Abstract
Microbial symbionts can be instrumental to the evolutionary success of their hosts. Here, we discuss medically significant tsetse flies (Diptera: Glossinidae), a group comprised of over 30 species, and their use as a valuable model system to study the evolution of the holobiont (i.e., the host and associated microbes). We first describe the tsetse microbiota, which, despite its simplicity, harbors a diverse range of associations. The maternally transmitted microbes consistently include two Gammaproteobacteria, the obligate mutualists Wigglesworthia spp. and the commensal Sodalis glossinidius, along with the parasitic Alphaproteobacteria Wolbachia. These associations differ in their establishment times, making them unique and distinct from previously characterized symbioses, where multiple microbial partners have associated with their host for a significant portion of its evolution. We then expand into discussing the functional roles and intracommunity dynamics within this holobiont, which enhances our understanding of tsetse biology to encompass the vital functions and interactions of the microbial community. Potential disturbances influencing the tsetse microbiome, including salivary gland hypertrophy virus and trypanosome infections, are highlighted. While previous studies have described evolutionary consequences of host association for symbionts, the initial steps facilitating their incorporation into a holobiont and integration of partner biology have only begun to be explored. Research on the tsetse holobiont will contribute to the understanding of how microbial metabolic integration and interdependency initially may develop within hosts, elucidating mechanisms driving adaptations leading to cooperation and coresidence within the microbial community. Lastly, increased knowledge of the tsetse holobiont may also contribute to generating novel African trypanosomiasis disease control strategies.
INTRODUCTION
Species interactions, across and within the domains of life, are ubiquitous in nature, fundamental in ecology, and pivotal with respect to evolutionary diversification. Throughout history, metazoans have formed intimate partnerships with microorganisms, some of which significantly contribute to the health and development of their host. Due to the important role of microbial symbionts in host fitness, research has increasingly focused on a more holistic examination of the biological system, encompassing the macroscopic host (i.e., animals or plants) and associated microbes, termed the holobiont (1) or the metaorganism (2), with the cumulative genetic material known as the hologenome (3). The hologenome theory of evolution has been used to examine the holobiont as a single unit undergoing evolution, adapting to persist in or expand its niche (3, 4).
Traditionally used in aquatic biology, applications of the holobiont concept have provided insights into marine microbiology such as coral health and the functioning of deep-sea hydrothermal vents (1, 3, 5). Heightened recognition of microbial symbionts as major contributors to host health has spurred the Human Microbiome Project, aimed at characterizing the microbial communities of several distinct spatial sites on the human body to better our understanding of their role in health and disease (6). These communities are highly complex (7) and can be composed of hundreds of species-level phylotypes, determined by ≥97% 16S rRNA sequence identity (8, 9). To fully understand processes occurring within complex holobionts, examination of simpler systems may aid in dissecting intimate interplay among the partners.
THE TSETSE FLY
The tsetse fly (Diptera: Glossinidae) provides a valuable model to study the evolution of a holobiont. The tsetse microbial symbionts range not only in the nature of their association (from mutualistic to parasitic) but also in their establishment times within the host (10). Thus, the system is distinct from other described insect symbioses, including sharpshooters, cicadas, spittlebugs, and mealybugs, where microbial symbionts have coevolved with one another and their host for a significant amount of time (11–16). A holobiont similar to tsetse is the pea aphid, Acyrthosiphon pisum, which harbors an obligate, nutrient-provisioning, mutualist Buchnera aphidicola (17), in addition to multiple facultative symbionts, such as Serratia symbiotica, Hamiltonella defensa (itself harboring a toxin-encoding phage protecting against parasitoid attacks), and Regiella insecticola, that provide defense against heat stress and parasites and have enabled an expansion of host-plant range (reviewed in reference 18). A notable distinction is that unlike the tsetse holobiont, which has evolved essential metabolic interdependency for both host and symbiont fitness (19, 20), S. symbiotica has been shown to partially recover host fitness upon Buchnera loss (21, 22), suggesting early steps in symbiont replacement or functional redundancy. Investigations regarding microbial community dynamics within tsetse will greatly contribute to the knowledge of how cooperation may evolve among players with differing establishment times.
In addition to its use as a model system for understanding the evolution of a holobiont, tsetse maintains significance as the sole and obligate vector of protozoan African trypanosomes (Trypanosoma spp.). These parasites (T. brucei rhodesiense and T. b. gambiense) are the causative agents of human African trypanosomiasis (HAT; commonly called “sleeping sickness”), a disease affecting the central nervous system that is lethal if left untreated. HAT threatens millions of people in approximately 36 countries and has been classified by Doctors Without Borders as a neglected tropical disease, impacting some of the poorest rural areas in Africa (23). Another African trypanosome, T. b. brucei, causes Nagana, a similar wasting disease in domesticated animals, particularly cattle, further impeding the economic development of affected areas (24). Disease relief is relatively nonexistent, as there are no vaccines available to prevent African trypanosomiasis, manual trapping is often unreliable due to the social unrest in many affected areas, diagnostics are limited, and the small arsenal of drugs available for treatment are associated with significant toxic side effects. Therefore, vector control is an alternative intervention to break the disease cycle (25), as advocated with other systems (reviewed in reference 26). For example, Wolbachia has been shown to inhibit the replication of multiple arboviruses and filarial nematodes within Aedes mosquitoes (27–30), as well as to shorten the host life span, not permitting cyclical pathogen development and transmission (31). Additionally, symbionts within the guts of mosquitoes and triatome bugs are being genetically modified to produce antiparasitic molecules, in efforts to block transmission of malaria (32) and Chagas disease (33), respectively. Knowledge of tsetse fly symbiosis not only stands to provide basic insight into how microbial partners adapt and respond to changes in ecological factors and parasite infections but may also be of applied value to generate novel modes of pest biocontrol (26).
Localized exclusively to sub-Saharan Africa, there are approximately 31 species and subspecies of tsetse flies (Diptera: Glossinidae), which can be divided into 3 groups: Glossina morsitans, G. palpalis, and G. fusca (34). The unique biology of tsetse contributes to the maintenance of a simple larval microbiota, consisting of only 3 maternally transmitted bacterial symbionts (35), in comparison to other Diptera, such as mosquitoes and fruit flies, which harbor a greater complexity of bacterial taxa (36, 37). One distinct feature of tsetse biology is that both sexes maintain a strictly hematophagous lifestyle, persisting solely on vertebrate blood, which limits the introduction of additional microbes through an oral/digestive route. Due to their restricted diet, tsetse rely on microbial symbionts for provisioning essential metabolites lacking in blood (38). Another contributing factor to microbiome simplicity is the tsetse reproductive strategy, known as adenotrophic viviparity. This reproductive strategy involves high maternal investment and a low reproductive output of only 6 to 8 offspring in their 3-to-4-month life span (34). Unlike many higher Dipteran, female tsetse have highly modified reproductive tracts (39), enabling the deposition of a single fertilized egg into a muscular uterus, which is connected to highly specialized accessory glands, referred to as milk glands. Milk secretions provide nourishment and a route through which microbial symbionts (40, 41) are transferred during intrauterine larval development. This form of reproduction transmits the microbiota with high fidelity while preventing exposure to transient microbes during early tsetse development (35).
