Abstract
Lysogenic bacteriophage D3 causes seroconversion of Pseudomonas aeruginosa PAO1 from serotype O5 to O16 by inverting the linkage between O-specific antigen (OSA) repeat units from α to β. The OSA units are polymerized by Wzy to modal lengths regulated by Wzz1 and Wzz2. A key component of the D3 seroconversion machinery is the inhibitor of α-polymerase (Iap) peptide, which is able to solely suppress α-linked long-chain OSA production in P. aeruginosa PAO1. To establish the target specificity of Iap for Wzyα, changes in OSA phenotypes were examined via Western immunoblotting for wzz1 and wzz2 single-knockout strains, as well as a wzz1 wzz2 double knockout, following the expression of iap from a tuneable vector. Increased induction of Iap expression completely abrogated OSA production in the wzz1 wzz2 double mutant, while background levels of OSA production were still observed in either of the single mutants. Therefore, Iap inhibition of OSA biosynthesis was most effective in the absence of both Wzz proteins. Sequence alignment analyses revealed a high degree of similarity between Iap and the first transmembrane segment (TMS) of either Wzz1 or Wzz2. Various topology prediction analyses of the Iap sequence consistently predicted the presence of a single TMS, suggesting a propensity for Iap to insert itself into the inner membrane (IM). The compromised ability of Iap to abrogate Wzyα function in the presence of Wzz1 or Wzz2 provides compelling evidence that inhibition occurs after Wzyα inserts itself into the IM and is achieved through mimicry of the first TMS from the Wzz proteins of P. aeruginosa PAO1.
INTRODUCTION
Lipopolysaccharide (LPS) is an integral structural component of the outer membrane of Gram-negative bacteria and is important for the survival of these bacteria in the environment or in a host. In Pseudomonas aeruginosa and many other opportunistic pathogens, LPS is a major virulence factor and is composed of a lipid A membrane anchor, a core oligosaccharide linker, and a distal polysaccharide termed O antigen (O-Ag) (1). P. aeruginosa simultaneously produces two forms of O-Ag, a homopolymeric common antigen (CPA) and an immunodominant heteropolymeric O-specific antigen (OSA) composed of repeating trisaccharide units (2).
In P. aeruginosa PAO1, OSA is synthesized via the Wzx/Wzy-dependent pathway (3), which requires the activity of several integral inner membrane (IM) proteins (4). Synthesis begins at the cytoplasmic leaflet of the IM on the lipid carrier undecaprenyl pyrophosphate (UndPP), with the formation of an OSA trisaccharide repeat constructed from newly synthesized nucleotide sugar precursors (5). The UndPP-linked OSA repeat is then transported through the cationic interior of the OSA flippase Wzx to the periplasmic leaflet of the IM (6, 7) and polymerized at the reducing terminus of the growing chain (8) by Wzy via a putative “catch-and-release” mechanism in an α-1-4 linkage (9, 10). The OSA chain length is regulated by the polysaccharide copolymerase (PCP) proteins Wzz1 and Wzz2, which interact with the nascent polysaccharides and confer long (12 to 16 and 22 to 30 repeats) and very long (40 to 50 repeats) modal lengths, respectively (11, 12). Full-length OSA is then ligated to the lipid A-core moiety by the O-Ag ligase WaaL (13, 14), forming the mature LPS molecule.
Variability in the OSA repeat sugar constituents, intra- and interglycosidic linkages of the OSA repeat residues, and the presence of side branch modifications are used to classify P. aeruginosa into 20 distinct serotypes according to the International Antigenic Typing Scheme (1). Similar OSA backbone sugar structures result in certain individual serotypes, for example, O2, O5, O16, O18, and O20, being classified into a single serogroup (serogroup O2). Immunochemical cross-reactivity of LPS of these serotypes with specific typing antisera or monoclonal antibodies (MAbs) substantiates their relatedness. In particular, the only difference between the OSA chemical structures of serotypes O5 and O16 is the α or β configuration of the interglycosidic bond at the reducing end, respectively (15, 16). Interestingly, the OSA biosynthesis cluster spanning the pa3160 to pa3145 (wzz1 to wbpL) genes of O5 is identical to that in serogroup O2. This suggests that genes located outside the wbp cluster are responsible for the chemical differences in the OSA structures. Therefore, genes responsible for the chemical differences in OSA structures were likely to have come from external sources such as lysogenic bacteriophage (17) or other ancestral microbial species.
Serotype conversion following bacteriophage infection has long been observed in diverse Gram-negative bacteria (18–22). Upon infection of P. aeruginosa PAO1 (serotype O5) by bacteriophage D3, the bacteria became resistant to future infections, illustrating the lysogenic property of this lambdoid D3 phage (23). D3 phage uses the O5 OSA as a receptor, leading to the downstream conversion of the bacteria to the O16 serotype. This conversion involved a switch from an α to a β bond linkage between OSA repeat units (24). Taking advantage of the annotated D3 phage genome (25), our group identified a 3.6-kb DNA fragment containing three open reading frames. Expression of the entire seroconverting unit in P. aeruginosa PAO1 resulted in the same observed loss of reactivity to O5-specific MAb MF15-4 (26) and a conversion to serotype O16 as described above. Characterization of this 3.6-kb fragment revealed a three-component system containing genes that encode a putative O-acetyltransferase, a putative β-polymerase (Wzyβ), and an α-polymerase inhibitor (Iap) peptide (27).
The Iap peptide is encoded by a 0.9-kb fragment of the three-gene seroconverting unit, which when supplied in trans, inhibits the production of α-linked long-chain OSA in serotypes O5, O18, and O20. Transformation of serotype O16 with iap did not show any inhibitory effect on OSA production, illustrating the specificity of Iap for α-linked OSA biosynthesis in the O2 serogroup (27). Our group performed Southern blot analysis and demonstrated that iap is present in the genomes of serotypes O2 and O16, which explains the lack of α-linked OSA in these strains. A subsequent study by our group showed that, in addition to iap, a chromosomal copy of wzyβ (responsible for the β-linked OSA of these serotypes) is actively expressed in the O2 and O16 serotypes (28).
The mechanism of Wzyα suppression by iap is unknown. The translated product of iap is a 3.1-kDa protein (27). In this study, we further investigated Iap-mediated inhibition of OSA in the P. aeruginosa PAO1 background by using previously generated chromosomal mutants deficient in the chain length regulator(s) wzz1 and/or wzz2. The observed differences in Iap-mediated inhibition were dependent on the presence or absence of both Wzz proteins. These observations suggested that inhibition due to Iap occurred downstream of Wzyα insertion into the IM. Strong sequence homology was detected between a putative transmembrane segment (TMS) in Iap and that of the first TMS of both PAO1 Wzz1 and Wzz2. Together, these findings indicate that the inhibitory activity of Iap is specifically targeted at Wzyα through mimicry of the Wzz TMS. Moreover, these data provide additional evidence to support direct interaction between Wzy and Wzz1/2 in the Wzx/Wzy-dependent LPS assembly pathway.
