Abstract
Iron is essential for pathogen survival, virulence, and colonization. Feo is suggested to function as the ferrous iron (Fe2+) transporter. The enterobacterial Feo system is composed of 3 proteins: FeoB is the indispensable component and is a large membrane protein likely to function as a permease; FeoA is a small Src homology 3 (SH3) domain protein that interacts with FeoB; FeoC is a winged-helix protein containing 4 conserved Cys residues in a sequence suitable for harboring a putative iron-sulfur (Fe-S) cluster. The presence of an iron-sulfur cluster on FeoC has never been shown experimentally. We report that under anaerobic conditions, the recombinant Klebsiella pneumoniae FeoC (KpFeoC) exhibited hyperfine-shifted nuclear magnetic resonance (NMR) and a UV-visible (UV-Vis) absorbance spectrum characteristic of a paramagnetic center. The electron paramagnetic resonance (EPR) and extended X-ray absorption fine structure (EXAFS) results were consistent only with the [4Fe-4S] clusters. Substituting the cysteinyl sulfur with oxygen resulted in significantly reduced cluster stability, establishing the roles of these cysteines as the ligands for the Fe-S cluster. When exposed to oxygen, the [4Fe-4S] cluster degraded to [3Fe-4S] and eventually disappeared. We propose that KpFeoC may regulate the function of the Feo transporter through the oxygen- or iron-sensitive coordination of the Fe-S cluster.
INTRODUCTION
Iron is an essential element for nearly all life forms (1–3). However, free cellular iron is toxic, and the solubility of ferric iron is poor. Thus, bacteria tightly regulate cellular iron levels through multiple iron transport pathways to achieve effective homeostasis (3–5). Feo, the ferrous iron (Fe2+) transport system, is likely a major route for transporting ferrous iron across the bacterial membrane under anaerobic or low-pH conditions, such as those in the gastrointestinal tract (6). Several systems have demonstrated the importance of Feo. Feo is critical for both survival and virulence in Helicobacter pylori (7). The Feo system is critical for virulence in Streptococcus suis (8) and for colonization in Escherichia coli and Salmonella enterica serovar Typhimurium (9, 10). Bacterial pathogens require the Feo system for enhanced colonization (1, 3, 6).
The feo operon was first identified in E. coli K-12, and its expression was shown to be under dual transcriptional control by the iron-sensing ferric uptake regulator (Fur) and oxygen-sensing fumarate nitrate reduction protein (FNR) regulators in response to different levels of iron and oxygen (6, 11). The feo operon from gammaproteobacteria encodes 3 proteins: FeoA, FeoB, and FeoC (11–13). FeoA, present in 90% of the feo operons, is a small Src homology 3 (SH3) domain protein necessary for ferrous iron transport (6, 12, 14–16). Recent enzymatic assays have suggested that FeoA may not act as a GTPase-activating protein as originally proposed (14). FeoB, an indispensable component of the Feo system, is a large protein consisting of an intracellular amino-terminal domain (NFeoB) and a transmembrane carboxyl-terminal domain presumed to form the Fe2+ pore function that functions as a permease. NFeoB consists of a small-GTPase domain (G domain) and a GDP dissociation inhibitor (GDI)-like helical domain. The crystal structures of several forms from E. coli and Klebsiella pneumoniae NFeoB assemble into a funnel-like trimer with a cytoplasmic pore that could facilitate gating and passage of unhydrated ferrous irons (17, 18). The G domain exhibits guanosine nucleotide-dependent conformational changes in the Switch I, Switch II, and G5 motifs of the G domain and changes in the distance between G and GDI-like domains. The conformational changes were suggested to trigger the opening (GTP bound) and closing (GDP bound) of the pore and to regulate Fe2+ transport (17).
FeoC is a small hydrophilic protein that is present in only 15% of the feo operons (14). The solution structures of FeoC from K. pneumoniae and E. coli possess a winged-helix structure often associated with DNA binding (19). The long, disordered wing loop 1 (W1) contains 4 conserved Cys residues in a sequence, CX4CXXCX5-8C, suitable for harboring a putative iron-sulfur (Fe-S) cluster (6, 19). Thus, FeoC was proposed to be an iron-sulfur cluster-dependent transcriptional regulator directly controlling the expression of the feo operon (transcriptional regulator model) (6). However, a recent study suggested that FeoC did not regulate the feo promoter in Yersinia pestis (20), and no report has confirmed the DNA binding activity of FeoC.
Two studies have recently suggested that FeoC may function at the posttranslational level. We showed that apo-KpFeoC binds to the N-terminal domain of KpFeoB (KpNFeoB) with high affinity (21). In the crystal, apo-KpFeoC binds to KpNFeoB at a site encompassing the Switch II region of the G domain and the C-terminal GDI-like domain such that the flexible W1 loop is potentially capable of interacting with residues in the nucleotide-binding site. We proposed that FeoC might coordinate the Fe-S cluster to regulate ferrous iron transport by modulating G-protein activity (G-protein modulator model). However, Kim et al. found that FeoC binds to FeoB, and the presence of FeoC prevents FeoB from proteolytic degradation by FtsH in Salmonella enterica under low-iron and low-oxygen conditions (22). This results in an elevated level of FeoB that enables Salmonella to take up Fe(II) under anaerobic and low-iron conditions (protease inhibitor model). The coordination of the Fe-S cluster on FeoC can play crucial roles in all 3 models. This study is the first to provide experimental evidence supporting the existence of an Fe-S cluster on FeoC. We present spectroscopic and mutational data proving its existence, the range of redox potentials, and the degradation of a [4Fe-4S] cluster on KpFeoC, with discussions regarding the models.
MATERIALS AND METHODS
Chemicals, bacterial strains, and vectors.
[99% 15N]H4Cl, [99% U-13C]-d-glucose, 99% D2O, and sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) were purchased from Cambridge Isotope Laboratories (Andover, MA). Basal medium Eagle (BME) vitamins, redox reagents, and corresponding antibiotics were purchased from Sigma-Aldrich (St. Louis, MO). Isopropyl-β-d-thiogalactoside (IPTG) was purchased from MDBio Inc. (Taipei, Taiwan). Air-tight Hellma UV-Vis cuvettes (114B-QS) were purchased from Sigma-Aldrich (St. Louis, MO). The E. coli strain BL21(DE3)/pLysS was purchased from Novagen (Madison, WI). The expression vector pGEX-6p-1 carrying the cDNA encoding a chimera fusion protein of the glutathione S-transferase (GST) tag and the FeoC from K. pneumoniae (subsp. pneumoniae NTUH-K2044) (pGEX-6p-1/GST-KpFeoC) was prepared in this lab. The fusion protein was first purified through the GST affinity column. The enzyme was digested to remove the GST tag and further purified through the size exclusion column (19). For preparing the holo-FeoC, the protein was purified through the GST column and concentrated aerobically, whereas the follow-up enzyme digestion was performed anaerobically (less than 5 ppm oxygen) in an anaerobic chamber from COY Lab (Grass Lake, MI) and purified through a Superdex-75 10/300 column attached to an ÄKTA purifier (GE Healthcare) inside the anaerobic chamber. Samples were kept at low temperature in a labtop cooler from Nalgene/Sigma-Aldrich (St. Louis, MO). All point mutants were cloned with the GST tag in this lab.
Protein expression and labeling.
The KpFeoC protein was prepared by growing BL21(DE3)/pLysS cells carrying the pGEX-6p-1/GST-KpFeoC vector in LB medium without supplementing additional iron. Uniformly 15N-labeled KpFeoC ([U-15N]-KpFeoC) protein was prepared by growing cells in the M9 medium supplemented with 1 g/liter 15NH4Cl in the presence of 50 mg/liter (wt/vol) FeCl3 and 1% (vol/vol) BME vitamins, as described previously (18, 19, 21), but with modifications: an additional 50 mg/liter FeCl3 (wt/vol) and 1% (vol/vol) BME vitamins were added to the growth medium. For preparing [U-13C, 15N]-KpFeoC protein, [U-13C]-glucose was added in 2 aliquots: 2 g/liter at the onset and 2 g/liter when the optical density at 600 nm (OD600) reached 2.5 (immediately after IPTG induction). Based on optical spectrum analysis, the holo-KpFeoC expressed in the LB medium is identical to that expressed in the M9 medium with iron. The addition of iron typically increases the yield and homogeneity.