TSETSE MICROBIAL COMMUNITY
The tsetse microbiota consists primarily of three vertically transmitted bacterial species (Fig. 1). These microbes include two enteric Gammaproteobacteria, obligate mutualists Wigglesworthia spp. (42) and the commensal Sodalis glossinidius (43). Tsetse can also harbor the Alphaproteobacteria Wolbachia (44), a facultative parasite infecting many different invertebrates (45, 46), which is typically restricted to the reproductive organs (47, 48). Field studies report a more complex diversity in adult flies (49–51), although these microbes are believed to be transient in nature. The tsetse holobiont provides opportunities to examine evolutionary aspects associated with adapting to microbial coresidence, as Wigglesworthia and Sodalis have drastically different times of establishment (10, 52). Moreover, interactions among microbes with various levels of host dependency and symbiotic roles can be empirically investigated.
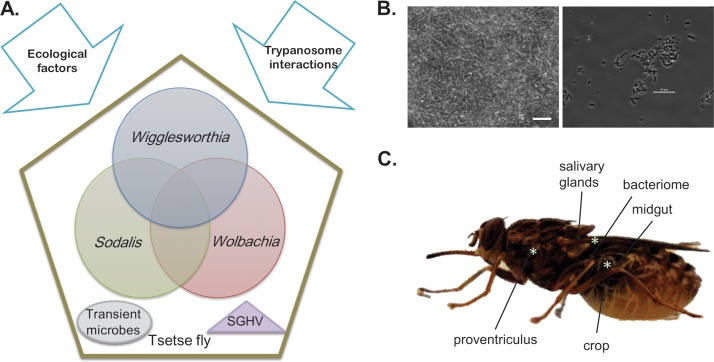
Fig 1.
The tsetse holobiont. (A) The holobiont is composed of the tsetse fly and its 3 vertically transmitted bacterial symbionts, Wigglesworthia, Sodalis, and Wolbachia, and may be influenced by intrinsic factors such as other transient microbes, salivary gland hypertrophy virus (SGHV), and trypanosomes as well as by abiotic factors. (B) The enteric microbiota (Wigglesworthia and Sodalis), whose genomes are both annotated, provide a natural model to examine the early evolution of cooperation and adaptations leading toward microbiome coresidency. Phase-contrast microscopy images of Wigglesworthia cells (left) within a G. morsitans bacteriome and Sodalis cells (right) within culture are shown. Scale bars signify 10 μm. (C) The protozoan parasite T. brucei subsp. potentially interacts with microbial symbionts throughout infection. Stages of infection (denoted by asterisks) signify colocalizations of microbes that include the midgut, moving in an anterior manner toward the proventriculus and culminating in the salivary glands.
Wigglesworthia spp. maintain an obligate mutualism with tsetse and display significant concordant evolution with its specific host species, dating back 50 to 80 million years (10). In both sexes, this symbiont is localized intracellularly in specialized host cells (bacteriocytes) at the anterior midgut, collectively comprising an organ known as the bacteriome (Fig. 1). An additional extracellular population is found in the female milk glands which is maternally transmitted to offspring (40, 41). Described roles of this symbiont include both nutrient provisioning, where Wigglesworthia supplements B vitamins lacking in the tsetse blood diet (38, 53–55), and contributions to the maturation of host immunity (35, 56, 57). Insight into these roles has been found by examining tsetse biology upon removal of the symbiont. For example, the bacteriome population is vital for nutrient provisioning during host reproduction, as flies lacking the Wigglesworthia bacteriome populations are sterile (19, 58, 59), with fecundity partially restored by B-vitamin or yeast extract supplementation (38, 59). Absence of the milk gland symbiont population does not inhibit reproduction (58), as these populations are believed to be dedicated to vertical transmission and the persistence of the symbiosis through evolutionary time. In addition, the presence of Wigglesworthia during larval stages is essential for proper immune development, as larvae that lack this symbiont are significantly compromised in the induction of pathways associated with cellular immunity (35). The immunocompromised phenotype of aposymbiotic larvae can be reversed by feeding their moms a diet supplemented with Wigglesworthia cell extracts (57). The presence of the tsetse's larval microbiota also contributes to the proper development of the adult peritrophic matrix, separating epithelial cells from the contents of the lumen, which regulates the timing of immune induction following parasite challenge (60). Wigglesworthia also impacts tsetse digestion, temperature sensitivity, and susceptibility to infection with trypanosomes (56, 58, 60). It is important to mention that removing Wigglesworthia also causes indirect effects on the host that are not related to the loss of symbiont function. Some examples include additional nutritional deficiencies due to inhibited blood meal digestion (58), transient microbial colonization arising from an altered immune state (35, 56, 57), and perturbations to the remaining tsetse microbiota (20).
The annotation of Wigglesworthia genomes, isolated from Glossina brevipalpis (Wgb) (53) and G. morsitans (Wgm) (54), revealed characteristics similar to those of other ancient insect symbionts (61–63), including a reduced size (∼0.7 Mb) and high adenine-thymine bias. Genome adaptations by Wigglesworthia, while in association with tsetse, are believed to have resulted in the loss of many capabilities required for a free-living lifestyle through reductive evolution (53, 54). Despite its small genome size, the majority of B-vitamin biosynthesis pathways remain intact, supporting the nutritional mutualism. Comparative analyses revealed that Wgm and Wgb maintain similar genomic repertoires with high synteny. Interestingly, some pockets of unique Wgm genes, potentially contributing to anabolic distinctions, were found (54). As this symbiont has undergone deep codiversification with the tsetse host (10), any unique capabilities of Wigglesworthia spp. influencing physiological and phenotypic differences between host species remain largely unknown.
In contrast to the ancient Wigglesworthia association, tsetse's commensal partner, Sodalis, is believed to have established recently within the tsetse host from a previously free-living progenitor. Evidence lies in its ability to be cultured (64), providing a tremendous benefit for empirical analyses, wide tsetse tissue tropism with both intra- and extracellular localization (41, 48), lack of coevolution with host species (10), and stochastic presence in the field (49, 51, 65). Similar to Wigglesworthia, Sodalis is vertically transmitted through the maternal milk glands (40, 41). Despite a more recent association, Sodalis displays genomic signatures indicating that adaption to the symbiosis has commenced. While the Sodalis genome (4.2 Mb) is larger than that of Wigglesworthia, it appears to be undergoing massive reduction, as it is composed of >50% pseudogenes (66, 67), indicative of relaxed selection on nonessential genes. Notably, Sodalis has modified its outer membrane protein A (OmpA), which represents a molecular adaptation that contributes to host immune tolerance (68). The OmpA protein is also utilized in biofilm production within the tsetse gut, further protecting Sodalis from host immune responses (69).
