencing suppressors were coinfiltrated with different combinations of the core proteins (Fig. 2). In agreement with the results shown in Fig. 1, isolated GFP-expressing foci appeared in infiltrated regions when pSYNV-MRrsGFP-CAT and the N, P, and L proteins were coinfiltrated (Fig. 2A), but as anticipated, GFP foci were not evident when bacteria harboring the N, P, or L plasmid were omitted from infiltration mixtures (Fig. 2A). In addition, Western blot analyses revealed the presence of GFP only in MR infiltrations containing the N, P, and L plasmids, and expression of the N and P proteins was also evident in the expected combinations (Fig. 2B). Similarly, CAT activity absolutely relied on coexpressed N, P, and L core proteins (Fig. 2C). These results demonstrate that all three core proteins are essential for reporter expression, and the results are also compatible with a scenario in which the transcribed SYNV MR agRNAs are encapsidated by the N, P, and L proteins to generate agNC intermediates needed for formation of gNCs that function in synthesis of GFP and CAT mRNAs.
Fig 2.
SYNV core protein requirements and gene silencing suppressor effects on GFP expression. (A) GFP foci in N. benthamiana leaves at 6 dpi with bacteria harboring the pSYNV-MReGFP-DsRed reporter cassette. The pGD panel shows a control pGD plasmid not encoding GFP. The −N, −P, and −L panels illustrate tissue infiltrated with pSYNV-MReGFP-DsRed and SYNV core protein mixtures lacking the N, P, or L plasmid, respectively. The +NPL panel shows fluorescence observed as single-cell foci scattered throughout the infiltrated regions of N. benthamiana leaves agroinfiltrated with mixtures containing the pSYNV-MReGFP-DsRed reporter cassette and the SYNV N, P and L plasmids. All experiments were conducted in the presence of p19 and γb gene silencing suppressors. The bar in the pGD panel represents 100 μm, and the remaining panels are the same magnification. (B) Western blot analysis showing GFP and SYNV N and P protein expression. Protein extracts recovered from leaf tissue were detected by immunoblotting with primary mouse GFP antibodies and secondary goat anti-mouse–horseradish peroxidase (HRP) antibodies or with rabbit antibodies elicited against the SYNV N and P proteins and secondary goat anti-rabbit–HRP antibodies. Lanes indicate uninfiltrated tissue (U), pGD-infiltrated tissue (pGD), and tissue infiltrated with pSYNV-MReGFP-DsRed lacking N (−N), P (−P), or L (−L), or pSYNV-MReGFP-DsRed containing N, P, and L (+NPL). mN, N protein monomer; dN, N protein dimer; other designations are as in Fig. 1. (C) Requirement of the N, P, and L proteins for CAT expression from SYNV plasmids in N. benthamiana tissue. Leaves were infiltrated with mixtures of bacteria containing various combinations of the N, P, and L plasmids for core protein expression, pSYNV-MRrsGFP-CAT, and the TBSV p19 and BSMV γb gene silencing suppressors. Designations along the side of the TLC plate are as in Fig. 1. (D) Effects of viral gene silencing suppressors on GFP expression from the pSYNV-MReGFP-DsRed reporter. N. benthamiana leaves were agroinfiltrated with plasmids for expression of the N, P, and L proteins and pSYNV-MReGFP-DsRed in the absence or presence of various combinations of the gene silencing suppressors TBSV p19, BSMV γb, and TEV HC-Pro. The numbers of GFP foci were substantially lower in the absence of gene silencing suppressors (−Suppressor) than in their presence, and a synergistic increase in the numbers of fluorescent cells occurred upon addition of two or more suppressors. The bar in the −Suppressor panel represents 100 μm, and the remaining panels are the same magnification.
Effects of RNA silencing suppressors on GFP expression.
Dolja and colleagues have shown previously that viral suppressors of host gene silencing greatly enhanced reporter gene expression from a Beet yellows closterovirus (BYV) minireplicon following Agrobacterium-mediated delivery (50), so experiments were carried out to determine the extent to which different gene silencing suppressors affected expression from the SYNV MR derivatives (Fig. 2D). To provide direct comparisons, we counted GFP foci from 1-mm2 fields (∼400 cells) of leaf tissue infiltrated with pSYNV-MReGFP-DsRed-L, the core proteins, and various combinations of the BSMV γb, TBSV p19, and TEV HC-Pro suppressor proteins. The results revealed that expression of each of the three suppressors increased the numbers of foci per 1-mm2 field (γb, 6.8 ± 2.3; p19, 10.8 ± 3.9; and HC-Pro, 5.5 ± 1.4) substantially over those of leaves lacking a suppressor (1.8 ± 0.6 foci/1-mm2 field). Moreover, combinations of γb and HC-Pro (17.7 ± 2.3), p19 and HC-Pro (11.6 ± 2.2), and γb and p19 (25.7 ± 5.4) enhanced GFP expression more dramatically (Fig. 2D). In addition, coinfiltration with agrobacteria containing plasmids encoding all three suppressors increased the fluorescence foci (35.9 ± 6.7 foci/1-mm2 field) more than 20-fold over infiltrations lacking suppressors (Fig. 2D). These experiments demonstrated that each of the individual suppressors and various combinations collectively increased the numbers of GFP foci. Therefore, suppression of host RNA interference is quite important for optimal biological activity of the SYNV MR derivatives in these experiments.
MR replication and mRNA synthesis.
