Abstract
Reactive nitrogen species (RNS), in particular nitric oxide (NO), are toxic to bacteria, and bacteria have mechanisms to allow growth despite this stress. Understanding how bacteria interact with NO is essential to understanding bacterial physiology in many habitats, including pathogenesis; however, many targets of NO and enzymes involved in NO resistance remain uncharacterized. We performed for the first time a metabolomic screen on NO-treated and -untreated bacteria to define broadly the effects of NO on bacterial physiology, as well as to identify the function of NnrS, a previously uncharacterized enzyme involved in defense against NO. We found many known and novel targets of NO. We also found that iron-sulfur cluster enzymes were preferentially inhibited in a strain lacking NnrS due to the formation of iron-NO complexes. We then demonstrated that NnrS is particularly important for resistance to nitrosative stress under anaerobic conditions. Our data thus reveal the breadth of the toxic effects of NO on metabolism and identify the function of an important enzyme in alleviating this stress.
INTRODUCTION
Vibrio cholerae causes cholera, a severe watery diarrhea responsible for millions of cases and thousands of deaths each year (Centers for Disease Control and Prevention [http://www.cdc.gov]). It is not, however, a member of the Enterobacteriaceae—its natural habitat is aquatic. It is thought that during most of its life cycle, when not infecting humans, V. cholerae resides in association with zooplankton, forming biofilms on the chitinous surfaces of crustaceans (1, 2). Thus, V. cholerae must display metabolic flexibility in order to thrive in these two different environments and respond to the different metabolic challenges therein.
A commonly encountered metabolic stress for pathogenic and nonpathogenic bacteria is the presence of reactive nitrogen species (RNS), in particular the well-studied molecule nitric oxide (NO). NO is formed as a by-product of nitrogen metabolism for many bacteria as an intermediate in denitrification (3), as well as from dedicated nitric oxide synthases (NOSs) in both bacteria and eukaryotes (4, 5), and is present in micromolar concentrations in some bacterial biofilms (6). NO can also be formed by chemical decomposition of nitrite in acid environments, such as the human stomach (7). NO is also a prominent component of the mammalian innate immune system, part of a battery of reactive oxygen species (ROS) and RNS produced by phagocytes when they encounter bacteria (7).
The mechanisms whereby NO inhibits bacterial growth are diverse, but one of the most important properties of NO is its ability to bind iron and form dinitrosyl iron complexes (DNICs) bound to iron-sulfur cluster proteins, inhibiting their function (8, 9). DNICs have also been shown to mediate the formation of nitrosothiols, another form of nitrosative stress that inhibits thiol-containing proteins (10). Through this mechanism and others, NO has been shown to inhibit a few enzymes in vitro, including such central metabolic enzymes as aconitase (11), dihydroxyacid dehydratase (12), alpha-ketoglutarate dehydrogenase (13), fructose-1,6-bisphosphate aldolase (14), argininosuccinate synthase (15), and components of the respiratory chain (16, 17). However, these enzymes have largely been studied in isolation, and there has not been any comprehensive study on the effects of NO on bacterial metabolism.
Bacteria possess several strategies for coping with nitrosative stress. The most obvious strategy is to directly remove the NO, and there are several enzymes known to convert NO into less toxic nitrogen oxides, such as nitrate (NO3−) or nitrous oxide (N2O). Another strategy is to alter carbon flux to bypass blockades and maintain redox homeostasis, a method used by Staphylococcus aureus by upregulating lactate dehydrogenase (18). The genes required for these responses are usually under the control of an NO-responsive transcription factor (19). One gene that is conserved throughout many Gram-negative bacteria, including such pathogens as Pseudomonas, Neisseria, and Brucella, and is also usually found under the control of one of these transcriptional regulators is nnrS. NnrS was initially described in Rhodobacter as a heme- and copper-containing transmembrane protein (20). Although the function of NnrS is unknown, we have previously shown that it contributes to nitrosative-stress tolerance in V. cholerae (21).
In this study, we performed a metabolomic screen with two goals: to identify more fully the effects of NO on bacterial metabolism by surveying the relative concentrations of metabolites from many different pathways in V. cholerae and to use these data to determine the function of NnrS. We found drastic changes in metabolic pathways in response to NO, suggesting that nitrosative stress forces bacteria to adapt dramatically. We also found that NnrS does not directly remove NO but instead protects the cellular iron pool from the formation of DNICs, thus protecting critical metabolic pathways from inhibition.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All strains of V. cholerae in this study were derived from O1 El Tor strain C6706. In-frame deletion strains were generated by sucrose counterselection, as described previously (22). The minimal media used contained 79 mM KH2PO4, 15 mM (NH4)2SO4, 0.65 mM MgSO4, 0.07 mM CaCl2, 0.018 mM FeSO4, and 0.013 mM MnSO4, with carbon sources added as indicated. For growth curves involving 2,2′-dipyridyl, FeSO4 was omitted and 0.2 mM 2,2′-dipyridyl was added. Yeast extract was added when indicated at a concentration of 0.5% (wt/vol). The plasmid used to complement the nnrS deletion was derived from pMal-c2x (New England BioLabs), in which the malE gene was replaced at the NdeI and SalI sites with nnrS tagged with six histidine codons at its 3′ end. Expression was induced by adding 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) to the growth media.
Growth curves.
