Abstract
When deprived of a combined-nitrogen source in the growth medium, the filamentous cyanobacterium Anabaena sp. PCC 7120 (Anabaena) can form heterocysts capable of nitrogen fixation. The process of heterocyst differentiation takes about 20 to 24 h, during which extensive metabolic and morphological changes take place. Guanosine tetraphosphate (ppGpp) is the signal of the stringent response that ensures cell survival by adjusting major cellular activities in response to nutrient starvation in bacteria, and ppGpp accumulates at the early stage of heterocyst differentiation (J. Akinyanju, R. J. Smith, FEBS Lett. 107:173–176, 1979; J Akinyanju, R. J. Smith, New Phytol. 105:117–122, 1987). Here we show that all1549 (here designated relana) in Anabaena, homologous to relA/spoT, is upregulated in response to nitrogen deprivation and predominantly localized in vegetative cells. The disruption of relana strongly affects the synthesis of ppGpp, and the resulting mutant, all1549Ωsp/sm, fails to form heterocysts and to grow in the absence of a combined-nitrogen source. This phenotype can be complemented by a wild-type copy of relana. Although the upregulation of hetR is affected in the mutant, ectopic overexpression of hetR cannot rescue the phenotype. However, we found that the mutant rapidly loses its viability, within a time window of 3 to 6 h, following the deprivation of combined nitrogen. We propose that ppGpp plays a major role in rebalancing the metabolic activities of the cells in the absence of the nitrogen source supply and that this regulation is necessary for filament survival and consequently for the success of heterocyst differentiation.
INTRODUCTION
The stringent response is primarily defined as inhibition of rRNA and protein synthesis in response to amino acid starvation in Escherichia coli (1). In addition to amino acid starvation, the stringent response is also involved in cell regulation under other stress conditions such as limitation of carbon (2), of iron (3), and of fatty acid (4), oxidative stress (5), or acid shock (6). This mechanism allows bacteria to divert cellular resources from growth toward survival and thus constitutes an important means for cellular adaptation under stress conditions. The stringent response is mediated by the signal molecules guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), collectively referred to as ppGpp. In E. coli, these signaling molecules are produced by two homologous enzymes, RelA and SpoT. RelA is involved in ppGpp synthesis, and SpoT is a bifunctional enzyme which contributes predominately to ppGpp degradation or slightly to ppGpp synthesis (1). However, by bioinformatics analysis, Mittenhuber found that most bacterial genomes encode only one dual-function enzyme which displays both synthesis and degradation activities (7). ppGpp signaling is widely present not only in bacteria but also in plants (8, 9). In Arabidopsis, the At-RSH1, a Rel/SpoT homolog, can complement the rel mutant phenotypes in E. coli and Streptomyces coelicolor A3 (2, 8). Recently, studies in metazoa provided evidence that ppGpp may also be present in animals; indeed, Mesh1 encoding ppGpp hydrolase was found in Drosophila and human (10). In Drosophila, deletion of Mesh1 led to body growth retardation and starvation resistance damage (10). These results imply that the control of cellular processes by ppGpp is evolutionarily conserved.
ppGpp acts as a global regulator in the control of a variety of developmental processes in bacteria such as biofilm formation in Listeria monocytogenes (11), quorum sensing in Pseudomonas aeruginosa (12), fruiting body development in Myxococcus xanthus (13), antibiotic production in Streptomyces (14, 15), and virulence in Legionella pneumophila (16). Anabaena sp. PCC 7120 (here Anabaena) is a filamentous nitrogen-fixing cyanobacterium that forms heterocysts when grown in a medium deprived of combined nitrogen. Heterocysts are specialized cells and provide a micro-oxic environment suitable for nitrogen fixation catalyzed by the oxygen-sensitive nitrogenase (17). Several hundreds of genes are involved in heterocyst differentiation (17, 18); among them, NtcA and HetR are key regulators required for the initiation of heterocyst development. NtcA is a global nitrogen-control transcription factor, and its DNA-binding activity is regulated by 2-oxoglutarate through an allosteric control mechanism (19, 20). HetR, also a DNA-binding protein, is a master regulator specifically required for the initiation of heterocyst development (21). A hetR mutant fails to form heterocysts, whereas overexpression of hetR increases heterocyst frequency (22–24). Expression of ntcA and hetR shows a mutual dependency during heterocyst development (25, 26).
Heterocyst development is induced upon deprivation of combined nitrogen, and this process lasts about 20 to 24 h before mature heterocysts can fix nitrogen to support filament growth. During this long period, extensive metabolic modification, such as the degradation of nitrogen reserves, occurs to ensure that the cells can survive during this period of nitrogen limitation (27). Since the stringent response plays critical roles in cell adaptation under nutrient limitation conditions in various bacteria, we wondered whether ppGpp signaling could be involved in the process of heterocyst development. In a previous study, Ning et al. reported that all1549 (ana-rsh), a spoT homolog in Anabaena, could complement the relA spoT mutant in E. coli. They were unable to inactivate all1549 and thus suggested that all1549 was essential for cell growth but with no link to heterocyst differentiation (28). On the basis of the present study, we report that the all1549 gene, here proposed as relana following the recent nomenclature (29), is required for diazotrophic growth of Anabaena and that the Relana protein is localized specifically in vegetative cells. Our results indicate that relana is involved in heterocyst development because it is required for cell survival during nitrogen step-down.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains and plasmids used in this study are described in Table 1. Anabaena and its derivatives were grown in BG11 medium (with nitrate) or BG110 medium (without combined nitrogen), as described previously (30). All cyanobacterial strains were grown at 30°C under conditions of illumination (∼40 μE m−2 s−1). Neomycin (50 μg ml−1) or spectinomycin (10 μg ml−1) was added to the culture medium when necessary. For the induction of amino acid starvation, serine hydroxamate (SHX; Sigma) was added to the culture at the final concentration of 1 g liter−1 (31). E. coli strains were grown in LB or minimal medium (MM). Plasmids were transferred from E. coli into Anabaena strains by conjugation as described previously (32).
Table 1.
