Abstract
Mycobacterium tuberculosis is one of the strongest reducers of nitrate among all mycobacteria. Reduction of nitrate to nitrite, mediated by nitrate reductase (NarGHJI) of M. tuberculosis, is induced during the dormant stage, and the enzyme has a respiratory function in the absence of oxygen. Nitrite reductase (NirBD) is also functional during aerobic growth when nitrite is the sole nitrogen source. However, the role of NirBD-mediated nitrite reduction during the dormancy is not yet characterized. Here, we analyzed nitrite reduction during aerobic growth as well as in a hypoxic dormancy model of M. tuberculosis in vitro. When nitrite was used as the sole nitrogen source in the medium, the organism grew and the reduction of nitrite was evident in both hypoxic and aerobic cultures of M. tuberculosis. Remarkably, the hypoxic culture of M. tuberculosis, compared to the aerobic culture, showed 32- and 4-fold-increased expression of nitrite reductase (NirBD) at the transcription and protein levels, respectively. More importantly, a nirBD mutant of M. tuberculosis was unable to reduce nitrite and compared to the wild-type (WT) strain had a >2-log reduction in viability after 240 h in the Wayne model of hypoxic dormancy. Dependence of M. tuberculosis on nitrite reductase (NirBD) was also seen in a human macrophage-based dormancy model where the nirBD mutant was impaired for survival compared to the WT strain. Overall, the increased expression and essentiality of nitrite reductase in the in vitro dormancy models suggested that NirBD-mediated nitrite reduction could be critical during the persistent stage of M. tuberculosis.
INTRODUCTION
Tuberculosis (TB) accounts for 1.4 million deaths annually and remains a serious health problem worldwide (1). The ability of the causative agent, Mycobacterium tuberculosis, to adapt to changing hostile environments within the host and shift into a dormant state has been recognized as one of the major reasons for its successful survival in humans (2). The nonreplicating persistent form not only helps the pathogen escape from the host defense mechanisms but also gives the bacilli the advantage of remaining unaffected by standard antitubercular drugs (3). Long-term persistence of M. tuberculosis in the latent stage, during antitubercular therapy, could also assist the pathogen to develop not just tolerance but resistance to currently used drugs (4). Therapeutic intervention which can target and kill the dormant tubercle bacilli therefore would not only provide a novel approach to combat TB but could also pave the way for complete eradication of the disease. A comprehensive knowledge of the metabolic state and physiology of M. tuberculosis during latent disease is required in order to discover such therapeutic options. The limited methods available for studying dormancy in M. tuberculosis, though not well characterized, have shown some degree of resemblance to actual in vivo latency (5–7). Based on the observation that human lung granulomas, where the TB bacilli reside, are hypoxic, an in vitro model in which oxygen depletion is induced gradually has been commonly used to study dormancy of M. tuberculosis (5, 8). In this model, replication ceases as the oxygen level decreases, and certain changes in energy metabolism are observed, including increased nitrate reductase activity (9). While the expression of nitrate reductase (NarGHJI) remained constant, the increase in the nitrate reductase activity during hypoxia was due to the increased expression of a nitrate transporter, NarK2 (10). Mycobacterium smegmatis was also reported to express a respiratory nitrate reductase and to have the ability to undergo hypoxia-induced in vitro dormancy, similar to M. tuberculosis (11, 12). Assimilatory reduction of nitrite produced by NarGHJI was also evident in this saprophytic strain, and a complete pathway of nitrate assimilation was discovered with the use of a minimal medium having nitrate, nitrite, or ammonium as the sole nitrogen source (13). By employing the defined nitrogen source for growth, the presence of functional nitrite reductase (NirBD) was confirmed in M. tuberculosis as well (14). However, the reduction of nitrite during the dormant stage of M. tuberculosis was not evaluated yet.
In this study, we characterized the reduction of nitrite and the expression of nirBD in the nonreplicating hypoxic stage, to evaluate the role of nitrite reductase (NirBD) during dormancy of M. tuberculosis. Survival of a nirBD mutant of M. tuberculosis in the Wayne hypoxic model of tuberculosis, as well as in a human macrophage-based dormancy model, was also compared with that of the wild-type (WT) strain to further determine the importance of nitrite reductase during persistent infection.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
M. tuberculosis H37Ra (ATCC 25177) was obtained from IMTECH, Chandigarh, India, and M. tuberculosis H37Rv (ATCC 27294) was obtained from the ATCC, Manassas, VA. The stock cultures of both the strains were first subcultured in 7H9 broth to an optical density at 620 nm (OD620) of ∼1.0 (14). The cultures were then washed three times with phosphate-buffered saline (PBS), and the OD620 was adjusted to 0.01 at the beginning of an experimental culture. Growth and reduction of nitrite in the culture were tested under nitrogen-limiting conditions using Mycobacterium phlei medium supplemented with NaNO3 (10 mM), NaNO2 (1 mM), or asparagine (7.5 mM) as the sole nitrogen source (13). One liter of this medium contained 5 g KH2PO4, 2.5 g sodium citrate, 0.60 g MgSO4, and 20 ml glycerol, and the pH was adjusted to 6.6 ± 0.2.
For aerobic cultivation, bacterial cultures were grown in 30 ml M. phlei medium in a 100-ml flask with an initial inoculum of ∼105 (OD620 ∼ 0.01) cells per ml. The flask was then kept under aerobic conditions in a shaker incubator (model 481; Thermo Electron Corporation) maintained at 150 rpm and 37°C. For cultivation of anaerobic dormant bacilli, the Wayne 0.5 HSR (headspace ratio) model was used with a starting inoculum of ∼105 cells/ml (5).
The number of viable bacilli at different time points in both aerobic and Wayne cultures was determined by first sonicating (Sonics VibraCell sonicator; 4 W for 60 s) the cultures to break up clumps and then spreading dilutions on 7H9 solid agar medium.