To monitor growth continuously, overnight saturated cultures were washed in phosphate-buffered saline (PBS), and 2 μl of washed culture was inoculated into 200 μl of the relevant growth medium in a 96-well plate in triplicate. (Z)-1-[N-(2-aminoethyl)-N-(2-ammonioethyl)amino]diazen-1-ium-1,2-diolate (DETA-NONOate) (Cayman Chemical) was included at a range of concentrations. The plate was covered with a transparent film, and growth was monitored every 4 min, using the absorbance at 600 nm after shaking for 2 min at each time point, with an automated plate reader (BioTek Synergy HT). Anaerobic growth curves were performed by inoculating 30 μl of washed, saturated culture into 3 ml minimal medium in individual test tubes and then placing the cultures in an anaerobic chamber (Coy Laboratories) equipped with a 37°C standing incubator, periodically withdrawing 200 μl, and measuring the absorbance at 600 nm. For aerobic growth curves, 10 mM glucose was used as a sole carbon source, and for anaerobic growth, 25 mM glucose was used, unless otherwise indicated.
Measurement of NO consumption.
Strains were inoculated into 120-ml serum flasks with 50 ml minimal medium containing 0.25% (wt/vol) glucose, Teflon magnetic bars, and crimp-sealed butyl septa. Prior to inoculation, the headspace atmosphere was replaced with helium by evacuation and refilling six times and then supplied with pure oxygen to reach 15 ml liter−1 and pure NO to 300 to 350 ppm (equivalent to ∼493 to 575 nM in the liquid). The flasks were then placed in a 37°C water bath. The initial pressure was adjusted to 1 atmosphere by releasing the overpressure through a 0.5-mm (inside diameter [ID]) cannula. The flasks were inoculated with 1 ml culture containing ∼3 × 108 cells and stirred while monitoring the oxygen and NO concentrations in the headspaces, which were then used to calculate the concentrations in the liquid. This incubation and measurement system has been described in detail previously (23).
Metabolomic study.
Overnight cultures of the three strains were inoculated at a ratio of 1:1,000 into 440 ml of LB in centrifuge bottles filled to the top and closed tightly. Twenty micromolar DETA-NONOate was added to the samples or 20 μM diethylamine triamine (DETA) to the control samples. All samples were incubated for 7 h at 37°C, after which 200 ml was discarded and the remainder was centrifuged at 6,000 rpm in a Sorvall SLA-3000 rotor for 10 min. The pellets were resuspended in 1 ml of PBS and then centrifuged again in a Nalgene cryovial for 5 min at 13,000 rpm in a tabletop centrifuge. The pellets were then flash-frozen in an ethanol-dry ice bath and stored at −80°C. This experiment was carried out five times on separate days and then analyzed in conjunction with Metabolon, Inc. (Durham, NC, USA). The extraction protocol, instrument settings, data processing, and quality control have been described in detail previously (24, 25). In brief, samples were extracted and analyzed by ultra-high-performance liquid chromatography–tandem mass spectrometry (UHPLC–MS-MS) and gas chromatography-mass spectrometry (GC-MS). Metabolites present in samples were identified by matching chromatographic and mass spectral data to an in-house library of chemical standards, and the relative abundances of metabolites were determined by area-under-the-peak analysis. Data were normalized to the protein concentration of the sample and further normalized so that the median concentration of each metabolite across all samples was 1.
Measurement of aconitase activity.
One milliliter of saturated overnight cultures was inoculated into 200-ml volumes of LB in 500-ml flasks with shaking at 37°C in the presence or absence of 100 μM DETA-NONOate, which had been freshly dissolved in 10 mM NaOH. After 3 h of growth, the bacteria were centrifuged and resuspended in 0.5 to 1 ml 50 mM Tris-HCl, pH 7.4, 0.6 mM MnCl2. Three hundred microliters of resuspended cells was lysed quickly by sonication at 400 W and spun in a tabletop centrifuge to remove cell debris. Thirty microliters of supernatant was immediately aliquoted in triplicate to a 48-well plate. Using a multichannel pipette, the aconitase reaction was started by adding 1 ml of reaction mixture (50 mM Tris-HCl, pH 7.4, 0.6 mM MnCl2, 0.2 mM NADP+, 1 U/ml porcine heart isocitrate dehydrogenase, 5 mM trisodium citrate) to the extracts. The citrate was added to the reaction mixture immediately before initiation. The activity was then calculated by monitoring the rate of formation of NADPH for approximately 10 min every 15 s in an automated plate reader (BioTek Synergy HT), using an extinction coefficient at 340 nm of 6,220 M−1 cm−1.
Measurement of ferrous iron.
One milliliter of saturated overnight cultures was inoculated into 200-ml volumes of LB broth in 500-ml flasks with shaking at 37°C in the presence or absence of 100 μM DETA-NONOate, which had been freshly dissolved in 10 mM NaOH. The cultures were centrifuged, washed twice, and resuspended in 50 mM Tris-HCl, pH 7.4. A 300-μl sample was sonicated briefly at 400 W. To avoid oxidation by oxygen, 100 μl was added within seconds to 10 μl of FerroZine reagent (3-[2-pyridyl]-5,6-diphenyl-1,2,4-triazine-p,p′-disulfonic acid disodium salt; 10 mM dissolved in 100 mM ammonium acetate). The samples were then centrifuged, and the absorbance at 562 nm was recorded, compared to a freshly prepared ferrous ammonium sulfate standard, and normalized to the protein concentration.
RESULTS
NnrS is important for resistance to NO but does not remove NO.
We previously reported that a strain of V. cholerae lacking the flavohemoglobin HmpA (ΔhmpA), which removes NO by conversion to nitrate, or lacking the transcriptional regulator NorR was hypersusceptible to growth inhibition by NO and was defective in colonizing the mouse gastrointestinal tract (21). Although a strain lacking only NnrS (ΔnnrS) displayed NO resistance comparable to that of the wild type, the deletion of nnrS in a ΔhmpA background (ΔhmpA ΔnnrS) resulted in severe sensitivity to NO compared to ΔhmpA (21). Thus, although HmpA is likely the dominant NO resistance protein of V. cholerae, NnrS plays an auxiliary role and may be important in environments in which HmpA is nonfunctional (such as strictly anaerobic conditions, as discussed below). To expand on our previous findings and begin to search for the function of NnrS, we performed growth curves over a range of concentrations of the NO donor DETA-NONOate and found that the ΔhmpA ΔnnrS strain was approximately 1 log unit more sensitive than the ΔhmpA strain (Fig. 1A). This phenotype could be complemented by expressing NnrS from a plasmid (Fig. 1A, ΔhmpA ΔnnrS/pNnrS).
