Abstract
Vibrio cholerae is motile by its polar flagellum, which is driven by a Na+-conducting motor. The stators of the motor, composed of four PomA and two PomB subunits, provide access for Na+ to the torque-generating unit of the motor. To characterize the Na+ pathway formed by the PomAB complex, we studied the influence of chloride salts (chaotropic, Na+, and K+) and pH on the motility of V. cholerae. Motility decreased at elevated pH but increased if a chaotropic chloride salt was added, which rules out a direct Na+ and H+ competition in the process of binding to the conserved PomB D23 residue. Cells expressing the PomB S26A/T or D42N variants lost motility at low Na+ concentrations but regained motility in the presence of 170 mM chloride. Both PomA and PomB were modified by N,N′-dicyclohexylcarbodiimide (DCCD), indicating the presence of protonated carboxyl groups in the hydrophobic regions of the two proteins. Na+ did not protect PomA and PomB from this modification. Our study shows that both osmolality and pH have an influence on the function of the flagellum from V. cholerae. We propose that D23, S26, and D42 of PomB are part of an ion-conducting pathway formed by the PomAB stator complex.
INTRODUCTION
Rotational nanomachines are membrane-embedded protein complexes which couple the flux of protons or sodium ions along a transmembrane electrochemical gradient to the generation of torque (1, 2). In F1Fo ATP synthases, this mechanism depends on reversible interactions of the coupling cation (H+ or Na+) with selected amino acid residues of the c subunits found in the membrane-embedded Fo complex (3, 4). In the Na+-translocating ATP synthase from Ilyobacter tartaricus, a ring of 11 c subunits rotates against the stator composed of a single a subunit and two b subunits. Rotation of the c ring is transmitted via a central shaft into the cytoplasmic F1 complex of the ATP synthase, enforcing ATP synthesis (2). The bacterial flagellum is another nanomachine, which transmits rotation of the basal body-motor complex in the cytoplasmic membrane to the extracellular flagellar filament. Torque is generated by several stator complexes which surround the rotor. They are composed of the subunits MotA and MotB in H+-dependent flagella or PomA and PomB in Na+-dependent flagella (5). Stator complexes which, depending on the environment of the cell, may use both Na+ and H+ (6) or Na+ and K+ (7) have also been described. A stator complex, or force-generating unit, is composed of four PomA (MotA) and two PomB (MotB) subunits (PomA4B2 or MotA4B2) and is anchored to peptidoglycan in the periplasm by a specific binding region of PomB (1, 8). The flux of the coupling cation (Na+, K+, or H+) along the electrochemical gradient through a putative channel formed by transmembrane helices III and IV of PomA and the single transmembrane helix of PomB (Fig. 1A) is proposed to alter the conformation of the FliG component of the rotor ring. This creates torque and forces the rotor to move with respect to the PomA4B2 (or MotA4B2) stator (9, 10). Conserved, charged amino acid residues within the transmembrane regions of the stator were studied with respect to their putative role as a ligand for Na+ in the access channel between PomA and PomB (11). It was shown that Asp24 in PomB of the flagellum of Vibrio alginolyticus (9, 12) or the homologous Asp23 in the highly conserved PomB from V. cholerae (13) (Fig. 1A and B) is required for flagellar function. This conserved aspartate was proposed to interact with Na+.
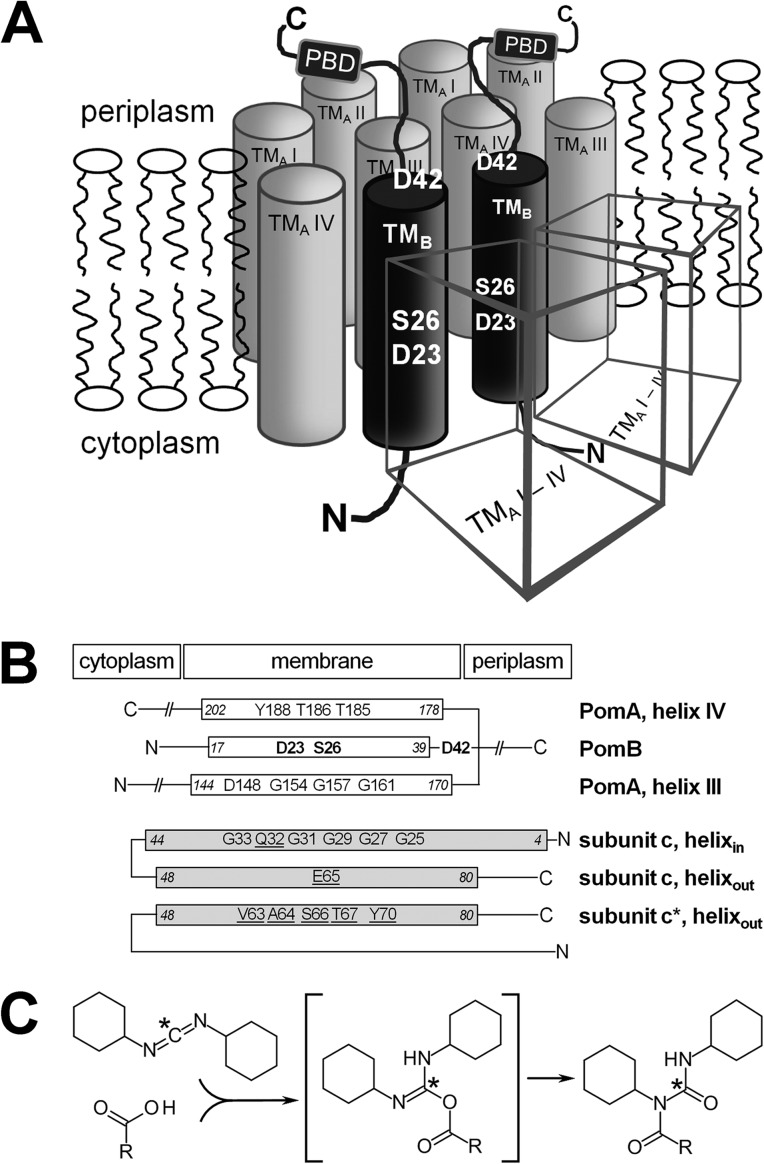
Fig 1.
Model of the V. cholerae PomA4B2 stator complex and comparison of its transmembrane helices with the inner and outer helices forming the membrane-embedded c ring of the Na+-translocating F1Fo ATPase from I. tartaricus. (A) Possible arrangement of transmembrane helices in a PomA4B2 complex adapted from the E. coli MotA4B2 model (39). Borders of transmembrane segments, as predicted by HMMTOP, are V7/V30 for transmembrane helix I of PomA (TMA I), V35/F56 for transmembrane helix II of PomA (TMA II), R144/D170 for transmembrane helix III of PomA (TMA III), A178/D202 for transmembrane helix IV of PomA (TMA IV), and W17/S39 for the transmembrane helix of PomB (TMB). The positions of the residues in PomB subjected to mutagenesis in this study are indicated (D23, S26, and D42). The cubic objects represent PomA subunits. PBD, peptidoglycan binding domain. (B) Transmembrane helices III and IV of PomA, together with the single transmembrane helix of PomB, likely provide interaction sites for the coupling cation (Na+) in the PomA4PomB2 stator complex of the flagellar motor. The carboxylate of D23 (PomB) is a putative ligand for Na+ which also might interact with S26 or D42 (shown in bold). In the membrane-embedded c ring of the Na+-translocating F1Fo ATPase from Ilyobacter tartaricus, seven residues (E65, Q32, Y70, T67, S66, V63, and A64, which are underlined) of three transmembrane helices from two neighboring c subunits (c and c*) are required to form a Na+ binding site. Due to conserved glycine residues in subunit c, the N-terminal ring of helices (helixin) exhibits a smaller diameter than the C-terminal ring of helices (helixout). Na+ engages in direct coordination with the carboxylate of Glu65 of the C-terminal ring of helices. The absence of Na+ and low pH promote protonation of E65, which is monitored from its modification with DCCD. (C) A carboxylic acid reacts with DCCD to form an intermediate (O-acylisocarbamide) which is unstable in the presence of water. In hydrophobic environments, such as membrane-spanning regions of proteins, a rearrangement occurs, and the stable N-acylcarbamide is formed. *, radioactive 14C, which, after modification, is covalently attached to the protein carrying the carboxyl group (E65 on subunit c in the case of the I. tartaricus ATPase). The structures were drawn with the Accelrys Draw (version 4.0) program (Accelrys).
The high-resolution structure of the c ring of the ATP synthase illustrates that the coordination of a sodium ion in the hydrophobic region of a membrane protein can involve one carboxylate per Na+ ion (3) (Fig. 1B). Comparing the mode of action of these two Na+-driven rotational machines, the flagellum and the F1Fo ATP synthase, reveals a remarkable difference. H+- and Na+-dependent systems are found with both types of machines, yet the Na+-driven ATPase may operate with H+ in the absence of Na+ and at low pH (14), whereas the Na+-dependent, polar flagellum obligatorily requires Na+ for its function (11). It seems likely that this functional difference is the result of a different interaction pattern of Na+ with the c ring or the flagellar stator complex. In the Na+-F1Fo ATP synthase from I. tartaricus, Na+ engages in direct coordination with a glutamate carboxyl group (Glu65) in a cation binding site constructed by several amino acid residues from one inner and two outer helices of two adjacent c subunits (3) (Fig. 1B). Consequently, the presence of the coordinated Na+ prevents modification of the carboxylate by N,N′-dicyclohexylcarbodiimide (DCCD) (15) (Fig. 1C).
We hypothesized that despite a potentially different Na+ interaction pattern, a similar Na+ protection mechanism in the DCCD reaction should be observable in the PomA4B2 flagellar stator complex due to the presence of conserved glutamate or asparate residues. Hence, we first investigated how the salt content and pH of the medium affect the in vivo motility of V. cholerae expressing variants of PomB, and next, we studied the accessibility of protons and Na+ to PomA and PomB in vitro using DCCD as a probe specific for protonated carboxylates in hydrophobic regions of these stator components. It is shown that both osmolality and pH govern the performance of the flagellum from V. cholerae and that PomB harbors at least one carboxyl group in a hydrophobic region with a pKa higher than that of a carboxyl group on PomA. The experimental observations can be explained by a perturbation of the arrangement of the structural water molecules within the ion pathway caused by a shift in the ionic composition of the periplasm of V. cholerae. We propose that the arrangement of water molecules in the pathway formed by PomB and PomA determines the access of Na+ to the flagellar motor.
MATERIALS AND METHODS
Sequence alignments.
Amino acid sequences for PomA/MotA and PomB/MotB homologues were obtained from NCBI BLAST using the algorithm blastp (protein-protein BLAST) with PomA (gi|51241595) or PomB (gi|51241597) from V. cholerae as query sequences. In most cases, the coupling cation specificity (Na+ or H+) has not been experimentally confirmed, although the proteins are designated PomA/PomB or MotA/MotB in the database. A multiple-sequence alignment for the PomA/MotA and PomB/MotB group of sequences was performed using the ClustalW algorithm (16) implemented in the BioEdit sequence alignment editor (version 7.0.5.3) (17) with default settings. Identical and similar residues were identified with BioEdit using an identity and similarity threshold of 83% and the BLOSUM62 scoring matrix (18). Putative transmembrane regions of PomA and PomB from V. cholerae were predicted using the HMMTOP server (19, 20).