Biotechnological advancements, notably culture-independent techniques, have accelerated the number of described host-associated bacterial species, stimulating interest in examination of the Enterobacteriaceae Sodalis-allied clade, as members are present in a diverse array of insect and environmental samples (43, 70–81) (Table 1). While this bacterial group appears to have an enhanced ability to establish within a variety of niches relative to most other characterized symbionts, much of the initial molecular phylogenetic analysis within the Sodalis-allied clade has utilized the conserved 16S rRNA gene, resulting in low resolution (70, 75, 82). Examination of surface-encoding loci from Sodalis isolates and related symbionts revealed early genomic host-specific modifications, likely aiding in their integration within different insects (82). Moreover, phylogenetic analyses of the internal transcribed spacer regions, which have an accelerated evolutionary rate (83), in conjunction with the adjacent 16S rRNA gene, have provided additional insights into the evolutionary divergence of this clade (84). The Sodalis isolates from various tsetse species formed a monophyletic clade, indicative of their divergence from additional known members of this group. Supporting the recent expansion of this clade, the phylogenies also demonstrated that symbionts from additional insect hosts were intertwined, suggesting either horizontal transfer between insects or the acquisition from a common environmental source. A recently described member obtained from an environmental source, known as strain HS, has provided novel insights into the progenitor of this clade (81). Comparative genomic analyses of strain HS, with other members of the Sodalis-allied clade, specifically, Sodalis and the Sitophilus oryzae primary symbiont (SOPE), revealed that both insect symbiont genomes were near-perfect subsets of the strain HS genome and yet each contained a unique set of pseudogenes (81). These results suggest that strain HS may be a representative environmental progenitor of the Sodalis-allied clade, which has independently formed symbioses with various insects. Continued examination of this clade will enhance knowledge of potential adaptations aiding in establishment within a broad range of niches, mechanisms facilitating host switching, and the impact of these host jumps on symbiont genome evolution.
Table 1.
Characterized members of the Enterobacteriaceae Sodalis-allied clade exhibiting ≥96% 16S rRNA identity
| Member of the Enterobacteriaceae Sodalis-allied clade as described in corresponding citation | Source | Insect order: family | Reference |
|---|---|---|---|
| Sodalis glossinidius | Tsetse fly, Glossina spp. | Diptera: Glossinidae | 43 |
| Symbiont | Hippoboscid fly, Craterina melbae | Diptera: Hippoboscidae | 70 |
| “Candidatus Sodalis melophagi” | Sheep ked, Melophagus ovinus | Diptera: Hippoboscidae | 71 |
| Sodalis-allied symbiont | Scutellerid stinkbug, Cantao ocellatus | Hemiptera: Scutelleridae | 72 |
| Sodalis-allied symbiont | Giant jewel stinkbug, Eucorysses grandis | Hemiptera: Scutelleridae | 73 |
| Symbiont | Long-tailed mealybug, Pseudococcus longispinus | Hemiptera: Pseudococcidae | 74 |
| Symbiont | Slender pigeon louse, Columbicola columbae | Phthiraptera: Philopteridae | 75 |
| Symbiont | Longhorn beetle, Tetropium castaneum | Coleoptera: Cerambycidae | 76 |
| Sitophilus primary symbiont | Grain weevil, Sitophilus spp. | Coleoptera: Curculionidae | 77 |
| Secondary symbiont | Chestnut weevil, Curculio sikkimensis | Coleoptera: Curculionidae | 78 |
| Secondary symbiont | Weevil, Archarius roelofsi | Coleoptera: Curculionidae | 79 |
| Secondary symbiont | Weevil, Curculio hachijoensis | Coleoptera: Curculionidae | 79 |
| Biostraticola tofi | Environmental, Tufa deposit biofilm isolate | n/aa | 80 |
| Strain HS | Environmental, hand wound | n/a | 81 |
n/a, not applicable.
The third bacterial member of the tsetse holobiont is an intracellular pathogen within the genus Wolbachia (44). While there is a high prevalence of Wolbachia infections in laboratory colonies (85), field populations are more stochastic and infection is also not detected in all tsetse species (86). This symbiont is transovarially transmitted through successive host generations and has recently been shown to induce cytoplasmic incompatibility within the tsetse host (59). The association may also have a long coevolutionary history with some tsetse species, as Wolbachia loci were found horizontally transferred into the host genome (86).
IMPACT OF ADDITIONAL MICROBES ON HOLOBIONT SUCCESS
The introduction of additional microbes can influence the fitness of the holobiont. For example, salivary gland hypertrophy virus (SGHV) infection is a well-characterized parasitic disease in tsetse (87). SGHV, a nuclear rod-shaped, enveloped DNA virus (88), has low infection rates in the field and can be both vertically and horizontally transmitted. This infection can quickly spread in laboratory colonies, driven by horizontal transmission through artificial feeding systems, which can harbor concentrated viral numbers in the blood that would otherwise quickly disseminate within a vertebrate host (87). Viral infection is associated with testicular degeneration and ovarian abnormalities (89–91), which can lead to decreased tsetse fertility and longevity (92, 93). This is just one example of how parasitic associations may impact the tsetse host and influence evolutionary adaptations by the bacterial symbionts. The interactions of SGHV and the tsetse microbiota remain largely unknown. SGHV infection, as well as other potential parasitic interactions, within tsetse populations should be considered when examining the success and evolution of the holobiont.
It should also be noted that recent field studies have found an unexpected diversity within the microbial community of tsetse which is dependent on host species and geographic region (49, 51). Differences in abiotic conditions and food sources may influence the composition of these transient microbial communities (49, 94). Although additional microbes have been found in association with tsetse in the field, only Wigglesworthia, Sodalis, and Wolbachia are maternally transmitted, as 16S rRNA clone libraries of 3rd instar larvae contain only these 3 bacterial species (35).
An additional major factor influencing the holobiont is trypanosome presence; once a fly becomes infected with trypanosomes, it remains infected for the duration of its life span (Fig. 1). Tsetse flies play an obligate role in the successful development and transmission of Trypanosoma spp. (reviewed in reference 95). A phenotypic characteristic differentiating tsetse species is their vector competency, i.e., their ability to support the development and transmission of trypanosomes to naive hosts (96–103). While in the fly, the parasites are also heavily bombarded by the tsetse immune system, including the synthesis of antimicrobial peptides and the production of reactive oxygen species (104–108). The presence of trypanosomes, as well as the related biological modifications within tsetse, such as heightened immune stimulation and increased competition for space and resources, may also impact their microbial symbionts.
THE TSETSE FLY AS A TOOL TO EXAMINE HOLOBIONT EVOLUTION
The hologenome theory of evolution has been used to describe how variation in the genetic material of symbiotic partners is an important factor in the adaptive evolution of the holobiont (3, 4). This theory is based on four assumptions, the first being that all metazoans are associated with microbial symbionts (4, 109). Second, the fitness of the holobiont requires cooperation among the partners, as conflict may prove detrimental to overall health. Within the tsetse holobiont, synergistic effects of coresidency were recently observed, as clearance of Wigglesworthia resulted in the loss of Sodalis over generations of the host (20). This phenomenon was possibly due to metabolic dependencies (110), as previously indicated by similar trends in population dynamics through tsetse development (111). Third, the hologenome can change through alterations of the genetic material of any partner. Symbionts can allow the holobiont to adapt more quickly to ecological disturbances, through the acquisition of novel capabilities by horizontal gene transfer or changes in population dynamics or community composition, thereby aiding in the persistence of the holobiont. Within the tsetse fly, an increase in the availability of nutrients, specifically, the supplementation of thiamine (B1) to blood meals, was shown to alter symbiont populations (55, 110) and decreased the transcription of corresponding biosynthetic loci by Wigglesworthia (55). Lastly, symbiotic associations must be passed on through generations of the host, maintaining the species composition of the holobiont. By applying the holobiont concept and hologenome theory of evolution to tsetse symbioses, research will elucidate novel mechanisms driving microbial species cooperation and adaptation, which enable successful cooccupancy within a specific niche. Because the Wigglesworthia, Sodalis, and T. b. brucei genomes are sequenced and annotated and tsetse's (G. morsitans) genome is soon to released (S. Aksoy, personal communication), comparative analyses have begun to spur empirical studies sure to advance our knowledge of microbial community evolution within hosts.