Fig 1.
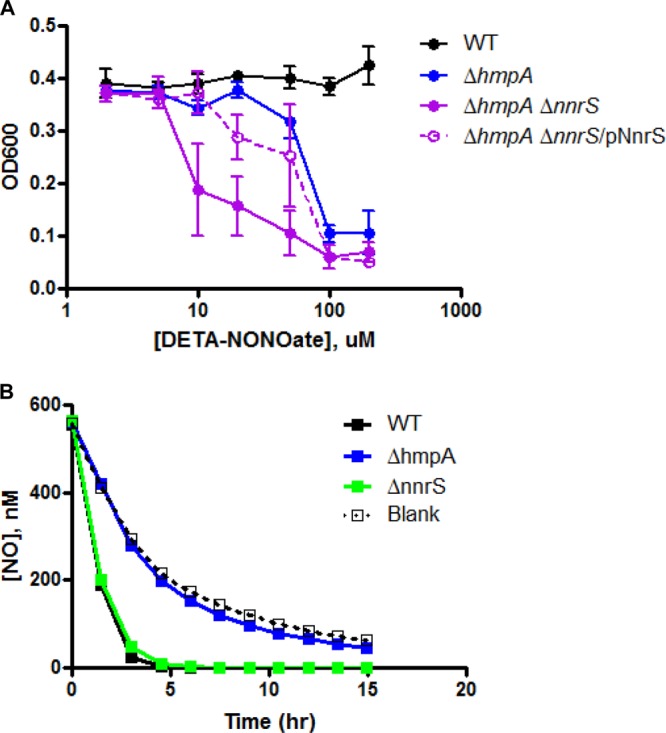
NnrS protects the cell from NO but does not remove it. (A) Strains of V. cholerae were inoculated into minimal medium with 10 mM glucose as the sole carbon source in the presence of increasing concentrations of the NO donor DETA-NONOate, and the optical density at 600 nm (OD600) was recorded over time to generate growth curves for each strain at each concentration. The graph shows the OD600 of each strain 6 h after inoculation, at the approximate time when the WT strain reached stationary phase. The error bars represent standard deviations. The data are representative of three independent experiments. (B) Strains of V. cholerae were inoculated into minimal medium, and the consumption of NO, which was added as a bolus of authentic NO gas into the headspace, was measured. “Blank” refers to a flask of medium without bacteria. Shown is an experiment done in single replicates, representative of at least three identical experiments. The concentration of NO represented on the graph is the concentration calculated in the liquid based on measurement of the concentration in the headspace (23, 26).
We next determined whether NnrS might remove NO directly. However, we were unable to detect any difference in the rate of NO consumption between the wild-type and ΔnnrS strains (Fig. 1B). In addition, we were unable to detect any metabolism of NO in the ΔhmpA strain above background auto-oxidation (26). These data suggest that HmpA is responsible for the removal of NO in V. cholerae and that NnrS protects against NO through a different mechanism.
A metabolomic study to identify pathways inhibited by NO.
Although several enzymes are known to be inhibited by NO, to date, no comprehensive study of central metabolism has been conducted to determine the breadth of its effects. By identifying molecules that increase or decrease in concentration in the cell, we reasoned that such a study could identify new enzymes inhibited by NO through the accumulation of intermediates upstream (and decrease downstream) of an NO-inhibited enzyme.
We therefore grew V. cholerae in the presence of DETA-NONOate or the control compound DETA and subjected bacterial pellets to analysis by mass spectrometry to identify the relative contents of a broad array of metabolites. We employed three strains: the wild type (WT) and ΔhmpA and ΔhmpA ΔnnrS strains. Intermediates from glycolysis, the tricarboxylic acid (TCA) cycle, amino acid synthesis, nucleotide synthesis, lipid metabolism, and various other pathways were quantified. The complete data set is available in the supplemental material. A large variety of these metabolites differed significantly between the NO-treated and untreated samples (Fig. 2A). Several enzymes previously shown to be inhibited by NO could be identified by the buildup of their substrates or upstream intermediates. For instance, fructose-1,6-bisphosphate aldolase is a zinc-dependent glycolytic enzyme demonstrated to be NO sensitive in Borrelia burgdorferi (14). In our study, upstream glycolytic intermediates, such as glucose, glucose-6-phosphate, and fructose-1,6-bisphosphate, all accumulated in the presence of NO, whereas downstream metabolites, such as 2-phosphoglycerate and 3-phosphoglycerate, decreased (see Fig. S1 in the supplemental material), thus confirming that this enzyme is likely inhibited in V. cholerae, too. Argininosuccinate synthetase, which converts citrulline to argininosuccinate in order to produce arginine, has been shown to be inhibited by NO in mitochondria (15). The 15-fold accumulation of citrulline in the presence of 20 μM DETA-NONOate in our study (see Fig. S2 in the supplemental material) suggests that the enzyme may be inhibited in bacteria, as well.
Fig 2.
