Abstract
The essential process of peptidoglycan synthesis requires two enzymatic activities, transpeptidation and transglycosylation. While the PBP2 and PBP3 transpeptidases perform highly specialized functions that are widely conserved, the specific roles of different glycosyltransferases are poorly understood. For example, Caulobacter crescentus encodes six glycosyltransferase paralogs of largely unknown function. Using genetic analyses, we found that Caulobacter glycosyltransferases are primarily redundant but that PbpX is responsible for most of the essential glycosyltransferase activity. Cells containing PbpX as their sole glycosyltransferase are viable, and the loss of pbpX leads to a general defect in the integrity of the cell wall structure even in the presence of the other five glycosyltransferases. However, neither PbpX nor any of its paralogs is required for the specific processes of cell elongation or division, while the cell wall synthesis required for stalk biogenesis is only partially disrupted in several of the glycosyltransferase mutants. Despite their genetic redundancy, Caulobacter glycosyltransferases exhibit different subcellular localizations. We suggest that these enzymes have specialized roles and normally function in distinct subcomplexes but retain the ability to substitute for one another so as to ensure the robustness of the peptidoglycan synthesis process.
INTRODUCTION
The peptidoglycan (PG) cell wall is essential for viability in the majority of bacterial species (reviewed in reference 1). It represents the load-bearing structure that protects bacteria from lysis induced by osmotic shock in the hypotonic environments where most prokaryotes thrive (1). Additionally, the cell wall is directly responsible for defining bacterial morphology. Degradation of PG with lysozyme under isotonic conditions avoids lysis but causes cells to adopt an amorphous shape (2); conversely, an isolated PG sacculus maintains the shape of the cell from which it was derived (3). PG is comprised of unbranched glycan strands containing alternating N-acetylglucosamine and N-acetyl-muramic acid sugars, with these strands cross-linked to each other by short peptide linkages (1, 4, 5). Thus, the overall cell wall structure is thought to incorporate a meshwork of stiff glycan chains and flexible peptide links.
The chemical composition of PG necessitates at least two chemical reactions for the incorporation of new subunits into the preexisting cell wall structure: the glycosyltransferase (GTase) reaction that polymerizes the sugars into glycan strands and the transpeptidation reaction that links the peptides of adjacent strands to generate the cross-linked meshwork. While the chemical composition of the PG and the chemical reactions involved in its synthesis are well characterized (reviewed in reference 6), the way these reactions are arranged in three-dimensional space to accomplish the overall architecture of the cell has remained enigmatic. Nevertheless, studies in rod-shaped bacteria have identified distinct modes of cell wall synthesis that occur at different stages of the cell cycle and rely on different subsets of biosynthetic enzymes and cytoskeletal elements (7, 8, 9, 10). The primary elongation mode is responsible for most of the increase in cell length and requires the MreB cytoskeleton and the PBP2 transpeptidase (TP) (11, 12, 13). The division mode is responsible for the creation of the septum that separates nascent daughter cells during cytokinesis; it relies on the FtsZ cytoskeleton and the PBP3 transpeptidase (11, 14, 15).
While the dedicated roles of the PBP2 and PBP3 TPs in elongation and division are known (reviewed in reference 16), there is conflicting evidence regarding the participation of specific GTases in the different modes of growth. In Escherichia coli, disruptions of PBP1A and PBP1B, the two principal GTases, are synthetically lethal (17, 18), but their individual deletions fail to strongly perturb either cell growth or morphology (19, 20, 21), indicating that both enzymes are capable of catalyzing transglycosylation involved in all modes of growth. However, in wild-type cells, only PBP1A interacts with the elongation-specific PBP2 and only PBP1B interacts with PBP3 (21, 22). Moreover, the roles of the other two E. coli GTases, PBP1C and MtgA, are unclear. In Bacillus subtilis, deletion of ponA, which encodes a PBP1 homolog, causes mild defects both in elongation and division, and PonA localizes to the lateral wall or to the division plane depending on the stage of the cell cycle (23).
Here, we examine the roles of the GTases in the vibrioid, stalked organism Caulobacter crescentus. Like all rod-like bacteria, Caulobacter cells require PG synthesis for elongation and septation (16). Moreover, these curved cells must also distinguish the convex and concave sides of the cell (24) and synthesize a thin, PG-containing appendage called the stalk (25). We show that PbpX acts as the primary Caulobacter GTase. The loss of PbpX leads to reduced cell wall integrity and stalk length and increases the sensitivity to PG-targeting antibiotics, while PbpX alone supports sufficient transglycosylation for growth. Additionally, while the distinct subcellular localizations of Caulobacter GTases imply different functions in the PG synthesis pathway, these functions appear to be redundant. We propose that subsets of GTases perform overlapping functions and that enzymes within a particular subset can substitute for each other to ensure stable continuation of cell wall synthesis.
MATERIALS AND METHODS
Strains, media, and DNA manipulations.
Caulobacter crescentus cells were grown in peptone-yeast extract (PYE) medium at 30°C, except for the PBP3Ts strains, which were grown at room temperature (∼22°C). The osmotic shock experiment was performed in M2G minimal medium with or without 150 mM sucrose (26). Stalk elongation was measured after >24 h of growth of cells inoculated from isolated colonies into Hutner base-imidazole-buffered-glucose-glutamate (HIGG) minimal medium (27) supplemented with 8.9 mM NH4Cl and with phosphate buffer (305 mM Na2HPO4, 195 mM KH2PO4, pH 7.0) to the final phosphate concentration of 30 μM. Antibiotics were used at the following concentrations: 2.5 μg/ml gentamicin (5 μg/ml on plates), 5 μg/ml kanamycin (25 μg/ml on plates), 1 μg/ml oxytetracycline (2 μg/ml on plates), 2.5 μg/ml A22, 9 μg/ml cephalexin, and 15 μg/ml amdinocillin.
