Abstract
ModE is the molybdate-sensing transcription regulator that controls the expression of genes related to molybdate homeostasis in Escherichia coli. ModE is activated by binding molybdate and acts as both an activator and a repressor. By genomic systematic evolution of ligands by exponential enrichment (SELEX) screening and promoter reporter assays, we have identified a total of nine operons, including the hitherto identified modA, moaA, dmsA, and napF operons, of which six were activated by ModE and three were repressed. In addition, two promoters were newly identified and direct transcription of novel genes, referred to as morA and morB, located on antisense strands of yghW and torY, respectively. The morA gene encodes a short peptide, MorA, with an unusual initiation codon. Surprisingly, overexpression of the morA 5′ untranslated region exhibited an inhibitory influence on colony formation of E. coli K-12.
INTRODUCTION
The transition metal molybdenum is essential for life. In nature, molybdenum is present in various oxidation states and transported into organisms in the form of the tetraoxyanion molybdate. In the case of Escherichia coli, molybdate is transported through an ABC-type transport system encoded by the modABC operon (1). The expression of the modABC operon is repressed by the molybdate-bound ModE transcription factor (2). The ModE protein functions as a homodimer and consists of two domains, the N-terminal DNA-binding domain containing a winged helix-turn-helix motif and the C-terminal molybdate-binding domain (3). Binding of molybdate to the C-terminal domain induces a conformational change of ModE so that it recognizes a palindromic sequence of its target promoters (4). In addition to repression of the molybdate transporter operon, the ModE-molybdate complex is involved in induction of three operons encoding molybdate-containing enzymes, dimethyl sulfoxide (DMSO) reductase (dmsABC), nitrate reductase (napFDAGHBC), and molybdenum cofactor synthase (moaABCDE) (5–7). The three known ModE-inducing targets are all involved in molybdenum metabolism and utilization. Since bacteria contain more than 50 species of the molybdate-containing enzyme, the repertoire of regulation targets of ModE could include more than the already-characterized operons.
In this study, attempts were made to identify the set of regulation targets of the E. coli ModE transcription factor. For this purpose, we employed the genetic systematic evolution of ligands by exponential enrichment (SELEX) screening system, which was developed for identification of the set of regulation targets recognized by DNA-binding transcription factors and was successfully used for the search of regulation targets by AscG (8), AllR (9), CitB (10), Cra (11, 12), cyclic AMP receptor protein (CRP) (13), Dan (14), LeuO (15), NemA (16), PdhR (17), RcdA (18), PgrR (19), RstA (20), RutR (21), and TyrR (22). After the genomic SELEX screening, we identified at least 10 binding sites of the ModE-molybdate complex. Transcription in vivo of the predicted promoters located next to these ModE-binding sites was analyzed using reporter assays in the presence and absence of ModE. Results indicated that ModE activates six promoters (dmsAp, napFp, moaAp, ybhKp, ynfEp, and ecop) and repressed the modA promoter. In addition, we identified two novel promoters that directed the transcription of an antisense sequence of yghX and cutC and hereby name them morA and morB, respectively. The morA gene encodes a small peptide, MorA, carrying an unusual N terminus of Pro, the most probable translation initiation amino acid. Interestingly, overexpression of the morA 5′ untranslated region (UTR) interfered with E. coli colony formation.
MATERIALS AND METHODS
E. coli strains and growth conditions.
Escherichia coli K-12 strains BW25113 (parent strain) and JW0744 (BW25113 with modE::Km) were previously constructed by Baba et al. (23). WJ0101 (W3110 type A with modE::Km) was isolated by P1 transduction with W3110 type A (24) using the P1 lysate prepared from JW0744. These E. coli cells were grown at 37°C in Luria broth (LB) medium.
Purification of ModE.
To construct a pModE plasmid for overproduction of His-tagged ModE, the DNA fragments were prepared by PCR using E. coli W3110 genome DNA as the template and a set of primer pairs, modEF and modER (for sequences, see Table S1 in the supplemental material). After digestion of the PCR-amplified fragments with NdeI and NotI, the PCR-amplified fragments were inserted into the pET21a(+) vector (Novagen) between the same restriction sites as those used for the preparation of insert DNA. The plasmids thus constructed were confirmed by DNA sequencing. pModE was transformed into E. coli BL21(DE3), and His-tagged ModE was overexpressed and purified as described by Yamamoto et al. (25). In brief, E. coli BL21(DE3) transformant was grown in 200 ml of LB medium at an optical density of 600 nm (OD600) of 0.6, and isopropyl-β-d-thiogalactopyranoside (IPTG) was added at the final concentration of 1 mM. After 3 h of incubation, cells were harvested by centrifugation, washed with a lysis buffer (50 mM Tris-HCl [pH 8.0, at 4°C] and 100 mM NaCl), and then stored at −80°C until use. For protein purification, frozen cells were suspended in 3 ml of lysis buffer containing 100 mM phenylmethylsulfonyl fluoride (PMSF). Cells were treated with lysozyme and then subjected to sonication for cell disruption. After centrifugation at 15,000 rpm for 20 min at 4°C, the resulting supernatant was mixed with 2 ml of 50% (wt/vol) Ni-nitrilotriacetic acid (NTA) agarose solution (Qiagen) and loaded onto a column. After being washed with 10 ml of the lysis buffer, the column was washed with 10 ml of washing buffer (50 mM Tris-HCl [pH 8.0, at 4°C] and 100 mM NaCl) and then 10 ml of washing buffer containing 10 mM imidazole. Proteins were eluted with 2 ml of an elution buffer (lysis buffer plus 200 mM imidazole), and peak fractions of transcription factors were pooled and dialyzed against a storage buffer (50 mM Tris-HCl [pH 7.6, at 4°C], 200 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol [DTT], and 50% [vol/vol] glycerol) and stored at −80°C until use. Protein concentration was measured by the Bradford method, and purity was checked on an SDS-PAGE gel.
Genomic SELEX.
The genomic SELEX method was carried out as previously described (10, 11). In brief, the genomic DNA fragments of E. coli K-12 W3110 type A (24) were regenerated by PCR by using the genomic library into pBR322 as the template, a pair of primers hybridized onto plasmid vector, and Ex Taq DNA polymerase (TaKaRa Bio). The mixture of DNA fragments (5 pmol) and His-tagged ModE (10 pmol) were mixed in a binding buffer containing sodium molybdate (1 mM) and incubated for 30 min at 37°C. The ModE-DNA mixture was applied to an Ni-NTA column, and after unbound DNA was washed out with the binding buffer containing 10 mM imidazole, the ModE-DNA complexes were eluted with 200 mM imidazole. DNA fragments recovered from the complexes were amplified by PCR. The concentrated PCR products were purified and labeled with Cy3. The fluorescent-labeled DNAs were hybridized to a DNA microarray consisting of 43,450 probes of a 60-base-long DNA, which were designed to cover the entire E. coli genome at 105-bp intervals (Oxford Gene Technology, Oxford, United Kingdom). The fluorescent intensity at each probe was measured by the array scanner and was indicated as the ratio to sum of the fluorescent intensity of each spot on a slide.
Construction of the lacZ reporter on plasmid.