Strains and plasmids
| Strain or plasmid | Descriptiona | Source |
|---|---|---|
| Strains | ||
| Anabaena sp. PCC 7120 | Wild type | Pasteur Culture Collection |
| Anabaena all1549Ωsp/sm | Disruption relana; Spr Smr | This study |
| Anabaena C1549 | Nmr Spr Smr; pRL25T-C1549 introduced into all1549Ωsp/sm strain | This study |
| E. coli MG1655 | Wild-type K-12 strain | 37 |
| E. coli CF1693 | MG1655 ΔrelA251 ΔspoT207 | 37 |
| Plasmids | ||
| pGEX-6p-1 | Expressing vector | GE Healthcare |
| pGEX-6p-1549 | pGEX-6p-1 carrying relana ORF | This study |
| pRL277 | Smr Spr; sacB-bearing cloning vector | 33 |
| pRL277-HRS | Smr Spr; pRL277 carrying an internal fragment (from position 277 to 1173 relative to the translational initiation codon) of the relana gene | This study |
| pRL25T | Kmr Nmr; pDU1-based shuttle vector | 32, 45 |
| pRL25T-p69-gfp | Kmr Nmr; pRL25T carrying relana promoter and gfp coding sequence | This study |
| pRL25T-C1549 | Kmr Nmr; pRL25T containing relana ORF and native promoter, used to complement mutant | This study |
| pRL25T-PpetE-hetR | Kmr Nmr; pRL25T carrying PpetE-hetR fusion | This study |
| pRL25N-1549-lgfp | Kmr Nmr; pRL25N-lgfp carrying relana under the control of native promoter | This study |
| pRL25N-ACT-lgfp | Kmr Nmr; pRL25N-lgfp carrying relana without ACT domain coding sequence under the control of native promoter | This study |
Km, kanamycin; Nm, neomycin; Sm, streptomycin; Sp, spectinomycin.
Plasmid construction.
E. coli strain TG1 was used for all cloning experiments. All constructs were verified by DNA sequencing. All primers used in this study are listed in Table 2.
Table 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) |
|---|---|
| HRS-1 | CCTCTGCAGGGATCCATGATGTAGTTGAAGATACAGA |
| HRS-2 | CCTAGATCTACTTTGCCATTCCAATAATT |
| oe1549-1 | CTTGAATTCATGAGCAGCATCGCTATTAGTTCCC |
| oe1549-2 | CTTCTCGAGTCATTCATCAATTTGACCAACACG |
| 1549gfp-1 | CTTGCGGCCGCGGATCCAAGAGTTGCTTTAACCCAGC |
| 1549gfp-2 | CCTCATATGACAATAGAAGAGGTGATCGG |
| Del_1549–2 | CTGCAGCTCGAGTTCATCAATTTGACCAACACGGCG |
| Del_ACT-2 | CTGCAGCTCGAGGACTTCGCCATAACCCATAC |
| PpetE-1 | CTTGGATCCTAAAGCCTGTGAAATTAACTG |
| petER-2 | GTTACTCATATGCGTTCTCCTAACCTGTAG |
| oehetR-1 | AGAACGCATATGAGTAACGACATCGATCTGA |
| oehetR-2 | CTTCTCGAGGCCGAGTCATTTGTCATCAC |
| rnpB-1 | CCAGTTCCGCTATCAGAGAG |
| rnpB-2 | GAGGAGAGAGTTGGTGGTAAG |
| hetR-1 | TACTCTGGCACGGTGACAAG |
| hetR-2 | AGGGCATAGAAGGGCATTCC |
| RT1549-1 | GATAGCCGTCGTCGTTCAG |
| RT1549-2 | TAATTCGCCAGATCCCTAAGC |
| cm1549-1 | CATTTGTATCTGGTGGCTTTTGTT |
| cm1549-2 | ACGCTATGTTCTCTTGCTTTTGTC |
Plasmid pRL277-HRS for relana gene disruption was constructed with the suicide vector pRL277 (33). Plasmid pRL277-HRS contained an internal fragment of relana (from position 277 to 1173 starting with the corresponding ATG codon) amplified by PCR using primers HRS-1 and HRS-2. The amplified fragment was digested with BglII and XhoI and subsequently ligated into the pRL277 suicide vector digested by the same enzymes.
To complement the all1549Ωsp/sm mutant, the relana gene and its promoter region were amplified by PCR using primers 1549gfp-1 and oe1549-2. The amplified fragment was digested with BamHI and XhoI and then cloned into the same sites of pRL25T. The final construct was named pRL25T-C1549. The pRL25T-p69-gfp plasmid was used to express the transcriptional fusion between relana and the green fluorescent protein (GFP) gene, gfp. The promoter region of relana was amplified by PCR using primers 1549gfp-1 and p69-2. The amplified fragment was digested with BamHI and PstI, cloned between the same sites of pBS-gfp, and then re-excised as a BamHI-XhoI fragment and inserted into the pRL25T shuttle vector between the BamHI and XhoI sites to give pRL25Tp69-gfp. The translational GFP fusion under the control of the relana promoter was amplified using primers 1549gfp-1 and Del_1549-2. This fragment was digested by BamHI and XhoI and then cloned into pRL25N-lgfp, which was derived from pRL25T bearing the gfp coding sequence (unpublished data) to produce pRL25N-1549-lgfp. Plasmid pRL25N-ACT-lgfp was a construct derived from Relana-GFP fusion but with the corresponding ACT domain deleted. DNA fragment was amplified by PCR using primers 1549gfp-1 and Del_ACT-2 and then digested by BamHI and XhoI and cloned into pRL25N-lgfp to produce pRL25N-ACT-lgfp. To construct plasmid pGEX-6p-1549, we amplified the relana gene by PCR using primers oe1549-1 and oe154-2. The 2,256-bp PCR product was digested by EcoRI and XhoI and cloned into vector pGEX-6p-1. The pRL25T-PpetE-hetR plasmid was made by overlapping extension PCR using primers PpetE-1 and petER-2 and primers oehetR-1 and oehetR-2. Subsequently, the PpetE-hetR fragment was digested by BamHI and XhoI and inserted into pRL25T at the same sites.
Construction of mutant.
For construction of the all1549Ωsp/sm mutant, plasmid pRL277-HRS was introduced into wild-type Anabaena by triparental mating. The mutant was selected by positive selection with spectinomycin. The single-crossover mutant, here called all1549Ωsp/sm, was confirmed by PCR using wild-type DNA as a positive control. For complementation, plasmid pRL25T-C1549 was introduced into the all1549Ωsp/sm mutant by conjugation, and the complemented strain was named C1549.
Measurement of ppGpp by high-performance liquid chromatography (HPLC).