The bacterial strains and plasmids used in this work are described in Tables S1 and S2 in the supplemental material, respectively. In addition, the primers used for constructing these strains and plasmids are listed in Table S3. The plasmids were introduced into Caulobacter by electroporation. The double deletion strains were obtained by ΦCr30 transduction, using a single mutant with a reintegrated deletion plasmid as the donor strain, or by double homologous recombination. The ΦCr30 transductions were performed using the standard protocol (28).
Microscopy.
Cells were immobilized on 1% agarose pads made with PYE medium and imaged with a Nikon 90i microscope equipped with a Nikon Plan Apo 100×/1.4 phase-contrast objective, a Rolera XR cooled charge-coupled-device (CCD) camera, and NIS Elements software. For the cell death assays, mid-log-phase cultures were grown in the presence of 5 μM propidium iodide (PI) for 1.5 h, and the cells were washed once with an equal volume of PYE prior to imaging. Membrane staining was achieved by adding 1 μg/ml of FM1-43 (Invitrogen) directly to the agarose pad. The images were processed with NIS Elements, Fiji, and Adobe Photoshop software. The stalk lengths were measured manually via Fiji.
Cell growth experiments.
Growth curve experiments were performed in a 96-well format using a Synergy HT microplate reader equipped with Gen5 software (BioTek). An overnight culture was diluted with fresh medium to an optical density at 660 nm (OD660) of 0.003. A 150-μl volume of each diluted sample was overlaid with 80 μl of mineral oil to prevent evaporation and allowed to grow at 30°C with continuous shaking at the medium setting. Optical density measurements at 660 nm were taken every 15 min in the course of 24 h. The doubling time was determined individually for every sample well, using a Matlab-based analysis program. The program subtracted the average blank absorbance value for each time point from each sample value for that time point and tested measurements in the time intervals ranging between 4 to 7 h and 2 to 10 h for the time interval that gave the best fit to an exponential curve, as determined by the greatest R2 value of the linear best-fit regression for the natural logarithm of optical density as a function of time. The doubling time was calculated from the corresponding slope of the linear regression line (doubling time = ln2/slope). The growth curves represent the average sample values minus the average blank values, and the error bars are the combined standard deviations for a sample and the blank.
RESULTS
Caulobacter encodes six PG glycosyltransferases.
A bioinformatic search of the Caulobacter crescentus genome revealed the presence of six proteins with a domain belonging to the biosynthetic transglycosylase superfamily (pfam00912). Two of these putative GTases have previously published names, PBP1A for CC1516 and PbpC for CC3277 (29, 30). Additionally, there are three other GTases belonging to the PBP1 family of proteins and thus possessing both a TG domain and a TP domain. We refer to these as PbpX (CC0252), PbpY (CC1875), and PbpZ (CC3570). The remaining GTase is the monofunctional transglycosylase MtgA (CC0325), which has only a TG domain. Primary sequence alignment of these GTases with the four found in Escherichia coli showed that Caulobacter MtgA bears close homology to E. coli MtgA (45% identity), PbpZ is homologous to E. coli PBP1C (34% identity), and PbpY is homologous to E. coli PBP1A (31% identity) (Fig. 1). However, the remaining Caulobacter TGs are more similar to each other than to any E. coli proteins, and Caulobacter lacks a PBP1B homolog (Fig. 1).
Fig 1.

Phylogenetic comparison of the glycosyltransferases. Horizontal line lengths are proportional to phylogenetic distances. The Caulobacter enzymes are shown in blue, and the E. coli (E.c.) enzymes are shown in pink. The phylogram was produced using ClustalW2.
Contributions of GTases to cell growth and cell shape.
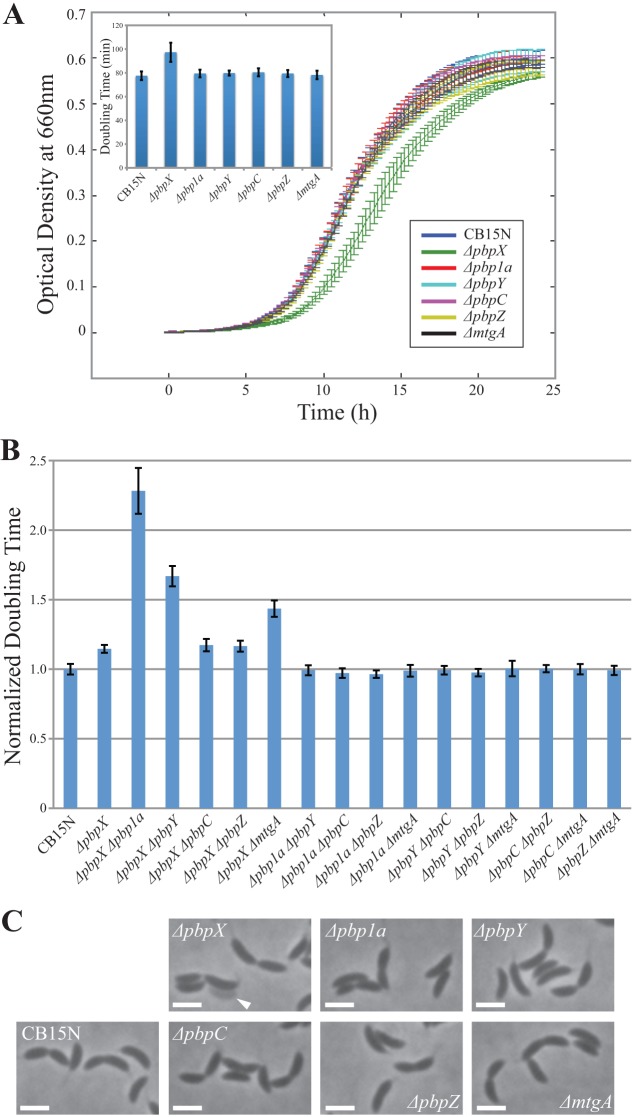
In order to characterize the specific contributions of the six GTases to cell wall synthesis, we constructed strains with in-frame deletions of each of the GTase-encoding genes. The doubling time of these mutants was analyzed in rich PYE medium, in which fast cellular growth requires the greatest rate of PG biosynthesis. While the growth rate of most single GTase mutants was indistinguishable from that of the wild-type strain, the ΔpbpX strain consistently produced an ∼15% delay in doubling time (Fig. 2A). Comparable growth rate reduction has previously been reported for mutants of pbp1b in E. coli and ponA (pbp1) in Bacillus subtilis (18, 31). We conclude that none of the Caulobacter GTases plays an essential role in PG synthesis that cannot be fulfilled by any of the others but that PbpX has the greatest contribution to cell growth.