To construct the lacZ fusion gene, pRS551 and pRS552 plasmids were used as vectors (26). The DNA fragment was amplified by PCR using the genome of the E. coli W3110 type A strain (24) as a template and a pair of primers, as follows: SC1_A_S and SC1_A_T for pSch1-anti-ybhK, SC1_B_S and SC1_B_T for pSch1-modA, SC2_A_S and SC2_A_T for pSch2-ybhK, SC2_B_S and SC2_B_T for pSch2-moaA, SC3_A_S-2 and SC3_A_T-2 for pSch3-anti-serS, SC3_B_S-2 and SC3_B_T-2 for pSch3-dmsA, C5_A_S and SC5_A_T for pSch5-anti-ynfD, SC5_B_S and SC5_B_T for pSch5-ynfE, SC6_A_S and SC6_A_T for pSch6-torY, SC6_B_S and SC6_B_T for pSch6-anti-cutC, SC7_A_S and SC7_A_T for pSch7-napF, SC7_B_S and SC7_B_T for pSch7-eco, SC9_A_S and SC9_A_T for pSch9-yghW, SC9_B_S and SC9_B_T for pSch9-anti-yghX, SC9_B_S and SC9_B_T-E1 for pSch9-anti-yghX fusion-1, SC9_B_S and SC9_B_T-E2 for pSch9-anti-yghX fusion-2, SC9_B_S and SC9_B_T-O1R65 for pORF1-R65, SC9_B_S and SC9_B_T-O2I2 for pORF2-I2, SC9_B_S and SC9_B_T-O2I10 for pORF2-I10, SC9_B_S and SC9_B_T-O2E11 for pORF2-E11, SC9_B_S and SC9_B_T-O2P12 for pORF2-P12, SC9_B_S and SC9_B_T-O2F13 for pORF2-F13, SC9_B_S and SC9_B_T-O2Q14 for pORF2-Q14, SC9_B_S and SC9_B_T-O2C15 for pORF2-C15, SC9_B_S and SC9_B_T-O2L17 for pORF2-L17, SC9_B_S and SC9_B_T-O2G22 for pORF2-G22, SC9_B_S and SC9_B_T-O2R30 for pORF2-R30, SC9_B_S and SC9_B_T-O2M34 for pORF2-M34, SC9_B_S and SC9_B_T-O2V37 for pORF2-V37, SC9_A_S and SC9_A_T-D6 for pmorA-19, SC9_A_S and SC9_A_T-D5 for pmorA+35, SC9_A_S and SC9_A_T-D4 for pmorA+78, SC9_A_S and SC9_A_T-D3 for pmorA+95, SC9_A_S and SC9_A_T-D2 for pmorA+118, SC9_A_S and SC9_A_T-D1 for pmorA+127, SC9_A_S and SC9_A_T-E1 for pmorA+444 (see Tables S1 and S2 in the supplemental material). The PCR product was digested with BamHI and EcoRI and then ligated into pRS551 or pRS552 at the corresponding sites. The DNA sequence of insertion on plasmids was confirmed by DNA sequencing using the lac30R primer complementary to lacZ in a vector (for sequences, see Table S1).
Measurement of β-galactosidase activity in E. coli.
E. coli cells grown in LB medium until an OD600 of 0.3 to 0.4 were subjected to measuring β-galactosidase activity with O-nitrophenyl-d-galactopyranoside as described by Miller (27).
Primer extension analysis.
The promoter-lacZ fusion plasmid was transformed into E. coli. Total RNA from transformants was extracted by the hot phenol method as described previously (28, 29). In brief, E. coli was grown in LB medium at 37°C until mid-log phase (OD600 = 0.3 to 0.4), cells were harvested, and total RNAs were prepared with the hot phenol method. After digestion with RNase-free DNase I (TaKaRa Bio), total RNA was reextracted with phenol, precipitated with ethanol, and dissolved with RNase-free water. The concentration of total RNA was determined by measuring the absorbance at 260 nm. The purity of total RNA was checked by agarose gel electrophoresis. Primer extension analysis was performed using fluorescent-labeled probes as described previously (30). In brief, total RNA (20 μg) and the 5′-fluorescein isothiocyanate (FITC)-labeled FITC-lac primer (for sequences, see Table S1 in the supplemental material) were mixed and the primer extension reaction was initiated by the addition of reverse transcriptase XL (Life Science). After incubation for 1 h at 50°C, DNA was extracted with phenol, precipitated with ethanol, and subjected to electrophoresis on a 6% polyacrylamide sequencing gel containing 8 M urea using DSQ-500L (Shimadzu).
DNase I footprinting analysis.
Probe was amplified by PCR using a pair of primers, 5′-FITC-labeled FITC-lac and SC9_A_T (for sequences, see Table S1 in the supplemental material), pSch9-yghW plasmid as the template, and Ex Taq DNA polymerase. DNase I footprinting assay was carried out under the standard reaction conditions (30). In brief, 1.0 pmol each of FITC-labeled probes was incubated at 37°C for 30 min with purified ModE (0.75 to 6 pmol) in 25 μl of binding buffer (10 mM Tris-HCl [pH 7.8], 150 mM NaCl, 3 mM magnesium acetate, 5 mM CaCl2, and 25 μg/ml bovine serum albumin [BSA]) containing 1 μM Na2MO4. After incubation for 30 min, DNA was digested by DNase I (TaKaRa Bio) for 30 s at 25°C, and then the reaction was terminated by the addition of phenol. DNA was precipitated by ethanol, dissolved in formamide dye solution, and analyzed by electrophoresis on a DNA analyzer DSQ-500L (Shimadu).
Western blotting.
To detect MorA-LacZ fusion protein, Western blotting was performed as previously described (31). In brief, E. coli cells grown in LB medium were harvested, washed, and resuspended in lysis buffer (50 mM Tris-HCl [pH 8.0, at 4°C] and 100 mM NaCl) containing 100 mM PMSF. After sonication, supernatant was recovered by centrifugation, subjected to 10% SDS-PAGE, and blotted onto polyvinylidene difluoride (PVDF) membranes using an iBlot semidry transfer apparatus (Invitrogen). Membranes were first immunodetected with anti-β-galactosidase (Promega) and anti-α-subunit of RNA polymerase (NeoClone) antibodies, followed by immunodetection with a horseradish peroxidase (HRP)-conjugated anti-mouse IgG (Nacalai Tesque) antibody and then development with a chemiluminescence kit (Nacalai Tesque). The image was analyzed with an LAS-4000 IR multicolor scanner (Fuji Film).
Affinity chromatography with APTG agarose.
APTG (4-aminophenyl-β-d-thiogalactopyranoside) agarose 4B (Sigma) was used as a resin for purification of the LacZ fusion protein (32). E. coli transformant JW0744/pORF2-M34 was grown in 400 ml of LB medium and at an OD600 of 1.0, and then cells were harvested by centrifugation, suspended in 12 ml of loading buffer (20 mM Tris-HCl [pH 7.4, at 4°C], 10 mM MgCl2, 10 mM DTT, and 1.6 M NaCl) containing 100 mM PMSF. Cells were treated with lysozyme and then subjected to sonication for cell disruption. After centrifugation at 15,000 rpm for 20 min at 4°C, the resulting supernatant was loaded onto a column filled with 0.5 ml of APTG agarose 4B. After being washed with 30 ml of loading buffer, proteins were eluted with 1 ml of an elution buffer (1 M borate and 10 mM DTT [pH 10.0, at 4°C]) four times. Elution fractions were neutralized by the addition of 1 ml of neutralization buffer (1 M Tris-HCl [pH 7.0, at 4°C]). Protein concentration was measured by the Bradford method, and purity was checked by SDS-PAGE.
Identification of proteins with a mass spectrometer.
