Abstract
Cells use complex mechanisms to regulate glucose transport and metabolism to achieve optimal energy and biomass production while avoiding accumulation of toxic metabolites. Glucose transport and glycolytic metabolism carry the risk of the buildup of phosphosugars, which can inhibit growth at high concentrations. Many enteric bacteria cope with phosphosugar accumulation and associated stress (i.e., sugar-phosphate stress) by producing a small RNA (sRNA) regulator, SgrS, which decreases phosphosugar accumulation in part by repressing translation of sugar transporter mRNAs (ptsG and manXYZ) and enhancing translation of a sugar phosphatase mRNA (yigL). Despite a molecular understanding of individual target regulation by SgrS, previously little was known about how coordinated regulation of these multiple targets contributes to the rescue of cell growth during sugar-phosphate stress. This study examines how SgrS regulation of different targets impacts growth under different nutritional conditions when sugar-phosphate stress is induced. The severity of stress-associated growth inhibition depended on nutrient availability. Stress in nutrient-rich media necessitated SgrS regulation of only sugar transporter mRNAs (ptsG or manXYZ). However, repression of transporter mRNAs was insufficient for growth rescue during stress in nutrient-poor media; here SgrS regulation of the phosphatase (yigL) and as-yet-undefined targets also contributed to growth rescue. The results of this study imply that regulation of only a subset of an sRNA's targets may be important in a given environment. Further, the results suggest that SgrS and perhaps other sRNAs are flexible regulators that modulate expression of multigene regulons to allow cells to adapt to an array of stress conditions.
INTRODUCTION
All organisms must produce biomass and generate energy from external substrates in order to grow. Microbes have evolved complex regulatory mechanisms to optimize uptake and metabolism of glucose, which is a preferred carbon source for many species, while avoiding accumulation of unnecessary and potentially toxic metabolic intermediates. In many bacteria, glucose is transported into cells mainly by the phosphoenolpyruvate phosphotransferase system (PTS), which consists of two general sugar transport proteins enzyme I (EI) and histidine protein (HPr), as well as two glucose-specific proteins glucose-specific enzyme IIA (EIIAGlc) and EIICBGlc (1). The expression of the ptsG gene encoding EIICBGlc is extensively regulated transcriptionally and posttranscriptionally. Transcription factor proteins responding to different environmental conditions regulate ptsG. For example, ptsG transcription is activated by the cyclic AMP (cAMP) receptor protein (CRP) (2), and negative control is exerted by the repressor Mlc, which inhibits ptsG transcription in the absence of glucose (3, 4). Posttranscriptional control of ptsG expression is mediated by the small RNA (sRNA) regulator SgrS (5). SgrS is produced in response to a metabolic stress known as sugar-phosphate or glucose-phosphate stress, which is characterized by cytoplasmic accumulation of certain phosphosugars and inhibition of cell growth. Escherichia coli cells require SgrS to resist stress—i.e., continue growing under stress conditions (6).
SgrS, like many other sRNA regulators, depends on the RNA chaperone Hfq for stability and to facilitate base pairing interactions with target mRNAs (7). SgrS forms base pairing interactions with ptsG mRNA that occlude the ptsG ribosome binding site (RBS), resulting in translation inhibition and subsequent RNase-E dependent degradation of the SgrS-ptsG duplex (5, 7, 8). We recently showed that SgrS represses translation of a second target mRNA, manXYZ (9, 10), which encodes the mannose (and auxiliary glucose) PTS transporter. SgrS-mediated inhibition of sugar transporter synthesis is believed to limit further uptake and accumulation of stressor phosphosugars during glucose-phosphate stress (Fig. 1) (10). SgrS was also recently demonstrated to act as a positive regulator of a novel target, yigL. SgrS-yigL mRNA base pairing selectively stabilizes a processed form of yigL mRNA by masking an RNase E cleavage site (38). SgrS-mediated stabilization of yigL mRNA allows for enhanced production of the encoded haloacid dehalogenase (HAD)-like phosphatase, which was previously shown to dephosphorylate glucose-6-phosphate and its analog 2-deoxyglucose-6-phosphate in vitro (11). This and other evidence suggests that SgrS enhances YigL production in order to promote dephosphorylation of sugar-phosphates so that the resulting uncharged sugars can be exported by an unknown efflux pump (Fig. 1) (38). In addition to regulating translation and stability of target mRNAs via base pairing interactions, we showed that sgrS encodes a functional protein, SgrT (12). Whereas SgrS base pairing activity affects new protein synthesis, SgrT inhibits the activity of extant sugar transporters (12). In a previous study, we showed that E. coli SgrS does not produce significant amounts of SgrT under typical glucose-phosphate stress conditions and that the base pairing function is sufficient for E. coli growth recovery (13).
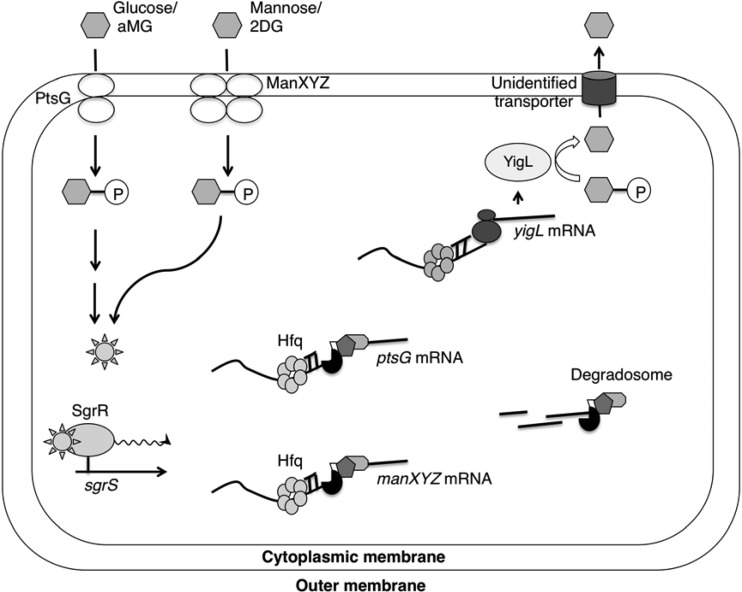
Fig 1.
Model for the SgrS-mediated glucose-phosphate (GP) stress response. During glucose-phosphate stress, SgrR activates transcription of sgrS. SgrS associates with Hfq and negatively regulates the ptsG and manXYZ mRNAs, which encode major PTS sugar transporters. In addition, SgrS positively regulates the yigL mRNA, encoding a phosphatase. Dephosphorylation of sugars is a prerequisite for their efflux through unknown transporters. The regulation of these target mRNAs by SgrS, in turn, helps cellular recovery from GP stress. aMG, αMG; P, phosphate group.
While there are unanswered questions regarding the roles of SgrS and the stress response in natural environments, it is clear that the response is broadly conserved among enteric bacteria (14, 15). Glucose-phosphate stress occurs in several different circumstances, all having in common perturbed glycolytic metabolism resulting in accumulation of nonmetabolizable phosphosugars. Wild-type E. coli and Salmonella enterica cells induce the stress response when the glucose analogs α-methyl glucoside (αMG) or 2-deoxyglucose (2DG) are taken up through the glucose or mannose PTSs, respectively, causing accumulation of α-methyl glucoside-6-phosphate or 2-deoxyglucose-6-phosphate (5, 10). Interestingly, unlike E. coli and Salmonella, some microbes, such as Klebsiella pneumoniae, possess enzymes for αMG catabolism (16), while other organisms can utilize 2DG as a carbon source (17), raising the possibility that these (or similar) compounds may be present in some natural environments. These two glucose analogs, αMG and 2DG, are ideal model stress inducers for our studies because they enter E. coli cells through different PTS transporters that are regulated by SgrS. PtsG (EIICBGlc) is the primary αMG transporter, whereas ManXYZ (mannose-specific enzyme IIABCD [EIIABCDMan]) transports 2DG (18, 19).
Most studies so far have focused on the molecular mechanisms of individual target regulation by SgrS, and these have revealed novel and interesting aspects of sRNA-mediated regulation. However, it is unknown how coordinated regulation of these multiple targets contributes to the physiology of the glucose-phosphate stress response. Importantly, the issue of multiple target regulation by bacterial sRNAs is relevant for dozens of other Hfq-binding sRNAs that have also been shown to control expression of many genes (20–22), and little is known about how this property contributes to sRNA-mediated stress responses. In the present study, we sought to establish SgrS as a model sRNA for exploring the consequences of coordinated regulation of multiple targets with regard to stress resistance. We hypothesized that regulation of some targets would be more important than others in terms of rescuing cell growth during stress and that this arrangement might change to match fluctuations in the environment. To address this hypothesis, we began by testing how SgrS regulation of different targets impacts growth in response to different inducers of stress (αMG or 2DG) in different nutrient environments. Our results demonstrate for the first time that regulation of individual sRNA targets can contribute differentially to a stress response depending upon the particular source of stress and other environmental conditions. With regard to glucose-phosphate stress, our results highlight the importance of different carbon sources in modulating the severity of sugar-phosphate-associated metabolic stress. We show that under less severe stress conditions (in nutrient-rich media), SgrS needs only to repress synthesis of the relevant sugar transporter in order to ensure stress resistance. When stress becomes more severe (in nutrient-poor media), regulation of additional SgrS targets becomes crucial for growth recovery. These results show that regulation of only a subset of SgrS targets is important for responding to a given stressor in a particular environment, suggesting a broad role in nature for SgrS-mediated responses to metabolic stress. Our results imply that sRNAs may have evolved as flexible regulators that adjust their regulons in accordance with environmental changes, thus providing bacteria with efficient and adaptable responses to different stressors.
MATERIALS AND METHODS
Bacterial strain and plasmid construction.