METABOLIC INTERACTIONS AMONG THE MICROBIOTA
While the genomic evolution and importance of individual symbiont species within tsetse have been examined, the community dynamics are only beginning to be explored. Although Wigglesworthia and Sodalis have different evolutionary histories with tsetse, they maintain parallel population dynamics through host development (111, 112), indicative of coordinated activities or a generalized level of host control. Unlike the extensive metabolic complementation observed within ancient coresident symbionts (11–16), comparative genomics reveals that Sodalis encodes a majority of Wigglesworthia genes (110). This genetic redundancy brings into question the factors contributing to the maintenance of both associations within the tsetse host and how cooperation, rather than competition, occurs among the symbionts. In addition to their colocalization within both the anterior midgut and milk glands, metabolic interplay among the partners has been previously described (55, 110). Specifically, a distinction between Wigglesworthia and Sodalis lies in their thiamine (vitamin B1) biosynthesis capabilities. The genome of Wigglesworthia encodes the complete pathway, while that of Sodalis does not. Thiamine, specifically in the form of thiamine monophosphate (TMP), the derivative putatively produced by Wigglesworthia, is required for Sodalis proliferation and intracellular infection (110). This nutrient may be imported by Sodalis through an ATP-driven thiamine ABC transporter (110). Moreover, Sodalis growth was dependent on TMP concentration, suggesting a role for TMP in the maintenance of homeostasis within the tsetse holobiont. TMP supplementation to the tsetse holobiont via blood meals also resulted in decreased transcription of the Wgm thiamine biosynthetic locus thiC and in reduced symbiont population density and cell viability (55). These alterations in symbiont dynamics may contribute to the efficiency and stability of the tsetse holobiont, with the reduction of the Wigglesworthia population likely representing a lower functional need. Metabolic interactions may be involved in symbiont population control and possibly aid in preventing antagonism among the partners. In support of this theory, a more intricate metabolic complementation has been observed among anciently coevolved microbial symbionts in other insect holobionts (11–16). For example, within the mealybug, essential amino acid synthesis necessitates a medley of gene products arising from both bacterial partners, and possibly the host, for completion (16).
UNDERSTANDING THE TSETSE HOLOBIONT FOR ENHANCED VECTOR CONTROL
Measures to prevent the spread of African trypanosomiasis, such as using mass insecticide spraying during outbreaks and the release of sterile males in restricted areas to reduce field populations, have been historically targeted at controlling the tsetse fly population (95). Such measures have been successful within their targeted locales (113). Nevertheless, the threat of trypanosomiasis remains relevant, as political instability (23) and decreased priority status implemented by local authorities, due to the reduced numbers of reported cases (114), impede the continuous efforts required to prevent the reestablishment and subsequent heightened disease incidence (115). By gaining a more holistic view of tsetse biology, insights into novel strategies for controlling the spread of the disease may be gained. In fact, a recent International Atomic Energy Agency (IAEA)-coordinated research project (CRP) aims to unravel the interactions between the tsetse host, Wigglesworthia, Sodalis, Wolbachia, and SGHV and the development of African trypanosomes to increase knowledge of ways to enhance refractoriness to trypanosome infection (116).
Past studies have demonstrated a positive correlation between Sodalis presence and trypanosome infections in field flies (65, 117). Sodalis is believed to contribute to the susceptibility of teneral flies (i.e., newly emerged unfed adults) through its endochitinase activity within the midgut which breaks down chitin (118), producing a byproduct of N-acetyl-d-glucosamine which inhibits the action of trypanocidal lectins (118, 119).
The role of Wigglesworthia in the tsetse fly's susceptibility to trypanosome infection remains largely unknown. A link between Wigglesworthia and trypanosome infection was suggested, as the removal of the symbiont resulted in higher susceptibility to midgut infection in older, nonteneral flies—typically a time point of low vector competency (58). Subsequently, the absence of Wigglesworthia was found to impair host immune system development (35). Thus, the higher susceptibility to trypanosome infection may be due to compromised immunity, although the effect of an altered nutritional state may also be a contributing factor. Genome comparisons between Wigglesworthia spp. revealed potential metabolome differences among the primary symbionts (54). One distinction lies in the complete retention of the chorismate (an intermediate in the production of aromatic compounds, including amino acids and vitamins) and downstream folate (vitamin B9) biosynthetic pathways by Wgm but not Wgb. Interestingly, the parasitic lifestyle of T. brucei subsp. has resulted in a highly restricted genomic repertoire, compensating for the absence of biosynthetic pathways by encoding transporters to sequester metabolites (including folate) from the environment (120, 121). This enhanced biosynthetic capability may contribute to the higher reported vector competency of the G. morsitans host relative to G. brevipalpis (96–98, 100), as trypanosomes necessitate exogenous folate for growth (122). Deeper investigation of this hypothesis is required. Enhanced understanding of the unique capabilities of specific Wigglesworthia spp. with respect to holobiont functioning may contribute to a more holistic view of factors that result in different levels of refractoriness between tsetse species.
Tsetse immune tolerance of symbionts may also influence the fly's susceptibility to trypanosome infection. For example, the contribution of the tsetse fly's immune system to the persistence of the Wigglesworthia symbiosis may also play a role in the fly's ability to transmit trypanosomes. The host pathogen recognition protein PGRP-LB, which scavenges peptidoglycan, thus preventing immune deficiency (IMD) signaling pathway stimulation, is intimately associated with maintaining the Wigglesworthia symbiosis (56, 123). PGRP-LB is maternally transmitted via milk secretions to developing offspring and is produced by adult flies only after their first blood meal (123). This protein also exhibits trypanocidal activity (123). Therefore, higher levels of PGRP-LB may aid in the refractory nature of older, nonteneral flies with respect to trypanosome infection.
Field studies examining the rates of microbial coinfections within tsetse may also provide insight into symbiont interactions. One study examining the association of coinfections in Glossina fuscipes fuscipes in Uganda found a negative correlation between the prevalences of Wolbachia and SGHV, while SGHV infection and trypanosome infection were positively correlated (124). This finding highlights the importance of examining the evolutionary and physiological effects of coinfection. One trypanosome control strategy that relies on symbiont interactions is known as paratransgenesis (for more details, see references 25, 26, and 125). Paratransgenesis involves manipulating Sodalis to express antitrypanosomal effector molecules and utilizes the cytoplasmic incompatibility properties of Wolbachia (59, 126) to drive the genetically modified symbiont into natural populations.
CONCLUSIONS
The low complexity of the tsetse holobiont and the annotated genomes of its members enable investigations into the evolutionary aspects of coresidence and holobiont adaptations when challenged with both intrinsic and ecological disturbances. Comparisons of the tsetse holobiont, in which members are still transitioning into the symbiotic lifestyle, to other anciently coevolved mutualisms will help describe mechanisms contributing to early integration and cooperation within a microbial community. Moreover, a more holistic and comprehensive understanding of the tsetse holobiont may identify additional factors promoting or inhibiting vector competency that may ultimately aid in controlling the spread of African trypanosomiasis.
ACKNOWLEDGMENTS
We thank Brian Weiss, Brittany Ott, and anonymous reviewers for critical review of the manuscript. We thank the Slovak Academy of Science and IAEA for providing tsetse pupae.
The Rio laboratory is supported by NIH NIAID and NSF-IOS funding.