Proteins in solution were digested with 2 ng/μl proteomic-grade trypsin (Roche) in aqueous 0.8 M urea buffer at 37°C overnight. Following digestion, tryptic peptides were then analyzed using a liquid chromatography-tandem mass spectrometry (LC-MS/MS) mass spectrometer (amaZon; Bruker). Data from mass spectrometry were analyzed using the MASCOT software (Matrix Science).
RESULTS
Search for ModE-binding sites on the E. coli K-12 genome.
To identify the set of regulation targets of E. coli ModE, we performed the genomic SELEX screening. For this purpose, a mixture of E. coli genome DNA fragments of 200 to 300 bp in length was incubated with the purified His-tagged ModE protein in the presence of sodium molybdate. The ModE-DNA complexes formed were isolated by affinity chromatography using Ni-NTA agarose. When the genomic SELEX was carried out in the absence of molybdate, specific SELEX DNA fragments were not recovered as judged by PAGE (the DNA formed a smear on the gel, as did the original DNA mixture). After two cycles of the genomic SELEX in the presence of molybdate, however, specific DNA fragments were recovered, which formed visible bands on the PAGE gel. The DNA fragments thus isolated were cloned into pT7Blue (Novagen) for sequencing, and the resulting sequences were mapped in the E. coli genome by using both SELEX-clos and SELEX-chip procedures (see Materials and Methods).
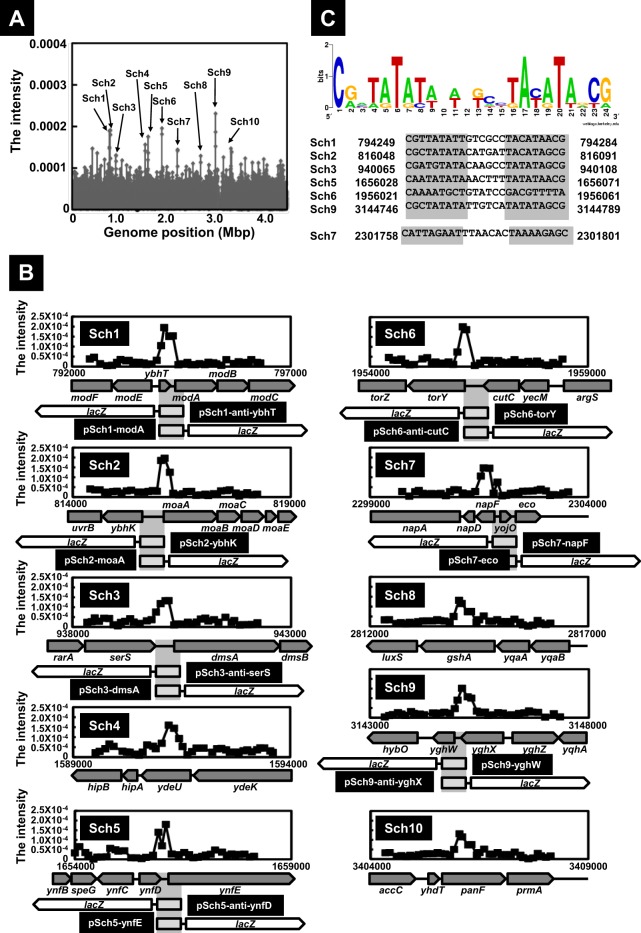
For identification of the set of the binding sites by ModE, we next performed SELEX-chip analysis. The same collection of genomic SELEX fragments as used for SELEX-clos was subjected, after fluorescent labeling, to hybridization with a DNA tilling microarray (Oxford Gene Technology, Oxford, United Kingdom) (11–14). The fluorescence intensity on each spot was represented as the ratio to sum of the fluorescent intensity of each spot on an array and plotted on the corresponding position on the E. coli genome. Since the 60-bp-long probes are aligned along the E. coli genome at 105-bp intervals, approximately 300-bp-long SELEX fragments should bind to two or more consecutive probes, and we employed this criterion for identification of positive peaks with a significant intensity (Fig. 1A). A total of 10 positive peaks, SELEX-chip 1 (Sch1) to Sch10, were identified (Fig. 1A and B). Seven ModE-binding sites, Sch1, Sch2, Sch3, Sch5, Sch6, Sch7, and Sch9, were all located within intergenic spacer regions between two neighboring genes. Since the four hitherto identified ModE-binding sites were included in this list, i.e., modABC on Sch1, moaABCDE on Sch2, dmsABC on Sch3, and napFDAGHBC on Sch7, we predicted that the remaining three ModE-binding sites, Sch5, Sch6, and Sch9, are involved in regulation of neighboring genes (Fig. 1B). The rest of three ModE-binding sites were located within coding regions, Sch4 on ydeU, Sch8 on gshA, and Sch10 on panF (Fig. 1B). In this study, we focused detailed analysis on the ModE targets located within spacer regions.
Fig 1.
Identification of ModE-binding sites on the E. coli genome. (A) SELEX-chip analysis for ModE was performed as described in Materials and Methods. SELEX fragments were mapped on the E. coli genome by using a DNA tilling microarray. The high intensity is indicated on the E. coli genome as an Sch number, indicating a ModE-binding site detected by SELEX-chip. (B) SELEX-chip detailed profiles on each ModE-binding position are shown with the organization of genes. At the center of each ModE-binding site, DNA fragments transcriptionally fused to the promoterless lacZ gene in both directions. The size of each insert is as follows: 498 bp for Sch1, 401 bp for Sch2, 613 bp for Sch3, 471 bp for Sch5, 502 bp for Sch6, 680 bp for Sch7, and 401 bp for Sch9 (see Table S2 in the supplemental material). (C) Using the set of six ModE-binding sequences identified after genomic SELEX screening, the consensus sequences reevaluated by using logo analysis (http://weblogo.berkeley.edu/) are shown. In the case of Sch7, an additional base was observed in the spacer of the consensus sequence.
Identification of the ModE-binding consensus sequence.
Within the ModE-molybdate complex-binding sequence on the modABC promoter, an inverted repeat of pentanucleotides, TATAT, with a spacer of 7 nucleotides (nt), was identified by the DNase I protection assay (33). Since we identified the ModE-molybdate binding sequences for six additional targets, we reexamined the ModE box sequence using the W-AlignACE program (34), which was successfully employed for identification of the MntR box sequence (35). After analysis of a set of 200-bp DNA sequences centered on each ModE-molybdate binding peak for six ModE target sequences (Sch1, Sch2, Sch3, Sch5, Sch6, and Sch9), we identified a conserved sequence of 24 bp in length (Fig. 1C). This revised ModE box is composed of an inverted repeat of 9 nucleotides, CGNTATATA, with a spacer of 6 nucleotides. This ModE box sequence includes the previously proposed consensus sequence of ModE binding (33). In the case of Sch7, the length of the spacer was a total of 7 nucleotides with the extent of one additional nucleotide (Fig. 1C).
Regulation of the newly identified promoters by ModE.