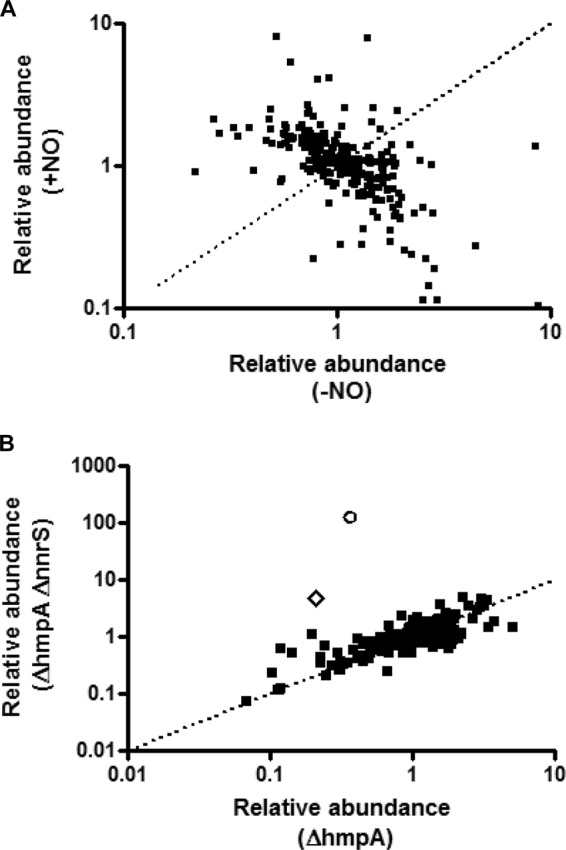
Scatter plots of metabolomic analyses of the effects of NO on V. cholerae. The relative concentrations of hundreds of metabolites were recorded for wild-type, ΔhmpA, and ΔhmpA ΔnnrS strains of V. cholerae. (A) Scatter plot of the relative abundances of metabolites in wild-type V. cholerae in the absence (x axis) and presence (y axis) of the NO donor DETA-NONOate. The area above the dotted line indicates relative accumulation in the presence of NO; the area below the dotted line indicates relative accumulation in the absence of NO. (B) Scatter plot of the relative abundances of metabolites in the presence of NO for the ΔhmpA strain (x axis) compared to the ΔhmpA ΔnnrS strain (y axis). The area above the dotted line indicates accumulation in the ΔhmpA ΔnnrS strain; the area below the dotted line indicates accumulation in the ΔhmpA strain. Circle, citrate; diamond, cis-aconitate.
Comparative metabolomics reveals a role for NnrS.
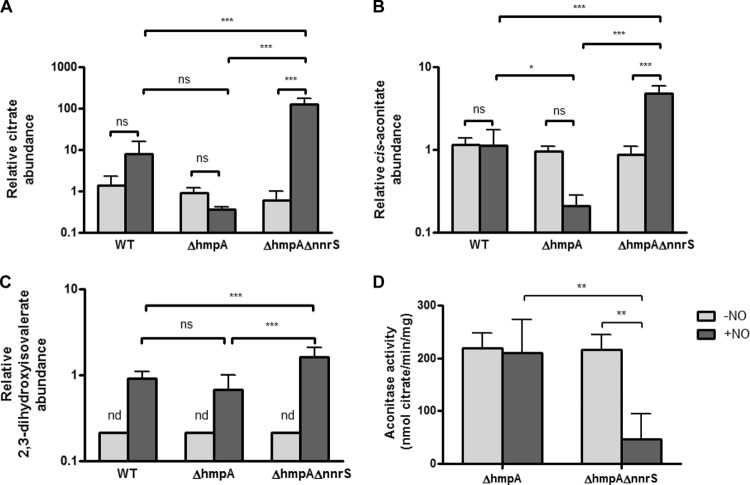
Knowing that NnrS is important for resistance to NO but that it does not remove NO, we hypothesized that there might be specific metabolic pathways protected by NnrS from nitrosative stress. Thus, we compared the results of the metabolomic study between the ΔhmpA and ΔhmpA ΔnnrS strains (Fig. 2B). We found that in the presence of NO, the ΔhmpA ΔnnrS strain accumulated more than 200-fold more citrate and 23-fold more cis-aconitate than the ΔhmpA strain (Fig. 2B and 3A and B). In addition, 2,3-dihydroxyisovalerate accumulated more in the ΔhmpA ΔnnrS strain than in the ΔhmpA strain, though only by 2.4-fold (Fig. 3C). Citrate and cis-aconitate are substrates of aconitase, and 2,3-dihydroxyisovalerate is a substrate of dihydroxyacid dehydratase, both enzymes of the dehydratase family known to be sensitive to NO (11, 12). The dehydratase family of enzymes is a unique family in which the iron-sulfur cluster reacts directly with its substrate. Dehydratases are thus exquisitely sensitive to NO due to the solvent-exposed nature of their iron-sulfur clusters: NO binds and forms dinitrosyl iron complexes at these sites, inactivating the enzyme (8). On the other hand, substrates of nondehydratase enzymes, such as citrulline and 1,6-fructose bisphosphate, that accumulated in all three strains did not accumulate any further in the absence of nnrS (see Fig. S1 and S2 in the supplemental material). This suggested to us that NnrS, although not removing NO directly, might serve some role specifically in protecting dehydratases or other iron-sulfur cluster-containing proteins from inhibition by NO. To test this hypothesis, we measured the aconitase activity in cell extracts of the ΔhmpA and ΔhmpA ΔnnrS strains (Fig. 3D). We found that in the absence of NO, the two strains had similar activities, but upon the addition of NO, aconitase activity in the ΔhmpA ΔnnrS strain dropped to approximately 25% of that of the ΔhmpA strain. This suggests that a decrease in the activity of dehydratases such as aconitase due to NO is prevented by NnrS.
Fig 3.
NnrS protects dehydratases from NO. (A to C) Relative concentrations of citrate (A), cis-aconitate (B), and 2,3-dihydroxyisovalerate (C) in the three strains from the metabolomic study in the presence (+NO) or absence (−NO) of DETA-NONOate. (D) Aconitase activity in cell extracts of V. cholerae grown in the presence or absence of DETA-NONOate. The error bars represent the standard deviations of the mean of three independent experiments. Statistical significance was determined by two-way analysis of variance (ANOVA) with Bonferroni's posttest. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant; nd, not detectable.