Chemicals and bacterial strains.
Unless stated otherwise, chemicals were purchased from Sigma-Aldrich Chemie GmbH, Germany. Oligonucleotides were custom synthesized by Eurofins MWG Operon, Germany. Yeast extract, tryptone, and Bacto agar were purchased from Becton, Dickinson. The V. cholerae strains used in this study were O395-N1 ΔpomAB (ΔctxA Strr ΔpomAB) (21) and O395-N1 toxT::lacZ ΔfliG (22). For standard cloning procedures, Escherichia coli strain XL-10 Gold (Stratagene) served as the host. To produce V. cholerae PomAB in E. coli, strain C41(DE3) (23) was used as the expression host.
Vectors for production of PomAB variants in V. cholerae.
The mutations resulting in the S26A, S26T, and D42N variants of PomB were introduced into plasmid pAB (13) by performing a two-step PCR procedure (24) using one pair of complementary oligonucleotides containing the codon for the desired mutation and two flanking oligonucleotides (VCmotAV-III and VCmotBR-III) matching the sequence of the wild-type (wt) genes (see Table S1 in the supplemental material). Plasmid pAB (Table 1) served as the template. Restriction of the resulting 1,762-bp pomAB PCR products with NdeI and XhoI and insertion into the NdeI/XhoI vector fragment of pAB yielded pAB-S26A, pAB-S26T, and pAB-D42N (Table 1). Plasmid pABGFP encoding PomA with an N-terminal polyhistidine tag and PomB fused to green fluorescent protein (GFP) at its N terminus and a Strep tag at its C terminus (Table 1) was constructed as follows. Restriction sites for NheI and AgeI were introduced into pAB upstream of pomB, resulting in pAB-NheI-AgeI (Table 1). Thereby, the complementary oligonucleotides VCmotBV-NheI-AgeI and VCmotBR-NheI-AgeI (see Table S1 in the supplemental material) contained the recognition sequences for NheI and AgeI, VCmotAV-III and VCmotBR-III served as flanking oligonucleotides, and pAB served as the template. The resulting 1,777-bp pomAB PCR product was restricted with NdeI and XhoI and inserted into the NdeI/XhoI vector fragment of pAB, yielding pAB-NheI-AgeI. The sf-gfp gene encoding an engineered superfolding (sf) GFP (25) was amplified using oligonucleotides GFP-V and GFP-R (see Table S1 in the supplemental material). Restriction of the PCR product with AgeI and NheI and subsequent ligation with the AgeI/NheI vector fragment of pAB-NheI-AgeI resulted in pABGFP. To introduce the PomB point mutations D23E, D23N, S26A, and S26T into plasmid pABGFP, the same PCR procedure (24) was used. Plasmid pABGFP was the template, and WS-013-GFPend-f and VCmotBR-III served as flanking oligonucleotides. Insertion of the resulting PCR products with a length of 1,059 bp into pABGFP via restriction sites for AgeI and XhoI yielded plasmids pABGFP-D23E, pABGFP-D23N, pABGFP-S26A, pABGFP-S26T, and pABGFP-D42N.
Table 1.
Plasmids
| Plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| pEC422 | Confers His6 tag to the N terminus of a target protein, Apr | 47 |
| pISC-2 | Para araC Apr | 48 |
| pET24b | PT7, confers His6 tag to the C terminus of a target protein, Kanr | Novagen |
| pET224-Streptag | PT7, confers Strep tag to the C-terminus of a target protein, Kanr | 49 |
| pISC-H | MCS of pISC-2 inserted into pEC422 | 13 |
| pAB | ParaBAD, codes for His6-PomA and PomB-Strep, Apr | 13 |
| pAB-D23N | ParaBAD, codes for His6-PomA and D23N variant of PomB-Strep, Apr | 13 |
| pAB-D23E | ParaBAD, codes for His6-PomA and D23E variant of PomB-Strep, Apr | 13 |
| pAB-S26A | ParaBAD, codes for His6-PomA and S26A variant of PomB-Strep, Apr | This work |
| pAB-S26T | ParaBAD, codes for His6-PomA and S26T variant of PomB-Strep, Apr | This work |
| pAB-D42N | ParaBAD, codes for His6-PomA and D42N variant of PomB-Strep, Apr | This work |
| pAB-NheI-AgeI | ParaBAD, codes for His6-PomA and PomB-Strep, NheI and AgeI restriction sites upstream of pomB, Apr | This work |
| pABGFP | ParaBAD, codes for His6-PomA and GFP-PomB-Strep, Apr | This work |
| pABGFP-D23N | ParaBAD, codes for His6-PomA and D23N variant of GFP-PomB-Strep, Apr | This work |
| pABGFP-D23E | ParaBAD, codes for His6-PomA and D23E variant of GFP-PomB-Strep, Apr | This work |
| pABGFP-S26A | ParaBAD, codes for His6-PomA and S26A variant of GFP-PomB-Strep, Apr | This work |
| pABGFP-S26T | ParaBAD, codes for His6-PomA and S26T variant of GFP-PomB-Strep, Apr | This work |
| pET18ABS | PT7, codes for PomA and PomB-Strep, Kanr | 13 |
| pET18ABS-D23N | PT7, codes for PomA and D23N variant of PomB-Strep, Kanr | This work |
Localization of GFP-PomB.
Overnight cultures of V. cholerae O395-N1 ΔpomAB transformed with plasmids pABGFP encoding His-PomA and PomB variants fused to GFP were added to 5 ml LB medium supplemented with 50 μg ml−1 streptomycin, 200 μg ml−1 ampicillin, and 0.4 mM, 1 mM, or 10 mM arabinose to give an optical density at 600 nm (OD600) of 0.01. After aerobic growth for 5 h at 30°C (180 rpm), 20 μl of the culture was spotted on a poly-l-lysine-coated glass slide (Sigma-Aldrich Chemie GmbH, Germany). V. cholerae toxT::lacZ ΔfliG strains transformed with plasmids coding for PomA and GFP-PomB wild type or variants served as negative controls. Cells were visualized using a fluorescence microscope (Zeiss Imager M1; Zeiss, Germany) equipped with a 38 HE filter set. Wavelengths were 470 nm for excitation (bandpass, 40 nm) and 525 nm for emission (bandpass, 50 nm). Pictures were analyzed with the help of the software AxioVision (Zeiss, Germany).
Swarming assays.
Swarming of cells was analyzed on swarming plates containing 0.25% (wt/vol) agar supplemented with 200 μg ml−1 ampicillin, 50 μg ml−1 streptomycin, and 1 mM arabinose to induce protein expression. The residual Na+ content of plates prepared with deionized H2O was 1.6 mM due to the sodium salts present in the agar. In M9 minimal medium (26), sodium salts were replaced with the corresponding potassium salts (10.3 g liter−1 K2HPO4·3H2O, 3.0 g liter−1 KH2PO4, 1.0 g liter−1 NH4Cl, 2 mM MgCl2, 0.1 mM CaCl2), and the residual Na+ concentration was 16 μM. As the carbon source, 0.2 M glucose was added (final concentration). LB medium without added NaCl (10 g liter−1 tryptone, 5 g liter−1 yeast extract) contained 11 mM Na+. NaCl or KCl (170 mM) was added as indicated. All media were buffered with 10 mM Tris, and the pH was adjusted with 5 M KOH or 5 M HCl. Single colonies of an overnight culture were stabbed on swarming plates using a round-tipped toothpick. Plates were incubated at 30°C for the indicated times.
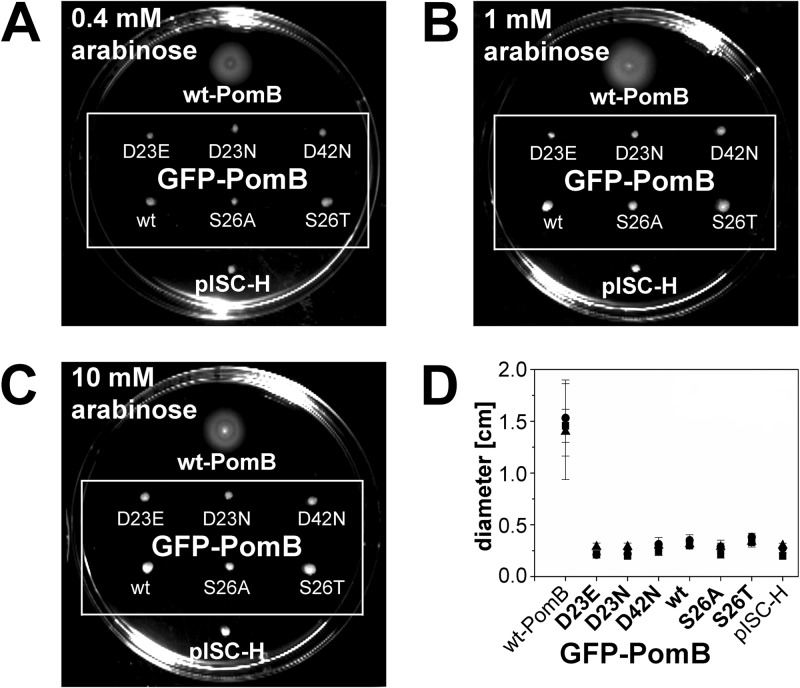
To investigate the impact of different arabinose concentrations on swarming, assays were performed with V. cholerae O395-N1 ΔpomAB transformed with plasmids coding for the GFP-PomB wild type and variants. Single colonies were stabbed on LB soft agar plates containing 0.25% (wt/vol) agar, 200 μg ml−1 ampicillin, 50 μg ml−1 streptomycin, and 0.4 mM, 1 mM, or 10 mM arabinose. Plates were incubated at 30°C for 22 h, and a total of 10 replicates for each condition were analyzed. The mean value and standard deviation of eight plates were calculated (the highest and lowest values were omitted). Strains transformed with either pAB encoding wild-type PomA and PomB or pISC-H for the empty vector were used as controls. Vector pISC-H comprises a promoter region which is similar to the promoter PBAD of pBAD/HisA (Invitrogen) but differs in four nucleotides in the segment between the −10 and the −35 regions (see Fig. S1 in the supplemental material). As a consequence, levels of target proteins expressed from the pISC or the pBAD vector at a given arabinose concentration are expected to differ.
Transmission electron microscopy.
Single colonies of V. cholerae O395-N1 ΔpomAB complemented with plasmids coding for His-PomA and the PomB wild type or its variants were inoculated at 30°C for 30 h in liquid M9 minimal medium with 10 mM arabinose buffered with 10 mM Tris-HCl and supplemented with 200 μg ml−1 ampicillin and 50 μg ml−1 streptomycin. Sodium salts were replaced by their corresponding potassium salts. Aliquots of the bacterial cultures (10 μl) were adsorbed on polyvinyl butyral (Pioloform)-coated copper grids (Plano, Germany) for 2 min. Unbound cells were removed using filter paper (Whatman, GE Healthcare, Germany). The grids were washed twice by incubation with a drop of distilled water for 1 min. Excess water was removed with filter paper. To stain the cells and their flagella, 2% uranyl acetate (5 μl) was applied onto the grids. After 1 min, excess stain was removed with filter paper and the grids were dried at room temperature. Micrographs of cells were recorded on a transmission electron microscope (LEO 912AB; Zeiss, Germany) at 80 kV and at a magnification of ×630.