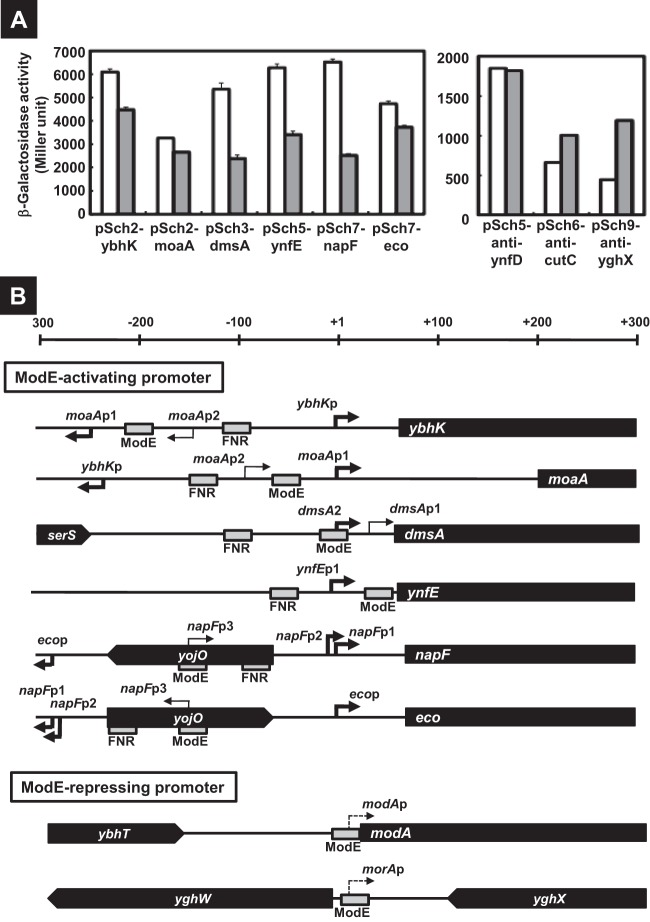
To examine the regulation of seven ModE targets, including three newly identified promoters, we next carried out the reporter assay in vivo in the presence and absence of ModE. For this purpose, we constructed a set of 14 promoter-lacZ transcription fusions. DNA fragments (400 to 700 bp in length) containing each ModE-binding region were amplified by PCR and then ligated into the pRS551 vector (26) in front of the lacZ open reading frame (ORF) in both directions (Fig. 1B; see also Table S2 in the supplemental material). A total of 14 promoter-lacZ fusion plasmids were constructed (see Fig. 1B), each carrying the spacer region of one of the seven ModE targets in both directions. All these plasmids could be successfully transformed into both BW25113 and modE-deficient mutant JW0744 except for pSch1-anti-ybhT and pSch9-yghW, which we failed to transform into the modE mutant (for details, see below). All these transformants were subjected to LacZ assay. β-Galactosidase activity was detected for transformants harboring pSch1-modA, pSch2-ybhK, pSch2-moaA, pSch3-dmsA, pSch5-ynfE, pSch7-napF, and pSch7-eco.
The activity of three known promoters, moaA (pSch2-moaA), dmsA (pSch3-dmsA), and napF (pSch7-napF), decreased in the modE-deficient mutant (Fig. 2A), indicating that ModE activates these promoters. This finding agrees with previous observations (5–7). Three newly identified ModE-regulated promoters, ybhK (pSch2-ybhK), ynfE (pSch5-ynfE), and eco (pSch7-eco), also showed decreased activity in the modE mutant (Fig. 2A), indicating that these promoters are also activated by ModE. To confirm regulation of the newly identified promoters by ModE, we next performed primer extension analysis. The transcription initiation site was identified for each promoter (Fig. 2B). cDNA signals of the primer extension were detected for all three promoters, ybhK, ynfE, and eco in BW25113, but the signals were weak in the modE mutant (data not shown), implying a positive role of ModE for these promoters.
Fig 2.
Promoters activated and repressed by ModE. (A) The lacZ reporter plasmids (see Table S2 in the supplemental material) were introduced into the BW25113 (white bars) and the modE-deficient JW0744 (gray bars), and then the transformants were subjected to β-galactosidase assay. The β-galactosidase activity was represented as a Miller unit in the graph. (B) Organization of the promoters controlled by ModE is shown. The locations of ModE- and FNR-binding sites (gray box) at relative positions to the transcription initiation site (bold and dot arrows represent ModE-activating and repressing sites, respectively) are shown for the promoters. The filled bars represent open reading frames of the target genes.
Identification of ModE-regulated promoters on the antisense strand of cutC and yghX.
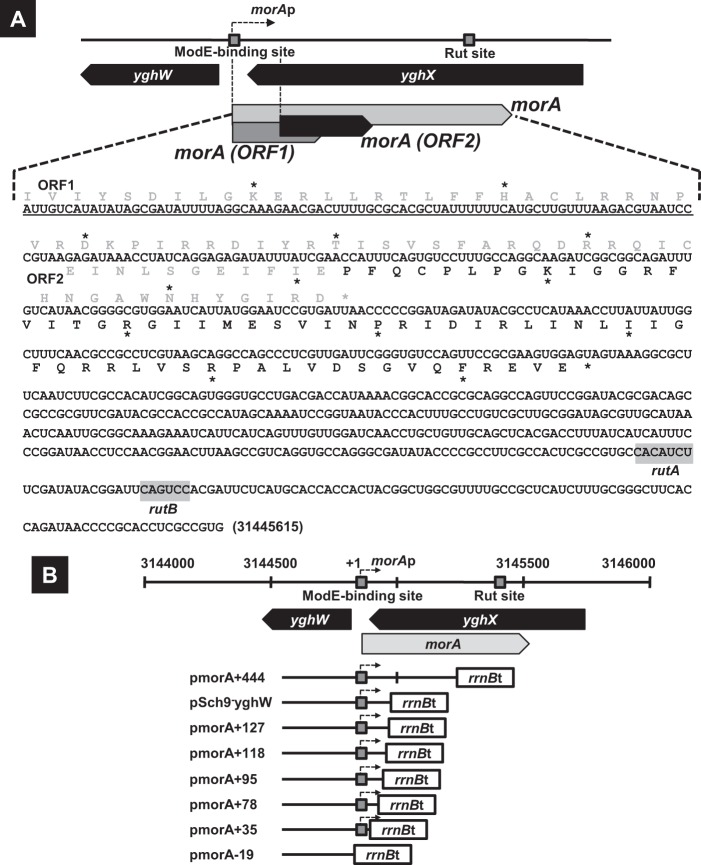
Among 14 promoter assay vectors, five constructs (pSch1-anti-ybhT, pSch3-anti-serS, pSch5-anti-ynfD, pSch6-anti-cutC, and pSch9-anti-yghX) carried the ModE-binding sequences that are located downstream of the respective ORF. Nevertheless, the promoter activity was detected for three constructs (pSch5-anti-ynfD, pSch6-anti-cutC, and pSch9-anti-yghX) (Fig. 2A, right), implying that these promoters transcribe the antisense sequence of the respective genes. In the case of pSch9-anti-yghX (see Fig. 1B), the ModE-binding sequence located downstream of yghX directed expression of LacZ, indicating the presence of a promoter on the antisense strand of yghX. This putative promoter directs transcription toward the opposite direction of that of yghX. The β-galactosidase activity directed by this unexpected promoter increased in the modE mutant, implying that this promoter is repressed by the ModE-molybdate complex. Sequence analysis of the antisense strand of yghX indicated the presence of two overlapping ORFs of 63 codons (ORF1) and 74 codons (ORF2) (Fig. 3A). Thus, we tentatively designated this putative gene as morA (ModE-regulated gene A). The presence of morA promoters was confirmed by measuring transcripts by primer extension analysis (Fig. 4A). The typical RpoD promoter −10 and −35 elements were identified. Electrophoretic mobility shift analysis indicated that dissociation constant (Kd) values for ModE-binding to the morA promoter were 65 nM and 186 nM with and without 1 μM molybdate, respectively (data not shown). By DNase I footprinting, a single ModE-binding site was identified between +23 and −5 of the morA promoter (Fig. 4), which overlapped with the binding site of the RNA polymerase (Fig. 2B).