NMR and EPR sample preparation.
Amicon (Millipore, Billerica, MA) tubes were used for protein concentration and buffer exchange following protein purification. Unless specified, the NMR (and EPR) buffers were 50 mM Tris and 100 mM NaCl, in 9% D2O at pH 7.8 for KpFeoC. We defined the native state of FeoC as freshly purified protein from E. coli in the anaerobic chamber. Unused samples were stored in bottles sealed with septum caps at −80°C. The DSS was added as the internal chemical shift standard. Excess molar ratios of sodium dithionite or dithiothreitol (DTT) were added to the reduced samples. Unless specified, the reduced state is the dithionite-reduced state (for NMR and EPR). NMR samples in the reduced state were prepared in an anaerobic chamber and transferred to NMR tubes with a J. Young valve (Wilmad, Vineland, NJ). EPR samples were prepared and frozen in the anaerobic chamber. Spectra were taken aerobically as solids. The pH values were measured before entering the chamber. apo-KpFeoC samples were prepared by dialyzing away irons from holo-KpFeoC, which had been exposed to oxygen for more than 5 days.
NMR spectroscopy, data processing, and analysis.
NMR spectra were acquired at specified temperatures on Bruker Avance 500- or 600-MHz spectrometers equipped with triple resonance cryogenic probes, as described previously (19). To observe the fast-relaxing hyperfine shifts, the repetition times of superWEFT sequences were set to 0.1 s for protons and 0.2 s for carbons and fine-tuned before measurements. Spectra were signal averaged for 160,000 scans for carbons and 40,000 scans for protons for a total acquisition time of 8 h for carbon and 2 h for protons. Spectra were obtained by subtracting 2 spectra with broad and desired line broadening (typically 60 Hz for protons and 600 Hz for carbons) and were baseline corrected using the spline protocol. Proton chemical shifts were referenced relative to internal DSS (taken as 0 ppm); carbon and nitrogen spectra were referenced indirectly by the canonical ratios (23). All spectra were processed by Topspin software (Bruker).
EPR.
Unless specified, the EPR sample buffer conditions were identical to those used in NMR experiments. The EPR spectra were acquired at 9.5390 GHz (measured using a Hewlett-Packard 5246L electronic counter) and 4G modulations with 20 mW power at a 4,096-point resolution, and the average from 4 scans was reported. To avoid loss of EPR signals caused by exposure to air, the sodium dithionite-reduced samples were transferred to the EPR tube and frozen within 10 s in the anaerobic chamber. To ensure full reduction, the reductant/protein molar ratios exceeded 4 and 8 for DTT and dithionite, respectively. Cavity signals were subtracted from the EPR spectra by exact g values using Origin (OriginLab, Northampton, MA). The simulated spectra were processed with SimFonia (Bruker BioSpin, Billerica, USA) and WINEPR (Bruker BioSpin, Billerica, USA).
X-ray absorption spectra.
We conducted the measurements of the Fe K-edge X-ray absorption spectroscopy at the wiggler beamline (BL17C) with a beam size of 2 by 2 mm at the National Synchrotron Radiation Research Center in Taiwan. Beam energy was calibrated to iron foil standards. The sample cell at a volume of 120 μl was sealed with thin Fe-free Kapton tapes. We performed sample loading in the anaerobic chamber and conducted data collection with the sleeping mode that halted the exposure by approximately 1 to 8 s after 2 to 24 s of radiation. The Lytle detector collected signals ranging from 6,912 to 7,912 eV with the florescence mode. Samples were maintained at 283 K throughout the experiments with an air-cooling device. Each scan ran for approximately 90 min. Based on collected spectra, the anaerobic samples were stable for 20 h, indicating that the effects of the photoreduction or radiation damage were negligible for the duration of the experiment. The data analysis and background subtraction were performed using the ATHENA program (24), a graphical interface in the IFEFFIT suite (version 1.2.12) (25). We used an average of 30 scans for the model fitting. Following baseline correction (with Rbkg = 1 using AUTOBK), the EXAFS data were analyzed and simulated using ARTEMIS to yield the fitting curves (24). Scattering paths of the [4Fe-4S] cluster were generated with the ATOMS (26) and FEFF (25) programs, in which the initial distance of Fe-Fe was set as 2.7 Å and that of Fe-S as 2.2 Å, based on measurements from the synthetic (27) and experimental Fe-S clusters (28). Two-shell models centered on iron atoms surrounded by (i) iron and sulfur atoms, such as [4Fe-4S]-(S-Cys)4 and [3Fe-4S]-(S-Cys)3, or (ii) iron, sulfur, and oxygen, [4Fe-4S]-(O-Ser)1-(S-Cys)3, were used in data analysis, and only atoms at distances within 1.8 ≤ R ≤ 3.3 Å were considered.
RESULTS
NMR evidence supports a paramagnetic center in KpFeoC.
Using relaxation-optimized sequences (see Materials and Methods), we observed a proton hyperfine shift of KpFeoC isolated directly from the cell culture (native state) at −14 ppm, indicating that KpFeoC possesses a paramagnetic center (Fig. 1A) (29). Upon addition of dithionite (reduced state), the resonance shifted upfield by 5 ppm to −19 ppm, indicating that the paramagnetic property of the paramagnetic center is redox state dependent. Hyperfine-shifted 13C resonances were also observed from the native state [U-13C, 15N]-KpFeoC (Fig. 1B). Although the assignments of the hyperfine-shifted proton and carbon resonances are currently unknown, the detection of these resonances supports the presence of a paramagnetic Fe-S cluster in KpFeoC. Upon raising the temperature from 293 K to 300 K, the proton resonance of the reduced state KpFeoC shifted upfield by 0.5 ppm, characteristic of the anti-Curie (increasing paramagnetic shifts with increasing temperature) hyperfine-shifted resonance (data not shown). However, the 13C resonances exhibited both Curie (resonance 1 and 3) and anti-Curie (resonance 4) behaviors (Fig. 1B). In comparison, the [1Fe] system of rubredoxin exhibited only the Curie behavior, suggesting that holo-KpFeoC may possess a higher-order Fe-S cluster (30–32), consistent with the X-ray absorption results (see below).
Fig 1.
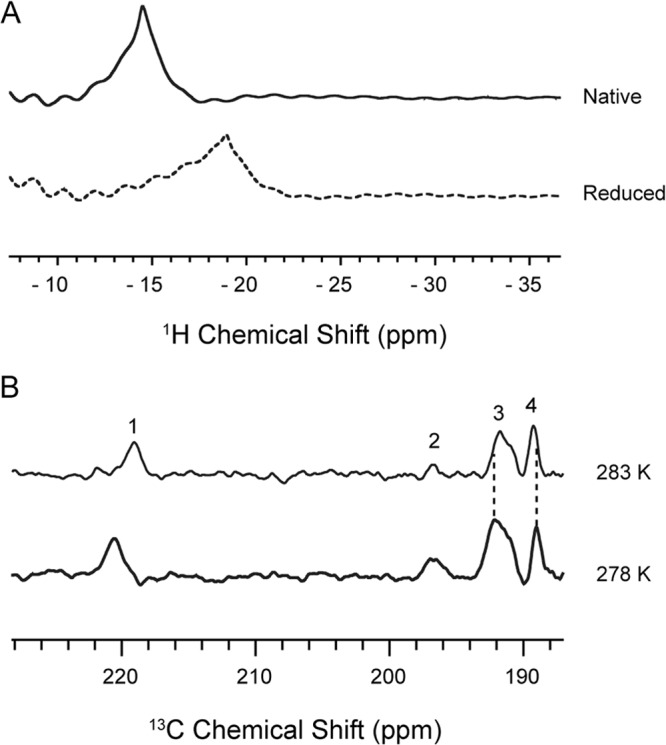
Hyperfine shifted NMR resonance of 1 mM unlabeled-KpFeoC detected by superWEFT pulse sequence. (A) 1H spectra of native state (mostly oxidized, top trace) and the dithionite-reduced state (bottom trace). (B) 13C spectra of 1.3 mM [U-13C, 15N]-KpFeoC at native state at 283 K (top trace) and 278 K (bottom trace). The recycle delay times for the superWEFT sequence were ∼0.1 and ∼0.2 s for proton and carbon spectra, respectively.