MATERIALS AND METHODS
DNA manipulations.
The strains and plasmids used in this study are outlined in Table 1. Cloning of iap into the arabinose-inducible expression vector pHERD20T (29) required amplification with iap-specific primers (see Fig. S1 in the supplemental material) containing EcoRI and PstI sites, respectively, from a previously generated pET28-His6-Iap construct (unpublished data). The His6 coding sequence was retained for use in future experiments. Ligation products were introduced into Escherichia coli DH10B by heat shock transformation. Plasmid DNA was extracted from positive clones with a plasmid purification kit (Life Technologies Inc., Burlington, ON, Canada) and identified by sequencing with the pBAD forward primer. QuikChange (Agilent) site-directed mutagenesis was used to remove the leader sequence present in frame at the 5′ end of the pHERD20T vector (see Fig. S1). Successful constructs were identified as described above. Positive clones and previously generated plasmids were introduced into electrocompetent P. aeruginosa PAO1 wzz1, wzz2, and wzz1 wzz2 chromosomal mutants with a Gene Pulser instrument (Bio-Rad).
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype, phenotype, or relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| O5 | Wild-type strain PAO1, IATS O5, A+ B+ | 61 |
| O16 | Wild-type strain, IATS serotype O16, A− B+ | ATCC 33363 |
| wzz1::Gmr | PAO1 derivative, A+ B+a | 12 |
| wzz2Δ | PAO1 derivative, A+ B+ | 11 |
| wzz2Δ wzz1::Gmr | PAO1 derivative, A+ B+ | 11 |
| wzyβ::Gmr | O16 derivative, A− B− | 28 |
| E. coli DH10B | F− araD139 Δ(ara leu)7697 ΔlacX74 galU galK rpsL deoR ϕ80dlacZΔM15 endA1 nupG recA1 mcrA Δ(mrr hsdRMS mcrBC) | Laboratory stock |
| Plasmids | ||
| pHERD20t | Pseudomonas expression vector (Plac); Apr | 29 |
| pHERD20tHis6iap | pHERD20t carrying His6-iap | This study |
| pET28His6iap | Escherichia expression vector | 27 |
| pUCP26 | 4.9-kb pUC18-based broad-host-range vector, Tcr | 43 |
| pUCP26iap | Broad-host-range vector containing iap | 27 |
| pUCP26wzz1 | Broad-host-range vector containing O5 wzz1 | 12 |
A superscript plus or minus after A or B denotes the presence or absence of the particular O polysaccharide, respectively. Resistance (superscript r) is shown for gentamicin (Gm), ampicillin (Ap), and tetracycline (Tc).
LPS preparation and visualization.
Each of the P. aeruginosa strains was grown overnight from a single colony with shaking (200 rpm, 37°C) in the presence of (i) 300 μg/ml of carbenicillin or 90 μg/ml of tetracycline to maintain the plasmids and (ii) different l-arabinose concentrations to induce expression from pHERD. In the Wzz investigations with pHERD20T-His6-Iap, 0, 0.1, 0.5, and 1% (wt/vol) l-arabinose concentrations were tested. The following morning, the cultures were equilibrated to the equivalent of 1 ml of culture at an optical density at 600 nm of 0.45. LPS was then prepared by the proteinase K digestion method (12, 30). The LPS samples (5 μl) were analyzed by Western immunoblotting probing with murine MAb MF15-4 (serotype O5 O-Ag specific) and MAb 5c-7-4 (inner core specific). To quantify OSA levels in each of the wzz mutants, independently isolated triplicate LPS samples were processed according to a previously described method (6).
Bioinformatic analyses.
The Iap sequence was analyzed by the following topology prediction algorithms to probe for the presence of a predicted TMS (31): TopCons (http://topcons.cbr.su.se/) (32), HMMTOP (http://www.enzim.hu/hmmtop) (33), TMHMM (http://www.cbs.dtu.dk/services/TMHMM) (34), PHOBIUS (http://phobius.sbc.su.se) (35), TMPred (http://www.ch.embnet.org/software/TMPRED_form.html) (36), Philius http://www.yeastrc.org/philius/pages/philius/runPhilius.jsp;jsessionid=6DD32D6307F337296FF09929EE7029D1) (37), TOPPRED (http://www.sbc.su.se/∼erikw/toppred2) (38), and PredictProtein (http://www.predictprotein.org) (39). Alignments of Iap with the PCP sequences were performed by ClustalW with a gap extension penalty of 1.0 (40, 41). A three-dimensional (3D) model of Iap was generated with the I-TASSER de novo platform, which produces a model structure based on sequence alignment with existing Protein Data Bank files (42).
RESULTS
The OSA chain length regulator proteins Wzz1 and Wzz2 are not the targets of Iap.
To determine whether Iap was inhibiting long-chain OSA biosynthesis through interference with either of the Wzz proteins in P. aeruginosa PAO1, the pD3 vector (i.e., pUCP26iap) was used to transform the wild-type (WT) strain, as well as the wzz1, wzz2, and wzz1 wzz2 knockout strains. Transformation of WT P. aeruginosa PAO1 with pD3 resulted in a reduction of OSA production (Fig. 1, lane 2), as detected through Western immunoblot analysis. Interestingly, OSA inhibition in either the wzz1 or the wzz2 single-mutant background occurred at the same observed amount as in the WT PAO1 background. This indicates that neither of the two Wzz proteins is the specific target of inhibition by Iap, as there was no restoration of long-chain OSA. In contrast, when iap was expressed in the wzz1 wzz2 double mutant, total abrogation of OSA production was observed (Fig. 1, lane 8).
Fig 1.
