Abstract
The foliar pathogen Pseudomonas syringae is a useful model for understanding the role of stress adaptation in leaf colonization. We investigated the mechanistic basis of differences in the osmotolerance of two P. syringae strains, B728a and DC3000. Consistent with its higher survival rates following inoculation onto leaves, B728a exhibited superior osmotolerance over DC3000 and higher rates of uptake of plant-derived osmoprotective compounds. A global transcriptome analysis of B728a and DC3000 following an osmotic upshift demonstrated markedly distinct responses between the strains; B728a showed primarily upregulation of genes, including components of the type VI secretion system (T6SS) and alginate biosynthetic pathways, whereas DC3000 showed no change or repression of orthologous genes, including downregulation of the T3SS. DC3000 uniquely exhibited improved growth upon deletion of the biosynthetic genes for the compatible solute N-acetylglutaminylglutamine amide (NAGGN) in a minimal medium, due possibly to NAGGN synthesis depleting the cellular glutamine pool. Both strains showed osmoreduction of glnA1 expression, suggesting that decreased glutamine synthetase activity contributes to glutamate accumulation as a compatible solute, and both strains showed osmoinduction of 5 of 12 predicted hydrophilins. Collectively, our results demonstrate that the superior epiphytic competence of B728a is consistent with its strong osmotolerance, a proactive response to an osmotic upshift, osmoinduction of alginate synthesis and the T6SS, and resiliency of the T3SS to water limitation, suggesting sustained T3SS expression under the water-limited conditions encountered during leaf colonization.
INTRODUCTION
Bacteria vary greatly in the ability to tolerate osmotic stress. Large differences in osmotolerance may be associated with highly divergent adaptation strategies, such as the maintenance of low versus high cytoplasmic salt levels. Similarly, small differences in osmotolerance may reflect subtle differences in osmoadaptation mechanisms, with these differences influencing the relative fitness of individual species and strains. In most terrestrial environments, water stress tolerance is an important component of bacterial fitness and is therefore relevant to understanding, predicting, and manipulating the ecological success of target organisms, such as for biocontrol (1, 2). Pseudomonas syringae is a foliar pathogen, as well as a common resident on leaves. It has been used extensively as a model to understand bacterial strategies of plant surface colonization and how they are impacted by water-mediated processes (e.g., references 3 to 5). Here, we explore how two of these well-studied strains, B728a and DC3000, differ in their responses to osmotic stress and in the ability to colonize leaves.
General osmotolerance mechanisms are similar across diverse bacteria and include the accumulation of compatible solutes by de novo synthesis and uptake. The compatible solutes synthesized de novo by Pseudomonas spp. vary among species and even strains but include the dipeptide N-acetylglutaminylglutamine amide (NAGGN), the disaccharide trehalose, and glutamate in P. syringae, P. aeruginosa, and P. putida (6–8); mannitol in P. putida (9); and glucosylglycerol, ectoine, and hydroxyectoine in halotolerant and halophilic species (10–12). Pseudomonas spp. can also import exogenous compounds, including glycine betaine and its precursor choline (6, 13, 14). Additional mechanisms of water stress tolerance include cellular aggregation (15) and the production of exopolymeric substances that encapsulate bacterial cells, as shown for alginate in P. putida and P. syringae cells subjected to matric, i.e., low water content, stress (16).
Low water availability is among the most significant obstacles faced by resident microbes on leaf surfaces. This prediction is supported by the identification of a P. syringae locus required for both fitness on leaves and osmotolerance in culture (17), the contribution of alginate to P. syringae fitness on leaves (18), the preferential survival of highly aggregated over nonaggregated P. syringae cells on drying leaf surfaces (15), and the strong induction of water stress tolerance genes in P. syringae cells on and in leaves (19). Interestingly, following leaf infiltration by P. syringae, endophytic population sizes were directly correlated with the availability of water in the leaf intercellular spaces (20), indicating a need to tolerate low water availability even at endophytic sites. Furthermore, P. syringae encountered particularly low water availability during a hypersensitive response (21), indicating a possible role for water limitation as a component of plant defense (5).
At present, we have little understanding of how P. syringae strains vary in the ability to cope with water limitation. P. syringae strains share many genes involved in osmoadaptation, including transporters for the uptake of osmoprotectants and putative biosynthetic loci for compatible solutes and alginate. P. syringae strains B728a, DC3000, and 1448A, which all have a complete genome sequence available, represent three distinct pathovars, namely, syringae, tomato, and phaseolicola, respectively. These pathovars have been classified into distinct genomospecies based on whole-genome DNA-DNA hybridization (22) and into distinct phylogroups based on the sequences of four housekeeping genes (23). B728a is thought to be better adapted for epiphytic survival than DC3000 and 1448A (24), which is suggestive of superior osmotolerance of B728a over the other strains. Here, we confirm the roles of several genes in P. syringae osmoadaptation and correlate a greater osmotolerance of strain B728a with superior epiphytic survival. By specifically comparing B728a to DC3000, we correlate the greater osmotolerance of B728a with greater activation of alginate genes and alginate production and higher osmoprotectant uptake activity. A comparison of the global transcriptomes of the two strains in response to an osmotic upshift shows primarily upregulation of genes in B728a but downregulation of genes in DC3000, with strain-dependent effects on transcripts for the type III secretion system (T3SS) and T6SS.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Bacteria were grown at 28°C in King's B medium (25) containing rifampin (KB-Rif), in the low-osmolarity medium ½21C (13) or ½21C amended with 25 mM succinate (½21CS) to evaluate bacterial osmotolerance or in MinAS medium (7), when indicated, to enable comparison with ½21CS medium in the evaluation of bacterial osmotolerance. Experiments evaluating the role of N in osmotolerance involved modified ½21CS medium that had the N content of MinAS medium [7.6 mM (NH4)2SO4] rather than that of ½21CS medium (9.4 mM NH4Cl), as well as ½21CS medium amended with glutamine to a final concentration of 4.7 mM. Antibiotics were added to the growth media as needed at the following concentrations (μg/ml): ampicillin (Ap), 100; kanamycin (Km), 50; rifampin (Rif), 100; spectinomycin (Sp), 60; tetracycline (Tet), 20; cycloheximide, 100.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description or relevant genotype | Reference |
|---|---|---|
| Strains | ||
| DC3000 | Wild-type P. syringae pv. tomato; Rifr | 54 |
| B728a | Wild-type P. syringae pv. syringae; Rifr | 55 |
| 1448A | Wild-type P. syringae pv. phaseolicola; Rifr | 56 |
| PAO1 | Wild-type P. aeruginosa; Rifr | 57 |
| DCΔtre | DC3000 with deletions ΔPSPTO_2760-2762 and ΔPSPTO_3125-3134; Rifr | 7 |
| DCΔggn | DC3000 ΔPSPTO_1630-1633; Rifr | This work |
| B7Δggn | B728a ΔPsyr_3747-3748 (ΔggnAB); Rifr | 28 |
| Plasmids | ||
| pTOK2T | Broad-host-range plasmid with functional lacZ activity; Tetr | 58 |
| pKD4 | Template for kan cassette flanked by FRT sites; Apr Kmr | 59 |
| pFlp2Ω | Flp recombinase-encoding plasmid; Apr Spr | 60 |
Characterization of bacterial responses to osmotic stress and survival and growth on plants.
Bacterial growth in culture was evaluated spectrophotometrically in microtiter plates as described by Chen et al. (13). Bacterial uptake of osmoprotectant compounds was evaluated by measuring the rates of uptake of [methyl-14C]choline and [methyl-14C]betaine at a final concentration of 10 μM in cells grown in ½21C. The sources of the radiochemicals and the transport assays were as described by Chen et al. (13). Bacterial production of polysaccharides was evaluated by growing cells in ½21CS or MinAS to an optical density at 600 nm (OD600) of 0.2 to 0.3, precipitating the polysaccharides with cold ethanol (ethanol/supernatant ratio, 4:1) at −20°C, and subjecting them to a meta-hydroxydiphenyl assay in which the absorbance at 520 nm was measured and d-glucuronic acid was used as the standard (26).
Bacterial survival following an osmotic upshift and an 8-h incubation with shaking was assessed by enumeration on KB-Rif medium. Bacterial survival following inoculation onto plant surfaces in the laboratory was evaluated by diluting ½21C-grown bacteria to a density of 108 CFU/ml in phosphate buffer (10 mM, pH 7) (PB) containing 0.01% Silwet L-77 (Lehle Seeds, Round Rock, TX), inoculating soybean (Glycine max cultivar Williams 82) and maize (Z. mays subsp. mays L. inbred B73) plants by leaf immersion in a bacterial suspension, and incubating the plants in a mist tent. Epiphytic bacterial populations were enumerated on individual leaves as described by Li et al. (27). Bacterial survival under field conditions was evaluated on tomato (Solanum lycopersicum cultivar Rio Grande PtoS), soybean Williams 82, and bean (Phaseolus vulgaris cultivar Bush Blue Lake 274) plants grown at the Horticultural Research Farm of Iowa State University near Gilbert, IA, with four replications of each strain-plant species combination arranged in a randomized complete block design. KB-Rif-grown bacteria were suspended in PB containing 0.01% Silwet L-77 (107 CFU/ml) and introduced onto plants with a handheld spray bottle with subsequent sampling and bacterial enumeration as described previously (7).