Estimation of nitrite in whole-cell culture.
Nitrite concentration in cultures was measured by the Griess method (15). Briefly, 1 ml of the culture was added to 1 ml of 1% sulfanilic acid (in 20% HCl) and 1 ml of 1% naphthylenediamine dihydrochloride (NEDD) solution. The reaction mixture was incubated for 15 min to develop a pink color. The absorbance of the supernatant was measured at 540 nm, and nitrite concentration was quantified; the values were compared to a standard curve of nitrite.
Generation of nirBD mutant of M. tuberculosis.
The nirBD operon was amplified from M. tuberculosis with the primers 5′-AGGGTCGAGCTCGACGTTGACGTCCTTGTC-3′ and 5′-GGTGATCTAGACCGCTACCCGCGCGACCTG-3′. The underlined bases indicate mismatches used to create SacI and XbaI restriction sites. The 3,387-bp fragment was cloned into pST-Blue (Novagen) vector, and sequenced. nirBD was subcloned into the suicide vector pJQ200SK (16) by cutting both plasmids with XbaI and SacI. Next, aph, a kanamycin resistance marker, was inserted into the NcoI site resulting in the deletion of part of nirB and nirD. This plasmid was electroporated into M. tuberculosis H37Rv, and knockout mutants were identified as previously described (10).
Expression analysis and quantification of mRNA levels.
To isolate total RNA, the spheroplast-based method was applied to aerobic and hypoxia-induced dormant cultures (17). Briefly, a spheroplast solution consisting of 0.002% lysozyme, 0.006% d-cycloserine, 1.4% glycine, 0.2% EDTA, and 0.1% lithium chloride (wt/vol) in distilled water was aseptically added to an M. tuberculosis culture grown aerobically to an OD620 of ∼1.0 or anaerobically in the Wayne model on the 7th day with different nitrogen sources. The treated cells were then used for total RNA isolation by the TRIzol extraction. A 1-μg portion of total RNA isolated from in vitro- and ex vivo-grown mycobacteria was treated with DNase I (Sigma) and then incubated at 70°C according to the manufacturers' instruction. DNase I-treated total RNA was used for cDNA synthesis using random primers and enhanced avian reverse transcriptase provided in the first-strand cDNA synthesis kit (Sigma) at 25°C for 10 min followed by incubation at 45°C for 50 min. The resulting cDNA was used as a template for PCR amplification.
PCR was carried out by using Taq DNA polymerase provided in the PCR core kit (Sigma) in a total volume of 50 μl. The amplification PCR product was first analyzed on a 2% agarose gel containing 1% (vol/vol) SafeView dye followed by nucleotide sequencing to verify the fragment.
Real-time quantitative PCR was performed with a brilliant SYBR green quantitative PCR (qPCR) master mix kit (Sigma, St. Louis, MO). Reactions were carried out in a volume of 25 μl, and the reaction mixtures consisted of a 0.05 μM concentration of forward and reverse primers, 12.5 μl of 2× master mix, and 2.5 μl of cDNA. Controls with no cDNA template were included in each run. The internal control for each reaction was 16S gene amplification.
The PCR parameters were as follows: (i) an initial denaturation step of 2 min at 95°C; (ii) 40 cycles of 30 s at 95°C, 30 s at the respective annealing temperature, and 30 s for extension at 72°C; and (iii) a final extension step of 7 min at 72°C. A melting curve analysis was then performed. All samples were run on a 2% agarose gel containing 1% (vol/vol) SafeView dye to verify that only a single band was produced. Each experiment was done three times with independent RNA samples isolated from similar conditions (primer sequences are given in Table 1).
Table 1.
Primer sequences, annealing temperatures, and amplification sizes
| Gene | Direction | Primer sequence | Annealing temp (°C) | Amplification size (bp) |
|---|---|---|---|---|
| narG | F | 5′-ACTACGCCGACAACACCAAGTTCGCCGACG-3′ | 68 | 158 |
| R | 5′-AGCGGCGCACATAGTCGACAAAGAACGGAA-3′ | |||
| nirB | F | 5′-GTCCCGGTTCGTTTCCTTCG-3′ | 68 | 155 |
| R | 5′- CGCGGGATACCAATGGACAC-3′ | |||
| glnA | F | 5′-CAACTTCTTTGTGCACGACCCGTT-3′ | 64 | 423 |
| R | 5′-AACTGGTAGTTGATCTCGGCCTGT-3′ | |||
| narK2 | F | 5′-TGCTTCGTGATGCACCCTACTTTCGGCCCA-3′ | 68 | 120 |
| R | 5′-CCGCCGAACACGATCGCGTACAGAAACGAC-3′ | |||
| 16Sa | F | 5′-ATGCATGTCTTGTGGTGGAAAGCG-3′ | 58 | 350 |
| R | 5′-TTCACGAACAACGCGACAAACCAC-3′ |
16S gene PCR was done for 25 cycles, while PCRs of other genes were done for 35 cycles.
Preparation of cell extracts and Nir enzyme assay.