Spectrophotometric evidence suggests a [2Fe-2S] or [4Fe-4S] cluster in KpFeoC.
UV-Vis spectra were diagnostic of the presence of iron-sulfur clusters (33). Figure 2 shows the UV-Vis spectra of GST-tagged KpFeoC (GST-KpFeoC) and 4 cysteine-to-serine mutants (C56S, C61S, C64S, and C71S; Fig. 2A) and their spectra without the GST tag (Fig. 2B). For comparison, we included the spectrum of Clostridium pasteurianum rubredoxin (CpRd), which contains a [1Fe] cluster resembling that of another winged-helix protein, PF0610 (34). The color of the (concentrated) native state GST-KpFeoC was dark red, and the UV-Vis spectrum contained peaks at 417 nm, 450 nm, and 550 nm. Upon enzymatic removal of the GST tag, the wild-type KpFeoC maintained a nearly identical UV-Vis spectrum, indicating that the absorption spectrum derived from KpFeoC (Fig. 2B). However, the peak intensity dropped 2-fold, likely because of cluster degradation during the prolonged enzyme digestion process (3 days). However, the UV-Vis spectrum of CpRd contained peaks at 384 nm, 490 nm, and 570 nm (Fig. 2C), suggesting that GST-KpFeoC likely contained a [2Fe-2S] (34, 35) or [4Fe-4S] (36–38) cluster but not a [1Fe] cluster.
Fig 2.
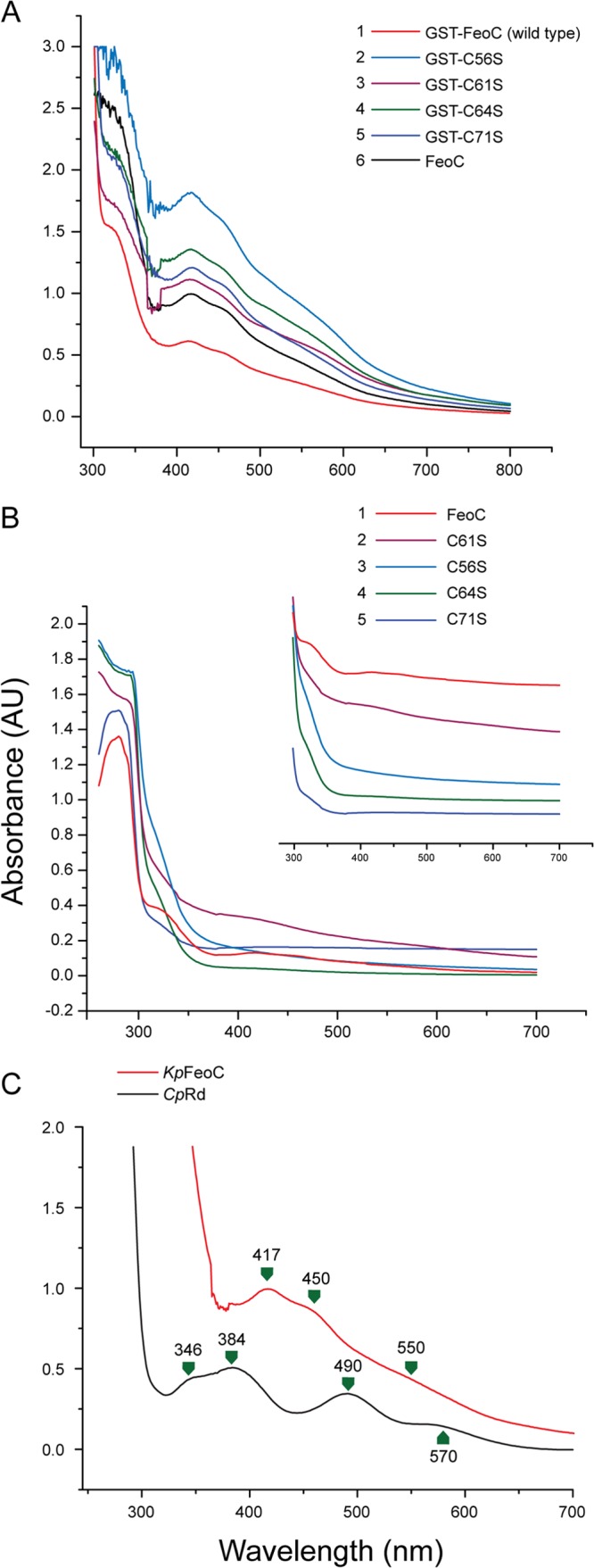
The UV-Vis spectra of various freshly prepared KpFeoC samples (0.5 to 3 mM) in native state. (A) The UV-Vis spectra of GST-tagged KpFeoC (GST-KpFeoC) and four cysteine-to-serine single-site KpFeoC mutants at concentrations of 2.5, 1.5, 2.6, 2.6, and 0.9 mM for C56S, C61S, C64S, C71S, and wild-type GST-FeoC, respectively. (B) The UV-Vis spectra of GST-tag-free KpFeoC and its single-site mutants at concentrations of 0.5, 0.9, 0.8, 0.5, and 0.15 mM for C56S, C61S, C64S, C71S, and wild-type FeoC, respectively. The 300-nm to 700-nm region, which exhibits the characteristic absorption of iron-sulfur clusters, is shown in the inset, shifted in the y axis direction for clarity. (C) Comparison of the UV-Vis spectrum of 3 mM KpFeoC (red) and 1.5 mM CpRd (black), which possesses [1Fe].
We further determined the fractional concentration of KpFeoC containing a [4Fe-4S] cluster by inductively coupled plasma mass spectrometry (ICP-MS) and spectrophotometry. For spectrophotometric measurements, we used the canonical extinction coefficient of 15,000 at 410 nm per [4Fe-4S] cluster (39). The results from both methods showed that 10% of native KpFeoC contain the [4Fe-4S] cluster.
EPR evidence supports a [4Fe-4S] cluster on KpFeoC.
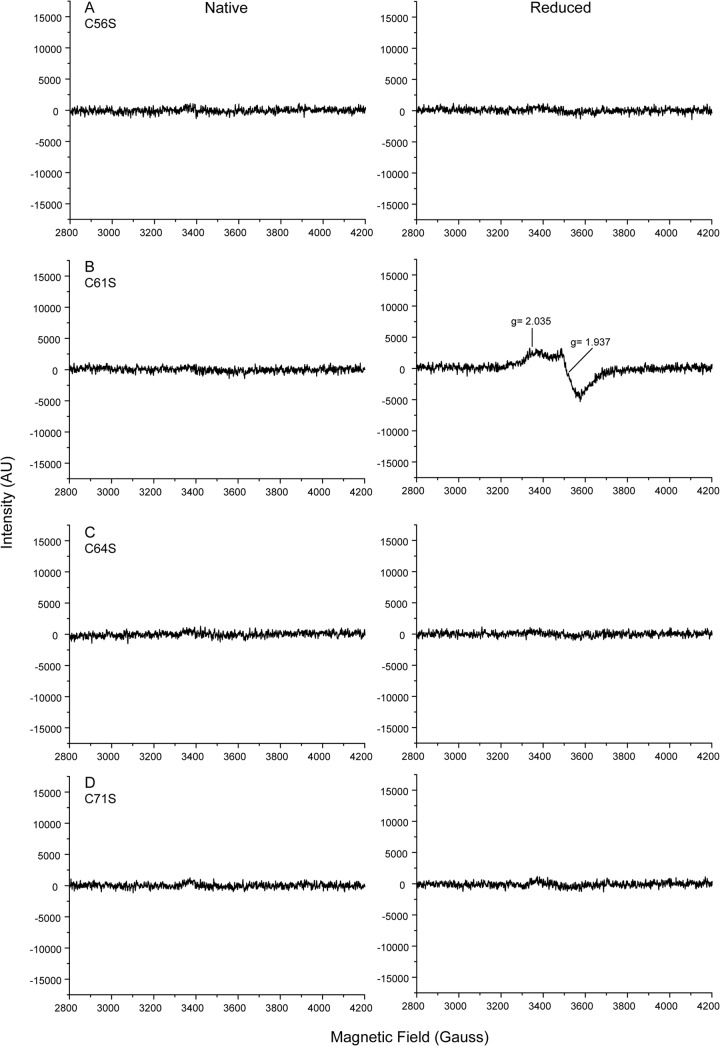
Iron sulfur clusters exhibit the EPR spectrum characteristic of the type of clusters (40–42). Thus, we employed EPR to assign the cluster type present in KpFeoC. At high temperature (77 K), we detected no signal within g values of 1.8 to 2.2 from native or reduced states of KpFeoC (data not shown). Upon lowering the temperature to 14 K, we observed 2 signals at g values of 2.060 and 2.007 from the native KpFeoC, characteristic of the [4Fe-4S]3+ state (Fig. 3A). Reducing KpFeoC by using DTT significantly reduced the resonance intensity, and we observed only residual resonances at g values of 2.05 and 2.008 (Fig. 3B), indicating that most of the protein had been reduced to the EPR-silent diamagnetic [4Fe-4S]2+ state. After further reducing KpFeoC with sodium dithionite, a stronger reducing agent, the EPR signals reappeared at g values of 2.038 and 1.937, reminiscent of those of the [4Fe-4S]1+ cluster (Fig. 3C) (36, 37). Thus, the redox potential of [4Fe-4S]2+/1+ lies between that of DTT and dithionite (43, 44). We also observed EPR signals at a g value of 4.3 for both native and DTT-reduced states (data not shown). We attribute this signal to free iron or nonspecifically bound iron on the protein (45). Table S1 in the supplemental material presents a summary of the EPR signal observed for KpFeoC under various conditions.