Growth of V. cholerae reference strain at pH 6.5 or 8.5 in the presence of 170 mM NaCl or KCl.
V. cholerae O395-N1 (wild type) was grown overnight at 37°C in 5 ml LB (10 g liter−1 tryptone, 5 g liter−1 yeast extract, and 10 g liter−1 NaCl supplemented with 50 mM KPi, pH 7.5, 100 mM Tris-HCl, pH 7.5, and 50 μg ml−1 streptomycin). Fifty microliters was used to inoculate 5 ml LB medium, pH 6.5 or 8.5 (10 g liter−1 tryptone, 5 g liter−1 yeast extract, 100 mM bis-Tris-HCl, pH 6.5, or Tris-HCl, pH 8.5), supplemented with 170 mM NaCl or KCl and 50 μg ml−1 streptomycin. Cells were grown aerobically for 15 h at 37°C and 180 rpm, and the resulting OD600 was determined. Mean values from three replicates are presented.
Determination of swimming speed.
V. cholerae ΔpomAB transformed with pAB was grown overnight in LB medium at 37°C and 180 rpm. Fresh LB medium without added NaCl buffered with 10 mM Tris-HCl, pH 7.0, 8.0, or 9.0, supplemented with 50 μg ml−1 streptomycin and 200 μg ml−1 ampicillin was inoculated with an overnight culture to an OD600 of 0.01. Arabinose at a final concentration of 10 mM was added immediately after inoculation. After incubation at 30°C and 180 rpm for 5 h, cultures were diluted 1:100 in LB medium buffered with 10 mM Tris, and the pH was adjusted with 5 M KOH or 5 M HCl. If indicated, 170 mM NaCl or KCl was added. The medium contained 50 μg ml−1 streptomycin, 200 μg ml−1 ampicillin, and 50 mM l-serine to enhance straight swimming of the bacteria (27). Thirty microliters was pipetted into one channel of a flat μ-Slide VI (Ibidi, Germany), and cells were immediately observed under ×200 magnification using a fluorescence microscope (Zeiss Imager M1; Zeiss, Germany) in its differential interference contrast (DIC) mode. After focusing in the z axis, video sequences were taken. Analyses of at least three sequences per condition and strain were performed by Wimasis GmbH, Germany, to achieve a minimum of 200 tracks per condition and strain. All bacteria showing a minimum speed of 4 μm s−1 were included in the analyses and represent 100% of the motile cells. The percentages of motile cells displaying speeds between 18 and 30 μm s−1 were cumulated.
Production of GFP-PomB wt and variants in V. cholerae ΔpomAB cells and membrane isolation.
V. cholerae ΔpomAB containing plasmid pABGFP or pABGFP-D23E, -D23N, -S26T, -S26A, or -D42N was grown in buffered LB medium (10 g tryptone, 5 g yeast extract, 10 g NaCl per 1 liter in 50 mM KPi, pH 8.0) supplemented with 200 μg/ml ampicillin and 50 μg/ml streptomycin. Single colonies were used to inoculate 10 ml medium. After 16 h at 37°C, the cultures were transferred to 1 liter fresh medium in 5-liter Erlenmeyer flasks, and aerobic growth was continued (37°C, 160 rpm) until an OD600 of 0.6 to 0.7 was reached. Protein expression was induced by adding 1 mM arabinose, and growth was continued for 3 h at 30°C. Cells (4 to 6 g [wet weight]) were harvested by centrifugation (5,000 × g, 30 min, 4°C) and washed with buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl). Pellets were resuspended in buffer A (50 mM Tris-HCl, pH 8.0, 300 mM KCl, 10% [vol/vol] glycerol; 5 ml buffer A per g of cells). After addition of a few crystals of DNase I, 0.5 mM MgCl2, and 0.2 mM diisopropylfluorophosphate (DFP), cells were broken by passages through an Emulsiflex cell disruptor at 20,000 lb/in2. Unbroken cells were removed by low-speed centrifugation (10,000 × g, 30 min, 4°C). Membranes were collected by ultracentrifugation of the supernatant (250,000 × g, 1 h, 4°C). The sedimented membranes were resuspended in buffer A (1 ml per g of cells) and stored at −80°C until use. Gradient centrifugation of membranes was performed with 30 to 60% sucrose gradients in buffer A in polycarbonate ultracentrifugation tubes (25 by 89 mm; Beckman). Ultracentrifugation of membranes for 2 h (4°C) in a type 70 Ti rotor (Beckman) at 110,000 × g yielded a brown band (see Fig. S2 in the supplemental material), which was used for further analysis by SDS-PAGE and Western blotting.
Production of PomAB and PomAB-D23N in E. coli.
Introduction of the PomB-D23N mutation into pET18ABS (13) using a QuikChange site-directed mutagenesis kit (Stratagene) with oligonucleotides VCb82V-D23N and VCb49R-D23N (see Table S1 in the supplemental material) yielded pET18ABS-D23N. E. coli C41(DE3) containing pET18ABS or pET18ABS-D23N was grown in buffered LB medium (10 g tryptone, 5 g yeast extract, 10 g NaCl per liter in 50 mM potassium phosphate, pH 8.0) supplemented with 50 μg ml−1 kanamycin. Single colonies were used to inoculate 10 ml medium. After 16 h at 37°C, the cultures were transferred to 1 liter of fresh medium in 5-liter Erlenmeyer flasks, and aerobic growth was continued (37°C, 160 rpm) until an OD600 of 0.7 was reached. Protein expression was induced by adding 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and growth was continued for 3.5 h. Cells (5 to 7 g [wet weight]) were harvested by centrifugation, washed with buffer (100 mM NaCl in 20 mM Tris-HCl, pH 8.0), frozen in liquid N2, and stored at −80°C. Cells were thawed on ice and resuspended in extraction buffer (0.2 M KCl and 10% [vol/vol] glycerol in 50 mM Tris-HCl, pH 8.0; 5 ml buffer per g of cells). After addition of a few crystals of DNase I, 0.5 mM MgCl2, and 0.2 mM DFP, cells were ruptured by three passages through a French pressure cell (Aminco) at 750 bars. Cell debris and unbroken cells were removed by centrifugation (12,000 × g, 20 min, 4°C), and the PomAB-containing membranes were collected by ultracentrifugation (250,000 × g, 1 h, 4°C). The membranes were resuspended in 1 ml extraction buffer per g of cells, and 2% (vol/vol) Triton X-100 or 3% (vol/vol) 5-cyclohexylpentyl β-d-maltoside (Cymal-5) were added. Membranes were stirred on ice for 10 min and subjected to ultracentrifugation (275,000 × g, 1 h, 4°C). The supernatant was loaded onto a Strep-Tactin column (gravity flow; bed volume, 1 ml; IBA GmbH, Germany) equilibrated with purification buffer (0.2 M KCl, 20 mM Tris-HCl, pH 8.0, 10% [vol/vol] glycerol, 0.1% [vol/vol] Triton X-100 or 0.3% [vol/vol] Cymal-5). The column was washed with 5 ml purification buffer. In the following, PomA refers to the wild-type protein, PomB refers to the wild-type protein with the Strep tag fused to its C terminus, and PomB-D23N refers to the PomB variant with the Strep tag carrying the D23N mutation. PomA was eluted together with PomB or PomB-D23N in a complex by adding 0.5-ml aliquots of purification buffer containing 2.5 mM desthiobiotin. The PomAB- or PomAB-D23N-containing fractions were combined and stored in liquid N2. This single chromatographic step yielded 0.2 to 0.3 mg partially purified PomAB or PomAB-D23N complex per gram of cells (wet weight). The identities of the prominent proteins at apparent molecular masses of below 45 kDa (PomB or PomB-D23N) and about 20 kDa (PomA) were confirmed by Edman sequencing and matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) (see Fig. S4 in the supplemental material). Inspection by SDS-PAGE revealed that PomA was copurified in similar amounts with wild-type PomB or PomB-D23N (see Fig. S5 in the supplemental material).
Analytical methods.
Protein was determined by the bicinchoninic acid method (28) using reagents from Pierce Biotechnology. Bovine serum albumin served as a standard. Na+ was determined by atomic absorption spectroscopy with a Shimadzu AA-6200 spectrometer. SDS-PAGE (29) was performed with 12% acrylamide/bisacrylamide (37.5:1.0; National Diagnostics). To 100 μl PomAB (12 μg) or PomAB-D23N (15 μg), 500 μl of ice-cold 12% trichloroacetic acid was added, and proteins were precipitated on ice for 15 min. After centrifugation (20,000 × g, 15 min, 4°C), the precipitate was washed with 500 μl ice-cold acetone and dissolved in 100 μl SDS-PAGE loading buffer (50 mM Tris-HCl, pH 6.8, 1% SDS, 10% [vol/vol] glycerol, 100 mM dithiothreitol, 1 mM bromophenol blue). Aliquots of 20 μl (2 to 3 μg protein) were separated on SDS-polyacrylamide gels, and proteins were stained with silver (30), since PomA is not efficiently stained with Coomassie blue. The silver-staining method allows estimation of the amount of protein loaded in the range from 11 to 300 ng (30). Transfer of proteins to membranes and immunostaining of the Strep tag of PomB were performed as described previously (13). GFP-PomB wild type and variants were visualized using a TyphoonTrio system (GE Healthcare) with excitation at 488 nm and a 526-nm emission filter. As molecular standards, Precision Plus Protein WesternC standards (Bio-Rad) were used.
Modification of PomAB and subunit c of ATPase with DCCD.
V. cholerae PomAB from the Strep-Tactin chromatographic step (30 to 100 μg) in purification buffer with 0.1% (vol/vol) Triton X-100 and 2.5 mM desthiobiotin was diluted in 3.5 ml buffer (50 mM MOPS [morpholinepropanesulfonic acid], 50 mM Tricine, pH 6.0, 7.0, or 8.0, adjusted with KOH) and concentrated to 100 μl using a centrifugal filter device (Millipore) with a molecular weight cutoff of 10,000. The c11 ring of I. tartaricus F1Fo ATP synthase was prepared from wild-type I. tartaricus cells as described previously (15).
Each PomAB aliquot contained 6 μg protein in 20 μl buffer, and pairs of aliquots were adjusted to the desired pH. Each of two c-ring aliquots contained 4 μg protein and 0.1% β-octylglucoside in 20 μl buffer (pH 7.0). To the first aliquots of PomAB and one aliquot of c ring, 1 μl 1 M NaCl (final concentration, 50 mM) and 2 μl 0.5 mM [14C]DCCD (in 25% ethanol, 50 μCi nmol−1; final concentration, 50 μM [14C]DCCD; Amersham, United Kingdom) were added. In the control reactions performed with the second aliquots, NaCl was omitted, and the residual Na+ concentration was less than 15 μM. After 1 h of incubation at room temperature (RT), SDS-PAGE loading buffer was added from a 5× stock solution (250 mM Tris-HCl, pH 6.8, 5% SDS, 50% glycerol [vol/vol], 500 mM dithiothreitol, 5 mM bromophenol blue) and the modified proteins were immediately separated by SDS-PAGE (29). After fixation of the gel in an aqueous solution containing 10% acetone and 30% isopropanol for 20 min and subsequent treatment of the gel with NAMP 100V Amplify enhancer solution (Amersham, United Kingdom) for 5 min, the gel was dried under vacuum on Whatman 3MM paper and exposed to a phosphor screen for 21 days. Radioactively labeled polypeptides were detected with a Molecular Dynamics PhosphorImager (Amersham, United Kingdom). The contrast of the autoradiograms was enhanced using the auto contrast function of Adobe Photoshop software. The volumes of the PomA and PomB bands were determined using Quantity One software (Bio-Rad).
