Abstract
The trace elements molybdenum and tungsten are essential components of cofactors of many metalloenzymes. However, in sulfate-reducing bacteria, high concentrations of molybdate and tungstate oxyanions inhibit growth, thus requiring the tight regulation of their homeostasis. By a combination of bioinformatic and experimental techniques, we identified a novel regulator family, tungstate-responsive regulator (TunR), controlling the homeostasis of tungstate and molybdate in sulfate-reducing deltaproteobacteria. The effector-sensing domains of these regulators are similar to those of the known molybdate-responsive regulator ModE, while their DNA-binding domains are homologous to XerC/XerD site-specific recombinases. Using a comparative genomics approach, we identified DNA motifs and reconstructed regulons for 40 TunR family members. Positional analysis of TunR sites and putative promoters allowed us to classify most TunR proteins into two groups: (i) activators of modABC genes encoding a high-affinity molybdenum and tungsten transporting system and (ii) repressors of genes for toluene sulfonate uptake (TSUP) family transporters. The activation of modA and modBC genes by TunR in Desulfovibrio vulgaris Hildenborough was confirmed in vivo, and we discovered that the activation was diminished in the presence of tungstate. A predicted 30-bp TunR-binding motif was confirmed by in vitro binding assays. A novel TunR family of bacterial transcriptional factors controls tungstate and molybdate homeostasis in sulfate-reducing deltaproteobacteria. We proposed that TunR proteins participate in protection of the cells from the inhibition by these oxyanions. To our knowledge, this is a unique case of a family of bacterial transcriptional factors evolved from site-specific recombinases.
INTRODUCTION
Molybdenum and tungsten are essential trace metals utilized by many living organisms in active centers of enzymes catalyzing different redox reactions (1). Enzymes containing molybdenum cofactors (molybdoenzymes), unlike tungstoenzymes, are widespread in aerobic organisms, including eukaryotes (2). In contrast to aerobic organisms, anaerobic microbes may use both tungstoenzymes and molybdoenzymes.
Some of these enzymes may incorporate either molybdenum or tungsten without loss of catalytic function, while other proteins require a specific metal for catalytic activity (3). For example, formate dehydrogenase 1 (FDH-1) from Desulfovibrio vulgaris Hildenborough was demonstrated to contain both metals, while isoenzyme FDH-3 incorporated only molybdenum (4).
A similar variability in metal specificity is known for molybdate and tungstate transporting systems. Specific high-affinity ATP-binding cassette (ABC) transporting systems uptake molybdenum and tungsten in the form of soluble oxyanions (molybdate and tungstate) (5). Among them are the bacterial TupABC transporting system, which is highly specific for tungstate and does not transport other anions (6), and the ModABC system, which can transport both molybdate and tungstate (7).
Regulation of the ModABC transporting system was extensively studied in Escherichia coli, where the transcriptional factor ModE was shown to negatively regulate the modABC operon in the presence of molybdate (8). In E. coli, ModE also induces expression of the molybdenum cofactor biosynthesis operon and molybdoenzymes in a molybdate-dependent manner (9, 10).
In sulfate-reducing bacteria (SRB), including the model bacterium D. vulgaris Hildenborough, a tight regulation of intracellular concentrations of molybdenum and tungsten is especially important, because these metals suppress SRB growth (11). Biocides that inhibit SRB growth are often used for prevention and control of the microbial biocorrosion process (12). Tetrahedral oxyanions, like molybdate and tungstate, inhibit ATP-dependent activation of sulfate by sulfate adenylyltransferase (SAT), the first enzyme of the sulfate reduction pathway (13). Evidence has also been obtained for inhibition of sulfate transport by molybdate in SRB (14). Despite the importance of this regulation, it has been poorly described in the literature, and no experimental verification of any regulatory mechanisms in SRB is known.
Attempts to find ModE-like regulatory mechanisms in Desulfovibrio spp. by comparing these bacteria with E. coli have not been successful since orthologs of modE and ModE binding motifs could not be found in Desulfovibrio spp. (15). However, a putative regulatory motif has been computationally inferred in D. vulgaris Hildenborough and Desulfovibrio alaskensis G20 upstream of molybdenum transport genes (15).
We carried out a comprehensive computational analysis of the putative molybdate transport regulation mechanism in Desulfovibrio spp. and related sulfate-reducing deltaproteobacteria. Here, we report our discovery of a novel family of transcriptional factors, named tungstate-responsive regulators (TunR), that control molybdenum and tungsten transport. These proteins evolved from site-specific recombinases to oxyanion-responsive regulators of transcription, thus forming a unique family of regulatory proteins. The regulatory function of a representative member of this TunR family from D. vulgaris Hildenborough was experimentally validated in vitro and in vivo, showing for the first time a mechanism of transcriptional control of molybdenum and tungsten transport in this bacterium.
MATERIALS AND METHODS
Data sources.
In this work, we used genome and protein sequences from MicrobesOnline (16) and NCBI GenBank (17) databases (Table 1).
Table 1.
Genomic data sourcesa
| Organism | DB source | Identifier(s) |
|---|---|---|
| Bilophila sp. 4_1_30 | NCBI | NZ_ADCO01000000 |
| Bilophila wadsworthia 3_1_6 | NCBI | NZ_ADCP00000000 |
| Desulfomicrobium baculatum DSM 4028 | MO | NC_013173 |
| Desulfovibrio aespoeensis Aspo-2 | NCBI | NC_014844 |
| Desulfovibrio alaskensis G20 | MO | NC_007519 |
| Desulfovibrio desulfuricans ND132 | NCBI | NC_016803 |
| Desulfovibrio desulfuricans subsp. desulfuricans strain ATCC 27774 | MO | NC_011883 |
| Desulfovibrio fructosovorans JJ | NCBI | NZ_AECZ00000000 |
| Desulfovibrio magneticus RS-1 | MO | NC_012795, NC_012796, NC_012797 |
| Desulfovibrio piger ATCC 29098 | MO | NZ_ABXU00000000 |
| Desulfovibrio salexigens DSM 2638 | MO | NC_012881 |
| Desulfovibrio sp. 3_1_syn3 | NCBI | NZ_ADDR01000000 |
| Desulfovibrio sp. 6_1_46AFAA | NCBI | NZ_ACWM01000000 |
| Desulfovibrio sp. A2 | NCBI | NZ_AGFG00000000 |
| Desulfovibrio sp. FW1012B | NCBI | NZ_ADFE00000000 |
| Desulfovibrio sp. U5L | NCBI | NZ_AHMC00000000 |
| Desulfovibrio vulgaris DP4 | MO | NC_008751, NC_008741 |
| Desulfovibrio vulgaris strain Hildenborough | MO | NC_002937, NC_005863 |
| Desulfovibrio vulgaris strain “Miyazaki F” | MO | NC_011769 |
| Desulfobacter postgatei 2ac9 | NCBI | NZ_AGJR00000000 |
| Desulfobulbus propionicus DSM 2032 | NCBI | NC_014972 |
| Desulfomonile tiedjei DSM 6799 | NCBI | NC_018025 |
| Desulfovibrio africanus strain Walvis Bay | NCBI | NC_016629 |
Identification of TunR family members.