Most strains used in this study are derivatives of E. coli DJ480 (D. Jin, National Cancer Institute), and all bacterial strains are listed in Table 1. The sequences of all oligonucleotides (Integrated DNA Technologies) used in the construction of mutant strains and plasmids are listed in Table 2.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Description or relevant characteristic(s) | Source or reference |
|---|---|---|
| Bacterial strains | ||
| MG1655 | Wild-type E. coli K-12 | D. Jin (NCI) |
| DJ480 | MG1655 ΔlacX74 | D. Jin (NCI) |
| CS104 | DJ480 ΔsgrS | 13 |
| CS123 | DJ480 sgrS1 | 13 |
| CS168 | DJ480 λattB::lacIq+ tetR Spr | 23 |
| CS194 | DJ480 ΔyigL::FRT λattB::lacIq+ tetR Spr | This study |
| CS195 | DJ480 ΔyigL::FRT ΔsgrS λattB::lacIq+ tetR Spr | This study |
| JH111 | DJ480 ΔsgrS λattB::lacIq+ tetR Spr | 10 |
| JH116 | DJ480 ΔsgrS manX′-′lacZ lacIq+ | 10 |
| JH171 | DJ480 ΔsgrS ptsG′-′lacZ lacIq+ | 10 |
| YS185 | DJ480 ΔyigL::FRT | This study |
| YS234 | DJ480 ΔsgrS yigL′-′lacZ λattB::lacIq+ tetR Spr | This study |
| YS236 | λattB::tet | This study |
| YS237 | DJ480 λattB::tet | This study |
| YS238 | DJ480 λattB::tet sgrS1 | This study |
| YS246 | ΔsgrS::kan-araC-PBAD-ccdB; mini λ | This study |
| YS247 | DJ480 λattB::tet ΔsgrS | This study |
| YS248 | DJ480 sgrS28 | This study |
| YS249 | DJ480 λattB::tet sgrS28 | This study |
| YS258 | DJ480 ΔsgrS cm-Plac-ptsG λattB::lacIq+ tetR Spr | This study |
| YS259 | DJ480 cm-Plac-ptsG λattB::lacIq+ tetR Spr | This study |
| YS265 | DJ480 cm-Plac-ptsG yigL::FRT ΔsgrS λattB::lacIq+ tetR Spr | This study |
| YS269 | DJ480 sgrS26 | This study |
| YS270 | DJ480 λattB::tet sgrS26 | This study |
| YS273 | DJ480 λattB::tet ΔyigL::FRT | This study |
| YS283 | ΔmanXYZ::kan ΔsgrS λattB::tet | This study |
| YS284 | ΔptsG::cm ΔsgrS λattB::tet | This study |
| YS285 | ΔmanXYZ::kan λattB::tet | This study |
| YS286 | ΔptsG::cm λattB::tet | This study |
| Plasmids | ||
| pBRCS12 | Vector control for pLCV1, pBRCS6, pBRJH19, pBR26 and pBRYS4 | 13 |
| pLCV1 | Plac-sgrS | 5 |
| pBRCS6 | Plac-sgrS1 | 13 |
| pBRJH26 | Plac-sgrS26 | 10 |
| pBRYS4 | Plac-sgrS28 | This study |
| pZE21 | Vector control for pZEYS2 | 23 |
| pZEYS2 | PLtetO-1-yigL | This study |
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′–3′) |
|---|---|
| O-YS156 | CCCAGCGGAAACCGCTCTACAGAGGTTTAAATTTCTTGTGTAGGCTGGAGCTGCTTCG |
| O-YS157 | GCGAAGTATCAGGTTGACAACTGACCAAATAAAGAACGAATTCCGGGGATCCGTCGAC |
| O-YS206 | CATACGTTATCCCCTTACGCCAAAGAACTCTGAAGCTGATTCCGGGGATCCGTCGA |
| O-YS207 | GTATCCATTGTAGCGAAGTATCAGGTTGACAACTGACCAGTGTAGGCTGGAGCTGCT |
| O-YS212 | TGAAAGTTGACTTGCCTGCATCATCACACACTGAGTATTGGTGTAAAATCACCCGC |
| O-YS213 | ACCTTCCCGTTTCGCTCAAGTTAGTATAAAAAAGCACTAGACATCATTAATTCCTA |
| O-YS214 | CCGGGCTATGAAATAGAAAAATGAATCCGTTGAAGCCGAAGCTAAATCTTCTTTATC |
| O-YS215 | CCCAAGCTTATTAAAGAGGAGAAATTAACTATGTACCAGGTTGTTGCGTCTGAT |
| O-YS216 | CCCGGATCCCCAAATAAAGAACGATTACGATAAATAGAGTTTACGCAGA |
| O-YS225 | CCTGTGACGGAAGATCACTTCGCAGAATAA |
| O-YS226 | CAGTGGGATGACCGCAATTCTGAAAGTTGACTTGCCTGCAATAGGAACTTCAAGATCC |
| O-YS227 | TACGGCGAGCCATCGTCATTATCCAGATCATACGTTCCTTATATTCCCCAGAACATCAGG |
| O-YS228 | CAGTGGGATGACCGCAATTCTGAAAGTTGACTTGCCTGCATCATCACACACTGAGTATT |
| O-YS230 | TACGGCGAGCCATCGTCATTATCCAGATCATACGTTCCC |
| O-YS238 | GTGCTCAGTATCTTGTTATCCGCTCACAATGTCAATGTTATCCGCTCACATTTATTTATCCGCTCACATTTATTTATCACTTAT |
| O-YS240 | AGCTCGTAATTAATGGCTAAAACGAGTAAAGTTCACCCCTGTGACGGAAGATCACTT |
| O-YS241 | CCTCGCCGTGTACAGGGCATCTAAGCGCCCTTTATTTATGTGCTCAGTATCTTGTTATC |
| O-YS261 | CAGTGGGATGACCGCAATTCTGAAAGTTGACTTGCCTGCATCATCTGTGACTGAGTATT |
Strains JH116 and JH171, which contain the manX′-′lacZ and ptsG′-′lacZ translational fusions, respectively, were described in a previous study (10). The yigL′-′lacZ translational fusion was created using a technique described previously (24). Briefly, a kanamycin cassette flanked by a FLP recombination target (FRT) site was amplified from template pKD13 using oligonucleotides O-YS206 and O-YS207 (Table 2) and integrated into the chromosome by λ Red recombination at the yigL locus. The kanamycin cassette was then removed using the helper plasmid pCP20 encoding the FLP recombinase, resulting in a strain carrying a single FRT site. Subsequently, translational fusion vector pCE40 (24) was integrated into the chromosome by FLP-dependent site-specific recombination, resulting in ′lacZ fused to the 17th codon of yigL and linked to the kanamycin cassette. The fusion was then transduced by P1 phage into a previously described strain, JH111 (10) (which is lacIq+ and ΔsgrS [Table 1]) to create strain YS234.
Strains CS104 and CS123, which carry the ΔsgrS and sgrS1 mutations, respectively, were described previously (13). The ΔmanXYZ::kan allele was moved into strain CS104 via P1 transduction to create strain CS184 by C. Wadler in our laboratory. The ΔptsG::cm allele was transduced into strain CS104 to create strain CV106. Chromosomal sgrS26 and sgrS28 alleles (Fig. 2A) were constructed using a strategy modified from the one described in a previous study (25), and a strain that carries the kanamycin cassette fused to the araC gene and the toxin gene ccdB under the control of the PBAD promoter (a gift from N. Majdalani, National Cancer Institute). The kan-araC-PBAD-ccdB region was PCR amplified by oligonucleotides O-YS226 and O-YS227 and inserted into the sgrS locus of strain NM300 (which carries a mini-λ encoding λ Red functions [5]), resulting in strain YS246. Mini-λ was maintained in strain YS246 by growth at 30°C. Subsequently, the sgrS26 allele was PCR amplified from plasmid pBRJH26, using oligonucleotides O-YS261 and O-YS230. The sgrS28 allele was amplified using genomic DNA from wild-type DJ480 as the template, and oligonucleotides O-YS228/O-YS230, which incorporated the desired point mutations. Following induction of λ Red functions in strain YS246, sgrS26 and sgrS28 PCR products were transformed by electroporation, yielding strains YS269 and YS248, respectively. Recombinants were obtained by counterselection against ccdB by plating cells on medium containing 1% l-arabinose.
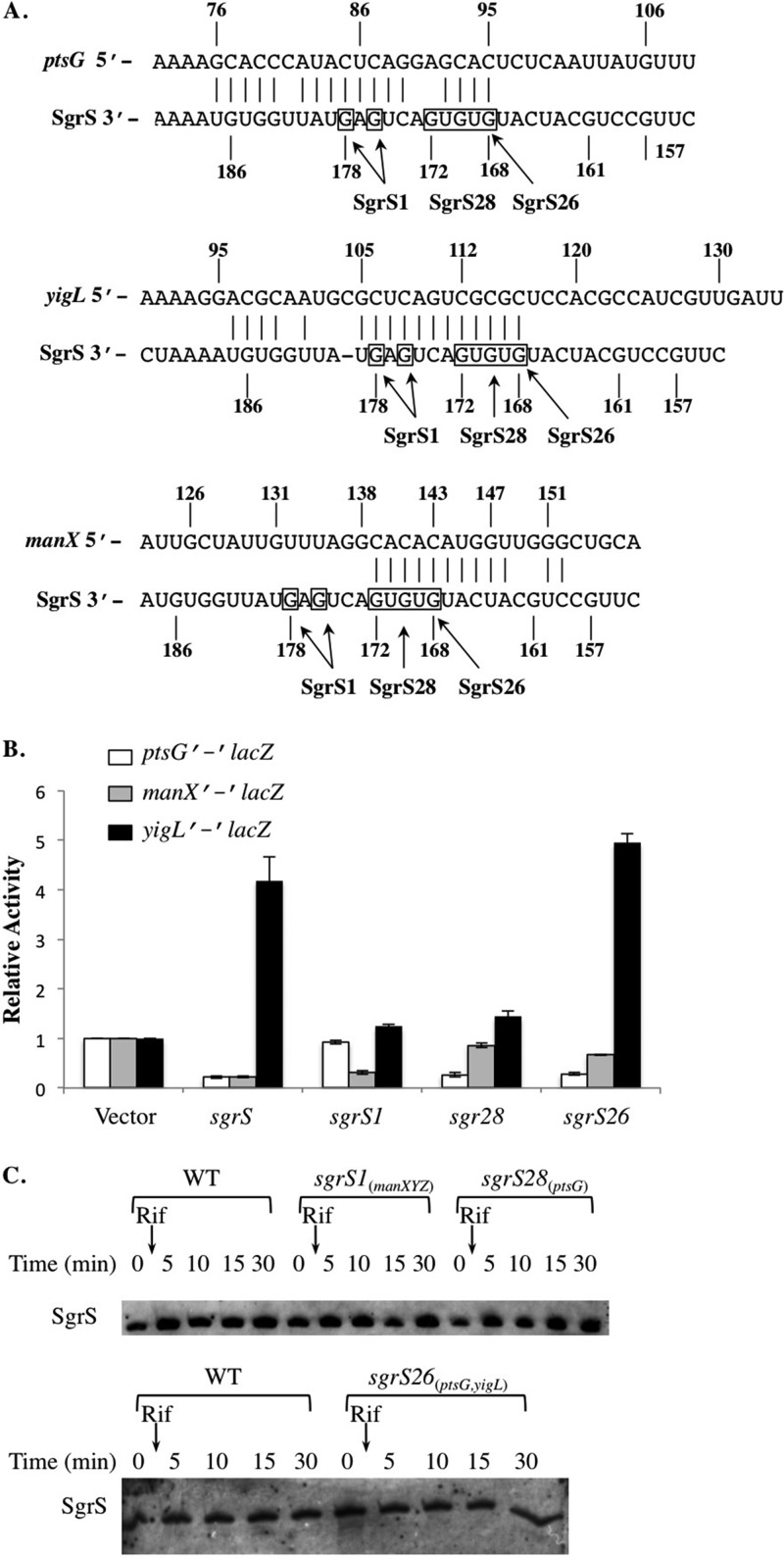
Fig 2.