The procedure for extraction of nucleotides with formic acid was adapted from those described previously (34, 35). For E. coli, 10 ml cells was collected from LB liquid culture by centrifugation. These cells were washed twice with MM and resuspended in an equal volume of MM (optical density at 600 nm [OD600] at 0.5). A 1-ml volume of this suspension was immediately transferred into a new tube containing 0.1 ml of 11 M formic acid and then vigorously mixed and incubated on ice for 45 min. In parallel, a 1-ml suspension was cultured for the indicated times at 37°C in the presence of serine hydroxamate (SHX; Sigma) at a final concentration of 1 g liter−1 followed by extraction with formic acid as described above. For Anabaena, 1.5 liters of cells (OD750 at 0.5) was collected by membrane filtration and then divided into three aliquots. One aliquot was resuspended in 100 ml BG11 medium and immediately extracted with 11 M formic acid. Another two parts were cultured for the indicated times in 100 ml BG11 medium plus 1 g liter−1 SHX followed by collection with filtration and extraction with 11 M formic acid. After extraction, all samples were centrifuged at 12,000 rpm for 10 min at 4°C. The supernatant was filtered through 0.22-μm-pore-size filters, adjusted to pH 4.0 with KOH, and stored at −20°C.
For the assay of ppGpp, a sample of 90 μl was subjected to processing using an anion-exchange column (Partisil SAX 10; Whatman) (4.6 by 250 mm) at a flow rate of 1 ml min−1. Nucleotides were separated by a gradient made of buffer A (7 mM KH2PO4; adjusted to pH 4.0 with H3PO4) and buffer B (0.5 M KH2PO4, 0.5 M Na2SO4; adjusted to pH 5.0 with KOH). The proportion of buffer B increased during 20 min from 0% (vol/vol) to 100% (vol/vol) and was then maintained for 20 min at 100% (vol/vol). Signals were detected by UV absorbance at 260 nm. The ppGpp standard was purchased from TriLink Biosciences. Standard curves were established using different concentrations of ppGpp, and the detection limit was approximately 100 nM.
RNA analysis.
To isolate RNA for real-time quantitative reverse transcription-PCR (qPCR) analysis, 30 ml of cells was filtered quickly on a membrane (5 μm pore size). Total RNA was extracted using a hot phenol method (36). RNA samples were treated with RNase-free DNase I (Invitrogen). Quantitative PCR (qPCR) was carried out as described previously (36) using specific primers (Table 2).
Cell viability test.
Cells were grown at the exponential phase and then collected and washed three times with BG110 medium. Cells were then adjusted to the initial optical density (OD750 = 0.12) and cultured into a medium deprived of combined nitrogen (BG110) at 30°C with shaking under conditions of illumination. To measure cell viability under such conditions over time, ammonium was added back to an aliquot at a different time point, and the aliquot was cultured to assess the ability to grow. Both optical density and chlorophyll a contents were monitored at various time points.
RESULTS
Relana is capable of ppGpp synthesis.
The Relana protein of Anabaena is a protein homologous to RelA/SpoT in E. coli. It was previously called Ana-Rsh (28), and we propose here to use the designation Relana, following the nomenclature of a recent review (29).To further confirm that relana is functionally related to Rel/SpoT as proposed by Ning et al. (28), plasmid pGEX-6p-1549, which carries the entire coding region of relana, was transformed into E. coli strain CF1693, a relA/spoT double mutant which cannot grow on minimal medium unless supplemented with multiple amino acids (37). The pGEX-6p-1 empty vector was used as a control. For growth analysis on LB and MM plates, the procedure was carried out as reported previously (31, 37). The CF1693 strain containing pGEX-6p-1 was unable to grow on a MM plate, while that containing pGEX-6p-1549 could grow under similar conditions, just as in the case of the wild-type MG1655 transformed with pGEX-6p-1 (Fig. 1). These results indicate that relana was expressed in E. coli and produced a functional protein capable of ppGpp synthesis.
Fig 1.
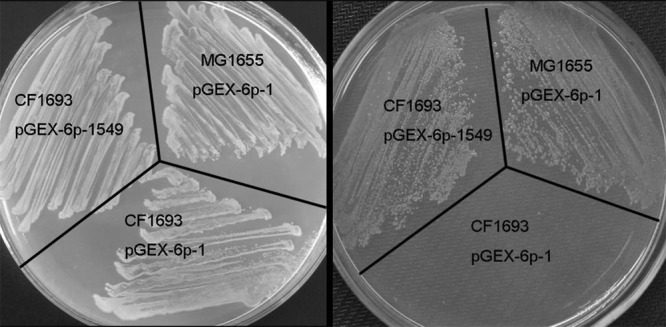
Complementation test of E. coli rel spoT mutant by relana of Anabaena. The images show the results of complementation of CF1693, a strain with mutations in the relA and spoT genes, on LB (left) and MM (right) plates. MG1655, a strain of wild-type E. coli, was transformed with empty vector. CF1693 was transformed with the plasmids as indicated.
The relana gene is required for diazotrophic growth and heterocyst differentiation.
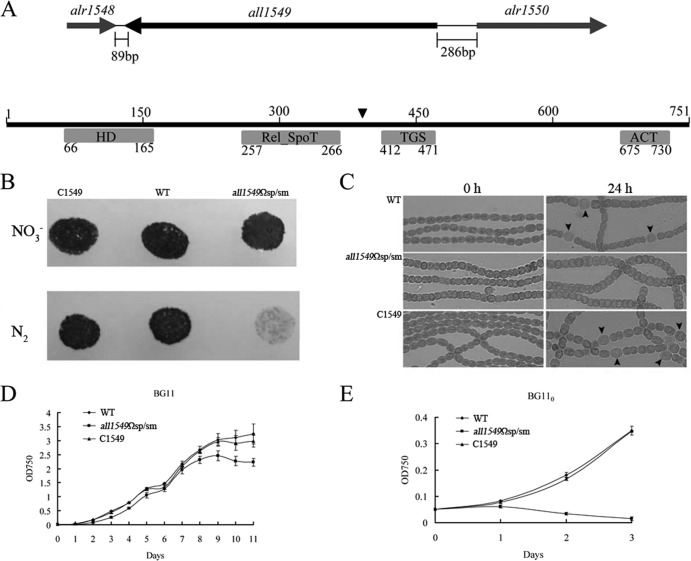
Relana is composed of four domains: HD (38), RelA_SpoT, TGS (39), and ACT (40–42). The structural conservation of Relana suggests that the corresponding gene plays an important role in the process of cell growth that is similar to that of SpoT as known in many other bacteria. In order to investigate the function of the relana gene, we attempted to inactivate this gene. Because relana forms a monocistronic unit (Fig. 2A), no polar effect on the transcription of its flanking genes was expected when the relana gene was disrupted. We first attempted to inactivate the coding region by double homologous recombination with insertion of an antibiotic resistance cassette but failed despite many attempts. The reason for this failure remains unknown, but the failure is consistent with the negative result reported by Ning et al. (28). We thus tried a second approach using an internal gene fragment borne on a suicide vector for gene inactivation. Plasmid pRL277-HRS, containing an internal 897-bp fragment (from position 277 to 1173 relative to the translational initiation codon) of the relana gene, was transferred into Anabaena by conjugation (43, 44). After selection, the insertion of the internal fragment on this suicide vector through homologous recombination would generate two partial alleles; neither of the alleles would be full, and thus, neither would be functional. After verification by PCR, the relana gene was found to be disrupted between the RelA_SpoT domain and the TGS domain (Fig. 2A). The resultant mutant, all1549Ωsp/sm, was fully segregated as confirmed by PCR using gene- and vector-specific primers 1549gfp-1/oe1549-2 and cm1549-1/cm1549-2 (data not shown).