TunR family proteins were identified in the NCBI database by comparison with the DVU0179 protein using BLAST (18) and protein domain annotations. Only those BLAST hits that had both predicted PF00589 and transport-associated oligonucleotide/oligosaccharide-binding fold (TOBE) domains were recognized as TunR family members. Annotations of protein domains were taken from MicrobesOnline and NCBI databases.
Phylogenetic analysis of transcription factors.
We used MUSCLE (19) for protein multiple alignment and ClustalX 2.1 (20) for phylogenetic analysis. Phylogenetic trees were constructed by the neighbor-joining method with default parameters, with calculation of bootstraps from 1,000 replications. Orthologous groups of transcription factors (TFs) on the phylogenetic tree were identified as high confidence clusters that have more than 40% pairwise protein identity within the cluster. Colocalization of regulatory genes with genes encoding different transporting systems was used as additional evidence for distinction of orthologous groups.
Motif reconstruction and regulon prediction.
We used the RegPredict Web server (21) for motif reconstruction. Sets of upstream sequences of TunR family genes and modABC genes (from −400 to +50 with respect to the translation start) were selected, and a common palindromic motif with the highest information content was identified in each set by using the “Discover profiles” tool of the RegPredict Web server.
For regulon reconstruction, we used a comparative genomics approach implemented in the RegPredict Web server (21) and the Genome Explorer software package (22). Briefly, a position-weight matrix was used for a whole-genome search in upstream regions of the coding genes (from −400 to +50 with respect to the translation start) with a threshold equal to a minimal score among all sites in a training set of the matrix. For promoter motif predictions, a position-weight matrix based on the sigma-70 motif identified in D. vulgaris Hildenborough (23) was used. A collection of TunR family regulons reconstructed with the RegPredict Web server is available in the RegPrecise database (24).
Growth conditions.
D. vulgaris Hildenborough strains (wild type [WT] and GZ6027) were grown from freezer stocks in MOYLS4 medium (25) at 30°C in an anaerobic chamber (COY, Grass Lake, MI). The antibiotic G418 (400 μg/ml) was always added to cultures of strain GZ6027. To deplete cells of Mo and W, cells were passaged three times using 2.5% inoculum through LS4D medium prepared with trace element solution lacking Mo and W (referred to hence as LS4D-Mo/W).
Construction of the GZ6027 strain.
The GZ6027 (tunR::mini-Tn5) strain was constructed using Tn5-RL27 transposon mutagenesis as described previously (26).
qRT-PCR.
WT and GZ6027 D. vulgaris cells depleted of molybdenum and tungsten were each grown in LS4D-Mo/W (40-ml cultures in triplicate) supplemented with and without 1 μM sodium molybdate or 0.3 μM sodium tungstate. Cells were harvested at mid-log phase (optical density at 600 nm [OD600] of ∼0.4 for WT and ∼0.3 for GZ6027) by centrifuging at 3,000 × g for 10 min at 4°C and homogenized in TRIzol LS (Life Technologies, Grand Island, NY) and stored at −80°C until ready for extraction. RNA was extracted with chloroform, and further RNA isolation was performed using the PureLink RNA minikit (Life Technologies) as per the manufacturer's instruction. On-column DNA digestion was performed using the PureLink DNase set (Life Technologies) as per the manufacturer's instruction. RNA was eluted in 50 μl RNase-free water, and a second round of DNase digestion was performed using 1 μl of Turbo DNA-free DNase (Life Technologies) at 37°C for 30 min, followed by DNase removal with DNase inactivation reagent. RNA was quantified spectrophotometrically using the NanoDrop ND-1000 (Thermo Fisher Scientific, Wilmington, DE), and 500 ng was reverse transcribed using iScript reverse transcription supermix (Bio-Rad, Hercules, CA) (25°C for 5 min, 42°C for 30 min, 85°C for 5 min). The iScript mix with no reverse transcriptase (RT) was used to set up parallel reactions to check for DNA contamination. Two microliters of the resulting cDNA was used as the template for quantitative PCRs (qPCRs) (in triplicate) using 2× SsoAdvanced SYBR green supermix (Bio-Rad) and 0.5 μM primers in a total reaction volume of 20 μl. qPCR primers were designed using PrimerQuest (Integrated DNA Technologies, Coralville, IO) to amplify 75- to 150-bp regions of target genes DVU0177 (modA), DVU0181 (modB), DVU0179 (first half of gene before transposon insertion), and rpoH (DVU1584). Primer sequences are in Table S1 in the supplemental material. Reactions were run on the Applied Biosystems Step One Plus PCR system (Life Technologies), with thermocycling conditions of 95°C for 1 min and 40 cycles of 95°C for 10 s, 59°C for 15 s, and 70°C for 35 s (data collection stage). Amplification efficiencies for the primer sets were determined using the standard curve method, where D. vulgaris genomic DNA was serially diluted 10-fold and used as the template in duplicate reactions. The slope of threshold cycle (ΔCT) versus log input between each target gene primer set and that of the reference gene (rpoH) was less than 0.1. Therefore, we used the 2−ΔΔCT method to calculate fold change in expression (27) using the rpoH gene as a reference. Standard deviations of the ΔΔCT value were incorporated into the fold change calculations to determine the error range.
DVU0179 cloning and purification.
DVU0179 was amplified from D. vulgaris Hildenborough genomic DNA, cloned into vector pEL01 (pSKB3 backbone with N-terminal 8×His and Strep II tags and a tobacco etch virus [TEV] protease cleavage site, Kanr) using the Gibson method (28) and transformed into E. coli BL21 Star (DE3) cells. Transformants were selected on LB-kanamycin plates and verified by sequencing. DVU0179 was overexpressed by growing 1 liter of the transformed E. coli in Terrific broth at 37°C until mid-log phase followed by growth at room temperature overnight with a few drops of antifoam B emulsion (Sigma-Aldrich, St. Louis, MO). Cells were lysed by sonication, and the protein was purified by fast protein liquid chromatography (FPLC) on the AKTA Explorer (GE Life Sciences, Pittsburgh, PA) with a HisTrap FF column (GE Life Sciences) as previously described (29). The protein was eluted directly onto a 5-ml StrepTrap FF column (GE Life Sciences, Pittsburgh, PA) for further purification. The column was washed with 20 mM Na-phosphate and 250 mM NaCl, pH 7.4, and the protein was eluted with wash buffer containing 2.5 mM desthiobiotin. The protein was quantified spectrophotometrically using the NanoDrop ND-1000 (Thermo Fisher Scientific, Wilmington, DE), and the tags were cleaved by incubation overnight with TEV-6×His protease in the presence of 50 mM Arg and 50 mM Glu at 4°C. NaCl was added to 500 mM and imidazole to 10 mM, and the protein prep was run on a fresh HisTrap column (5 ml) to remove the TEV, cleaved tags, and uncleaved DVU0179. A final buffer exchange was performed to remove excess salt and imidazole using a 26/10 desalting column (GE Life Sciences, Pittsburgh, PA), and the protein was concentrated using an Amicon stirred cell (EMD Millipore, Billerica, MA).
EMSA.
The DNA fragments for electrophoretic mobility shift assays (EMSA) were prepared by annealing the top biotinylated and bottom unlabeled oligonucleotides (IDT, San Diego, CA) as described previously (29) (see Table S1 in the supplemental material for primer sequences). Purified DVU0179 protein (33 pmol) was mixed with 200 fmol of biotinylated DNA (WT or mutated) in 10 mM Tris HCl (pH 7.5), 50 mM KCl, 5 mM MgCl2, and 1 μg/ml poly(dI·dC) in a total reaction volume of 20 μl and incubated at room temperature for 20 min. Electrophoresis, blotting, and chemiluminescent detection were performed as described previously (29). Final imaging of the blot was done using the Fluor Chem Q system (Protein Simple, Santa Clara, CA).