NnrS protects the cellular iron pool from NO.
The inhibition of dehydratases by DNIC formation occurs through the reaction of NO with the “chelatable-iron pool” (CIP), which is not a chemically defined mixture but is thought to be the cellular iron that is loosely coordinated and can thus be bound by chelators (9). It is thought that the chelatable-iron pool is composed of both free and protein-bound iron (8, 9). Other groups have shown that chelation of iron with 2,2′-dipyridyl prevents the formation of DNICs (8). Thus, we hypothesized that chelation of iron might complement the NO-dependent toxic effect of the deletion of nnrS by depleting the free iron available to react with NO. When we added both yeast extract and 2,2′-dipyridyl to minimal medium with no added iron, severely restricting cellular iron, there was no growth defect in the ΔhmpA ΔnnrS strain compared to the ΔhmpA strain (Fig. 4A). The addition of yeast extract alone only partially complemented the defect (compare Fig. 4A to 1A). This result is to be expected, since many of the pathways dependent on iron-sulfur clusters are biosynthetic, and thus, their inhibition might be overcome by supplementation with yeast extract. To further test the hypothesis that NnrS protects against iron-NO complex formation, we measured the chelatable ferrous iron content of cells treated with NO. It has been demonstrated that addition of NO to cellular systems depletes chelatable iron by causing it to form macromolecule-bound DNICs (9). In addition, the decomposition of DNICs by oxygen and l-cysteine causes the release of ferrous iron (27). Thus, a cell with an increased number of DNICs, or one defective in decomposing them, would have a lower ferrous iron concentration detectable by reagents such as FerroZine. Indeed, we found that the ΔhmpA ΔnnrS strain had a lower chelatable ferrous iron content than the ΔhmpA strain (Fig. 4B), again supporting the hypothesis that NnrS prevents the formation of iron-NO complexes.
Fig 4.
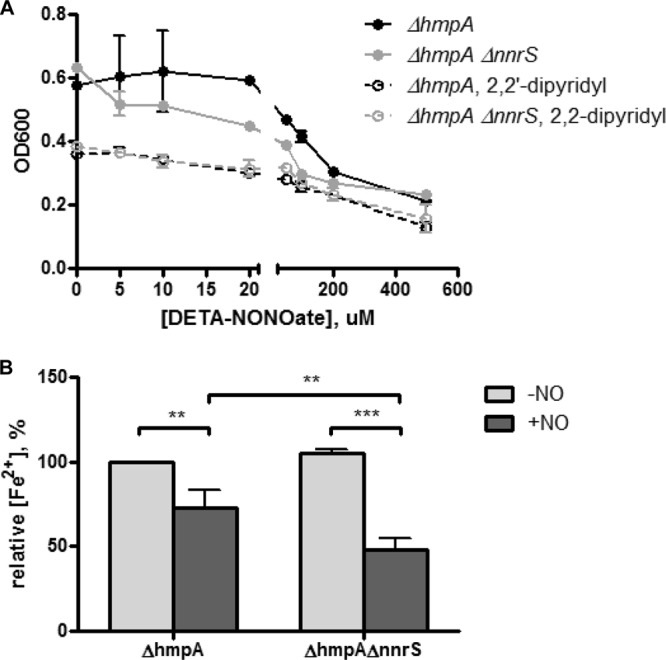
NnrS protects the cellular iron pool from NO. (A) V. cholerae ΔhmpA and ΔhmpA ΔnnrS strains were inoculated into minimal medium with glucose and yeast extract in the presence of increasing concentrations of DETA-NONOate with (dashed lines) or without (solid lines) the iron chelator 2,2′-dipyridyl. Shown is the OD600 at 10 h of growth, representative of two independent experiments. (B) Relative ferrous iron contents of ΔhmpA and ΔhmpA ΔnnrS strains in the presence (+NO) or absence (−NO) of DETA-NONOate. The error bars represent the standard deviations of the mean of three independent experiments. Statistical significance was determined by two-way ANOVA with Bonferroni's posttest to compare individual groups. **, P < 0.01; ***, P < 0.001.
NnrS is important during anaerobic nitrosative stress.
To this point, all the effects of deleting nnrS were examined only in the genetic background lacking hmpA. To determine the physiological relevance of NnrS, we sought to find a condition under which the single deletion of nnrS might have effects on resistance to NO. Previous work had shown that the primary mechanism of action of flavohemoglobins, such as HmpA, is through dinitrosylation of NO, a reaction that is dependent on O2 (28). Hmp of Escherichia coli also possesses an O2-independent NO reductase activity in vitro (29), but the activity is slow, and its physiological relevance is uncertain (30). Thus, we hypothesized that under anaerobic conditions, the effect of HmpA in V. cholerae might be less dominant and NnrS might become more important.
We found that in a strictly anoxic environment, even wild-type V. cholerae was highly sensitive to NO: growth was inhibited at micromolar concentrations of DETA-NONOate under anaerobic conditions (Fig. 5), whereas millimolar concentrations had no effect in the presence of oxygen (Fig. 1A). This heightened sensitivity is probably due to multiple factors, including the absence of nonenzymatic clearance of NO by O2, but may in part be explained by the oxygen dependence of HmpA. Interestingly, the ΔnnrS strain was more sensitive than the wild type under anoxic conditions (Fig. 5). We observed this phenotype during fermentation (Fig. 5) and during anaerobic respiration on fumarate (see Fig. S5 in the supplemental material). This suggests that NnrS may play an important role in anaerobic environments.
Fig 5.