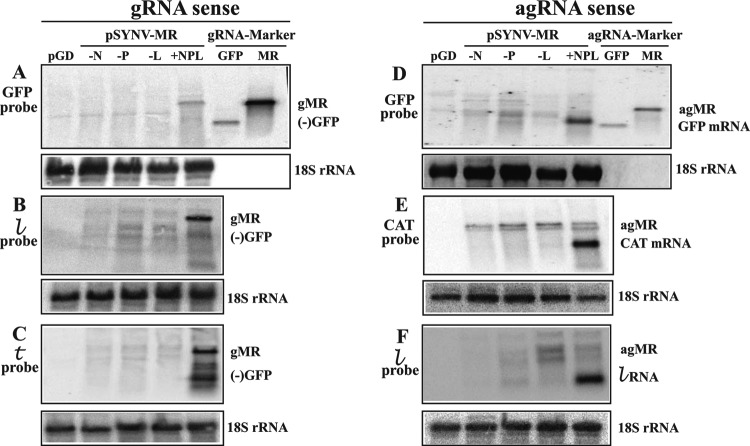
To identify RNAs that might participate in SYNV-MR replication and reporter gene expression, Northern blotting was conducted with N. benthamiana tissue infiltrated for 10 dpi with pSYNV-MRrsGFP-CAT (Fig. 3). In these experiments, RNAs extracted from infiltrated tissue were first hybridized with 32P-labeled probes to detect genomic-sense RNAs that might represent replication products (Fig. 3A, B, and C). As anticipated, the GFP probe revealed a distinct band corresponding in size to the 1,791-nt MR gRNA marker only when leaves were infiltrated with a mixture of bacteria harboring pSYNV-MRrsGFP-CAT and the SYNV N, P and L plasmids (Fig. 3A, lane +NPL). Similar RNAs corresponding in size to the gRNA were also evident in  and
and  probe hybridizations with RNAs from tissue coexpressing pSYNV-MRrsGFP-CAT and the SYNV N, P, and L proteins (Fig. 3B and C, lanes +NPL). In addition, lower-molecular-weight RNAs that are thought to be premature termination products were detected in tissue expressing SYNV-MRrsGFP-CAT and the N, P, and L proteins after hybridization with the antigenomic
probe hybridizations with RNAs from tissue coexpressing pSYNV-MRrsGFP-CAT and the SYNV N, P, and L proteins (Fig. 3B and C, lanes +NPL). In addition, lower-molecular-weight RNAs that are thought to be premature termination products were detected in tissue expressing SYNV-MRrsGFP-CAT and the N, P, and L proteins after hybridization with the antigenomic  probe. The
probe. The  probe was designed to hybridize primarily to 208 nt at the 3′ end of the gRNA and should have recognized RNAs corresponding to nearly complete, but not to incomplete, RNAs (Fig. 3B). The
probe was designed to hybridize primarily to 208 nt at the 3′ end of the gRNA and should have recognized RNAs corresponding to nearly complete, but not to incomplete, RNAs (Fig. 3B). The  probe was complementary to the 5′ end of the gRNA and should have detected incomplete replication products as well as the full-length gRNA (Fig. 3C). This appears to be the case, as the major gRNA sequence identified with the
probe was complementary to the 5′ end of the gRNA and should have detected incomplete replication products as well as the full-length gRNA (Fig. 3C). This appears to be the case, as the major gRNA sequence identified with the  probe comigrated with the MR marker RNA (Fig. 3B, lane +NPL). In contrast, four discrete RNAs migrating more slowly than the MR marker RNA were evident in the
probe comigrated with the MR marker RNA (Fig. 3B, lane +NPL). In contrast, four discrete RNAs migrating more slowly than the MR marker RNA were evident in the  probe blots of tissue infiltrated with the N, P, and L proteins (Fig. 3C, lane +NPL). However, these products are substantially larger than the 160-nt
probe blots of tissue infiltrated with the N, P, and L proteins (Fig. 3C, lane +NPL). However, these products are substantially larger than the 160-nt  sequence, which does not accumulate to detectable levels in infected plants (32). Hence, the latter results indicate that these “strong-stop” components represent accumulation of incomplete SYNV MR gRNA replication products.
sequence, which does not accumulate to detectable levels in infected plants (32). Hence, the latter results indicate that these “strong-stop” components represent accumulation of incomplete SYNV MR gRNA replication products.
Fig 3.
RNA blot analysis of pSYNV-MR transcription and replication products. (A) Northern blot detection of genomic-sense MR RNAs hybridizing to a positive-sense rsGFP probe. Total RNA extracts from N. benthamiana leaves at 10 dpi with bacteria harboring pSYNV-MRrsGFP-CAT, combinations of the N, P, and L proteins, plus the p19 and γb suppressors were used. RNA from mock-infiltrated tissue (pGD) was used as a negative control. A genomic sense RNA corresponding in size to the 1,791-nt genomic-sense MR (gMR) size marker hybridized to the probe only in tissue infiltrated with derivatives harboring pSYNV-MRrsGFP-CAT and plasmids for expression of SYNV N, P, and L proteins. Sample loading in all panels was evaluated by hybridization with an 18S rRNA probe (bottom). The 720-nt negative-sense complement to the GFP mRNA (−GFP) and the gMR size markers are identical in panels A, B, and C. (B) Presence of gRNAs hybridizing to the positive-sense leader ( ) probe. (C) Hybridization of genomic-sense RNAs in N. benthamiana extracts to a positive-sense trailer (
) probe. (C) Hybridization of genomic-sense RNAs in N. benthamiana extracts to a positive-sense trailer ( ) probe complementary to the 3′ end of the gRNAs. (D) Antigenomic RNAs hybridizing to a negative-sense rsGFP probe. A hybridizing RNA appearing slightly above the 720-nt GFP ORF size marker (GFP mRNA) was observed only in N. benthamiana leaves infiltrated with a complete mixture of plasmids expressing pSYNV-MRrsGFP-CAT and the SYNV N, P, and L proteins. The location of a marker corresponding in size to the 1,791-nt “positive-sense” (antigenomic) RNA is shown along the side of the panel. Only minor amounts of a hybridizing band corresponding to this RNA were detected in any of the hybridizations, and these are variable, suggesting that progeny agRNA transcripts are not amplified. (E) Detection of SYNV-MR agRNAs hybridizing to the negative-sense CAT probe. A 660-nt hybridizing band, corresponding to the CAT mRNA, hybridized to the probe only in tissue infiltrated with derivatives harboring pSYNV-MR and plasmids for expression of the N, P, and L proteins. (F) Analysis of SYNV agRNAs hybridizing to a negative-sense leader (
) probe complementary to the 3′ end of the gRNAs. (D) Antigenomic RNAs hybridizing to a negative-sense rsGFP probe. A hybridizing RNA appearing slightly above the 720-nt GFP ORF size marker (GFP mRNA) was observed only in N. benthamiana leaves infiltrated with a complete mixture of plasmids expressing pSYNV-MRrsGFP-CAT and the SYNV N, P, and L proteins. The location of a marker corresponding in size to the 1,791-nt “positive-sense” (antigenomic) RNA is shown along the side of the panel. Only minor amounts of a hybridizing band corresponding to this RNA were detected in any of the hybridizations, and these are variable, suggesting that progeny agRNA transcripts are not amplified. (E) Detection of SYNV-MR agRNAs hybridizing to the negative-sense CAT probe. A 660-nt hybridizing band, corresponding to the CAT mRNA, hybridized to the probe only in tissue infiltrated with derivatives harboring pSYNV-MR and plasmids for expression of the N, P, and L proteins. (F) Analysis of SYNV agRNAs hybridizing to a negative-sense leader ( ) probe. The intense hybridization corresponding to the 144-nt
) probe. The intense hybridization corresponding to the 144-nt  RNA was present only in tissue infiltrated with derivatives harboring the pSYNV MR and plasmids for expression of the core proteins.
RNA was present only in tissue infiltrated with derivatives harboring the pSYNV MR and plasmids for expression of the core proteins.