RESULTS
Identification of a candidate regulatory gene for the ModABC transport system in Desulfovibrio spp.
We collected several lines of computational evidence supporting a regulatory role for the DVU0179 protein from D. vulgaris Hildenborough and its homologs in Desulfovibrio spp. Major lines of evidence are based on protein sequence comparisons, inspection of the evolutionary conservation of the genomic neighborhood or synteny, and a transcription factor (TF)-binding motif analysis.
In our search for transcriptional regulators in the neighborhood of the modABC genes, we could not find a protein with a DNA-binding domain typical for transcriptional regulatory functions. This regulation is normally carried out by proteins that contain both a TF-specific DNA-binding domain and a domain that senses signal molecules. However, we found a protein, DVU0179, which could possibly play a regulatory role because it contains two tandem transport-associated oligonucleotide/oligosaccharide-binding fold (TOBE) domains similar to molybdate-binding domains of the ModE protein that regulates the mod operon in E. coli (8). In contrast to ModE, the protein we found has no helix-turn-helix DNA-binding domain. Instead, this protein contains a phage integrase family domain (PF00589 in the PFAM database [30]). The PF00589 domain is similar to the catalytic domains of the XerC and XerD proteins, site-specific tyrosine recombinases that resolve concatenated dimer chromosomes after replication (31). However, DVU0179 homologs probably lack recombinase activity, as shown by their comparison with the PF00589 integrase model that we present in Table S2 in the supplemental material. More than half of DVU0179 homologs have mutations of the key tyrosine residue, and all of them have mutations in one or several other active-site residues annotated in the UniProt database (32) for XerD recombinase (UniProt entry P0A8P8). Assuming that mutations in active-site residues do not disrupt DNA-binding ability of the protein, we would expect that DVU0179 homologs are capable of site-specific DNA binding. Thus, we hypothesize that DVU0179 and its homologs had evolved from a recombinase to carry out transcriptional regulation.
Another piece of evidence for the involvement of DVU0179 in the regulation of modABC genes is the fact that a regulatory motif found in regions upstream of modABC genes in D. vulgaris Hildenborough and D. alaskensis G20 (15) contained some similarity (positions 3, 4, 5, and 7 in Fig. 1) to a well-known XerC-binding site in E. coli (33). The XerC-XerD complex of E. coli binds the 28-bp dif sequence, which is located in the genome replication termination region (31). It is important to note that the dif motif lacks symmetry due to differences in the binding sites of XerC and XerD, which form a heterotetrameric complex. A palindromic structure of the proposed DVU0179 motif suggests that it may be recognized by a homodimeric protein complex rather than a heteromultimeric complex like XerC-XerD (Fig. 1). Thus, we hypothesized that the DVU0179 protein recognizes the identified palindromic motif and regulates the expression of the modABC genes.
Fig 1.
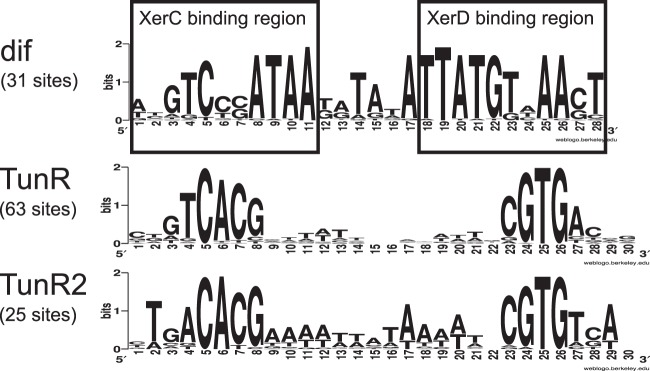
Motif logos of deltaproteobacterial dif and Desulfovibrio TunR- and TunR2-binding sites.
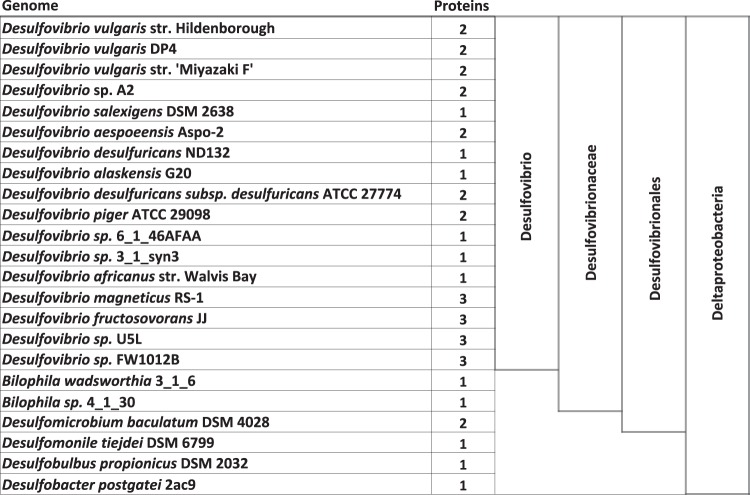
A functional linkage between DVU0179 and modABC genes was strengthened by the observed evolutionary conservation of the colocalization of orthologs of these genes. Comparative analysis of modABC loci identified DVU0179 homologs in 14 Desulfovibrio sp. genomes (Fig. 2). In eight genomes, DVU0179 homologs are transcribed divergently from modABC operons. In four other genomes, DVU0179 homologs are transcribed divergently from modA genes, and the modBC operons are located downstream from the regulatory genes. In Desulfovibrio desulfuricans ND132, the DVU0179 homolog is predicted to be transcribed divergently from an operon that encodes ModA, ModB, and a hypothetical protein. In Desulfovibrio salexigens, the DVU0179 homolog is transcribed divergently from a six-gene operon that is annotated to contain modABCAB and a gene encoding a hypothetical protein. Only in the genomes of Desulfovibrio africanus, Desulfovibrio sp. FW1012B, and Desulfovibrio sp. U5L is the DVU0179 homolog not linked with the corresponding modABC loci. These observations are consistent with a predicted functional linkage of DVU0179 and modABC genes.
Fig 2.
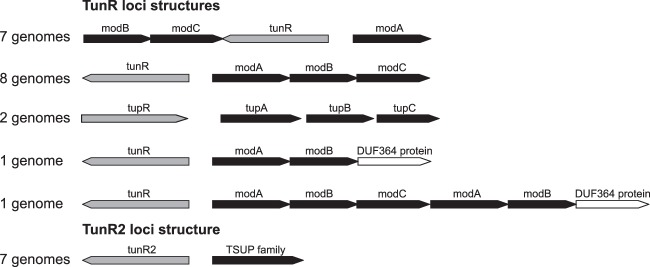
Genomic context of genes associated with TunR family members.
We expanded our search for evolutionary conservation to the putative regulatory motif upstream of modABC genes in Desulfovibrio sp. genomes to provide additional evidence for a regulatory role for DVU0179. This analysis demonstrated that the 30-bp palindromic motif is present upstream of modABC genes in all studied genomes containing DVU0179 homologs (see Table S3 in the supplemental material).