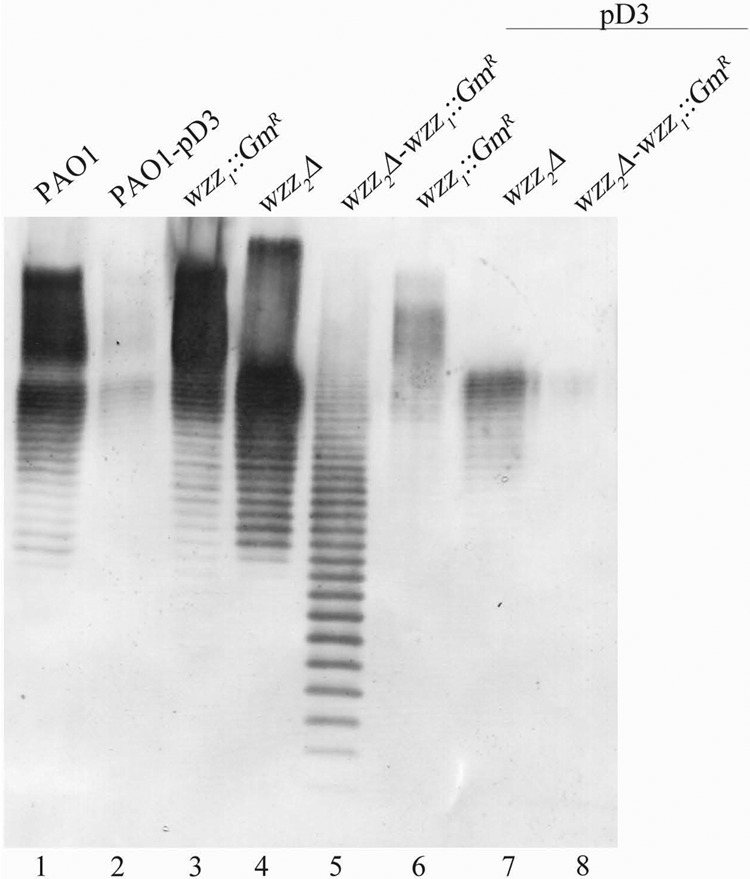
Effect of iap expression in P. aeruginosa PAO1 wzz chromosomal mutant strains. Shown is a Western immunoblot assay probed with OSA-specific MAb MF15-4. The control lanes include P. aeruginosa PAO1, PAO1 transformed with pD3, each wzz mutant construct (wzz1, wzz2, and wzz1 wzz2), and the wzz mutants transformed with pD3.
OSA inhibition by Iap is influenced by the presence of Wzz proteins.
To ensure that the complete abrogation of OSA production observed in the wzz1 wzz2 double mutant transformed with pD3 was not simply a high-dosage artifact due to constitutive expression from the multicopy pUCP26 backbone (43), iap was subcloned into the tuneable pHERD20T plasmid. Titratable iap expression was attained through the addition of increasing amounts of l-arabinose. To rule out the possibility that any observed variations in OSA levels were simply due to differences in the amounts of LPS being produced by the three Wzz mutant strains, the OSA levels from all three strains were quantified via densitometry following Western immunoblotting analysis. The density of OSA was normalized by comparing the intensity of the OSA band to that of its respective inner core oligosaccharide, which is synthesized via an unrelated pathway and therefore levels would remain constant despite changes in OSA production. Thus, these lower-molecular-weight bands detected in the Western immunoblot assays were used as a loading control. Equivalent amounts of OSA were detected in all three wzz mutants (see Fig. S3 in the supplemental material).
In either the wzz1 or the wzz2 single-knockout strain, OSA production was inhibited to levels similar to WT, retaining background amounts equivalent to each other when induced with up to 1% l-arabinose (Fig. 2A and B). In contrast, uninduced expression of iap in the wzz1 wzz2 double mutant was sufficient to inhibit OSA production. Total abrogation was observed at 0.1% l-arabinose induction, a 10-fold lower concentration than that required to observe changes in OSA production in either of the single-wzz mutants (Fig. 2C).
Fig 2.
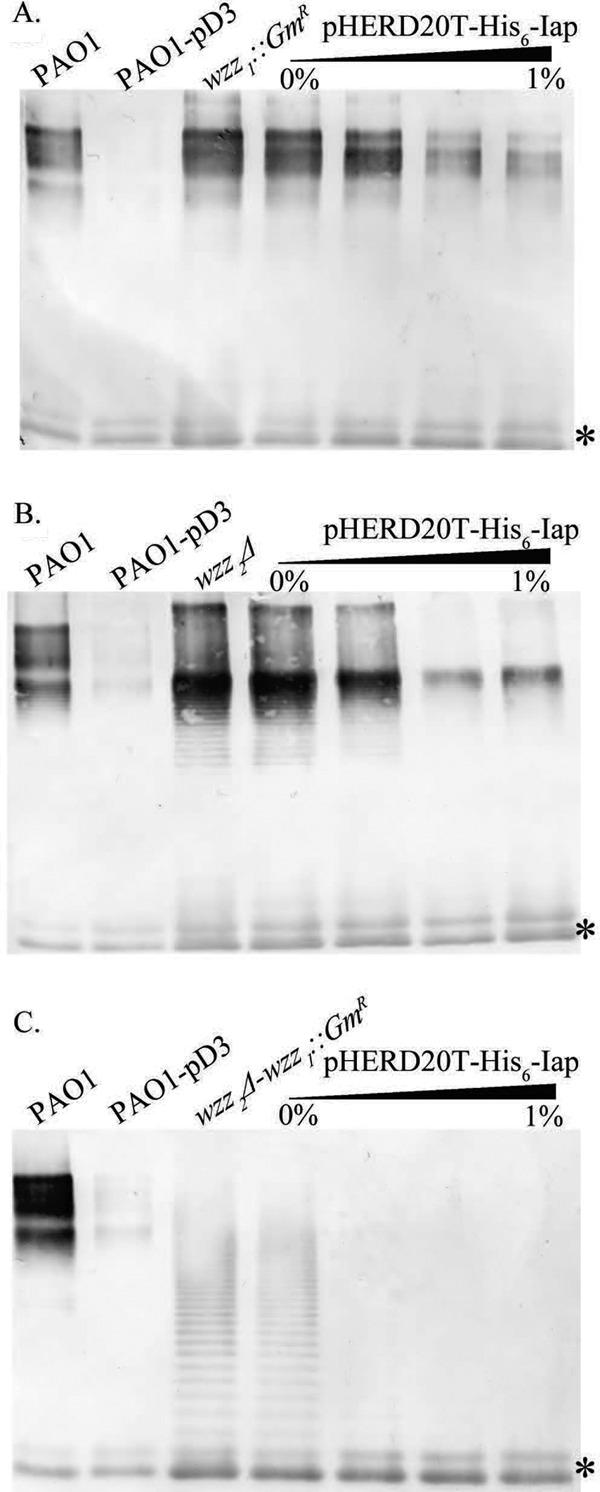
Effects of iap titration on OSA levels in various wzz knockout strains. Shown are Western immunoblot assays of LPS prepared from P. aeruginosa PAO1 wzz chromosomal mutants transformed with pHERD20T-His6-Iap. Expression levels were titrated via increasing concentrations of l-arabinose (0, 0.1, 0.5, and 1.0%). Immunoblot assays were subsequently probed with MAbs specific to O5 OSA (MAb MF15-4) and inner core oligosaccharide (MAb 5c-7-4) (*). WT P. aeruginosa PAO1 and PAO1 transformed with pUCP26iap were used as WT and OSA− controls in each set of blots. (A) LPS harvested from the wzz1 chromosomal mutant background. (B) LPS harvested from the wzz2 chromosomal mutant background. (C) LPS harvested from the wzz1 wzz2 double chromosomal mutant.