Fig 3.
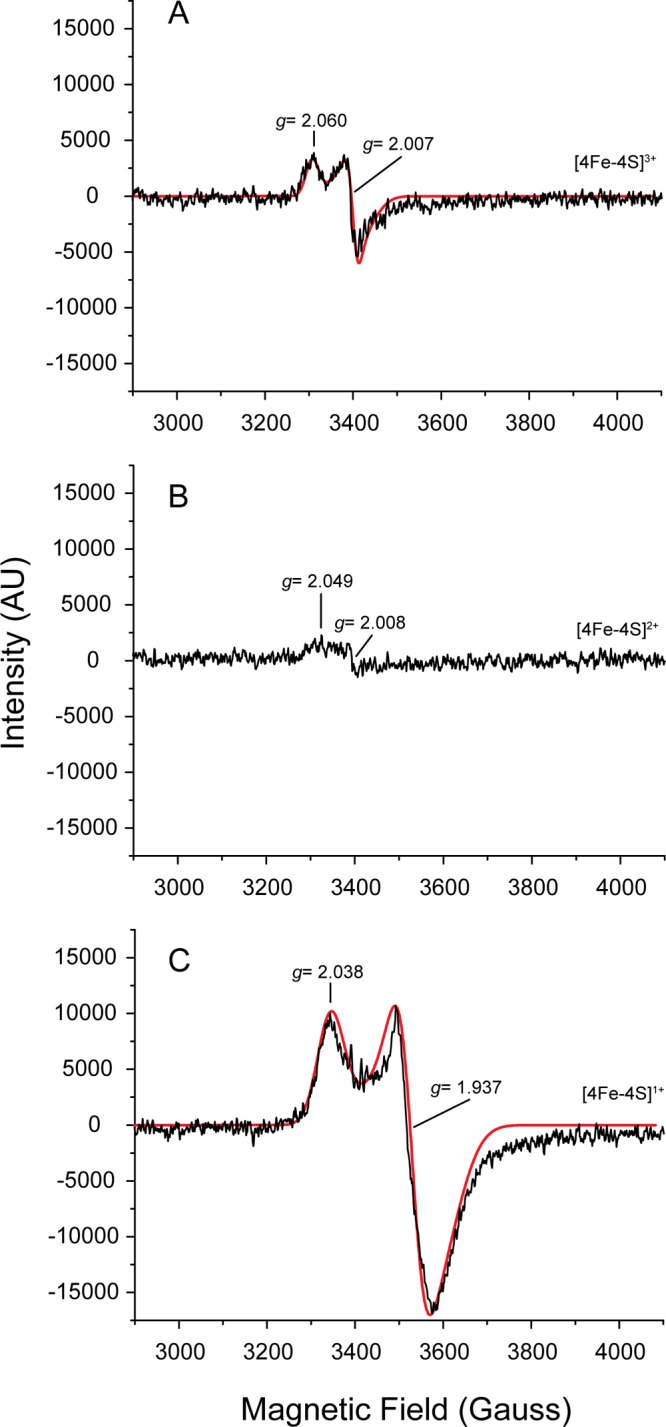
EPR spectra (first derivatives) of KpFeoC at different redox states at 14 K. (A) Total of 2.1 mM KpFeoC at the native state; (B) 2.7 mM KpFeoC at the DTT-reduced state; (C) 2.7 mM KpFeoC fully reduced by sodium dithionite. A SimFonia simulated spectrum was overlaid (red solid line). The assigned redox states of the [4Fe-4S] clusters were labeled on the right. EPR properties are summarized in Table S1 in the supplemental material.
Fe K-edge X-ray absorption structures support the [4Fe-4S] cluster.
We further investigated holo-KpFeoC using Fe K-edge X-ray absorption spectra. We conducted our initial attempt under air, and the Fe K-edge X-ray absorption near-edge structure (XANES) features of the rising edge appeared at approximately 7,125.9 eV (Fig. 4A), suggesting oxygen degradation, which was confirmed by extended X-ray absorption fine structure (EXAFS; data not shown). To protect the cluster from oxygen degradation, we repeated the experiment with samples prepared in anaerobic conditions (Fig. 4A). Consequently, the near-edge downshifted to 7,118.7 eV and the preedge (corresponding to 1s to 3d orbital transitions) peak downshifted to 7,112.7 eV, suggesting that anoxic samples have fewer positive charges to the irons because of a reduced oxidation state (46). We then collected the EXAFS to verify the cluster and ligands (Fig. 4B). We chose the EXAFS of k ranging from 3.12 to 12.00 Å−1 for analysis (Fig. 4B, inset) and applied the Fourier transform to yield the distances of the ligating atoms (iron and sulfur; Fig. 4B). We selected three models for simulation: [4Fe-4S]-(S-Cys) × 4, [3Fe-4S]-(S-Cys) × 3 (degradation intermediate), and [4Fe-4S]-(O-Ser) × 1-(S-Cys) × 3 (serine replacement). Among them, [4Fe-4S]-(S-Cys) × 4 was the most optimal model, which yielded the lowest Rf value of 0.07%. According to the model, the average Fe-S distances were reported as 2.26 ± 0.05 Å and the Fe-Fe distance was 2.71 ± 0.09 Å, consistent with canonical [4Fe-4S] clusters (27, 28). The EXAFS results were in good agreement with the EPR results, and a higher coordinated number of Fe-Fe bonds excluded the [2Fe-2S], supporting the temperature-dependent EPR results. Therefore, both the EPR and EXAFS results suggest that the [4Fe-4S] is the native cluster in FeoC.
Fig 4.