SgrS mutant alleles differentially regulate expression of ptsG, manX, and yigL. (A) Base pairing between SgrS and the three targets, the ptsG, manX, and yigL mRNAs, are indicated by vertical lines. The sequence directly above SgrS and allele names (SgrS1, SgrS26, and SgrS28) indicate the mutated bases and their positions in different SgrS mutants. (B) The ΔsgrS strains with ptsG′-′lacZ, manX′-′lacZ, or yigL′-′lacZ carrying an empty vector, Plac-sgrS, Plac-sgrS1, Plac-sgrS26, or Plac-sgrS28 were grown to early log phase and exposed to 0.1 mM IPTG. Samples were collected 60 min after IPTG addition and assayed for β-galactosidase activity. Specific activities were normalized to the levels in the strains carrying the empty vector to yield relative activity (fold). Three independent experiments were performed; results reported are averages plus standard deviations (error bars). (C) Strains were grown to early log phase and exposed to 0.5% αMG for 10 min. Rifampin (Rif) (250 μg/ml) was then added to all cultures, and RNA samples were harvested at the indicated time points and subjected to Northern blot analysis. Blots shown are representative of three independent experiments. WT, wild type.
To create the ΔyigL::FRT-kan-FRT allele, a kanamycin cassette flanked by FLP recombination target sites was amplified from template pKD13 (24) using oligonucleotides O-YS156/O-YS157. The ΔyigL::FRT-kan-FRT allele was then moved into the previously described strains CS168 and JH111 (10, 13) by P1 transduction. Subsequently, the kanamycin resistance cassettes in these strains were eliminated using pCP20, a plasmid that expresses FLP recombinase (24); the resulting strains are YS184, CS194, and CS195.
The attB::tet allele in strain YS236 was created using primers O-YS213/O-YS214, with homology to the attB locus to amplify the tetracycline resistance cassette, followed by λ Red recombination (26). This mutant allele was then transduced into strains DJ480, CV106, CS104, CS123, CS184, YS185, YS208, YS248, and YS269 to yield strains YS237, YS284, YS247, YS238, YS83, YS273, YS285, YS249, and YS270, respectively. The ΔptsG::cm allele was moved into strain YS237 by P1 transduction to produce strain YS286.
To insert the Plac promoter on the chromosome upstream of ptsG, we first amplified a chloramphenicol cassette from strain CV700, using oligonucleotides O-YS225 and O-YS238 that contain sequences homologous to the cat gene and the Plac promoter. The resulting PCR product, which has the chloramphenicol cassette linked to the Plac promoter, then served as the template for the next round of PCR amplification. Using oligonucleotides O-YS240 and O-YS241, we obtained a new PCR product, which contains the cat gene-linked Plac promoter that is flanked by the region from −177 to −140 relative to the ptsG start codon, as well as the first 30 nucleotides (nt) of ptsG coding sequence. Using this PCR product and the λ Red recombination system (26), we created the cm-Plac-ptsG allele, which was then transduced into strains JH111, CS168, and CS195, resulting in strains YS258, YS259, and YS265, respectively.
Strains DH5α (Invitrogen) and XL10 (Stratagene) were used for cloning and QuikChange mutagenesis, respectively. To construct plasmid pZEYS2, the yigL gene was amplified by PCR using the forward primer O-YS215, which contains a HindIII site and a 24-nt fragment from the pQE80L vector (Qiagen) carrying the ribosome binding site, followed by yigL sequence. The reverse primer, O-YS216, contains a BamHI site and sequences homologous to the region downstream of the predicted yigL terminator. The HindIII- and BamHI-digested PCR product was then cloned into the previously described vector pZE21 (23). Plasmids pLCV1, pBRCS6, and pBRJH26 that carry wild-type sgrS, sgrS1, and sgrS26, respectively, were described in previous studies (10, 13). Plasmid pBRYS4, containing the sgrS28 allele, was created using the QuikChange II site-directed mutagenesis kit (Agilent Technologies) with oligonucleotide O-YS212 and the previously described plasmid pBRJH19 (10) as the template.
β-Galactosidase assays.
Strains containing translational fusions were grown overnight in TB (Bacto tryptone) medium (BD, Franklin Lakes, NJ) with 100 μg/ml ampicillin and subcultured 1:200 to fresh medium. Cultures were grown to an optical density at 600 nm (OD600) of ∼0.5 and exposed to 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Samples were taken 1 h later and assayed for β-galactosidase activity as described previously (23). The β-galactosidase activity (measured in Miller units) produced by cells carrying the vector control was set at 1.0. Activity values for other strains were normalized to the vector control to give relative activity for experimental samples.
RNA extraction and Northern blot analyses.
To examine the stabilities of SgrS variants, strains were grown in LB medium to an OD600 of ∼0.5 and exposed to 0.5% αMG (Sigma) for 10 min. Rifampin (250 μg/ml) was then added to the cultures, and RNA was extracted at the indicated time intervals by the previously described hot phenol method (5). Northern blot analysis with probe sgrS-1bio (5) was used to detect the SgrS RNA.
Growth experiments.
For growth competition experiments, cells were grown overnight in LB or minimal MOPS (morpholinepropanesulfonic acid) medium (Teknova) supplemented with 0.4% glycerol or 0.2% fructose as indicated. Two competing strains were mixed at 1:1 ratios (based on OD600), inoculated in fresh media, and grown to an OD600 of ∼0.03 (minimal MOPS medium with glycerol) or to an OD600 of ∼0.1 (LB and minimal MOPS medium with fructose). Cultures were then exposed to 0.5% αMG or 2DG (Sigma) or kept under nonstress conditions. Due to the overall lower growth rate in glycerol, cells grown with glycerol were exposed to αMG until they reached a lower OD600 and incubated longer. Culture samples were collected right after the initial mixing, as well as 3 h (LB), 17 h (minimal MOPS medium with fructose), or 19 h (minimal MOPS medium with glycerol) after exposure of the cells to the stress inducer. Serial dilutions were made from the collected samples and plated on LB agar with or without 100 μg/ml tetracycline.
For the experiments involving removal of inducers, strains were grown overnight in LB or minimal MOPS medium supplemented with 0.4% glycerol (25 μg/ml kanamycin was added to the medium when working with strains carrying plasmids pZE21 and pZEYS2) in the presence of 0.1 mM IPTG and then subcultured 1:200 in fresh media. Anhydrotetracycline (aTc) (25 ng/ml) was added to the subcultures when working with strains carrying plasmids. Cells were harvested at an OD600 of ∼0.1 by filtration, washed, and resuspended in fresh media with 0.5% αMG and in the presence or absence of 0.1 mM IPTG. aTc at 25 ng/ml was added back to these cultures when working with strains carrying plasmids pZE21 and pZEYS2. In other regular growth experiments, strains were grown in minimal MOPS medium supplemented with 0.4% glycerol, 25 μg/ml kanamycin, and 25 ng/ml aTc to an OD600 of ∼0.1 and then exposed to 0.5% αMG.
For growth experiments involving the supplementation of Casamino Acids, strains were grown in minimal MOPS medium with 0.4% glycerol or 0.2% fructose overnight and subcultured to an OD600 of ∼0.05 in fresh medium. Casamino Acids (final concentration of 0.1%) and/or 0.5% αMG were also added to the medium to the subcultures as indicated. The growth of strains was monitored using a FLUOstar Omega multimode microplate reader (BMG Labtech).
RESULTS
Mutations in SgrS base pairing determinants have differential effects on regulation of three targets.