Modification of PomAB and its D23N variant with DCCD after prolonged incubation with Na+.
To promote binding of Na+ to PomAB prior to modification with DCCD, pairs of aliquots, each containing 6 μg PomAB or PomAB-D23N equilibrated at pH 7.5 or 8.5, were allowed to react with 50 mM Na+ for 10 min. Controls contained less than 15 μM Na+. Subsequently, 50 μM [14C]DCCD was added, and the reaction (at RT) was stopped after 30 min by the addition of SDS-PAGE loading buffer and separation of PomAB (the wt and D23N variant) on SDS-polyacrylamide gels.
Time-dependent modification of PomAB with DCCD without added Na+.
Each of eight aliquots of 20 μl contained 20 μg PomAB in 50 mM MOPS-Tricine (pH 7.5 or 8.5, adjusted with KOH), 0.1% Triton X-100. Two aliquots of 20 μl each contained 4 μg c ring in 50 mM MOPS-Tricine, pH 7.0 (adjusted with KOH), 0.1% β-octylglucoside. At 0, 30, 50, or 55 min, DCCD modification of PomAB was initiated by adding 2 μl 0.5 mM [14C]DCCD (in 25% ethanol, 50 μCi nmol−1; final concentration, 50 μM [14C]DCCD; Amersham, United Kingdom). The reaction of the c ring with DCCD was started at 50 or 55 min. At 60 min, SDS-PAGE loading buffer was added to each aliquot, and the reaction mixtures were immediately loaded on the SDS-polyacrylamide gel to separate the reactants and stop the reaction. Autoradiograms of proteins modified with [14C]DCCD were obtained as described above.
RESULTS
Sequence comparison of transmembrane helices of the PomA4B2 stator complex with the inner and outer helices of the c ring of the Na+-translocating F1Fo ATPase.
Transmembrane helices III and IV of PomA, together with the single transmembrane helix of PomB in the flagellar stator complex, provide access of Na+ to flagellar motor components (12). The question arises whether the stator subunits PomA and PomB also offer a specific Na+ binding site. The aspartate residue D24 of PomB in V. alginolyticus is critical for the function of the flagellum and was proposed to act as a ligand for Na+ in a binding site of the stator built by residues from both PomA and PomB (9). Of particular interest are charged or polar amino acid residues in transmembrane regions of PomA and PomB. To describe the positions of these residues with respect to the hydrophobic stretches in PomA and PomB, we performed multiple-sequence alignments of a set of 62 (PomA and MotA) and 49 (PomB and MotB) nonredundant sequences (see Fig. S6 and S7 in the supplemental material). The amino acid residues which are predicted to be situated at the end of transmembrane helices in PomA are R144 and D170 (helix III) and A178 and D202 (helix IV). In PomB, the N-terminal helix ranges from W17 to S39 (Fig. 1B; see Fig. S6 and S7 in the supplemental material). Helix III of PomA comprises the conserved glycine residues G154, G157, and G161 (Fig. 1B; see Fig. S6 in the supplemental material). Subunits c of the Fo part of the F1Fo ATP synthase form a membrane-embedded ring with several binding sites for the coupling cation, which is Na+ in the case of the Ilyobacter tartaricus enzyme. This ring is formed by 11 c subunits, each contributing an inner and an outer helix. Together they form two rings of alpha helices, both concentrically oriented but in staggered positions with respect to each other. Tight packing of the individual c subunits is enabled since the outer ring of helices exhibits a larger diameter than the inner helix ring with its conserved glycine residues G25, G27, G29, G31, and G33 (Fig. 1B) (31, 32).
The c ring of the I. tartaricus ATP synthase harbors 11 Na+ binding sites. Each site is formed by ligands from the inner helix and outer helix from one c subunit and from the outer helix of an adjacent c subunit. A conserved glutamate (E65 in I. tartaricus) on one of the outer helices directly coordinates Na+. Besides that, Y70 of the same outer helix, Q32 of the inner helix (same c subunit), and S66 and T67, as well as the backbone carbonyls of V63 and A64 from the neighboring c subunit, together with a water molecule, are (directly and indirectly) involved in Na+ coordination (33). The ion-binding E65 carboxylate conformation is referred to as the ion-locked conformation (3, 34, 35). We compared the amino acid sequences of these three helices of the c ring with the three helices (helices III and IV from PomA, the N-terminal helix of PomB) shown to be critical for stator function in the PomAB complex. In the PomAB complex, a strictly conserved D23 is situated in the transmembrane helix of PomB, followed by the conserved residues S26 (within the membrane) and D42 (predicted to be located in the periplasm). Helix IV of PomA harbors one strictly conserved tyrosine residue and two strictly conserved threonine residues. Does this similar arrangement of amino acid residues in the three helices of the c ring and the PomAB stator complex reflect a similar function of the two membrane protein complexes with respect to Na+ binding? We addressed this question by studying the motility of V. cholerae expressing PomB and its D23E, D23N, S26A/T, and D42N variants under different conditions of salt and pH.
GFP-PomB fusion proteins carrying mutations in the N-terminal helix of PomB localize to the pole of V. cholerae cells.
To study the in vivo effect of pH on motility on selected variants of PomB, the polar localization of PomB variants in V. cholerae cells must be confirmed. The production of PomAB-D23N in V. cholerae carrying the wild-type pomAB genes on its chromosome exerts a dominant negative effect on flagellar function (13). This demonstrates that the PomAB variant competes with the endogenous PomAB and is assembled into the flagellum. For the closely related V. alginolyticus, Homma and coworkers proposed that replacing the corresponding D24 of PomB by N prevented the assembly of the stator complex of the polar flagellum (36). In that study, a PomB variant carrying a GFP fusion at the N terminus was used to detect V. alginolyticus PomB in the cytoplasmic membrane by fluorescence microscopy. We followed a similar approach using GFP-PomB expressed together with PomA in V. cholerae ΔpomAB. It is important to exclude the possibility that fluorescent signals observed at the poles of V. cholerae cells expressing PomB variants result from nonfunctional GFP fusion proteins which may aggregate at the cellular membrane. To this end, we transformed the aflagellate ΔfliG V. cholerae strain with plasmids encoding PomA and variants of PomB. The gene product of fliG is essential for the assembly of the flagellar complex in V. cholerae (21). Expression of PomA together with GFP-PomB variants in the pomAB deletion strain resulted in prominent, fluorescent spots at one or both poles of the cells, indicating the presence of GFP-PomB variants at cellular sites which correlated with the position of the flagellar stator (Fig. 2). In addition, we observed fluorescent signals at or in the cellular membrane which could represent preassembled PomAB complexes (5). For induction of expression, 10 mM arabinose was added, as lower concentrations (0.4 and 1 mM arabinose) did not result in detectable amounts of the fluorescent proteins. In the ΔfliG background, we did not detect fluorescent signals at the poles of the cells at arabinose concentrations of 0.4 mM or 10 mM (Fig. 3). The cells were producing the PomB-GFP fusion proteins, as can be seen from the slight fluorescence of the cytoplasm, which was the highest with the GFP-PomB-D42N variant at 10 mM arabinose (Fig. 3). It is concluded that fluorescent proteins detected in the pomAB deletion strain transformed with plasmids encoding PomA together with PomB-GFP fusion proteins represent PomB-GFP variants assembled into the stator.
Fig 2.
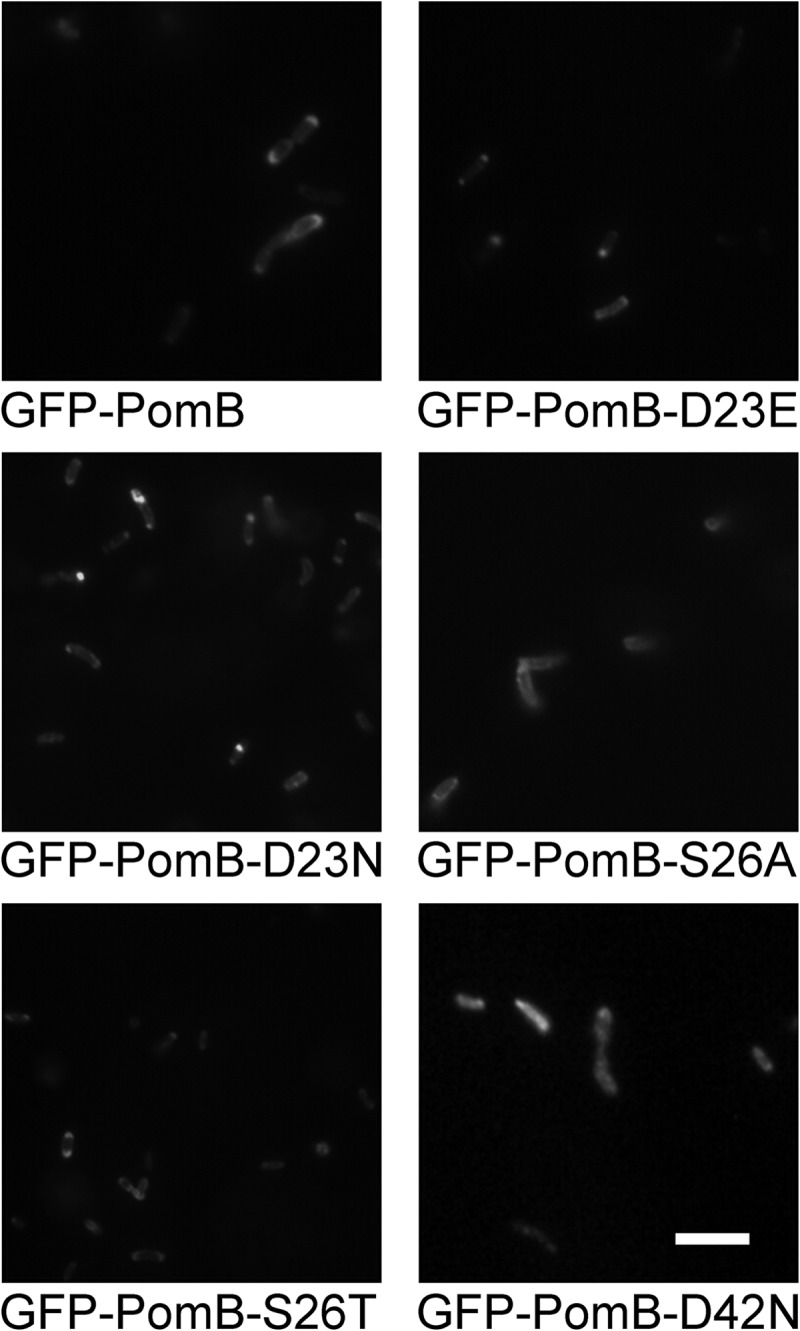
Localization of GFP-PomB variants coexpressed with wild-type PomA in a pomAB deletion strain of V. cholerae. Expression of PomA and GFP-PomB wt and variants was started using 10 mM arabinose. After 5 h, the polar localization (signals on one or both poles) of GFP-PomB was analyzed by counting a minimum of 322 cells. Bar, 5 μm.