Fig 2.
Phenotype of the all1549Ωsp/sm mutant. (A) Genomic context of relana from Anabaena. The black triangle indicates the position of disruption following single-crossover recombination. (B) Growth on solid agar plates with nitrogen (NO3−) or without nitrogen (N2). (C) Micrographs of the wild type (WT), the all1549Ωsp/sm mutant, and the complemented strain (C1549) after nitrogen deprivation. The arrowheads point to heterocysts. (D and E) Measurement of growth rate. Strains were grown in BG11 (+N) (D) or in BG110 (−N) (E). Error bars indicate standard deviations of the results determined from triplicate cultures.
We checked the growth of the all1549Ωsp/sm mutant on either nitrogen-containing (BG11) or nitrogen-free (BG110) medium. The all1549Ωsp/sm mutant grew slightly more slowly than the wild-type strain when nitrate was used as a nitrogen source but completely failed to grow in the absence of a combined-nitrogen source (Fig. 2D and E). Furthermore, microscopic observation showed that the all1549Ωsp/sm mutant did not form heterocysts after induction by deprivation of combined nitrogen. In comparison, the wild-type strain formed heterocysts, as expected (Fig. 2C). To check if the phenotype of the all1549Ωsp/sm mutant was due to the disruption of the relana gene, a complementation test was carried out. Plasmid pRL25T-C1549, constructed using a pDU1-based shuttle vector (32, 45) harboring the entire open reading frame (ORF) of relana as well as its promoter region, was transferred into mutant strain all1549Ωsp/sm. The complemented strain, called C1549, grows at a rate slightly slower than that of the wild type when nitrate is present in the growth medium. When cultured in the absence of a combined nitrogen source, C1549 recovered the ability to grow as well as to form heterocysts just like the wild type (Fig. 2). We therefore conclude that relana is required for heterocyst development.
Cellular and subcellular localization of Relana protein.
It was previously shown that relana was downregulated in heterocysts by using a transcriptional fusion to gfp (28). We could confirm this result using a similar fusion, the pRL25T-p69-gfp plasmid (data not shown). We further tested the expression of relana using qPCR and primers RT1549-1 and RT1549-2, following the deprivation of combined nitrogen. hetR, upregulated in response to nitrogen depletion in the wild-type strain (22), was used as a positive control. As shown in Fig. 3A, the transcriptional level of relana was enhanced approximately 2-fold at 24 h after the culture was moved from nitrogen-replete to nitrogen-depleted medium. This result was similar to the data obtained by transcriptome analysis (46).
Fig 3.
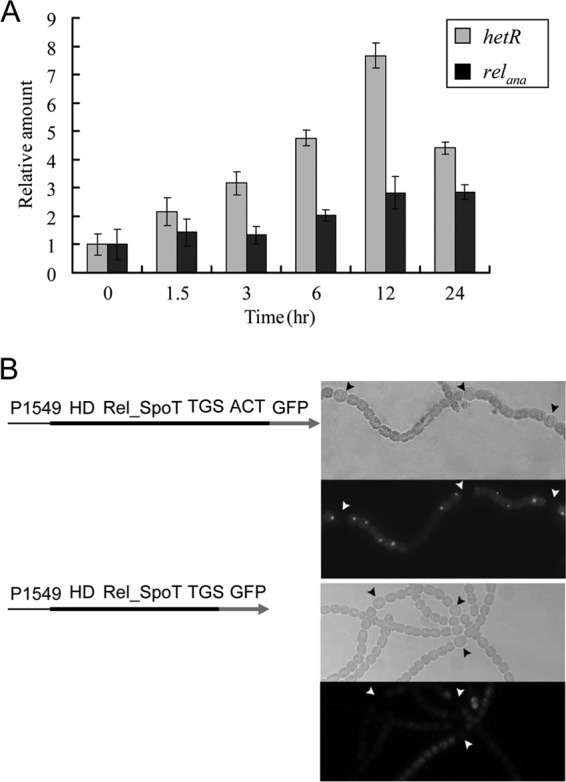
Spatiotemporal expression of relana. (A) Transcriptional levels of relana gene in wild type (WT). The result determined with the hetR gene was used as the positive control. The rnpB gene was selected as the reference gene for normalizing the data obtained by qPCR. (B) Localization of full-length and truncated Relana in the wild-type strain bearing plasmids pRL25N-1549-lgfp and pRL25N-ACT-lgfp, respectively. P1549 represents the native promoter. The results were visualized under bright-field and fluorescence microscopy at 24 h after nitrogen starvation. Arrowheads indicate heterocysts.
To test the localization of Relana in the filament in the wild-type background, we constructed a Relana-GFP translational fusion in which the gfp gene was fused at the 3′ terminus of relana with its native promoter. Fluorescence foci, often one per cell, were observed in the vegetative cells under the nitrogen-deprivation condition, but such foci were absent from heterocysts (Fig. 3B). Previous reports suggested that SpoT homologous protein is aggregated in E. coli (47) and forms trimers in Mycobacterium tuberculosis (48), but the physiological significance remains unclear. When the ACT domain at the C-terminal end of Relana was removed from the translational fusion, the fusion product was still localized in vegetative cells but the GFP-dependent fluorescence became diffuse compared to that from the full-length Relana-GFP fusion (Fig. 3B), although both the gfp fusion constructs could complement the all1549Ωsp/sm mutant.
Disruption of relana gene prevents accumulation of ppGpp.
As the relana gene was interrupted at the position corresponding to the region just downstream of the RelA_SpoT domain of the whole protein (Fig. 2A), we wondered what would be the effect on the level of ppGpp produced in the cells. We first examined if the disrupted relana gene was still transcribed and would thus produce a truncated form of mRNA. Using qPCR analysis with primers RT1549-1 and RT1549-2, the transcriptional level of relana was determined in both the wild type and the mutant. While expression of relana in the wild type increased after the deprivation of combined nitrogen, expression of the truncated form of relana in the all1549Ωsp/sm mutant was not upregulated and was detected at a much lower level than in the wild-type strain (Fig. 4A). This result further confirmed the disruption of relana in the all1549Ωsp/sm mutant and demonstrated that the truncated form of this gene was expressed only weakly.