Complementary hybridization experiments were carried out to detect GFP and CAT mRNAs and agRNAs (Fig. 3D, E, and F). The GFP probe revealed the presence of a product corresponding in size to GFP mRNA in tissue infiltrations containing the N, P, and L genes (Fig. 3D, lane +NPL). This RNA migrates slightly more slowly and is broader than the 720-nt GFP marker RNA, which lacks the 5′ UTR and poly(A) sequences anticipated for the SYNV MR GFP mRNA. The requirement for the N, P, and L proteins (Fig. 3D, lanes −N, −P, and −L) provides additional evidence that the MR GFP mRNA is derived from gNCs. Hybridizations with CAT probes were also conducted to determine whether sequences corresponding to the CAT mRNA in pSYNV-MRrsGFP-CAT were present in infiltrated tissue (Fig. 3E). These hybridizations revealed an intense RNA of ∼660 nt that migrated near the position expected for the CAT mRNA (Fig. 3E, lane +NPL). These results are also consistent with the observed CAT activities in tissue infiltrated with pSYNV-MRrsGFP-CAT and the N, P, and L plasmids (Fig. 1C and 2C). In addition, a probe designed to detect the 144-nt antigenomic sense  RNA revealed a lower-molecular-weight RNA in infiltrations containing the N, P, and L plasmids. This RNA corresponded in size to the
RNA revealed a lower-molecular-weight RNA in infiltrations containing the N, P, and L plasmids. This RNA corresponded in size to the  RNA (Fig. 3F, lane +NPL) and was absent in tissue infiltrations lacking the N, P, or L plasmid (Fig. 3F, lanes −N, −P, and −L).
RNA (Fig. 3F, lane +NPL) and was absent in tissue infiltrations lacking the N, P, or L plasmid (Fig. 3F, lanes −N, −P, and −L).
We had anticipated that full-length agRNAs originating from 35S2 promoter-mediated transcription of the SYNV-MR derivatives would accumulate in tissue coinfiltrated with bacteria harboring only pSYNV-MRrsGFP-CAT and the γb and p19 genes. However, tissue infiltrations lacking N-, P-, or L-expressing bacteria contained only minor doublet RNA products of a size approximating that of agRNAs, and even these products were variable in different extracts (Fig. 3D, E, and F). We also had envisioned that agRNA replication products would accumulate in tissue infiltrated with bacteria harboring pSYNV-MRrsGFP-CAT and the N, P, and L plasmids, but we were unable to detect accumulation of enhanced RNAs migrating more slowly than the GFP mRNAs with any of the hybridization probes (Fig. 3D, E, and F, lanes +NPL). These results thus indicate that detectable amounts of full-length agRNAs do not accumulate even under conditions that support replication of gRNAs.
Polar expression of reporter genes from the N and P ORF positions.
It has been documented that gene expression of nonsegemented NSR viruses involves progressive transcriptional attenuation of genes from the 3′ to the 5′ ends of the gRNA templates (51). To investigate whether the SYNV MRs recapitulate the polar transcription mechanism, two pairs of MR constructs were engineered to facilitate comparisons of the relative reporter expression levels from the N and P ORF positions (Fig. 4A). Both the SYNV MReGFP-CAT and MRCAT-eGFP express eGFP and CAT reporter genes that allow visual inspection of eGFP expression by fluorescence microscopy and enzymatic quantification of CAT activities, but the constructs differ in the order of reporter genes relative to the N and P ORFs. Upon infiltration into N. benthamiana leaves with bacteria containing the N, P, and L plasmids, eGFP expression was evident for both MR constructs starting at 5 dpi (Fig. 4B). Time course fluorescence microscopy analyses showed that eGFP was generally brighter when expressed from SYNV-MReGFP-CAT than when expressed from SYNV-MRCAT-eGFP at 5, 7, and 10 dpi (Fig. 4B), although the numbers of GFP foci appeared to be similar (data not shown). In addition, Western blot analyses confirmed the presence of higher levels of eGFP proteins in leaf tissues infiltrated with SYNV-MReGFP-CAT than in those infiltrated with SYNV-MRCAT-eGFP (Fig. 4C). In contrast, analyses of CAT enzymatic activities revealed that SYNV-MReGFP-CAT-infiltrated leaf extracts had lower levels of CAT expression than SYNV-MRCAT-eGFP-infiltrated leaf extracts (Fig. 4D). A second pair of MR derivatives, SYNV-MReGFP-DsRed and SYNV-MRDsRed-eGFP, was constructed to provide additional fluorescent reporter genes for visual analysis. In the presence of N, P, and L core proteins, intense eGFP and DsRed fluorescent foci were observed for both constructs under confocal microscopy (Fig. 4E). In these cases, eGFP and DsRed foci colocalized in individual cells, and fluorescent reporters for both constructs were more intense when substituted into the N ORF than when substituted into the P ORF (Fig. 4E). It is worth noting that, in order to visually evaluate the relative fluorescence intensity, the same microscope settings were applied for expression of the two constructs. Western blot analyses also verified the polar expression of the reporter genes observed in the microscopy studies (Fig. 4F). Similar to the results of our experiments with SYNV-MRrsGFP-CAT described above (Fig. 2A and B), fluorescent foci were not observed in leaf tissues infiltrated with any of the MR constructs when any of the N, P, or L plasmids were omitted from the agrobacterium mixtures (data not shown). These data thus convincingly show that higher levels of reporter expression from the N ORF position of SYNV MRs are due to positional effects rather than to intrinsic defects associated with a particular construct or reporter gene. Thus, the SYNV MR derivatives recapitulate the polar transcription process generally found in NSR viruses (2) and provide an ideal platform for studies of the regulation of mRNA transcription.
Fig 4.
Analysis of the polarity of reporter gene expression from SYNV MR derivatives. (A) Schematic illustrations of SYNV MR derivatives expressing either eGFP, DsRed, or CAT reporter genes. For each pair of MRs, the constructs differ only in the orders of reporter genes relative to each other. (B to D) Expression of eGFP and CAT in leaves infiltrated with pSYNV-MReGFP-CAT or pSYNV-MRCAT-eGFP, along with equal mixtures of agrobacteria harboring N, P, or L plasmids and gene silencing suppressors. (B) GFP expression monitored at 5, 7, and 10 dpi under an epifluorescence microscope. (C) Tissue extracts prepared from infiltrated leaves shown in panel B at 10 dpi were blotted with anti-GFP antibody for Western blots. Coomassie blue stains of the small subunit of Rubisco (Rubisco L) shown in panels C and F were used as controls to assess protein loading. The designations mGFP and dGFP indicate the monomer and dimer forms of GFP protein, respectively. (D) CAT activities analyzed by TLC. CAT products are as designated in Fig. 1. (E and F) Simultaneous visualization of eGFP and DsRed and evaluation of their relative expression levels. (E) Leaves infiltrated with pSYNV-MReGFP-DsRed or pSYNV-MRDsRed-eGFP as described for panel B. The expression of fluorescent reporters was monitored at 7 dpi under a Zeiss LSM 780 confocal microscope. The same microscopy setting was used for both constructs to facilitate equal evaluations of the relative fluorescence levels. (F) Levels of eGFP and DsRed expression in infiltrated leaves at 5 and 10 dpi were analyzed in Western blots with specific antibodies.
Effects of N protein mutants on reporter gene expression.