Previous studies have demonstrated that duplex formation between SgrS and its target mRNAs is essential for regulation. For each of the targets, the residues of SgrS involved in pairing partially overlap and are partially distinct (10). For example, though two G-C base pairs formed by SgrS residues G176 and G178 (Fig. 2A) are critical for translational repression of ptsG by SgrS (7), they are not required for inhibition of manX translation (10). To begin to elucidate how SgrS coordinates regulation of multiple targets, we sought to identify additional mutations in sgrS that would result in differential target regulation. Regulation by wild-type and mutant SgrS was monitored using translational lacZ fusions to the three known SgrS targets. As shown in Fig. 2B, wild-type SgrS (expressed from a plasmid under the control of the Plac promoter) repressed ptsG′-′lacZ (∼3-fold repressed compared to the vector control) and manX′-′lacZ (∼3-fold repressed) and activated yigL′-′lacZ (∼4.4-fold increase compared to the vector control). These results were consistent with previous reports (10; Papenfort et al., submitted). The sgrS allele with G176C and G178C mutations (Fig. 2A), which we refer to as sgrS1, repressed manX′-′lacZ almost as efficiently as wild-type SgrS but no longer regulated the activities of ptsG′-′lacZ (as observed previously [7, 10]) or yigL′-′lacZ (Fig. 2B). Thus, with regard to these three targets, SgrS1 specifically regulated only manXYZ and is hereafter referred to as SgrS1manXYZ. SgrS28 carries five point mutations (G172C, T171A, G170C, T169A, and G168C [Fig. 2A]). When expressed from the Plac plasmid, SgrS28 repressed ptsG′-′lacZ to a degree similar to wild-type SgrS (Fig. 2B) but failed to regulate either manX′-′lacZ or yigL′-′lacZ (Fig. 2B). Since SgrS28 specifically regulated only ptsG, it is referred to as SgrS28ptsG. A previous study in our laboratory identified the sgrS26 allele (Fig. 2A) with a G168C mutation as being defective for regulation of manX′-′lacZ (10). We confirmed this finding and further showed that SgrS26 repressed ptsG′-′lacZ activity and enhanced yigL′-′lacZ activity to a degree similar to wild-type SgrS (Fig. 2B). Since SgrS26 regulated both ptsG and yigL but was deficient in regulation of manXYZ, it is now referred to as SgrS26ptsG,yigL. Altogether, we defined three mutants with distinct target repertoires, two that each regulate only one of the three known SgrS targets (SgrS1manXYZ and SgrS28ptsG) and the third, which regulates two of the three known targets (SgrS26ptsG,yigL).
Regulation of ptsG, but not manXYZ or yigL, is crucial for recovery from αMG-induced stress in nutrient-rich medium.
In order to verify that differences in regulation were not due to decreased stability of mutant sRNAs, we tested the induction of chromosomally encoded sgrS mutant alleles in response to αMG and monitored stability using a rifampin chase. After αMG treatment, all three mutant SgrS molecules were present at levels similar to that of the wild type and also showed similar stabilities (Fig. 2C). These results indicated that the observed differences in regulation of targets were not due to effects on SgrS stability.
To examine how strains expressing target-specific mutant alleles cope with stress in the presence of various levels of nutrients, we examined stress induced by αMG under nutrient-rich and nutrient-poor conditions. Growth competition experiments were used to monitor the relative fitness of strains grown under stress conditions; these experiments were conducted as described in Materials and Methods. Briefly, the two strains were mixed at a 1:1 ratio. The mixed culture was exposed to αMG at early log phase and then was grown to saturation. The numbers of viable cells at both the initial mixing and the end of the experiment were determined by plating for CFU, and a competition index (CI) (27) was calculated as follows: (log10 strain A recovered/log10 strain B recovered)/(log10 strain A inoculated/log10 strain B inoculated). A CI equal to 1 indicates that the two strains compete evenly for resources in mixed culture; a CI that is less than 1 suggests that strain B outcompetes strain A, whereas a CI that is greater than 1 shows that strain A outcompetes strain B.
For a control, we first compared the growth of a strain marked with tetracycline resistance at a neutral genomic location (attB::tet) with the wild-type parent. The CI for the attB::tet strain versus the wild-type parent was ∼1.0 in both the absence and presence of αMG when cells were grown in nutrient-rich LB medium (Table 3). This result demonstrated that the tetracycline resistance gene does not cause a growth defect under these conditions. As shown in Table 3, in the absence of stress (LB medium without αMG), all strains competed equally, indicating that SgrS does not play a significant role in E. coli growth in rich medium. In contrast, under αMG-induced stress conditions, growth of the ΔsgrS mutant was attenuated when it competed with the wild-type strain (CI = 0.6), which is consistent with the important role of SgrS in the glucose-phosphate stress response, as reported previously (6). The sgrS1manXYZ mutant, which specifically regulates manXYZ but not ptsG or yigL (Fig. 2), also exhibited a growth defect in competition with the wild-type strain (CI = 0.5 [Table 3]). The CI for the sgrS1manXYZ mutant versus the ΔsgrS mutant was 1.0 (Table 3), indicating that SgrS regulation of manXYZ alone gives no growth advantage when the cells are stressed by αMG. In contrast, the strain with the sgrS28ptsG allele that allows regulation of ptsG but not manXYZ or yigL, performed well in growth competition with the wild-type strain (the CI, at 0.8, was not statistically different from 1.0 [Table 3]). Moreover, the sgrS28ptsG strain outcompeted the other two sgrS mutants with CIs of ∼3 (Table 3). These results strongly suggested that during stress with αMG in rich medium, regulation of ptsG, which encodes the major transporter of αMG, is a crucial function of SgrS in the stress response, whereas regulation of manXYZ and yigL is not required.
Table 3.
Competition assays to measure the effects of sgrS mutations on growth with αMG in LB medium
| E. coli strain A genotype | E. coli strain B genotype | Without αMG |
With 0.5% αMG |
||
|---|---|---|---|---|---|
| CIa | P valueb | CI | P value | ||
| λattB::tet | WT | 1.08 ± 0.06 | NS | 1.0 ± 0.3 | NS |
| ΔsgrS λattB::tet | WT | 0.82 ± 0.01 | NS | 0.6 ± 0.1 | 0.024 |
| sgrS1manXYZ λattB::tet | WT | 0.98 ± 0.21 | NS | 0.5 ± 0.04 | 0.01 |
| sgrS28ptsG λattB::tet | WT | 0.93 ± 0.08 | NS | 0.8 ± 0.07 | NS |
| sgrS1manXYZ λattB::tet | ΔsgrS | 1.02 ± 0.15 | NS | 1.0 ± 0.01 | NS |
| sgrS28ptsG λattB::tet | ΔsgrS | 0.97 ± 0.12 | NS | 3.5 ± 0.3 | 0.03 |
| sgrS28ptsG λattB::tet | sgrS1manXYZ | 1.03 ± 0.08 | NS | 2.9 ± 0.1 | 0.013 |
See Materials and Methods for detailed procedures. The competition index (CI) is calculated as follows: (log10 strain A output/log10 strain B output)/(log10 strain A input/log10 strain B input). The results presented here are the averages ± standard deviations of three independent experiments.
Student's t test was used to compare the output and inoculum. NS, not significant (P ≥ 0.05).
To further test the roles of manXYZ and yigL in growth during stress, strains with deletions in each locus were tested in growth competition assays in rich LB medium with αMG (Table 4). The ΔmanXYZ mutant (CI = 1.05) competed evenly with the wild-type strain under these conditions, and the ΔmanXYZ ΔsgrS double mutant competed evenly with its ΔsgrS parent (CI = 1.06 [Table 4]), indicating that the absence of the ManXYZ sugar transporter provides no protection from αMG stress. Likewise, the ΔyigL mutant performed well in competition with the wild-type strain (CI = 1.02 [Table 4]), consistent with the results above and demonstrating that regulation of yigL is dispensable for growth recovery when cells are stressed with αMG in rich medium. In contrast, a ΔptsG mutant outcompeted the wild-type strain (CI = 2.6), and a ΔptsG ΔsgrS double mutant outcompeted its ΔsgrS parent strain (CI = 2.26) during growth in LB with αMG (Table 4). These results support the conclusions from analyses of sgrS mutant alleles (Table 3) and are consistent with the notion that PtsG is the major αMG transporter and that repression of PtsG synthesis is important for growth recovery in the presence of αMG.
Table 4.
Competition assays to measure the effects of mutations in the three sgrS targets on growth with αMG in LB medium
| E. coli strain A genotype | E. coli strain B genotype | Without αMG |
With 0.5% αMG |
||
|---|---|---|---|---|---|
| CIa | P valueb | CI | P value | ||
| ΔptsG λattB::tet | WT | 0.96 ± 0.05 | NS | 2.6 ± 0.1 | 2.0E−5 |
| ΔmanXYZ λattB::tet | WT | 1.01 ± 0.1 | NS | 1.05 ± 0.04 | NS |
| ΔsgrS ΔptsG λattB::tet | ΔsgrS | 1.02 ± 0.03 | NS | 2.26 ± 0.18 | 0.0025 |
| ΔsgrS ΔmanXYZ λattB::tet | ΔsgrS | 0.98 ± 0.04 | NS | 1.06 ± 0.11 | NS |
| ΔyigL λattB::tet | WT | 0.98 ± 0.07 | NS | 1.02 ± 0.14 | NS |
See Materials and Methods for detailed procedures. The competition index (CI) is calculated as follows: (log10 strain A output/log10 strain B output)/(log10 strain A input/log10 strain B input). The results presented here are the averages ± standard deviations of three independent experiments.
Student's t test was used to compare the output and inoculum. NS, not significant (P ≥ 0.05).
Repression of ptsG by an SgrS-independent mechanism promotes recovery from αMG-induced stress in nutrient-rich medium.