Footnotes
Published ahead of print 8 July 2013
REFERENCES
- 1.Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1–10 [Google Scholar]
- 2.Bosch TCG, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology 114:185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5:355–362 [DOI] [PubMed] [Google Scholar]
- 4.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32:723–735 [DOI] [PubMed] [Google Scholar]
- 5.Beinart RA, Sanders JG, Faure B, Sylva SP, Lee RW, Becker EL, Gartman A, Luther GW, Seewald JS, Fisher CR, Girguis PR. 2012. Evidence for the role of endosymbionts in regional-scale habitat partitioning by hydrothermal vent symbioses. Proc. Natl. Acad. Sci. U. S. A. 109:E3241–E3250. 10.1073/pnas.1202690109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.NIH HMP Working Group, Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, Baker CC, Di Francesco V, Howcroft TK, Karp RW, Lunsford RD, Wellington CR, Belachew T, Wright M, Giblin C, David H, Mills M, Salomon R, Mullins C, Akolkar B, Begg L, Davis C, Grandison L, Humble M, Khalsa J, Little AR, Peavy H, Pontzer C, Portnoy M, Sayre MH, Starke-Reed P, Zakhari S, Read J, Watson B, Guyer M. 2009. The NIH Human Microbiome Project. Genome Res. 19:2317–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ibrahim M, Anishetty S. 2012. A meta-metabolome network of carbohydrate metabolism: Interactions between gut microbiota and host. Biochem. Biophys. Res. Commun. 428:278–284 [DOI] [PubMed] [Google Scholar]
- 8.Grice EA, Segre JA. 2011. The skin microbiome. Nat. Rev. Microbiol. 9:244–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen XA, Li S, Aksoy S. 1999. Concordant evolution of a symbiont with its host insect species: molecular phylogeny of genus Glossina and its bacteriome-associated endosymbiont, Wigglesworthia glossinidia. J. Mol. Evol. 48:49–58 [DOI] [PubMed] [Google Scholar]
- 11.McCutcheon JP, Moran NA. 2007. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. U. S. A. 104:19392–19397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takiya DM, Tran PL, Dietrich CH, Moran NA. 2006. Co-cladogenesis spanning three phyla: leafhoppers (Insecta: Hemiptera: Cicadellidae) and their dual bacterial symbionts. Mol. Ecol. 15:4175–4191 [DOI] [PubMed] [Google Scholar]
- 13.Wu D, Daugherty SC, Van Aken SE, Pai GH, Watkins KL, Khouri H, Tallon LJ, Zaborsky JM, Dunbar HE, Tran PL, Moran NA, Eisen JA. 2006. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 4:e188. 10.1371/journal.pbio.0040188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCutcheon JP, McDonald BR, Moran NA. 2009. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. U. S. A. 106:15394–15399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2:708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCutcheon JP, von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr. Biol. 21:1366–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 18.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55:247–266 [DOI] [PubMed] [Google Scholar]
- 19.Nogge G. 1976. Sterility in tsetse flies (Glossina morsitans Westwood) caused by loss of symbionts. Experientia 32:995–996 [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Brelsfoard C, Wu Y, Aksoy S. 2013. Intercommunity effects on microbiome and GpSGHV density regulation in tsetse flies. J. Invertebr. Pathol. 112:S32–S39 http://dx.doi.org/10.1016/j.jip.2012.03.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koga R, Tsuchida T, Fukatsu T. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. Biol. Sci. 270:2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koga R, Tsuchida T, Sakurai M, Fukatsu T. 2007. Selective elimination of aphid endosymbionts: effects of antibiotic dose and host genotype, and fitness consequences. FEMS Microbiol. Ecol. 60:229–239 [DOI] [PubMed] [Google Scholar]
- 23.Medecins Sans Frontieres 2012. Human African trypanosomiasis (sleeping sickness), p 63–77 In Zabriskie P. (ed), Fighting neglect. Medecins Sans Frontieres, Geneva, Switzerland: http://www.msf.es/fighting-neglect/Fighting%20Neglect_ENG.pdf [Google Scholar]
- 24.Budd LT. 1999. DFID-funded tsetse and trypanosome research and development since 1980: economic analysis, vol 2 Natural Resources International Limited, Aylesford, United Kingdom [Google Scholar]
- 25.Aksoy S, Weiss B, Attardo G. 2008. Paratransgenesis applied for control of tsetse transmitted sleeping sickness. Adv. Exp. Med. Biol. 627:35–48 [DOI] [PubMed] [Google Scholar]
- 26.Weiss B, Aksoy S. 2011. Microbiome influences on insect host vector competence. Trends Parasitol. 27:514–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frentiu FD, Robinson J, Young PR, McGraw EA, O'Neill SL. 2010. Wolbachia-mediated resistance to Dengue virus infection and death at the cellular level. PLoS One 5:e13398. 10.1371/journal.pone.0013398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O'Neill SL. 2009. A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139:1268–1278. 10.1016/j.cell.2009.11.042 [DOI] [PubMed] [Google Scholar]
- 29.Bian G, Xu Y, Lu P, Xie Y, Xi Z. 2010. The endosymbiotic bacterium Wolbachia induces resistance to Dengue virus in Aedes aegypti. PLoS Pathog. 6:e1000833. 10.1371/journal.ppat.1000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kambris Z, Cook PE, Phuc HK, Sinkins SP. 2009. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326:134–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cook PE, Hugo LE, Iturbe-Ormaetxe I, Williams CR, Chenoweth SF, Ritchie SA, Ryan PA, Kay BH, Blows MW, O'Neill SL. 2006. The use of transcriptional profiles to predict adult mosquito age under field conditions. Proc. Natl. Acad. Sci. U. S. A. 103:18060–18065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Ghosh AK, Bongio N, Stebbings KA, Lampe DJ, Jacobs-Lorena M. 2012. Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc. Natl. Acad. Sci. U. S. A. 109:12734–12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durvasula RV, Sundaram RK, Kirsch P, Hurwitz I, Crawford CV, Dotson E, Beard CB. 2008. Genetic transformation of a Corynebacterial symbiont from the Chagas disease vector Triatoma infestans. Exp. Parasitol. 119:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leak S. 1999. Tsetse biology and ecology. CABI Publishing, Nairobi, Kenya [Google Scholar]
- 35.Weiss BL, Wang JW, Aksoy S. 2011. Tsetse immune system maturation requires the presence of obligate symbionts in larvae. PLoS Biol. 9:e1000619. 10.1371/journal.pbio.1000619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osei-Poku J, Mbogo CM, Palmer WJ, Jiggins FM. 2012. Deep sequencing reveals extensive variation in the gut microbiota of wild mosquitoes from Kenya. Mol. Ecol. 21:5138–5150 [DOI] [PubMed] [Google Scholar]