The requirement of the N, P, and L genes for MR replication should permit analyses of biological activity of core protein mutants that we have been previously able to characterize only by their biochemical properties and cytological effects. Among these, we have shown that mutations within the N protein are important for N and P protein interactions and for nuclear localization (30, 40). To determine the transcriptional activities of these mutants, bacteria containing individual N protein mutant plasmids were mixed with bacteria harboring plasmids for expression of the P and L proteins, the pSYNV-MReGFP-DsRed derivative, and the p19 and γb proteins. The combinations were infiltrated into N. benthamiana leaves, and the numbers of GFP foci were compared with those of the wild-type N protein (NWT) (Fig. 5A). The N protein mutants when expressed along with the P and L proteins had varied effects, but all resulted in lower numbers of foci than infiltrations containing the NWT protein. For example, the NF42A mutant, containing site-specific alterations in the helix-loop-helix region, resulted in numbers of GFP foci that were reduced by about half (14.7 ± 4.6 foci/50-mm2 field, compared to 26.5 ± 12.8 foci in tissue infiltrated with NWT combinations), and Western blotting also indicated lower accumulation of GFP protein in the infiltrated tissue (Fig. 5A). Our earlier study (30) revealed that NF42A impairs homologous N protein interactions, has reductions in N-P protein interactions, and fails to form discernible subnuclear foci. However, NF42A was able to form subnuclear foci when expressed in conjunction with the P protein (30). More substantial reductions in GFP foci were observed with the NY51A (6.5 ± 3.8 foci/50-mm2 field) and NYIY (9.4 ± 3.8 foci) mutants (Fig. 5A). Interestingly, the NY51A mutant was previously shown to be compromised to a lesser extent in N-N and N-P binding interactions than the NF42A mutant, it but failed to form subnuclear foci when coexpressed with the P protein (30). In contrast, the NYIY mutant, which has alanine substitutions for tyrosine, isoleucine, and tyrosine at residues 48, 50, and 51, functions in homologous N-N and heterologous N-P interactions, although less efficiently than NWT, and also appears to be compromised in the formation of subnuclear foci and in P protein nuclear import (30). However, the most substantial mutant effects in which GFP foci were not present occurred with the C-terminal NKKRR mutant (40), which destroys the bipartite nuclear localization signal and results in total cytosolic localization. Therefore, the N protein mutant activities shown in Fig. 5A indicate that N and P protein interactions and the ability of the N protein to form subnuclear foci affect expression of MR reporter genes and suggest that the ability of the N protein to enter the nucleus is absolutely critical for MR replication. Thus, our results in toto show that the SYNV MR derivatives provide valuable reporters for use in conjunction with experiments to assess biochemical activities and cytological effects of mutations in the SYNV core proteins.
Fig 5.
Reporter gene expression to assess the functions of N protein mutants and cis-acting effects of MR mutations. (A) Effects of SYNV N protein mutants on GFP expression from pSYNV-MReGFP-DsRed. Expression of GFP at 6 dpi in leaves infiltrated with mixtures of bacteria containing plasmids for expression of pSYNV-MReGFP-DsRed, the P and L proteins, the p19 and γb suppressors, and various N protein mutants. Western blots show GFP (top) and N protein accumulation. Nwt, wild-type N protein; NF42A and NY51A, proteins with site-specific mutations in the helix-loop-helix region of the SYNV N gene; NYIY, triple mutant N protein containing alanine substitutions for tyrosine, isoleucine, and tyrosine at residues 48, 50, and 51, respectively. The NKKRR mutant contains alanine substitutions targeting lysine and arginine residues 469, 470, 480, and 481 that destroy the bipartite nuclear localization signal and disrupt nuclear localization. Immunoblotting of GFP elicited by the N protein derivatives is shown at the bottom right. See the text for various effects that these mutations have on N and P protein interactions and N and P protein colocalization in subnuclear foci. The designations along the side of the blot identify positions of the GFP monomers (mGFP) and dimer (dGFP) and the N protein monomers and dimers (mN and dN, respectively). NKKRR forms a high-molecular-weight multimer that does not migrate into the depicted region of the gel. (B) cis-acting effects of minireplicon mutants on GFP expression in infiltrated tissue. pGD, pGD lacking GFP; GFP, pGD encoding GFP; MRwt, pSYNV-MRGFP-DsRed; Δ , 18-nt deletion at the 5′ terminus of the
, 18-nt deletion at the 5′ terminus of the  sequence of pSYNV-MReGFP-DsRed; Δ
sequence of pSYNV-MReGFP-DsRed; Δ , 18-nt deletion at the 3′ terminus of pSYNV-MRGFP-DsRed; ΔHRz, HRz deletion preceding pSYNV-MReGFP-DsRed to prevent transcript processing. The lower panel depicts a Western blot probe of GFP accumulation in tissue infiltrated with the derivatives shown in the other panels.
, 18-nt deletion at the 3′ terminus of pSYNV-MRGFP-DsRed; ΔHRz, HRz deletion preceding pSYNV-MReGFP-DsRed to prevent transcript processing. The lower panel depicts a Western blot probe of GFP accumulation in tissue infiltrated with the derivatives shown in the other panels.
cis-acting effects of minireplicon mutants on GFP expression.
Partial inverse complementarity  and
and  terminal sequences are one of the general hallmarks of the Mononegavirales (2). These sequences serve as promoter recognition sites for synthesis of agRNAs and gRNAs during the replication cycle and form terminal “panhandle” regions of gRNAs and defective interfering RNAs (52, 53). In the plant rhabdoviruses, the
terminal sequences are one of the general hallmarks of the Mononegavirales (2). These sequences serve as promoter recognition sites for synthesis of agRNAs and gRNAs during the replication cycle and form terminal “panhandle” regions of gRNAs and defective interfering RNAs (52, 53). In the plant rhabdoviruses, the  and
and  sequences are considerably longer than those of the rhabdovirus model, VSV, and they vary substantially in length and sequence (24). Comparison of the 3′ and 5′ termini of the SYNV genome reveals that except for two mismatches, the terminal 18 nucleotides are complementary. To evaluate the requirements of these sequences for GFP expression, deletions in the
sequences are considerably longer than those of the rhabdovirus model, VSV, and they vary substantially in length and sequence (24). Comparison of the 3′ and 5′ termini of the SYNV genome reveals that except for two mismatches, the terminal 18 nucleotides are complementary. To evaluate the requirements of these sequences for GFP expression, deletions in the  and
and  regions were generated in pSYNV-MReGFP-DsRed to produce the Δ
regions were generated in pSYNV-MReGFP-DsRed to produce the Δ and Δ
and Δ mutants. As anticipated, fluorescent foci were not observed when either of these mutant derivatives was infiltrated into N. benthamiana leaves, and GFP was not detected in Western blots (Fig. 5B, panels Δ
mutants. As anticipated, fluorescent foci were not observed when either of these mutant derivatives was infiltrated into N. benthamiana leaves, and GFP was not detected in Western blots (Fig. 5B, panels Δ and Δ
and Δ ). To further assess the requirement of precise end termini for replication of the MR, the HRz at the 5′ end of pSYNV-MReGFP-DsRed was deleted to produce a ΔHRz MR transcript with a 16-nt sequence preceding the 5′ terminus of the
). To further assess the requirement of precise end termini for replication of the MR, the HRz at the 5′ end of pSYNV-MReGFP-DsRed was deleted to produce a ΔHRz MR transcript with a 16-nt sequence preceding the 5′ terminus of the  sequence. Interestingly, in this case leaves infiltrated with ΔHRz MR had reduced numbers of GFP foci (6.0 ± 1.5 foci/50-mm2 field) compared to those infiltrated with MRwt (20.6 ± 3.3) and much lower GFP expression in Western blots (Fig. 5B, ΔHRz). Taken together, these results indicate that the first 18 nucleotides of the
sequence. Interestingly, in this case leaves infiltrated with ΔHRz MR had reduced numbers of GFP foci (6.0 ± 1.5 foci/50-mm2 field) compared to those infiltrated with MRwt (20.6 ± 3.3) and much lower GFP expression in Western blots (Fig. 5B, ΔHRz). Taken together, these results indicate that the first 18 nucleotides of the  and
and  sequences are required for reporter gene expression and suggest that HRz processing is important but not obligatory for generation of functional NCs.
sequences are required for reporter gene expression and suggest that HRz processing is important but not obligatory for generation of functional NCs.