Mutant construction.
A DC3000 deletion mutant lacking the ggn locus (PSPTO_1630-1633) was constructed by amplifying two 1- to 2-kb fragments flanking this locus from the DC3000 genome and a kanamycin resistance-encoding (kan) cassette with flanking FLP recombination target (FRT) sites from pKD4 with the primer pairs listed in Table S1 in the supplemental material. The three fragments were ligated together by splice overlap extension PCR with the F1 and R2 primers. The resulting fragment was cloned into SmaI-digested pTOK2T and introduced into DC3000 by transformation; deletion mutants were identified as Rifr Kmr Tets. The pFlp2Ω plasmid was introduced to excise the kan cassette and later cured with 20% sucrose for counterselection. This DCΔggn mutant was confirmed to be deficient in the production of NAGGN by 13C nuclear magnetic resonance analysis (see Fig. S1 in the supplemental material). We also used a B728a deletion mutant lacking ggnAB that was previously constructed (28) and confirmed to be deficient in the production of NAGGN (27).
Preparation of osmotically stressed cells for RNA extraction.
To prepare RNA for transcriptome studies, B728a and DC3000 cells were each grown in ½21CS broth with three sequential subcultures. Mid-log-phase cells were harvested from the final subculture by centrifugation at 5,000 × g for 10 min, and the pellets were resuspended in 3.6 ml of prewarmed ½21CS to a final OD600 of 0.4 (approximately 8 × 108 cells/ml). Aliquots of 0.3 ml were transferred to 10-ml tubes, incubated at 28°C with shaking for 15 min, and then amended with 0.8 ml of prewarmed ½21CS medium lacking NaCl (−0.2 MPa) or supplemented with NaCl to a final concentration of 219 mM NaCl (−1 MPa). The cells were incubated at 28°C with shaking for 15 min, at which time 2 ml of RNA-stabilizing agent (RNAProtect Bacteria Reagent; Qiagen Inc., Valencia, CA) was added. This procedure was performed with two replicate cultures of each strain on a single day, and the cells from the two cultures were combined and harvested by centrifugation at 5,000 × g for 10 min. The supernatants were removed, and the cell pellets were stored at −20°C. The same procedure was repeated on a second day. The RNA isolated from each of the two cell pellets was pooled, and this pooled RNA, which was therefore derived from four independent cultures, served as a single biological replicate. In this manner, two independent biological replicates were prepared for each strain-treatment combination.
RNA extraction, hybridization, and microarray design.
RNA was purified with a Qiagen RNeasy minikit, and DNA was removed by on-column DNase I digestion, with subsequent DNase I removal with the RNeasy column purification kit (Qiagen Inc.). RNA integrity was evaluated with an Agilent 2100 bioanalyzer. RNA samples were sent to Roche NimbleGen Inc. (Madison, WI) for conversion into cDNA, labeling with U-CYA-3 fluorophore, and hybridization to the appropriate microarrays. Open reading frame (ORF)-based microarrays were designed by using complete genome sequence information for DC3000, including both plasmids (accession numbers NC_004578, NC_004632, and NC_004633), and a draft-phase sequence for B728a (accession numbers NZ_AABP02000001 to NZ_AABP02000026), as this microarray was generated before the sequence was finalized. Two complete microarrays were present on each slide and were considered technical replicates. Each gene was represented by a minimum of 17 (DC3000) or 19 (B728a) 24-mer nucleotide probes, with each technical replicate containing a complete set of perfect-match and mismatch probes. The fluorescence intensity of each probe was measured, and the fluorescent intensities were subjected to robust multiarray averaging, which included adjustment for the background intensity, quantile normalization, and median polishing. A robust estimated mean value for each gene in the array was determined for each technical replicate, and the two values for each gene were averaged.
Although the data are not included here, the microarray experiment with DC3000 included three additional treatments and the results of these treatments were included in the analysis. The microarray data were analyzed as described previously (19). Briefly, linear models for microarray data analysis (29) were applied to share information across genes when estimating error variances. The adjusted variance term was used to calculate Welch t statistics for pairwise comparison of the osmotic stress versus basal medium treatments, and Q values, and thus the associated false-discovery rates (FDRs), were estimated from the corresponding distribution of P values (30). Average fold changes across selected genes were calculated by exponentiating the arithmetic average of the estimated log fold changes of the individual genes.
Identification and annotation of orthologs in B728a, DC3000, and PAO1.
Previous work identified putative orthologs in B728a, DC3000, and PAO1 by applying Ortholuge analysis (31) to the genomes of these strains (32). For B728a or DC3000 genes with multiple predicted orthologs, orthology was evaluated on the basis of synteny; orthologs were assigned only for genes with a single probable ortholog in both strains. Orthologs in PAO1 were not assigned when different PAO1 genes were predicted as orthologs to orthologous genes in B728a and DC3000. Gene annotation was checked manually by comparing gene names and functions among the orthologs of all of the Pseudomonas sp. strains subjected to the Ortholuge program in the Pseudomonas Genome Database (32).
qRT-PCR.
Quantitative reverse transcription-PCR (qRT-PCR) analysis of selected genes in DC3000 was performed for comparison to the microarray data. Bacterial cells were grown in ½21CS medium containing 0.22 M NaCl (−1 MPa) and 0.44 M NaCl (−1.9 MPa). Cells were harvested after 15 and 60 min of incubation with hyperosmolarity, and RNA extraction and purification were performed as described above. One-step RT conversion of RNA to cDNA and amplification of selected targets was performed with SYBR green I-based Stratagene Full Velocity master mix (Stratagene, Cedar Creek, TX) in an Opticon thermocycler (Opticon Inc., Orangeburg, NY). The housekeeping gene hemD (PSPTO_0129) was used as an internal control to normalize the induction values of all genes. The fold change for each ORF was calculated by the 2(−ΔΔCT) method (33) by using two replicate samples for each treatment per time point. The sequences of the primers used are shown in Table S1 in the supplemental material.
RESULTS
P. syringae B728a exhibited superior osmotolerance and epiphytic survival compared to two other P. syringae strains.
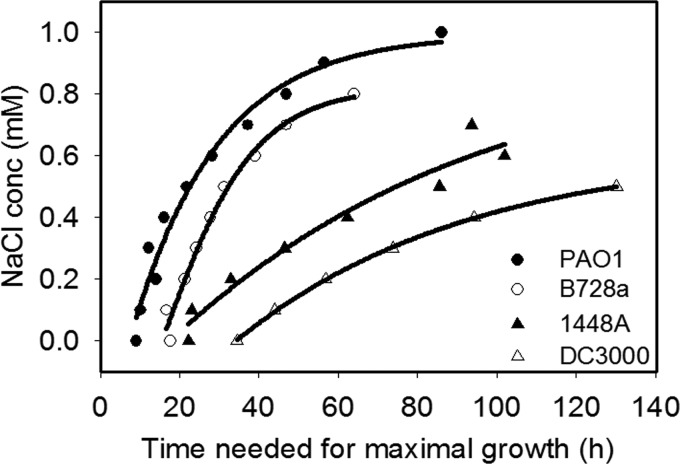
Three P. syringae strains were examined for tolerance to osmotic stress during growth, with P. aeruginosa strain PAO1 included for comparison. The strains, B728a, DC3000, and 1448A, differed greatly in their growth dynamics in media amended with NaCl, with B728a exhibiting shorter lag periods than the other P. syringae strains prior to growth (see Fig. S2 in the supplemental material). When osmotolerance was evaluated on the basis of the time required to attain a maximal growth rate, a term that reflected a combination of the growth rate and the length of the lag period, P. aeruginosa PAO1was more osmotolerant than B728a, which in turn was more osmotolerant than 1448A and DC3000 (Fig. 1). DC3000, 1448A, and B728a did not grow at NaCl concentrations greater than 0.7, 0.8, and 0.9 M, respectively (see Fig. S2).
Fig 1.
Impact of NaCl on the growth of three P. syringae strains, B728a, 1448A, and DC3000, and one P. aeruginosa strain, PAO1. Bacteria were grown in ½21C medium amended with 0 to 1 M NaCl at 0.1-M intervals (see Fig. S2 in the supplemental material). The time required for each strain to reach its maximal growth rate at a given NaCl concentration was determined, and these values were fitted into a modified Gompertz model.