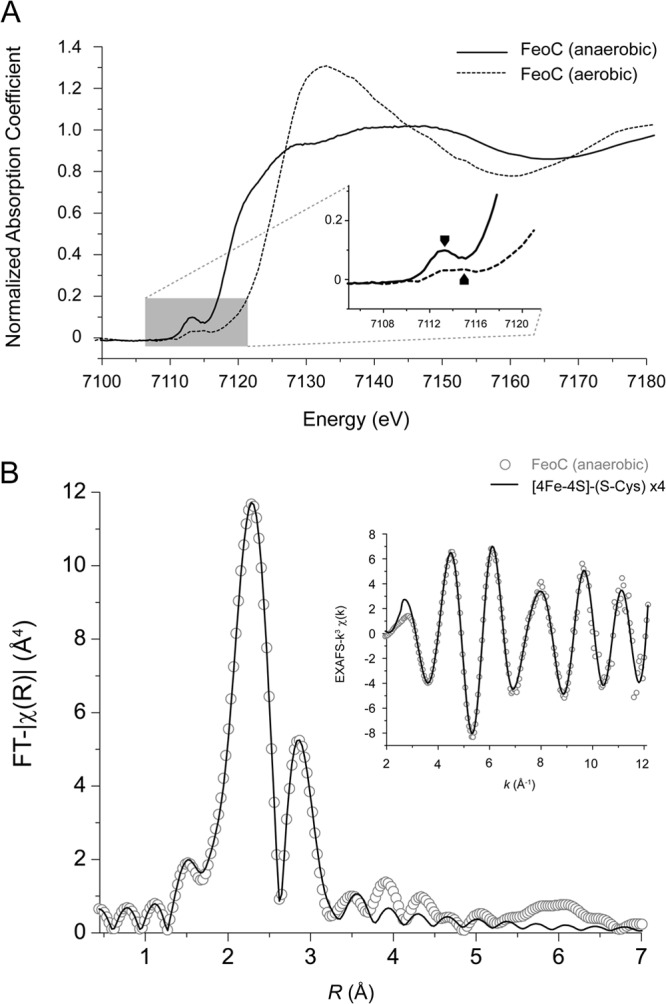
X-ray absorption spectra. (A) The normalized XANES of holo-FeoC at the Fe K-edge. Both experiments done in anaerobic and aerobic conditions were overlaid. The preedge region was enlarged, with absorption peaks labeled with black arrow at 7,113 eV and red arrow at 7,115 eV. (B) The EXAFS data of holo-FeoC. The experimental data (open circles) and simulated spectra (solid lines) according to model [4Fe-4S]-(S-Cys) × 4 were overlaid. The k3-weighted Fourier transform over a k range of ∼3.12 to 12 Å−1. The raw data of EXAFS (k3χ) used for the analysis are shown in the inset.
The [4Fe-4S] cluster of KpFeoC is sensitive to oxygen.
Similar to other iron-sulfur proteins, the holo-KpFeoC was oxygen sensitive (40). When exposed to oxygen at 4°C, native KpFeoC gradually lost its characteristic absorption peaks (Fig. 5). The change in peak height was fitted to a single exponential decay function of OD = c + A × 0.5t/t1/2, and the results yielded a half-life (t1/2) of 16.0 ± 1.0, 15.8 ± 1.0, and 18.3 ± 1.3 h for absorbance at 417 nm, 450 nm, and 550 nm, respectively, with an average half-life of 17 h. We monitored the protein using SDS-PAGE analysis, which indicated that the proteins were nondegraded (data not shown). Our data suggested that the [4Fe-4S] cluster in KpFeoC was oxygen labile. Proteins purified from a size exclusion column in air for 2 h at 4°C were depleted of the Fe-S cluster, suggesting that the degradation rate is likely less than 1 h under the chromatography conditions. The limited oxygen availability in the 1-mm by 10-mm by 10-mm cuvette without stirring might slow the degradation rate measured by the spectrophotometric experiments.
Fig 5.
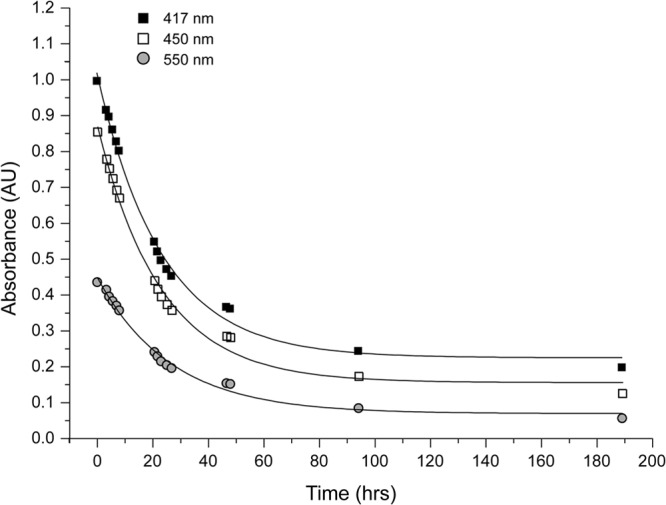
Kinetics of the O2-induced degradation of the [4Fe-4S] cluster in KpFeoC (0.1 mM). The optical absorbance at 417 nm (filled squares), 450 nm (empty squares), and 550 nm (filled circles) was monitored at various time points of O2 exposure. A total of 100 μl of sample was kept at 4°C in 50 mM Tris, 100 mM NaCl (pH 7.8), without stirring. Fittings of the curves (solid lines) yielded half-lives of 16.0 ± 1.0, 15.8 ± 1.0, and 18.3 ± 1.3 h, as monitored at 417 nm, 450 nm, and 550 nm, respectively.
To identify the degraded products, we initially treated the freshly prepared native KpFeoC sample with 1 mM DTT anaerobically and then exposed it to air at 4°C. We used a series of EPR spectra at 14 K at various exposure times (see Fig. S1 in the supplemental material). Because DTT-reduced KpFeoC was diamagnetic, the initial spectrum showed only a signal from the residual nonreduced protein. Further exposure to oxygen resulted in the appearance of a peak at a g value of 2.010, which gradually increased in intensity and peaked at 20 h. Further exposure to oxygen resulted in a gradual loss of the EPR signal. The time course of the oxidation process is consistent with the initial buildup of the [3Fe-4S]1+ cluster, the only iron-sulfur cluster with an isotropic g value at 2.01. The end product of the oxidation process is the loss of the iron-sulfur cluster and the generation of apo-FeoC and thus the loss of the EPR signal.
To assess the final state of the oxidization product, we added DTT or dithionite to reduce the cluster after 25 h of exposure to oxygen. DTT did not produce any change in the EPR spectrum (see Fig. S1D in the supplemental material), whereas dithionite greatly reduced the EPR signal (see Fig. S1E). These results suggested that 25-h O2-exposed KpFeoC did not contain a sufficient concentration of [2Fe-2S]2+ (EPR silent), because dithionite should produce [2Fe-2S]1+ (EPR active). The absence of a g value of 1.96 was indicative of the depleted [4Fe-4S]1+ cluster (the product of the dithionite reduction of the [4Fe-4S] cluster) in the final oxidation product, indicating that [4Fe-4S] was completely degraded. We concluded that [3Fe-4S] is likely the intermediate oxidative degradation process of the [4Fe-4S] cluster in KpFeoC.
Cysteines in the W1 loop are the ligands for the [4Fe-4S] cluster.
KpFeoC contains 4 conserved cysteine residues, which are all located in the W1 loop. To assess the roles of these cysteines in cluster formation, we generated 4 single-site Cys-to-Ser mutants (C56S, C61S, C64S, and C71S) and analyzed them with EPR (Fig. 6). Optical spectra (Fig. 2) of all GST-tagged mutants were similar to those of the wild-type KpFeoC. However, upon enzymatic removal of the GST tag, only the C61S mutant maintained the characteristic absorption spectrum of the [4Fe-4S] cluster (Fig. 2) and EPR signals similar to those of the wild-type KpFeoC (Fig. 6B, right). The intensity of [4Fe-4S]1+ of C61S was approximately 30% that of the wild type at a similar concentration, indicating a less stable cluster. These results indicated that Cys56, Cys64, and Cys71 are crucial for the formation and stability of the iron-sulfur cluster, whereas Cys61 also plays a role in stabilizing the cluster but is less essential. We suggest that Cys61 is the fourth ligand with the support from EXAFS, but it is possible to substitute Cys61 with other nearby glutamic acid or serine residues in the C61S mutant. In certain cases, serine can also serve as a ligand for the iron-sulfur cluster (35).
Fig 6.
EPR spectra (first derivatives) of four KpFeoC mutant proteins at 14 K at native (left) and dithionite-reduced (right) states. (A) C56S, 1.5 mM; (B) C61S, 2.9 mM; (C) C64S, 2.4 mM; (D) C71S, 1.1 mM.