Fig 2.
Genetic analysis of GTase deletions. (A) The growth curves of single GTase deletion mutants and wild-type CB15N cells are presented. The plot renders the averaged results of 12 replicates per strain. The inset represents the calculated doubling times from the same experiment. (B) The plot shows the calculated doubling times of the double GTase mutants normalized to wild-type CB15N. CB15N and ΔpbpX are presented as controls. All error bars represent standard deviations (SD). (C) Phase-contrast images of the single-deletion mutants are presented. The arrowhead indicates a lysed ghost cell. All scale bars are 2 μm.
We also constructed GTase double-deletion strains in all 15 possible combinations. Although Δpbp1a, ΔpbpY, and ΔmtgA failed to significantly affect the growth rate when present as single deletions, when coupled with ΔpbpX, these mutations caused a synthetic growth delay, exacerbating the growth perturbation of the single ΔpbpX mutant (Fig. 2B). None of the other combinations of deletions had any synthetic phenotypes, suggesting that PBP1A, PbpY, and MtgA are mostly redundant and become important only in the absence of PbpX, while the contributions of PbpC and PbpZ to the overall growth are comparatively negligible.
In order to assess the effect of the GTases on cell shape, we examined the morphology of the deletion mutants. Neither cell width nor cell length was appreciably perturbed in any of the single or double GTase mutants. Moreover, all mutants exhibited the characteristic Caulobacter curved morphology (Fig. 2C; see also Fig. S1 in the supplemental material). Coupled with the growth analysis, these results show that PbpX is the only GTase whose loss affects growth and that none of the GTases is required for cell elongation, division, or curvature formation.
PbpX plays a general role in the process of cell wall assembly.
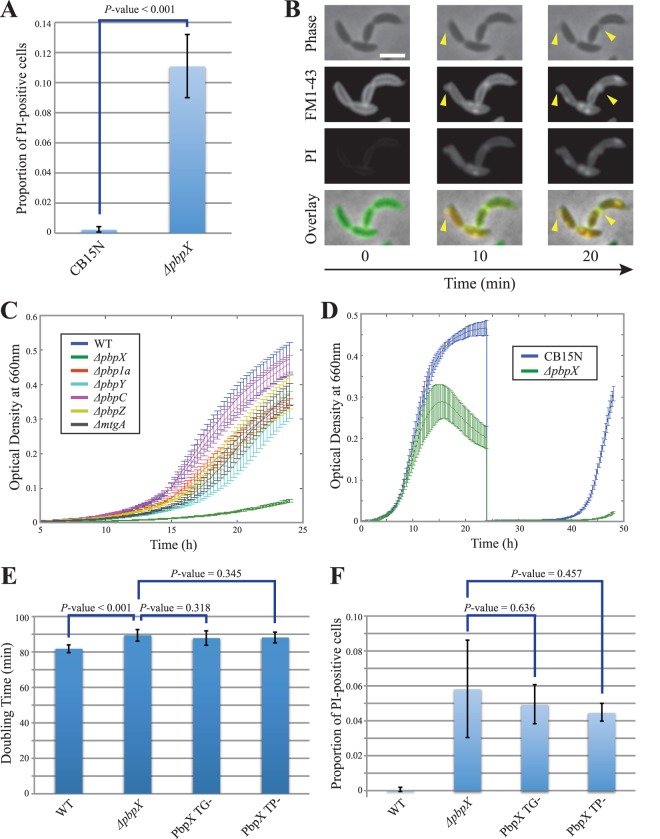
Growth analysis suggested that of the six GTases, PbpX has the greatest contribution to the process of PG synthesis. Interestingly, while ΔpbpX cells exhibited proper cell shape, we noticed that ΔpbpX cultures also contained lysed “ghost” cells in every imaging field (Fig. 2C, arrowhead). These ghost cells were extremely rare in wild-type Caulobacter cultures or in the cultures of any of the other GTase single mutants. To confirm that the loss of PbpX causes sporadic cell death, we treated wild-type and ΔpbpX cells with PI, a membrane-impermeative DNA-binding dye. In exponentially growing cultures, the incidence of PI-positive cells was >10-fold higher for ΔpbpX populations than for those of the wild-type CB15N strain (Fig. 3A). In order to determine the cause of this sporadic cell death, we imaged ΔpbpX cells in real time in the presence of a membrane-labeling dye, FM1-43. We observed that some of the cells developed membrane bulges characteristic of cell wall lesions (Fig. 3B). Such lesions were never observed in wild-type cells. The membrane bulges occurred at the division plane, near a pole, or anywhere along the cylindrical portion of the cell, thus failing to specifically correlate with either division or elongation modes of PG synthesis. Instead, it seems that pbpX deletion causes a general weakening of the cell wall structure. Supporting this conclusion, ΔpbpX cells showed increased sensitivity to osmotic pressure (see Fig. S2A in the supplemental material).
Fig 3.