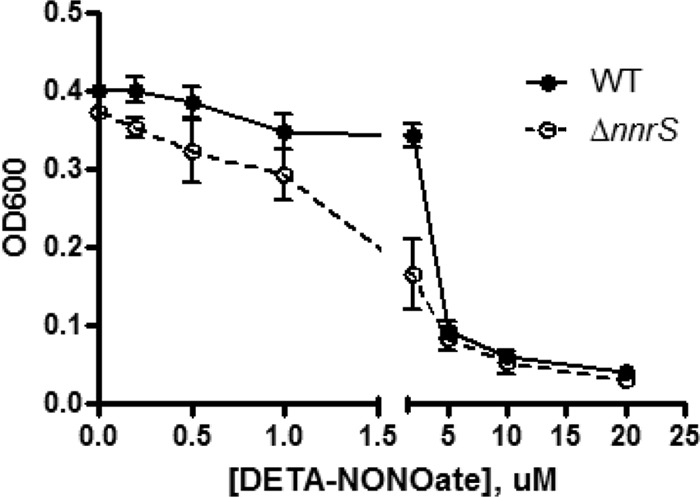
NnrS is important during nitrosative stress under anaerobic conditions. Shown is the growth of the wild-type and ΔnnrS strains in the presence of increasing concentrations of DETA-NONOate in an anaerobic environment in minimal medium with 25 mM glucose as the sole carbon source. The graph shows the OD600 at 12 h of growth. The error bars represents standard deviations. The data are representative of three independent experiments.
DISCUSSION
Nitrosative stress, derived from reactive nitrogen species such as NO, is a ubiquitous challenge for bacteria. During infection, pathogenic bacteria encounter high concentrations of NO released by phagocytes (31). NO is also formed inorganically when nitrite from the mouth reaches the low pH of the stomach (7). NO can also be generated by other bacteria through denitrification or by nitric oxide synthase (5, 32). Furthermore, NO has been found to reach high concentrations in polymicrobial biofilms (6). In other words, bacteria are constantly encountering NO and must adapt metabolically. To date, there had been no study detailing the scope of these metabolic effects, so we performed a metabolomic study on NO-treated and untreated cells. We found a wide breadth of effects, most pronounced in central carbon metabolism: an accumulation of upstream glycolytic intermediates pointed to a block in glycolysis at the fructose-1,6-bisphosphate aldolase step, and an accumulation of citrate indicated a block in the TCA cycle. In addition, high citrulline concentrations implied a defect in arginine synthesis, all validating studies in various other prokaryotic and eukaryotic systems that identified these pathways as targets of NO. We also found that the concentrations of the polyamines 1,3-aminodipropane (DAP) and spermidine were increased 9- and 3-fold, respectively (see Fig. S3 in the supplemental material). In uropathogenic E. coli, nitrosative stress has been shown to increase polyamine production, which was linked to RNS resistance (33), suggesting that such a mechanism might exist in V. cholerae, too. Polyamines have also been linked to biofilm production in V. cholerae (34). Furthermore, NO sensing has been shown to influence biofilm formation in other bacterial species through H-NOX domain proteins and cyclic-di-GMP production (35–37). The increase in polyamines thus suggests an additional possible link between NO and biofilms, which we are currently investigating.
This study also identified some metabolic pathways that may be affected by NO but have not been reported to be. We observed an accumulation of cysteine and glycine, as well as a decrease in both oxidized and reduced glutathione concentrations (see Fig. S4 in the supplemental material). Taken together, this may indicate a block in glutathione synthesis that occurs from the ligation of cysteine, glutamate, and glycine. Glutathione is a critical molecule in maintaining the proper functional redox state of many intracellular enzymes by regenerating the active form of thiol-dependent active sites. Glutathione is formed by two enzymes, gamma-glutamylcysteine synthetase and glutathione synthetase; we are currently investigating the inhibition of these enzymes by NO. We did not observe an increase in glutamate, as one might expect (in fact, there was a slight decrease). However, glutamate is a critical branch point for many pathways in central carbon metabolism, so the interruption of glutathione synthesis may not necessarily result in a detectable accumulation of glutamate.
All these data suggest that the effects of NO on bacterial physiology are quite broad. It is no wonder, then, that bacteria have evolved multiple mechanisms to cope with this stress. One obvious strategy is to simply remove the NO itself directly. There are multiple enzymes known to perform this task, including nitric oxide reductase (NOR), flavorubredoxin (NorV), and flavohemoglobin (Hmp), as well as the hybrid cluster protein (Hcp) thought to remove the related compound hydroxylamine (38). These compounds are nearly always under the control of an NO-responsive transcriptional regulator, such as NnrR, NsrR, NorR, or HcpR (19), all of which bind NO and alter gene expression. In many gammaproteobacteria, however, there is another gene, nnrS, that is also under the control of one of these regulators but whose function was previously unknown (20). Unlike most of the other factors under the control of these regulators, NnrS does not appear to remove NO directly. Instead, we found that it relieves a major stress caused by NO: formation of iron-NO complexes. Mutants lacking nnrS were significantly inhibited for growth in the presence of NO, mainly due to the sequestration of the cellular iron pool by NO. One of the most toxic effects of NO is the formation of protein-bound DNICs, which are directly inhibitory to iron-sulfur cluster proteins (9). We found that NnrS protects against this effect, allowing critical enzymes, such as aconitase, to function in the presence of NO.
We noticed similarities between our findings regarding NnrS and another protein involved in NO tolerance, YtfE. In E. coli, ytfE is under the control of the regulator NsrR and has been shown to protect iron-sulfur cluster-containing proteins, such as aconitase and fumarase, from damage due to NO or hydrogen peroxide (39–41). YtfE is a member of a putative family of nonheme di-iron proteins that includes ScdA from S. aureus and DnrN from Neisseria gonorrhoeae (41). Interestingly, we noticed that this family of proteins (Pfam family PF04405 [http://pfam.sanger.ac.uk]) is distributed primarily among the order Enterobacteriales and is absent from the Vibrionales, whereas NnrS (Pfam family PF05940) is absent from the Enterobacteriales but is found widely within the Vibrionales. Although both are present within the Alteromonadales, particularly within the genus Shewanella, the phylogenetic distribution suggests there may be some convergent evolution between the two different proteins fulfilling similar functions. On the other hand, parallels between NnrS and YtfE are not perfect. The growth defect in the ytfE deletion mutant of E. coli was found to worsen in the presence of 2,2′-dipyridyl (39), whereas it improved growth of the nnrS deletion strain (Fig. 4A). Further work on both NnrS and YtfE will hopefully shed light on how these proteins protect iron-sulfur clusters from NO.