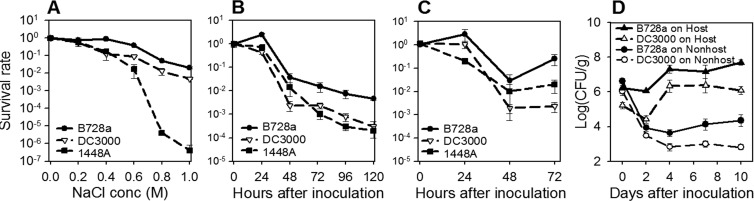
The ability of the strains to tolerate an osmotic upshift, as well as a period of drying on leaves, was examined. Following an upshift to NaCl concentrations greater than 0.2 M, a greater number of B728a cells than DC3000 and 1448A cells remained viable (Fig. 2A). Similarly, a greater proportion of B728a than DC3000 and 1448A cells survived on leaves of the two nonhost plant species soybean and maize under conditions that promoted drying of leaf surfaces (Fig. 2B and C). When B728a and DC3000 were examined on plants in a field environment, B728a established and maintained larger populations than DC3000 on the nonhost and host plant species during a 10-day period of monitoring (Fig. 2D). B728a and DC3000 were selected for further studies on the basis of their large difference in osmotolerance in culture (Fig. 1) and their fitness on leaves (Fig. 2).
Fig 2.
Survival of P. syringae strains following 8 h of exposure to hyperosmotic stress (A), during exposure to drying on soybean leaves (B) and maize leaves (C) in a growth chamber, and on leaves of host and nonhost plant species in the field (D). For panel A, strains were grown in ½21C medium and exposed to 0 to 1 M NaCl for 8 h; the survival rate was calculated as the proportion of cells that were present at 0 M NaCl. For panel B, strains were grown in ½21C medium and introduced onto plant leaves; cells were enumerated by plating, and the proportion of cells present at 0 h was determined. For panel D, strains were grown in King's B medium, DC3000 cells were introduced onto tomato (host) and soybean (nonhost) leaves, and B728a cells were introduced onto bean (host) and soybean (nonhost) leaves; plants for panel D were grown and inoculated under field conditions. The values shown are means ± standard errors (n = 6 [A to C] or 12 [D]).
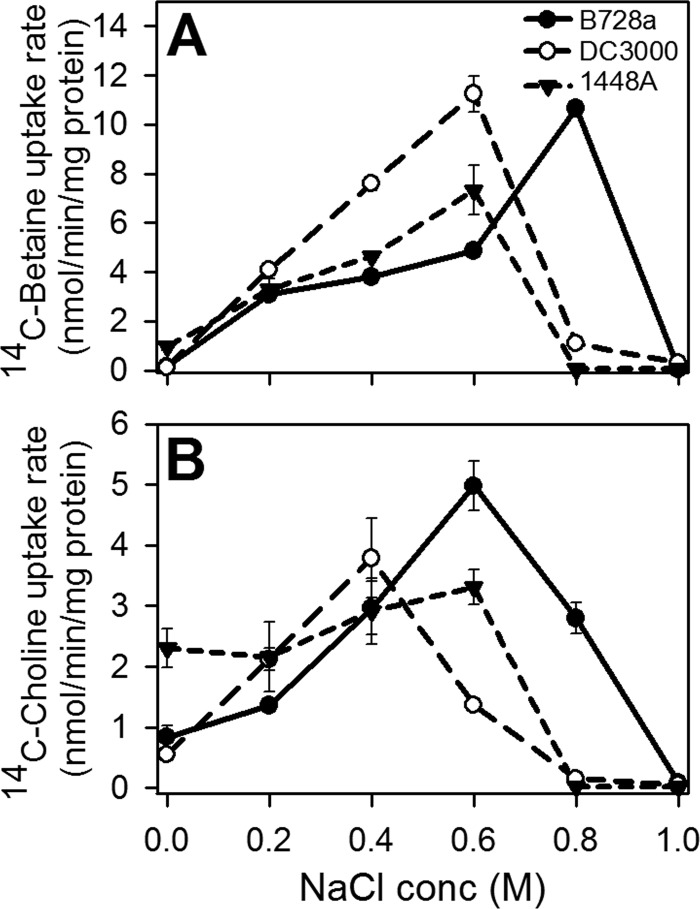
B728a exhibited higher rates of glycine betaine and choline uptake than DC3000 at high osmolarities.
We have previously reported that choline and glycine betaine provide high levels of osmoprotection for B728a, DC3000, and 1448A (28). To evaluate if these strains differ in the rate at which they transport these two osmoprotectants, we measured the rate of uptake of 14C-labeled derivatives (Fig. 3). DC3000 and 1448A exhibited higher glycine betaine uptake rates than B728a at NaCl concentrations of ≤0.6 M, but B728a exhibited a much higher uptake rate at 0.8 M NaCl (Fig. 3A). Similarly, B728a exhibited higher rates of choline uptake than the other strains at osmolarities of >0.4 M NaCl (Fig. 3B).
Fig 3.
Effect of NaCl on the initial rates of radiolabeled glycine betaine (A) and choline (B) uptake by P. syringae strains. Glycine betaine and choline were each provided at a final concentration of 10 μM. The values shown are means ± standard errors (n = 3) and are representative of two replicate experiments.
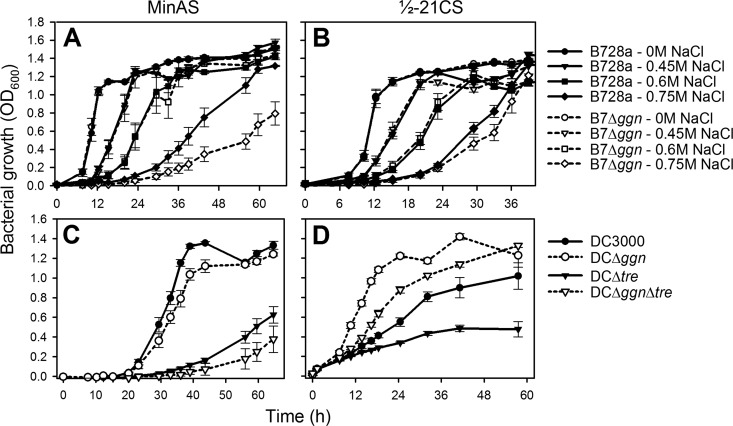
B728a consistently benefited from de novo synthesis of the compatible solute NAGGN, whereas DC3000 surprisingly did not.
In the absence of exogenous osmoprotectants, these P. syringae strains accumulated NAGGN, trehalose, and glutamate in response to osmotic stress (see Fig. S3 in the supplemental material) (7). A B728a mutant that lacked the ggnAB genes required for NAGGN synthesis (27, 34), designated B7Δggn, exhibited impaired osmotolerance but only at high NaCl concentrations (>0.6 M) (Fig. 4A and B). A DC3000 mutant that lacked the ggn operon, designated DCΔggn, exhibited reduced osmotolerance in the low-osmoticum medium MinAS but also only at high NaCl concentrations (≥0.4 M) (Fig. 4C; see Fig. S4A); the operon contained two putative genes downstream of ggnAB that are not required for NAGGN synthesis (34). A trehalose-deficient DC3000 mutant, designated DCΔtre (Table 1), showed a much greater growth delay than DCΔggn in MinAS medium, with a mutant that lacked the loci for both NAGGN and trehalose production showing the most severe reduction in osmotolerance (Fig. 4C; see Fig. S4A). These results demonstrate that trehalose and NAGGN perform complementary protective roles in promoting osmotolerance in DC3000, with trehalose contributing more under these conditions.
Fig 4.
Contribution of compatible solute biosynthetic loci to growth of wild-type and mutant strains of B728a (A, B) and DC3000 (C, D) at high osmolarity in MinAS (A, C) and ½21CS (B, D) media. The B728a cultures were grown in media amended with various NaCl concentrations, as indicated, whereas the DC3000 cultures were grown in media amended with 0.4 M NaCl. Values represent the mean OD600 ± the standard error (n = 4) and are representative of three independent experiments.
Interestingly, the DCΔggn mutant showed enhanced growth under high osmolarity in ½21CS medium, demonstrating that the ggn locus detrimentally impacted DC3000 osmotolerance in this medium (Fig. 4D; see Fig. S4B). The magnitude of the growth benefit of DCΔggn was similar in the presence and absence of the loci for trehalose production (Fig. 4D). In contrast, B7Δggn showed reduced growth in both MinAS and ½21CS media (Fig. 4A and B). Since NAGGN synthesis is a nitrogen-demanding process, we hypothesized that the 1.6-fold lower N content of ½21CS than MinAS may have caused a depletion of cellular N reserves in DC3000 during NAGGN synthesis. Modifying the N content of ½21CS to that of MinAS by amending it with (NH4)2SO4, however, did not increase the growth of DC3000 at high osmolarity (data not shown), but interestingly, amending ½21CS with glutamine dramatically increased the growth of DC3000 and DCΔtre at high osmolarity while having little to no impact on the growth of DCΔggn and DCΔggnΔtre (see Fig. S5 in the supplemental material).