DISCUSSION
The minimal infectious unit of NSR viruses is the NC, which consists of the viral gRNA and associated N, P, and L core proteins required for transcription and replication (52, 53). Early in infection, all NSR viruses rely on gNCs for transcription of viral mRNAs, and agRNAs required for agNC assembly. The agNC then participates in formation of progeny gRNAs that can form gNCs that participate in secondary rounds of mRNA transcription and replication. Therefore, in order to conduct reverse genetic studies of NSR viruses, recombinant RNAs must be assembled into NCs (7, 10, 15). Although reverse genetic systems have been described for most mammalian NSR viruses, most of these have involved transformation of tissue culture lines generated from the viral hosts that produce well-defined plaques from which recombinant virus can be recovered (10, 13, 14, 49, 52).
Several substantial problems have constrained reverse genetic applications with plant NSR viruses, which include the nucleorhabdoviruses and cytorhabdoviruses in the Rhabdoviridae, tospoviruses in the Bunyaviridae, ophioviruses in the Ophioviridae, and several nonenveloped viruses in the genus Tenuivirus (23–28, 53). Except for members of the fungus-transmitted Ophioviridae, other plant NSR viruses replicate in insect vectors, but unfortunately, cell lines of most of these vectors are not available, and those that have been developed are quite fastidious in their growth and maintenance requirements (54, 55). Moreover, infected insect cells generally fail to form plaques or display other easily distinguishable infection phenotypes. Another major constraint is that NSR virus generation in plant cells requires circumvention of the cell wall to enable efficient delivery of the numbers of plasmids needed to express the core proteins and viral genome derivatives required for generation of infectious nucleocapsids. Moreover, plant NSR infections do not result in cell lysis, and thus reliable detection of primary infection foci in plant cells is difficult because of the lack of easily discernible cytopathologies. Therefore, reverse genetic applications to plant NSR viruses require approaches substantially different from those used for animal NSR viruses.
We have devised an alternative approach with the nucleorhabdovirus SYNV to circumvent most of the technical issues that have previously hampered reverse genetic applications to plant NSR viruses. This strategy relies on coinfiltration of N. benthamiana leaves with Agrobacterium derivatives separately harboring pGD plasmids encoding the SYNV N, P, and L core proteins and an SYNV MR derivative containing reporter genes flanked by SYNV  and
and  sequences. These SYNV MR derivatives initiate transcription of agRNAs in which nearly authentic SYNV
sequences. These SYNV MR derivatives initiate transcription of agRNAs in which nearly authentic SYNV  and
and  termini are generated by cis cleavage with two ribozymes (HRz and ΔRz). Biological activities of the resulting agRNA transcripts were confirmed by reporter gene expression only in leaves that had been coinfiltrated with bacteria containing plasmids encoding all three SYNV core protein genes, by the presence of mRNA transcripts corresponding to the reporter genes and the
termini are generated by cis cleavage with two ribozymes (HRz and ΔRz). Biological activities of the resulting agRNA transcripts were confirmed by reporter gene expression only in leaves that had been coinfiltrated with bacteria containing plasmids encoding all three SYNV core protein genes, by the presence of mRNA transcripts corresponding to the reporter genes and the  RNA sequence, and by the appearance of gRNAs complementary to the agRNAs. These results also show that the recombinant N, P, and L core proteins are functionally active and indicate that they can participate in nucleocapsid assembly and in transcription and replication processes needed for expression of reporter genes substituted for the SYNV N and P ORFs.
RNA sequence, and by the appearance of gRNAs complementary to the agRNAs. These results also show that the recombinant N, P, and L core proteins are functionally active and indicate that they can participate in nucleocapsid assembly and in transcription and replication processes needed for expression of reporter genes substituted for the SYNV N and P ORFs.
GFP expression from SYNV-MRrsGFP-CAT exhibited several interesting variations in the time course of appearance and duration of expression compared to transient GFP expression from pGD-GFP. In the presence of γb and p19, bright pGD-GFP fluorescence appeared throughout infiltrated tissue by 2 to 3 days after infiltration, but the intensity declined during the next week. Although the appearance of GFP expression from SYNV MRs was slower, intense GFP foci began to appear within the infiltrated regions at about 5 dpi of the MR derivatives and the SYNV N, P, and L proteins. The numbers of foci then increased for the next few days, and intense GFP expression from the foci was maintained until the infiltrated areas began to senesce between 2 and 3 weeks after infiltration. These observations suggest that assembly of biologically active SYNV MR derivatives is inefficient, like that of animal NSR viruses (17), and this may account for the relatively low proportion of cells expressing the reporters in infiltrated leaves.
It is important to note that the biological activities of the SYNV MRs were enhanced substantially by viral suppressors of RNA silencing. In these experiments, relatively low numbers of fluorescent GFP foci occurred in tissue infiltrated only with SYNV MR derivatives and the N, P, and L proteins. However, the numbers of foci increased markedly when pGD vectors harboring BSMV γb, TBSV p19, or TEV HC-Pro genes were added individually to the infiltration mixtures. Larger numbers of fluorescent foci appeared when two RNA silencing suppressors were included, and the prevalence of the foci increased in the presence of all three suppressors. The γb, p19, and HC-Pro suppressors disrupt various steps in plant RNA silencing pathways (44), which likely accounts for their additive positive effects on SYNV MR expression. A previous report with relevance to the SYNV MR results has shown that expression of a minireplicon of a complex plus-strand RNA virus, Beet yellows closterovirus (BYV), is enhanced by diverse RNA silencing suppressors (50). In that study, Agrobacterium-delivered BYV minireplicons had negligible expression of GFP reporter genes in the absence of RNA silencing suppressors, whereas each of several suppressor proteins resulted in dramatic increases in GFP expression. Thus, our results complement those of the Dolja lab (50) and suggest that reverse genetic recovery of functional NSR virus derivatives may be improved by coexpression of suppressors of host RNA interference pathways.