Phenotypic analysis of pbpX perturbation. (A) The plot shows the proportions of wild-type (CB15N) and mutant cells stained positively with propidium iodide (PI). A representative result from 5 experiments is presented. (B) ΔpbpX cells were stained with FM1-43 membrane dye (green) and PI death marker dye (red) and subjected to time-lapse microscopy. The arrowheads indicate the sites of membrane bulging. The scale bar is 2 μm. (C) Growth curves of GTase single mutants in the PBP3Ts background are presented. The curve for the PBP3Ts parent strain (WT) is shown as a control. Growth was assayed at the permissive temperature of 30°C. Each curve represents the averaged results from 12 replicates. (D) CB15N and ΔpbpX strains grown for 24 h in the presence of 2.5 μg/ml A22, diluted into fresh medium without A22 to OD660 = 0.003, and allowed to grow for another 24 h (24 to 48 h on the graph). The presented growth curve renders the averaged results from 12 replicates (0 to 24 h) or 6 replicates (24 to 48 h). (E) Calculated growth rates of the TG− and TP− point mutants of PbpX are shown. CB15N and ΔpbpX growth rates are provided for comparison. (F) The proportions of PI-positive cells in the cultures of TG-deficient (TG−) and TP-deficient (TP−) mutants of PbpX are shown. P values are provided above the strain data. All error bars represent SD.
In order to examine PbpX participation in the elongation and division processes more directly, we analyzed the growth of the ΔpbpX mutant in the presence of A22, an MreB cytoskeleton-targeting drug that specifically perturbs elongation (32, 33), or a PBP3 temperature-sensitive allele, which specifically perturbs division at 37°C (14, 34). The ΔpbpX mutant showed increased sensitivity to A22 compared with the wild-type CB15N strain (Fig. 3D). Similarly, the ΔpbpX PBP3Ts double mutant grew much slower at the permissive temperature than either the wild-type strain or any other GTase single mutant (Fig. 3C). We also observed increased susceptibility of the ΔpbpX mutant to PBP2-targeting amdinocillin and PBP3-targeting cephalexin (see Fig. S2B and C in the supplemental material), but the interpretation of these data is complicated by the binding of both antibiotics to multiple Caulobacter penicillin-binding proteins (PBPs) (14, 35) and the hypersensitivity of the deletion mutant to the non-growth mode-specific transpeptidation inhibitor ampicillin (data not shown). Thus, the ΔpbpX mutant is hypersensitive to inhibition of either elongation or division, which is consistent with PbpX contributing to the general process of cell wall synthesis required for both of these modes of growth.
PbpX localizes to all known sites of Caulobacter cell wall insertion.
Since cell growth, viability, and antibiotic sensitivity assays pointed to PbpX as the primary GTase in Caulobacter, we sought to examine whether its subcellular localization is consistent with participation in the general process of PG assembly. In Caulobacter, cell wall insertion occurs in a least three patterns: distributed patches that mediate cell elongation, cell cycle-dependent midcell growth that mediates cell division, and stalked-pole growth that mediates stalk biogenesis (9, 10, 36). To determine the localization of PbpX, we constructed a native-site N-terminal fusion of PbpX to monomeric superfolder green fluorescent protein (msfGFP-PbpX). This fusion was fully functional, as a strain that carried it as the sole copy of pbpX had wild-type doubling time, did not experience significant cell lysis, and was not hypersensitive to cephalexin (see Fig. S3 in the supplemental material). msfGFP-PbpX exhibited a patchy, uneven localization around the cell periphery consistent with the pattern of PG insertion that occurs during elongation (see Fig. 5A). Dividing cells that had initiated constriction also accumulated msfGFP-PbpX at the division plane, consistent with a role in septation. Finally, a fraction of the cells accumulated msfGFP-PbpX as a focus that localized to the stalked pole whenever the stalked pole could be clearly identified. msfGFP-PbpX fluorescence was also often observed in the stalk itself. Thus, the pattern of PbpX localization is similar to that of all known sites of Caulobacter PG insertion, consistent with its general function in cell wall assembly.
Fig 5.
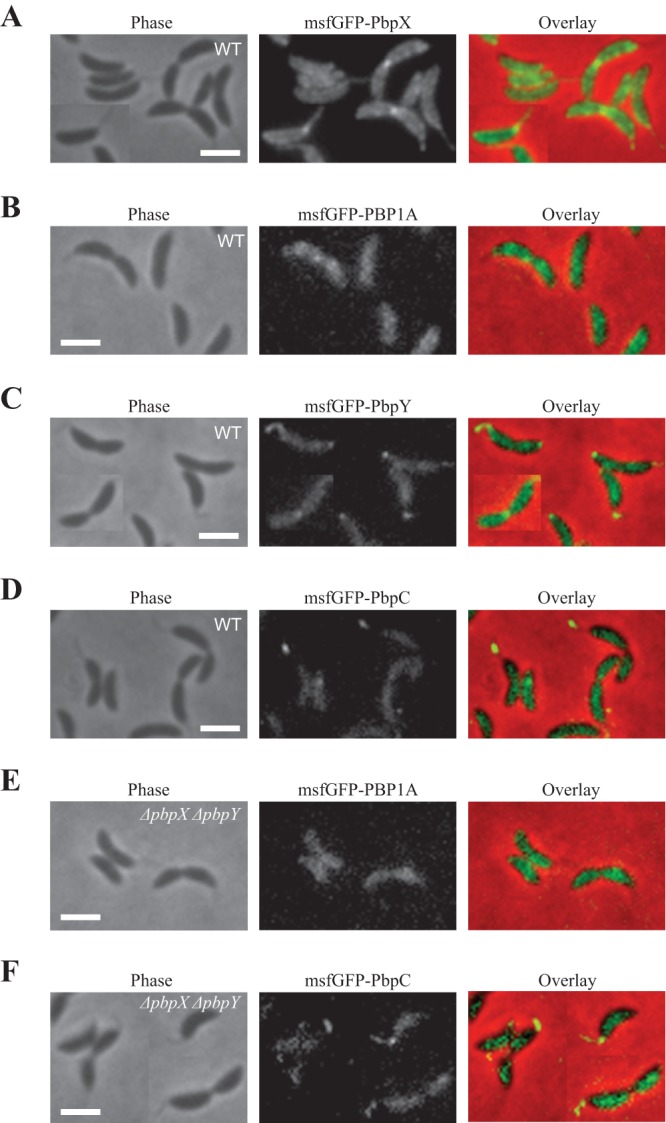
GTases exhibit different localization patterns. (A) Phase, msfGFP-PbpX fluorescence, and overlay images are shown. (B) Phase, msfGFP-PBP1A fluorescence, and overlay images are shown. (C) Phase, msfGFP-PbpY fluorescence, and overlay images are shown. (D) Phase, msfGFP-PbpC fluorescence, and overlay images are shown. (E) Phase, msfGFP-PBP1A fluorescence, and overlay images of cells expressing native-site msfGFP-PBP1A fusion in the ΔpbpX ΔpbpY genetic background are shown. (F) Phase, msfGFP-PbpC fluorescence, and overlay images of cells expressing native-site msfGFP-PbpC fusion in the ΔpbpX ΔpbpY genetic background are shown. All scale bars are 2 μm.