B728a and DC3000 differed in their responses to NaCl at the transcriptional level.
We characterized the global transcriptome of these strains following a 15-min exposure to a water potential of −1 MPa imposed by NaCl (0.22 M). The treatments included not only B728a and DC3000 with and without exposure to osmotic stress but also DC3000 exposed to three additional treatments that, although not discussed here, provided greater statistical power to the DC3000 analysis. Consequently, to assess if compensation for the lower statistical power of the B728a analysis was needed, we evaluated the B728a data set at 1% and 10% FDRs and evaluated the DC3000 results only at a 1% FDR. We found that using FDRs of 1% for DC3000 and 10% for B728a yielded similar proportions of differentially expressed genes in both strains. In DC3000, 5,582 genes were analyzed and 551, or 10%, differed in transcript abundance in response to NaCl (FDR, ≤1%). In B728a, 4,418 genes were analyzed and 411, or 9%, differed in transcript abundance in response to NaCl (FDR, ≤10%). Greater than 60% of the differentially expressed genes in B728a were increased in transcript abundance regardless of the selected FDR cutoff; in contrast, 58% of the differentially expressed genes in DC3000 were decreased in transcript abundance. We used ortholog predictions (32) and synteny to identify 3,861 genes that had single orthologs in B728a and DC3000. Among these 3,861 genes, the majority with altered transcript abundance showed increased abundance in B728a and decreased abundance in DC3000 (Fig. 5).
Fig 5.
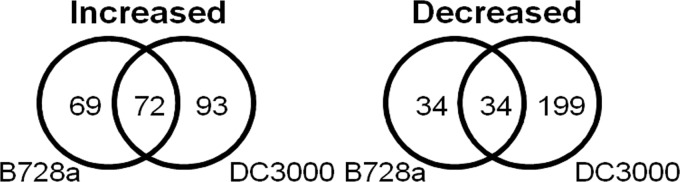
Numbers of B728a and DC3000 genes that were increased or decreased in transcript abundance in response to NaCl among the 3,861 genes that had orthologs in both strains (FDR, ≤1%). The use of an FDR of ≤10% to identify B728a transcripts that were altered in abundance changed the values in the diagram, from left to right, to 135, 99, and 66 for those that were increased and to 85, 65, and 168 for those that were decreased.
We assigned Gene Ontology (GO) terms to the P. syringae strains based on the GO terms assigned in P. aeruginosa PAO1. By using ortholog predictions and synteny to identify P. syringae orthologs of PAO1 genes, we were able to assign GO terms to 2,034 genes in B728a and 2,060 genes in DC3000. Fisher's exact test was used to identify the GO functions that were overrepresented among the genes exhibiting differential transcript abundance compared with their representation in the genome (see Fig. S6 in the supplemental material). In this analysis, individual genes could belong to multiple GO categories. Only two GO categories of differentially abundant transcripts were shared by the strains, betaine biosynthetic processes and betaine metabolic processes, both of which relate to the potential to accumulate the compatible solute betaine. The NaCl-responsive stimulon of B728a fell into 30 GO categories that were overrepresented, with 26 containing mostly genes with increased transcript abundance and 15 involving cell wall and/or polysaccharide metabolic processes. In contrast, the NaCl-responsive stimulon of DC3000 fell into 50 GO categories that were overrepresented, with 45 containing mostly genes with decreased transcript abundance and 46 involving primarily cytosolic processes. Thus, the two strains responded differently to the osmotic stress, with these differences suggesting that B728a was active in modifying cell envelope components whereas DC3000 slowed some of its metabolic processes, which generally dominate the cytoplasm.
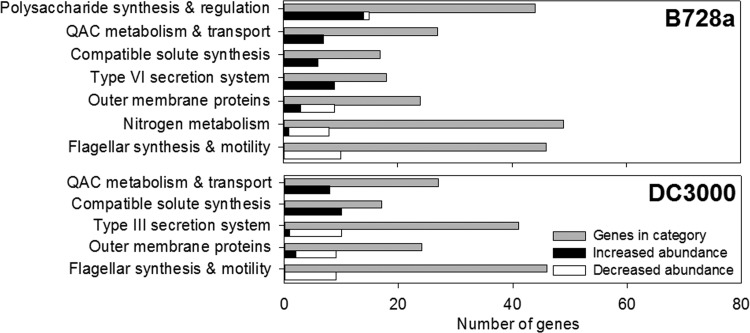
We classified the 3,861 orthologous genes in B728a and DC3000 into functional groups based on the primary function predicted for each gene. Genes with multiple primary functions were not classified, and each gene was assigned to only one functional group. A chi-square test was performed to compare the proportion of differentially expressed genes in each of these functional groups to those in the genome, with a permutation test performed when the groups were small. Similar results were obtained whether an FDR of 1% (Fig. 6) or 10% (data not shown) was used to identify differentially expressed transcripts in B728a. Both strains exhibited increases in the abundance of transcripts involved in the transport and metabolism of quaternary ammonium compounds such as betaine and choline (13, 14) and the synthesis of compatible solutes (see Table S2 in the supplemental material), supporting known mechanisms of osmoadaptation and the results of the gene ontology analysis described above. We did not observe strain specificity in the changes in the abundances of the transcripts for compatible solute synthesis but did find that the increases in the NAGGN transcripts were larger than those in the trehalose transcripts. The B728a ectC gene encodes an enzyme that is functional in the synthesis of the compatible solute ectoine but requires ectAB, which are missing from B728a (8). We found no evidence of ectC induction by osmotic stress; thus, if ectC is a remnant of a prior osmoresponsive ectoine synthetic pathway in B728a, it no longer exhibits osmoresponsiveness. DC3000 lacked orthologs to these ectoine synthetic genes. The strains also showed variable changes in outer membrane proteins, showing decreases for at least a third of the 12 porin-encoding genes (see Table S2), consistent with the outer membrane as a major site of alteration by osmotic stress. Lastly, both strains had decreased transcript levels of 22 to 44% of the genes involved in flagellar synthesis and motility (see Table S2), consistent with reduced movement as a response to water limitation. Functional groups that differed between the strains included polysaccharide synthesis and regulation, the T6SS and T3SS, and nitrogen metabolism (Fig. 6). We further discuss differences within these groups below.
Fig 6.
Number of genes in each of the functional categories that were significantly overrepresented in the NaCl-responsive transcriptome compared to the genome (FDR, ≤5% for the overrepresentation analysis). The values shown are the total number of genes in each category and the number genes that were increased or decreased in transcript abundance, with differential expression based on an FDR of ≤1%. QAC, quaternary ammonium compound.
Polysaccharide synthesis and regulation.
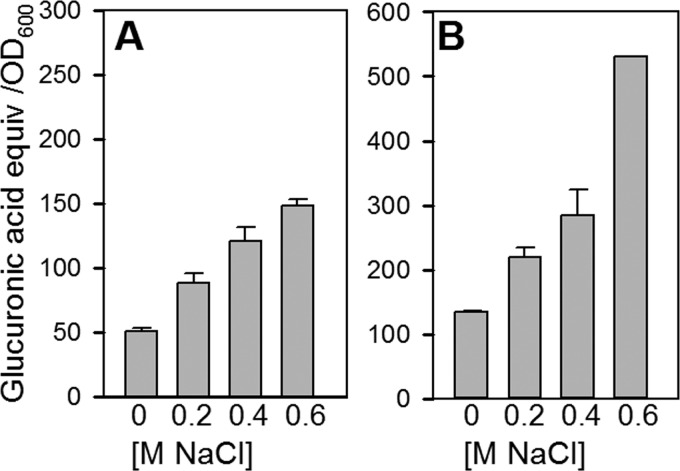
Transcripts for the alginate regulatory genes were increased in both strains, as demonstrated by average increases for mucB, algR, and algZ of 3.2- and 2.3-fold for B728a and DC3000, respectively (see Table S2 in the supplemental material). However, transcripts for the alginate biosynthetic genes were strongly increased only in B728a, as demonstrated by average increases of 6.4- and 1.4-fold for these genes in B728 and DC30000, respectively (see Table S2). Given the importance of alginate to water stress tolerance (16), greater alginate production by B728a under hyperosmolarity may contribute to its greater osmotolerance. In support of this prediction, we found that B728a produced a uronic acid-containing polysaccharide that resembled alginate (see Fig. S7 in the supplemental material), produced more with increasing osmolarity (Fig. 7), and lost its production upon the inactivation of algD (see Fig. S7). DC3000, in contrast, did not show osmotic stimulation of a uronic acid-containing polysaccharide resembling alginate (see Fig. S7).