Several features related to the structural properties of the SYNV MR constructs argue against spurious expression of reporter genes from the primary 35S2 MR agRNA transcripts. First, HRz- and ΔRz-processed MR transcripts are unlikely to be translated because they should lack both a 5′ cap structure and a poly(A) tail. Even if ribozyme processing is incomplete and capped agRNA transcripts accumulate in the cytoplasm, a second translation impediment is the presence of an 84-nt ORF present in the 144-nt  sequence followed by three in-frame termination codons that should abrogate translation of RNAs containing the
sequence followed by three in-frame termination codons that should abrogate translation of RNAs containing the  ORF. Moreover, if ribosome readthrough past the three termination codons occurred, the first ORF in the SYNV N protein is in a different reading frame from the
ORF. Moreover, if ribosome readthrough past the three termination codons occurred, the first ORF in the SYNV N protein is in a different reading frame from the  ORF. Third, internal ORFs are not readily translatable in eukaryotic cells, and this should especially be the case with reporter genes substituted into the SYNV P ORF position. Finally, strict SYNV N, P, and L protein requirements for accumulation of viral mRNA and reporter gene expression (Fig. 2) provide additional evidence that the primary 35S2 agRNA transcripts do not function in translation of reporter genes. These results collectively provide a compelling argument that the primary agRNA transcripts are encapsidated by the core proteins to form agNCs that participate as intermediates in synthesis of functional gNCs and that the gNCs function in synthesis of reporter mRNAs.
ORF. Third, internal ORFs are not readily translatable in eukaryotic cells, and this should especially be the case with reporter genes substituted into the SYNV P ORF position. Finally, strict SYNV N, P, and L protein requirements for accumulation of viral mRNA and reporter gene expression (Fig. 2) provide additional evidence that the primary 35S2 agRNA transcripts do not function in translation of reporter genes. These results collectively provide a compelling argument that the primary agRNA transcripts are encapsidated by the core proteins to form agNCs that participate as intermediates in synthesis of functional gNCs and that the gNCs function in synthesis of reporter mRNAs.
To evaluate transcription from the SYNV MR plasmids, hybridization experiments were carried out with SYNV-MRrsGFP-CAT. We had anticipated that irrespective of the presence of the N, P, and L proteins, a limited amount of 35S2 promoter-driven agRNA transcripts corresponding in size to the SYNV MR would accumulate. Nevertheless, we were unable to consistently detect agRNA species of this size in infiltrations lacking bacteria harboring plasmids for core protein expression. This may have been a consequence of low transcription of the agRNAs in limited numbers of foci, combined with rapid RNA turnover. However, when infiltrations included bacteria with plasmids encoding the N, P, and L proteins, the hybridizations reveal the presence of agRNAs (positive sense) corresponding in size and sequence to the 5′  RNA and to the GFP and CAT mRNAs, but full-length agRNAs were not evident. The three observed agRNA species theoretically should have been transcribed only from gNCs, so additional experiments were carried out to identify progeny gRNAs that might be replicated from very-low-abundance full-length agRNA transcripts. These results revealed a hybridizing species corresponding to the size and specificity expected for the full-length SYNV MR gRNAs, but only in tissue expressing the N, P, and L proteins. This suggests that primary agRNAs that are below our limits of detection participate in replication activities. In other hybridization results, the
RNA and to the GFP and CAT mRNAs, but full-length agRNAs were not evident. The three observed agRNA species theoretically should have been transcribed only from gNCs, so additional experiments were carried out to identify progeny gRNAs that might be replicated from very-low-abundance full-length agRNA transcripts. These results revealed a hybridizing species corresponding to the size and specificity expected for the full-length SYNV MR gRNAs, but only in tissue expressing the N, P, and L proteins. This suggests that primary agRNAs that are below our limits of detection participate in replication activities. In other hybridization results, the  sequence probe, which was designed to hybridize to the 5′ ends of nascent gRNAs, revealed the presence of three truncated gRNAs designated “strong-stop” species. These species might result from selective degradation of agRNA transcripts, or possibly low levels of one or more of the core proteins needed for gNC assembly could have resulted in accumulation of truncated gNCs that were able to protect only the encapsidated portions of the replicating gRNAs from degradation.
sequence probe, which was designed to hybridize to the 5′ ends of nascent gRNAs, revealed the presence of three truncated gRNAs designated “strong-stop” species. These species might result from selective degradation of agRNA transcripts, or possibly low levels of one or more of the core proteins needed for gNC assembly could have resulted in accumulation of truncated gNCs that were able to protect only the encapsidated portions of the replicating gRNAs from degradation.
Since NSR RNAs generally require precise 5′ and 3′ termini for nucleocapsid formation and replication (51, 52, 53), we investigated the requirement for an authentic 5′ agRNA terminus for SYNV-MReGFP-DsRed recovery by inactivating the HRz to eliminate precise processing at the 5′ ends of the 35S2 transcripts. In these experiments, the number of foci and the extent of GFP expression after HRz inactivation were reduced substantially by the HRz mutant, but a few foci still appeared. Similar results have been reported in ribozyme mutant experiments to investigate generation of recombinant Measles virus and Borna disease virus (56). As was the case with SYNV, that study showed that recombinant virus rescue was reduced but not entirely eliminated by mutations that destroyed autocatalytic ribozyme processing. Interestingly, progeny virus analyses also revealed that an authentic 5′ terminus had been regenerated during virus replication. Because previous studies with VSV have shown that the initiation site for NC assembly by the N and N-P protein complexes resides within 19 nucleotides at the 5′ of the agRNA (57), our results suggest that NC formation may be generated internally at initiation sites on the unprocessed agRNA transcripts or that the protein complexes may scan to the initiation sites from an upstream location. Although these findings have mechanistic significance for nucleocapsid formation during replication of mononegaviruses, an important practical consideration is that there may be more flexibility in engineering reverse genetic systems than previously recognized.
In addition to yielding valuable clues about conditions necessary for engineering biologically active reverse genetic derivatives, minireplicons have provided bioassays to evaluate nucleocapsid protein functions and requirements for cis-acting elements present on genomic RNAs (10). We therefore used SYNV-MReGFP-DsRed to evaluate the functional effects of N protein mutations that compromise nuclear import, subnuclear localization, and protein-protein interactions (30, 40). The resulting bioassay experiments clearly revealed that NLS amino acid substitutions that eliminate nuclear entry completely eliminate the ability of the N protein to function in expression of pSYNV-MReGFP-DsRed. Other mutations that affect subnuclear localization and protein interactions resulted in reduced numbers of foci, but the mutant proteins still retained some activity. Hence, these results show that the SYNV MRs provide a valuable resource to assess the biological activities of SYNV core protein mutants that affect biochemical or cellular functions.