For the nitrite reductase enzyme assay, spheroplast solution was added to M. tuberculosis cultures grown aerobically (up to an OD620 of ∼1.0) and anaerobically in the 0.5 HSR (headspace ratio) Wayne model (up to 7 days) (5). After incubation for 1 h, 50 ml of bacterial cells were concentrated using centrifugation at 10,000 rpm for 10 min at 4°C. The pellet obtained was washed twice with 2 ml of potassium phosphate buffer (50 mM, pH 6.6 ± 0.2) and resuspended in 2 ml of potassium phosphate buffer (50 mM, pH 7.0) containing protease inhibitor (protease cocktail; Sigma). The prepared mixture was sonicated in a water bath for 5 min at 50 kHz. The lysate was obtained after removal of unbroken cells by centrifugation at 10,000 rpm for 5 min at 4°C. The lysate was transferred to another tube and ultracentrifuged at 100,000 rpm for 1 h at 4°C. After centrifugation, the supernatant was again transferred to another tube while the pellet, including the membrane fraction, was resuspended in 2 ml of potassium phosphate buffer containing protease inhibitor. After the total protein concentrations in the supernatant and membrane fractions had been determined using the Bradford method, the nitrite reductase assay was done as described previously (18, 19). Briefly, the final assay mixture contained 1.6 ml of sodium phosphate buffer (50 mM, pH 7.2), 100 μl of methyl viologen (0.01%), 50 μl of enzyme (50 μg/ml), 50 μl of sodium nitrite (1 mM), and 200 μl sodium dithionite-sodium bicarbonate solution (0.8%). The tube was sealed with Parafilm and kept in a 37°C water bath incubator for 30 min. After 30 min of incubation, the assay reaction was stopped by vigorous shaking till the dark blue color disappeared. The amount of nitrite remaining as the substrate in the enzyme assay mixture was measured using the Griess method and compared with that in a control assay where whole-cell extract was not added.
THP-1 monocytes based in vitro model of intracellular dormancy of tubercle bacilli.
Human THP-1 monocytoid cell line (TIB3456; ATCC, Manassas, VA) was maintained at 37°C and 5% CO2 in HEPES-buffered RPMI 1640 (Sigma-Aldrich, St. Louis, MO) medium with 10% heat-inactivated fetal bovine serum (FBS), 50 μg/ml gentamicin, and 100 μg/ml penicillin (pH 7.2). Cells were seeded at 106/well in a 24-well plate free of antibiotics and were activated with 1 μM retinoic acid and vitamin D (cholecalceferol) (RAVD). After 72 h treatment with RAVD, cells were infected with M. tuberculosis or the nirBD mutant at a multiplicity of infection (MOI) of 1:10. At 24 h postinfection, the cells were washed with medium to remove unphagocytosed bacteria. Cells were then incubated in a CO2 incubator at 37°C and treated with RAVD every 72 h. The intracellular bacterial burden was determined at different time points by lysing the macrophages with a 0.05% SDS solution, plating dilutions on 7H11 agar plates, and counting the colonies after 21 days of incubation at 37°C. A stable number of viable bacilli along with the formation of multinucleated giant cells (MNGC) was used as an indicator of onset of intracellular dormancy of M. tuberculosis (20).
Data and statistical analysis.
Data were analyzed using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA). Results are expressed as means ± standard deviations (SD) unless otherwise stated. Student's t test and standard one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparison test were used to determine statistical significance. A P value of <0.05 was considered significant.
RESULTS
Reduction of nitrite by M. tuberculosis during aerobic growth and hypoxic dormancy in vitro.
An assimilatory function of nitrite reductase (NirBD) of M. tuberculosis during its active replicating stage was previously reported, when WT M. tuberculosis but not the nirBD mutant grew on nitrite as the sole nitrogen source (14). However, the reduction of nitrite by nitrite reductase (NirBD) remained unknown during the dormant stage of the pathogen, when it does not multiply. We used the Wayne model of dormancy, which uses gradual oxygen depletion in vitro, to analyze the reduction of nitrite during the nonreplicating persistence of M. tuberculosis (5). Since nitrite reduction was detected when nitrate or nitrite was used as the nitrogen source in the medium, we used an M. phlei medium having nitrite as the sole nitrogen source to cultivate M. tuberculosis (13). We used avirulent M. tuberculosis H37Ra in this experiment, whose NirBD protein has 100% amino acid similarity with the NirBD of M. tuberculosis H37Rv.
The time-dependent reduction of nitrite was measured along with the CFU in the Wayne model as well as in the aerobic culture (Fig. 1). In the aerobic culture, the growth of M. tuberculosis on nitrite as the sole nitrogen source, with a concurrent time-dependent depletion of nitrite from the medium, confirmed that the organism reduced nitrite for assimilation (Fig. 1). The observed growth is due to assimilation of nitrite and not because of formation of ammonia or other contaminants in the medium.
Fig 1.
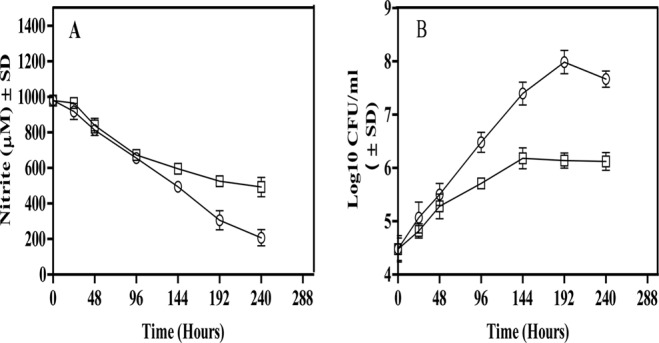
Nitrite reduction and CFU of M. tuberculosis H37Ra during aerobic growth and in the Wayne model. Bacteria were cultured with 1 mM nitrite as the sole nitrogen source. (A) Nitrite reduction was determined by testing aliquots at the indicated time points for depletion of nitrite from the medium. (B) Growth of the organism was determined by testing aliquots at the indicated time points by plating on the 7H11 agar plates to enumerate CFU. Circles represent the aerobic cultures, whereas rectangles represent the hypoxic cultures. The results are averages ± SD from three identical experiments.