DISCUSSION
KpFeoC forms an oxygen-sensitive [4Fe-4S] cluster.
The Fe-S proteins contain inorganic iron and sulfur as cofactors (40, 47, 48). Iron and sulfur are redox active, and the cluster can undergo redox reactions under physiological conditions. Studies have documented several types of biological Fe-S clusters, including the simplest [1Fe] cluster in rubredoxin to the complex [8Fe-7S] cluster in nitrogenase (42, 49). A change in cluster oxidation states is associated with characteristic magnetic properties, and thus EPR spectra are effective fingerprints of the cluster type and redox state of the Fe-S cluster (40, 41). For the [4Fe-4S] cluster at near-liquid helium temperature, the [4Fe-4S]3+ and [4Fe-4S]1+ states are paramagnetic and thus can be distinguished from the diamagnetic [4Fe-4S]2+ state (41, 50). The 2 paramagnetic states can be distinguished from the g factors in the EPR spectra, which can be detected only at near-liquid helium temperature because of fast relaxation at ambient temperature (50). However, in NMR spectroscopy, the paramagnetic effect from [2Fe-2S]2+ and [4Fe-4S]2+ clusters causes a hyperfine shift in the NMR resonances, which can be detected (32, 51–53). A summary of the EPR results in the literature, combined with our results of the holo-KpFeoC, is presented in Table S1 in the supplemental material for comparison.
Based on UV-Vis spectrophotometry, NMR, and EPR evidence, we demonstrated that KpFeoC forms an Fe-S cluster. The characteristic UV-Vis spectrum and NMR spectra suggested that the iron-sulfur cluster is not the simple [1Fe] type. The presence of the EPR signal at a g value of 1.937 in the dithionite-reduced state ruled out the possibility of the [3Fe-4S]1+/0+ cluster, because the reduced state of [3Fe-4S]0+ is diamagnetic (41, 50). As suggested in previous studies, [2Fe-2S] and [4Fe-4S] clusters can be distinguished by temperature-dependent EPR intensities (32, 50, 54). The EPR signals of [2Fe-2S] are observable above 77 K, whereas the EPR signal of the [4Fe-4S]1+ cluster is observable only at a temperature below 30 K. This was the case in KpFeoC (resonances disappeared above 37 K; data not shown), confirming the identity of the [4Fe-4S] cluster in KpFeoC. The EXAFS results further support the conclusion of [4Fe-4S], excluding the possibility of [2Fe-2S], and confirm that [4Fe-4S] is ligated by 4 cysteines. Protein exposure to oxygen generated the [3Fe-4S] intermediate, further supporting the [4Fe-4S] identity in native KpFeoC. In summary, UV-Vis, NMR, EPR, and EXAFS provide unequivocal evidence of the presence of the [4Fe-4S] cluster on KpFeoC.
Redox states and oxygen-induced degradation of holo-FeoC.
Cysteine typically coordinates each tetrahedral Fe site in the form of thiolate (RS−). However, other residues, such as aspartate (RCO2−), histidine (N=), and serine (R-O−), are occasionally encountered in clusters, and these ligands were shown to modify redox potential (55). The protein environment also affects the redox potential of an iron-sulfur cluster. We showed that the [4Fe-4S] cluster on KpFeoC is coordinated to the cysteine residues in the W1 loop. We estimated the redox potential of the [4Fe-4S] in the 3 oxidation states by examining the effect of DTT or dithionite on the native KpFeoC sample. We detected that most KpFeoC protein in the [4Fe-4S]3+ state reduced to the [4Fe-4S]2+ state by DTT and further reduced to the [4Fe-4S]1+ state by dithionite. Because the redox potential of DTT is −0.33 V and that of dithionite is −0.66 V (43, 44), the results indicated that the redox potential of [4Fe-4S]3+/2+ of FeoC is higher than −0.33 V and that of [4Fe-4S]2+/1+ is between −0.33 V and −0.66 V. However, we did not determine the precise redox potential of holo-KpFeoC.
The loss and gain of the iron-sulfur cluster is a common sensing mechanism for the Fe-S proteins to exhibit their biological activity. The cluster-assembling machinery assembles the iron-sulfur cluster, and degradation by oxidative agents removes the cluster. The ICP-MS results showed that 20% of the freshly prepared GST-KpFeoC contained the Fe-S cluster, indicating that the Fe-S cluster-assembling machinery is capable of assembling at least 20% of the overexpressed GST-KpFeoC. We examined the degradation of holo-KpFeoC by exposing the protein to oxygen and detected the presence of a [3Fe-4S]2+ intermediate. To assess whether the [2Fe-2S] state is the final cluster degradation product of holo-KpFeoC, we added dithionite to the final product to reduce the diamagnetic [2Fe-2S]2+ state, if present, to the paramagnetic [2Fe-2S]1+ state. We did not detect the EPR signal near a g value of 2.0, suggesting that the [2Fe-2S]2+ state is not the final product. The pathway is similar to that of FNR, but the rates differ, suggesting various sensory/regulatory mechanisms (56–58).
Biological implications.
Iron-sulfur proteins play key roles in catalytic reactions, in electron transfer in both oxidative phosphorylation and photosynthesis, and in gene regulation (41, 42, 48, 59, 60). The roles of bacterial iron-sulfur regulatory proteins as sensors/switches have been extensively reviewed (42, 60–62). Iron-sulfur clusters sense environmental changes by interacting with small molecules to exhibit rich chemistries and regulate cellular events. Iron-sulfur proteins acting as transcriptional regulators, such as the E. coli fumarate-nitrate reduction regulator protein (EcFNR), can alter binding affinities to specific DNA sequences by various cluster states. EcFNR is activated only when the O2-labile [4Fe-4S] cluster is assembled; holo-FNR recognizes specific binding sites in excess of 100 promoters (63, 64). The iron-sulfur cluster regulator (IscR) exhibits different DNA-binding properties in its apo and holo states, controlling different subsets of gene expressions (65–69). The cytoplasmic aconitase also regulates gene expression through differential binding affinities of the apo and holo protein to the iron regulatory elements within the mRNA of genes related to iron metabolism (70, 71). The NreB contains a [4Fe-4S]2+ cluster, but it does not bind to nucleotides, acting as a transcriptional activator by interacting with the response regulator, NreC, to regulate the expression of the nreABC operon (72, 73). Thus, the iron-sulfur proteins regulate transcription through direct binding to the DNA or RNA. They also exhibit transcriptional activity indirectly by affecting the activity of other proteins that interact with DNA activators or repressors.
In the literature, 3 models have been proposed for FeoC function: the transcriptional regulator model (6), the G-protein modulator model (21), and the protease inhibitor model (22). Available data does not currently support the transcriptional regulator model, and we could not detect KpFeoC binding to DNA using the gel-shift or SELEX experiments (20, 22); evidence supporting the G-protein modulator model is lacking. The protease inhibitor model appeared to be the only model supported by in vivo data. However, it is likely too early to discount any of these models, and it is conceivable that FeoC may possess dual functions or function differently in various systems. The presence of the Fe-S cluster on FeoC can have a substantial effect on its function, regardless of its role as a transcriptional regulator, a G-protein modulator, or a protease inhibitor. Our present work confirming the existence of the Fe-S cluster on FeoC should facilitate future studies in defining the roles and function of the Fe-S cluster. Future works are necessary to confirm their biological role. The low yield and oxygen sensitivity of holo-KpFeoC hampers current studies focused on clarifying the role of the Fe-S cluster. Advanced understanding requires the development of methods for generating high-yield holo-KpFeoC. However, the oxygen sensitivity of the [Fe-S] cluster on FeoC in vitro does not necessarily indicate that the cluster functions as an oxygen sensor in vivo.
In summary, by using spectrophotometric, NMR, EPR, and X-ray absorption methods, we showed that KpFeoC contains a [4Fe-4S] cluster that can be degraded by oxygen. Using single-site mutation and EXAFS techniques, we identified the crucial cysteine residues in the W1 loop as the ligands of the Fe-S cluster. Detection of the oxygen-sensitive Fe-S cluster in FeoC raises the possibility that the Fe-S cluster might play a role in regulating Feo activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank John L. Markley (the University of Wisconsin-Madison) for providing the plasmid encoding CpRd and Ping-Yu Chen (National Chung Hsing University) for providing the EPR service. We thank Jyh-Fu Lee, Chih-Wen Pao, and Jeng-Lung Chen for their help on the data collection of X-ray absorption at the beamline BL17C in NSRRC. We also thank Feng-Chun Lo for the discussion of the EXAFS data.