Sequence conservation between Iap and the predicted N-terminal TMS of Wzz1/2 suggests a mechanism of specificity.
Upon the alignment of the Iap amino acid sequence with the full-length sequence of either Wzz1 or Wzz2, a high degree of sequence conservation was observed between Iap and the N termini of both Wzz proteins in a region corresponding to the predicted first TMS of either Wzz (Fig. 3; see Fig. S4 in the supplemental material). Conversely, alignments of Iap with annotated full-length Wzz protein sequences from a heterologous strain, P. aeruginosa PA7 (serotype O12), and from other bacteria (E. coli K-12, Salmonella enterica serovar Typhimurium, and Vibrio cholerae) did not display homology to their respective TMS regions; instead, the Iap sequence was more favorably aligned with tracts present in the soluble periplasmic domains of these heterologous Wzz proteins. This difference in alignment regardless of amino acid characteristics demonstrates an association with the OSA biosynthesis machinery of the O5 serotype of Iap to WzzO5 (see Fig. S4).
Fig 3.
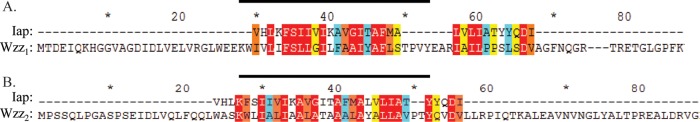
Sequence alignment of Iap with Wzz1 (A) and Wzz2 (B) of P. aeruginosa PAO1. The alignment was produced in ClustalW and colored according to conservation score (out of 10) as represented by Jalview, where red is 10, orange is 9, yellow is 8, and blue is 7. The black line denotes the predicted transmembrane region of either Wzz protein. Both alignments demonstrate strong similarities between the Iap sequence and Wzz from P. aeruginosa strain PAO1, alluding to substrate specificity.
In silico prediction of Iap topology suggests a single TMS-spanning domain.
The Iap sequence is composed of 31 amino acids, wherein 52% of the residues are hydrophobic, supporting the notion of membrane localization. The Iap amino acid sequence was subjected to in silico topology prediction analyses by nine distinct algorithms (31, 44), which consistently predicted a single TMS-spanning domain. On the basis of these outputs, Iap was predicted to contain a TMS between residues 6 and 28. In addition, the N and C termini were predicted to reside in the cytoplasm and periplasm, respectively (Fig. 4).
Fig 4.
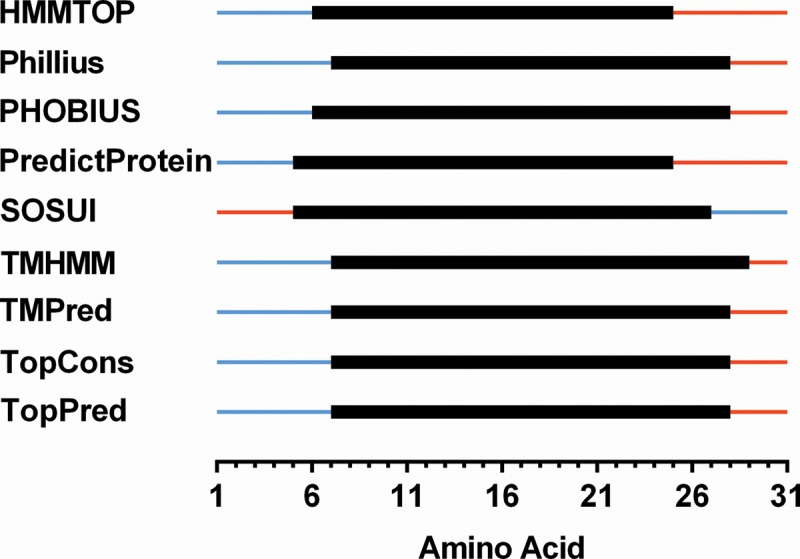
Topology prediction algorithm analysis of Iap. The amino acid sequence of Iap was analyzed by nine different algorithms to determine whether a conserved single TMS-spanning domain is predicted for a putative TMS. The sequence of Iap amino acids 1 to 31 is depicted. Color key: blue line, cytoplasmic localization; black box, range of amino acids predicted to form a TMS; red line, periplasmic localization.
To further support these findings, a de novo 3D structure of Iap was generated by the alignment modeling platform I-TASSER. Upon superimposition of the three highest-scoring models, a conserved α-helical region between residues 12 and 29 was displayed. This corresponds to the putative TMS of Iap (Fig. 5). Surface electrostatic analysis of the Iap structure revealed hydrophobic properties of the α-helical segment, while the N-terminal tail was shown to be cationic. These data fit with the proposed orientation of Iap in the IM (Fig. 4) and are consistent with the demonstrated “positive-inside rule,” which states that the cytoplasmic domains of TMS-containing proteins or peptides retain a positive net charge (45, 46) (see Fig. S6 in the supplemental material).
Fig 5.