Fig 3.
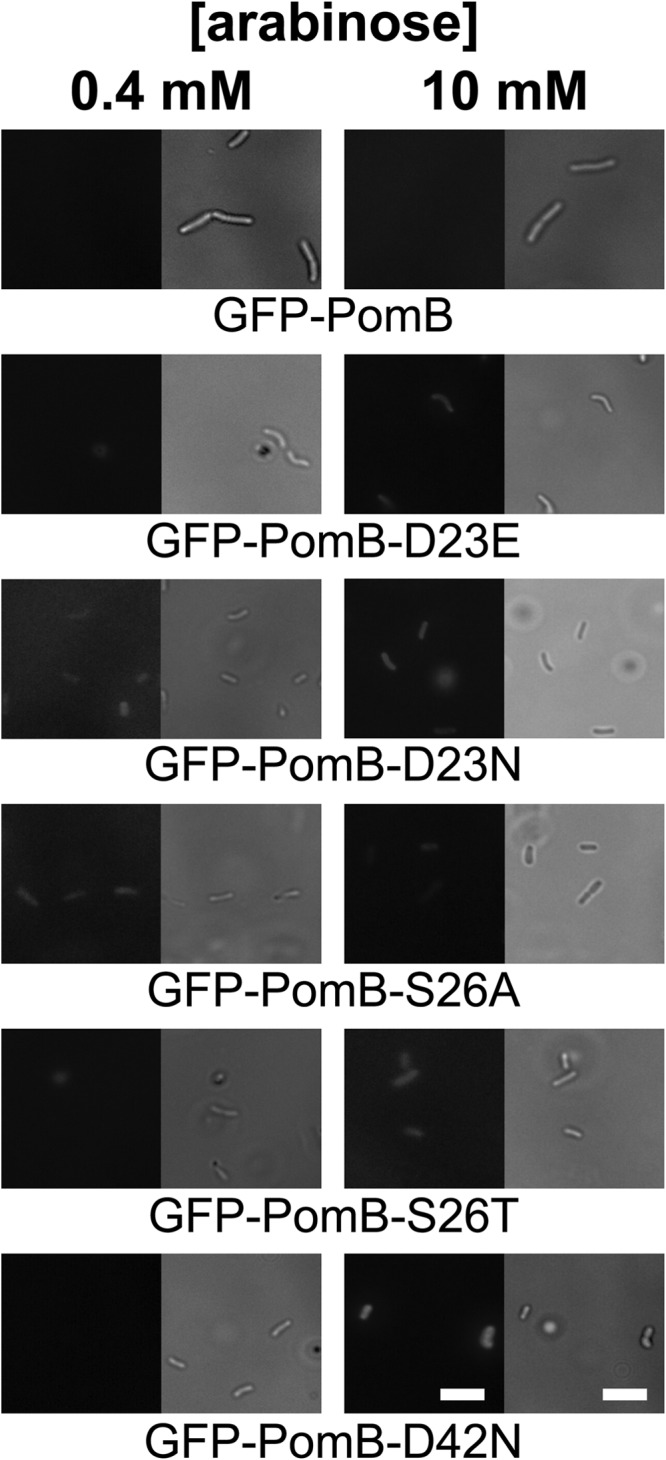
Localization of GFP-PomB variants coexpressed with wild-type PomA in a fliG deletion strain of V. cholerae. Cells were grown aerobically in liquid medium containing 0.4 mM (left column) or 10 mM (right column) arabinose. After 5 h, 20 μl culture was spotted on poly-l-lysine-covered glass slides and the slides were inspected using a fluorescence microscope to observe GFP-PomB (left pictures in each panel). The same section of the microscopic preparation was also observed in the DIC mode of the microscope (right pictures in each panel). Bars, 5 μm.
To address whether the insertion of a mutation in the transmembrane helix of PomB affected the localization of GFP-PomB, we performed a quantitative analysis. Of a total of 500 V. cholerae cells producing wild-type GFP-PomB, 197 showed prominent GFP signals at one pole, 222 showed prominent GFP signals at two poles, and 81 cells exhibited distinct GFP signals in the cellular membrane (Table 2). With the S26T and S26A variants of GFP-PomB, the corresponding numbers (signals at one pole/signals at two poles/membrane-associated signals) were 259/191/50 and 232/161/107, respectively. In a total of 322 cells producing GFP-PomB-D23E, the corresponding numbers were 128 (signals at one pole), 184 (signals at two poles), and 10 (membrane-associated signals). In a total of 342 cells producing GFP-PomB-D23N, 101 cells exhibited GFP signals at one pole, 225 cells exhibited GFP signals at two poles, and 16 cells showed membrane-associated GFP signals. In a total of 305 cells producing GFP-PomB-D42N, 117 cells showed GFP signals at one pole, 106 cells showed GFP signals at both poles, and 82 cells showed membrane-associated GFP signals (Table 2). We conclude that in V. cholerae, the insertion of mutations at position 23, 26, or 42 in PomB has no apparent influence on the localization of the GFP-PomB variant versus that of wild-type GFP-PomB. In membranes from V. cholerae ΔpomAB, wild-type GFP-PomB and the D23E, D23N, D42N, S26A, and S26T variants were present in similar amounts, as judged from the intensity of bands in the SDS-polyacrylamide gels (Fig. 4A) and Western blots (Fig. 4B). It should be noted that the motility of the V. cholerae ΔpomAB strain producing PomA and GFP-PomB was greatly diminished (Fig. 5). Swarming rings reached only approximately 15% of the diameter of the strains transformed with the vector producing PomA and PomB. For comparison, swarming rings of V. cholerae producing the nonmotile PomB-D23N variant (13) or control cells transformed with the empty vector lacking pomA and pomB exhibited swarming rings with diameters that were 7 to 10% of those of cells producing wild-type PomA and PomB (Fig. 5). It is therefore concluded that the N-terminally attached GFP interferes with the function of PomB in the stator complex.
Table 2.
Cellular localization of GFP-PomB in V. choleraea
| V. cholerae strain | No. (%) of cells with GFP fluorescence signal |
|||
|---|---|---|---|---|
| At one pole | At two poles | Membrane associated | Total | |
| GFP-PomB (wt) | 197 (39.4) | 222 (44.4) | 81 (16.2) | 500 (100) |
| GFP-PomB-D23E | 128 (39.8) | 184 (57.1) | 10 (3.1) | 322 (100) |
| GFP-PomB-D23N | 101 (29.5) | 225 (65.8) | 16 (4.7) | 342 (100) |
| GFP-PomB-S26A | 232 (46.4) | 161 (32.2) | 107 (21.4) | 500 (100) |
| GFP-PomB-S26T | 259 (51.8) | 191 (38.2) | 50 (10.0) | 500 (100) |
| GFP-PomB-D42N | 117 (38.4) | 106 (34.8) | 82 (26.9) | 305 (100) |
Wild-type PomA and GFP-PomB (wild type and variants) were coexpressed in V. cholerae ΔpomAB. The total number of cells of each strain which were included in the analysis represents 100%.
Fig 4.
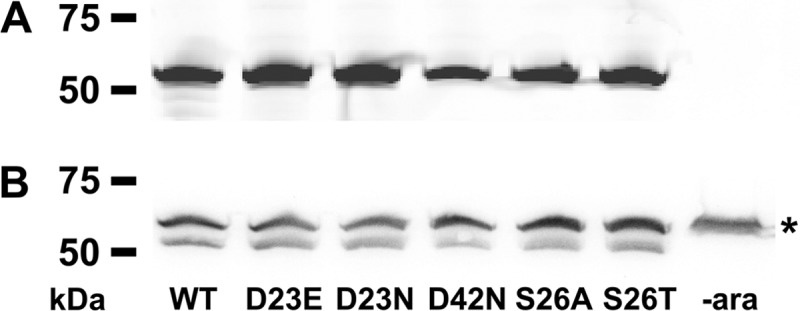
Detection of GFP-PomB variants in membranes from V. cholerae purified by sucrose density centrifugation. Cells were grown in the presence of 1 mM arabinose to induce protein expression, and membranes were isolated and purified by sucrose density centrifugation. A total of 200 μg protein per lane was loaded on an SDS-polyacrylamide gel and subjected to electrophoresis. (A) In-gel fluorescence of GFP-PomB wt and variants recorded with a TyphoonTrio system (GE Healthcare) with excitation at 488 nm and a 526-nm emission filter. The control (lane to the very right) was uninduced whole cells. As a molecular size standard, Precision Plus Protein WesternC standards (Bio-Rad) were used. (B) Western blot analysis of the gel from panel A blotted on a nitrocellulose membrane. GFP-PomB (with a C-terminal Strep tag) was visualized using Strep-Tactin-horseradish peroxidase and enhanced chemiluminescence. The upper bands (*) result from detection of the biotinylated subunit α of V. cholerae oxaloacetate decarboxylase by the Strep-Tactin-horseradish peroxidase conjugate. −ara, lacking arabinose.
Fig 5.
Swarming of V. cholerae O395-N1 ΔpomAB transformed with plasmids coding for PomA and GFP-PomB (wt and variants). Swarming assays were performed on LB soft agar plates (0.25% agar), pH 7.0, supplemented with 0.4 mM (A), 1 mM (B), or 10 mM (C) arabinose and incubated at 30°C for 22 h. (D) After 22 h, diameters were measured. In total, 10 experiments per condition were performed, and the average of 8 experiments was calculated (the highest and the lowest values were omitted). ■, 0.4 mM arabinose; •, 1 mM arabinose; ▲, 10 mM arabinose. V. cholerae ΔpomAB expressing PomA and wt PomB from plasmid pAB and V. cholerae ΔpomAB transformed with the empty vector pISC-H served as controls.
Effect of pH and osmolality on motility of V. cholerae expressing PomB (wild type and variants).
When the Na+-translocating ATP synthase operates at saturating Na+ concentrations, increasing the pH from 8.0 to 9.0 results in the 2-fold activation of enzymatic activity (37). At very low Na+ concentrations, ATPase activity is drastically diminished and the enzyme translocates protons. Lowering the proton concentration from pH 7.0 to pH 9.0 decreases activity by 70% (38). To find out whether a similar dependence of the activity of the flagellum on the concentration of Na+ and H+ could be observed, we investigated the motility of V. cholerae ΔpomAB coexpressing wild-type PomA and variants of PomB at pH 7.0, 8.0, and 9.0 in rich or minimal medium and at different Na+ concentrations. The flagellation of V. cholerae was not affected by the chosen experimental conditions. Representative transmission electron microscopic pictures of V. cholerae cells grown in M9 minimal medium at pH 8.0 and with low Na+ concentrations are presented in Table S2 and Fig. S3 in the supplemental material.
Strikingly, lowering the proton concentration did not result in the stimulation of the motility (followed as swarming in soft agar) of cells expressing wild-type PomB under any of the conditions tested (Fig. 6 and 7), but diminished motility of the cells was observed at both low and high Na+ concentrations. At the lowest Na+ concentration tested (1.6 mM in minimal medium) and at pH 9.0, the motility of cells expressing wild-type PomB was lost. Lowering the pH to 7.0 under these conditions of a low Na+ concentration led to an increase in motility which reached roughly 50% of the motility observed at high Na+ concentrations at pH 7.0 (Fig. 7). In marked contrast to the Na+-translocating ATPase, the performance of the Na+ gradient-driven flagellum decreased when the proton concentration of the external medium was lowered.