In the Wayne model, reduction of nitrite was clearly visible in the replicating phase (0 to 144 h) of M. tuberculosis H37Ra, with a steady depletion of nitrite from the culture medium. However, a slow depletion of nitrite was also evident during the nonreplicating phase (144 to 240 h). Although the rate of nitrite depletion during the nonreplicating phase seemed lower, the number of cells was also more than 10-fold lower than that in the aerobically grown culture. Interestingly, when nitrite reduction was calculated in terms of micromoles of nitrite depleted per 106 cells per day, the nonreplicating dormant cultures had an 8- to 12-fold-higher rate of nitrite reduction at hours 144, 192, and 240 than the aerobic culture (Table 2). Reduction of nitrite by nonreplicating M. tuberculosis confirmed that nitrite reductase (NirBD) was active during dormancy. Second, the increased reduction rate of nitrite by the hypoxic culture of M. tuberculosis compared to the aerobic culture also suggested a potentially vital role of the NirBD in hypoxic metabolism of the pathogen.
Table 2.
Rate of nitrite reduction by M. tuberculosis H37Ra under aerobic and Wayne hypoxic culture conditions
| Time (h) | Reduction rate (mean ±SD)a |
|
|---|---|---|
| Aerobic (+O2) | Hypoxic (−O2) | |
| 144 | 2.37 ± 0.28 | 19.57 ± 2.13 |
| 192 | 1.89 ± 0.17 | 21.73 ± 1.68 |
| 240 | 1.74 ± 0.13 | 17.38 ± 2.021 |
Micromoles nitrite reduced/48 h/106 cells of M. tuberculosis.
Comparative expression of nirB at the transcriptional and translational level during aerobic and hypoxic conditions by M. tuberculosis.
Since we observed an increased rate of nitrite reduction by nonreplicating cultures compared to the aerobic replicating cultures, we investigated whether the expression of nitrite reductase (NirBD) at the transcriptional and translational levels also differed between these two cultures.
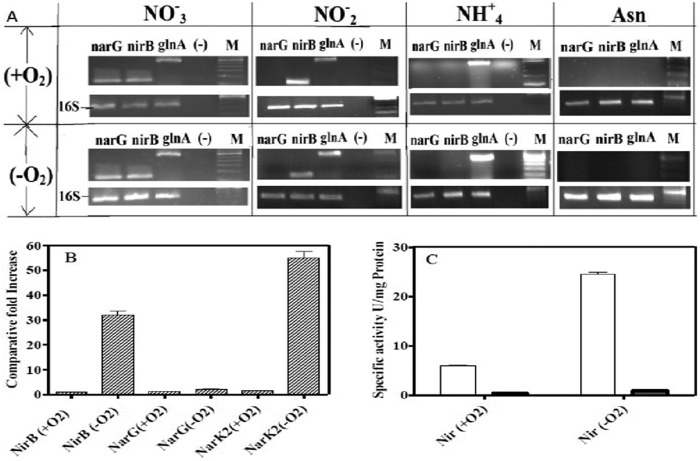
To analyze nitrite reductase gene expression, RT-PCR was done using cDNA synthesized from total RNA isolated from bacilli grown in different nitrogen sources (Fig. 2A). The expression of nitrate reductase MRA_1172 (narG), nitrite reductase MRA_261 (nirB), and glutamine synthetase MRA_2230 (glnA1) in aerobic as well as hypoxic dormant cultures was seen when nitrate was used as the sole nitrogen source. Expression of nirB and glnA1 but not narG was detected when nitrite was used as a nitrogen source, indicating that substrate-dependent transcription of the genes is required for nitrate/nitrite metabolism. In the presence of ammonium, only glnA1 expression was detected, while none of the nitrate- or nitrite-metabolizing genes were expressed when asparagine was the nitrogen source. This further confirmed that in both aerobic and dormant cultures of M. tuberculosis H37Ra, the transcription of nirB is seen only when nitrite is present.
Fig 2.
(A) Expression of M. tuberculosis genes involved in nitrate/nitrite metabolism under aerobic and Wayne hypoxic conditions with different nitrogen sources. cDNA was prepared from total RNA isolated from bacilli grown in the presence of nitrate, nitrite, ammonium, or asparagine either aerobically (+O2) or in the Wayne model (−O2). cDNA was used as the template for PCR amplification using gene-specific primers for narG (nitrate reductase), nirB (nitrite reductase), and glnA1 (glutamine synthetase), and the amplified products are shown after electrophoresis on agarose gels. Lane (-), negative control without cDNA; lane M, 100-bp marker (Invitrogen). (B) Quantitative PCR analysis of nirB, narG, and narK2 expression in bacilli grown in the presence of nitrite as the nitrogen source under different conditions. cDNA was prepared from the M. tuberculosis culture grown under aerobic (+O2) or hypoxic (−O2) conditions. The relative levels of the transcripts are shown as the difference from that obtained under aerobic conditions, while the 16S gene was used as an internal control. (C) Specific activity of nitrite reductase enzymes in the cytoplasm and membrane fractions of M. tuberculosis cells grown in the presence of nitrite as the nitrogen source. Specific enzyme activity was measured during mid-logarithmic phase for aerobic cultures (+O2) and on the 7th day in Wayne model cultures (−O2). White bars represent the Nir activity measured in cytoplasmic fractions, whereas gray bars represent Nir activity measured in membrane fractions. One unit of specific activity of enzyme was defined as 1 μM NO2− depleted per min/mg of total protein during the in vitro Nir enzyme assay.
Quantitative PCR of nirB was done to compare the transcript levels of nirB gene between aerobic replicating and hypoxic dormant cultures of M. tuberculosis (Fig. 2B). It was observed that the level of nirB expression during the nonreplicating dormancy was ∼32-fold higher than that during aerobic growth. The transcript levels of narK2 were increased 62-fold during the nonreplicating stage and served as positive controls for this study due to their known induction during dormancy (10).