Fig 7.
Effect of osmolarity on the production of uronic acid-containing polysaccharides by B728a with cells grown in ½21CS (A) and MinAS (B) media. Media were amended with the molar concentrations of NaCl indicated on the x axis, and the ethanol-precipitated polysaccharides were quantified by the meta-hydroxydiphenyl assay and expressed as micrograms of glucuronic acid equivalent per OD600 unit. The polysaccharide from B728a resembled the uronic acid-containing polysaccharide alginate on the basis of its color and absorption spectrum, whereas the polysaccharide from DC3000 did not (see Fig. S7 in the supplemental material). The values are means ± standard errors (n = 5 [A] or 2 [B]); the error bars are present but not visible for all samples.
T6SS.
The genes encoding T6SSs have been shown to fall into three distinct clusters, designated Hcp secretion islands (HSI) because of the ability of the T6SS to secrete an Hcp protein. Of these clusters, B728a has only HSI-I whereas DC3000 has both HSI-I and HSI-II gene clusters (35). None of the genes in either of the DC3000 clusters were impacted by osmotic stress, whereas 10 of the 21 genes in the B728a T6SS HSI-I cluster were induced, and 9 of these 10 had orthologs in DC3000 (see Table S2). These 10 genes include impK, clpV, and icmF, which are predicted to encode components of the secretion system itself (36), but not hcp1 (Psyr_4965), which was not induced although it encodes a secreted protein (36).
T3SS.
Genes categorized as T3SS associated included those for the T3SS components, as well as the secreted helper proteins, effectors, and their chaperones. None of the 52 T3SS-associated genes in B728a exhibited altered transcript levels in response to osmotic stress. In contrast, 41% of the 66 T3SS-associated genes in DC3000 exhibited altered levels, with all of these showing a decrease in transcript abundance (see Table S2). The ½21CS medium selected for these studies was not designed as an inducing medium for the T3SS genes; however, the average transcript level of these genes in this medium was higher than 25% of the genes in B728a and 54% of the genes in DC3000, suggesting that there was a sufficient level of transcription in both strains in the medium to detect reductions in transcript abundance.
Nitrogen metabolism.
Out of approximately 55 genes classified as N metabolism genes in the two strains, 9 genes in B728a and 3 genes in DC3000 showed changes in transcript abundance in response to osmotic stress. Among these, only one, a glutamine amidotransferase-encoding gene, was increased under stress, and it was increased only in B728a (see Table S2). Although the substrate of this glutamine amidotransferase is not known, this enzyme could potentially supplement the glutamine amidotransferase activity of GgnA during NAGGN synthesis. In contrast, both strains showed decreased levels of the glutamine synthetase-encoding glnA1 transcript (see Table S2), no change in the transcript levels of four other putative glutamine synthetase genes, and decreased levels of the glnK transcript, which encodes a regulator of glutamine synthetase (see Table S2). The reduced glnA1 levels may indicate a pathway by which glutamate is accumulated in response to osmotic upshock, namely, by reducing glutamate depletion due to glutamine synthesis; this is consistent with the lack of induction of the gltD and gltB genes involved in glutamate synthesis.
The B728a and DC3000 genomes encode 12 putative hydrophilins.
Hydrophilins are a class of small, highly hydrophilic proteins that can enhance tolerance to water limitation (37, 38). We identified 12 putative hydrophilin orthologs in both B728a and DC3000 on the basis of the criteria of small size, high glycine content, and high hydrophilicity. We designated these hfn (hydrophilin function) genes (Table 2). Transcripts for five of these putative hydrophilins were increased by osmotic stress. The proteins encoded by Psyr_3782, its DC3000 ortholog PSPTO_1596, and the putative hydrophilin-encoding, osmoinduced PAO1 gene PA4738 (39) exhibit 40, 36, and 55% amino acid identity to the Escherichia coli hydrophilin YjbJ, respectively. The hydrophilin YjbJ has been demonstrated to rescue enzymatic activity from the negative effects of osmotic stress in vitro (37).
Table 2.
Osmotic-stress-induced changes in transcript levels of genes predicted to encode hydrophilins
| Gene | B728aa |
DC3000a |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Locus | AA | Gly | Hyd | Fold changeb | Locus | AA | Gly | Hyd | Fold changeb | |
| hfnA | Psyr_3782 | 63 | 12.7 | 0.89 | 8.83c | PSPTO_1596 | 63 | 12.7 | 0.95 | 2.85d |
| hfnB | Psyr_0768 | 51 | 3.9 | 1.74 | 2.43c | PSPTO_0891 | 76 | 6.6 | 1.14 | 1.76d |
| hfnC | Psyr_0858 | 135 | 10.4 | 0.97 | 2.07c | PSPTO_0993 | 135 | 8.1 | 0.96 | 2.52d |
| hfnD | Psyr_3971 | 156 | 20.5 | 1.22 | Mc | PSPTO_4237 | 152 | 19.7 | 1.16 | 2.50d |
| hfnE | Psyr_0128 | 121 | 8.3 | 0.99 | 1.81c | PSPTO_0281 | 121 | 9.1 | 0.99 | 1.57d |
| hfnF | Psyr_3320 | 49 | 8.2 | 1.97 | Mc | PSPTO_3546 | 47 | 6.4 | 1.93 | (1.17)d |
| hfnG | Psyr_1879 | 51 | 8.9 | 1.83 | 1.70 | PSPTO_2069 | 61 | 8.2 | 1.91 | (1.10) |
| hfnH | Psyr_1972 | 103 | 7.8 | 1.64 | (1.27)c | PSPTO_2162 | 164 | 4.9 | 1.38 | (1.19) |
| hfnI | Psyr_4518 | 183 | 10.4 | 1.14 | (1.13) | PSPTO_0656 | 189 | 10.1 | 1.12 | (−1.24) |
| hfnJ | Psyr_0132 | 82 | 11.0 | 1.29 | M | PSPTO_0274 | 84 | 9.5 | 1.09 | (1.11) |
| hfnK | Psyr_0032 | 176 | 16.5 | 0.98 | (1.09) | PSPTO_0161 | 171 | 17.0 | 1.00 | (−1.03) |
| hfnL | Psyr_4262 | 117 | 9.4 | 0.95 | (−1.15) | PSPTO_4600 | 117 | 9.4 | 0.99 | (1.02) |
Hydrophilins were predicted on the basis of protein size (<200 amino acids), glycine content (≥6% of amino acid content), and estimated hydrophilicity (≥0.95) based on ExPASy's ProtScale (61). Values that did not meet a selection criterion in one of the two strains are in italics. The orthologs of HfnB exhibit distinct sizes because of the absence of an N-terminal domain in Psyr_0768. AA, number of amino acids; Gly, percent glycine content; Hyd, estimated hydrophilicity.
The fold change values in bold had significantly different transcript levels in the presence of NaCl versus in its absence on the basis of an FDR of 1%. For B728a, the fold changes not in bold had significantly different transcript levels on the basis of an FDR of 10%. Values in parentheses did not show significant differences in their transcript levels in response to NaCl and are included for comparison. M, the gene is missing from the B728a microarray.
This gene showed increased transcript abundance in response to osmotic stress in a subsequent transcriptome analysis (19).
This gene showed increased transcript abundance in response to a matric stress (polyethylene glycol [molecular weight, 8,000]), which represents a distinct form of water stress (62).
Transcript levels of selected genes exhibited similar qualitative changes by qRT-PCR and microarray analyses but were differentially influenced by osmotic stress level and exposure time.
The transcript levels of selected genes were evaluated by qRT-PCR following osmotic upshock with a moderate stress level similar to that used for the microarray, namely, −1 MPa osmotic upshock for 15 min, as well as for a longer time (60 min) and at a higher stress level (−1.9 MPa) for 15 and 60 min for comparison. The changes in transcript level, as evaluated by qRT-PCR, were qualitatively similar to those from the microarray for all of the genes examined (Table 3), although the quantitative fold change values were not correlated. Whereas the transcript levels of many genes such as ggnA were not strongly impacted by either stress level or exposure time, the glgA gene in the trehalose synthesis operon showed increased expression with increased stress and exposure time, whereas the hydrophilin gene hfnA showed the opposite (Table 3). The glnA1 gene was affected by stress and exposure time but in a more complex manner (Table 3).
Table 3.