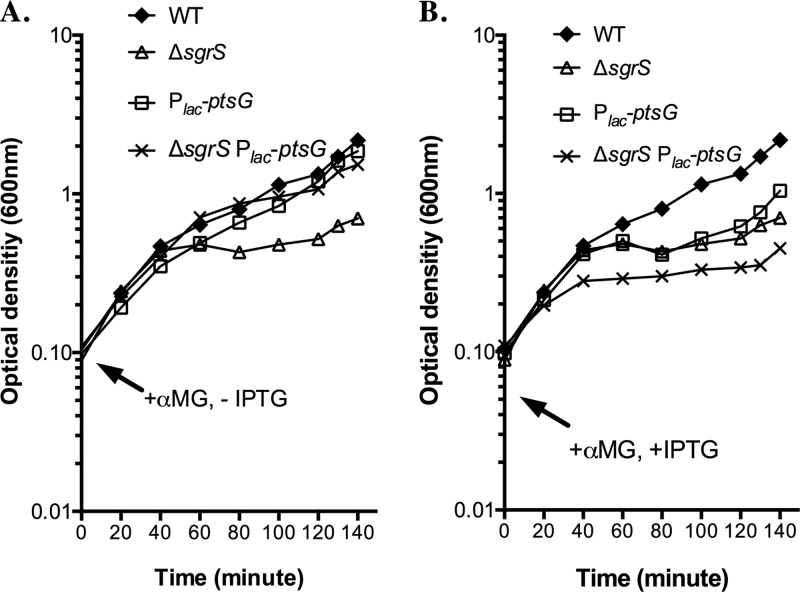
The sgrS base pairing mutations may affect the ability of SgrS to regulate other as-yet-unknown mRNA targets. Transcriptome analyses conducted in our laboratory have suggested that in addition to ptsG, manXYZ, and yigL, there are other mRNAs whose levels are altered upon SgrS induction (M. Bobrovskyy, G. Richards, D. Balasubramanian, and C. K. Vanderpool, unpublished data). While we do not yet know which (if any) of these are direct targets of SgrS, we wondered whether the sgrS mutations we constructed might affect recovery from stress by altering regulation of other targets. To address this issue, we tested whether cell growth could be rescued (in LB with αMG) by specific downregulation only of ptsG by a mechanism not dependent on SgrS. To this end, a Plac promoter was inserted upstream of ptsG on the chromosome to control its expression at the level of transcription. By removing the inducer IPTG, new synthesis of PtsG (EIICBGlc) protein could be stopped via turning off ptsG transcription in a manner that is independent of SgrS function. If stopping new synthesis of PtsG were sufficient for recovery from stress, we expected that a ΔsgrS mutant in which ptsG transcription had been turned off (ΔsgrS Plac-ptsG mutant [Fig. 3A]) would show better growth than its ΔsgrS parent with ptsG under the control of the native promoter (ΔsgrS mutant [Fig. 3A]). Cells that were wild type for sgrS grew well under stress conditions, regardless of the transcriptional control of ptsG (Fig. 3A, compare the wild type to Plac-ptsG mutant). This indicated that in the presence of SgrS, growth recovery under αMG stress conditions was not significantly affected by turning off transcription of Plac-ptsG (by removal of IPTG). In contrast, in the ΔsgrS mutant background, cells with Plac-ptsG recovered significantly better than cells expressing ptsG from its native promoter (Fig. 3A, compare ΔsgrS strain to ΔsgrS Plac-ptsG strain). This result suggested that in the absence of SgrS, cell growth in the presence of αMG can be rescued by turning off new PtsG synthesis by an SgrS-independent mechanism, i.e., stopping ptsG transcription. On the other hand, when IPTG was persistently present in the culture media, strains harboring Plac-ptsG strains grow worse under stress than their corresponding parental strains with ptsG expressed from its native promoter (Fig. 3B), presumably because the Plac promoter drives ptsG overexpression, resulting in higher αMG-6-phosphate accumulation. Collectively, these results are consistent with the idea that inhibition of PtsG synthesis is one of the primary adaptive effects mediated by SgrS under αMG stress conditions in rich medium.
Fig 3.
Regulation of ptsG by SgrS is crucial for recovery from αMG-induced stress. Strains were grown in LB medium overnight and then subcultured 1:200 in fresh medium, both in the presence of 0.1 mM IPTG. Cells were harvested at an optical density at 600 nm of ∼0.1 by filtration, washed, and resuspended in fresh medium with 0.5% αMG and in the absence (A) or presence (B) of 0.1 mM IPTG. Growth of all cultures was monitored by OD600 throughout the whole procedure, but only the measurements following resuspension of cells are reported in the graphs. Results shown are representative of at least three independent trials.
SgrS-mediated regulation of ptsG and yigL, but not manXYZ, is required for recovery from αMG stress in certain minimal media.
We reported previously that sgrS mutant strains have more pronounced growth defects when glucose-phosphate stress is induced in minimal medium compared with rich medium (23). Therefore, we were interested in determining how regulation of different SgrS targets contributed to growth recovery in the more stringent stress induced in minimal medium. To address this, we tested growth competition between wild-type and sgrS mutant strains grown with αMG as the stressor in minimal MOPS medium with glycerol (Table 5). (We also performed competition assays in minimal MOPS medium with fructose and αMG, and found that, as for competitions in LB with αMG, regulation of ptsG mRNA was crucial, whereas regulation of manXYZ and yigL appeared dispensable [Table 3; data not shown].) In minimal MOPS medium with glycerol, the attB::tet marker had no effect on growth without or with αMG (Table 5), and all strains competed evenly in the absence of the stressor (Table 5, without αMG). With αMG, both the ΔsgrS mutant (CI = 0.38 [Table 5]) and the sgrS1manXYZ mutant (CI = 0.42 [Table 5]) were at a significant growth disadvantage compared to the wild-type cells, whereas they competed evenly with one another (CI = 1.01 [Table 5]). In addition, the ΔmanXYZ mutant (CI = 1.02 [Table 6]) and the ΔmanXYZ ΔsgrS mutant (CI = 0.97 [Table 6]) competed equally with their respective parent strains during growth in minimal medium with αMG. These results (together with those in Tables 3 and 4) are consistent with the notion that the regulatory action of SgrS on manXYZ does not play a significant role in the response to αMG regardless of the nutrient content of the growth medium. In contrast, the competitiveness of strains with sgrS28ptsG was very different between rich and minimal media containing glycerol and αMG. The sgrS28ptsG mutant was severely attenuated in competition with the wild-type strain in minimal medium containing glycerol and αMG (CI = 0.34 [Table 5]). In fact, under these severe stress conditions, the sgrS28ptsG mutant competed evenly with the ΔsgrS mutant (CI = 1.11 [Table 5]) and the sgrS1manXYZ mutant (CI = 1.02 [Table 5]). On the other hand, under the same conditions, the ΔptsG mutant outcompeted the wild-type strain (CI = 2.87 [Table 6]), and the ΔptsG ΔsgrS mutant grew much better than its ΔsgrS parent strain (CI = 2.23 [Table 6]), suggesting that blocking αMG uptake by eliminating its major transporter PtsG can protect cells from αMG stress in minimal medium with glycerol. We interpret these results to mean that when cells are stressed while growing in minimal medium containing glycerol and αMG, regulating ptsG is not the only crucial contribution of SgrS to the stress response, and regulation of other targets is also required.
Table 5.
Competition assays to measure the effects of sgrS mutations on growth with αMG in minimal MOPS medium with glycerol
| E. coli strain A genotype | E. coli strain B genotype | Without αMG |
With 0.5% αMG |
||
|---|---|---|---|---|---|
| CIa | P valueb | CI | P value | ||
| λattB::tet | WT | 0.94 ± 0.04 | NS | 1.04 ± 0.06 | NS |
| ΔsgrS λattB::tet | WT | 0.98 ± 0.1 | NS | 0.38 ± 0.11 | <0.0001 |
| sgrS1manXYZ λattB::tet | WT | 1.03 ± 0.12 | NS | 0.42 ± 0.06 | 0.014 |
| sgrS28ptsG λattB::tet | WT | 0.96 ± 0.3 | NS | 0.34 ± 0.02 | 0.022 |
| sgrS26ptsG,yigL λattB::tet | WT | 0.92 ± 0.07 | NS | 0.5 ± 0.03 | 0.015 |
| sgrS1manXYZ λattB::tet | ΔsgrS | 0.96 ± 0.13 | NS | 1.01 ± 0.19 | NS |
| sgrS26ptsG,yigL λattB::tet | ΔsgrS | 1.1 ± 0.51 | NS | 1.86 ± 0.64 | 0.023 |
| sgrS28ptsG λattB::tet | ΔsgrS | 0.93 ± 0.21 | NS | 1.11 ± 0.16 | NS |
| sgrS26ptsG,yigL λattB::tet | sgrS1manXYZ | 1.03 ± 0.06 | NS | 1.75 ± 0.13 | 0.019 |
| sgrS28ptsG λattB::tet | sgrS1manXYZ | 0.91 ± 0.12 | NS | 1.02 ± 0.1 | NS |
| sgrS26ptsG,yigL λattB::tet | sgrS28ptsG | 1.03 ± 0.11 | NS | 2.09 ± 0.25 | 0.02 |
See Materials and Methods for detailed procedures. The competition index (CI) is calculated as follows: (log10 strain A output/log10 strain B output)/(log10 strain A input/log10 strain B input). The data presented are the averages ± standard deviations of three independent experiments.
Student's t test was used to compare the output and inoculum. NS, not significant (P ≥ 0.05).
Table 6.
Competition assays to measure the effects of mutations in the three sgrS targets on growth with αMG in minimal MOPS medium with glycerol
| E. coli strain A genotype | E. coli strain B genotype | Without αMG |
With 0.5% αMG |
||
|---|---|---|---|---|---|
| CIa | P valueb | CI | P value | ||
| ΔptsG λattB::tet | WT | 1.02 ± 0.15 | NS | 2.87 ± 0.12 | 0.0008 |
| ΔmanXYZ λattB::tet | WT | 0.98 ± 0.11 | NS | 1.02 ± 0.008 | NS |
| ΔsgrS ΔptsG λattB::tet | ΔsgrS | 0.97 ± 0.02 | NS | 2.23 ± 0.46 | 0.04 |
| ΔsgrS ΔmanXYZ λattB::tet | ΔsgrS | 0.97 ± 0.12 | NS | 0.97 ± 0.07 | NS |
| ΔyigL λattB::tet | WT | 1.01 ± 0.02 | NS | 0.26 ± 0.06 | 0.031 |
| ΔyigL λattB::tet | ΔsgrS | 0.98 ± 0.04 | NS | 1.0 ± 0.11 | NS |
See Materials and Methods for detailed procedures. The competition index (CI) is calculated as follows: (log10 strain A output/log10 strain B output)/(log10 strain A input/log10 strain B input). The results presented here are the averages ± standard deviations of three independent experiments.
Student's t test was used to compare the output and inoculum. NS, not significant (P ≥ 0.05).