This project was supported by the National Science Council of the Republic of China, grant NSC100-2311-B-001-023. The NMR experiments were conducted on NMR spectrometers at the High-Field Nuclear Magnetic Resonance Center (HFNMRC), supported by the National Research Program for Biopharmaceuticals, the National Science Council of the Republic of China.
Footnotes
Published ahead of print 16 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00687-13.
REFERENCES
- 1.Schaible UE, Kaufmann SH. 2004. Iron and microbial infection. Nat. Rev. Microbiol. 2:946–953 [DOI] [PubMed] [Google Scholar]
- 2.Conrad ME, Umbreit JN. 2002. Pathways of iron absorption. Blood Cells Mol. Dis. 29:336–355 [DOI] [PubMed] [Google Scholar]
- 3.Andrews SC, Robinson AK, Rodriguez-Quinones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237 [DOI] [PubMed] [Google Scholar]
- 4.Krewulak KD, Vogel HJ. 2008. Structural biology of bacterial iron uptake. Biochim. Biophys. Acta 1778:1781–1804 [DOI] [PubMed] [Google Scholar]
- 5.Clarke TE, Tari LW, Vogel HJ. 2001. Structural biology of bacterial iron uptake systems. Curr. Opin. Med. Chem. 1:7–30 [DOI] [PubMed] [Google Scholar]
- 6.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143–157 [DOI] [PubMed] [Google Scholar]
- 7.Velayudhan J, Hughes NJ, McColm AA, Bagshaw J, Clayton CL, Andrews SC, Kelly DJ. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274–286 [DOI] [PubMed] [Google Scholar]
- 8.Aranda J, Cortes P, Garrido ME, Fittipaldi N, Llagostera M, Gottschalk M, Barbe J. 2009. Contribution of the FeoB transporter to Streptococcus suis virulence. Int. Microbiol. 12:137–143 [PubMed] [Google Scholar]
- 9.Stojiljkovic I, Cobeljic M, Hantke K. 1993. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol. Lett. 108:111. [DOI] [PubMed] [Google Scholar]
- 10.Tsolis RM, Baumler AJ, Heffron F, Stojiljkovic I. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64:4549–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hantke K. 1987. Ferrous iron transport mutants in Escherichia coli K12. FEMS Microbiol. Lett. 44:53–57 [Google Scholar]
- 12.Kammler M, Schon C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hantke K. 2003. Is the bacterial ferrous iron transporter FeoB a living fossil? Trends Microbiol. 11:192–195 [DOI] [PubMed] [Google Scholar]
- 14.Lau CK, Ishida H, Liu Z, Vogel HJ. 2013. Solution structure of Escherichia coli FeoA and its potential role in bacterial ferrous iron transport. J. Bacteriol. 195:46–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reference deleted.
- 16.Kim H, Lee H, Shin D. 2012. The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem. Biophys. Res. Commun. 423:733–738 [DOI] [PubMed] [Google Scholar]
- 17.Guilfoyle A, Maher MJ, Rapp M, Clarke R, Harrop S, Jormakka K. 2009. Structural basis of GDP release and gating in G protein coupled Fe2+ transport. EMBO J. 28:2677–2685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hung KW, Chang YW, Eng Chen ETJH, Chen YC, Sun YJ, Hsiao CD, Dong G, Spasov KA, Unger VM, Huang TH. 2010. Structural fold, conservation and Fe(II) binding of the intracellular domain of prokaryote FeoB. J. Struct. Biol. 170:501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hung KW, Juan TH, Hsu YL, Huang TH. 2012. NMR structure note: the ferrous iron transport protein C (FeoC) from Klebsiella pneumoniae. J. Biomol. NMR 53:161–165 [DOI] [PubMed] [Google Scholar]
- 20.Fetherston JD, Mier I, Jr, Truszczynska H, Perry RD. 2012. The Yfe and Feo transporters are involved in microaerobic growth and virulence of Yersinia pestis in bubonic plague. Infect. Immun. 80:3880–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung K-W, Tsai J-Y, Juan T-H, Hsu Y-L, Hsiao C-D, Huang T-H. 2012. Crystal structure of the Klebsiella pneumoniae NFeoB/FeoC complex and roles of FeoC in regulation of Fe2+ transport by the bacterial Feo system. J. Bacteriol. 194:6518–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H, Lee H, Shin D. 2013. The FeoC protein leads to high cellular levels of the Fe(II) transporter FeoB by preventing FtsH protease regulation of FeoB in Salmonella enterica. J. Bacteriol. 195:3364–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. 1998. Recommendations for the presentation of NMR structures of proteins and nucleic acids. J. Mol. Biol. 280:933–952 [DOI] [PubMed] [Google Scholar]
- 24.Ravel B, Newville M. 2005. ATHENA, ARTEMIS, HEPHAESTUS: data analysis for X-ray absorption spectroscopy using IFEFFIT. J. Synchrotron Radiat. 12:537–541 [DOI] [PubMed] [Google Scholar]
- 25.Newville M. 2001. IFEFFIT: interactive XAFS analysis and FEFF fitting. J. Synchrotron Radiat. 8:322–324 [DOI] [PubMed] [Google Scholar]
- 26.Ravel B. 2001. ATOMS: crystallography for the X-ray absorption spectroscopist. J. Synchrotron Radiat. 8:314–316 [DOI] [PubMed] [Google Scholar]
- 27.Venkateswara Rao P, Holm RH. 2004. Synthetic analogues of the active sites of iron-sulfur proteins. Chem. Rev. 104:527–559 [DOI] [PubMed] [Google Scholar]
- 28.Mulder DW, Ortillo DO, Gardenghi DJ, Naumov AV, Ruebush SS, Szilagyi RK, Huynh B, Broderick JB, Peters JW. 2009. Activation of HydA(DeltaEFG) requires a preformed [4Fe-4S] cluster. Biochemistry 48:6240–6248 [DOI] [PubMed] [Google Scholar]
- 29.Inubushi T, Becker ED. 1983. Efficient detection of paramagnetically shifted NMR resonances by optimizing the WEFT pulse sequence. J. Magn. Reson. 51:128–133 [Google Scholar]
- 30.Lin IJ, Xia B, King DS, Machonkin TE, Westler WM, Markley JL. 2009. Hyperfine-shifted (13)C and (15)N NMR signals from Clostridium pasteurianum rubredoxin: extensive assignments and quantum chemical verification. J. Am. Chem. Soc. 131:15555–15563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng H, Markley JL. 1995. NMR spectroscopic studies of paramagnetic proteins: iron-sulfur proteins. Annu. Rev. Biophys. Biomol. Struct. 24:209–237 [DOI] [PubMed] [Google Scholar]
- 32.Sweeney WV, Rabinowitz JC. 1980. Proteins containing 4Fe-4S clusters: an overview. Annu. Rev. Biochem. 49:139–161 [DOI] [PubMed] [Google Scholar]
- 33.Orme-Johnson WH. 1973. Iron-sulfur proteins: structure and function. Annu. Rev. Biochem. 42:159–204 [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Lee H-S, Sugar FJ, Jenney FEJ, Adams MWW, Prestegard JH. 2007. PF0610, a novel winged helix-turn-helix variant possessing a rubredoxin-like Zn ribbon motif from the hyperthermophilic archaeon, Pyrococcus furiosus. Biochemistry 46:752–761 [DOI] [PubMed] [Google Scholar]
- 35.Cheng H, Xia B, Reed GH, Markley JL. 1994. Optical, EPR, and 1H NMR spectroscopy of serine-ligated [2Fe-2S] ferredoxins produced by site-directed mutagenesis of cysteine residues in recombinant Anabaena 7120 vegetative ferredoxin. Biochemistry 33:3155–3164 [DOI] [PubMed] [Google Scholar]
- 36.Berndt C, Lillig CH, Wollenberg M, Bill E, Mansilla MC, de Mendoza D, Seidler A, Schwenn JD. 2004. Characterization and reconstitution of a 4Fe-4S adenylyl sulfate/phosphoadenylyl sulfate reductase from Bacillus subtilis. J. Biol. Chem. 279:7850–7855 [DOI] [PubMed] [Google Scholar]