- 37.Broderick NA, Lemaitre B. 2012. Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3:307–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nogge G. 1981. Significance of symbionts for the maintenance of an optimal nutritional state for successful reproduction in hematophagous arthropods. Parasitology 82:101–104 [Google Scholar]
- 39.Tobe SS, Langley PA. 1978. Reproductive physiology of Glossina. Annu. Rev. Entomol. 23:283–307 [DOI] [PubMed] [Google Scholar]
- 40.Ma WC, Denlinger DL. 1974. Secretory discharge and microflora of milk gland in tsetse flies. Nature 247:301–303 [Google Scholar]
- 41.Balmand S, Lohs C, Aksoy S, Heddi A. 2013. Tissue distribution and transmission routes for the tsetse fly endosymbionts. J. Invertebr. Pathol. 112:S116–S122 http://dx.doi.org/10.1016/j.jip.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aksoy S. 1995. Wigglesworthia gen. nov. and Wigglesworthia glossinidia sp. nov., taxa consisting of the mycetocyte-associated, primary endosymbionts of tsetse flies. Int. J. Syst. Bacteriol. 45:848–851 [DOI] [PubMed] [Google Scholar]
- 43.Dale C, Maudlin I. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int. J. Syst. Bacteriol. 49(Pt 1):267–275 [DOI] [PubMed] [Google Scholar]
- 44.O'Neill SL, Gooding RH, Aksoy S. 1993. Phylogenetically distant symbiotic microorganisms reside in Glossina midgut and ovary tissues. Med. Vet. Entomol. 7:377–383 [DOI] [PubMed] [Google Scholar]
- 45.Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. 2008. How many species are infected with Wolbachia?—A statistical analysis of current data. FEMS Microbiol. Lett. 281:215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6:741–751 [DOI] [PubMed] [Google Scholar]
- 47.Zhou WG, Rousset F, O'Neill S. 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. Biol. Sci. 265:509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng Q, Aksoy S. 1999. Tissue tropism, transmission and expression of foreign genes in vivo in midgut symbionts of tsetse flies. Insect Mol. Biol. 8:125–132 [DOI] [PubMed] [Google Scholar]
- 49.Geiger A, Fardeau ML, Grebaut P, Vatunga G, Josenando T, Herder S, Cuny G, Truc P, Ollivier B. 2009. First isolation of Enterobacter, Enterococcus, and Acinetobacter spp. as inhabitants of the tsetse fly (Glossina palpalis palpalis) midgut. Infect. Genet. Evol. 9:1364–1370 [DOI] [PubMed] [Google Scholar]
- 50.Geiger A, Fardeau ML, Falsen E, Ollivier B, Cuny G. 2010. Serratia glossinae sp. nov., isolated from the midgut of the tsetse fly Glossina palpalis gambiensis. Int. J. Syst. Evol. Microbiol. 60:1261–1265 [DOI] [PubMed] [Google Scholar]
- 51.Lindh JM, Lehane MJ. 2011. The tsetse fly Glossina fuscipes fuscipes (Diptera: Glossina) harbours a surprising diversity of bacteria other than symbionts. Antonie Van Leeuwenhoek 99:711–720 [DOI] [PubMed] [Google Scholar]
- 52.Weiss BL, Mouchotte R, Rio RVM, Wu YN, Wu Z, Heddi A, Aksoy S. 2006. Interspecific transfer of bacterial endosymbionts between tsetse fly species: infection establishment and effect on host fitness. Appl. Environ. Microbiol. 72:7013–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402–407 [DOI] [PubMed] [Google Scholar]
- 54.Rio RVM, Symula RE, Wang J, Lohs C, Wu YN, Snyder AK, Bjornson RD, Oshima K, Biehl BS, Perna NT, Hattori M, Aksoy S. 2012. Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: Glossinidae) obligate symbiont Wigglesworthia. mBio 3:e00240–11. 10.1128/mBio.00240-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snyder AK, McLain C, Rio RVM. 2012. The tsetse fly obligate mutualist Wigglesworthia morsitans alters gene expression and population density via exogenous nutrient provisioning. Appl. Environ. Microbiol. 78:7792–7797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang JW, Wu YN, Yang GX, Aksoy S. 2009. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. U. S. A. 106:12133–12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiss BL, Maltz M, Aksoy S. 2012. Obligate symbionts activate immune system development in the tsetse fly. J. Immunol. 188:3395–3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pais R, Lohs C, Wu YN, Wang JW, Aksoy S. 2008. The obligate mutualist Wigglesworthia glossinidia influences reproduction, digestion, and immunity processes of its host, the tsetse fly. Appl. Environ. Microbiol. 74:5965–5974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alam U, Medlock J, Brelsfoard C, Pais R, Lohs C, Balmand S, Carnogursky J, Heddi A, Takac P, Galvani A, Aksoy S. 2011. Wolbachia symbiont infections induce strong cytoplasmic incompatibility in the tsetse fly Glossina morsitans. PLoS Pathog. 7:e1002415. 10.1371/journal.ppat.1002415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss BL, Wang J, Maltz MA, Wu Y, Aksoy S. 2013. Trypanosome infection establishment in the tsetse fly gut is influenced by microbiome-regulated host immune barriers. PLoS Pathog. 9:e1003318. dio:10.1371/journal.ppat.1003318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wernegreen JJ. 2002. Genome evolution in bacterial endosymbionts of insects. Nat. Rev. Genet. 3:850–861 [DOI] [PubMed] [Google Scholar]
- 62.Wernegreen JJ. 2012. Strategies of genomic integration within insect-bacterial mutualisms. Biol. Bull. 223:112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10:13–26 [DOI] [PubMed] [Google Scholar]
- 64.Welburn SC, Maudlin I, Ellis DS. 1987. In vitro cultivation of Rickettsia-like-organisms from Glossina spp. Ann. Trop. Med. Parasitol. 81:331–335 [DOI] [PubMed] [Google Scholar]
- 65.Farikou O, Njiokou F, Mbida Mbida JA, Njitchouang GR, Djeunga HN, Asonganyi T, Simarro PP, Cuny G, Geiger A. 2010. Tripartite interactions between tsetse flies, Sodalis glossinidius and trypanosomes—an epidemiological approach in two historical human African trypanosomiasis foci in Cameroon. Infect. Genet. Evol. 10:115–121 [DOI] [PubMed] [Google Scholar]
- 66.Toh H, Weiss BL, Perkin SAH, Yamashita A, Oshima K, Hattori M, Aksoy S. 2006. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 16:149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Belda E, Moya A, Bentley S, Silva FJ. 2010. Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics 11:449. 10.1186/1471-2164-11-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiss BL, Wu YN, Schwank JJ, Tolwinski NS, Aksoy S. 2008. An insect symbiosis is influenced by bacterium-specific polymorphisms in outer-membrane protein A. Proc. Natl. Acad. Sci. U. S. A. 105:15088–15093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maltz MA, Weiss BL, O'Neill M, Wu Y, Aksoy S. 2012. OmpA-mediated biofilm formation is essential for the commensal bacterium Sodalis glossinidius to colonize the tsetse fly gut. Appl. Environ. Microbiol. 78:7760–7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nováková E, Hypsa V. 2007. A new Sodalis lineage from bloodsucking fly Craterina melbae (Diptera, Hippoboscoidea) originated independently of the tsetse flies symbiont Sodalis glossinidius. FEMS Microbiol. Lett. 269:131–135 [DOI] [PubMed] [Google Scholar]