Fig 3.
The structure of the morA gene and construction of reporter plasmids for analysis of morA expression. (A) The speculated RNA sequence of the morA transcript was shown. Underlined segments indicate the minimum nucleotide sequence for inhibition of colony formation by overexpression of morA in the modE mutant (see Results). Two potential open reading frames, ORF1 and ORF2, and predicted terminators rutA and rutB are indicated in the morA RNA sequence. Actual amino acid residues to express the fused LacZ protein (see Fig. 4) are represented by black symbols in the peptide sequence of ORF2. (B) To analyze growth inhibition of E. coli by overexpression of morA (see Results), the rrnB terminator was followed by several lengths of morA genes in pRS551: −257 to +444 (pmorA+444), −257 to +144 (pSch9-yghW), −257 to +127 (pmorA+127), −257 to +118 (pmorA+118), −257 to +95 (pmorA+95), −257 to +78 (pmorA+78), −257 to +35 (pmorA+35), and −257 to −19 (pmorA-19) (see Table S2 in the supplemental material).
Fig 4.
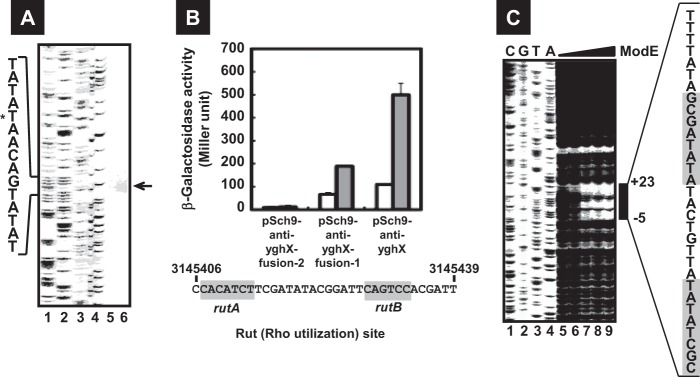
The structure of the morA gene repressed by ModE. (A) E. coli BW25113 (lane 5) and the modE mutant (lane 6) harboring pSch9-anti-yghX (morA-lacZ) were grown in LB medium until an OD600 of 0.3 to 0.4, and total RNAs were prepared as described in Materials and Methods and subjected to primer extension assay. Sanger ladders are synthesized using FITC-lac primer and each plasmid as a template and A, T, G, and C ladders are indicated in lanes 1, 2, 3, and 4, respectively. (B) The transcriptionally fused lacZ genes of several lengths of the morA promoter were constructed with pRS551: 257 to +744 (pSch9-anti-yghX fusion-2), −257 to +444 (pSch9-anti-yghX fusion-1), and −257 to +144 (pSch9-anti-yghX) (see Table S2 in the supplemental material). Three lacZ reporter plasmids introduced into BW25113 (white bars) and the modE-deficient JW0744 (gray bars). The transformants were subjected to β-galactosidase assay (top). The detected Rho utilization site (Rut), rutA and rutB, is shown in the bottom panel. The numbers represent the positions in the E. coli MG1655 genome (GenBank no. U00096.2). (C) An FITC-labeled probe (0.04 μM) prepared as described in Materials and Methods was incubated with 0 μM (lane 5), 0.03 μM (lane 6), 0.06 μM (lane 7), 0.12 μM (lane 8), and 0.24 μM (lane 9) of ModE and then digested by DNase I. Sanger ladders are synthesized using FITC-lac primer and pSch9-yghW plasmid as a template (lanes 1 to 4). Bars indicate the regions protected from DNase I digestion. The numbers represent the position of protection from DNase I relative to the morA transcription start site, and the highlighted sequence indicates ModE-binding consensus (see Fig. 1C).
The ModE-binding site within pSch6-anti-cutC is located downstream of cutC (see Fig. 1B) but was found to be functional as detected by expression of reporter LacZ (Fig. 2A, right). Even though a long reading frame was not detected in the antisense strand of cutC, we predicted that this promoter directs a yet-to-be-identified gene, tentatively designated here as morB. The β-galactosidase activity directed by this morB promoter also increased in the modE mutant, suggesting the negative regulation of the morB promoter by ModE.
Organization of the ModE-regulated morA gene.
To identify the transcription unit of the newly identified morA gene, we examined the length of transcript directed by the morA promoter. For this purpose, we constructed two types of the morA-lacZ transcription fusion plasmids, pSch9-anti-yghX fusion-1 (lacZ gene fused at +444 from the morA transcription start site) and pSch9-anti-yghX fusion-2 (lacZ gene fused at +744) (see Table S2 in the supplemental material). The β-galactosidase activity from pSch9-anti-yghX fusion-1 increased in the modE mutant, as in the case of original pSch9-anti-yghX (Fig. 4B). In contrast, the LacZ activity was not detected for pSch9-anti-yghX fusion-2 in both BW25113 and the modE mutant (Fig. 4B). Taking all this together, we predicted that morA transcription is terminated between +444 and +744. In this region, we found the Rut site, rutA and rutB, for the Rho-dependent termination signal at λtR1 (36) between +634 and +660 from the start site of morA transcription (Fig. 3A and 4B). In the case of λtR1, transcription is terminated downstream, less than 100 bp from the Rut site (36). The size of the morA transcript was thus predicted to be ∼744 nt in length.
Characterization of ORFs encoded by the morA gene.
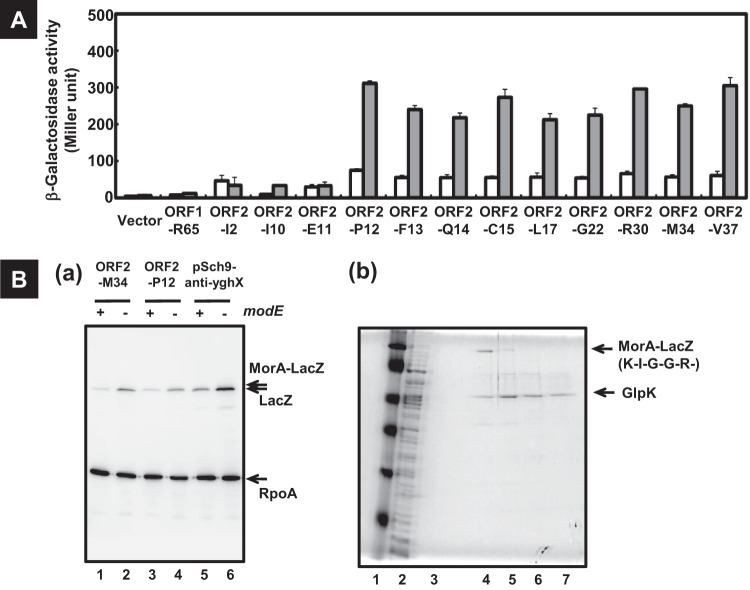
Next, we examined whether two open reading frames, ORF1 and ORF2, within the morA gene are expressed in vivo. For this purpose, we constructed a series of morA-lacZ protein fusion plasmids, each containing portions of the morA gene (Fig. 3A and Fig. 4B; see also Table S2 in the supplemental material). Since the β-galactosidase activity directed by plasmids carrying morA(ORF1)-lacZ fusions was not detected, we concluded that ORF1 was not expressed. In contract, morA(ORF2)-lacZ fusions directed LacZ expression in the modE mutant (Fig. 5), indicating that morA(ORF2) is expressed in the absence of repression by ModE.