It was recently reported that yigL also plays a critical role in the glucose-phosphate stress response (38). Consistently, we found that a ΔyigL mutant growing in competition with its wild-type parent in minimal MOPS medium containing glycerol and αMG was at a significant disadvantage (CI = 0.26 [Table 6]). Under these conditions, the ΔyigL mutant competed evenly with the ΔsgrS mutant (CI = 1.0 [Table 6]), implying that yigL plays an essential role in recovery from αMG stress in minimal medium with glycerol. This result led us to hypothesize that cells expressing sgrS28ptsG fail to grow well in minimal medium containing glycerol and αMG because SgrS28ptsG cannot regulate yigL. We tested this hypothesis—that SgrS regulation of both ptsG and yigL is required for growth recovery during αMG stress in minimal medium—by competing a strain expressing sgrS26ptsG,yigL, which expresses SgrS capable of regulating ptsG and yigL (Fig. 2A and B) with sgrS mutant strains. Consistent with our hypothesis, the sgrS26ptsG,yigL strain outcompeted the ΔsgrS (CI = 1.86 [Table 5]), sgrS28ptsG (CI = 2.09 [Table 5]), and sgrS1manXYZ (CI = 1.75 [Table 5]) mutants. These results strongly suggest that SgrS regulation of ptsG and yigL together protects cells from αMG stress under nutrient-poor conditions. However, while necessary, regulation of these two targets alone was not sufficient to promote full growth recovery, since the sgrS26ptsG,yigL mutant still displayed a growth deficit in competition with the wild-type strain (CI = 0.5 [Table 5]). More broadly, these results suggest that the importance of regulating different subsets of mRNA targets varies depending on specific stress conditions.
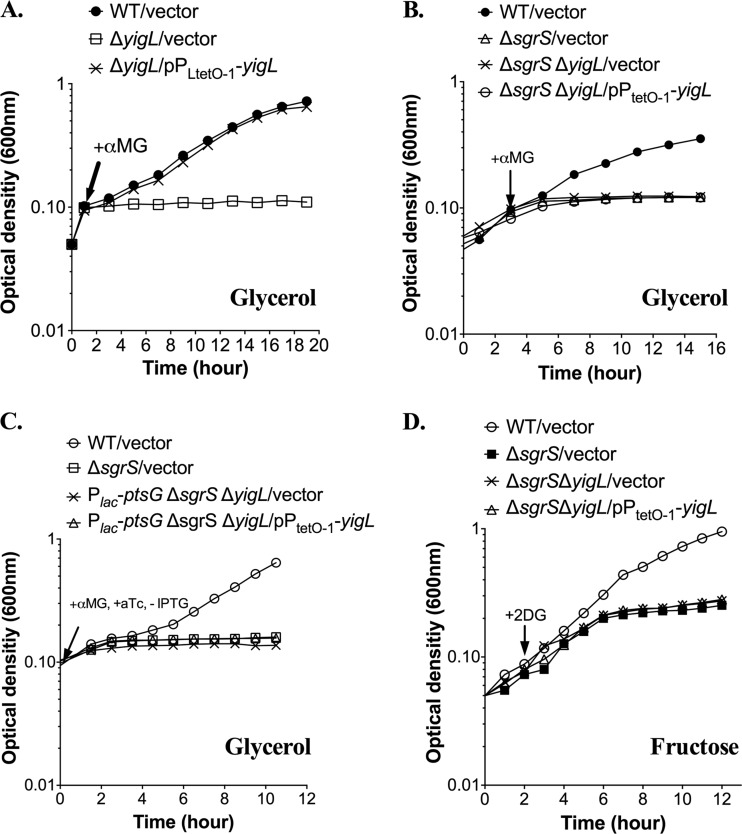
To further test the contributions of ptsG and yigL regulation to recovery from αMG-induced stress, we again employed the ΔsgrS Plac-ptsG strain, where the expression of ptsG can be manipulated in an SgrS-independent fashion. In this strain background, yigL was deleted from the chromosome and expressed in trans from the inducible PLtetO-1 promoter. Strains were grown and stressed in minimal MOPS medium supplemented with glycerol and αMG. The ΔyigL strain carrying a vector control or the PLtetO-1-yigL plasmid behaved as expected: the yigL mutant was immediately and strongly inhibited by the addition of αMG to cultures grown on minimal MOPS medium with glycerol (Fig. 4A), and induction of the plasmid-borne copy of yigL restored a wild-type pattern of growth to the ΔyigL mutant, confirming that the plasmid complements the yigL growth defect in an sgrS+ host (Fig. 4A). The ΔsgrS ΔyigL double mutant experienced immediate growth inhibition similar to the ΔsgrS and ΔyigL parent strains (Fig. 4B). However, yigL carried on a plasmid failed to restore growth during stress in the ΔsgrS ΔyigL double mutant background (Fig. 4B), confirming the importance of SgrS-mediated regulation of targets in addition to yigL under these conditions.
Fig 4.
SgrS-mediated regulation of multiple targets, including ptsG, yigL, and additional targets, is required for recovery from αMG-induced stress. (A and B) Strains were grown in minimal MOPS medium supplemented with 0.4% glycerol in the presence of 25 ng/ml aTc to an OD600 of ∼0.1 and then exposed to 0.5% αMG. (C) Strains were grown overnight in minimal MOPS medium supplemented with 0.4% glycerol and 25 μg/ml kanamycin and then subcultured 1:200 in fresh medium, both in the presence of 0.1 mM IPTG. aTc (25 ng/ml) was also present in all the subcultures. Cells were harvested at an OD600 of ∼0.1 by filtration, washed, and resuspended in fresh medium with 0.5% αMG and 25 ng/ml aTc. Growth of all cultures was monitored by OD600 throughout the whole procedure, but only the measurements following resuspension of cells were reported in the graphs. (D) Strains were grown in minimal MOPS medium supplemented with 0.2% fructose in the presence of 25 ng/ml aTc to an OD600 of ∼0.1 and then exposed to 0.5% αMG. All results are representative of at least three independent experimental trials.
By controlling ptsG transcriptional repression and yigL induction, independent of SgrS, we further validated the results of competition assays (sgrS26ptsG,yigL [Table 5]), suggesting that regulation of these two targets is necessary but not sufficient for growth rescue during αMG stress in minimal medium. Wild-type cells recovered from αMG stress, whereas ΔsgrS cells were severely growth inhibited (Fig. 4C). The growth of ΔsgrS ΔyigL Plac-ptsG cells carrying the vector control was similarly inhibited, even though ptsG transcription was turned off (by removal of IPTG), validating the results of growth competition experiments (sgrS28ptsG [Table 5]) that showed that repression of ptsG alone did not provide a growth advantage in minimal medium with αMG. Introduction of the PLtetO-1-yigL plasmid in the ΔsgrS ΔyigL Plac-ptsG strain with simultaneous regulation of yigL and ptsG (by addition or removal of the appropriate inducers) did not rescue cells from stress caused by αMG (Fig. 4C).
SgrS-mediated regulation of manXYZ mRNA becomes crucial under different stress conditions.
The experiments described so far establish that when glucose-phosphate stress is induced by αMG, the regulatory action of SgrS on ptsG plays a prominent role in stress recovery, whereas regulation of manXYZ does not contribute to growth (Tables 3, 4, and 5 and Fig. 3). These results are consistent with the substrate preferences of these two PTS transporters: EIICBGlc (PtsG) is more specific for αMG, and EIIABCDMan has higher specificity for the glucose analog 2-deoxyglucose (2DG) (28, 29). We previously demonstrated that PtsG plays a bigger role in induction of the stress response, as measured by increased sgrS transcription, when cells are exposed to αMG, whereas ManXYZ is required for induction in response to 2DG (5, 10, 19). We therefore theorized that regulation of manXYZ by SgrS would be important for growth recovery during 2DG-induced stress. To test this hypothesis, growth competition was performed with wild-type and sgrS mutant strains stressed in minimal MOPS medium with fructose (Table 7), because stress and growth inhibition of E. coli cells were previously observed under this condition (30). With 2DG, the ΔsgrS mutant was at a significant growth disadvantage in competition with the wild-type strain (CI = 0.46 [Table 7]), highlighting the crucial role of SgrS in mitigating 2DG-induced stress. Interestingly, the sgrS1manXYZ mutant, which specifically regulates manXYZ, but not ptsG or yigL (Fig. 2), competed equally with the wild-type strain (CI = 1.03 [Table 7]) and outcompeted the ΔsgrS mutant (CI = 2.25 [Table 7]). In contrast, the sgrS28ptsG strain (regulation of ptsG but not manXYZ or yigL) was at a growth disadvantage compared with both the wild type (CI = 0.68 [Table 7]) and the sgrS1manXYZ strain (CI = 0.59 [Table 7]). (All strains competed evenly in the absence of stress, and the selective marker [attB::tet] did not affect growth with or without 2DG [Table 7].) Similar results were observed when glycerol was used as the sole carbon source (data not shown). Collectively, these data indicate that, as predicted, regulation of manXYZ by SgrS becomes essential when the stressor is 2DG, a ManXYZ substrate, whereas regulation of ptsG and yigL does not contribute to growth recovery under these conditions.
Table 7.
Competition assays to measure the effects of sgrS mutations on growth with 2DG in minimal MOPS medium with fructose
| E. coli strain A genotype | E. coli strain B genotype | Without 2DG |
With 0.5% 2DG |
||
|---|---|---|---|---|---|
| CIa | P valueb | CI | P value | ||
| λattB::tet | WT | 0.84 ± 0.11 | NS | 0.84 ± 0.3 | NS |
| ΔsgrS λattB::tet | WT | 1.09 ± 0.14 | NS | 0.46 ± 0.1 | 0.023 |
| sgrS1manXYZ λattB::tet | WT | 0.97 ± 0.16 | NS | 1.03 ± 0.13 | NS |
| sgrS28ptsG λattB::tet | WT | 1.00 ± 0.15 | NS | 0.68 ± 0.04 | 0.01 |
| sgrS1manXYZ λattB::tet | ΔsgrS | 0.98 ± 0.22 | NS | 2.25 ± 0.59 | 0.015 |
| sgrS28ptsG λattB::tet | ΔsgrS | 0.96 ± 0.11 | NS | 1.12 ± 0.34 | NS |
| sgrS28ptsG λattB::tet | sgrS1manXYZ | 1.01 ± 0.06 | NS | 0.59 ± 0.08 | 0.019 |
See Materials and Methods for detailed procedures. The competition index (CI) is calculated as follows: (log10 strain A output/log10 strain B output)/(log10 strain A input/log10 strain B input). The results presented here are the averages ± standard deviations of three independent experiments.