Fig 4.
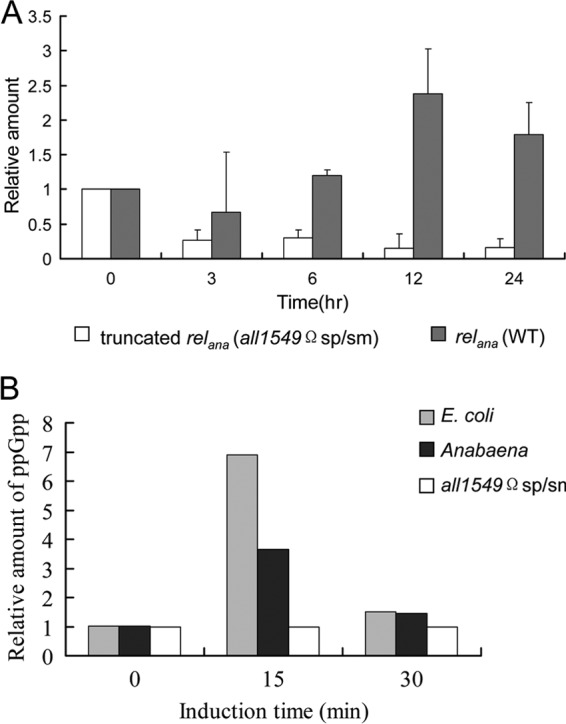
Determination of ppGpp level. (A) Transcriptional levels of the truncated form of relana in the mutant all1549Ωsp/sm compared to that in the wild type were analyzed at the indicated times after nitrogen step-down. (B) ppGpp level in Anabaena induced by amino acid starvation. SHX was used at the final concentration 1 g liter−1. Data shown are representative of the results of two independent experiments.
Since the truncated form of relana may still contain a functional catalytic domain, we thus checked the level of ppGpp in both the wild type and the mutant by using the HPLC method (34). An E. coli strain was included as a positive control. Serine hydroxamate (SHX), which has been shown to elicit a strong amino acid starvation response in various organisms, was used to trigger the accumulation of ppGpp (31, 49, 50). For E. coli, we could show a pattern of ppGpp accumulation after the treatment with SHX, as previously reported (51). As shown in Fig. 4B, ppGpp accumulation reaches the level of 32 pmol ml−1 OD−1 in the wild-type Anabaena strain at 15 min after treatment by SHX, followed by a drop after 30 min of treatment, but remained higher than the basal level found at time zero. In contrast, ppGpp accumulation was not observed under similar conditions in the all1549Ωsp/sm mutant and remained at the basal level with or without treatment by SHX (Fig. 4B). Taken together, these results indicated that the all1549Ωsp/sm mutant produced a truncated form of mRNA, expressed at a very low level, and had a strong defect in ppGpp synthesis.
hetR induction is affected in the all1549Ωsp/sm mutant.
In Anabaena, the hetR gene is essential for the initiation of heterocyst development and is upregulated during the developmental process (22). To understand the reason why the all1549Ωsp/sm mutant fails to form heterocysts, we checked the transcript level of hetR by qPCR in the all1549Ωsp/sm mutant as well as in the wild type. In the wild-type strain, the expression of hetR increased about 7-fold at 12 h after deprivation of combined nitrogen. However, in the all1549Ωsp/sm mutant, the transcript level of hetR was increased only slightly under similar conditions (Fig. 5A). We also used transcriptional fusion to gfp as a reporter to examine the expression of hetR. In the wild-type strain, GFP fluorescence derived from the transcriptional fusion of hetR-gfp was detected after nitrogen step-down and mainly focused in the heterocyst after 24 h. In contrast, no GFP fluorescence was observed in the all1549Ωsp/sm mutant under similar conditions (data not shown). These results indicated that the inactivation of relana impaired the regulation of hetR during heterocyst differentiation.
Fig 5.
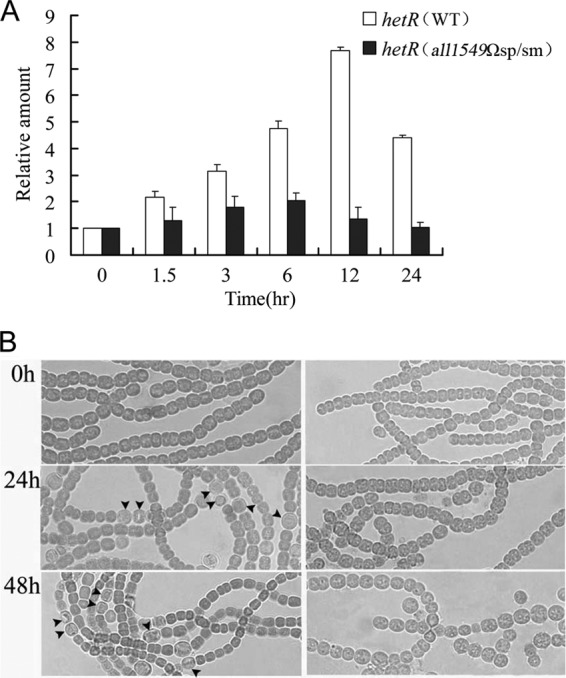
Ectopic expression of hetR in wild type (WT) and mutant. (A) Transcriptional levels of hetR were analyzed at the indicated times after nitrogen step-down. (B) Phenotypes after ectopic expression of hetR in the WT and all1549Ωsp/sm. Left panel, wild-type strain; right panel, all1549Ωsp/sm mutant strain. Arrowheads indicate the positions of heterocysts.
Since the upregulation of hetR was affected and heterocysts are not formed, we examined whether an ectopic expression of hetR could restore the capacity of all1549Ωsp/sm to form heterocysts. Plasmid pRL25T-PpetE-hetR containing hetR under the control of the copper-inducible promoter PpetE was transferred into both the wild type and the mutant. In the wild-type strain, overexpression of hetR increased the frequency of heterocysts and produced contiguous heterocysts as reported previously (Fig. 5B) (24). In contrast, no heterocyst was observed under similar conditions in the all1549Ωsp/sm mutant (Fig. 5B), even though the plasmid was well present and the overexpression of hetR occurred well in this strain as examined by qPCR (data not shown). Therefore, ectopic expression of hetR cannot bypass the need for ppGpp in heterocyst differentiation. The inactivation of relana, and the drop in ppGpp levels may affect the cellular activities of which the upregulation of hetR is only part, suggesting that this mutation has a much broader effect (see below).