Moreover, even in two genomes (Desulfovibrio sp. FW1012B and Desulfovibrio sp. U5L) where DVU0179 homolog genes are not linked with modABC loci, putative binding sites were identified upstream of both the modABC operon and the DVU0179 homologs. Taken together, these observations provided the logical basis to the hypothesis that this motif is the binding site for DVU0179 and its homologs.
Expression of TunR-dependent genes in D. vulgaris Hildenborough is regulated by tungstate.
The detailed reconstruction of the TunR family regulons revealed that all transporter systems tightly associated with each TunR family subgroup (modABC, toluene sulfonate uptake [TSUP] family gene, and tupABC) are under the control of the TunR family regulators. Additional members of regulons include enzymes containing a molybdenum cofactor and molybdopterin-binding proteins. Thus, all these regulons would be involved in homeostasis of molybdenum and tungsten in sulfate-reducing deltaproteobacteria. To confirm these predictions, we analyzed expression of modABC and tunR genes in wild-type and TunR mutant cultures of D. vulgaris Hildenborough under different conditions.
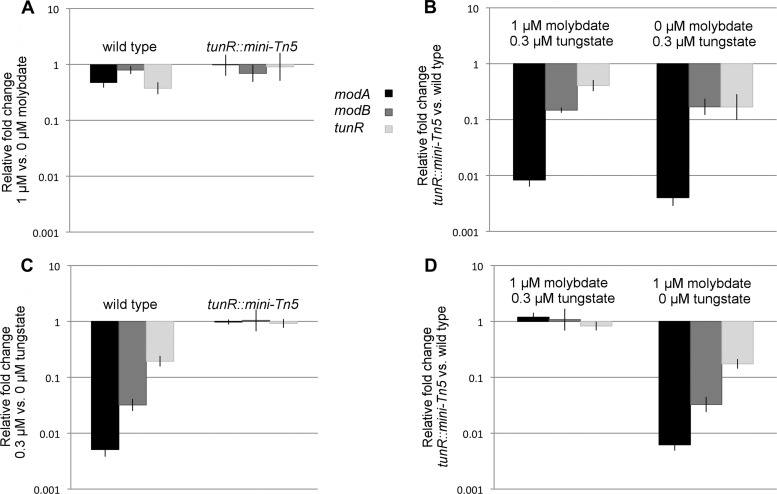
We constructed a transposon insertion mutant of the tunR gene (DVU0179) in D. vulgaris Hildenborough (strain GZ6027). We then examined the expression of modA (DVU0177), modB (DVU0181), and tunR genes in wild-type and GZ6027 strains and determined the effect of metal supplementation using quantitative reverse transcriptase PCR (qRT-PCR). When wild-type cells were grown in medium with 1 μM sodium molybdate, the expression of modA, modB, or tunR did not change relative to cells grown in unamended medium (Fig. 3A). However, the expression of modA and modB were strongly repressed (∼200- and 30-fold, respectively) and that of tunR was modestly repressed (∼5-fold) in the presence of 0.3 μM sodium tungstate (Fig. 3C). In GZ6027, the TunR regulon genes were not significantly affected by the addition of either molybdate or tungstate (Fig. 3A and C). GZ6027 also showed highly reduced expression of the three genes relative to the wild type in the absence of Mo and W amendments or in the presence of Mo (Fig. 3B and D). However, when tungstate was present, TunR regulon expression was similar to that of the wild type (Fig. 3D). The obtained results suggest that TunR functions as an activator of the modA, modBC, and tunR genes in the absence of tungstate and that the addition of 0.3 μM tungstate disrupts the TunR-dependent gene activation.
Fig 3.
Transcription of modA, modB, and tunR is activated by tunR and repressed by tungstate. (A) Effect of addition of molybdate in wild type and GZ6027 (tunR::mini-Tn5); (B) effect of tunR knockout in the presence and absence of molybdate; (C) effect of addition of tungstate in wild type and GZ6027 (tunR::mini-Tn5); (D) effect of tunR knockout in the presence and absence of tungstate.
TunR binds the predicted DNA motif in vitro.
In order to validate the computationally predicted DNA-binding motif of TunR, we heterologously expressed and purified the TunR/DVU0179 protein and further assessed it using electrophoretic mobility shift assay (EMSA). The recombinant TunR protein was able to shift a 46-bp biotin-labeled DNA fragment that contains the predicted TunR-binding site (30 bp) from the upstream region of the modA gene in D. vulgaris Hildenborough (Fig. 4) in the presence of nonspecific competitor DNA [poly(dI·dC)]. With a modified substrate, where specific substitutions were made in the conserved base pairs within the two half sites, the shift was eliminated, thus confirming the specificity of TunR binding to this site (Fig. 4).
Fig 4.
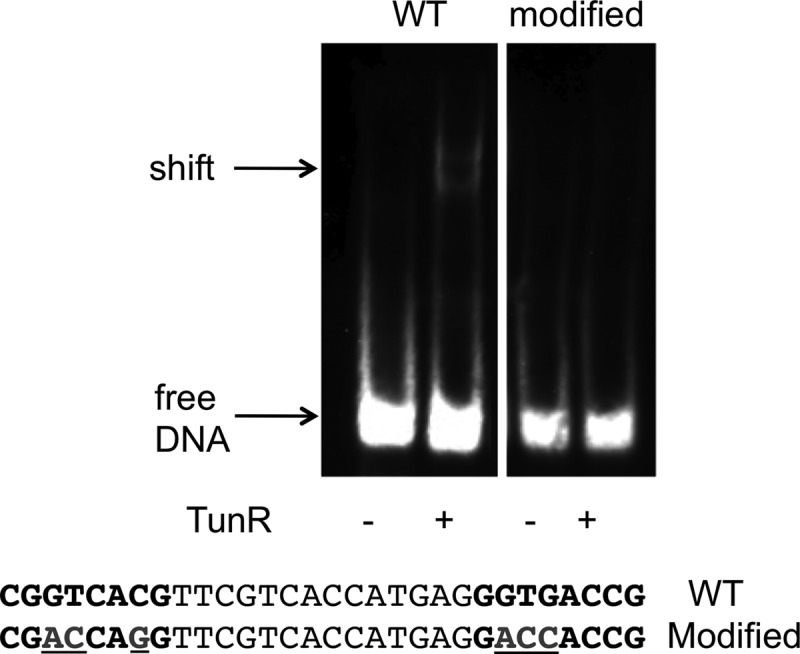
TunR binds to the predicted DNA motif. Electrophoretic mobility shift assay where TunR (33 pmol) was tested for binding to a DNA fragment (200 fmol) containing the predicted binding site motif. Lanes 1 and 2, wild-type-binding motif; lanes 3 and 4, modified motif; the sequences of the 30-nt motif (total DNA size of 46 bp) are shown with the 8-bp conserved half sites in bold, and the base changes in the modified substrate are underlined.
Identification of a novel family of transcription factors.
Starting with a newly found DVU0179 regulator, we found a novel TunR family of related regulators by carrying out a comprehensive search of its members across an entire phylogenetic tree of prokaryotic genomes.