Fig 5.
Expression of short peptide MorA. (A) The protein-fused lacZ genes of several lengths of the potential peptide-coding region in the morA gene were constructed with pRS552 (vector). The amino acid residue to fuse to the LacZ protein expressed from each plasmid is as follows: 65th arginine of ORF1 in pORF1-R65, 37th valine of ORF2 in pORF2-V37, 34th methionine of ORF2 in pORF2-M34, 30th arginine of ORF2 in pORF2-R30, 22nd glycine of ORF2 in pORF2-G22, 17th leucine of ORF2 in pORF2-L17, 15th cysteine of ORF2 in pORF2-C15, 14th glutamine of ORF2 in pORF2-Q14, 13th phenylalanine of ORF2 in pORF2-F13, 12th proline of ORF2 in pORF2-P12, 11th glutamate of ORF2 in pORF2-E11, 10th isoleucine of ORF2 in pORF2-I10, and 2nd leucine of ORF2 in pORF2-I2. These lacZ plasmids were introduced into the BW25113 (white bars) and the modE-deficient JW0744 (gray bars). The transformants were subjected to β-galactosidase assay as described for Fig. 2. (Ba) E. coli BW25113 (odd lanes) and the modE mutant (even lanes) harboring pORF2-M34 (lanes 1 and 2), pORF2-P12 (lanes 3 and 4), and pSch9-anti-yghX (morA-lacZ) (lanes 5 and 6) were grown, and the soluble fractions of total proteins were subjected to Western blotting with antibody of both β-galactosidase and the α-subunit of E. coli RNA polymerase. Arrows indicate the detected proteins. (Bb) The cell lysate of E. coli transformant JW0744/pORF2-M34 was prepared. The resulting supernatant (lane 2 is whole extracts) was subjected to affinity chromatography with an APTG agarose 4B column. Samples were analyzed by SDS-PAGE (lane 3 is the flowthrough, lane 4 is the first elution, lane 5 is the second elution, lane 6 is the third elution, lane 7 is the fourth elution). Lane 1 represents marker proteins (Nacalai Tesque), 117 kDa, 88 kDa, 47.2 kDa, 37 kDa, 28.3 kDa, and 22.2 kDa. Arrows indicate the proteins identified by mass spectrometry. The N-terminal sequence of MorA(ORF2), predicted by analysis with LC-MS/MS, is shown.
To identify the initiation site of ORF2 expression, a series of morA(ORF2)-lacZ fusions were constructed, each including a junction segment between the Sch9 spacer and 3′-terminal sequence of morA. To our surprise, LacZ expression was not detectable in both BW25113 and the modE mutant for the morA(ORF2)-lacZ fusions with junction at the 2nd Ile, 10th Ile, and 11th Gln of ORF2 (Fig. 3A and 5A). Upstream from the 11th Gln of ORF2, the typical initiation codons do not exist. These results suggested that translation of ORF2 initiated at the 12th Pro, an unusual amino acid residue for translation initiation. Western blotting indicated that MorA(ORF2)-LacZ from pORF2-M32 and pORF2-P12 increased in the modE-deficient mutant (Fig. 5Ba). The migration of MorA(ORF2)-LacZ proteins was slightly slower than that of LacZ from the morA-lacZ transcription fusion gene on pSch9-anti-yghX (Fig. 5Ba).
For identification of the N-terminal residue of ORF2, we affinity purified MorA(ORF2)-LacZ and identified its N-terminal sequence. MorA(ORF2)-LacZ was expressed from pORF2-M34 (see Fig. 3A and 5B) in the modE mutant, subjected to APTG agarose, and then eluted with an alkaline buffer. A high level of MorA(ORF2)-LacZ and glycerol kinase (GlpK) was detected in the eluate (Fig. 5Bb). Purified MorA(ORF2)-LacZ formed a single band on a PAGE gel (Fig. 5Bb). The purified MorA(ORF2)-LacZ was digested with trypsin and subjected to LC-MS mass spectrometry. Based on the LC-MS analysis, the N-terminal proximal tryptic segment was found to carry the 20th Lys of ORF2 (Fig. 5Bb). These results indicate that MorA(ORF2) seem to be translated at the 12th Pro.
Inhibition of cell growth by overexpression of the morA 5′ UTR.
As noted above, we failed to transform the pSch9-yghW plasmid containing the spacer between yghW and yghX, including a portion of the morA gene into the modE-deficient mutant, JW0744. One possible explanation is that the expression of morA in the absence of repressor ModE is toxic for cell growth as measured by colony formation. Transcription of morA by pSch9-yghW should terminate at the rrnB terminator within the vector, leading to production of a 144-nucleotide-long RNA (Fig. 3B). The inhibitory effect of cell growth by pSch9-yghW transformation might be due to the expression of this truncated morA transcript in the absence of ModE. To examine this possibility, we constructed a series of plasmids, each carrying different lengths of the morA transcript (Fig. 3B). We failed to transform pmorA+444 harboring a 444-bp sequence from the morA transcription initiation site, whereas pmorA-19 lacking the morA promoter and sequence could be transformed in the modE mutant. To determine the minimum region of morA transcript for transformation inhibition, a series of constructs, pmorA+127 (127 nt of morA), pmorA+118 (118 nt of morA), pmorA+95 (95 nt of morA), pmorA+78 (78 nt of morA), and pmorA+35 (35 nt of morA), was tested for transformation into the modE mutant (Fig. 3B). Transformants of the modE mutant were obtained for plasmids pmorA+78 and pmorA+35, producing short transcripts, but not with plasmids pmorA+127, pmorA+118, and pmorA+95, producing transcripts more than 95 nt long. The inhibition of transformation by a series of plasmids was also observed using WJ0101 (ΔmodE), the derivative of W3110 (see Table S1 in the supplemental material). These results indicate that a 95-nt sequence of morA transcript is inhibitory for the growth of E. coli. The mechanism of cell growth by morA RNA remains to be examined.
DISCUSSION
Regulon.
The essential transition metal molybdenum is present in various oxidation states in nature and transported into organisms in the form of the tetraoxyanion molybdate. The transcription factor ModE of E. coli is a dual regulator. In the presence of molybdate, ModE repressed transcription of the modABC operon encoding the transporter of molybdate (2) and activates the synthesis of molybdoenzymes and proteins for Mo metabolism, including dmsABC encoding dimethyl sulfoxide reductase, napFDAGHBC encoding nitrate reductase, and moaABCDE coding for molybdenum cofactor synthase (5–7) (see Fig. 2). Since bacteria contain more than 50 species of the molybdate-containing enzyme, the repertoire of regulation targets by ModE should include more than the hitherto characterized operons. In fact, after genomic SELEX screening, we identified three novel targets (ybhK, ynfEFGHI, and eco) in addition to four known targets (modABC, dmsABC, napFDAGHBC, and moaABCDE). The ynfEFGHI operon encodes the paralogue of the dimethyl sulfoxide reductase DmsABC, a molybdenum-dependent enzyme, and supports anaerobic growth in the presence of DMSO in a dmsABC deletion mutant (37). Another newly identified ModE target, ybhK, encodes a homologue of YvcK, which is required for a normal cell shape and is involved in carbon metabolism in Bacillus subtilis, and restores the growth of the B. subtilis yvcK mutant (38). eco encodes a periplasmic protein of 142 amino acids that is a potent inhibitor of serine protease (39). The ynfEFGHI operon is induced under anaerobic conditions in a fumarate nitrate reduction regulator (FNR)-dependent manner (40). It is noteworthy that the ModE-activated promoters are always associated with FNR-binding sites within a distance of less than 100 bp (see Fig. 2), suggesting a cooperative function between ModE and FNR for induction of promoters by ModE.