The requirements of the  and
and  regions for the SYNV MR recovery also confirmed our expectations, because deletion of the first 16 nt of either terminus completely destroyed the ability to generate GFP foci. This finding provides a unique opportunity for more refined sequence and structural evaluations of
regions for the SYNV MR recovery also confirmed our expectations, because deletion of the first 16 nt of either terminus completely destroyed the ability to generate GFP foci. This finding provides a unique opportunity for more refined sequence and structural evaluations of  and
and  requirements for SYNV MR recovery. It is also likely that the cis-acting requirements of the GJ regions for regulation of mRNA transcription and replication of SYNV MR derivatives will be amenable to a similar mutagenesis strategy.
requirements for SYNV MR recovery. It is also likely that the cis-acting requirements of the GJ regions for regulation of mRNA transcription and replication of SYNV MR derivatives will be amenable to a similar mutagenesis strategy.
In conclusion, our construction of SYNV MR plasmids now permits direct analysis of the biological requirements of the core proteins and cis-acting elements encoded by the SYNV genome. We have also conducted experiments to assess the activities of mutations known to affect P protein cellular localization and protein interactions, and we plan to present these findings in an upcoming communication. In addition, the L protein has a number of conserved motifs common to other rhabdovirus L proteins (41) that are ripe for mutagenesis studies. Moreover, a number of characterized nucleorhabdoviruses appear to be closely related to SYNV (24), and it would be informative to determine whether substitution of the conserved motifs of these viruses into the SYNV core proteins will affect specific replication steps and recovery of SYNV MR derivatives. More importantly from a practical perspective, the SYNV MR constructs provide platforms for future engineering of autonomously replicating minigenomes encoding the N, P, and L proteins and for eventual construction of full-length SYNV derivatives suitable for reverse genetic analyses of replication, pathogenesis, and vector transmission. Thus, we anticipate that our SYNV MR findings will initiate future biological studies of SYNV and that the agroinfiltration strategy we have devised will be applicable to engineering reverse genetic systems for other plant NSR viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Steven Ruzin and Denise Schichnes for advice and support with microscopy carried out in conjunction with this research and the Biological Imaging Facility at UC—Berkeley for access to the Zeiss Lumar epifluorescence dissecting microscope and the Zeiss LSM 510 confocal microscope used during the research. Andrew Ball kindly provided the transcription-ΔRz vector (pTV2.0) used for the ΔRz constructions.
This research was supported in part by NSF competitive grant MCB-03, 16907, National Institutes of Health award RO3 AI059178-01, and U.S. Department of Agriculture competitive grant award 96-35303-3563 to A.O.J., NSF competitive grant IOS-07, 49519 to M.M.G., grant Y12C140049 from the Zhejiang Provincial Foundation for Natural Science of China to M.S., and China National Science Funds for Excellent Young Scientists grant 31222004 and Zhejiang Provincial Natural Science Foundation of China under grant no. LR12C14001 to Z.L.
Footnotes
Published ahead of print 24 July 2013
This article is dedicated to the late Richard Francki and Lindsay Black, who were instrumental in helping initiate early rhabdovirus research in the Jackson lab.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01397-13.
REFERENCES
- 1.Ahlquist P, French R, Janda M, Loesch-Fries LS. 1984. Multicomponent RNA plant virus infection derived from cloned viral cDNA. Proc. Natl. Acad. Sci. U. S. A. 81:7066–7070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pringle CR. 2005. Order Mononegavirales, p 609–614 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses Academic Press [Google Scholar]
- 3.Tordo N, Benmansour A, Calisher C, Dietzgen RG, Fang RX, Jackson AO, Kurath G, Nadin-Davis S, Tesh RB, Walker PJ. 2005. Family Rhabdoviridae, p 626–644 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA [Google Scholar]
- 4.Luytjes W, Krystal M, Enami M, Parvin JD, Palese P. 1989. Amplification, expression, and packaging of foreign gene by influenza virus. Cell 59:1107–1113 [DOI] [PubMed] [Google Scholar]
- 5.Pattnaik AK, Wertz GW. 1991. Cells that express all five proteins of vesicular stomatitis virus from cloned cDNAs support replication, assembly, and budding of defective interfering particles. Proc. Natl. Acad. Sci. U. S. A. 88:1379–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawson ND, Stillman EA, Whitt MA, Rose JK. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. U. S. A. 92:4477–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann G, Whitt MA, Kawaoka Y. 2002. A decade after the generation of a negative-sense RNA virus from cloned cDNA—what have we learned? J. Gen. Virol. 83:2635–2662 [DOI] [PubMed] [Google Scholar]
- 8.Whelan SP, Ball LA, Barr JN, Wertz GT. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. U. S. A. 92:8388–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnell MJ, Mebatsion T, Conzelmann KK. 1994. Infectious rabies viruses from cloned cDNA. EMBO J. 13:4195–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walpita P, Flick R. 2005. Reverse genetics of negative-stranded RNA viruses: a global perspective. FEMS Microbiol. Lett. 244:9–18 [DOI] [PubMed] [Google Scholar]
- 11.Roberts A, Rose JK. 1999. Redesign and genetic dissection of the rhabdoviruses. Adv. Virus Res. 53:301–319 [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cornu TI, de la Torre JC. 2001. Ring finger Z protein of lymphocytic choriomeningitis virus (LCMV) inhibits transcription and RNA replication of an LCMV S-segment minigenome. J. Virol. 75:9415–9426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conzelmann KK. 2004. Reverse genetics of Mononegavirales. Curr. Top. Microbiol. Immunol. 283:1–41 [DOI] [PubMed] [Google Scholar]
- 15.de la Torre JC. 2006. Reverse-genetic approaches to the study of Borna disease virus. Nat. Rev. Microbiol. 4:777–783 [DOI] [PubMed] [Google Scholar]
- 16.Emonet SE, Urata S, de la Torre JC. 2011. Arenavirus reverse genetics: new approaches for the investigation of arenavirus biology and development of antiviral strategies. Virology 411:416–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pekosz A, He B, Lamb RA. 1999. Reverse genetics of negative-strand RNA viruses: closing the circle. Proc. Natl. Acad. Sci. U. S. A. 96:8804–8806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flanagan EB, Zamparo JM, Ball LA, Rodriguez LL, Wertz GW. 2001. Rearrangement of the genes of vesicular stomatitis virus eliminates clinical disease in the natural host: new strategy for vaccine development. J. Virol. 75:6107–6114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schnell MJ, Tan GS, Dietzschold B. 2005. The application of reverse genetics technology in the study of rabies virus (RV) pathogenesis and for the development of novel RV vaccines. J. Neurovirol. 11:76–81 [DOI] [PubMed] [Google Scholar]