Fig 6.
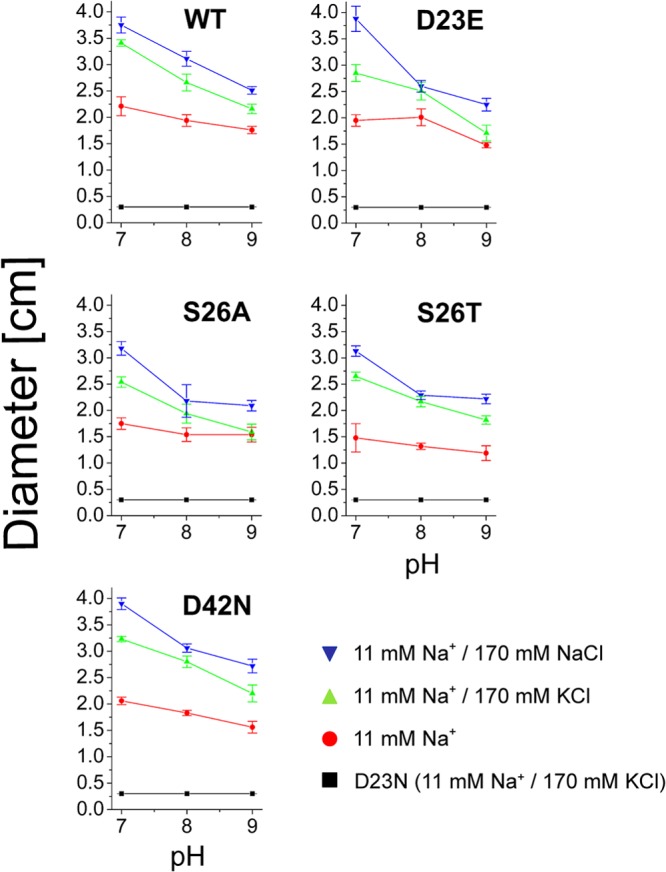
Influence of pH and salt on swarming of V. cholerae cells expressing PomB variants in LB medium. V. cholerae lacking the chromosomal pomA and pomB genes was transformed with plasmids encoding His6-PomA (wt) together with PomB-Strep (wt) or its D23E, S26A, S26T, and D42N variants. The diameter of the swarming rings was determined after 21 h. Mean values from 10 experiments are shown. No swarming was observed with cells transformed with the empty expression vector or with cells expressing the D23N variant of PomB (negative control, black trace).
Fig 7.
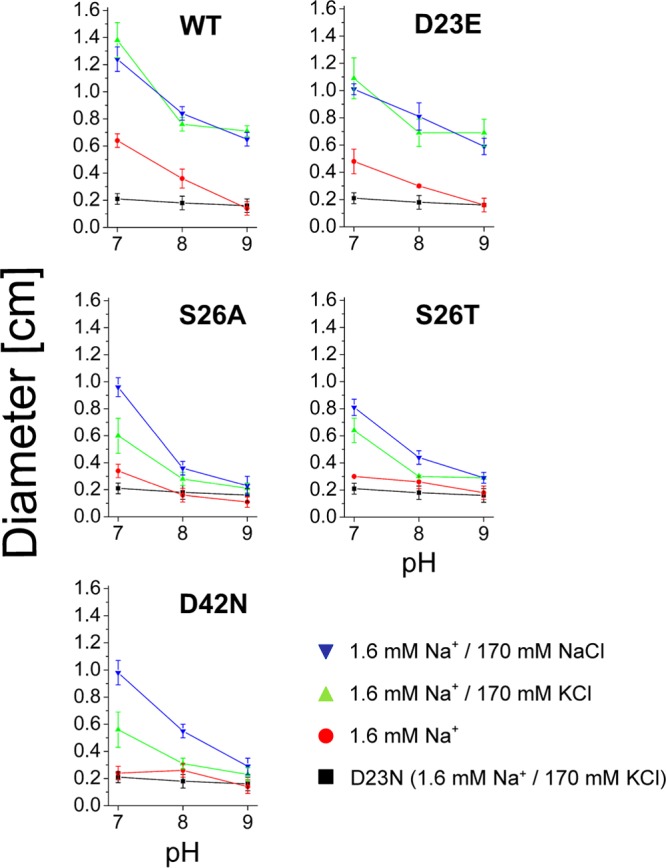
Influence of pH and salt on swarming of V. cholerae cells expressing PomB variants in minimal medium. V. cholerae lacking the chromosomal pomA and pomB genes was transformed with plasmids encoding His6-PomA (wt) together with PomB-Strep (wt) or its D23E, S26A, S26T, and D42N variants. The diameter of the swarming rings was determined after 45 h. Mean values from 10 experiments are shown. No swarming was observed with cells transformed with the empty expression vector or with cells expressing the D23N variant of PomB (negative control, black trace).
The stimulation of motility of cells expressing wild-type PomB by Na+ (170 mM, added as NaCl) was observed on both rich medium and minimal medium at pH 7.0, 8.0, and 9.0. However, this increase in motility was also achieved when NaCl was replaced by KCl. Under extreme conditions (pH 9.0 and 1.6 mM Na+ in minimal medium), when cells were immotile, addition of 170 mM KCl rescued motility to the same extent as that observed with 170 mM NaCl (Fig. 7). Similar results were obtained when the carboxyl group at position 23 in PomB was maintained in the D23E variant. In a previous study, we observed a slightly increased motility of cells expressing wild-type PomB compared to that of cells expressing PomB-D23E at pH 7.0. Those studies were performed using LB medium buffered with 10 mM potassium phosphate, and the diameters of swarming rings were determined after 7.5 h of incubation at 37°C (13). In the present study, when M9 minimal medium buffered with 10 mM Tris and glucose as the carbon source was used, we again observed a slight stimulation of the motility of cells expressing wild-type PomB compared to that of cells expressing PomB-D23E at pH 7.0 (Fig. 7). In these experiments, the diameters of swarming rings were measured after 45 h at 30°C. The diminished swarming motility observed under alkaline conditions could be caused by a decrease in energy conversion or membrane potential in the V. cholerae reference strain. We compared the final yield of cells grown in LB medium at pH 6.5 or 8.5 with 170 mM KCl or NaCl added. Stationary phase was reached after 15 h under all conditions tested, and the final OD600s were 1.48 ± 0.12 (pH 6.5) and 1.49 ± 0.22 (pH 8.5) in the presence of 170 mM KCl. With 170 mM NaCl added, the final OD600s were 1.33 ± 0.09 (pH 6.5) and 1.29 ± 0.14 (pH 8.5). The decreased swarming motility observed at alkaline pH is therefore not caused by energy limitation of the V. cholerae cells.
We also determined the swimming speeds of V. cholerae cells producing wild-type PomAB in liquid LB medium at different proton and salt concentrations (Fig. 8). In accord with the observed pH-dependent swarming behavior on solid medium (Fig. 6), there was a decrease in motility with increasing pH at all salt concentrations investigated. Notably, the percentage of motile cells at low ionic strength (11 mM Na+ added) and pH 8.0 was higher than that under conditions with 170 mM NaCl or KCl, and the addition of salts did not stimulate the motility of cells at pH 9.0. In summary, the results show that the osmolality of the external medium rather than its Na+ concentration determines the performance of the flagellum.
Fig 8.
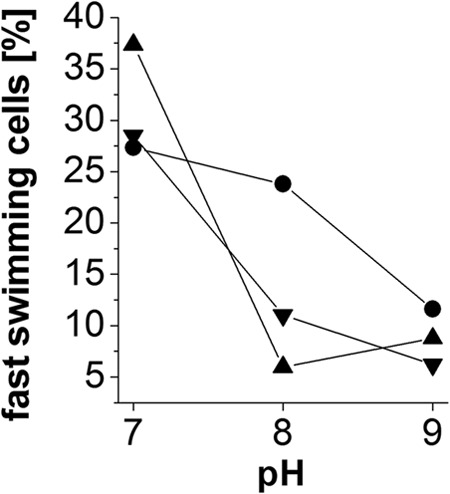
Influence of external pH and salt on swimming speed of a V. cholerae ΔpomAB strain producing PomAB. The swimming speed of single cells was determined by recording their movement. A minimum of 200 tracks of individual cells was analyzed for each condition of pH and salt. Only bacteria exhibiting a swimming speed of ≥4 μm s−1 were included in the analysis, and these represent 100% of the motile cells under the indicated condition. Data points represent the fraction of fast-swimming cells exhibiting motilities between 18 and 30 μm s−1 as a percentage of the total number of motile cells at the indicated condition. ▼, 11 mM Na+ and 170 mM NaCl; ▲, 11 mM Na+ and 170 mM KCl; •, 11 mM Na+.
Critical role for S26 and D42 of PomB for motility of V. cholerae.
One could argue that the observed effects of pH and osmolality on flagellar performance on minimal medium do not result from the altered function of PomB but are due to processes preceding the formation of a functional flagellum, such as synthesis, membrane insertion, or assembly of its components. This possibility can be excluded since introducing single-site mutations at positions S26 and D42 in PomB influenced the motility of V. cholerae in a salt- and pH-dependent manner. The D23N variant of PomB completely prevented the motility of V. cholerae cells on both rich and minimal media (Fig. 6 and 7). The PomB-S26A/T and PomB-D42N variants had no significant effect on motility when analyzed on rich medium, but they drastically altered the pH profile of motility on minimal medium. Residues D23 and S26 (or T26) are conserved in homologues of PomB and MotB, whereas D42 represents an amino acid residue which is exclusively conserved in PomB homologues (see Fig. S7 in the supplemental material). The activity of the flagellum at pH 9.0 clearly depended on serine at position 26 and on aspartate at position 42, irrespective of the osmolality of the minimal medium. This is in contrast to the findings for wild-type PomB and the D23E variant, which regained their function at pH 9.0 by addition of 170 mM KCl or NaCl. The difference in function of wild-type PomB and the D23E variant from that of the S26A/T and D42N variants was also apparent at pH 7.0 and a low Na+ concentration (1.6 mM). Here, cells containing PomB-S26A/T or D42N were nonmotile, whereas the presence of wild-type PomB or its D23E variant resulted in 50% of the motility observed in the presence of 170 mM KCl or NaCl (Fig. 7).
Modification of PomAB with DCCD.
In the c11 ring from the I. tartaricus ATP synthase, the interaction of Na+ with E65 is evident from the protection of modification of E65 with DCCD in the presence of Na+ (15). At physiological pH, only the protonated carboxyl group of the glutamate residue undergoes covalent modification with the carbodiimide (Fig. 1C). Coordination of Na+ by the carboxylate of E65 prevents protonation and therefore labeling with radioactive DCCD (15). We asked whether PomA4B2, like the c subunits, contains a critical carboxylate which binds Na+ or H+ and studied the influence of Na+ on the modification of the PomAB or the PomAB-D23N complex by DCCD. The observed influence of pH on the motility of V. cholerae suggested that the critical Asp23 of PomB may undergo protonation even in the presence of Na+. The I. tartaricus c11 ring was used as a control.