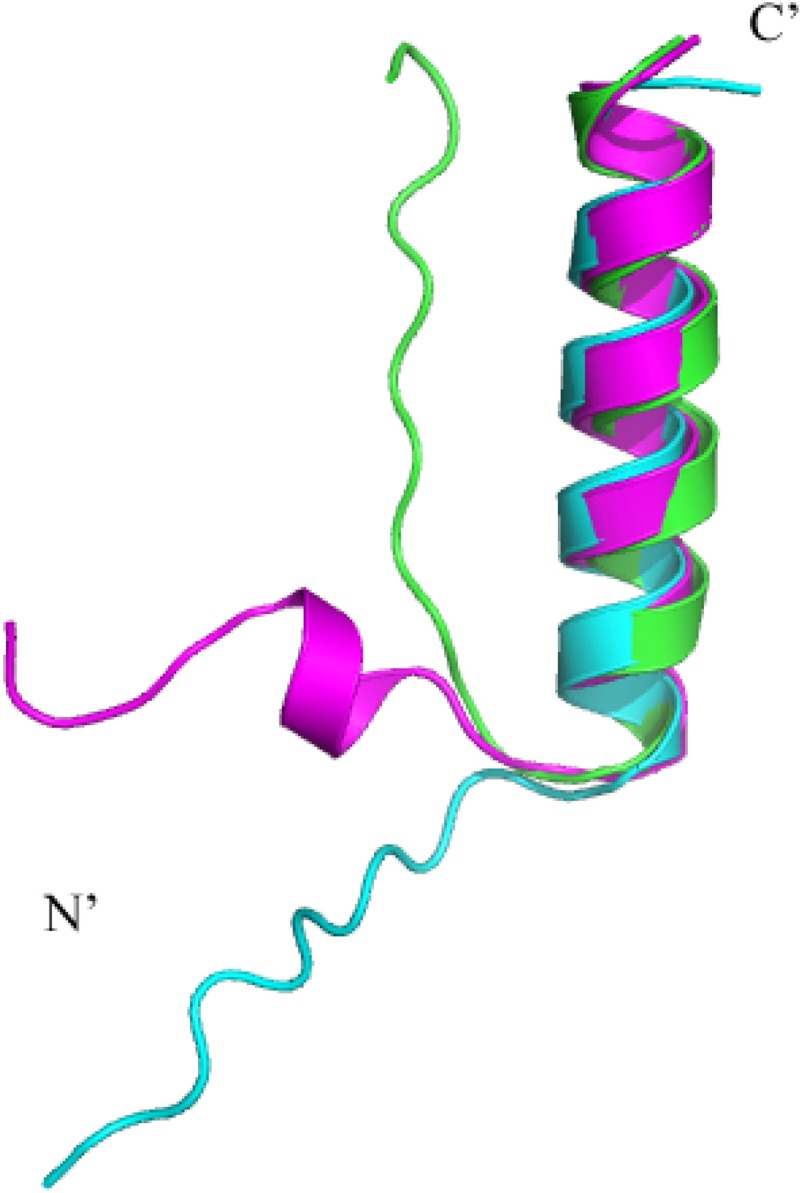
I-TASSER three-dimensional modeling of Iap. The three highest-quality models for the Iap structure were superimposed with PyMOL. Residues 1 to 13 are predicted to form a disordered coil, while residues 14 to 31 are predicted to form an α-helix.
Overexpression of Wzz in P. aeruginosa serotype O16 results in the recovery of α-linked OSA.
As mentioned in the introduction, serotype O16 has previously been shown to be unable to produce detectable α-linked OSA. If Iap in this background is indeed mimicking a Wzz TMS, then overexpression of Wzz should be able to outcompete Iap for access to Wzyα and restore the synthesis of α-linked OSA. To test this hypothesis, Western immunoblotting of triplicate LPS samples obtained from the previously generated P. aeruginosa serotype O16 wzyβ::Gmr strain transformed with the pUCP26wzz1 plasmid showed LPS bands that reacted with O5-specific antibodies (Fig. 6). This indicates that α-linked OSA similar to that seen in strain PAO1 was produced by this transformant.
Fig 6.
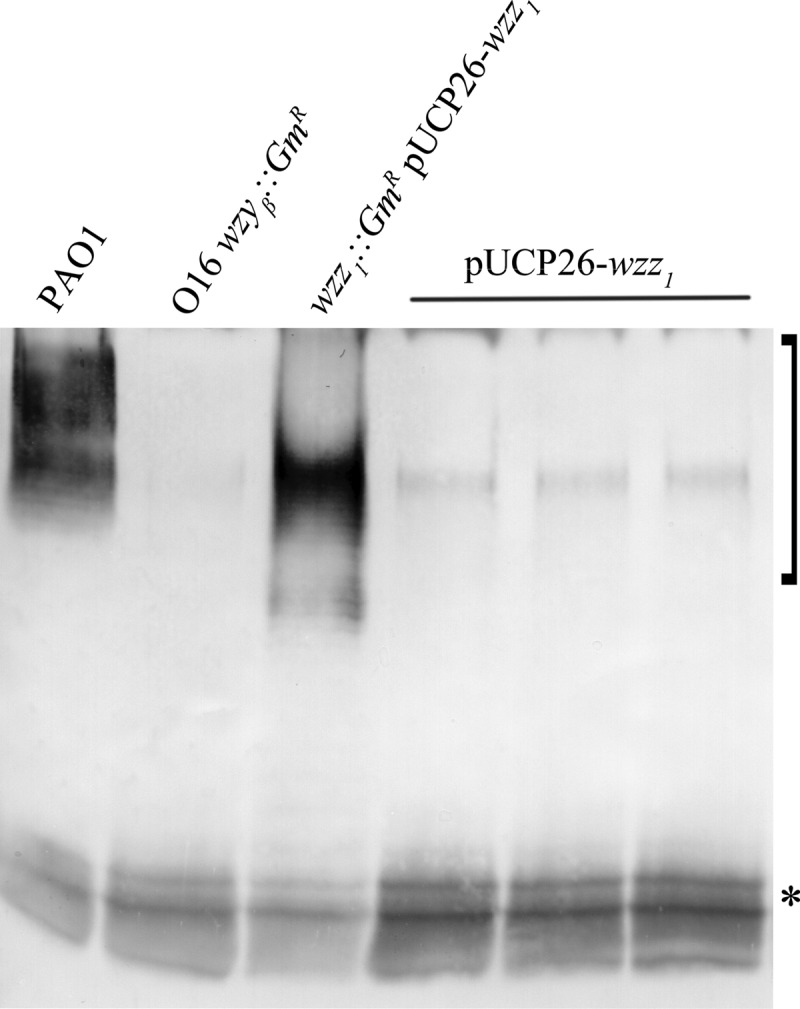
Effect of wzz1 overexpression in P. aeruginosa serotype O16 wzyβ::Gmr. Shown is a Western immunoblot assay probed with OSA-specific MAb MF15-4 (bracket) and MAb 5c7-4 (asterisk). The control lanes include P. aeruginosa PAO1, O16 wzyβ::Gmr, and the PAO1 wzz1 wzz2 double mutant transformed with pUCP26wzz1. The last three lanes contain O16 wzyβ::Gmr transformed with pUCP26wzz1.
DISCUSSION
Seroconversion in P. aeruginosa PAO1 caused by bacteriophage D3 was observed earlier by our group after transformation of the bacterium with the aforementioned “seroconverting unit” (consisting of three genes from the D3 chromosome), resulting in the abrogation of long-chain OSA production. One of these three genes, iap, was identified as the cause of inhibition of long- and very-long-chain OSA biosynthesis (27).