Recent studies have shown that both hetP and hetZ genes, acting at the early stages of heterocyst differentiation, are directly regulated by HetR and were able bypass the requirement of hetR when ectopically expressed (52, 53). Since overexpression of hetR failed to rescue the failure of heterocyst differentiation in the all1549Ωsp/sm mutant, these two genes were in turn overexpressed in the all1549Ωsp/sm mutant. Again, none of them could rescue the defect of heterocyst differentiation of the all1549Ωsp/sm mutant (data not shown).
Loss of ppGpp affects cell viability under conditions of nitrogen deprivation.
Heterocyst differentiation takes about 20 h, during which no external nitrogen source is available to sustain cell growth. Heterocysts become mature enough to fix N2 to ensure the growth of the filament only at the end of the differentiation process. The low expression level of the master regulator of heterocyst development, hetR alone, cannot provide a rational explanation for the phenotypes of the all1549Ωsp/sm mutant, since ectopic expression of hetR failed to rescue the phenotypes. We thus explored whether the viability of the filaments could be affected early enough so that heterocyst differentiation might not be initiated or sustained. For this experiment, filaments were cultured first under conditions of combined-nitrogen sufficiency and then deprived of the combined-nitrogen source. During different times following the step-down of combined nitrogen, ammonium was added back as a nitrogen source, and cell viability was tested by following the optical density or chlorophyll content (Fig. 6). Similar results were obtained with measurement of either the optical density or the chlorophyll content; thus, only the data corresponding to the optical density measurement are shown here. For the wild type, the growth of the cells became faster when ammonium was added back, as expected, and this was true during the whole process of this experiment till 36 h after the combined-nitrogen step-down (Fig. 6A). In the all1549Ωsp/sm mutant, in contrast, cells could not grow under conditions of combined-nitrogen deprivation, as demonstrated earlier. When ammonium was added back 3 h after the step-down, cell growth resumed just as in the wild type. However, when ammonium was added back at or after 6 h, the filaments failed to recover their growth (Fig. 6B). Taken together, these results demonstrated that the inactivation of relana affected the ability of the filaments to sustain their viability between 3 and 6 h after the induction of heterocyst differentiation.
Fig 6.
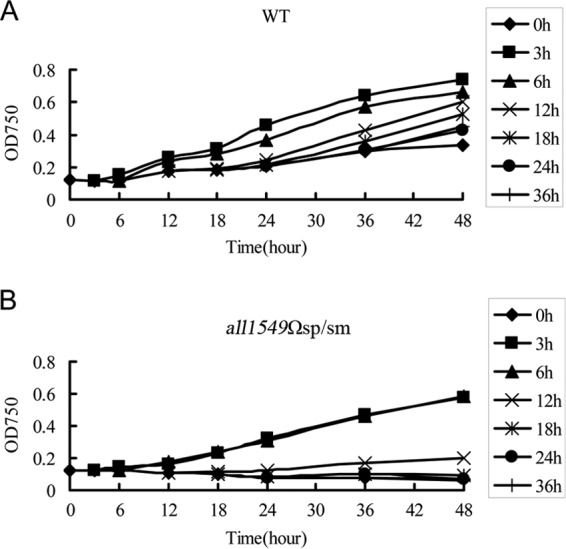
Cell viability test. (A) Wild-type strain. (B) all1549Ωsp/sm mutant. Data shown are representative of the results of two independent experiments. After nitrogen starvation, samples were taken and ammonium was added back at different time points as indicated by each curve: 0 h, 3 h, 6 h, 12 h, 18 h, 24 h, and 36 h.
DISCUSSION
ppGpp functions as a second messenger and is involved in various processes of cell growth and development. When the rel gene in Rhizobium etli is disrupted, both the morphology and the size of bacteroid are altered and nodules fail to fix nitrogen (54, 55), showing the importance of the stringent response in symbiosis. In Sorangium cellulosum So ce56, the Srel mutant is unable to form multicellular fruiting bodies under conditions of nutrient starvation (56). ppGpp also affected secondary metabolite biosynthesis (56–58).
In this study, we characterized a mutant in which the relana gene was disrupted in Anabaena. The phenotype of the mutant can be complemented by a wild-type copy provided on a plasmid. Although a portion of the 5′-terminal end is still present and thus may encode a partial protein capable of ppGpp synthesis, we have two arguments indicating that even if this is true, the level of ppGpp present in this mutant must be very low. First, the 5′ portion of the relana gene present in the chromosome gives only a low level of expression that is even lower, after induction of heterocyst differentiation, than the basal level detected at time zero; second, after treatment with SHX, the ppGpp accumulates rapidly in the wild type but not in the mutant. Thus, we conclude that the interruption of Relana results in a very low level of ppGpp synthesis in the mutant. It was reported a long time ago that ppGpp levels increase at the early stage of heterocyst differentiation (59, 60). Our data further indicate that this ppGpp synthesis activity is required for heterocyst development. Indeed, no heterocysts are formed in the mutant that cannot grow under nitrogen-fixing conditions. The enhanced expression of hetR does not occur in the mutant compared to the wild type. While this may account for the failure in the initiation of heterocyst differentiation in many mutants, it is not the case for the all1549Ωsp/sm mutant since the defect cannot be rescued by overexpression of either hetR or its direct targets hetZ and hetP. Our data indicate that the inactivation of relana produced a profound physiological defect, affecting the survival ability of Anabaena at the early stage of the transfer to nitrogen-fixing conditions. This effect makes the filament unable to respond properly to combined-nitrogen deprivation and thus to initiate heterocyst differentiation.
Heterocyst differentiation is a relatively long process, taking about 20 h before heterocysts can provide fixed nitrogen for filament growth. Therefore, during this long period of time, the cells must readjust their metabolism, in the absence of external nitrogen input, to maintain the viability of the filaments. We propose that ppGpp plays a role in the regulation of such metabolic rebalancing. Such a role mirrors what happens in E. coli under conditions of amino acid starvation or other nutrient starvation, during which major nutrient-consuming activities are shut down so that the cellular resources are redirected toward those activities essential for cell survival (5, 61). A similar situation may occur in Anabaena, where ppGpp accumulation at the early stage of heterocyst differentiation allows the cells to reprogram their metabolic activities so that the differentiation can proceed; otherwise, uncontrolled, nutrient-demanding activities may make the cells unable to keep pace with the demand for nutrients. The failure to properly activate expression of hetR, as well as that of several other genes (hetP, hetZ, hetC, and ntcA) involved in heterocyst development (data not shown), can be regarded as a consequence of such a misregulation. Our studies reveal a new mechanism involved in heterocyst development that ensures filament survival during the early stage of the differentiation process. Interestingly, we found that the amount of cyanophycin, an important nitrogen reserve for cyanobacteria (62), fell rapidly following nitrogen starvation in the all1549Ωsp/sm mutant, in contrast to the wild type, in which it remains stable or even increases, possibly as a safeguard response to nitrogen starvation and cell survival strategy (data not shown). This observation is consistent with the metabolic rebalancing necessary for the proper functioning of the developmental process.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 31170048 and 30970088) and the Fundamental Research Funds for the Central Universities (program no. 2011QC070 and 2011PY090).