- 37.Netz DJ, Stith CM, Stumpfig M, Kopf G, Vogel D, Genau HM, Stodola JL, Lill R, Burgers PM, Pierik AJ. 2012. Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8:125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yukl ET, Elbaz MA, Nakano MM, Moënne Loccoz P. 2008. Transcription factor NsrR from Bacillus subtilis senses nitric oxide with a 4Fe-4S cluster. Biochemistry 47:13084–13092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duin EC, Lafferty ME, Crouse BR, Allen RM, Sanyal I, Flint DH, Johnson MK. 1997. [2Fe-2S] to [4Fe-4S] cluster conversion in Escherichia coli biotin synthase. Biochemistry 36:11811–11820 [DOI] [PubMed] [Google Scholar]
- 40.Beinert H, Holm RH, Munck E. 1997. Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277:653–659 [DOI] [PubMed] [Google Scholar]
- 41.Beinert H. 2000. Iron-sulfur proteins: ancient structures, still full of surprises. J. Biol. Inorg. Chem. 5:2–15 [DOI] [PubMed] [Google Scholar]
- 42.Johnson DC, Dean DR, Smith AD, Johnson MK. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247–281 [DOI] [PubMed] [Google Scholar]
- 43.Cleland WW. 1964. Dithiothreitol, a new protective reagent for Sh groups. Biochemistry 3:480–482 [DOI] [PubMed] [Google Scholar]
- 44.Mayhew SG. 1978. The redox potential of dithionite and SO-2 from equilibrium reactions with flavodoxins, methyl viologen and hydrogen plus hydrogenase. Eur. J. Biochem. 85:535–547 [DOI] [PubMed] [Google Scholar]
- 45.Bou-Abdallah F, Chasteen ND. 2008. Spin concentration measurements of high-spin (g' = 4.3) rhombic iron(III) ions in biological samples: theory and application. J. Biol. Inorg. Chem. 13:15–24 [DOI] [PubMed] [Google Scholar]
- 46.Shulman GR, Yafet Y, Eisenberger P, Blumberg WE. 1976. Observations and interpretation of x-ray absorption edges in iron compounds and proteins. Proc. Natl. Acad. Sci. U. S. A. 73:1384–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qi W, Cowan JA. 2011. Structural, mechanistic and coordination chemistry of relevance to the biosynthesis of iron-sulfur and related iron cofactors. Coord. Chem. Rev. 255:688–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleischhacker AS, Kiley PJ. 2011. Iron-containing transcription factors and their roles as sensors. Curr. Opin. Chem. Biol. 15:335–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peters JW, Stowell MHB, Soltis SM, Finnegan MG, Johnson MK, Rees DC. 1997. Redox-dependent structural changes in the nitrogenase P-cluster. Biochemistry 36:1181–1187 [DOI] [PubMed] [Google Scholar]
- 50.Cammack R, Patil DS, Fernandez VM. 1985. Electron-spin-resonance/electron-paramagnetic-resonance spectroscopy of iron-sulphur enzymes. Biochem. Soc. Trans. 13:572–578 [DOI] [PubMed] [Google Scholar]
- 51.Lin IJ, Chen Y, Fee JA, Song J, Westler WM, Markley JL. 2006. Rieske protein from Thermus thermophilus: 15N NMR titration study demonstrates the role of iron-ligated histidines in the pH dependence of the reduction potential. J. Am. Chem. Soc. 128:10672–10673 [DOI] [PubMed] [Google Scholar]
- 52.Hsueh KL, Westler WM, Markley JL. 2010. NMR investigations of the Rieske protein from Thermus thermophilus support a coupled proton and electron transfer mechanism. J. Am. Chem. Soc. 132:7908–7918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arnesano F, Banci L, Piccioli M. 2005. NMR structures of paramagnetic metalloproteins. Q. Rev. Biophys. 38:167–219 [DOI] [PubMed] [Google Scholar]
- 54.Cammack R. 1975. Effects of solvent on the properties of ferredoxins. Biochem. Soc. Trans. 3:482–488 [DOI] [PubMed] [Google Scholar]
- 55.Link TA. 1999. The structures of Rieske and Rieske-type proteins. Adv. Inorg. Chem. 47:83–157 [Google Scholar]
- 56.Khoroshilova N, Popescu C, Munck E, Beinert H, Kiley PJ. 1997. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4s] to [2Fe-2s] conversion with loss of biological activity. Proc. Natl. Acad. Sci. U. S. A. 94:6087–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Green J, Bennett B, Jordan P, Ralph ET, Thomson AJ, Guest JR. 1996. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crack JC, Jervis AJ, Gaskell AA, White GF, Green J, Thomson AJ, Le Brun NE. 2008. Signal perception by FNR: the role of the iron-sulfur cluster. Biochem. Soc. Trans. 36:1144–1148 [DOI] [PubMed] [Google Scholar]
- 59.Kiley PJ, Beinert H. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6:181–185 [DOI] [PubMed] [Google Scholar]
- 60.Crack JC, Green J, Hutchings MI, Thomson AJ, Le Brun NE. 2012. Bacterial iron-sulfur regulatory proteins as biological sensor-switches. Antioxid. Redox Sign. 17:1215–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. 2009. Metalloproteins and metal sensing. Nature 460:823–830 [DOI] [PubMed] [Google Scholar]
- 62.Ayala-Castro C, Saini A, Outten FW. 2008. Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 72:110–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moore LJ, Kiley PJ. 2001. Characterization of the dimerization domain in the FNR transcription factor. J. Biol. Chem. 276:45744–45750 [DOI] [PubMed] [Google Scholar]
- 64.Scott C, Partridge JD, Stephenson JR, Green J. 2003. DNA target sequence and FNR-dependent gene expression. FEBS Lett. 541:97–101 [DOI] [PubMed] [Google Scholar]
- 65.Outten FW, Djaman O, Storz G. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52:861–872 [DOI] [PubMed] [Google Scholar]
- 66.Nesbit AD, Giel JL, Rose JC, Kiley PJ. 2009. Sequence-specific binding to a subset of IscR-regulated promoters does not require IscR Fe-S cluster ligation. J. Mol. Biol. 387:28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yeo WS, Lee JH, Lee KC, Roe JH. 2006. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 61:206–218 [DOI] [PubMed] [Google Scholar]
- 68.Fleischhacker AS, Stubna A, Hsueh KL, Guo Y, Teter SJ, Rose JC, Brunold TC, Markley JL, Munck E, Kiley PJ. 2012. Characterization of the [2Fe-2S] cluster of Escherichia coli transcription factor IscR. Biochemistry 51:4453–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajagopalan S, Teter SJ, Zwart PH, Brennan RG, Phillips KJ, Kiley PJ. 2013. Studies of IscR reveal a unique mechanism for metal-dependent regulation of DNA binding specificity. Nat. Struct. Mol. Biol. 20:740–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hentze MW, Kuhn LC. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 93:8175–8182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haile DJ, Rouault TA, Harford JB, Kennedy MC, Blondin GA, Beinert H, Klausner RD. 1992. Cellular regulation of the iron-responsive element binding protein: disassembly of the cubane iron-sulfur cluster results in high-affinity RNA binding. Proc. Natl. Acad. Sci. U. S. A. 89:11735–11739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kamps A, Achebach S, Fedtke I, Unden G, Gotz F. 2004. Staphylococcal NreB: an O-2-sensing histidine protein kinase with an O-2-labile iron-sulphur cluster of the FNR type. Mol. Microbiol. 52:713–723 [DOI] [PubMed] [Google Scholar]
- 73.Müllner M, Hammel O, Mienert B, Schlag S, Bill E, Unden G. 2008. A PAS domain with an oxygen labile [4Fe-4S]2+ cluster in the oxygen sensor kinase NreB of Staphylococcus carnosus. Biochemistry 47:13921–13932 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.