Student's t test was used to compare the output and inoculum. NS, not significant (P ≥ 0.05).
Consistent with the observations described above, the ΔmanXYZ mutant outcompeted the wild-type strain (CI = 2.04 [Table 8]), and the ΔmanXYZ ΔsgrS mutant had a growth advantage over its ΔsgrS parent (CI = 1.90 [Table 8]) during growth with 2DG. Both the ΔptsG (CI = 0.98) and ΔptsG ΔsgrS (CI = 1.08) mutant strains competed evenly with their respective parent strains under the same conditions (Table 8), which is consistent with the notion that PtsG does not contribute significantly to 2DG uptake. In addition, the ΔyigL mutant competed well against the wild-type strain (CI = 0.92 [Table 8]), indicating that YigL is unlikely to play a significant role in the cellular response to 2DG under these conditions. We further tested the effect of YigL on growth with 2DG in minimal media, using the PtetO-1-yigL+ plasmid. While the wild-type strain recovered from 2DG stress, the plasmid-borne copy of YigL failed to improve the growth of the ΔsgrS ΔyigL mutant in the presence of 2DG (Fig. 4D). Together, these results strongly suggested that recovery from stress induced by 2DG requires SgrS-mediated regulation of manXYZ, whereas regulation of ptsG and yigL is dispensable for the response to 2DG.
Table 8.
Competition assays to measure the effects of mutations in the three sgrS targets on growth with 2DG in minimal MOPS medium with fructose
| E. coli strain A genotype | E. coli strain B genotype | Without 2DG |
With 0.5% 2DG |
||
|---|---|---|---|---|---|
| CIa | P valueb | CI | P value | ||
| ΔptsG λattB::tet | WT | 1.03 ± 0.15 | NS | 0.98 ± 0.008 | NS |
| ΔmanXYZ λattB::tet | WT | 0.99 ± 0.06 | NS | 2.04 ± 0.12 | 0.003 |
| ΔsgrS ΔptsG λattB::tet | ΔsgrS | 0.96 ± 0.14 | NS | 1.08 ± 0.07 | NS |
| ΔsgrS ΔmanXYZ λattB::tet | ΔsgrS | 1.02 ± 0.11 | NS | 1.90 ± 0.27 | 0.04 |
| ΔyigL λattB::tet | WT | 0.97 ± 0.10 | NS | 0.92 ± 0.08 | NS |
See Materials and Methods for detailed procedures. The competition index (CI) is calculated as follows: (log10 strain A output/log10 strain B output)/(log10 strain A input/log10 strain B input). The results presented here are the averages ± standard deviations of three independent experiments.
Student's t test was used to compare the output and inoculum. NS, not significant (P ≥ 0.05).
Nutrient supplementation in minimal media improves growth during GP stress.
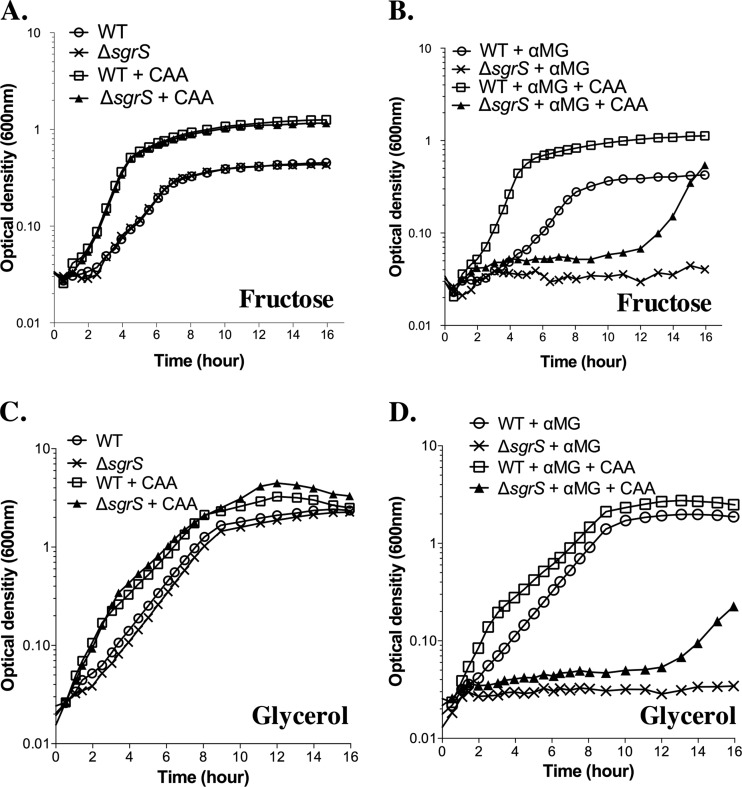
Our results so far have demonstrated that variations in nutrient content of the growth medium can influence glucose-phosphate (GP) stress-associated growth phenotypes, as well as the requirements for the regulatory activities of SgrS. When growing in LB medium, E. coli cells use amino acids as the carbon source (31, 32), whereas under our minimal medium growth conditions, cells were given glycerol or fructose as a carbon source and had to synthesize their own amino acids. Another study from our laboratory (an accompanying article [33]) revealed that one underlying cause of glucose-phosphate stress is depletion of central glycolytic metabolites. Given this, we reasoned that E. coli cells may be more stressed during growth in minimal media compared to rich media, because in minimal media, already low pools of central metabolites would be further reduced because of the need to draw on these metabolites for precursors of amino acid biosynthesis. To investigate whether supplementation of amino acids reduced the severity of glucose-phosphate stress-associated growth inhibition, we compared growth of wild-type and sgrS mutant strains in minimal media with fructose or glycerol in the presence and absence of Casamino Acids (CAA) (Fig. 5). As expected, CAA supplementation enhanced the growth rates of wild-type and mutant strains in these minimal media in the absence of stress (Fig. 5A and C). Similarly, in both media, the presence of CAA improved the growth of wild-type cells stressed with αMG (Fig. 5B and D, compare the wild type [WT] plus αMG to the WT plus αMG plus CAA). The growth improvement conferred by CAA on the stressed wild-type strain resembled that observed for nonstressed cells, suggesting that the growth potential of wild-type cells stressed in minimal medium is not drastically limited by depleted pools of central metabolites. We expected this because wild-type cells induce SgrS to reduce αMG uptake and subsequent metabolite depletion. The sgrS mutant strain, on the other hand, is much more severely inhibited by αMG, yet added CAA also improved growth of this strain (Fig. 5B and D, compare the ΔsgrS mutant plus αMG to the ΔsgrS mutant plus αMG plus CAA), albeit after a much longer lag (approximately 12 h following exposure of the cells to αMG). These results are consistent with the idea that at least one factor accounting for lack of growth of sgrS mutant cells stressed in nutrient-poor conditions is a lack of central metabolites available to divert to amino acid biosynthesis. These results provide a rationale for the less severe growth inhibition experienced by cells growing in rich media containing amino acids compared with cells growing in minimal media.
Fig 5.
Supplementation with Casamino Acids improves growth during glucose-phosphate stress. Strains were grown in minimal MOPS medium supplemented with 0.2% fructose (A and B) or 0.4% glycerol (C and D). In addition, 0.5% αMG and/or 0.1% Casamino Acids (CAA) were present in the media as indicated. Results shown are representative of at least three independent experimental trials.
DISCUSSION
In recent years, hundreds of novel sRNAs have been identified in E. coli, Salmonella, and many other bacterial species. However, SgrS is one of only a few base pairing sRNA regulators for which we have detailed knowledge concerning its regulation, targets, and perhaps most importantly, a clearly associated growth phenotype. These features make SgrS an excellent model for unraveling molecular mechanisms of sRNA-mediated coordinate regulation of multiple targets and unifying these mechanisms with their physiological relevance. While many studies have demonstrated that Hfq-dependent sRNAs regulate multiple mRNA targets (34–36), how regulation of individual targets or target subsets contributes specifically to growth physiology under different conditions has not been well studied. Here, we began to investigate this issue by studying the physiological impact of SgrS regulation of its multitarget regulon.
We report some of the first evidence supporting the idea that coordinated regulation of multiple genes in an sRNA's regulon directly contributes to cell growth potential during stress. Identification of SgrS mutants with altered target specificities allowed us to assess the importance of SgrS-mediated regulation of different targets under a variety of conditions. One stress variable that we manipulated was the stress-inducing phosphosugar. The two sugars we used, αMG and 2DG, are both glucose analogs, but are taken up via distinct PTS transporters, PtsG and ManXYZ, respectively (30, 37). We found that when αMG was the stressor, SgrS regulation of ptsG was crucial for continued growth, both in the context of growth competition (Table 3) and when strains were growing in pure culture (Fig. 3). In contrast, regulation of manXYZ by SgrS conferred no growth advantage during αMG-induced stress (Table 3). On the other hand, when cells were stressed by uptake of 2DG, regulation of manXYZ by SgrS was crucial, whereas regulation of ptsG was dispensable (Table 7). These specific results track with the known substrate specificities of the two PTS transporters and make perfect biological sense based on what we know about the glucose-phosphate stress response. However, prior to our study, it had not been demonstrated that regulation of different subsets of sRNA target genes could allow cells to respond effectively to changing stress conditions. Thus, our experimental approaches and results have helped to shed light on a broader issue: how sRNA-mediated regulation of multiple mRNA targets can provide a flexible stress response that promotes optimal cell physiology under fluctuating environmental conditions.