Comparison of osmoinduced changes in transcript levels based on microarray versus qRT-PCR analyses for selected DC3000 genes
| Locus | Genec | Fold change in transcript levela after: |
||||
|---|---|---|---|---|---|---|
| 15-min osmotic upshock |
60-min osmotic upshock |
|||||
| Microarray | Moderate NaClb | High NaClb | Moderate NaClb | High NaClb | ||
| PSPTO_1633 | ggnA | 5.24 | 3.2 (0.7) | 3.8 (0.4) | 2.3 (0.1) | 4.8 (2.3) |
| PSPTO_4578 | opuCD | 4.68 | 3.1 (0.2) | 1.5 (0.1) | 4.7 (0.7) | 7.0 (2.6) |
| PSPTO_4906 | 4.62 | 5.8 (3.7) | 2.5 (1.7) | 8.5 (0.7) | 4.2 (1.7) | |
| PSPTO_1596 | hfnA | 2.85 | 9.3 (1.1) | 4.2 (0.4) | 5.8 (3.7) | 2.2 (0.3) |
| PSPTO_3125 | glgA | 2.16 | 1.6 (0.0) | 2.9 (1.2) | 2.7 (0.5) | 7.5 (4.6) |
| PSPTO_0359 | glnA1 | −2.94 | −8.3 (1.7) | −2.9 (0.7) | −2.4 (0.9) | −25.3 (2.2) |
| PSPTO_1274 | cspC | −3.29 | −2.5 (1.2) | −1.3 (0.4) | −2.0 (0.4) | −7.6 (0.4) |
Fold change in transcript level relative to conditions without hyperosmolarity, as described in Materials and Methods. The values are averages of two replicates (standard deviation).
DC3000 cells in ½21CS were subjected to a 15- or 60-min osmotic upshock with a moderate (0.22 M) or high (0.44 M) NaCl concentration.
The gene functions are as defined in Table 2 (see also Table S3 in the supplemental material), with the addition of PSPTO_4906, which encodes a putative ferritin/DPS gene, and PSPTO_1274, which encodes a putative cold shock protein.
DISCUSSION
In this study, we demonstrated markedly different survival of two related foliar bacterial pathogens in water-stressed environments, and these differences were correlated with gene expression profiles that suggest differences in their coping strategies and responses to osmotic stress. P. syringae strain B728a exhibits strong epiphytic competency and is thought to be better adapted to epiphytic survival than other P. syringae strains, including DC3000 (24, 40). Our side-by-side comparison of the fitness of B728a with that of DC3000 confirmed the superior fitness of B728a on nonhost plant species (Fig. 2B to D), where all or a majority of the bacteria reside in epiphytic sites (41), and on host species (Fig. 2D), where at least some of the growth occurs endophytically (41). Moreover, the greater osmotolerance of B728a than DC3000 is consistent with a contribution of osmotolerance to epiphytic survival. In a global transcriptome analysis, we found that B728a exhibited a proactive response to osmotic stress based on primarily increases in gene expression, whereas DC3000 exhibited primarily decreases in gene expression. This response of DC3000 was not due to a general reduction in transcriptional activity because only 233 of the 3,861 genes examined were detectably decreased in expression (Fig. 5).
B728a and DC3000 differ in their osmoprotectant uptake properties (Fig. 3). The higher rates of uptake of choline and glycine betaine by B728a at high osmolarities may have resulted from higher expression of genes for the glycine betaine/choline transporter OpuC in B728a than in DC3000 (see Table S2 in the supplemental material) and/or from greater osmotic activation of the BetT and OpuC transporters (13, 14). High osmoprotectant uptake rates under water-limited conditions on leaves may provide a particularly strong fitness benefit because of the presence of plant-derived choline (28) coupled with the significant energy savings of compatible solute accumulation via uptake rather than synthesis.
NAGGN synthesis contributed to the osmotolerance of B728a, as expected, but only at a much higher NaCl concentration than in a previous study (8). This difference was probably because of the added stress imposed by the presence of glycerol in the medium in that study. NAGGN synthesis also contributed to the osmotolerance of DC3000 but surprisingly did so in a medium-specific manner, with loss of NAGGN synthesis associated with a dramatic increase in DC3000 osmotolerance in ½21CS medium. NAGGN requires nitrogen from three glutamine molecules per NAGGN molecule (34); thus, the detrimental impact of NAGGN synthesis may be due to its depletion of glutamine reserves in DC3000 in ½21CS medium. This prediction is supported by the ability of exogenous glutamine to restore strong growth to DC3000 and a trehalose-deficient DC3000 mutant in this medium while not affecting the growth of NAGGN-deficient DC3000 mutants. The basis for the distinct impact of the loss of NAGGN synthesis on DC3000 growth in the two media remains unclear, however, because neither medium contains glutamine.
Changes in the transcript levels of genes involved in nitrogen metabolism suggested a mechanism by which glutamate accumulates as a compatible solute. In osmotically stressed E. coli and Salmonella enterica cells, glutamate accumulates through pathways other than direct synthesis, presumably through small changes in reactions involving glutamate, but these have not been identified (42, 43). Osmotic stress decreased the expression of the glutamine synthetase gene glnA1 in B728a and DC3000, thus enabling the accumulation of glutamate by reducing the rate at which it is converted to glutamine. Although reduced glutamine synthesis could interfere with the glutamine requirement for NAGGN, glutamate may accumulate early following an osmotic upshift, thus allowing it to function as a counterion to K+ prior to a transition to greater glutamine synthesis. Our data on the dynamics of glnA1 expression at two time points following an osmotic upshift support this model at a moderate osmotic stress but suggest a sustained repression of glnA1 at a high osmotic stress (Table 3).
We previously identified the loci for trehalose biosynthesis in DC3000 and demonstrated that trehalose synthesis contributes to osmotolerance in DC3000 (7). Here, we found that trehalose was a much larger contributor to DC3000 osmotolerance than NAGGN; that is, trehalose synthesis could fully or nearly fully compensate for the loss of NAGGN synthesis, but the reverse was not true. Unfortunately, we did not have pyramided deletion mutants to perform a similar analysis of B728a, but the reduced growth of the B728aΔggn mutant only at high osmotic stress suggests that trehalose was similarly a large contributor to B728a osmotolerance.
We were surprised to find evidence of osmoinduction of the alginate biosynthesis genes in B728a but not in DC3000 and for increased production of a uronic acid-containing polysaccharide, likely alginate, with increasing osmolarity in B728a but not in DC3000 (Fig. 7; see Fig. S7 in the supplemental material). Alginate production is stimulated by high osmolarity in many Pseudomonas spp. (44) and contributes to the epiphytic fitness of P. syringae (18). Alginate production, however, has not yet been shown to protect cells from osmotic stress. In fact, Chang et al. (16) recently provided the first clear demonstration that alginate protects bacterial cells against water stress, although the protection was effective against matric but not osmotic stress for P. putida and B728a. Their use of a yeast extract-containing medium, however, may have suppressed an osmotic response due to exogenous choline, which suppresses the induction of osmoregulated genes (28). The identity of the B728a uronic acid-containing polysaccharide as alginate was confirmed on the basis of its loss in an algD insertional mutant (see Fig. S7). This mutant was not altered in its osmotolerance when grown in ½21CS or MinAS at NaCl concentrations of up to 0.75 M (data not shown), suggesting that alginate did not contribute to B728a osmotolerance under these conditions; however, this result does not exclude the possibility of compensatory osmoadaptation mechanisms in B728a. If alginate is involved in osmoadaptation, then the lack of osmoinduction of the alginate biosynthesis genes and alginate production in DC3000 may be associated with the lower osmotolerance of this strain.
Protein translocation machinery, including the T3SS and T6SS, is used by diverse bacterial species to deliver protein effectors to plant host cells or bacterial prey cells, respectively. An unexpected finding was the osmoinduction of transcripts involved in the T6SS in B728a but not in DC3000. Hcp secretion provided evidence of functionality of the T6SS of B728a (45). The T6SS of DC3000 has not been shown to be functional and may not function given the presence of an ISPssy transposase in both the HSI-I and HSI-II gene clusters (35). On the basis of the role of T6SSs in protein transfer during antagonistic bacterial cell-cell interactions (46, 47), osmoinduction of the core T6SS machinery may be relevant to the interaction of B728a with other epiphytic organisms rather than to B728a osmoadaptation per se and may contribute to epiphytic fitness by antagonizing other microbes. Although osmoinduction may signal the presence of the cell in a phyllosphere habitat (19), the ecological benefit of osmotic stress as a T6SS-inducing signal is not clear.
The decreased abundance of T3SS transcripts during osmoadaptation in DC3000 but not in B728a is consistent with the downregulation of several T3SS-associated genes observed in P. syringae pv. phaseolicola (48) and P. aeruginosa (39) by hyperosmolarity. The lack of such downregulation in B728a may reflect the greater resilience of T3SS expression when confronted with water-limited conditions. Previous studies have shown that DC3000 expresses the T3SS mainly inside leaves (49) and that following invasion, DC3000 eventually encounters water-limited conditions in the leaf intercellular spaces, with the kinetics and extent of water limitation dependent on the nature of the DC3000-host interaction (20, 21). Osmoregulation of the T3SS may therefore reflect the integration of signals influencing the interaction of DC3000 with plants rather than a role for the T3SS in DC3000 osmoadaptation per se.