To characterize the TunR transcription factor family in full detail, we identified all representatives of this family in the GenBank database. We considered only proteins with similarity in both the N-terminal recombinase-like domain and C-terminal TOBE domains, representing the true TunR homologs. As a result, 40 proteins from 23 bacterial species were identified (see Table S4 in the supplemental material). All bacterial species considered belong to sulfate-reducing deltaproteobacteria. Most of the TunR family proteins were identified in 17 Desulfovibrio sp. genomes (32 proteins), four proteins in three Desulfovibrionales species, and three proteins in other deltaproteobacteria. For all these TunR family members, we carried out a sequence analysis of amino acid variants in all active-site positions in the recombinase-like domain. We found that one or more amino acids in these active positions are altered in all 40 proteins (see Table S2 in the supplemental material). Thus, we argue that all TunR family proteins are not active as DNA cleavage enzymes but function as transcription factors that specifically bind to their target DNA sites and control gene expression.
Phylogenetic analysis and genome context analysis identified two distinct groups of orthologs within the TunR family (see Fig. S1 in the supplemental material). Genes encoding proteins from both ortholog groups were found to be colocalized with the genes encoding transport systems. Proteins from the largest TunR group that includes all orthologs of DVU0179 are encoded by genes that are positionally linked with modABC genes. The second group, named TunR2, includes seven genes colocalized with orthologous genes encoding TSUP family transporters. For other tunR2 genes, no conserved gene neighborhood was observed. The TunR2 group is the most phylogenetically diverse.
Reconstruction of the TunR family regulons.
To fully characterize the novel TunR family, we reconstructed regulons for all members of this family by searching for the associated TF-binding motifs in both subgroups. A known motif upstream of modABC genes found to be associated with DVU0179 was a starting point for this analysis.
We searched for good matches of the putative binding motif in upstream regions of modABC genes in all Desulfovibrio genomes and built a novel regulatory motif for the TunR family proteins with these matches as a training set. By comparison with this motif, TunR-binding sites were found in all 19 genomes that contain tunR genes. The binding sites were located upstream of the modABC genes and were typically proximal to the tunR genes.
We discovered additional binding sites of TunR in nine of these genomes (Table 2). In D. alaskensis G20, the TunR-binding site was found upstream of the operon encoding ModA and ModB proteins and a hypothetical protein. Other TunR-binding sites were identified upstream of phsAB operons encoding a putative thiosulfate reductase (in six genomes) and upstream of a four-gene operon encoding an ABC-type transporting system and a TonB-dependent receptor (in four genomes).
Table 2.
Occurrence and functional roles of genes regulated by TunR family members
| Group | Genome | Occurrence of genea |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| tunR | tunR2 | modABC | modAB-DUF364 genes | ABC-mop | phsAB | fmdE | modE | fdhE | TSUP family gene | TonB-ABC genes | SulP family gene | mop | modABCAB-DUF364 genes | tupBAC | ||
| TunR | Bilophila sp. 4_1_30 | + | + | |||||||||||||
| Bilophila wadsworthia 3_1_6 | + | + | ||||||||||||||
| Desulfomicrobium baculatum DSM 4028 | + | + | ||||||||||||||
| Desulfovibrio aespoeensis Aspo-2 | + | + | ||||||||||||||
| Desulfovibrio alaskensis G20 | + | + | + | |||||||||||||
| Desulfovibrio desulfuricans ND132 | + | + | + | + | ||||||||||||
| Desulfovibrio desulfuricans subsp. desulfuricans strain ATCC 27774 | + | + | + | + | + | |||||||||||
| Desulfovibrio fructosovorans JJ | + | + | + | + | ||||||||||||
| Desulfovibrio magneticus RS-1 | + | + | + | + | ||||||||||||
| Desulfovibrio piger ATCC 29098 | + | + | + | |||||||||||||
| Desulfovibrio salexigens DSM 2638 | + | + | ||||||||||||||
| Desulfovibrio sp. 3_1_syn3 | + | + | ||||||||||||||
| Desulfovibrio sp. 6_1_46AFAA | + | + | ||||||||||||||
| Desulfovibrio sp. A2 | + | + | ||||||||||||||
| Desulfovibrio sp. FW1012B | + | + | + | + | ||||||||||||
| Desulfovibrio sp. U5L | + | + | + | + | ||||||||||||
| Desulfovibrio vulgaris DP4 | + | + | ||||||||||||||
| Desulfovibrio vulgaris strain Hildenborough | + | + | ||||||||||||||
| Desulfovibrio vulgaris strain “Miyazaki F” | + | + | + | + | + | + | + | + | ||||||||
| TunR2 | Desulfobacter postgatei 2ac9 | + | + | |||||||||||||
| Desulfobulbus propionicus DSM 2032 | + | |||||||||||||||
| Desulfomicrobium baculatum DSM 4028 | + | |||||||||||||||
| Desulfomonile tiedjei DSM 6799 | + | + | ||||||||||||||
| Desulfovibrio aespoeensis Aspo-2 | + | + | ||||||||||||||
| Desulfovibrio africanus strain Walvis Bay | + | + | ||||||||||||||
| Desulfovibrio fructosovorans JJ | + | + | ||||||||||||||
| Desulfovibrio magneticus RS-1 | + | + | + | + | ||||||||||||
| Desulfovibrio sp. A2 | + | + | ||||||||||||||
| Desulfovibrio sp. FW1012B | + | + | ||||||||||||||
| Desulfovibrio sp. U5L | + | + | ||||||||||||||
| Desulfovibrio vulgaris DP4 | + | + | ||||||||||||||
| Desulfovibrio vulgaris strain Hildenborough | + | + | ||||||||||||||
| Desulfovibrio vulgaris strain “Miyazaki F” | + | + | ||||||||||||||
Regulons of different TunR family members are shown. The presence of genes for the respective functional roles in the regulon is shown by a plus sign. Genes and their predicted functional roles are as follows: tunR, TunR group regulator; tunR2, TunR2 group regulator; modABC, ModABC transporter; modAB-DUF364 genes, ModA and ModB components of ModABC transporter and hypothetical DUF364 family protein; ABC-mop, ABC transporter and molybdate-binding protein; phsAB, thiosulfate reductase; fmdE, formylmethanofuran dehydrogenase subunit E; modE, ModE-like transcriptional regulator; fdhE, formate dehydrogenase accessory protein; TSUP family gene, TSUP family transporter; TonB-ABC genes, ABC transporter and TonB-dependent receptor; SulP family gene, SulP family transporter; mop, molybdate-binding protein; modABCAB-DUF364 genes, ModABC transporter and hypothetical DUF364 family protein; tupBAC, TupABC transporter.
In D. desulfuricans ND132, predicted TunR-binding sites were found upstream of two four-gene operons encoding ABC transport systems. The first ND132 operon appears to encode subunits of an Fe(III) ABC transporter and an ModA-like periplasmic molybdate-binding protein. Proteins encoded by the second ND132 operon are similar to three subunits of a sulfonate transporting system and a molybdopterin-binding protein. Both operons have no close homologs in other deltaproteobacteria.
Unusual expansion of the TunR regulon was observed in D. vulgaris Miyazaki. In addition to TunR-binding sites upstream of modABC genes and the phsAB operon, TunR-binding sites were also found upstream of an operon encoding a tungstate-specific transporter, TupABC, and four other genes. One of these genes encodes a truncated transcriptional regulator, ModE, that possesses only a DNA-binding domain. Other genes possibly regulated by TunR are annotated to encode a molybdate/tungstate-binding protein, a formate dehydrogenase accessory protein, and the formylmethanofuran dehydrogenase subunit E.