Both enzymatic activities of PbpX are required for proper growth.
We found that of the six predicted Caulobacter GTases, PbpX plays the greatest role in cell growth and the maintenance of PG integrity. Because PbpX contains both TG and TP domains, we sought to determine whether its cellular function requires its glycosyltransferase or transpetidase activity. A TP-deficient (TP−) mutant was constructed by substituting the chromosomal copy of pbpX with a mutant variant that replaced the TP site catalytic serine residue (6) with an alanine (S366A). A TG-deficient (TG−) mutant was similarly constructed by mutating the invariably conserved TG domain glutamates (E101Q E158Q), which are predicted to be involved in catalysis based on the crystal structure and genetic analyses of PBP1 homologs (6, 37). These mutant variants of PbpX protein were stable, as evidenced by a Western blot of endogenous msfGFP-PbpX fusions of the wild-type and mutant proteins with anti-GFP antibodies (data not shown). Both TG− and TP− mutants grew slower than the wild-type strain, with a doubling time close to that of the ΔpbpX mutant (Fig. 3E). The growth perturbation was due to sporadic lysis, as the cultures of both mutants exhibited a high incidence of PI-positive cells (Fig. 3F). Additionally, both mutants were hypersensitive to cephalexin (see Fig. S4A in the supplemental material). It is also noteworthy that the introduced mutations specifically affected the catalytic activity of only one domain, since the TG− and TP− mutants could complement each other upon coexpression, presumably due to heterodimerization (Fig. S4B). Consequently, we conclude that both enzymatic activities are critical for PbpX function. This result is consistent with the observation that TG activity of E. coli PBP1A is susceptible to TP inhibition (21) and with the fact that transpeptidation relies on the glycosyltransferase activity for substrates.
PbpX can function as the sole glycosyltransferase in Caulobacter.
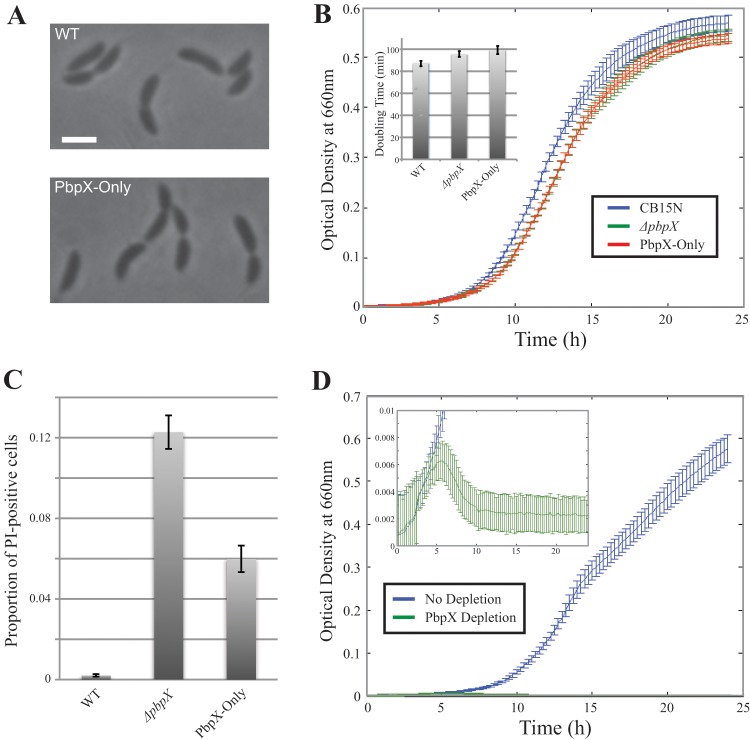
Since the PbpX localization suggested that it could participate in all modes of new PG synthesis, we tested whether it is sufficient to catalyze the transglycosylation reactions needed for growth by constructing a mutant strain with deletions in all TG-encoding genes except for pbpX. The cell shape of the PbpX-Only quintuple deletion mutant (Δpbp1a ΔpbpY ΔpbpC ΔpbpZ ΔmtgA) was indistinguishable from that of wild-type cells (Fig. 4A). While the doubling time and the incidence of death in PbpX-Only cultures were greater than in CB15N, the death rate was lower than that of the ΔpbpX mutant and the growth rate was only marginally lower (Fig. 4B and C). Moreover, depletion of PbpX in the PbpX-Only background led to the loss of viability (Fig. 4D), indicating the absence of unaccounted, cryptic biosynthetic PG GTases in the Caulobacter genome. Thus, PbpX is sufficient for catalysis of PG transglycosylation in Caulobacter, and its efficiency is comparable to that of the rest of the GTases combined.
Fig 4.
PbpX is sufficient for growth in Caulobacter. (A) Phase-contrast images of cells encoding PbpX as the sole GTase (PbpX-Only) and wild-type (CB15N) cells are presented. The scale bar is 2 μm. (B) Growth curves of CB15N, ΔpbpX, and PbpX-Only cells are shown. The plot renders the averaged results of 28 replicates per strain. The inset represents the calculated doubling times from the same experiment. (C) The proportions of PI-positive PbpX-Only and wild-type (CB15N) cells are shown. The ΔpbpX value is provided as a control. (D) Cells expressing xylose-inducible PbpX as the sole GTase were grown in PYE supplemented with either xylose (No depletion) or glucose (PbpX Depletion). The presented growth curve shows the averaged results of 42 replicates per condition. The inset renders a zoomed-in region of the same plot. All error bars are SD.