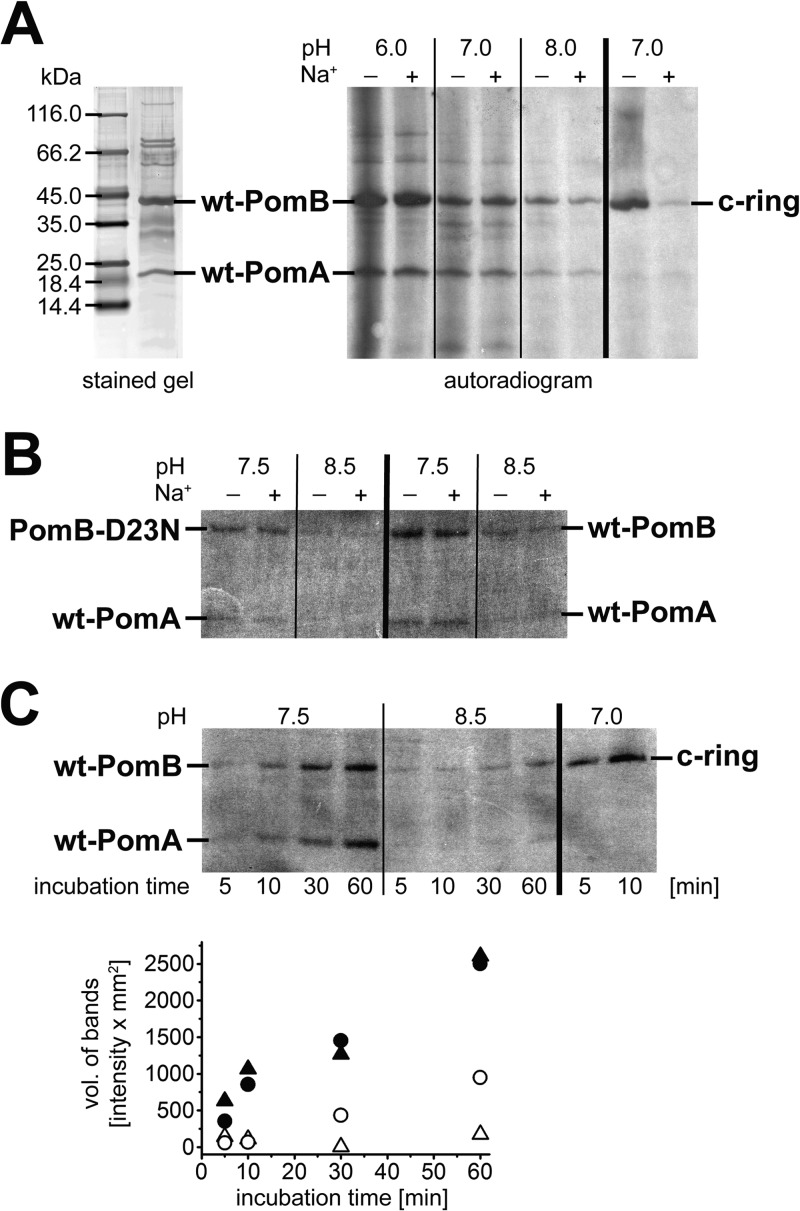
Figure 9A shows the autoradiogram of the PomAB complex after modification with [14C]DCCD at pH 6.0, 7.0, or 8.0 in the presence of 15 μM or 50 mM Na+ and subsequent separation of the subunits by SDS-PAGE. Both proteins were labeled, demonstrating the presence of at least one protonated carboxyl group per stator subunit in a hydrophobic environment. The modification of the PomAB complex was not decreased by adding 50 mM Na+, while in a control reaction, the presence of Na+ completely protected the c11 ring from the carbodiimide modification (Fig. 9A). This indicates that Na+ does not prevent the protonation of carboxyl groups in hydrophobic regions of the PomAB stator subunits, such as D23 in the N-terminal, membrane-spanning helix of PomB. To promote binding of Na+ to the PomAB complex and protection against DCCD modification, Na+ (50 mM) was added to PomAB 10 min prior to the addition of [14C]DCCD, and the pH was raised to 8.5, facilitating the deprotonation of carboxyl groups (Fig. 9B). As expected, the overall modification of the proteins with [14C]DCCD was reduced at alkaline pH, but clearly, Na+ did not have any observable effects on the rates of the modification. Interestingly, modification of wild-type PomB was enhanced compared to that of the PomB-D23N variant at pH 7.5 and 8.5 (Fig. 9B), suggesting that D23 contributed to the reaction with [14C]DCCD. We next investigated the influence of pH on the rates of [14C]DCCD modification of PomA and PomB. The kinetic analysis revealed that in the wild-type PomAB complex equilibrated at pH 7.5, both PomA and PomB were modified by [14C]DCCD at similar rates (Fig. 9C). At pH 8.5, PomB, but not PomA, was still modified by DCCD.
Fig 9.
SDS-PAGE of the PomAB complex and modification with [14C]DCCD. (A) (Left) SDS-PAGE of PomAB (2.2 μg in 5-cyclohexylpentyl β-d-maltoside) stained with silver; (right) autoradiogram of PomAB and the c ring separated by SDS-PAGE after modification with [14C]DCCD in the presence or absence of Na+. PomAB (in Triton X-100, 6 μg per lane) was allowed to react with [14C]DCCD at pH 6.0, 7.0, or 8.0 for 1 h in the presence of 50 mM Na+ (lanes +) or without added Na+ (lanes −). The reaction was also performed with the c ring (4 μg per lane) under the same conditions at pH 7.0. (B) Comparison of modification of PomAB and the PomAB-D23N variant with [14C]DCCD at pH 7.5 or 8.5. Each reaction mixture contained 6 μg wild-type PomAB or 6 μg PomAB-D23N and 50 mM Na+ (lanes +) or no added Na+ (lanes −). The pH was adjusted with MOPS-Tricine. After 10 min of incubation at room temperature, [14C]DCCD was added and incubation was prolonged for 30 min. Reaction mixtures were diluted in SDS loading buffer and subjected to SDS-PAGE. The autoradiogram of the SDS-polyacrylamide gel is shown. (C) Time course of [14C]DCCD modification of PomA and PomB at pH 7.5 or 8.5 without added Na+. (Top) Autoradiogram of the corresponding SDS-polyacrylamide gel. Six micrograms of wild-type PomAB was loaded per lane after reaction with [14C]DCCD for the indicated times. The pH was adjusted with MOPS-Tricine. The residual Na+ concentration in the reaction mixtures was less than 15 μM. As a control, 4 μg of c ring was incubated for 5 or 10 min with [14C]DCCD. (Bottom) Quantification of the radioactivity incorporated into PomA at pH 7.5 (▲) or pH 8.5 (Δ) and into PomB at pH 7.5 (•) or pH 8.5 (○).
Taken together, we conclude that PomB harbors at least one carboxyl group in a hydrophobic region which has a higher pKa than a carboxyl group on PomA.
DISCUSSION
Rotation of the flagellum depends on the torque generated by its stator complexes, composed of four PomA (or MotA) and two PomB (or MotB) subunits embedded in the cytoplasmic membrane (39). It was suggested that a prerequisite for flagellar rotation is the direct coordination of Na+ with the conserved D24 in PomB from V. alginolyticus (9). Like the conserved D23 in the highly related PomB from V. cholerae (13), replacing this aspartate with asparagine results in an inactive flagellum, yet this critical carboxyl group does not represent the sole determinant of coupling cation selectivity in Na+-driven flagellar motors. By comparing various investigations which described the function of chimeric motors built from subunit domains of Na+- or H+-gradient-driven motors, Sowa and Berry concluded “that there is no single determining component for ion selectivity” (40). This is in marked contrast to another rotational nanomachine, the Na+-translocating F1Fo ATP synthase, with its conserved, critical amino acid residues of subunits c which determine the coupling cation selectivity (Na+ versus H+) of the rotor (Fig. 1) (3, 41). In the present study, we investigated the functional role of the conserved D23, S26, and D42 residues in PomB from V. cholerae.
The D23N variant of PomB was assembled into the polar flagellum of V. cholerae, indicating that the nonmotile phenotype was caused by the lack of this critical carboxylate embedded within the membrane-bound helix of PomB but not by the failure of PomB-D23N to integrate into the stator. For the D24N variant of V. alginolyticus, Homma and coworkers found that introduction of the D24N mutation prevented assembly of GFP-PomB-D24N into the stator (36). Assembly was also confirmed for the S26A/T and D42N variants of PomB. Assuming that both Na+ and H+ have access to the conserved aspartate 23 in the PomAB stator complex by a channel opening toward the periplasmic side of the membrane, we would expect that there is competition between Na+ and H+ for binding to specific, functional sites on PomA and/or PomB. As a consequence, the pH profile of flagellar activity should reveal an increase from pH 7.0 toward pH 9.0. With the pomAB deletion strain from V. cholerae coexpressing wild-type PomA and PomB, the contrary was observed. Cells exhibited the highest motility at pH 7.0 on both rich (LB) and minimal medium with glucose as the carbon source. One would also expect that the protonation of carboxyl groups in membrane-embedded regions of PomA and PomB, which act as ligands for the coupling cation, is prevented or diminished by adding Na+. The fact that we did not observe protection from modification with [14C]DCCD in the presence of Na+ suggests that in the PomAB complex, Na+ is not directly coordinated with carboxyl groups. The observed effect could also indicate that the actual Na+ binding affinity in the PomAB complex is lower than that in the Na+ binding c rings (4, 33), although the Na+-to-proton specificity in PomAB obviously remains high, since the V. cholerae flagellum motor cannot be operated with a proton (instead of a Na+) motif force. Alternatively, our failure to detect protection of [14C]DCCD modification by Na+ could be caused by the inaccessibility of a Na+ binding site within the stator complex purified in a closed (or plugged) conformation (42, 43). Kandori and colleagues studied the access of Na+ to D23 of PomB in the stator complex from V. alginolyticus using attenuated total reflectance-Fourier transform infrared (ATR-FTIR) spectroscopy. By comparing the properties of wild-type PomB with those of the D23N variant of PomB, they concluded that Na+ binds to the carboxylate of D23 with an apparent Kd (dissociation constant) of 85 or 98 mM (9). We cannot compare the results of their study with those of our investigations since the ATR-FTIR spectra were recorded at pH 5.5. At this pH, V. cholerae does not grow and cannot be studied with respect to motility. It should also be noted that at pH 5.5, the critical carboxyl group required for Na+ transport by the ATP synthase undergoes modification by DCCD, despite saturating concentrations of Na+ (41).