ModE has been reported to activate hyc encoding a formate hydrogenlyase, narXL encoding a two-component regulatory system, and deoCABD operons encoding the deoxyribose utilization enzyme by transcriptome analysis of an modE and moeA double mutant (1, 41). Under the genomic SELEX screening conditions employed in this study, we failed to detect these promoters as ModE-binding sites, suggesting that the ModE-binding affinity to these promoters might be too low to detect in vitro.
By promoter assay, we detected two ModE-repressing promoters that direct transcription of antisense strands. The newly identified morA gene is located on the antisense strand of the yghX gene and is transcribed toward the opposite direction of yghX (see Fig. 3). Likewise, the morB gene is located downstream of cutC and directs transcription of the antisense strand of cutC toward the opposite direction of cutC. These two promoters, morA and morB, were found to be repressed by ModE, as in the case of the modA promoter. In concert with the repression mode, the ModE-binding site in the modA promoter overlaps with the binding site of RNA polymerase, suggesting competitive binding of ModE with RNA polymerase. We constructed a single-knockout mutant of both morA and morB. Growth in these strains, however, were affected neither by growth in the presence nor absence of molybdate (data not shown). Also, the intracellular level of molybdenum in both mutants did not change in comparison to that of a parent strain (data not shown).
Nature of the newly identified morA.
Within the morA transcript of more than 700 nucleotides in length, there are two ORFs, morA(ORF1) and morA(ORF2), of which ORF2 was found to be expressed as detected by LacZ fusion assay and Western blotting (see Fig. 3 and 5). No homologous peptide of sequence similarity with MorA(ORF1) and MorA(ORF2) was detected by BLAST search. The morA(ORF2) lacks the typical organization for translation initiation. Nevertheless, the morA(ORF2)-lacZ fusion was found to be expressed (see Fig. 3 and 5). LC-MS/MS analysis of tryptic digests of MorA(ORF2) indicated the presence of a peptide with the 20th Lys at its N terminus. Translation of LacZ fusion proteins was detected when lacZ was fused downstream of the 12th Pro within a total of 74 codons of morA(ORF2). The canonical translation initiation region of E. coli consists of the initiation codon, a Shine-Dalgarno (SD) sequence, and an adequate spacer between them (42). Translation initiation in E. coli occurs on the three canonical codons, AUG (Met codon), GUG (Val codon), or UUG (Leu codon). The complete E. coli genomic sequence indicates that AUG, GUG, and UUG are, respectively, used for 3,542, 612, and 130 genes (43). Genes starting with noncanonical codons are rare in E. coli, but there are some exceptions starting with noncanonical codons, such as AUC (Ile codon) (44), AUA (Ile codon) (45), AUU (Ile codon) (46), GCG (Val codon), and UUC (Phe codon) (47). Translation initiation factor IF3 is involved in the discrimination against initiation on noncanonical codons, but this discrimination is neutralized by generating mutations within the initiator tRNAs so as to disrupt the complementary between codon-anticodon pairs (48). Up to the present time, however, translation initiation from Pro or Lys codons has not been reported. Translation from the 12th Pro codon experimentally detected may also be mediated through continuation (or trans-translation) of a nascent polypeptide initiated at another gene, leading to the formation of a chimeric protein. Actually, we failed to complete the translation of MorA(ORF2) in vitro using the reconstitution system of E. coli translation, PURESYSTEM classic II (BioComber) (data not shown), suggesting that other factors are required for the complete translation of MorA(ORF2).
The high-level expression of morA was found to inhibit E. coli cell growth as measured by colony formation. Deletion analysis of morA showed that the 95-nucleotide-long transcript of morA is enough for expression of the inhibition activity of modE-deficient colony formation. DNA random mutagenesis showed that a C-to-T substitution at +47 suppressed the inhibitory activity of cell growth (data not shown). Noncoding RNAs (ncRNAs) are known to play critical regulatory roles in bacteria. Most ncRNAs act as regulators of gene expression by base pairing with mRNA of target genes (49). However, we failed to find the complementary sequences of the 95-nucleotide-long transcript of morA on the E. coli genome. For the expression of inhibitory function of some ncRNA species, the RNA chaperone protein Hfq is needed (49). We tried to isolate modE and hfq double mutants by P1 transduction but failed, implying that small amounts of transcripts under the control of ModE might be stable in the absence of Hfq. The E. coli transformant with the arabinose-inducible morA plasmid showed normal growth in LB liquid medium with and without arabinose (data not shown). It is possible that overexpression of morA could inhibit colony formation on solid medium but not E. coli growth in a liquid medium. It is not clear, however, how morA RNA affects E. coli colony formation.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2008-2012.
We are grateful to Antonio Tsuneshige of Hosei University for critical reading of the manuscript. We also thank the National BioResource Project (NBRP) of Japan for providing E. coli strains and Ayako Kori, Mie Kanda, and Kayoko Yamada for preparation of proteins and technical support.
Footnotes
Published ahead of print 2 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00304-13.