GTases may have specialized functions.
If PbpX is sufficient for transglycosylation and acts as the primary GTase in Caulobacter, are the remaining GTases merely redundant or might they accomplish different functions? In order to address this question, we attempted to construct native-site fluorescent fusions to each of the other GTases. We were unable to produce functional fusions to PbpZ and MtgA. Intriguingly, all of the remaining GTases had different localization patterns. As previously reported for a fusion of PBP1A to eGFP (29), msfGFP-PBP1A localized around the cell periphery with no particular enrichment either at the division plane or at the stalked pole (Fig. 5B), suggesting that the primary function of PBP1A is elongation. msfGFP-PbpY also exhibited a patchy localization along the periphery but additionally had midcell ring localization in dividing cells and bipolar foci and stalk localization (Fig. 5C). While the patchy, division plane, and stalk localization patterns are consistent with PbpY contributing to elongation, division, and stalk biogenesis, respectively, the function of the bipolar localization is unclear, especially since the nonstalked pole is thought to be inert (9). In contrast to PBP1A and PbpY, msfGFP-PbpC primarily localized to the stalk (Fig. 5D), as has been previously reported (30). Interestingly, deletions of pbpX and pbpY failed to relocalize msfGFP-PBP1A to the stalk or to the division plane or to relocalize msfGFP-PbpC to the main body of the cell or the septum (Fig. 5E and F), indicating that the different localization patterns are an inherent property of the GTases themselves. Thus, although none of the GTases appear to be essential for any of the specific PG synthesis modes, they may perform specialized and only partially overlapping functions in wild-type cells.
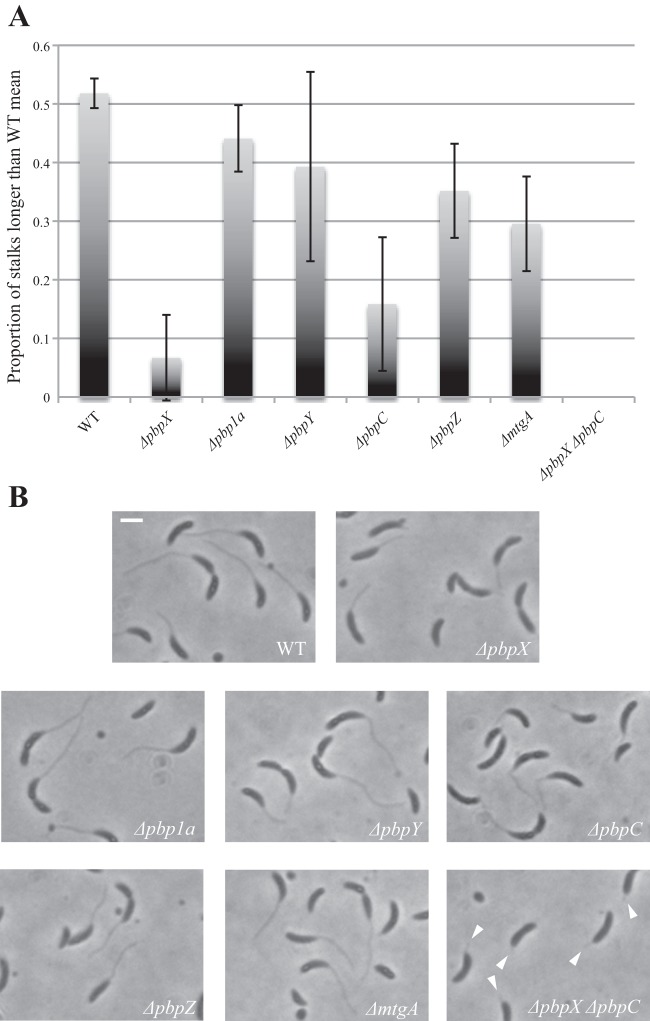
Based on the different localization patterns of the GTases with respect to the stalk, we examined whether they played different roles in stalk biogenesis. All of the single GTase mutants retained the ability to form a stalk (Fig. 2C). However, while wild-type cells could elongate their stalks in response to phosphate-limiting growth conditions, this stalk elongation was severely curtailed in some of the mutant cells, particularly those of the ΔpbpX and ΔpbpC mutants (Fig. 6). The ΔpbpX ΔpbpC double mutant showed greater inhibition of stalk elongation than either of the two single mutants (Fig. 6). Thus, stalk biogenesis greatly relies on PbpX and PbpC, which is consistent with the localization of these two proteins to the stalked pole.
Fig 6.
Stalk length analysis of the GTase mutants. (A) The proportions of stalks with length exceeding the mean stalk length of wild-type CB15N are plotted. The average results of 3 independent experiments are presented. None of the ΔpbpX ΔpbpY cells had stalk length above the threshold. The error bars show SD. (B) Single GTase deletion mutants grown in HIGG medium with low phosphate content and imaged with phase-contrast microscopy. The arrowheads indicate severely shortened stalks of the ΔpbpX ΔpbpC double mutant. Wild-type CB15N is provided as a control. The scale bar is 2 μm.
DISCUSSION
The presence of multiple genes encoding PG glycosyltransferases in bacterial genomes raises the issue of the evolutionary incentive to maintain these paralogous enzymes. This issue is particularly pertinent to Caulobacter crescentus, which encodes six GTases in contrast to the four GTases encoded by most related Alphaproteobacteria, such as Asticcacaulis biprosthecum, Hyphomonas neptunium, Rhodobacter sphaeroides, and Sinorhizobium meliloti. The synthetic lethality of pbp1a and pbp1b mutations in E. coli points to the importance of redundancy to ensure robustness of the essential process of PG synthesis (17, 18). Consistent with the importance of redundancy in GTase systems, we find that all single GTase mutants in Caulobacter are viable. However, robustness does not require six or even four paralogs, particularly since the presence of several GTases, such as E. coli PBP1C and MtgA, is insufficient for growth (17, 18, 38). Additionally, many essential processes are mediated by nonredundant enzymes (see reference 39, for instance), suggesting that essentiality itself is not necessarily a strong driver of the emergence of redundancy.