The D23 and S26 residues of PomB from V. cholerae are set apart by one helical turn and therefore are likely to face the same side of the transmembrane helix. In the closely related organism V. alginolyticus, Terauchi and coworkers demonstrated that a mutation in S27 diminished motility compared to that of the wild type (11). We propose that D23 and S26 in the transmembrane helix of PomB could contribute to the arrangement of water molecules or a hydrogen bond network which is important for proper function of the Na+-conducting stator complex or that these residues contribute to Na+ coordination, even though the affinity of the putative Na+ binding sites in the PomAB ion pathway is lower but, nevertheless, still highly specific for Na+ under physiological ion concentrations. Support for this hypothesis is given by our observation that chloride, which acts as a chaotrope (44), enhances the motility of V. cholerae cells. Anions may distort structural water molecules in close proximity to D23 and S26 and thereby facilitate the access of Na+ to PomA and PomB. In accord with this notion, we observed that the loss of motility at low ionic strength in the S26A/T or D42N variant of PomB was partially compensated for by externally added chloride at pH 7.0 but not at pH 9.0. We propose that D23 and S26 of PomB are of functional importance for the PomAB stator complex, particularly if the H+ concentration decreases. Under these alkaline conditions, D42, predicted to be located in the periplasm of V. cholerae, is also required for motility. In summary, our study demonstrated that the motility of V. cholerae is strongly influenced by external pH and osmolality. The function of D23, S26, and D42 of PomB is impaired in a salt- and H+-dependent manner. This suggests an ion-conducting pathway through the PomAB stator complex involving these conserved residues of PomB.
At a concentration of at least 0.1 M, the Na+ concentration in diarrheal stools from persons infected with V. cholerae is not expected to limit the performance of the flagellum of V. cholerae (45), yet V. cholerae is a versatile bacterium which is able to proliferate outside the human host in many different environments. These include freshwater habitats with salinity much lower than that found in estuaries or coastal areas (46). Our study suggests that in these environmental habitats, it is the overall salinity rather than the absolute Na+ concentration which determines the motility of V. cholerae.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christoph von Ballmoos (University of Stockholm) for performing the MALDI-MS analyses, Claudia Häse (Oregon State University) for providing the V. cholerae ΔfliG strain, Georg Kaim (PCRlab GbR, Achern, Germany) for helpful discussions, and Yasmin Kolar (University of Hohenheim, Stuttgart, Germany) for experimental assistance.
This work was supported by a grant from the Velux Foundation (to J.S.). T.M. was supported by the Cluster of Excellence Macromolecular Complexes at the Goethe University Frankfurt (project EXC 115).
Footnotes
Published ahead of print 23 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00353-13.
REFERENCES
- 1.Berg HC. 2008. Bacterial flagellar motor. Curr. Biol. 18:R689–R691 [DOI] [PubMed] [Google Scholar]
- 2.von Ballmoos C, Wiedenmann A, Dimroth P. 2009. Essentials for ATP synthesis by F1F0 ATP synthases. Annu. Rev. Biochem. 78:649–672 [DOI] [PubMed] [Google Scholar]
- 3.Meier T, Polzer P, Diederichs K, Welte W, Dimroth P. 2005. Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308:659–662 [DOI] [PubMed] [Google Scholar]
- 4.Krah A, Pogoryelov D, Meier T, Faraldo-Gómez JD. 2010. On the structure of the proton-binding site in the Fo rotor of chloroplast ATP synthases. J. Mol. Biol. 395:20–27 [DOI] [PubMed] [Google Scholar]
- 5.Thormann KM, Paulick A. 2010. Tuning the flagellar motor. Microbiology 156:1275–1283 [DOI] [PubMed] [Google Scholar]
- 6.Terahara N, Krulwich TA, Ito M. 2008. Mutations alter the sodium versus proton use of a Bacillus clausii flagellar motor and confer dual ion use on Bacillus subtilis motors. Proc. Natl. Acad. Sci. U. S. A. 105:14359–14364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terahara N, Sano M, Ito M. 2012. A Bacillus flagellar motor that can use both Na+ and K+ as a coupling ion is converted by a single mutation to use only Na+. PLoS One 7:e46248. 10.1371/journal.pone.0046248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yonekura K, Maki-Yonekura S, Homma M. 2011. Structure of the flagellar motor protein complex PomAB: implications for the torque-generating conformation. J. Bacteriol. 193:3863–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudo Y, Kitade Y, Furutani Y, Kojima M, Kojima S, Homma M, Kandori H. 2009. Interaction between Na+ ion and carboxylates of the PomA-PomB stator unit studied by ATR-FTIR spectroscopy. Biochemistry 48:11699–11705 [DOI] [PubMed] [Google Scholar]
- 10.Paul K, Gonzalez-Bonet G, Bilwes AM, Crane BR, Blair D. 2011. Architecture of the flagellar rotor. EMBO J. 30:2962–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terauchi T, Terashima H, Kojima S, Homma M. 2011. The critical role of a conserved residue, PomB-F22, in the transmembrane segment of the flagellar stator complex for conducting ions and generating torque. Microbiology 157:2422–2432 [DOI] [PubMed] [Google Scholar]
- 12.Terashima H, Kojima S, Homma M. 2010. Functional transfer of an essential aspartate for the ion-binding site in the stator proteins of the bacterial flagellar motor. J. Mol. Biol. 397:689–696 [DOI] [PubMed] [Google Scholar]
- 13.Vorburger T, Stein A, Ziegler U, Kaim G, Steuber J. 2009. Functional role of a conserved aspartic acid residue in the motor of the Na+-driven flagellum from Vibrio cholerae. Biochim. Biophys. Acta 1787:1198–1204 [DOI] [PubMed] [Google Scholar]
- 14.Laubinger W, Dimroth P. 1989. The sodium ion translocating adenosinetriphosphatase of Propionigenium modestum pumps protons at low sodium ion concentrations. Biochemistry 28:7194–7198 [DOI] [PubMed] [Google Scholar]
- 15.Meier T, Matthey U, von Ballmoos C, Vonck J, Krug von Nidda T, Kühlbrandt W, Dimroth P. 2003. Evidence for structural integrity in the undecameric c-rings isolated from sodium ATP synthases. J. Mol. Biol. 325:389–397 [DOI] [PubMed] [Google Scholar]
- 16.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 18.Henikoff S, Henikoff JG. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U. S. A. 89:10915–10919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tusnády GE, Simon I. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489–506 [DOI] [PubMed] [Google Scholar]
- 20.Tusnády GE, Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850 [DOI] [PubMed] [Google Scholar]
- 21.Gosink KK, Häse CC. 2000. Requirements for conversion of the Na+-driven flagellar motor of Vibrio cholerae to the H+-driven motor of Escherichia coli. J. Bacteriol. 182:4234–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Häse CC, Mekalanos JJ. 1999. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 96:3183–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298 [DOI] [PubMed] [Google Scholar]
- 24.Barik S. 1993. Site-directed mutagenesis by double polymerase chain reaction: megaprimer method. Methods Mol. Biol. 15:277–286 [DOI] [PubMed] [Google Scholar]
- 25.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88 [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27.Yoshida S, Sugiyama S, Hojo Y, Tokuda H, Imae Y. 1990. Intracellular Na+ kinetically interferes with the rotation of the Na+-driven flagellar motors of Vibrio alginolyticus. J. Biol. Chem. 265:20346–20350 [PubMed] [Google Scholar]
- 28.Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76–85 [DOI] [PubMed] [Google Scholar]
- 29.Schägger H, von Jagow G. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368–379 [DOI] [PubMed] [Google Scholar]
- 30.Nesterenko MV, Tilley M, Upton SJ. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J. Biochem. Biophys. Methods 28:239–242 [DOI] [PubMed] [Google Scholar]
- 31.Vonck J, Krug von Nidda T, Meier T, Matthey U, Mills DJ, Kühlbrandt W, Dimroth P. 2002. Molecular architecture of the undecameric rotor of a bacterial Na+-ATP synthase. J. Mol. Biol. 321:307–316 [DOI] [PubMed] [Google Scholar]
- 32.Pogoryelov D, Klyszejko AL, Krasnoselska GO, Heller EM, Leone V, Langer JD, Vonck J, Müller DJ, Faraldo-Gómez JD, Meier T. 2012. Engineering rotor ring stoichiometries in the ATP synthase. Proc. Natl. Acad. Sci. U. S. A. 109:E1599–E1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meier T, Krah A, Bond PJ, Pogoryelov D, Diederichs K, Faraldo-Gómez JD. 2009. Complete ion-coordination structure in the rotor ring of Na+-dependent F-ATP synthases. J. Mol. Biol. 391:498–507 [DOI] [PubMed] [Google Scholar]
- 34.Pogoryelov D, Yildiz Ö, Faraldo-Gomez JD, Meier T. 2009. High-resolution structure of the rotor ring of a proton-dependent ATP synthase. Nat. Struct. Mol. Biol. 16:1068–1073 [DOI] [PubMed] [Google Scholar]
- 35.Preiss L, Yildiz Ö, Hicks DB, Krulwich TA, Meier T. 2010. A new type of proton coordination in an F1Fo-ATP synthase rotor ring. PLoS Biol. 8:e1000443. 10.1371/journal.pbio.1000443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. 2009. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71:825–835 [DOI] [PubMed] [Google Scholar]
- 37.Neumann S, Matthey U, Kaim G, Dimroth P. 1998. Purification and properties of the F1Fo ATPase of Ilyobacter tartaricus, a sodium ion pump. J. Bacteriol. 180:3312–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kluge C, Dimroth P. 1993. Kinetics of inactivation of the F1Fo ATPase of Propionigenium modestum by dicyclohexylcarbodiimide in relationship to H+ and Na+ concentration: probing the binding site for the coupling ions. Biochemistry 32:10378–10386 [DOI] [PubMed] [Google Scholar]
- 39.Kim EA, Price-Carter M, Carlquist WC, Blair DF. 2008. Membrane segment organization in the stator complex of the flagellar motor: implications for proton flow and proton-induced conformational change. Biochemistry 47:11332–11339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sowa Y, Berry RM. 2008. Bacterial flagellar motor. Q. Rev. Biophys. 41:103–132 [DOI] [PubMed] [Google Scholar]
- 41.Krah A, Pogoryelov D, Langer JD, Bond PJ, Meier T, Faraldo-Gómez JD. 2010. Structural and energetic basis for H+ versus Na+ binding selectivity in ATP synthase Fo rotors. Biochim. Biophys. Acta 1797:763–772 [DOI] [PubMed] [Google Scholar]
- 42.Li N, Kojima S, Homma M. 2011. Characterization of the periplasmic region of PomB, a Na+-driven flagellar stator protein in Vibrio alginolyticus. J. Bacteriol. 193:3773–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu S, Homma M, Kojima S. 2012. Intragenic suppressor of a plug deletion nonmotility mutation in PotB, a chimeric stator protein of sodium-driven flagella. J. Bacteriol. 194:6728–6735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collins KD. 2004. Ions from the Hofmeister series and osmolytes: effects on proteins in solution and in the crystallization process. Methods 34:300–311 [DOI] [PubMed] [Google Scholar]
- 45.Swerdlow DL, Ries AA. 1992. Cholera in the Americas: guidelines for the clinician. JAMA 267:1495–1499 [PubMed] [Google Scholar]
- 46.Colwell RR. 2004. Infectious disease and environment: cholera as a paradigm for waterborne disease. Int. Microbiol. 7:285–289 [PubMed] [Google Scholar]
- 47.Schulz H, Fabianek RA, Pellicioli EC, Hennecke H, Thöny-Meyer L. 1999. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc. Natl. Acad. Sci. U. S. A. 96:6462–6467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arslan E, Schulz H, Zufferey R, Kunzler P, Thöny-Meyer L. 1998. Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251:744–747 [DOI] [PubMed] [Google Scholar]
- 49.Schneider K, Dimroth P, Bott M. 2000. Biosynthesis of the prosthetic group of citrate lyase. Biochemistry 39:9438–9450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.