The expression of nirB was also measured at the translational level by determining the specific activity of nitrite reductase in cell extracts obtained from aerobic and hypoxic cultures of M. tuberculosis H37Ra (Fig. 2C). The cytoplasmic fraction of cell extract obtained from the Wayne model showed a 4-fold increase in the specific activity of nitrite reductase compared to that in the extracts from aerobically grown M. tuberculosis H37Ra. The membrane fraction of cell extracts did not show any discernible nitrite reductase activity, which also confirmed the cytoplasmic location of NirBD. The increased expression of nirB at both the transcriptional and translational levels correlated with the increased rate of nitrite reduction during the nonreplicating stage and suggested that the nitrite reductase (NirBD) could have an important function during dormancy of M. tuberculosis.
Growth and survival of a nirBD mutant of M. tuberculosis in the Wayne model.
The inability of nirBD-deficient M. tuberculosis to grow and assimilate nitrite was previously observed only under aerobic culture condition (14). Since the assimilation of nitrite may not be required when the organism is not multiplying, it was of interest to examine whether M. tuberculosis could survive without a functional nitrite reductase during hypoxia. Survival of the nirBD mutant compared to WT M. tuberculosis was therefore evaluated in the Wayne model. Given that the nirBD mutant was unable to grow when nitrite was used as the sole nitrogen source, asparagine was also added to the medium to allow growth.
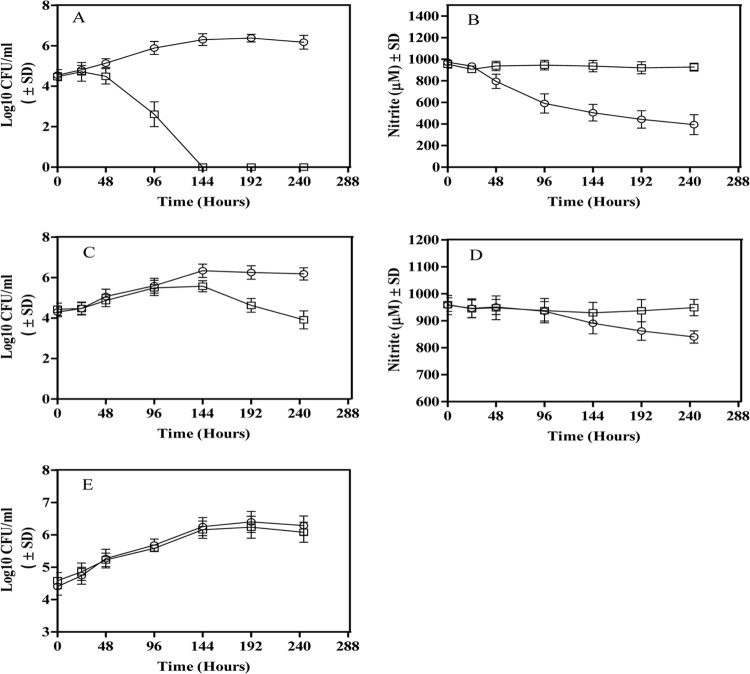
The reduction of nitrite and the viability of the nirBD mutant were then compared to those of WT M. tuberculosis H37Rv in the Wayne model (Fig. 3). The genetic similarity of nitrite reductases in virulent and avirulent M. tuberculosis was corroborated at the functional level when H37Rv showed a pattern of growth and nitrite reduction similar to that in the Wayne model with nitrite as the sole nitrogen source (Fig. 1; Fig. 3A and B). The nirBD mutant was unable to assimilate nitrite with no indication of growth or nitrite reduction in the Wayne model when nitrite was used as the sole nitrogen source.
Fig 3.
Viability and nitrite reduction by the nirBD mutant and WT M. tuberculosis in the Wayne model with different nitrogen sources. Viability (A, C, and E) and nitrite levels (B and D) in M. tuberculosis H37Rv (○) and the nirBD mutant (□) when nitrite (A and B) or nitrite and asparagine (C and D) or asparagine only (E) was the nitrogen source. The results are averages ± SD from three identical experiments.
Nonetheless, in a medium where asparagine was added with nitrite, the mutant showed a growth rate comparable to that of WT M. tuberculosis H37Rv in the replicating phase of the Wayne hypoxic culture (Fig. 3C). Reduction of nitrite by either the nirBD mutant or WT M. tuberculosis was not detected during the replicating phase of Wayne cultures, when nitrite and asparagine were present in the medium (Fig. 3D). However, once cultures shifted down into the nonreplicating phase in the Wayne model, a slow reduction of nitrite by WT was detected. The nirBD mutant did not reduce any discernible amount of nitrite and was unable to maintain viability during this nonreplicating phase in the Wayne model. The viability of the nirBD mutant was reduced by more than 2 logs compared to that of the WT M. tuberculosis at hour 240 in the Wayne hypoxic culture (Fig. 3C). The results thus indicated that the function of NirBD of M. tuberculosis under hypoxic conditions not only could be essential but also could be different from that during the aerobic stage of growth. However, it remained unknown whether NirBD is essential for the survival of M. tuberculosis during the dormant stage when nitrite is not present in the medium. In order to address this question, asparagine was used as the sole nitrogen source in the Wayne cultures, and the survival of the nirBD mutant was examined. With asparagine as the nitrogen source, the viability of nirBD mutant remained comparable to that of WT M. tuberculosis in the Wayne model for at least 240 h (Fig. 3E). Thus, it could be concluded that the nirBD is indispensable when nitrite as a substrate for the enzyme is available.
Survival of the nirBD mutant of M. tuberculosis during intracellular dormancy.
Evidence from the earlier experiment suggested that the expression and functional role of nitrite reductase of M. tuberculosis are dependent on the nitrogen source (Fig. 2 and 3). Since the nitrogen source(s) present in the axenic dormant culture may not mimic what is available to M. tuberculosis during its actual persistence within a cell, we examined the role of nitrite reductase in an intracellular model of dormancy.