Another possibility is that naturally occurring PG-targeting antibiotics with different spectra of activity drive the duplication and diversification of PBPs, selecting for multiple-PBP-encoding organisms that thereby increase the probability of at least one PBP escaping inhibition by an encountered antibiotic. However, Caulobacter is resistant to β-lactam antibiotics due to the action of a β-lactamase (40), thus reducing such antibiotic-induced selective pressures. Additionally, we show that ΔpbpX mutation renders cells hypersensitive to several antibiotics, including amdinocillin, cephalexin, A22, and ampicillin, so the multiplicity of GTases would be practically important only as long as PbpX were to remain uninhibited.
A third possibility is that different GTases perform specialized, albeit partially overlapping, functions. This idea is analogous to the long-established participation of different PG transpeptidases in specific modes of bacterial growth (11). We report that Caulobacter GTases exhibit different localization patterns, and deletions of the GTase-encoding genes produce different phenotypes with respect to stalk length, implying the existence of specialized functions for these enzymes. Of the six GTases, PbpX is the only one whose loss is detrimental to growth rate, producing membrane bulging, sporadic lysis, and sensitivity to osmotic pressure. Its deletion also has synthetic growth phenotypes with Δpbp1a, ΔpbpY, and ΔmtgA mutants. Conversely, PbpX alone is sufficient for transglycosylation. Thus, PbpX appears to act as the primary GTase in Caulobacter. Accordingly, it localizes similarly to all sites of PG synthesis, including the lateral cell wall, the division septum, and the stalk/stalked pole, while pbpX deletion sensitizes cells to antibiotics that affect either division or elongation and hinders the ability of the stalk to lengthen under low-phosphate conditions. PbpY also localizes to the lateral wall, septum, and stalk/stalked pole, but its deletion does not produce an overt effect on any of these processes, suggesting that its role is comparatively minor. PBP1A localizes only to the lateral wall (29) (Fig. 5B) and thus may participate only in cellular elongation. In contrast, PbpC localizes primarily to the stalk (30) (Fig. 5D), and the ΔpbpC mutant produces considerably shorter stalks in low-phosphate media (30) (Fig. 6), also exhibiting a synthetic stalk length phenotype when coupled with ΔpbpX. Thus, PbpC appears to be specialized for stalk biogenesis. Our inability to construct functional fluorescent fusions to PbpZ and MtgA obscures the functions of these enzymes. However, considering the inability of PBP1A and PbpC to localize to the division plane in the ΔpbpX ΔpbpY mutant, it is reasonable to hypothesize that either PbpZ or MtgA is responsible for division under these circumstances, particularly since MtgA localizes to the division plane in a pbp1aTs Δpbp1b double mutant of E. coli (41). The contribution of PbpZ to overall PG synthesis is likely negligible since an ortholog of the gene is absent from the genome of a close Caulobacter crescentus relative, Caulobacter segnis.
Since localization and deletion analyses suggest specialized roles for the GTases, what determines these roles? Based on the observed redundancy of these enzymes, one plausible hypothesis is that different GTases have different binding affinities for the growth mode-specific members of the PG remodeling complex (such as PBP2 or PBP3) or for the cytoskeleton-based localization-determining structures. In that case, GTases would be expected to relocalize to the sites with lower-affinity interaction partners in the absence of competing GTases that normally localize there, thus compensating for the lost function and lowering the selection cost of single mutations. Such relocalization has been reported for MtgA in E. coli (41), but we have not observed it for either PBP1A or PbpC in the ΔpbpX ΔpbpY double-mutant background. An alternative model is that interaction with unique localization determinants allows only particular subsets of GTases to accomplish particular functions, so PBP1A alone, for instance, would not be able to support either division or stalk biogenesis, having an exclusive function in elongation.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Gitai laboratory and Ned Wingreen for helpful discussions and comments and Noriko Ohta for providing strains.
This work was supported in part by an HFSP Young Investigator award and NIH New Innovator Award (DP2OD004389) to Z.G.
Footnotes
Published ahead of print 9 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00600-13.