REFERENCES
- 1.Self WT, Grunden AM, Hasona A, Shanmugam KT. 1999. Transcriptional regulation of molybdoenzyme synthesis in Escherichia coli in response to molybdenum: ModE-molybdate, a repressor of the modABCD (molybdate transport) operon is a secondary transcriptional activator for the hyc and nar operons. Microbiology 145:41–55 [DOI] [PubMed] [Google Scholar]
- 2.Grunden AM, Ray RM, Rosentel JK, Healy FG, Shanmugam KT. 1996. Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J. Bacteriol. 178:735–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall DR, Gourley DG, Leonard GA, Duke EM, Anderson LA, Boxer DH, Hunter WN. 1999. The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds. EMBO J. 18:1435–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schüttelkopf AW, Boxer DH, Hunter WN. 2003. Crystal structure of activated ModE reveals conformational changes involving both oxyanion and DNA-binding domains. J. Mol. Biol. 326:761–767 [DOI] [PubMed] [Google Scholar]
- 5.McNicholas PM, Rech SA, Gunsalus RP. 1997. Characterization of the ModE DNA-binding sites in the control regions of modABCD and moaABCDE of Escherichia coli. Mol. Microbiol. 23:515–524 [DOI] [PubMed] [Google Scholar]
- 6.McNicholas PM, Gunsalus RP. 2002. The molybdate-responsive Escherichia coli ModE transcriptional regulator coordinates periplasmic nitrate reductase (napFDAGHBC) operon expression with nitrate and molybdate availability. J. Bacteriol. 184:3253–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNicholas PM, Chiang RC, Gunsalus RP. 1998. Anaerobic regulation of the Escherichia coli dmsABC operon requires the molybdate-responsive regulator ModE. Mol. Microbiol. 27:197–208 [DOI] [PubMed] [Google Scholar]
- 8.Ishida Y, Kori A, Ishihama A. 2009. Participation of regulator AscG of the beta-glucoside utilization operon in regulation of the propionate catabolism operon. J. Bacteriol. 191:6136–6144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasegawa A, Ogasawara H, Kori A, Teramoto J, Ishihama A. 2008. The transcription regulator AllR senses both allantoin and glyoxylate and controls a set of genes for degradation and reutilization of purines. Microbiology 154:3366–3378 [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto K, Matsumoto F, Oshima T, Fujita N, Ogasawara N, Ishihama A. 2008. Anaerobic regulation of citrate fermentation by CitAB in Escherichia coli. Biosci. Biotechnol. Biochem. 72:3011–3014 [DOI] [PubMed] [Google Scholar]
- 11.Shimada T, Fujita N, Maeda M, Ishihama A. 2005. Systematic search for the Cra-binding promoters using genomic SELEX system. Genes Cells 10:907–918 [DOI] [PubMed] [Google Scholar]
- 12.Shimada T, Yamamoto K, Ishihama A. 2011. Novel members of the Cra regulon involved in carbon metabolism in Escherichia coli. J. Bacteriol. 193:649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shimada T, Fujita N, Yamamoto K, Ishihama A. 2011. Novel roles of cAMP receptor protein (CRP) in regulation of transport and metabolism of carbon sources. PLoS One 6:e20081. 10.1371/journal.pone.0020081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teramoto J, Yoshimura SH, Takeyasu K, Ishihama A. 2010. A novel nucleoid protein of Escherichia coli induced under anaerobiotic growth conditions. Nucleic Acids Res. 38:3605–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimada T, Yamamoto K, Ishihama A. 2009. Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux. J. Bacteriol. 191:4562–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umezawa Y, Shimada T, Kori A, Yamada K, Ishihama A. 2008. The uncharacterized transcription factor YdhM is the regulator of the nemA gene, encoding N-ethylmaleimide reductase. J. Bacteriol. 190:5890–5897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogasawara H, Ishida Y, Yamada K, Yamamoto K, Ishihama A. 2007. PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J. Bacteriol. 189:5534–5541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimada T, Katayama Y, Kawakita S, Ogasawara H, Nakano M, Yamamoto K, Ishihama A. 2012. A novel regulator RcdA of the csgD gene encoding the master regulator of biofilm formation in Escherichia coli. Microbiologyopen 1:381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimada T, Yamazaki K, Ishihama A. 2013. Novel regulator PgrR for switch control of peptidoglycan recycling in Escherichia coli. Genes Cells 18:123–134 [DOI] [PubMed] [Google Scholar]
- 20.Ogasawara H, Hasegawa A, Kanda E, Miki T, Yamamoto K, Ishihama A. 2007. Genomic SELEX search for target promoters under the control of the PhoQP-RstBA signal relay cascade. J. Bacteriol. 189:4791–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada T, Hirao K, Kori A, Yamamoto K, Ishihama A. 2007. RutR is the uracil/thymine-sensing master regulator of a set of genes for synthesis and degradation of pyrimidines. Mol. Microbiol. 66:744–757 [DOI] [PubMed] [Google Scholar]
- 22.Yang J, Ogawa Y, Camakaris H, Shimada T, Ishihama A, Pittard AJ. 2007. folA, a new member of the TyrR regulon in Escherichia coli K-12. J. Bacteriol. 189:6080–6084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jishage M, Ishihama A. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli strain W3110. J. Bacteriol. 179:959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto K, Hirao K, Oshima T, Aiba H, Utsumi R, Ishihama A. 2005. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J. Biol. Chem. 280:1448–1456 [DOI] [PubMed] [Google Scholar]
- 26.Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 27.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28.Yamamoto K, Ishihama A. 2005. Transcriptional response of Escherichia coli to external zinc. J. Bacteriol. 187:6333–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamamoto K, Ishihama A. 2005. Transcriptional response of Escherichia coli to external copper. Mol. Microbiol. 56:215–227 [DOI] [PubMed] [Google Scholar]
- 30.Ogasawara H, Yamamoto K, Ishihama A. 2011. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 193:2587–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto K, Oshima T, Nonaka G, Ito H, Ishihama A. 2011. Induction of the Escherichia coli cysK gene by genetic and environmental factors. FEMS Microbiol. Lett. 323:88–95 [DOI] [PubMed] [Google Scholar]
- 32.Steers E, Jr, Cuatrecasas P. 1974. Isolation of beta-galactosidase by chromatography. Methods Enzymol. 34:350–358 [DOI] [PubMed] [Google Scholar]
- 33.Self WT, Grunden AM, Hasona A, Shanmugam KT. 2001. Molybdate transport. Res. Microbiol. 152:311–321 [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Guo L, Fan Z, Jiang T. 2008. W-AlignACE: an improved Gibbs sampling algorithm based on more accurate position weight matrices learned from sequence and gene expression/ChIP-chip data. Bioinformatics 24:1121–1128 [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto K, Ishihama A, Busby SJW, Grainger DC. 2011. The Escherichia coli K-12 MntR miniregulon includes dps which encodes the major stationary-phase DNA-binding protein. J. Bacteriol. 193:1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CY, Richardson JP. 1987. Sequence elements essential for rho-dependent transcription termination at lambda tR1. J. Biol. Chem. 262:11292–11299 [PubMed] [Google Scholar]
- 37.Lubitz SP, Weiner JH. 2003. The Escherichia coli ynfEFGHI operon encodes polypeptides which are paralogues of dimethyl sulfoxide reductase (DmsABC). Arch. Biochem. Biophys. 418:205–216 [DOI] [PubMed] [Google Scholar]
- 38.Görke B, Foulquier E, Galinier A. 2005. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 151:3777–3791 [DOI] [PubMed] [Google Scholar]
- 39.McGrath ME, Gillmor SA, Fletterick RJ. 1995. Ecotin: lessons on survival in a protease-filled world. Protein Sci. 4:141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu M, Busby SJ, Browning DF. 2009. Activation and repression at the Escherichia coli ynfEFGHI operon promoter. J. Bacteriol. 191:3172–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao H, Hasona A, Do PM, Ingram LO, Shanmugam KT. 2005. Global gene expression analysis revealed an unsuspected deo operon under the control of molybdate sensor, ModE protein, in Escherichia coli. Arch. Microbiol. 184:225–233 [DOI] [PubMed] [Google Scholar]
- 42.Kozak M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187–208 [DOI] [PubMed] [Google Scholar]
- 43.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 44.Chalut C, Eglay JM. 1995. AUC is used as a start codon in Escherichia coli. Gene 156:43–45 [DOI] [PubMed] [Google Scholar]
- 45.Köpke AK, Leggatt PA. 1991. Initiation of translation at an AUA codon for an archaebacterial protein gene expressed in E. coli. Nucleic Acids Res. 19:5169–5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romero A, García P. 1991. Initiation of translation at AUC, AUA and AUU codons in Escherichia coli. FEMS Microbiol. Lett. 68:325–330 [DOI] [PubMed] [Google Scholar]
- 47.Chattapadhyay R, Pelka H, Schulman LH. 1990. Initiation of in vivo protein synthesis with non-methionine amino acids. Biochemistry 29:4263–4268 [DOI] [PubMed] [Google Scholar]
- 48.Meinnel T, Sacerdot C, Graffe M, Blanquet S, Springer M. 1999. Discrimination by Escherichia coli initiation factor IF3 against initiation on non-canonical codons relies on complementarity rules. J. Mol. Biol. 290:825–837 [DOI] [PubMed] [Google Scholar]
- 49.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.