A recently developed human macrophage (THP-1)-based model of intracellular dormancy of tuberculosis was employed to analyze the survival of the nirBD mutant (20). Briefly, in this model, the dormancy of M. tuberculosis in THP-1 macrophages is induced by constant treatment with retinoic acid (RA) and vitamin D3 (VD). Combined treatment with RA and VD enhances the expression of DC-SIGN (dendritic cell-specific intercellular adhesion molecule-3-grabbing nonintegrin) and mannose receptors on THP-1, which inhibits the subsequent intracellular growth of M. tuberculosis and induces the dormancy phenotype of the pathogen. After treatment of infected macrophages with RA and VD for a certain period of time, formation of multinucleated giant cells (MNGCs) confirms the establishment of dormancy of M. tuberculosis.
The viability of the nirBD mutant and WT M. tuberculosis was monitored in this model to determine the contribution of nitrite reductase during intracellular dormancy (Fig. 4). After the initial time point, both the WT and the nirBD mutant showed decreasing viability. After 15 days, the tubercle bacilli entered dormancy (as evidenced by the formation of MNGC), and the viable count remained constant for WT for the next 25 days (Fig. 4A). In contrast, the number of viable nirBD mutants continued to decrease, although at a reduced rate. At day 25 after the onset of dormancy, the number of viable bacilli of nirBD mutant was reduced by more than 1.5 logs compared to the WT M. tuberculosis. The inability of nirBD mutant to maintain its viability in this intracellular dormancy model of tuberculosis further strengthened the idea that nitrite reductase is involved in supporting the survival of M. tuberculosis during latency. More interestingly, the dependence of M. tuberculosis on nitrite reductase could be seen even without external addition of nitrite in this intracellular dormancy.
Fig 4.
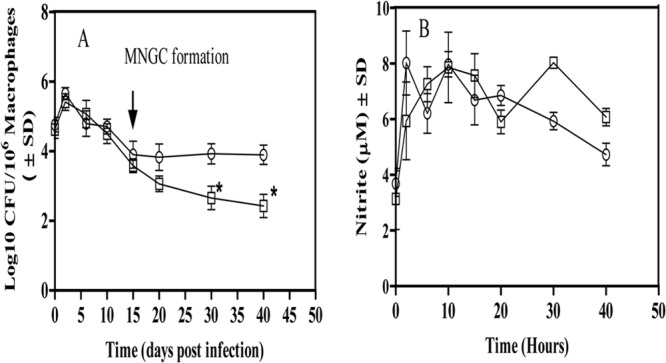
(A) Survival of the nirBD mutant and WT M. tuberculosis H37Rv in THP-1 macrophages treated with retinoic acid and vitamin D (RAVD). Macrophages were lysed at the indicated time points and plated on 7H11 agar to determine the intracellular burden of bacilli. *, P < 0.05. ↓, MNGC formation, which indicates the induction of dormancy in intracellular M. tuberculosis. (B) Presence of nitrite within THP-1 macrophages infected with the nirBD mutant (□) and WT M. tuberculosis H37Rv (○). The results are averages ± SD from three identical experiments.
In order to know whether the substrate for nitrite reductase is made available by endogenous production of nitrite by M. tuberculosis-infected macrophages, we examined the nitrite levels in the cells harboring dormant bacilli (Fig. 4B). Macrophages infected with the nirBD mutant as well as those infected with WT M. tuberculosis maintained a stable level of ∼5 μM nitrite per 106 cells during the intracellular dormancy. Although it was expected that if nitrite reductase (NirBD) was functional during this intracellular dormancy, the amount of nitrite produced by macrophages should have decreased with time for WT M. tuberculosis, a constant production of nitrite by intrinsic nitric oxide generation could be the possible reason for stable levels of nitrite detection (21).
DISCUSSION
One of the key factors in the success of M. tuberculosis as a pathogen is its ability to persist for extended periods within the human host in a dormant state. The understanding of metabolic pathways operative during latent tuberculosis is an active area of research. Inhibition of aerobic respiration by either hypoxia, nitric oxide, or carbon monoxide is proposed to be the key signal driving M. tuberculosis into a nonreplicating persistent state. The discovery of microbial factors which allow the pathogen to adapt to this stress could identify vital drug targets.
The response of M. tuberculosis to low oxygen is one of the best characterized of all responses to environmental triggers which induce dormancy. The reduction of nitrate to nitrite increases during hypoxia as the bacteria use nitrate as an alternative terminal electron acceptor (9, 10). The enzyme responsible, NarGHJI, is representative of the bacterial respiratory nitrate reductases. However, it can also play an assimilatory role during aerobic condition and allow M. tuberculosis to replicate when nitrate is the sole nitrogen source (14, 22). In this case, nitrite is further reduced to ammonium by the NirBD nitrite reductase, as was confirmed when the nirB mutant failed to grow on either nitrate or nitrite (14). Although it was known that NarGHJI could play a respiratory and assimilatory role, whether the same is true of for NirBD was not known.
In this study, we discovered that NirBD-mediated reduction of nitrite also occurred during the hypoxic dormant stage. Indeed, nitrite reduction was at a higher rate than in the aerobic growth phase when nitrite was the nitrogen source (Fig. 1 and Table 2). The addition of asparagine to the medium resulted in a decrease in the rate of nitrite reduction (Fig. 3).
The fact that the reduction in the viability of nirBD mutant compared to the WT was seen only during the hypoxic stage suggested that the function of NirBD of M. tuberculosis might be different under aerobic and anaerobic conditions. The high level of asparagine (7.5 mM) in comparison to nitrite (1 mM) suggests that the asparagine was not depleted as the cultures entered dormancy, which is further supported by the persistence of both strains in the Wayne model with asparagine only (Fig. 3E).