- 71.Chrudimsky T, Husnik F, Novakova E, Hypsa V. 2012. Candidatus Sodalis melophagi sp. nov.: phylogenetically independent comparative model to the tsetse fly symbiont Sodalis glossinidius. PLoS One 7:e40354. 10.1371/journal.pone.0040354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, Ito M, Fukatsu T. 2010. Primary gut symbiont and secondary, Sodalis-allied symbiont of the Scutellerid stinkbug Cantao ocellatus. Appl. Environ. Microbiol. 76:3486–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, Ito M, Fukatsu T. 2011. Bacterial symbionts of the giant jewel stinkbug Eucorysses grandis (Hemiptera: Scutelleridae). Zool. Sci. 28:169–174 [DOI] [PubMed] [Google Scholar]
- 74.Duron O, Bouchon D, Boutin S, Bellamy L, Zhou L, Engelstadter J, Hurst GD. 2008. The diversity of reproductive parasites among arthropods: Wolbachia do not walk alone. BMC Biol. 6:27. 10.1186/1741-7007-6-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fukatsu T, Koga R, Smith WA, Tanaka K, Nikoh N, Sasaki-Fukatsu K, Yoshizawa K, Dale C, Clayton DH. 2007. Bacterial endosymbiont of the slender pigeon louse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl. Environ. Microbiol. 73:6660–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grünwald S, Pilhofer M, Holl W. 2010. Microbial associations in gut systems of wood- and bark-inhabiting longhorned beetles [Coleoptera: Cerambycidae]. Syst. Appl. Microbiol. 33:25–34. 10.1016/j.syapm.2009.10.002 [DOI] [PubMed] [Google Scholar]
- 77.Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P. 1998. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G + C content of an endocytobiotic DNA. J. Mol. Evol. 47:52–61 [DOI] [PubMed] [Google Scholar]
- 78.Toju H, Hosokawa T, Koga R, Nikoh N, Meng XY, Kimura N, Fukatsu T. 2010. “Candidatus Curculioniphilus buchneri,” a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl. Environ. Microbiol. 76:275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toju H, Tanabe AS, Notsu Y, Sota T, Fukatsu T. 2013. Diversification of endosymbiosis: replacements, co-speciation and promiscuity of bacteriocyte symbionts in weevils. ISME J. 7:1378–1390. 10.1038/ismej.2013.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Verbarg S, Frühling A, Cousin S, Brambilla E, Gronow S, Lünsdorf H, Stackebrandt E. 2008. Biostraticola tofi gen. nov., spec. nov., a novel member of the family Enterobacteriaceae. Curr. Microbiol. 56:603–608 [DOI] [PubMed] [Google Scholar]
- 81.Clayton AL, Oakeson KF, Gutin M, Pontes A, Dunn DM, von Niederhausern AC, Weiss RB, Fisher M, Dale C. 2012. A novel human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect-bacterial symbioses. PLoS Genet. 8:e1002990. 10.1371/journal.pgen.1002990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Snyder AK, McMillen CM, Wallenhorst P, Rio RVM. 2011. The phylogeny of Sodalis-like symbionts as reconstructed using surface-encoding loci. FEMS Microbiol. Lett. 317:143–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gürtler V, Stanisich VA. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3–16 [DOI] [PubMed] [Google Scholar]
- 84.Snyder AK, Adkins KZ, Rio RVM. 2011. Use of the internal transcribed spacer (ITS) regions to examine symbiont divergence and as a diagnostic tool for Sodalis-related bacteria. Insects 2:515–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cheng Q, Ruel TD, Zhou W, Moloo SK, Majiwa P, O'Neill SL, Aksoy S. 2000. Tissue distribution and prevalence of Wolbachia infections in tsetse flies, Glossina spp. Med. Vet. Entomol. 14:44–50 [DOI] [PubMed] [Google Scholar]
- 86.Doudoumis V, Tsiamis G, Wamwiri F, Brelsfoard C, Alam U, Aksoy E, Dalaperas S, Abd-Alla A, Ouma J, Takac P, Aksoy S, Bourtzis K. 2012. Detection and characterization of Wolbachia infections in laboratory and natural populations of different species of tsetse flies (genus Glossina). BMC Microbiol. 12(Suppl 1):S3 http://www.biomedcentral.com/1471-2180/12/S1/S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abd-Alla AMM, Parker AG, Vreysen MJB, Bergoin M. 2011. Tsetse salivary gland hypertrophy virus: hope or hindrance for tsetse control? PLoS Negl. Trop. Dis. 5:e1220. 10.1371/journal.pntd.0001220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jaenson TG. 1978. Virus-like rods associated with salivary gland hyperplasia in tsetse, Glossina pallidipes. Trans. R. Soc. Trop. Med. Hyg. 72:234–238 [DOI] [PubMed] [Google Scholar]
- 89.Jura WG, Odhiambo TR, Otieno LH, Tabu NO. 1988. Gonadal lesions in virus-infected male and female tsetse, Glossina pallidipes (Diptera: Glossinidae). J. Invertebr. Pathol. 52:1–8 [DOI] [PubMed] [Google Scholar]
- 90.Sang RC, Jura W, Otieno LH, Ogaja P. 1996. Ultrastructural changes in the milk gland of tsetse Glossina morsitans centralis (Diptera; Glossinidae) female infected by a DNA virus. J. Invertebr. Pathol. 68:253–259 [DOI] [PubMed] [Google Scholar]
- 91.Sang RC, Jura WG, Otieno LH, Mwangi RW, Ogaja P. 1999. The effects of a tsetse DNA virus infection on the functions of the male accessory reproductive gland in the host fly Glossina morsitans centralis (Diptera; Glossinidae). Curr. Microbiol. 38:349–354 [DOI] [PubMed] [Google Scholar]
- 92.Sang RC, Jura WG, Otieno LH, Tukei PM, Mwangi RW. 1997. Effects of tsetse DNA virus infection on the survival of a host fly, Glossina morsitans centralis (Diptera; Glossinidae). J. Invertebr. Pathol. 69:253–260 [Google Scholar]
- 93.Sang RC, Jura WG, Otieno LH, Mwangi RW. 1998. The effects of a DNA virus infection on the reproductive potential of female tsetse flies, Glossina morsitans centralis and Glossina morsitans morsitans (Diptera: Glossinidae). Mem. Inst. Oswaldo Cruz 93:861–864 [DOI] [PubMed] [Google Scholar]
- 94.Farikou O, Njiokou F, Simo G, Asonganyi T, Cuny G, Geiger A. 2010. Tsetse fly blood meal modification and trypanosome identification in two sleeping sickness foci in the forest of southern Cameroon. Acta Trop. 116:81–88 [DOI] [PubMed] [Google Scholar]
- 95.Walshe DP, Ooi CP, Lehane MJ, Haines LR. 2009. The enemy within: interactions between tsetse, trypanosomes and symbionts. Adv. Insect Phys. 37:119–175. 10.1016/S0065-2806(09)37003-4 [DOI] [Google Scholar]
- 96.Harley JM. 1971. Comparison of the susceptibility of infection with Trypanosoma rhodesiense of Glossina pallidipes, G. morsitans, G. fuscipes and G. brevipalpis. Ann. Trop. Med. Parasitol. 65:185–189 [DOI] [PubMed] [Google Scholar]
- 97.Moloo SK, Kutuza SB. 1988. Comparative study on the susceptibility of different Glossina species to Trypanosoma brucei brucei infection. Trop. Med. Parasitol. 39:211–213 [PubMed] [Google Scholar]
- 98.Moloo SK, Kabata JM, Sabwa CL. 1994. A study on the maturation of procyclic Trypanosoma brucei brucei in Glossina morsitans centralis and G. brevipalpis. Med. Vet. Entomol. 8:369–374 [DOI] [PubMed] [Google Scholar]