REFERENCES
- 1.Höltje JV. 1998. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev. 62:181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henning U, Rehn K, Hoehn B. 1973. Cell envelope and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. U. S. A. 70:2033–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weibull C. 1953. The isolation of protoplasts from Bacillus megaterium by controlled treatment with lysozyme. J. Bacteriol. 66:688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labischinski H, Barnickel G, Bradaczek H, Giesbrecht P. 1979. On the secondary and tertiary structure of murein. Low and medium-angle X-ray evidence against chitin-based conformations of bacterial peptidoglycan. Eur. J. Biochem. 95:147–155 [DOI] [PubMed] [Google Scholar]
- 5.Gan L, Chen S, Jensen GJ. 2008. Molecular organization of Gram-negative peptidoglycan. Proc. Natl. Acad. Sci. U. S. A. 105:18953–18957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sauvage E, Kerff F, Terrak M, Ayala JA, Charlier P. 2008. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32:234–258 [DOI] [PubMed] [Google Scholar]
- 7.de Pedro MA, Quintela JC, Höltje JV, Schwarz H. 1997. Murein segregation in Escherichia coli. J. Bacteriol. 179:2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniel RA, Errington J. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113:767–776 [DOI] [PubMed] [Google Scholar]
- 9.Aaron M, Charbon G, Lam H, Schwarz H, Vollmer W, Jacobs-Wagner C. 2007. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 64:938–952 [DOI] [PubMed] [Google Scholar]
- 10.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, Vannieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew. Chem. Int. Ed. Engl. 51:12519–12523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spratt BG. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. U. S. A. 72:2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wachi M, Doi M, Tamaki S, Park W, Nakajima-Iijima S, Matsuhashi M. 1987. Mutant isolation and molecular cloning of mre genes, which determine cell shape, sensitivity to mecillinam, and amount of penicillin-binding proteins in Escherichia coli. J. Bacteriol. 169:4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitai Z, Dye N, Shapiro L. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 101:8643–8648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costa T, Priyadarshini R, Jacobs-Wagner C. 2008. Localization of PBP3 in Caulobacter crescentus is highly dynamic and largely relies on its functional transpeptidase domain. Mol. Microbiol. 70:634–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Errington J, Daniel RA, Scheffers DJ. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 32:321–344 [DOI] [PubMed] [Google Scholar]
- 17.Suzuki H, Nishimura Y, Hirota Y. 1978. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 75:664–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yousif SY, Broome-Smith JK, Spratt BG. 1985. Lysis of Escherichia coli by β-lactam antibiotics: deletion analysis of the role of Penicillin-Binding Proteins 1A and 1B. J. Gen. Microbiol. 131:2839–2845 [DOI] [PubMed] [Google Scholar]
- 19.García del Portillo F, de Pedro MA. 1990. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. J. Bacteriol. 172:5863–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laubacher ME, Melquist AL, Chandramohan L, Young KD. 2013. Cell sorting enriches Escherichia coli mutants that rely on peptidoglycan endopeptidases to suppress highly aberrant morphologies. J. Bacteriol. 195:855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banzhaf M, van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J, Zapun A, Breukink E, Nguyen-Distèche M, den Blaauwen T, Vollmer M. 2012. Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Mol. Microbiol. 85:179–194 [DOI] [PubMed] [Google Scholar]
- 22.Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman ME, Kannenberg K, von Rechenberg M, Nguyen-Distèche M, den Blaauwen T, Höltje JV, Vollmer W. 2006. Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Mol. Microbiol. 61:675–690 [DOI] [PubMed] [Google Scholar]
- 23.Claessen D, Emmins R, Hamoen LW, Daniel RA, Errington J, Edwards DH. 2008. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 68:1029–1046 [DOI] [PubMed] [Google Scholar]
- 24.Cabeen MT, Charbon G, Vollmer W, Born P, Ausmees N, Weibel DB, Jacobs-Wagner C. 2009. Bacterial cell curvature through mechanical control of cell growth. EMBO J. 28:1208–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poindexter JS, Hagenzieker JG. 1982. Novel peptidoglycans in Caulobacter and Asticcacaulis spp. J. Bacteriol. 150:332–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foreman R, Fiebig A, Crosson S. 2012. The LovK-LovR two-component system is a regulator of the general stress pathway in Caulobacter crescentus. J. Bacteriol. 194:3038–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poindexter JS. 1978. Selection for nonbuoyant morphological mutants of Caulobacter crescentus. J. Bacteriol. 135:1141–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ely B, Johnson RC. 1977. Generalized transduction in Caulobacter crescentus. Genetics 87:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hocking J, Priyadarshini R, Takacs CN, Costa T, Dye NA, Shapiro L, Vollmer W, Jacobs-Wagner C. 2012. Osmolarity-dependent relocation of penicillin-binding protein PBP2 to the division site in Caulobacter crescentus. J. Bacteriol. 194:3116–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kühn J, Briegel A, Mörschel E, Kahnt J, Leser K, Wick S, Jensen GJ, Thanbichler M. 2010. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 29:327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Popham DL, Setlow P. 1995. Cloning, nucleotide sequence, and mutagenesis of the Bacillus subtilis ponA operon, which codes for Penicillin-Binding Protein (PBP) 1 and a PBP-related factor. J. Bacteriol. 177:326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329–341 [DOI] [PubMed] [Google Scholar]
- 33.Bean GJ, Flickinger ST, Westler WM, McCully ME, Sept D, Weibel DB, Amann KJ. 2009. A22 disrupts the bacterial actin cytoskeleton by directly binding and inducing a low-affinity state in MreB. Biochemistry 48:4852–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohta N, Ninfa AJ, Allaire A, Kulick L, Newton A. 1997. Identification, characterization, and chromosomal organization of cell division cycle genes in Caulobacter crescentus. J. Bacteriol. 179:2169–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koyasu S, Fukuda A, Okada Y. 1982. Penicillin-Binding Proteins in the soluble fraction of Caulobacter crescentus. J. Gen. Microbiol. 128:1117–1124 [Google Scholar]
- 36.Divakaruni AV, Baida C, White CL, Gober JW. 2007. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 66:174–188 [DOI] [PubMed] [Google Scholar]
- 37.Terrak M, Sauvage E, Derouaux A, Dehareng D, Bouhss A, Breukink E, Jeanjean S, Nguyen-Distèche M. 2008. Importance of the conserved residues in the peptidoglycan glycosyltransferase module of the class A penicillin-binding protein 1b of Escherichia coli. J. Biol. Chem. 283:28464–28470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schiffer G, Höltje JV. 1999. Cloning and characterization of PBP1C, a third member of the multimodular class A penicillin-binding proteins of Escherichia coli. J. Biol. Chem. 274:32031–32039 [DOI] [PubMed] [Google Scholar]
- 39.Papanikou E, Karamanou S, Economou A. 2007. Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 5:839–851 [DOI] [PubMed] [Google Scholar]
- 40.West L, Yang D, Stephens C. 2002. Use of the Caulobacter crescentus genome sequence to develop a method for systematic genetic mapping. J. Bacteriol. 184:2155–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Derouaux A, Wolf B, Fraipont C, Breukink E, Nguyen-Distèche M, Terrak M. 2008. The monofunctional glycosyltransferase of Escherichia coli localizes to the cell division site and interacts with penicillin-binding protein 3, FtsW, and FtsN. J. Bacteriol. 190:1831–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.