A putative TunR-type-binding site was also found in D. desulfuricans ATCC 27774 upstream of the tupBAC operon encoding a high-specificity tungstate transporting system. We associate this binding site with an additional copy of the tunR gene directly upstream. In Desulfovibrio piger, an additional copy of the tunR gene was also found upstream of the tupBAC operon, but no candidate TunR site was found in this locus. A possible explanation is a deletion in the tupBAC regulatory region in D. piger, because the tunR-tupA intergenic region in this genome is only 67 bp long, in contrast to 182 bp in D. desulfuricans ATCC 27774.
The motif specific for the TunR2 regulators was identified by the analysis of noncoding regions of loci where tunR2 genes are colocalized with TSUP family transporter genes (associated with the TunR2 group). The identified TunR2-specific motif (a 30-nucleotide [nt] conserved inverted repeat) is very similar to the TunR motif, but it differs in one pair of the palindromic positions, which allows the two motifs to be distinguished (Fig. 1). Namely, the T4/A27 pair is specific for the TunR-binding motif, and the A4/T27 pair is specific for the TunR2-binding motif. In species that have both regulators (nine Desulfovibrio sp. genomes), we utilized this notable distinction for the assignment of putative binding sites to either TunR or TunR2 during the whole-genome regulon reconstruction.
Additional members of the TunR2 regulon were identified by scanning all genomes for the TunR2-binding motif. The TunR2 regulon typically includes one or two regulated genes (Table 2) that encode permeases from either the TSUP or SulP family. Only Desulfovibrio magneticus has TunR2-binding sites upstream of both transporters. In four genomes where TunR2 regulates SulP family transporters, a TunR2-binding site was also found upstream of a gene annotated to encode a molybdate/tungstate-binding protein.
Two copies of tunR2 were found in genomes of D. magneticus, Desulfovibrio fructosovorans, Desulfovibrio sp. FW1012B, and Desulfovibrio sp. U5L. In each of these four genomes, additional TunR2-binding sites were identified upstream of predicted molybdopterin-binding proteins. These additional sites are highly similar to the sites upstream of SulP family transporter genes, so we expect that both TunR2 paralogs can bind all these binding sites.
Promoter prediction.
The position of a transcription factor-binding site relative to a promoter may determine whether the regulatory role is to activate or repress transcription of the downstream genes (34). Using the sigma-70 promoter motif reconstructed in D. vulgaris Hildenborough (23), we predicted putative promoters upstream of target genes of the TunR family TFs (see Table S3 in the supplemental material). For modABC genes, TunR-binding sites are located just upstream of −35 promoter sequences in most cases, and they were predicted to be activating sites. However, the TunR sites upstream of the phsAB operons overlap −10 sequences, a typical position for repressor binding. Predicted binding sites of TunR2 proteins upstream of the TSUP and SulP family transporter genes usually overlap −10 promoter elements and the predicted transcriptional start points, so we hypothesize that TunR2 functions as a repressor.
DISCUSSION
In this study, we discovered a novel family of tungstate-responsive transcriptional factors which was found only in sulfate-reducing deltaproteobacteria. A striking feature of this family of regulators is that their DNA-binding domains are not similar to any known transcriptional regulator and appear to have been evolved from a protein of a totally different function that belongs to the phage integrase/recombinase family (PF00589). We named this novel family TunR since our experimental data confirm that a member of the family in D. vulgaris Hildenborough regulates a transcriptional response to a change in concentration of tungstate. We thoroughly investigated this family using comparative genomics analysis in all genomes where it exists. This analysis resulted in computational reconstruction of complete regulons for all known TunR family members. To verify regulon prediction in a model bacterium, D. vulgaris Hildenborough, TunR-dependent expression of the molybdate transporting system ModABC was demonstrated in vivo. The expression studies suggest that tungstate is an effector of the TunR protein. Specific binding of the TunR protein to the predicted binding site was also confirmed in vitro. TunR protein would be able to bind tungstate directly, because C-terminal domains of TunR and ModE proteins are similar and binding of molybdate and tungstate to TOBE domains of the E. coli ModE protein has been previously demonstrated in vitro (35). A binding of molybdate with C-terminal domains of ModE induces conformational change of the ModE protein that affects its dimerization status (35), and we suppose that TunR quaternary structure may change upon molybdate binding as well. Though SRB are known to be very susceptible to trace metal oxyanions, regulators responsive to tungstate have not yet been studied in these bacteria. A unique feature of TunR protein from D. vulgaris Hildenborough is an ability to differentiate between molybdate and tungstate. Such specificity has not been shown for other oxyanion-sensing bacterial transcription factors.
Evolutionary scenario of the TunR family.
TunR family proteins have a unique domain architecture that includes an N-terminal recombinase-like domain that is involved in DNA binding and two C-terminal TOBE domains sensing oxyanions. We performed phylogenetic analysis for each TunR protein domain to elucidate possible scenarios of evolution and origin of this novel TF family. The phylogenetic tree based on the DNA-binding domain only (see Fig. S2 in the supplemental material) clusters all TunR proteins into a single group. Similarly, these proteins form a distinct monophyletic group on the phylogenetic tree based on the first TOBE domain (see Fig. S3). Thus, the most parsimonious explanation of TunR family origin in deltaproteobacteria is that it emerged as a result of a single event of fusion of a recombinase-like protein and a molybdate/tungstate-binding protein.
Further analysis revealed that N-terminal and C-terminal domains of TunR have different evolutionary origins. The closest homologs of N-terminal domains of TunR were found in three sulfate-reducing bacteria, two deltaproteobacteria, Desulfohalobium retbaense (Dret_2529) and Syntrophobacter fumaroxidans (Sfum_2367), and Nitrospira Thermodesulfovibrio yellowstonii (THEYE_A0306) (Fig. 5). Though Nitrospira protein seems to be fully functional since it possesses all required active-site residues in its recombinase domain, two proteins from deltaproteobacteria miss two of these residues. Thus, we assume that the ancestor of the DNA-binding domain of TunR lost its original recombinase activity by acquiring point mutations in active-site residues prior to the fusion event. In contrast to the N-terminal DNA-binding domain, TunR TOBE domains do not have close homologs in deltaproteobacteria. Despite some deltaproteobacteria having modE genes with the TOBE domain (15), the closest TunR homologs (∼50% identity) are putative molybdate-binding proteins from Pseudomonas spp. and ModE regulators from alphaproteobacteria (see Fig. S3 in the supplemental material).
Fig 5.
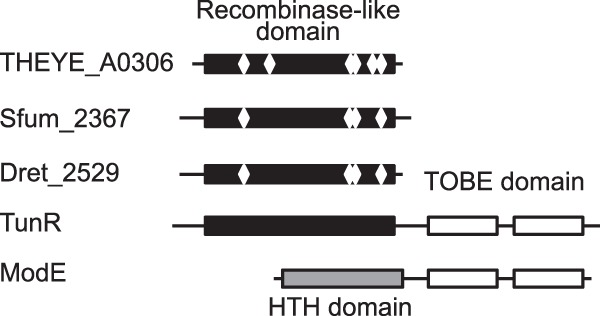
Domain structure of TunR, ModE, and TunR homologs. Rectangles represent protein domains, and white diamonds show approximate positions of active site residues in PFAM [29] annotations.