- 99.Moloo SK, Okumu IO, Kuria NM. 1998. Comparative susceptibility of Glossina longipennis and G. brevipalpis to pathogenic species of Trypanosoma. Med. Vet. Entomol. 12:211–214 [DOI] [PubMed] [Google Scholar]
- 100.Moloo SK, Orinda GO, Sabwa CL, Minja SH, Masake RA. 1999. Study on the sequential tsetse-transmitted Trypanosoma congolense, T. brucei brucei and T. vivax infections to African buffalo, eland, waterbuck, N′Dama and Boran cattle. Vet. Parasitol. 80:197–213 [DOI] [PubMed] [Google Scholar]
- 101.Distelmans W, D'Haeseleer F, Kaufman L, Rousseeuw P. 1982. The susceptibility of Glossina palpalis palpalis at different ages to infection with Trypanosoma congolense. Ann. Soc. Belg. Med. Trop. 62:41–47 [PubMed] [Google Scholar]
- 102.Dale CC, Welburn SC, Maudlin I, Milligan PJ. 1995. The kinetics of maturation of trypanosome infections in tsetse. Parasitology 111:187–191 [DOI] [PubMed] [Google Scholar]
- 103.Peacock L, Ferris V, Bailey M, Gibson W. 2012. The influence of sex and fly species on the development of trypanosomes in tsetse flies. PLoS Negl. Trop. Dis. 6:e1515. 10.1371/journal.pntd.0001515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hao Z, Kasumba I, Lehane MJ, Gibson WC, Kwon J, Aksoy S. 2001. Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc. Natl. Acad. Sci. U. S. A. 98:12648–12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hao Z, Kasumba I, Aksoy S. 2003. Proventriculus (cardia) plays a crucial role in immunity in tsetse fly (Diptera: Glossinidiae). Insect Biochem. Mol. Biol. 33:1155–1164 [DOI] [PubMed] [Google Scholar]
- 106.Lehane MJ, Aksoy S, Gibson W, Kerhornou A, Berriman M, Hamilton J, Soares MB, Bonaldo MF, Lehane S, Hall N. 2003. Adult midgut expressed sequence tags from the tsetse fly Glossina morsitans morsitans and expression analysis of putative immune response genes. Genome Biol. 4:R63. 10.1186/gb-2003-4-10-r63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hu C, Aksoy S. 2006. Innate immune responses regulate trypanosome parasite infection of the tsetse fly Glossina morsitans morsitans. Mol. Microbiol. 60:1194–1204 [DOI] [PubMed] [Google Scholar]
- 108.Nayduch D, Aksoy S. 2007. Refractoriness in tsetse flies (Diptera: Glossinidae) may be a matter of timing. J. Med. Entomol. 44:660–665 [DOI] [PubMed] [Google Scholar]
- 109.Rosenberg E, Zilber-Rosenberg I. 2011. Symbiosis and development: the hologenome concept. Birth Defects Res. C Embryo Today 93:56–66 [DOI] [PubMed] [Google Scholar]
- 110.Snyder AK, Deberry JW, Runyen-Janecky L, Rio RVM. 2010. Nutrient provisioning facilitates homeostasis between tsetse fly (Diptera: Glossinidae) symbionts. Proc. Biol. Sci. 277:2389–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rio RV, Wu YN, Filardo G, Aksoy S. 2006. Dynamics of multiple symbiont density regulation during host development: tsetse fly and its microbial flora. Proc. Biol. Sci. 273:805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hamidou Soumana I, Berthier D, Tchicaya B, Thevenon S, Njiokou F, Cuny G, Geiger A. 2013. Population dynamics of Glossina palpalis gambiensis symbionts, Sodalis glossinidius, and Wigglesworthia glossinidia, throughout host-fly development. Infect. Genet. Evol. 13:41–48 [DOI] [PubMed] [Google Scholar]
- 113.Gechere G, Terefe G, Belihu K. 2012. Impact of tsetse and trypanosomiasis control on cattle herd composition and calf growth and mortality at Arbaminch District (Southern Rift Valley, Ethiopia). Trop. Anim. Health Prod. 44:1745–1750 [DOI] [PubMed] [Google Scholar]
- 114.Simarro PP, Jannin J, Cattand P. 2008. Eliminating human African trypanosomiasis: where do we stand and what comes next? PLoS Med. 5:e55. 10.1371/journal.pmed.0050055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Welburn SC, Maudlin I, Simarro PP. 2009. Controlling sleeping sickness—a review. Parasitology 136:1943–1949 [DOI] [PubMed] [Google Scholar]
- 116.Van Den Abbeele J, Bourtzis K, Weiss B, Cordon-Rosales C, Miller W, Abd-Alla AM, Parker A. 2013. Enhancing tsetse fly refractoriness to trypanosome infection—a new IAEA coordinated research project. J. Invertebr. Pathol. 112:S142–S147 http://dx.doi.org/10.1016/j.jip.2012.07.020 [DOI] [PubMed] [Google Scholar]
- 117.Maudlin I, Welburn SC, Mehlitz D. 1990. The relationship between rickettsia-like-organisms and trypanosome infections in natural populations of tsetse in Liberia. Trop. Med. Parasitol. 41:265–267 [PubMed] [Google Scholar]
- 118.Welburn SC, Arnold K, Maudlin I, Gooday GW. 1993. Rickettsia-like organisms and chitinase production in relation to transmission of trypanosomes by tsetse-flies. Parasitology 107:141–145 [DOI] [PubMed] [Google Scholar]
- 119.Welburn SC, Maudlin I. 1992. The nature of the teneral state in Glossina and its role in the acquisition of trypanosome infection in tsetse. Ann. Trop. Med. Parasitol. 86:529–536 [DOI] [PubMed] [Google Scholar]
- 120.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme W, Hannick L, Aslett MA, Shallom J, Marcello L, Hou LH, Wickstead B, Alsmark UCM, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetter J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, et al. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309:416–422 [DOI] [PubMed] [Google Scholar]
- 121.Jackson AP, Sanders M, Berry A, McQuillan J, Aslett MA, Quail MA, Chukualim B, Capewell P, MacLeod A, Melville SE, Gibson W, Barry JD, Berriman M, Hertz-Fowler C. 2010. The genome sequence of Trypanosoma brucei gambiense, causative agent of chronic human african trypanosomiasis. PLoS Negl. Trop. Dis. 4:e658. 10.1371/journal.pntd.0000658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sienkiewicz N, Jaroslawski S, Wyllie S, Fairlamb AH. 2008. Chemical and genetic validation of dihydrofolate reductase-thymidylate synthase as a drug target in African trypanosomes. Mol. Microbiol. 69:520–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang J, Aksoy S. 2012. PGRP-LB is a maternally transmitted immune milk protein that influences symbiosis and parasitism in tsetse's offspring. Proc. Natl. Acad. Sci. U. S. A. 109:10552–10557. 10.1073/pnas.1116431109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alam U, Hyseni C, Symula RE, Brelsfoard C, Wu YN, Kruglov O, Wang JW, Echodu R, Alioni V, Okedi LM, Caccone A, Aksoy S. 2012. Implications of microfauna-host interactions for trypanosome transmission dynamics in Glossina fuscipes fuscipes in Uganda. Appl. Environ. Microbiol. 78:4627–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rio RV, Hu Y, Aksoy S. 2004. Strategies of the home-team: symbioses exploited for vector-borne disease control. Trends Microbiol. 12:325–336 [DOI] [PubMed] [Google Scholar]
- 126.Doudoumis V, Alam U, Aksoy E, Abd-Alla AMM, Tsiamis G, Brelsfoard C, Aksoy S, Bourtzis K. 2013. Tsetse-Wolbachia symbiosis: comes of age and has great potential for pest and disease control. J. Invertebr. Pathol. 112:S94–S103. 10.1016/j.jip.2012.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]