The nirBD mutant had reduced survival in macrophages (Fig. 4). This was analyzed by utilizing a macrophage cell culture system that allowed longer experimental times, up to 40 days (20). The indispensability of NirBD could indicate that M. tuberculosis is subjected to hypoxia in the macrophage. Indeed, prior studies have shown that the intraphagosomal oxygen levels in M. tuberculosis-infected macrophages are lower than the cytosolic oxygen concentration, supporting the notion that hypoxia could be associated universally with dormancy (23, 24).
There are several possible roles that nitrite reduction may play. One is the assimilation of nitrite as a nitrogen source. This occurs in vitro in culture with nitrite or nitrate as the only nitrogen source. The reduction of nitrite to ammonium consumes 6 electrons and is energetically costly, which may explain why M. tuberculosis prefers asparagine over nitrite. There was no induction of nirB in the presence of ammonium or asparagine (Fig. 2A).
There is very little information on the nitrogen sources available to M. tuberculosis within macrophages. Amino acid biosynthesis mutants of M. tuberculosis and M. bovis do not replicate in macrophages (25–27) and are attenuated in mice (28, 29), suggesting that the phagosome is limiting for amino acids. Free amino acids and peptides are not thought to be the main source of nitrogen. Nitrite and nitrate, the main breakdown products of nitric oxide, may serve as significant nitrogen sources for M. tuberculosis at times. However, the medium used to grow macrophages in vitro, RPMI, contains high levels of glutamine, which results in abnormally high levels of glutamine in the phagosome (27). This makes the detection of an assimilatory role for NirBD difficult.
The increased rate of nitrite reduction when the pathogen is not replicating suggests a possible role in respiration. The attenuation of the M. tuberculosis nirBD mutant during hypoxic dormancy even when other nitrogen sources besides nitrite were available further supports a nonassimilatory function. Under these conditions, the expression of nirBD was dependent on the presence of nitrite and lack of oxygen (Fig. 2). The M. tuberculosis nirBD gene product is not predicted to be membrane bound and hence was not detected in the membrane (Fig. 2B), unlike the Nrf nitrite reductase system of Escherichia coli, and thus would not contribute to the proton gradient (30). Nitrite reduction is a NAD(P)H-dependent process, and a role for NirBD in this process to maintain the NADH/NAD balance during an interruption in aerobic respiration is possible. The higher cost in electrons for the reduction of nitrite would be a benefit under these conditions. A similar role has been proposed for nitrate reductase in M. tuberculosis, which is also required for survival in human macrophages (10, 31).
A role in nitrite and nitric oxide detoxification is also possible. Nitrite has been shown to be toxic to M. tuberculosis and therefore must be exported or converted to ammonium, and under hypoxia conditions, it is exported (32). The nitrite reductase NirBD of E. coli is involved mainly in detoxification of internal nitrite (33). Nitric oxide can also be produced from the dismutation of nitrite. Nitric oxide is bactericidal at high levels and more stable in the absence of oxygen (34). This nitric oxide may be responsible for the decreased viability seen with the nirBD mutant in the Wayne model (Fig. 3C).
The production of ammonium to reduce the acidity of the environment of the phagosome is another possible role for nitrite reductase. It has been reported that secretion of ammonium by M. tuberculosis inhibits the phagosome-lysosome fusion (35). NirBD could be one source of this ammonium production from nitrite.
Nitrite could be readily available by the spontaneous degradation of nitric oxide in mammalian tissues (21). Organs where the infection of M. tuberculosis is most common, including lungs, liver, and kidneys, can produce 0.1 to 1 mM nitrate and nitrite. Tuberculosis patients have higher levels of these NO breakdown products (nitrate and nitrite) in their serum than control patients (36). Here, a steady level of nitrite was detected in macrophage cultures, suggesting that nitrite would be available during the persistent stage (Fig. 4B).
Nitrite reductase is an important virulence factor for other intracellular pathogens. NO production by macrophages, while toxic in the short term, actually increased long-term survival for the intracellular pathogen Brucella abortus (37). This was proposed to be due to the production of nitrate and nitrite, which were subsequently used as terminal electron acceptors. Both nitrate and nitrite reductase mutants of Campylobacter jejuni showed reduced colonization of chickens (38).
With the development of animal models representing a true latent infection which closely resembles human tuberculosis, it should now be possible to confirm whether the in vitro role of nitrite reductase (NirBD) correlates with the function of the enzyme during in vivo persistence of M. tuberculosis.
ACKNOWLEDGMENTS
We are grateful to CSIR, New Delhi, India, for providing the financial support to carry out the research.
We are also grateful to Sheetal Singh, Halliburton, Pune, India, for significant contributions in designing and editing the manuscript. We are grateful to the Microbial Type Culture Collection (MTCC) Chandigarh, India, for providing M. tuberculosis H37Ra (ATCC 25177).