Results from the present study indicate that the specificity of the Iap inhibitory activity is for native Wzyα of serotype O5. We initially attempted colocalization studies to directly examine Iap-Wzyα interaction; however, because of the hydrophobic nature of the Iap peptide, such evidence could not be obtained. Subsequently, we decided to take advantage of the well-defined wzz1, wzz2, and wzz1 wzz2 chromosomal mutants and the unique panel of MAbs from our laboratory to elucidate the target of Iap inhibition. Our results show that the target of Iap-mediated OSA inhibition is Wzyα, a key component of the putative membrane complex involved in the Wzx/Wzy-dependent LPS assembly pathway.
The phenotype resulting from Iap expression was the loss of long- and very-long-chain OSA in P. aeruginosa PAO1, especially when the peptide was expressed in multicopy plasmid pUCP26. Iap expressed in the wzz1 and wzz2 single-PCP mutants did not display observable changes in OSA production compared to the effect of Iap in the WT PAO1 background; therefore, the chain length regulators are not the target of Iap function. This is consistent with the previous findings of our group that expression from the pD3 plasmid (consisting of the three-gene seroconverting segment of bacteriophage D3) in P. aeruginosa PAO1 resulted in β-linked OSA subunits being polymerized by phage-encoded Wzyβ to form LPS of defined modal lengths (despite the presence of Iap) and that PCP proteins are not the target of Iap inhibition (27, 28).
In order to be able to draw conclusions about the effect of Iap activity in the different wzz mutant backgrounds, it is essential to determine whether equivalent levels of OSA are being produced. To our knowledge, this is the first study to investigate whether the absence of Wzz1, Wzz2, or both would affect the total amount of OSA on the cell surface (see Fig. S3 in the supplemental material). With equivalent levels of OSA being synthesized by the three wzz mutant strains, the low Iap levels required for suppression of LPS production in the wzz1 wzz2 double mutant suggest that in the absence of Wzz proteins, Iap has unencumbered access to Wzyα. This is in contrast to either of the single wzz mutants, where a larger amount of Iap would be required to outcompete Wzz for Wzyα by the presence of the remaining Wzz protein.
In addition, this suggests that Iap-mediated inhibition of Wzyα function occurs after the α-polymerase has inserted itself into the IM. If the Iap were to interfere with the wzyα RNA or disrupt the transport of Wzyα or its insertion into the cell membrane, then use of the pHERD system to express iap in P. aeruginosa would have resulted in a consistent inhibition phenotype regardless of the WT or copolymerase mutant background strain. This is regardless of whether a WT or copolymerase mutant background was used to test the effect of iap.
In previous investigations, neither WT O16 nor O16 wzyβ::Gmr was able to produce α-linked OSA because of the presence of the seroconverting unit, particularly iap, within the genome. Previous work in our lab demonstrated that Wzyα from serotype O16 is indeed functional; however, it is being actively inhibited. To further investigate the Iap-Wzy-Wzz interaction, Wzz was overexpressed in the O16 wzyβ::Gmr background to determine whether any amount of α-linked OSA can be restored. Several advantages were exploited through the use of this particular strain. First, the lack of CPA allowed a more clear observation of changes in the OSA. In addition, iap expressed from the chromosome precluded the use of two separate plasmids. Finally, with the absence of the β-polymerase, any restoration of α-linked OSA was more easily detected. As reported previously (28), no background O5 OSA is polymerized in the absence of Wzyβ. When Wzz was overexpressed, partial restoration of the α-linked OSA phenotype was restored, indicating that Wzz was able to outcompete Iap to interact with Wzyα. Future characterization of the binding kinetics and affinity of Iap for Wzyα will help us to better understand, at the molecular level, the events that lead to the abrogation of OSA biosynthesis.
Wzz proteins exist throughout all Gram-negative organisms and are required not only for the biosynthesis of LPS but also for the biosynthesis of enterobacterial common antigen (47) and capsule polysaccharide (48). Wzz proteins are characterized by two TMSs, a large periplasmic domain, and the preference to organize in a higher-ordered homo-oligomeric state (11, 12, 49–53). The sequence similarity between PCP proteins is limited to approximately 19 to 20%; however, the secondary structures of these proteins are quite conserved (12). The sequence similarity between Iap and the proposed N-terminal TMS of either P. aeruginosa PAO1 Wzz suggests a mechanism of specificity to the O5 OSA machinery. Simply on the basis of hydrophobicity, it would not be unexpected for a hydrophobic IM-spanning peptide to align with other TMS domains. However, alignment of Iap with heterologous Wzz protein sequences does not yield the same alignment positioning as that detected with the P. aeruginosa PAO1 Wzz1 or Wzz2 template. This similarity might provide a clue to substrate specificity, explaining the targeted inhibition of OSA production in serotype O5. Recently, high-resolution X-ray crystal structures of the periplasmic domain of Wzz from Shigella flexneri and from other bacterial species have been obtained (53, 54). These structures revealed a conserved structural motif, and functional investigation through site-directed mutagenesis identified residues proposed to be involved in substrate binding and oligomerization (49, 50, 55).
A leading hypothesis to describe the interplay between proteins in the Wzx/Wzy-dependent pathway, including Wzy and Wzz, is based on their loose proximity to one another and supposes that the resulting interaction is mediated through a mutually shared OSA substrate bridging the two proteins (11, 56, 57). However, genetic data indirectly support interaction of the Wzx flippase with the corresponding Wzy and Wzz proteins (58). In addition, a new investigation that used heterologous bacterial two-hybrid screening has provided evidence to support interactions between Wzy proteins and their corresponding PCP (59).