V. cholerae is an aquatic organism and frequently lives on the molts of microscopic crustaceans (1), where the carbon sources are likely more limited. We have previously shown (21) that NnrS probably does not play a significant role during growth in the mammalian intestine, where carbon sources are likely more diverse than on a crustacean molt, which is made primarily of chitin, a polymer of the amino sugar N-acetylglucosamine. In fact, V. cholerae can use chitin as its sole carbon source (42, 43), a situation resembling minimal medium. Thus, we suspect that in “minimal-medium-like” environments, such as chitinous surfaces, NnrS might play a more prominent role in resistance to NO, as demonstrated by the more pronounced growth defect in minimal medium (Fig. 1A) than in rich medium (Fig. 4A). Interestingly, there is one bacterial species, Saccharophagus degradans, that has been reported to possess nnrS as its only gene under the control of a dedicated NO-responsive transcription factor (19). This species of bacteria is found in a habitat in which its only carbon source is agar, which is another sugar polymer (44). Thus, the phylogenetic distribution of NnrS, as well as the data in this study, support the conclusion that NnrS is important in resisting nitrosative stress, particularly in environments with low carbon diversity, abundant iron, or low oxygen, in order to protect the cell against inhibition of iron-containing proteins by NO.
In summary, this work employed metabolomics for the first time to identify new targets of NO, a common source of metabolic stress for bacteria. We also found that one of the most important targets of NO, the cellular iron pool and iron-sulfur cluster enzymes, is protected from damage by NnrS, an NO-regulated protein with a previously unknown function.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mark Goulian and Fevzi Daldal (University of Pennsylvania) and Aaron Landry (Louisiana State University) for their thoughtful suggestions.
This study was supported by NIH grant R01AI080654 to J.Z.
A.M.S. designed, performed, and interpreted experiments and wrote the manuscript. B.L. designed, performed, and interpreted experiments and revised the manuscript. L.R.B., J.P.S., and J.Z. designed and interpreted experiments and revised the manuscript.
We declare no competing financial interests.
Footnotes
Published ahead of print 9 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00836-13.
REFERENCES
- 1.Huq A, Small EB, West PA, Huq MI, Rahman R, Colwell RR. 1983. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl. Environ. Microbiol. 45:275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rawlings TK, Ruiz GM, Colwell RR. 2007. Association of Vibrio cholerae O1 El Tor and O139 Bengal with the copepods Acartia tonsa and Eurytemora affinis. Appl. Environ. Microbiol. 73:7926–7933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffith OW, Stuehr DJ. 1995. Nitric oxide synthases: properties and catalytic mechanism. Annu. Rev. Physiol. 57:707–734 [DOI] [PubMed] [Google Scholar]
- 5.Gusarov I, Starodubtseva M, Wang Z-Q, McQuade L, Lippard SJ, Stuehr DJ, Nudler E. 2008. Bacterial nitric-oxide synthases operate without a dedicated redox partner. J. Biol. Chem. 283:13140–13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber F, Polerecky L, de Beer D. 2008. Nitric oxide microsensor for high spatial resolution measurements in biofilms and sediments. Anal. Chem. 80:1152–1158 [DOI] [PubMed] [Google Scholar]
- 7.Lundberg JO, Weitzberg E, Gladwin MT. 2008. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 7:156–167 [DOI] [PubMed] [Google Scholar]
- 8.Landry AP, Duan X, Huang H, Ding H. 2011. Iron-sulfur proteins are the major source of protein-bound dinitrosyl iron complexes formed in Escherichia coli cells under nitric oxide stress. Free Radic. Biol. Med. 50:1582–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toledo JC, Bosworth CA, Hennon SW, Mahtani HA, Bergonia HA, Lancaster JR. 2008. Nitric oxide-induced conversion of cellular chelatable iron into macromolecule-bound paramagnetic dinitrosyliron complexes. J. Biol. Chem. 283:28926–28933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosworth CA, Toledo JC, Zmijewski JW, Li Q, Lancaster JR. 2009. Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc. Natl. Acad. Sci. U. S. A. 106:4671–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner PR, Costantino G, Szabó C, Salzman AL. 1997. Nitric oxide sensitivity of the aconitases. J. Biol. Chem. 272:25071–25076 [DOI] [PubMed] [Google Scholar]
- 12.Hyduke DR, Jarboe LR, Tran LM, Chou KJY, Liao JC. 2007. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:8484–8489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, Becker LA, Navarre WW, Castor ME, Libby SJ, Fang FC. 2011. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar Typhimurium. Cell Host Microbe 10:33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourret TJ, Boylan JA, Lawrence KA, Gherardini FC. 2011. Nitrosative damage to free and zinc-bound cysteine thiols underlies nitric oxide toxicity in wild-type Borrelia burgdorferi. Mol. Microbiol. 81:259–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao G, Xie L, Gross SS. 2004. Argininosuccinate synthetase is reversibly inactivated by S-nitrosylation in vitro and in vivo. J. Biol. Chem. 279:36192–36200 [DOI] [PubMed] [Google Scholar]
- 16.Clementi E, Brown GC, Feelisch M, Moncada S. 1998. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc. Natl. Acad. Sci. U. S. A. 95:7631–7636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husain M, Bourret TJ, McCollister BD, Jones-Carson J, Laughlin J, Vázquez-Torres A. 2008. Nitric oxide evokes an adaptive response to oxidative stress by arresting respiration. J. Biol. Chem. 283:7682–7689 [DOI] [PubMed] [Google Scholar]