Footnotes
Published ahead of print 9 August 2013
REFERENCES
- 1.WHO 2012. Tuberculosis. Fact sheet no. 104 WHO, Geneva, Switzerland: http://www.who.int/mediacentre/factsheets/fs104/en/index.html. [Google Scholar]
- 2.Tufariello JM, Chan J, Flynn JL. 2003. Latent tuberculosis: mechanisms of host and bacillus that contribute to persistent infection. Lancet Infect. Dis. 3:578–590 [DOI] [PubMed] [Google Scholar]
- 3.Cosma CL, Sherman DR, Ramakrishnan L. 2003. The secret lives of the pathogenic mycobacteria. Annu. Rev. Microbiol. 57:641–676 [DOI] [PubMed] [Google Scholar]
- 4.Gomez JE, McKinney JD. 2004. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis. 84:29–44 [DOI] [PubMed] [Google Scholar]
- 5.Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717–731 [DOI] [PubMed] [Google Scholar]
- 7.McCune RM, Tompsett R. 1957. Fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. I. The persistence of drug-susceptible tubercle bacilli in the tissues despite prolonged antimicrobial therapy. J. Exp. Med. 104:737–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, Lee EG, Park SK, Raleigh JA, Cho SN, McMurray DN, Flynn JL, Barry CE. 2008. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76:2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wayne LG, Hayes LG. 1998. Nitrate reduction as a marker for hypoxic shiftdown of Mycobacterium tuberculosis. Tuber. Lung Dis. 79:127–132 [DOI] [PubMed] [Google Scholar]
- 10.Sohaskey CD, Wayne LG. 2003. Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J. Bacteriol. 185:7247–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khan A, Sarkar D. 2006. Identification of a respiratory-type nitrate reductase and its role for survival of Mycobacterium smegmatis in Wayne model. Microb. Pathog. 41:90–95 [DOI] [PubMed] [Google Scholar]
- 12.Dick T, Lee BH, Murugasu-Oei B. 1998. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 163:159–164 [DOI] [PubMed] [Google Scholar]
- 13.Khan A, Akhtar S, Ahmad NA, Sarkar D. 2008. Presence of a functional nitrate assimilation pathway in Mycobacterium smegmatis. Microb. Pathog. 44:71–77 [DOI] [PubMed] [Google Scholar]
- 14.Malm S, Tiffert Y, Micklinghoff J, Schultze S, Joost I, Weber I, Horst S, Ackermann B, Schmidt M, Wohlleben W, Ehlers S, Geffers R, Reuther J, Bange FC. 2009. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155:1332–1339 [DOI] [PubMed] [Google Scholar]
- 15.Nicholas DJD, Nason A. 1957. Determination of nitrate and nitrite. Methods Enzymol. 3:1981–1984 [Google Scholar]
- 16.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram negative bacteria. Gene 127:15–21 [DOI] [PubMed] [Google Scholar]
- 17.Akhtar S, Sarkar S, Mishra A, Sarkar D. 2011. A method to extract intact and pure RNA from mycobacteria. Anal. Biochem. 417:286–288 [DOI] [PubMed] [Google Scholar]
- 18.Bradford MM. 1976. A rapid and sensitive for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 19.Sengupta S, Melkote S, Shaila MS, Rao GR. 1996. Purification and characterization of assimilatory nitrite reductase from Candida utilis. Biochem. J. 317:147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estrella JL, Kan-Sutton C, Gong X, Rajagopalan M, Lewis DE, Hunter RL, Eissa NT, Jagannath C. 2011. A novel in vitro human macrophage model to study the persistence of Mycobacterium tuberculosis using vitamin D3 and retinoic acid activated THP-1 macrophages. Front. Microbiol. 67:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelm M. 1999. Nitric oxide metabolism and breakdown. Biochim. Biophys. Acta 1411:273–289 [DOI] [PubMed] [Google Scholar]
- 22.Khan A, Sarkar D. 2012. Nitrate reduction pathways in mycobacteria and their implications during latency. Microbiology 158:301–307 [DOI] [PubMed] [Google Scholar]
- 23.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James PE, Grinberg OY, Michaels G, Swartz HM. 1995. Intraphagosomal oxygen in stimulated macrophages. J. Cell. Physiol. 163:241–247 [DOI] [PubMed] [Google Scholar]
- 25.Bange FC, Brown AM, Jacobs WR., Jr 1996. Leucine auxotrophy restricts growth of Mycobacterium bovis BCG in macrophages. Infect. Immun. 64:1794–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAdam RA, Weisbrod TR, Martin J, Scuderi JD, Brown AM, Cirillo JD, Bloom BR, Jacobs WR., Jr 1995. In vivo growth characteristics of leucine and methionine auxotrophic mutants of Mycobacterium bovis BCG generated by transposon mutagenesis. Infect. Immun. 63:1004–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tullius MV, Harth G, Horwitz MA. 2003. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 71:3927–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hondalus MK, Bardarov S, Russell R, Chan J, Jacobs WR, Jr, Bloom BR. 2000. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect. Immun. 68:2888–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith DA, Parish T, Stoker NG, Bancroft GJ. 2001. Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infect. Immun. 69:1142–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Einsle O. 2011. Structure and function of formate-dependent cytochrome c nitrite reductase, NrfA. Methods Enzymol. 496:399–422 [DOI] [PubMed] [Google Scholar]
- 31.Jung JY, Ranjna ML, Maria G, Rengarajan J, Sohaskey CD, Bange FC, Robinson CM. 2013. The intracellular environment of human macrophages that produce nitric oxide promotes growth of mycobacteria. Infect. Immun. 81:3198–3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giffin MM, Raab RW, Morganstern M, Sohaskey CD. 2012. Mutational analysis of the respiratory nitrate transporter NarK2 of Mycobacterium tuberculosis. PLoS One 7:e45459. 10.1371/journal.pone.0045459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page L, Griffiths L, Cole JA. 1990. Different physiological roles of two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch. Microbiol. 154:349–354 [DOI] [PubMed] [Google Scholar]
- 34.Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. 2011. The response of mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front. Microbiol. 2:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gordon AH, Hart PD, Young MR. 1980. Ammonia inhibits phagosome-lysosome fusion in macrophages. Nature 286:79–80 [DOI] [PubMed] [Google Scholar]
- 36.Dubaniewicz A, Holownia A, Kalinowski L, Wybieralska M, Dobrucki IT, Singh M. 2013. Is mycobacterial heat shock protein 16kDa, a marker of the dormant stage of Mycobacterium tuberculosis, a sarcoid antigen? Hum. Immunol. 74:45–51 [DOI] [PubMed] [Google Scholar]
- 37.Wang M, Qureshi N, Soeurt N, Splitter G. 2001. High levels of nitric oxide production decrease early but increase late survival of Brucella abortus in macrophages. Microb. Pathog. 31:221–230 [DOI] [PubMed] [Google Scholar]
- 38.Weingarten RA, Grimes JL, Olson JW. 2008. Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl. Environ. Microbiol. 74:1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]