Although considerable efforts have been made to investigate the periplasmic domain of Wzz proteins, the importance of the two TMS regions has been largely overlooked. Conserved motifs spanning both the N- and C-terminal TMSs of S. flexneri have been investigated by site-directed mutagenesis (60). Of particular relevance to this study are the residues substituted in the N-terminal TMS contained within the conserved “KTMII” motif. Complete loss of high-molecular-weight O-Ag was observed in the K31A mutant, whereas an M32T mutation resulted in altered O-Ag modal lengths approximately 10 to 15 residues shorter than the WT length, thus demonstrating the importance of the N-terminal TMS regions in O-Ag chain length regulation (60). In a recent study by our group (S. T. Islam, S. M. Huszczynksi, T. Nugent, A. C. Gold, and J. S. Lam, submitted for publication), substitutions of cytoplasmic amino acid residues in Wzyα from P. aeruginosa PAO1 were found to eliminate OSA bands at modal lengths corresponding to those regulated by Wzz1 (but not those regulated by Wzz2); these cytoplasmic amino acid substitutions in Wzyα have led to altered predicted packing of the C-terminal TMS of the polymerase. These observations suggest that a specific interaction between Wzy and Wzz1 had been disrupted, as OSA bands corresponding to Wzz2-mediated very-long-chain regulation were not affected. If Wzyα and Wzz1/2 were simply localized in close proximity, the site-directed mutations introduced into the cytoplasmic amino acid residues of Wzyα should not have exerted an effect on the downstream process of OSA chain length regulation. These observations are consistent with the result shown in the present study, providing the first evidence of a direct interaction between homologously expressed Wzy and Wzz proteins, with the TMS of Wzz proteins likely playing an integral role in this process.
On the basis of our present observations and the documented relationship between Wzy and Wzz, a possible mechanism of Iap-mediated inhibition of OSA polymerization would involve Iap outcompeting Wzz for interaction with Wzy. Hence, the bacteria would exhibit a phenotype lacking long-chain polysaccharide with α-linked O units. Herein, we have used Iap as a unique tool with which to examine the biosynthesis of O-Ag via the Wzx/Wzy-dependent pathway. Further investigations of the molecular mechanism of Iap-mediated O-Ag inhibition would shed more light on the assembly of various virulence-associated cell surface polysaccharides via this assembly scheme.
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by operating grants from Cystic Fibrosis Canada (CFC) and the Canadian Institutes of Health Research (CIHR) (grant MOP-14687) to J.S.L. V.L.T. and S.T.I. are recipients of CFC doctoral studentships, and J.S.L. holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology.
Footnotes
Published ahead of print 16 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00903-13.
REFERENCES
- 1.Lam JS, Taylor VL, Islam ST, Hao Y, Kocíncová D. 2011. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front. Microbiol. 2:118. 10.3389/fmicb.2011.00118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King JD, Kocíncová D, Westman EL, Lam JS. 2009. Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 15:261–312 [DOI] [PubMed] [Google Scholar]
- 3.Whitfield C. 1995. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 3:178–185 [DOI] [PubMed] [Google Scholar]
- 4.Islam ST, Taylor VL, Qi M, Lam JS. 2010. Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures. MBio 1:e00189–10. 10.1128/mBio.00189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocchetta HL, Burrows LL, Pacan JC, Lam JS. 1998. Three rhamnosyltransferases responsible for assembly of the A-band d-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A-band and B-band lipopolysaccharide synthesis. Mol. Microbiol. 28:1103–1119 [DOI] [PubMed] [Google Scholar]
- 6.Islam ST, Fieldhouse RJ, Anderson EM, Taylor VL, Keates RAB, Ford RC, Lam JS. 2012. A cationic lumen in the Wzx flippase mediates anionic O-antigen subunit translocation in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 84:1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Islam ST, Lam JS. 2013. Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ. Microbiol. 15:1001–1015 [DOI] [PubMed] [Google Scholar]
- 8.Robbins PW, Bray D, Dankert M, Wright A. 1967. Direction of chain growth in polysaccharide synthesis. Science 158:1536–1542 [DOI] [PubMed] [Google Scholar]
- 9.Islam ST, Gold AC, Taylor VL, Anderson EM, Ford RC, Lam JS. 2011. Dual conserved periplasmic loops possess essential charge characteristics that support a catch-and-release mechanism of O-antigen polymerization by Wzy in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 286:20600–20605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Kievit TR, Dasgupta T, Schweizer H, Lam JS. 1995. Molecular cloning and characterization of the rfc gene of Pseudomonas aeruginosa (serotype O5). Mol. Microbiol. 16:565–574 [DOI] [PubMed] [Google Scholar]
- 11.Daniels C, Griffiths C, Cowles B, Lam JS. 2002. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ. Microbiol. 4:883–897 [DOI] [PubMed] [Google Scholar]
- 12.Burrows LL, Chow D, Lam JS. 1997. Pseudomonas aeruginosa B-band O-antigen chain length is modulated by Wzz (Ro1). J. Bacteriol. 179:1482–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abeyrathne PD, Daniels C, Poon KKH, Matewish MJ, Lam JS. 2005. Functional characterization of WaaL, a ligase associated with linking O-antigen polysaccharide to the core of Pseudomonas aeruginosa lipopolysaccharide. J. Bacteriol. 187:3002–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abeyrathne PD, Lam JS. 2007. WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O antigen onto lipid A-core. Mol. Microbiol. 65:1345–1359 [DOI] [PubMed] [Google Scholar]
- 15.Knirel YuA, Vinogradov EV, Kocharova NA, Paramonov NA, Kochetkov NK, Dmitriev BA, Stanislavsky ES, Lányi B. 1988. The structures of O-specific polysaccharides and serological classification of Pseudomonas aeruginosa. Acta Microbiol. Hung. 35:3–24 [PubMed] [Google Scholar]
- 16.Lam JS, Handelsman MY, Chivers TR, MacDonald LA. 1992. Monoclonal antibodies as probes to examine serotype-specific and cross-reactive epitopes of lipopolysaccharides from serotypes O2, O5, and O16 of Pseudomonas aeruginosa. J. Bacteriol. 174:2178–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burrows LL, Charter DF, Lam JS. 1996. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol. Microbiol. 22:481–495 [DOI] [PubMed] [Google Scholar]
- 18.Simmons DAR, Romanowska E. 1987. Structure and biology of Shigella flexneri O antigens. J. Med. Microbiol. 23:289–302 [DOI] [PubMed] [Google Scholar]
- 19.Uetake H, Luria SE, Burrous JW. 1958. Conversion of somatic antigens in Salmonella by phage infection leading to lysis or lysogeny. Virology 5:68–91 [DOI] [PubMed] [Google Scholar]
- 20.Losick R. 1969. Isolation of a trypsin-sensitive inhibitor of O-antigen synthesis involved in lysogenic conversion by bacteriophage ε15. J. Mol. Biol. 42:237–246 [DOI] [PubMed] [Google Scholar]