We are very grateful to Emmanuelle Bouveret for the gift of E. coli strains.
Footnotes
Published ahead of print 9 August 2013
REFERENCES
- 1.Potrykus K, Cashel M. 2008. (p) ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51 [DOI] [PubMed] [Google Scholar]
- 2.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980–5990 [PubMed] [Google Scholar]
- 3.Vinella D, Albrecht C, Cashel M, D'Ari R. 2005. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 56:958–970 [DOI] [PubMed] [Google Scholar]
- 4.Seyfzadeh M, Keener J, Nomura M. 1993. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 90:11004–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang DE, Smalley DJ, Conway T. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289–306 [DOI] [PubMed] [Google Scholar]
- 6.Wells DH, Gaynor EC. 2006. Helicobacter pylori initiates the stringent response upon nutrient and pH downshift. J. Bacteriol. 188:3726–3729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mittenhuber G. 2001. Comparative genomics and evolution of genes encoding bacterial (p) ppGpp synthetases/hydrolases (the Rel, RelA and SpoT proteins). J. Mol. Microbiol. Biotechnol. 3:585–600 [PubMed] [Google Scholar]
- 8.van der Biezen EA, Sun J, Coleman MJ, Bibb MJ, Jones JD. 2000. Arabidopsis RelA/SpoT homologs implicate (p)ppGpp in plant signaling. Proc. Natl. Acad. Sci. U. S. A. 97:3747–3752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K, Kasai K, Ochi K. 2004. Identification of the bacterial alarmone guanosine 5′-diphosphate 3′-diphosphate (ppGpp) in plants. Proc. Natl. Acad. Sci. U. S. A. 101:4320–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun D, Lee G, Lee JH, Kim HY, Rhee HW, Park SY, Kim KJ, Kim Y, Kim BY, Hong JI, Park C, Choy HE, Kim JH, Jeon YH, Chung J. 2010. A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat. Struct. Mol. Biol. 17:1188–1194 [DOI] [PubMed] [Google Scholar]
- 11.Taylor CM, Beresford M, Epton HA, Sigee DC, Shama G, Andrew PW, Roberts IS. 2002. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J. Bacteriol. 184:621–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Delden C, Comte R, Bally AM. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris BZ, Kaiser D, Singer M. 1998. The guanosine nucleotide (p) ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chakraburtty R, Bibb M. 1997. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. J. Bacteriol. 179:5854–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M. 2007. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2). Genome Biol. 8:R161. 10.1186/gb-2007-8-8-r161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zusman T, Gal-Mor O, Segal G. 2002. Characterization of a Legionella pneumophila relA insertion mutant and toles of RelA and RpoS in virulence gene expression. J. Bacteriol. 184:67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar K, Mella-Herrera RA, Golden JW. 2010. Cyanobacterial heterocysts. Cold Spring Harb. Perspect. Biol. 2:a000315. 10.1101/cshperspect.a000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Golden JW, Yoon HS. 2003. Heterocyst development in Anabaena. Curr. Opin. Microbiol. 6:557–563 [DOI] [PubMed] [Google Scholar]
- 19.Zhao MX, Jiang YL, He YX, Chen YF, Teng YB, Chen Y, Zhang CC, Zhou CZ. 2010. Structural basis for the allosteric control of the global transcription factor NtcA by the nitrogen starvation signal 2-oxoglutarate. Proc. Natl. Acad. Sci. U. S. A. 107:12487–12492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurent S, Chen H, Bedu S, Ziarelli F, Peng L, Zhang CC. 2005. Nonmetabolizable analogue of 2-oxoglutarate elicits heterocyst differentiation under repressive conditions in Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. U. S. A. 102:9907–9912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X, Dong Y, Zhao J. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. U. S. A. 101:4848–4853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buikema WJ, Haselkorn R. 1991. Characterization of a gene controlling heterocyst differentiation in the cyanobacterium Anabaena 7120. Genes Dev. 5:321–330 [DOI] [PubMed] [Google Scholar]
- 23.Khudyakov IY, Golden JW. 2004. Different functions of HetR, a master regulator of heterocyst differentiation in Anabaena sp. PCC 7120, can be separated by mutation. Proc. Natl. Acad. Sci. U. S. A. 101:16040–16045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buikema WJ, Haselkorn R. 2001. Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. U. S. A. 98:2729–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frias JE, Flores E, Herrero A. 1994. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 14:823–832 [DOI] [PubMed] [Google Scholar]
- 26.Muro-Pastor AM, Valladares A, Flores E, Herrero A. 2002. Mutual dependence of the expression of the cell differentiation regulatory protein HetR and the global nitrogen regulator NtcA during heterocyst development. Mol. Microbiol. 44:1377–1385 [DOI] [PubMed] [Google Scholar]
- 27.Wolk CP, Ernst A, Elhai J. 2004. Heterocyst metabolism and development, p 769–823 In Bryant D. (ed), The molecular biology of cyanobacteria, vol 1 Springer, Dordrecht, Netherlands [Google Scholar]
- 28.Ning D, Qian Y, Miao X, Wen C. 2011. Role of the all1549 (ana-rsh) gene, a relA/spoT homolog, of the cyanobacterium Anabaena sp. PCC7120. Curr. Microbiol. 62:1767–1773 [DOI] [PubMed] [Google Scholar]
- 29.Boutte CC, Crosson S. 2013. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 21:174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang CC. 1993. A gene encoding a protein related to eukaryotic protein kinases from the filamentous heterocystous cyanobacterium Anabaena PCC 7120. Proc. Natl. Acad. Sci. U. S. A. 90:11840–11844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nanamiya H, Kasai K, Nozawa A, Yun CS, Narisawa T, Murakami K, Natori Y, Kawamura F, Tozawa Y. 2008. Identification and functional analysis of novel (p) ppGpp synthetase genes in Bacillus subtilis. Mol. Microbiol. 67:291–304 [DOI] [PubMed] [Google Scholar]
- 32.Elhai J, Wolk CP. 1988. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 167:747–754 [DOI] [PubMed] [Google Scholar]
- 33.Black TA, Cai Y, Wolk CP. 1993. Spatial expression and autoregulation of hetR, a gene involved in the control of heterocyst development in Anabaena. Mol. Microbiol. 9:77–84 [DOI] [PubMed] [Google Scholar]