In summary, we suggest a possible evolutionary scenario of gradual conversion of a site-specific recombinase into an oxyanion-responsive transcriptional regulator. To our knowledge, this is the first known case of a novel transcription factor family emerged from site-specific recombinases.
TunR function determines its phylogenetic distribution.
Remarkably, all proteins of the TunR family were found only in sulfate-reducing deltaproteobacteria (Fig. 6), which can be explained by special features of both TunR proteins and energy metabolism of sulfate reducers. TunR was demonstrated to be a tungstate-responsive protein (Fig. 3) and is expected to sense tungstate concentration directly. ModABC is the most conserved member of TunR regulons in Desulfovibrionales. It is generally recognized as a molybdate transporter, but it can also transport tungstate since it was demonstrated in E. coli that the ModA subunit of the ModABC binds both molybdate and tungstate with similar affinities (7). The substrate specificity of the ModABC transporter was not determined in D. vulgaris, although its tungstate-dependent regulation suggests it may be involved in the uptake of tungstate as well as molybdate. In accordance with our results, a strong repression of modB transcription by tungstate was observed in D. alaskensis NCIMB 13491, whereas a negative effect of molybdate was less pronounced (36). However, several sulfate-reducing deltaproteobacteria have no TunR family members, i.e., Desulfatibacillum alkenivorans, Desulfobacterium autotrophicum, Desulfococcus oleovorans, and Desulfotalea psychrophila. We assume an existence of other transcriptional factors that control expression of molybdate and tungstate transporters in these bacteria.
Fig 6.
Phylogenetic distribution of the TunR family.
An importance of the strict control of Mo and W homeostasis for SRB comes from metabolic peculiarities of sulfate reduction. The first step in the sulfate reduction pathway is ATP-dependent activation of sulfate by SAT that provides adenosine-5′-phosphosulfonate (APS) for APS reductase. SAT can also convert chromate, molybdate, and tungstate into unstable analogs of APS (13). It leads to depletion of the ATP pool, and, in turn, to inhibition of SRB growth. Experimental studies demonstrated that molybdate inhibits sulfate reduction and growth of SRB even at micromolar concentrations (37), and the addition of tungstate had a similar effect (38). In contrast, molybdate and tungstate stimulate growth of the SRB at nanomolar concentrations, because these metals are indispensable for enzymes involved in energy metabolism, such as formate dehydrogenases (4) and aldehyde oxidoreductase (39). Thus, tight control of intracellular concentrations of these metals in a narrow range is vital for SRB.
At the same time, bioavailability of tungstate oxyanions for SRB under sulfide-rich conditions may be greater than that of molybdate oxyanions. SRB live in anoxic environments and produce sulfide as an end product of their metabolism. Desulfovibrio spp. are able to reduce Mo(VI) to Mo(IV) that results in the removal of molybdate from solution (40) due to the formation of less soluble molybdenum sulfides (41), which are not suitable for uptake. In contrast to molybdate, the tungstate would be more stable because of the lower reduction potential of the W(IV)-W(VI) redox couple than the reduction potential of the Mo(IV)-Mo(VI) couple (42). Higher solubility of the tungsten disulfide than the molybdenum disulfide may also influence its stability (43). Thus, the tungstate-dependent regulation of molybdate and tungstate transport is important for the physiology of SRB.
TunR2 is a putative protection factor against sulfate reduction inhibition.
The second largest group of TunR family regulators, TunR2, has two remarkable differences from the first group of TunR proteins. First, the TunR2 regulons do not include any known molybdate or tungstate transporters. Instead, genes regulated by TunR2 encode transporters from the TSUP and SulP families. Second, the positional analysis of binding sites in promoter regions of target genes suggests that TunR and TunR2 proteins may function in opposite modes. TunR-binding sites are located upstream of predicted promoters, which is a typical location for transcriptional activators (34). In contrast, TunR2 is characterized by binding sites that overlap putative transcription start sites, suggesting a transcriptional repression mechanism.
We propose that, similarly to TunR, TunR2 binds to DNA in the absence of an effector (possibly molybdate and/or tungstate ions). The proposed negative regulatory effect of TunR2 proteins may be influenced by their dissociation from DNA upon oxyanion binding. We hypothesize that TunR2 represses its target genes in the absence of the effector and derepresses the genes in response to an excess of molybdate and/or tungstate; however, this hypothesis awaits further experimental validation.
The biological role of TunR2-dependent regulation may be linked with a function of protecting SRB from an inhibitory effect of a high intracellular concentration of molybdate and tungstate on sulfate reduction. Two hypothetical mechanisms of such protection may be proposed: TunR2 may regulate either molybdate/tungstate efflux or uptake of a substance that overcomes the inhibiting effect of molybdate and tungstate. The former mechanism is supported by a wide spread of efflux systems for a metal resistance in the bacterial world (44). Molybdate or tungstate efflux proteins have not been described yet, but most well-studied bacteria are much less susceptible to these oxyanions than SRB. The latter mechanism is supported by a finding that the SulP family contains sulfate uptake transporters (45), and the TSUP family members are involved in transport of sulfur-based compounds (46). An excess of molybdate and tungstate is known to inhibit the sulfate reduction in Desulfovibrio spp. (11). A plausible explanation of the TunR2-dependent upregulation of sulfur-related transporters is that it would supply more sulfate for adenylation in Mo/W-replete conditions and thus overcome inhibition of sulfate reduction.
Supplementary Material
ACKNOWLEDGMENTS
This work was conducted by Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA; http://enigma.lbl.gov), a Scientific Focus Area Program at Lawrence Berkeley National Laboratory, and was supported by the Office of Science, Office of Biological and Environmental Research, of the U.S. Department of Energy under contracts no. DE-AC02-05CH11231 with Lawrence Berkeley National Laboratory and DE-SC0004999 with Sanford-Burnham Medical Research Institute and Lawrence Berkeley National Laboratory.
Footnotes
Published ahead of print 2 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00679-13.