- 21.Sun Q, Lan R, Wang Y, Wang J, Wang Y, Li P, Du P, Xu J. 2013. Isolation and genomic characterization of SfI, a serotype-converting bacteriophage of Shigella flexneri. BMC Microbiol. 13:39. 10.1186/1471-2180-13-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark CA, Beltrame J, Manning PA. 1991. The oac gene encoding a lipopolysaccharide O-antigen acetylase maps adjacent to the integrase-encoding gene on the genome of Shigella flexneri bacteriophage Sf6. Gene 107:43–52 [DOI] [PubMed] [Google Scholar]
- 23.Holloway BW, Cooper GN. 1962. Lysogenic conversion in Pseudomonas aeruginosa. J. Bacteriol. 84:1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuzio J, Kropinski AM. 1983. O-antigen conversion in Pseudomonas aeruginosa PAO1 by bacteriophage D3. J. Bacteriol. 155:203–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kropinski AM. 2000. Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J. Bacteriol. 182:6066–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam JS, MacDonald LA, Lam MY, Duchesne LG, Southam GG. 1987. Production and characterization of monoclonal antibodies against serotype strains of Pseudomonas eruginosa. Infect. Immun. 55:1051–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton GJ, Daniels C, Burrows LL, Kropinski AM, Clarke AJ, Lam JS. 2001. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol. Microbiol. 39:1237–1247 [DOI] [PubMed] [Google Scholar]
- 28.Kaluzny K, Abeyrathne PD, Lam JS. 2007. Coexistence of two distinct versions of O-antigen polymerase, Wzy-α and Wzy-β, in Pseudomonas aeruginosa serogroup O2 and their contributions to cell surface diversity. J. Bacteriol. 189:4141–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 74:7422–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hitchcock PJ, Brown TM. 1983. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Islam ST, Lam JS. 2013. Topological mapping methods for α-helical bacterial membrane proteins—an update and a guide. Microbiologyopen 2:350–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernsel A, Viklund H, Hennerdal A, Elofsson A. 2009. TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37:W465–W468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tusnády GE, Simon I. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489–506 [DOI] [PubMed] [Google Scholar]
- 34.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markova model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 35.Käll L, Krogh A, Sonnhammer ELL. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027–1036 [DOI] [PubMed] [Google Scholar]
- 36.Hofmann K, Stoffel W. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166 [Google Scholar]
- 37.Reynolds SM, Käll L, Riffle ME, Bilmes JA, Noble WS. 2008. Transmembrane topology and signal peptide prediction using dynamic Bayesian networks. PLoS Comput. Biol. 4:e1000213. 10.1371/journal.pcbi.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Heijne G. 1992. Membrane protein structure prediction: hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487–494 [DOI] [PubMed] [Google Scholar]
- 39.Rost B, Yachdav G, Liu J. 2004. The PredictProtein server. Nucleic Acids Res. 32:W321–W326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 42.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West SEH, Schweizer HP, Dall C, Sample AK, Runyen-Janecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86 [DOI] [PubMed] [Google Scholar]
- 44.Nilsson J, Persson B, von Heijne G. 2000. Consensus predictions of membrane protein topology. FEBS Lett. 486:267–269 [DOI] [PubMed] [Google Scholar]
- 45.Fontaine F, Fuchs RT, Storz G. 2011. Membrane localization of small proteins in Escherichia coli. J. Biol. Chem. 286:32464–32474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heijne G. 1986. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 5:3021–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barr K, Klena J, Rick PD. 1999. The modality of enterobacterial common antigen polysaccharide chain lengths is regulated by o349 of the wec gene cluster of Escherichia coli K-12. J. Bacteriol. 181:6564–6568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75:39–68 [DOI] [PubMed] [Google Scholar]
- 49.Papadopoulos M, Morona R. 2010. Mutagenesis and chemical cross-linking suggest that Wzz dimer stability and oligomerization affect lipopolysaccharide O-antigen modal chain length control. J. Bacteriol. 192:3385–3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kintz EN, Goldberg JB. 2011. Site-directed mutagenesis reveals key residue for O antigen chain length regulation and protein stability in Pseudomonas aeruginosa Wzz2. J. Biol. Chem. 286:44277–44284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larue K, Kimber MS, Ford R, Whitfield C. 2009. Biochemical and structural analysis of bacterial O-antigen chain length regulator proteins reveals a conserved quaternary structure. J. Biol. Chem. 284:7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larue K, Ford RC, Willis LM, Whitfield C. 2011. Functional and structural characterization of polysaccharide co-polymerase proteins required for polymer export in ATP-binding cassette transporter-dependent capsule biosynthesis pathways. J. Biol. Chem. 286:16658–16668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tocilj A, Munger C, Proteau A, Morona R, Purins L, Ajamian E, Wagner J, Papadopoulos M, Van Den Bosch L, Rubinstein JL, Fethiere J, Matte A, Cygler M. 2008. Bacterial polysaccharide co-polymerases share a common framework for control of polymer length. Nat. Struct. Mol. Biol. 15:130–138 [DOI] [PubMed] [Google Scholar]
- 54.Kalynych S, Yao D, Magee J, Cygler M. 2012. Structural characterization of closely related O-antigen lipopolysaccharide (LPS) chain length regulators. J. Biol. Chem. 287:15696–15705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalynych S, Ruan X, Valvano MA, Cygler M. 2011. Structure-guided investigation of lipopolysaccharide O-antigen chain length regulators reveals regions critical for modal length control. J. Bacteriol. 193:3710–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carter JA, Jiménez JC, Zaldívar M, Álvarez SA, Marolda CL, Valvano MA, Contreras I. 2009. The cellular level of O-antigen polymerase Wzy determines chain length regulation by WzzB and WzzpHS-2 in Shigella flexneri 2a. Microbiology 155:3260–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalynych S, Valvano MA, Cygler M. 2012. Polysaccharide co-polymerases: the enigmatic conductors of the O-antigen assembly orchestra. Protein Eng. Des. Sel. 25:797–802 [DOI] [PubMed] [Google Scholar]
- 58.Marolda CL, Tatar LD, Alaimo C, Aebi M, Valvano MA. 2006. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O-antigen. J. Bacteriol. 188:5124–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marczak M, Dźwierzyńska M, Skorupska A. 2013. Homo- and heterotypic interactions between Pss proteins involved in the exopolysaccharide transport system in Rhizobium leguminosarum bv. trifolii. Biol. Chem. 394:541–559 [DOI] [PubMed] [Google Scholar]
- 60.Daniels C, Morona R. 1999. Analysis of Shigella flexneri Wzz (Rol) function by mutagenesis and cross-linking: Wzz is able to oligomerize. Mol. Microbiol. 34:181–194 [DOI] [PubMed] [Google Scholar]
- 61.Hancock RE, Carey AM. 1979. Outer membrane of Pseudomonas aeruginosa: heat-2-mercaptoethanol-modifiable proteins. J. Bacteriol. 140:902–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