Both strains decreased transcripts encoding porins and flagellar proteins in response to osmotic stress. Although osmotic stress did not alter porin gene expression in P. aeruginosa cells (39), it decreased the levels of porin protein in P. fluorescens (50), including three proteins encoded by genes that were downregulated in B728a and DC3000, OprD, OprE, and OprQ (see Table S2 in the supplemental material). The fact that at least half of the porin-encoding genes and most of the outer membrane protein-encoding genes were not altered by an osmotic upshift in the two P. syringae strains supports a directed effect on a few porins, potentially reducing permeability to specific molecules, rather than a nonspecific effect on outer membrane proteins due to osmoinduced membrane perturbations. Similar to the porins, osmotic stress did not alter flagellar gene expression in P. aeruginosa cells (39) but reduced the expression and protein levels of the flagellar subunit FliC and reduced the motility of P. fluorescens cells (50). Whereas osmotic stress reduced many flagellar gene transcripts in P. syringae (see Table S2) and matric stress reduced at least flgC and fliE in P. putida (51), suggesting reduced motility of pseudomonads in response to water stress, osmotic stress did not affect flagellar gene transcripts in Desulfovibrio vulgaris (52), indicating that this is not a generalized bacterial response to water limitation.
The accumulation of small, glycine-rich hydrophilic proteins can enhance cellular tolerance to water limitation (38). Such proteins were first identified in plants and called late embryogenesis abundant (LEA) proteins but were later discovered in a range of prokaryotes and eukaryotes and renamed hydrophilins (38). These proteins share a highly unstructured state but little sequence similarity (38). Their intrinsic disorder is thought to be central to their abilities to physically interfere with protein aggregation (53) and provide hydrophilic surfaces that order water molecules around other proteins, thus stabilizing them (37). In contrast to LEA proteins in plants, relatively little is known of hydrophilins in bacteria. The few putative hydrophilins that have been identified include 14 from among the bacterial proteins in the Swiss Protein Database and 5 predicted from the E. coli genome (38); genes for four of the E. coli proteins were induced by osmotic stress (38). The other bacterial hydrophilins identified to date are three P. aeruginosa proteins that were identified on the basis of similarity to the E. coli hydrophilins and induction by osmotic upshock (39). Here, we identified 12 P. syringae proteins as putative hydrophilins. Genes for five of these were induced by osmotic stress in both P. syringae strains, providing good evidence that they are hydrophilins. Interestingly, only 1 of these 12 was an ortholog of the predicted hydrophilins in P. aeruginosa, and the gene encoding this hydrophilin, designated hfnA, exhibited the greatest increase in transcript abundance in response to osmotic stress of the 12 putative hydrophilin-encoding genes; furthermore, hfnA encodes an ortholog of the only hydrophilin shown thus far to have hydrophilin activity, E. coli YjbJ (37).
In summary, these results provide a detailed model of P. syringae osmoadaptation and insight into strain differences that are correlated with osmotolerance. Collectively, the results suggest that following exposure of P. syringae to a moderate osmotic upshift, P. syringae upregulates genes not only for compatible solute synthesis and osmoprotectant transport but also for hydrophilin proteins, which may provide an additional mechanism for osmoadaptation. A comparison of P. syringae strains B728a and DC3000 showed that superior osmotolerance and epiphytic fitness were associated with (i) higher osmoprotectant transport rates; (ii) stronger metabolic support for NAGGN production; (iii) global upregulation of genes, suggesting a proactive response to osmotic stress; (iv) osmotic induction of the alginate biosynthetic genes and alginate production; (v) osmotic induction of the T6SS, a system whose ecological role is not yet fully understood; and (vi) greater resiliency of the T3SS upon water limitation, a property that could ensure continued expression of this critical secretion system in the face of water limitation during leaf colonization and infection.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the National Research Initiative of the USDA-CSREES (grants 2005-35319-15300 and 2008-35600-18786).
We thank Dan Nettleton, Steve Lund, and Justin Recknor for help with the statistical analysis; Nicole O'Tool for her many hours of organizing data files; and Larry Halverson and Shanshan Li for reviewing the manuscript.
Footnotes
Published ahead of print 16 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00787-13.
REFERENCES
- 1.Cabrefiga J, Francés J, Montesinos E, Bonaterra A. 2011. Improvement of fitness and efficacy of a fire blight biocontrol agent via nutritional enhancement combined with osmoadaptation. Appl. Environ. Microbiol. 77:3174–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagen MJ, Stockwell VO, Whistler CA, Johnson KB, Loper JE. 2009. Stress tolerance and environmental fitness of Pseudomonas fluorescens A506, which has a mutation in rpoS. Phytopathology 99:679–688 [DOI] [PubMed] [Google Scholar]
- 3.Dulla G, Lindow SE. 2008. Quorum size of Pseudomonas syringae is small and dictated by water availability on the leaf surface. Proc. Natl. Acad. Sci. U. S. A. 105:3082–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schenk A, Weingart H, Ullrich MS. 2008. The alternative sigma factor AlgT, but not alginate synthesis, promotes in planta multiplication of Pseudomonas syringae pv. glycinea. Microbiology 154:413–421 [DOI] [PubMed] [Google Scholar]
- 5.Beattie GA. 2011. Water relations in the interaction of foliar bacterial pathogens with plants. Annu. Rev. Phytopathol. 49:533–555 [DOI] [PubMed] [Google Scholar]
- 6.D'Souza-Ault MR, Smith LT, Smith GM. 1993. Roles of N-acetylglutaminylglutamine amide and glycine betaine in adaptation of Pseudomonas aeruginosa to osmotic stress. Appl. Environ. Microbiol. 59:473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freeman BC, Chen C, Beattie GA. 2010. Identification of the trehalose biosynthetic loci of Pseudomonas syringae and their contribution to fitness in the phyllosphere. Environ. Microbiol. 12:1486–1497 [DOI] [PubMed] [Google Scholar]
- 8.Kurz M, Burch AY, Seip B, Lindow SE, Gross H. 2010. Genomic-driven investigation of compatible solute biosynthesis pathways of Pseudomonas syringae pv. syringae and their contribution to water stress tolerance. Appl. Environ. Microbiol. 76:5452–5462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kets EPW, Galinski EA, de Wit M, de Bont JAM, Heipieper HJ. 1996. Mannitol, a novel bacterial compatible solute in Pseudomonas putida S12. J. Bacteriol. 178:6665–6670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mikkat S, Galinski EA, Berg G, Minkwitz A, Schoor A. 2000. Salt adaptation in pseudomonads: characterization of glucosylglycerol-synthesizing isolates from brackish coastal waters and the rhizosphere. Syst. Appl. Microbiol. 23:31–40 [DOI] [PubMed] [Google Scholar]
- 11.Pocard JA, Smith LT, Smith GM, Le Rudulier D. 1994. A prominent role for glucosylglycerol in the adaptation of Pseudomonas mendocina Skb70 to osmotic stress. J. Bacteriol. 176:6877–6884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seip B, Galinski EA, Kurz M. 2011. Natural and engineered hydroxyectoine production based on the Pseudomonas stutzeri ectABCD-ask gene cluster. Appl. Environ. Microbiol. 77:1368–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Beattie GA. 2007. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-β-synthase domains are required for its osmoregulatory function. J. Bacteriol. 189:6901–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C, Beattie GA. 2008. Pseudomonas syringae BetT is a low-affinity choline transporter that is responsible for superior osmoprotection by choline over glycine betaine. J. Bacteriol. 190:2717–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monier JM, Lindow SE. 2003. Differential survival of solitary and aggregated bacterial cells promotes aggregate formation on leaf surfaces. Proc. Natl. Acad. Sci. U. S. A. 100:15977–15982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang WS, van de Mortel M, Nielsen L, de Guzman GN, Li XH, Halverson LJ. 2007. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J. Bacteriol. 189:8290–8299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beattie GA, Lindow SE. 1994. Survival, growth, and localization of epiphytic fitness mutants of Pseudomonas syringae on leaves. Appl. Environ. Microbiol. 60:3790–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Peñaloza-Vázquez A, Chakrabarty AM, Bender CL. 1999. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol. Microbiol. 33:712–720 [DOI] [PubMed] [Google Scholar]