- 20.Roberts A, Buonocore L, Price R, Forman J, Rose JK. 1999. Attenuated vesicular stomatitis viruses as vaccine vectors. J. Virol. 73:3723–3732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reuter JD, Vivas-Gonzalez BE, Gomez D, Wilson JH, Brandsma JL, Greenstone HL, Rose JK, Roberts A. 2002. Intranasal vaccination with a recombinant vesicular stomatitis virus expressing cottontail rabbit papillomavirus L1 protein provides complete protection against papillomavirus-induced disease. J. Virol. 76:8900–8909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Moller P, Wagner R, Volchkov V, Klenk HD, Feldmann H, Stroher U. 2004. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J. Virol. 78:5458–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falk BW, Tsai JH. 1998. Biology and molecular biology of viruses in the genus Tenuivirus. Annu. Rev. Phytopathol. 36:139–163 [DOI] [PubMed] [Google Scholar]
- 24.Jackson AO, Dietzgen RG, Goodin MM, Bragg JN, Deng M. 2005. Biology of plant rhabdoviruses. Annu. Rev. Phytopathol. 43:623–660 [DOI] [PubMed] [Google Scholar]
- 25.Whitfield AE, Ullman DE, German TL. 2005. Tospovirus-thrips interactions. Annu. Rev. Phytopathol. 43:459–489 [DOI] [PubMed] [Google Scholar]
- 26.Jackson AO, Dietzgen RG, Fang RX, Goodin MM, Hogenhout SA, Deng M, Bragg JN. 2008. Plant rhabdoviruses, p 187–196 In Mahy BWJ, van RegenmortelMHV. (ed), Encyclopedia of virology, 3rd ed. Academic Press, Oxford, United Kingdom [Google Scholar]
- 27.Ramirez BC. 2008. Tenuivirus, p 320–323 In Mahy BWJ, van RegenmortelMHV. (ed), Encyclopedia of virology, 3rd ed. Academic Press, Oxford, United Kingdom [Google Scholar]
- 28.Tsompana M, Moyer JW. 2008. Tospovirus, p 157–163 In Mahy BWJ, van RegenmortelMHV. (ed), Encyclopedia of virology, 3rd ed. Academic Press, Oxford, United Kingdom [Google Scholar]
- 29.Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. 2002. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. Plant J. 31:375–383 [DOI] [PubMed] [Google Scholar]
- 30.Deng M, Bragg JN, Ruzin S, Schichnes D, King D, Goodin MM, Jackson AO. 2007. Role of the sonchus yellow net virus N protein in formation of nuclear viroplasms. J. Virol. 81:5362–5374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zuidema D, Heaton LA, Hanau R, Jackson AO. 1986. Detection and sequence of plus-strand leader RNA of sonchus yellow net virus, a plant rhabdovirus. Proc. Natl. Acad. Sci. U. S. A. 83:5019–5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi TJ, Wagner JD, Jackson AO. 1994. Sequence analysis of the trailer region of sonchus yellow net virus genomic RNA. Virology 202:33–40 [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34.Herold J, Andino R. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394–6400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrotta AT, Been MD. 1990. The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res. 18:6821–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pattnaik AK, Ball LA, LeGrone AW, Wertz GW. 1992. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell 69:1011–1020 [DOI] [PubMed] [Google Scholar]
- 37.Heaton LA, Hillman BI, Hunter BG, Zuidema D, Jackson AO. 1989. Physical map of the genome of sonchus yellow net virus, a plant rhabdovirus with six genes and conserved gene junction sequences. Proc. Natl. Acad. Sci. U. S. A. 86:8665–8668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou H, Jackson AO. 1996. Expression of the barley stripe mosaic virus RNA beta “triple gene block.” Virology 216:367–379 [DOI] [PubMed] [Google Scholar]
- 39.Heaton LA, Zuidema D, Jackson AO. 1987. Structure of the M2 protein gene of sonchus yellow net virus. Virology 161:234–241 [DOI] [PubMed] [Google Scholar]
- 40.Goodin MM, Austin J, Tobias R, Fujita M, Morales C, Jackson AO. 2001. Interactions and nuclear import of the N and P proteins of sonchus yellow net virus, a plant nucleorhabdovirus. J. Virol. 75:9393–9406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi TJ, Kuwata S, Koonin EV, Heaton LA, Jackson AO. 1992. Structure of the L (polymerase) protein gene of sonchus yellow net virus. Virology 189:31–39 [DOI] [PubMed] [Google Scholar]
- 42.An G, Ebert P, Mitra A, Ha S. 1989. Binary vectors, p 29–47 In Gelvin S, Schilperoort R, Verma D. (ed), Plant molecular biology manual. Springer, Dordrecht, Netherlands [Google Scholar]
- 43.Bragg JN, Jackson AO. 2004. The C-terminal region of the Barley stripe mosaic virus gammab protein participates in homologous interactions and is required for suppression of RNA silencing. Mol. Plant Pathol. 5:465–481 [DOI] [PubMed] [Google Scholar]
- 44.Burgyan J, Havelda Z. 2011. Viral suppressors of RNA silencing. Trends Plant Sci. 16:265–272 [DOI] [PubMed] [Google Scholar]
- 45.Pruss G, Ge X, Shi XM, Carrington JC, Bowman Vance V. 1997. Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9:859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim HS, Bragg JN, Ganesan U, Lawrence DM, Yu J, Isogai M, Hammond J, Jackson AO. 2008. Triple gene block protein interactions involved in movement of Barley stripe mosaic virus. J. Virol. 82:4991–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martins CR, Johnson JA, Lawrence DM, Choi TJ, Pisi AM, Tobin SL, Lapidus D, Wagner JD, Ruzin S, McDonald K, Jackson AO. 1998. Sonchus yellow net rhabdovirus nuclear viroplasms contain polymerase-associated proteins. J. Virol. 72:5669–5679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith AJ, Struhl K. 1989. Current protocols in molecular biology. Greene Publishing Associates, New York, NY [Google Scholar]
- 49.Neumann G, Kawaoka Y. 2004. Reverse genetics systems for the generation of segmented negative-sense RNA viruses entirely from cloned cDNA. Curr. Top. Microbiol. Immunol. 283:43–60 [DOI] [PubMed] [Google Scholar]
- 50.Chiba M, Reed JC, Prokhnevsky AI, Chapman EJ, Mawassi M, Koonin EV, Carrington JC, Dolja VV. 2006. Diverse suppressors of RNA silencing enhance agroinfection by a viral replicon. Virology 346:7–14 [DOI] [PubMed] [Google Scholar]
- 51.Whelan SP, Barr JN, Wertz GW. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:61–119 [DOI] [PubMed] [Google Scholar]
- 52.Lyles DS, Rupprecht CE. 2007. Rhabdoviridae, p 1364–1408, In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE.Fields virology, 5th ed. Lippincott Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 53.Kormelink R, Garcia ML, Goodin M, Sasaya T, Haenni AL. 2011. Negative-strand RNA viruses: the plant-infecting counterparts. Virus Res. 162:184–202 [DOI] [PubMed] [Google Scholar]
- 54.Black LM. 1979. Vector cell monolayers and plant viruses. Adv. Virus Res. 25:191–271 [DOI] [PubMed] [Google Scholar]
- 55.Sylvester E, Richardson J. 1992. Aphid-borne rhabdoviruses-relationship with their aphid vectors, p 313–341 In Harris KF. (ed), Advances in disease vector research, vol 9, Springer-Verlag, New York, NY [Google Scholar]
- 56.Martin A, Staeheli P, Schneider U. 2006. RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication. J. Virol. 80:5708–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moyer SA, Smallwood-Kentro S, Haddad A, Previc L. 1991. Assembly and transcription of synthetic vesicular stomatitis virus nucleocapsids. J. Virol. 65:2170–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.