- 18.Richardson AR, Libby SJ, Fang FC. 2008. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science 319:1672–1676 [DOI] [PubMed] [Google Scholar]
- 19.Rodionov DA, Dubchak IL, Arkin AP, Alm EJ, Gelfand MS. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. Plos Comput. Biol. 1:e55. 10.1371/journal.pcbi.0010055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartnikas TB, Wang Y, Bobo T, Veselov A, Scholes CP, Shapleigh JP. 2002. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a haem-copper protein. Microbiology 148:825–833 [DOI] [PubMed] [Google Scholar]
- 21.Stern AM, Hay AJ, Liu Z, Desland FA, Zhang J, Zhong Z, Zhu J. 2012. The NorR regulon is critical for Vibrio cholerae resistance to nitric oxide and sustained colonization of the intestines. MBio 3:e00013–12. 10.1128/mBio.00013-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metcalf WW, Jiang W, Daniels LL, Kim SK, Haldimann A, Wanner BL. 1996. Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13 [DOI] [PubMed] [Google Scholar]
- 23.Molstad L, Dörsch P, Bakken LR. 2007. Robotized incubation system for monitoring gases (O2, NO, N2O N2) in denitrifying cultures. J. Microbiol. Methods 71:202–211 [DOI] [PubMed] [Google Scholar]
- 24.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. 2009. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem. 81:6656–6667 [DOI] [PubMed] [Google Scholar]
- 25.Reitman ZJ, Jin G, Karoly ED, Spasojevic I, Yang J, Kinzler KW, He Y, Bigner DD, Vogelstein B, Yan H. 2011. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc. Natl. Acad. Sci. U. S. A. 108:3270–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nadeem S, Dörsch P, Bakken LR. 2013. Autoxidation and acetylene-accelerated oxidation of NO in a 2-phase system: implications for the expression of denitrification in ex situ experiments. Soil Biol. Biochem. 57:606–614 [Google Scholar]
- 27.Yang J, Duan X, Landry AP, Ding H. 2010. Oxygen is required for the l-cysteine-mediated decomposition of protein-bound dinitrosyl-iron complexes. Free Radic. Biol. Med. 49:268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gardner PR, Gardner AM, Martin LA, Salzman AL. 1998. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. U. S. A. 95:10378–10383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SO, Orii Y, Lloyd D, Hughes MN, Poole RK. 1999. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 445:389–394 [DOI] [PubMed] [Google Scholar]
- 30.Gonçalves VL, Nobre LS, Vicente JB, Teixeira M, Saraiva LM. 2006. Flavohemoglobin requires microaerophilic conditions for nitrosative protection of Staphylococcus aureus. FEBS Lett. 580:1817–1821 [DOI] [PubMed] [Google Scholar]
- 31.Ischiropoulos H, Zhu L, Beckman JS. 1992. Peroxynitrite formation from macrophage-derived nitric oxide. Arch. Biochem. Biophys. 298:446–451 [DOI] [PubMed] [Google Scholar]
- 32.Choi PS, Naal Z, Moore C, Casado-Rivera E, Abruña HD, Helmann JD, Shapleigh JP. 2006. Assessing the impact of denitrifier-produced nitric oxide on other bacteria. Appl. Environ. Microbiol. 72:2200–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bower JM, Mulvey MA. 2006. Polyamine-mediated resistance of uropathogenic Escherichia coli to nitrosative stress. J. Bacteriol. 188:928–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGinnis MW, Parker ZM, Walter NE, Rutkovsky AC, Cartaya-Marin C, Karatan E. 2009. Spermidine regulates Vibrio cholerae biofilm formation via transport and signaling pathways. FEMS Microbiol. Lett. 299:166–174 [DOI] [PubMed] [Google Scholar]
- 35.Henares BM, Higgins KE, Boon EM. 2012. Discovery of a nitric oxide responsive quorum sensing circuit in Vibrio harveyi. ACS Chem. Biol. 7:1331–1336 [DOI] [PubMed] [Google Scholar]
- 36.Carlson HK, Vance RE, Marletta MA. 2010. H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol. Microbiol. 77:930–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plate L, Marletta MA. 2012. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol. Cell 46:449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poole RK. 2005. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 33:176–180 [DOI] [PubMed] [Google Scholar]
- 39.Justino MC, Almeida CC, Gonçalves VL, Teixeira M, Saraiva LM. 2006. Escherichia coli YtfE is a di-iron protein with an important function in assembly of iron-sulphur clusters. FEMS Microbiol. Lett. 257:278–284 [DOI] [PubMed] [Google Scholar]
- 40.Justino MC, Almeida CC, Teixeira M, Saraiva LM. 2007. Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J. Biol. Chem. 282:10352–10359 [DOI] [PubMed] [Google Scholar]
- 41.Overton TW, Justino MC, Li Y, Baptista JM, Melo AMP, Cole JA, Saraiva LM. 2008. Widespread distribution in pathogenic bacteria of di-iron proteins that repair oxidative and nitrosative damage to iron-sulfur centers. J. Bacteriol. 190:2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reguera G, Kolter R. 2005. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J. Bacteriol. 187:3551–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nalin DR, Daya V, Reid A, Levine MM, Cisneros L. 1979. Adsorption and growth of Vibrio cholerae on chitin. Infect. Immun. 25:768–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chi W-J, Chang Y-K, Hong S-K. 2012. Agar degradation by microorganisms and agar-degrading enzymes. Appl. Microbiol. Biotechnol. 94:917–930 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.