- 19.Yu X, Lund SP, Scott RA, Greenwald JW, Records AH, Nettleton D, Lindow SE, Gross DC, Beattie GA. 2013. Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. U. S. A. 110:E425–E434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright CA, Beattie GA. 2004. Pseudomonas syringae pv. tomato cells encounter inhibitory levels of water stress during the hypersensitive response of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 101:3269–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman BC, Beattie GA. 2009. Bacterial growth restriction during host resistance to Pseudomonas syringae is associated with leaf water loss and localized cessation of vascular activity in Arabidopsis thaliana. Mol. Plant Microbe Interact. 22:857–867 [DOI] [PubMed] [Google Scholar]
- 22.Gardan L, Shafik H, Belouin S, Broch R, Grimont F, Grimont PAD. 1999. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int. J. Syst. Bacteriol. 49:469–478 [DOI] [PubMed] [Google Scholar]
- 23.Hwang MSH, Morgan RL, Sarkar SF, Wang PW, Guttman DS. 2005. Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 71:5182–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindeberg M, Myers CR, Collmer A, Schneider DJ. 2008. Roadmap to new virulence determinants in Pseudomonas syringae: insights from comparative genomics and genome organization. Mol. Plant Microbe Interact. 21:685–700 [DOI] [PubMed] [Google Scholar]
- 25.King EO, Ward MK, Raney DE. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301–307 [PubMed] [Google Scholar]
- 26.Blumenkrantz N, Asboe-Hansen G. 1973. New method for quantitative determination of uronic acids. Anal. Biochem. 54:484–489 [DOI] [PubMed] [Google Scholar]
- 27.Li S, Yu X, Beattie GA. 2013. Glycine betaine catabolism contributes to Pseudomonas syringae tolerance to hyperosmotic stress by relieving betaine-mediated suppression of compatible solute synthesis. J. Bacteriol. 195:2415–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C, Li S, McKeever DR, Beattie GA. 13 June 2013, posting date. The widespread plant-colonizing bacterial species Pseudomonas syringae detects and exploits an extracellular pool of choline in hosts. Plant J. (Epub ahead of print.) 10.1111/tpj.12262 [DOI] [PubMed] [Google Scholar]
- 29.Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Gen. Mol. Biol. 3:Article 3. [DOI] [PubMed] [Google Scholar]
- 30.Nettleton D, Hwang JTG, Caldo RA, Wise RP. 2006. Estimating the number of true null hypotheses from a histogram of P values. J. Agric. Biol. Environ. Stat. 11:337–356 [Google Scholar]
- 31.Fulton DL, Li YY, Laird MR, Horsman BGS, Roche FM, Brinkman FSL. 2006. Improving the specificity of high-throughput ortholog prediction. BMC Bioinform. 7:270. 10.1186/1471-2105-7-270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winsor GL, Lam DKW, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock REW, Brinkman FSL. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res. 39:D596–D600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 34.Sagot B, Gaysinski M, Mehiri M, Guigonis J-M, Le Rudulier D, Alloing G. 2010. Osmotically induced synthesis of the dipeptide N-acetylglutaminylglutamine amide is mediated by a new pathway conserved among bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:12652–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarris PF, Skandalis N, Kokkinidis M, Panopoulos NJ. 2010. In silico analysis reveals multiple putative type VI secretion systems and effector proteins in Pseudomonas syringae pathovars. Mol. Plant Pathol. 11:795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Records AR. 2011. The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol. Plant Microbe Interact. 24:751–757 [DOI] [PubMed] [Google Scholar]
- 37.Reyes JL, Rodrigo M-J, Colmenero-Flores JM, Gil J-V, Garay-Arroyo A, Campos F, Salamini F, Bartels D, Covarrubias AA. 2005. Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ. 28:709–718 [Google Scholar]
- 38.Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. 2000. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J. Biol. Chem. 275:5668–5674 [DOI] [PubMed] [Google Scholar]
- 39.Aspedon A, Palmer K, Whiteley M. 2006. Microarray analysis of the osmotic stress response in Pseudomonas aeruginosa. J. Bacteriol. 188:2721–2725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feil H, Feil WS, Chain P, Larimer F, DiBartolo G, Copeland A, Lykidis A, Trong S, Nolan M, Goltsman E, Thiel J, Malfatti S, Loper JE, Lapidus A, Detter JC, Land M, Richardson PM, Kyrpides NC, Ivanova N, Lindow SE. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 102:11064–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabaratnam S, Beattie GA. 2003. Differences between Pseudomonas syringae pv. syringae B728a and Pantoea agglomerans BRT98 in epiphytic and endophytic colonization of leaves. Appl. Environ. Microbiol. 69:1220–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLaggan D, Naprstek J, Buurman ET, Epstein W. 1994. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J. Biol. Chem. 269:1911–1917 [PubMed] [Google Scholar]
- 43.Botsford JL, Alvarez M, Hernandez R, Nichols R. 1994. Accumulation of glutamate by Salmonella typhimurium in response to osmotic stress. Appl. Environ. Microbiol. 60:2568–2574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh S, Koehler B, Fett WF. 1992. Effect of osmolarity and dehydration on alginate production by fluorescent pseudomonads. Curr. Microbiol. 25:335–339 [Google Scholar]
- 45.Records AR, Gross DC. 2010. Sensor kinases RetS and LadS regulate Pseudomonas syringae type VI secretion and virulence factors. J. Bacteriol. 192:3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Basler M, Ho BT, Mekalanos JJ. 2013. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152:884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carruthers MD, Nicholson PA, Tracy EN, Munson RS., Jr 2013. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 8:e59388. 10.1371/journal.pone.0059388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahme LG, Mindrinos MN, Panopoulos NJ. 1992. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J. Bacteriol. 174:3499–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boureau T, Routtu J, Roine E, Taira S, Romantschuk M. 2002. Localization of hrpA-induced Pseudomonas syringae pv. tomato DC3000 in infected tomato leaves. Mol. Plant Pathol. 3:451–460 [DOI] [PubMed] [Google Scholar]
- 50.Guyard-Nicodème M, Bazire A, Hémery G, Meylheuc T, Mollé D, Orange N, Fito-Boncompte L, Feuilloley M, Haras D, Dufour A, Chevalier S. 2008. Outer membrane modifications of Pseudomonas fluorescens MF37 in response to hyperosmolarity. J. Proteome Res. 7:1218–1225 [DOI] [PubMed] [Google Scholar]
- 51.van de Mortel M, Halverson LJ. 2004. Cell envelope components contributing to biofilm growth and survival of Pseudomonas putida in low-water-content habitats. Mol. Microbiol. 52:735–750 [DOI] [PubMed] [Google Scholar]
- 52.He Z, Zhou A, Baidoo E, He Q, Joachimiak MP, Benke P, Phan R, Mukhopadhyay A, Hemme CL, Huang K, Alm EJ, Fields MW, Wall J, Stahl D, Hazen TC, Keasling JD, Arkin AP, Zhou J. 2010. Global transcriptional, physiological, and metabolite analyses of the responses of Desulfovibrio vulgaris Hildenborough to salt adaptation. Appl. Environ. Microbiol. 76:1574–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakrabortee S, Tripathi R, Watson M, Schierle GSK, Kurniawan DP, Kaminski CF, Wise MJ, Tunnacliffe A. 2012. Intrinsically disordered proteins as molecular shields. Mol. Biosyst. 8:210–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore RA, Starratt AN, Ma SW, Morris VL, Cuppels DA. 1989. Identification of a chromosomal region required for biosynthesis of the phytotoxin coronatine by Pseudomonas syringae pv. tomato. Can. J. Microbiol. 35:910–917 [Google Scholar]
- 55.Loper JE, Lindow SE. 1987. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology 77:1449–1454 [Google Scholar]
- 56.Mansfield J, Jenner C, Hockenhull R, Bennett MA, Stewart R. 1994. Characterization of avrPphE, a gene for cultivar-specific avirulence from Pseudomonas syringae pv. phaseolicola which is physically linnked to hrpY, a new hrp gene identified in the halo-blight bacterium. Mol. Plant Microbe Interact. 7:726–739 [DOI] [PubMed] [Google Scholar]
- 57.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FSL, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Foger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK-S, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock REW, Lory S, Olson MV. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 58.Chen C, Malek AA, Wargo MJ, Hogan DA, Beattie GA. 2010. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 75:29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu L, McGrane RS, Beattie GA. 2013. Light regulation of swarming motility in Pseudomonas syringae integrates signaling pathways mediated by a bacteriophytochrome and a LOV protein. MBio 4:e00334–00313. 10.1128/mBio.00334-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607 In Walker JM. (ed), The proteomics protocols handbook. Humana Press Inc, Totowa, NJ [Google Scholar]
- 62.Peterson KP. 2009. Evaluation of the expression of water stress-responsive Pseudomonas syringae genes during plant infection and in the presence of low osmotic versus low matric potential in culture. M.S. thesis Iowa State University, Ames [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.