- 34.Ochi K, Kandala JC, Freese E. 1981. Initiation of Bacillus subtilis sporulation by the stringent response to partial amino acid deprivation. J. Biol. Chem. 256:6866–6875 [PubMed] [Google Scholar]
- 35.Washio K, Lim SP, Roongsawang N, Morikawa M. 2010. Identification and characterization of the genes responsible for the production of the cyclic lipopeptide arthrofactin by Pseudomonas sp. MIS38. Biosci. Biotechnol. Biochem. 74:992–999 [DOI] [PubMed] [Google Scholar]
- 36.Shi L, Li JH, Cheng Y, Wang L, Chen WL, Zhang CC. 2007. Two genes encoding protein kinases of the HstK family are involved in synthesis of the minor heterocyst-specific glycolipid in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 189:5075–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Battesti A, Bouveret E. 2006. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62:1048–1063 [DOI] [PubMed] [Google Scholar]
- 38.Aravind L, Koonin EV. 1998. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 23:469–472 [DOI] [PubMed] [Google Scholar]
- 39.Sankaranarayanan R, Dock-Bregeon AC, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D. 1999. The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell 97:371–381 [DOI] [PubMed] [Google Scholar]
- 40.Chipman DM, Shaanan B. 2001. The ACT domain family. Curr. Opin. Struct. Biol. 11:694–700 [DOI] [PubMed] [Google Scholar]
- 41.Grant GA. 2006. The ACT domain: a small molecule binding domain and its role as a common regulatory element. J. Biol. Chem. 281:33825–33829 [DOI] [PubMed] [Google Scholar]
- 42.Hsieh MH, Goodman HM. 2002. Molecular characterization of a novel gene family encoding ACT domain repeat proteins in Arabidopsis. Plant Physiol. 130:1797–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ning D, Xu X. 2004. alr0117, a two-component histidine kinase gene, is involved in heterocyst development in Anabaena sp. PCC 7120. Microbiology 150(Pt 2):447–453 [DOI] [PubMed] [Google Scholar]
- 44.Awai K, Wolk CP. 2007. Identification of the glycosyl transferase required for synthesis of the principal glycolipid characteristic of heterocysts of Anabaena sp. strain PCC 7120. FEMS Microbiol. Lett. 266:98–102 [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Huang XZ, Wang L, Risoul V, Zhang CC, Chen WL. 2013. Phenotypic variation caused by variation in the relative copy number of pDU1-based plasmids expressing the GAF domain of Pkn41 or Pkn42 in Anabaena sp. PCC 7120. Res. Microbiol. 164:127–135 [DOI] [PubMed] [Google Scholar]
- 46.Flaherty BL, Van Nieuwerburgh F, Head SR, Golden JW. 2011. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genomics 12:332. 10.1186/1471-2164-12-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gentry DR, Cashel M. 1995. Cellular localization of the Escherichia coli SpoT protein. J. Bacteriol. 177:3890–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Avarbock A, Avarbock D, Teh JS, Buckstein M, Wang ZM, Rubin H. 2005. Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44:9913–9923 [DOI] [PubMed] [Google Scholar]
- 49.Brockmann-Gretza O, Kalinowski J. 2006. Global gene expression during stringent response in Corynebacterium glutamicum in presence and absence of the rel gene encoding (p) ppGpp synthase. BMC Genomics 7:230. 10.1186/1471-2164-7-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemos JA, Lin VK, Nascimento MM, Abranches J, Burne RA. 2007. Three gene products govern (p)ppGpp production by Streptococcus mutans. Mol. Microbiol. 65:1568–1581 [DOI] [PubMed] [Google Scholar]
- 51.Battesti A, Bouveret E. 2009. Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J. Bacteriol. 191:616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higa KC, Callahan SM. 2010. Ectopic expression of hetP can partially bypass the need for hetR in heterocyst differentiation by Anabaena sp. strain PCC 7120. Mol. Microbiol. 77:562–574 [DOI] [PubMed] [Google Scholar]
- 53.Du Y, Cai Y, Hou S, Xu X. 2012. Identification of the HetR recognition sequence upstream of hetZ in Anabaena sp. strain PCC 7120. J. Bacteriol. 194:2297–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Calderón-Flores A, Du Pont G, Huerta-Saquero A, Merchant-Larios H, Servin-Gonzalez L, Duran S. 2005. The stringent response is required for amino acid and nitrate utilization, nod factor regulation, nodulation, and nitrogen fixation in Rhizobium etli. J. Bacteriol. 187:5075–5083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moris M, Braeken K, Schoeters E, Verreth C, Beullens S, Vanderleyden J, Michiels J. 2005. Effective symbiosis between Rhizobium etli and Phaseolus vulgaris requires the alarmone ppGpp. J. Bacteriol. 187:5460–5469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Knauber T, Doss SD, Gerth K, Perlova O, Muller R, Treuner-Lange A. 2008. Mutation in the rel gene of Sorangium cellulosum affects morphological and physiological differentiation. Mol. Microbiol. 69:254–266 [DOI] [PubMed] [Google Scholar]
- 57.Jin W, Ryu YG, Kang SG, Kim SK, Saito N, Ochi K, Lee SH, Lee KJ. 2004. Two relA/spoT homologous genes are involved in the morphological and physiological differentiation of Streptomyces clavuligerus. Microbiology 150:1485–1493 [DOI] [PubMed] [Google Scholar]
- 58.Gomez-Escribano JP, Martin JF, Hesketh A, Bibb MJ, Liras P. 2008. Streptomyces clavuligerus relA-null mutants overproduce clavulanic acid and cephamycin C: negative regulation of secondary metabolism by (p)ppGpp. Microbiology 154:744–755 [DOI] [PubMed] [Google Scholar]
- 59.Akinyanju J, Smith RJ. 1979. Accumulation of ppGpp and pppGpp during nitrogen deprivation of the cyanophyte Anabaena cylindrica. FEBS Lett. 107:173–176 [DOI] [PubMed] [Google Scholar]
- 60.Akinyanju J, Smith RJ. 1987. The accumulation of phosphorylated guanosine nucleotides in Anabaena cylindrica. New Phytol. 105:117–122 [DOI] [PubMed] [Google Scholar]
- 61.Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T. 2008. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68:1128–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon RD. 1973. Measurement of the cyanophycin granule polypeptide contained in the blue-green alga Anabaena cylindrica. J. Bacteriol. 114:1213–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]