REFERENCES
- 1.Zhang Y, Gladyshev VN. 2008. Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 379:881–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kletzin A, Adams MW. 1996. Tungsten in biological systems. FEMS Microbiol. Rev. 18:5–63 [DOI] [PubMed] [Google Scholar]
- 3.Sevcenco A-M, Bevers LE, Pinkse MWH, Krijger GC, Wolterbeek HT, Verhaert PDEM, Hagen WR, Hagedoorn P-L. 2010. Molybdenum incorporation in tungsten aldehyde oxidoreductase enzymes from Pyrococcus furiosus. J. Bacteriol. 192:4143–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Da Silva SM, Pimentel C, Valente FMA, Rodrigues-Pousada C, Pereira IAC. 2011. Tungsten and molybdenum regulation of formate dehydrogenase expression in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 193:2909–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilar-Barajas E, Díaz-Pérez C, Ramírez-Díaz MI, Riveros-Rosas H, Cervantes C. 2011. Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 24:687–707 [DOI] [PubMed] [Google Scholar]
- 6.Makdessi K, Andreesen JR, Pich A. 2001. Tungstate uptake by a highly specific ABC transporter in Eubacterium acidaminophilum. J. Biol. Chem. 276:24557–24564 [DOI] [PubMed] [Google Scholar]
- 7.Rech S, Wolin C, Gunsalus RP. 1996. Properties of the periplasmic ModA molybdate-binding protein of Escherichia coli. J. Biol. Chem. 271:2557–2562 [DOI] [PubMed] [Google Scholar]
- 8.Anderson LA, Palmer T, Price NC, Bornemann S, Boxer DH, Pau RN. 1997. Characterisation of the molybdenum-responsive ModE regulatory protein and its binding to the promoter region of the modABCD (molybdenum transport) operon of Escherichia coli. Eur. J. Biochem. 246:119–126 [DOI] [PubMed] [Google Scholar]
- 9.Anderson LA, McNairn E, Lubke T, Pau RN, Boxer DH, Leubke T. 2000. ModE-dependent molybdate regulation of the molybdenum cofactor operon moa in Escherichia coli. J. Bacteriol. 182:7035–7043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McNicholas PM, Gunsalus RP. 2002. The molybdate-responsive Escherichia coli ModE transcriptional regulator coordinates periplasmic nitrate reductase (napFDAGHBC) operon expression with nitrate and molybdate availability. J. Bacteriol. 184:3253–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranade D. 1999. Evaluation of the use of sodium molybdate to inhibit sulphate reduction during anaerobic digestion of distillery waste. Bioresour. Technol. 68:287–291 [Google Scholar]
- 12.Videla HA, Herrera LK. 2005. Microbiologically influenced corrosion: looking to the future. Int. Microbiol. 8:169–180 [PubMed] [Google Scholar]
- 13.Taylor BF, Oremland RS. 1979. Depletion of adenosine triphosphate in Desulfovibrio by oxyanions of group VI elements. Curr. Microbiol. 3:101–103 [Google Scholar]
- 14.Newport PJ, Nedwell DB. 1988. The mechanisms of inhibition of Desulfovibrio and Desulfotomaculum species by selenate and molybdate. J. Appl. Microbiol. 65:419–423 [Google Scholar]
- 15.Rodionov DA, Dubchak I, Arkin A, Alm E, Gelfand MS. 2004. Reconstruction of regulatory and metabolic pathways in metal-reducing delta-proteobacteria. Genome Biol. 5:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, Dubchak IL, Alm EJ, Arkin AP. 2010. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 38:D396–D400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res. 41:D36–D42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boratyn GM, Camacho C, Cooper PS, Coulouris G, Fong A, Ma N, Madden TL, Matten WT, McGinnis SD, Merezhuk Y, Raytselis Y, Sayers EW, Tao T, Ye J, Zaretskaya I. 22 April 2013. BLAST: a more efficient report with usability improvements. Nucleic Acids Res. [Epub ahead of print.] 10.1093/nar/gkt282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics Chapter 2:Unit 2.3 [DOI] [PubMed] [Google Scholar]
- 21.Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, Gelfand MS, Arkin AP, Mironov AA, Dubchak I. 2010. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 38:W299–W307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mironov AA, Vinokurova NP, Gel'fand MS. 2000. Software for analyzing bacterial genomes. Mol. Biol. (Mosk.) 34:253–262 [PubMed] [Google Scholar]
- 23.Price MN, Deutschbauer AM, Kuehl JV, Liu H, Witkowska HE, Arkin AP. 2011. Evidence-based annotation of transcripts and proteins in the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 193:5716–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novichkov PS, Laikova ON, Novichkova ES, Gelfand MS, Arkin AP, Dubchak I, Rodionov DA. 2010. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 38:D111–D118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zane GM, Yen HB, Wall JD. 2010. Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 76:5500–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193–201 [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 28.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6:343–345 [DOI] [PubMed] [Google Scholar]
- 29.Rajeev L, Luning EG, Dehal PS, Price MN, Arkin AP, Mukhopadhyay A. 2011. Systematic mapping of two component response regulators to gene targets in a model sulfate reducing bacterium. Genome Biol. 12:R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer ELL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blakely G, May G, McCulloch R, Arciszewska LK, Burke M, Lovett ST, Sherratt DJ. 1993. Two related recombinases are required for site-specific recombination at dif and cer in E. coli K-12. Cell 75:351–361 [DOI] [PubMed] [Google Scholar]
- 32.UniProt Consortium 2012. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res. 40:D71–D75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kono N, Arakawa K, Tomita M. 2011. Comprehensive prediction of chromosome dimer resolution sites in bacterial genomes. BMC Genomics 12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Hijum SAFT, Medema MH, Kuipers OP. 2009. Mechanisms and evolution of control logic in prokaryotic transcriptional regulation. Microbiol. Mol. Biol. Rev. 73:481–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gourley DG, Schuttelkopf AW, Anderson LA, Price NC, Boxer DH, Hunter WN. 2001. Oxyanion binding alters conformation and quaternary structure of the C-terminal domain of the transcriptional regulator mode. Implications for molybdate-dependent regulation, signaling, storage, and transport. J. Biol. Chem. 276:20641–20647 [DOI] [PubMed] [Google Scholar]
- 36.Mota CS, Valette O, González PJ, Brondino CD, Moura JJG, Moura I, Dolla A, Rivas MG. 2011. Effects of molybdate and tungstate on expression levels and biochemical characteristics of formate dehydrogenases produced by Desulfovibrio alaskensis NCIMB 13491. J. Bacteriol. 193:2917–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nemati M, Mazutinec TJ, Jenneman GE, Voordouw G. 2001. Control of biogenic H(2)S production with nitrite and molybdate. J. Ind. Microbiol. Biotechnol. 26:350–355 [DOI] [PubMed] [Google Scholar]
- 38.Lie TJ, Godchaux W, Leadbetter ER. 1999. Sulfonates as terminal electron acceptors for growth of sulfite-reducing bacteria (Desulfitobacterium spp.) and sulfate-reducing bacteria: effects of inhibitors of sulfidogenesis. Appl. Environ. Microbiol. 65:4611–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hensgens CMH, Nienhuis-Kuiper ME, Hansen TA. 1994. Effects of tungstate on the growth of Desulfovibrio gigas NCIMB 9332 and other sulfate-reducing bacteria with ethanol as a substrate. Arch. Microbiol. 162:143–147 [Google Scholar]
- 40.Tucker MD, Barton LL, Thomson BM. 1997. Reduction and immobilization of molybdenum by Desulfovibrio desulfuricans. J. Environ. Quality 26:1146 [Google Scholar]
- 41.Biswas KC, Woodards NA, Xu H, Barton LL. 2009. Reduction of molybdate by sulfate-reducing bacteria. Biometals 22:131–139 [DOI] [PubMed] [Google Scholar]
- 42.Holm RH, Solomon EI, Majumdar A, Tenderholt A. 2011. Comparative molecular chemistry of molybdenum and tungsten and its relation to hydroxylase and oxotransferase enzymes. Coord. Chem. Rev. 255:993–1015 [Google Scholar]
- 43.Hille R. 2002. Molybdenum and tungsten in biology. Trends Biochem. Sci. 27:360–367 [DOI] [PubMed] [Google Scholar]
- 44.Nies DH. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313–339 [DOI] [PubMed] [Google Scholar]
- 45.Alper SL, Sharma AK. 2013. The SLC26 gene family of anion transporters and channels. Mol. Aspects Med. 34:494–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shlykov MA, Zheng WH, Chen JS, Saier MH., Jr 2012. Bioinformatic characterization of the 4-toluene sulfonate uptake permease (TSUP) family of transmembrane proteins. Biochim. Biophys. Acta 1818:703–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.