Changing the nutrients available to cells by culturing in rich or minimal media allowed us to discern that factors other than the sugar stressor can modulate glucose-phosphate stress-associated growth phenotypes. Cells stressed with αMG show different patterns of growth depending on the nutrient content of the medium. In rich (LB) medium, wild-type and sgrS mutant cells continue growing for ∼2 generations after αMG exposure. After that, growth of wild-type cells is unaffected, while sgrS mutant growth slows dramatically (5) (Fig. 3). With plentiful nutrients available in LB, restoring regulation of a single target, ptsG mRNA, was sufficient to rescue growth of sgrS mutant cells (Table 3 and Fig. 3), suggesting that simply reducing αMG uptake relieves stress under these conditions. Results from another study in our laboratory showed that one important cause of glucose-phosphate stress-associated growth inhibition of sgrS mutant strains is depletion of glycolytic metabolites, including phosphoenolpyruvate (PEP) (33).We propose that in wild-type cells growing in nutrient-rich media, SgrS-mediated repression of PtsG synthesis reduces uptake of the nonmetabolizable sugar and consequently reduces PEP consumption by the PTS. This activity of SgrS allows cells to continue growing using the amino acids available in LB as a carbon source.
In contrast, when growing in minimal medium, both wild-type and sgrS mutant cells experience almost immediate inhibition after exposure to αMG (23) (Fig. 4B). Wild-type cells subsequently recover, but sgrS mutant growth remains inhibited (Fig. 4B). We postulate that reduced levels of central metabolites is more growth limiting in minimal medium (compared to rich medium), because cells have to draw on central metabolite pools for amino acid biosynthetic precursors. Restoring regulation of ptsG alone failed to rescue growth of sgrS mutant cells in minimal medium containing glycerol and αMG (Table 5 and Fig. 4). Regulation of both ptsG and yigL provided some relief of growth inhibition but failed to fully restore sgrS mutant growth to wild-type levels (Table 5 and Fig. 4). These observations indicate that both reducing αMG uptake (via repression of ptsG) and enhancing αMG efflux (via activation of the sugar phosphatase yigL mRNA, a prerequisite for efflux [38]) are necessary but not sufficient for the stress response in nutrient-poor conditions. Our results imply that other as-yet-uncharacterized SgrS target mRNAs must also be regulated for full recovery under these conditions. We speculate that these other SgrS target mRNAs may encode metabolic enzymes or regulators that help reroute metabolism in order to replenish these metabolites. Ongoing studies in our laboratory are testing this hypothesis.
Consistent with the idea that reduced levels of central metabolites (including amino acid biosynthetic precursors) are limiting for sgrS mutant growth (33), we found that supplementation of minimal media with amino acids mitigates growth inhibition associated with αMG stress (Fig. 5). We postulate that when stressed cells growing in minimal media are provided with exogenous amino acids, they are spared from utilizing the already limited central metabolites for amino acid biosynthesis. In addition, the amino acids may help rescue the growth of sgrS mutant cells by serving as the substrates for gluconeogenesis that allow cells to make more PEP and other upstream metabolites. The lag we observe in growth recovery of the αMG-stressed sgrS mutant provided with amino acids may reflect the time it takes for cells to replenish the limiting metabolites through gluconeogenesis.
This study provides insight into two separate aspects of the glucose-phosphate stress response. First, our demonstration that cells require SgrS regulation of different target subsets depending on the nature of the environmental conditions when stress is induced suggests that SgrS and perhaps other sRNAs have evolved to be flexible regulators that modulate expression of multigene regulons in order to allow cells to adapt to an array of related stress conditions. Second, our analysis here of growth and competitiveness of wild-type and mutant strains stressed under different nutritional conditions, combined with our other study (33) is fully consistent with our model that glucose-phosphate stress is caused by an imbalance of central metabolites. In sum, our work shows that the SgrS-mediated response to stress has three main components that vary in importance depending upon the nutrients available. The first arm of the stress response, repression of sugar transport protein synthesis, is all that is required if stress occurs in a nutrient-rich environment with available amino acids, perhaps because this reduces the drain on central metabolites for biosynthesis and provides cells a route to replenish these limiting metabolites. Under nutrient-poor conditions, the cell needs at least two additional SgrS-dependent functions in order to recover from stress—activation of sugar efflux (through activation of yigL and subsequent dephosphorylation and efflux of accumulated sugars [38]) and another unknown activity. These studies set the stage for future work aimed at answering two important questions. (i) What is the exact nature of metabolic defects of cells experiencing stress in different nutritional environments? (ii) What are the other SgrS-mediated cellular responses that are important for overcoming stress under nutrient-limiting conditions? Answering the first question will require detailed analyses of intracellular metabolite levels and changes in metabolic fluxes in response to glucose-phosphate stress. Resolving the second will involve identification of other members of the SgrS regulon and assessing how their regulation contributes to stress-associated growth phenotypes. Both of these are active areas of investigation in this laboratory, and we anticipate they will lead to new insights into the regulation of central metabolism as well as physiology of glucose-phosphate stress.
ACKNOWLEDGMENTS
We sincerely thank John Cronan for providing the plasmid pZE21. We also thank Jennifer Rice and Caryn Wadler for construction of several strains used in our study. We are grateful to James Imlay and James Slauch for fruitful discussions and Greg Richards, Divya Balasubramanian, and Chelsea Lloyd for critical reading of the manuscript and helpful comments, as well as to members of J. Slauch's laboratory and the members of C. K. Vanderpool's laboratory for moral support, useful discussions, and suggestions.
This work was supported by the University of Illinois at Urbana-Champaign and the American Cancer Society Scholar Grant (research scholar grant ACS2008-01868).
Footnotes
Published ahead of print 19 July 2013
REFERENCES
- 1.Deutscher J, Francke C, Postma PW. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimata K, Takahashi H, Inada T, Postma P, Aiba H. 1997. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 94:12914–12919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plumbridge J. 1998. Expression of ptsG, the gene for the major glucose PTS transporter in Escherichia coli, is repressed by Mlc and induced by growth on glucose. Mol. Microbiol. 29:1053–1063 [DOI] [PubMed] [Google Scholar]
- 4.Tanaka Y, Kimata K, Inada T, Tagami H, Aiba H. 1999. Negative regulation of the pts operon by Mlc: mechanism underlying glucose induction in Escherichia coli. Genes Cells 4:391–399 [DOI] [PubMed] [Google Scholar]
- 5.Vanderpool CK, Gottesman S. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54:1076–1089 [DOI] [PubMed] [Google Scholar]
- 6.Vanderpool CK. 2007. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 10:146–151 [DOI] [PubMed] [Google Scholar]
- 7.Kawamoto H, Koide Y, Morita T, Aiba H. 2006. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 61:1013–1022 [DOI] [PubMed] [Google Scholar]
- 8.Morita T, Mochizuki Y, Aiba H. 2006. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl. Acad. Sci. U. S. A. 103:4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rice JB, Balasubramanian D, Vanderpool CK. 2012. Small RNA binding site multiplicity involved in translational regulation of a polycistronic mRNA. Proc. Natl. Acad. Sci. U. S. A. 109:E2691–E2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice JB, Vanderpool CK. 2011. The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res. 39:3806–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Carmel I, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, Yakunin AF. 2006. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J. Biol. Chem. 281:36149–36161 [DOI] [PubMed] [Google Scholar]
- 12.Wadler CS, Vanderpool CK. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. U. S. A. 104:20454–20459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wadler CS, Vanderpool CK. 2009. Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res. 37:5477–5484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Englesberg E, Anderson RL, Weinberg R, Lee N, Hoffee P, Huttenhauer G, Boyer H. 1962. l-Arabinose-sensitive, l-ribulose 5-phosphate 4-epimerase deficient mutants of Escherichia coli. J. Bacteriol. 84:137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kukasawa T, Nikaido H. 1961. Galactose-sensitive mutants of Salmonella. II. Bacteriolysis induced by galactose. Biochim. Biophys. Acta 48:470–483 [DOI] [PubMed] [Google Scholar]
- 16.Pikis A, Hess S, Arnold I, Erni B, Thompson J. 2006. Genetic requirements for growth of Escherichia coli K12 on methyl-alpha-D-glucopyranoside and the five alpha-D-glucosyl-D-fructose isomers of sucrose. J. Biol. Chem. 281:179000–179008 [DOI] [PubMed] [Google Scholar]
- 17.Eichhorn MM, Cynkin MA. 1965. Microbial metabolism of 2-deoxyglucose; 2-deoxygluconic acid dehydrogenase. Biochemistry 4:159–165 [DOI] [PubMed] [Google Scholar]
- 18.Postma PW, Lengeler JW, Jacobson GR. 1993. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol. Rev. 57:543–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richards GR, Vanderpool CK. 2011. Molecular call and response: the physiology of bacterial small RNAs. Biochim. Biophys. Acta 1809:525–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massé E, Vanderpool CK, Gottesman S. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187:6962–6971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. 2011. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfa-dependent GcvB small RNA. Mol. Microbiol. 81:1144–1165 [DOI] [PubMed] [Google Scholar]
- 22.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Vanderpool CK. 2011. Regulation and function of Escherichia coli sugar efflux transporter A (SetA) during glucose-phosphate stress. J. Bacteriol. 193:143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ellermeier CK, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161 [DOI] [PubMed] [Google Scholar]
- 25.Le Roux F, Binesse J, Saulnier D, Mazel D. 2007. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel counterselectable suicide vector. Appl. Environ. Microbiol. 73:777–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu DG, Ellis HM, Lee EC, Jenkins NA, Poerland NG, Court DL. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim B, Richards SM, Gunn JS, Slauch JM. 2010. Protecting against antimicrobial effectors in the phagosome allows SodCII to contribute to virulence in Salmonella enterica serovar Typhimurium. J. Bacteriol. 192:2140–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stock JB, Waygood EB, Meadow ND, Postma PW, Roseman S. 1982. Sugar transport by the bacterial phosphotransferase system. The glucose receptors of the Salmonella typhimurium phosphotransferase system. J. Biol. Chem. 257:14543–14552 [PubMed] [Google Scholar]
- 29.Rephaeli AW, Saier MH., Jr 1980. Substrate specificity and kinetic characterization of sugar uptake and phosphorylation, catalyzed by the mannose enzyme II of the phosphotransferase system in Salmonella typhimurium. J. Biol. Chem. 255:8585–8591 [PubMed] [Google Scholar]
- 30.Amaral D, Kornberg HL. 1975. Regulation of fructose uptake by glucose in Escherichia coli. J. Gen. Microbiol. 90:157–168 [DOI] [PubMed] [Google Scholar]
- 31.Prüss BM. 1994. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol. Microbiol. 12:973–984 [DOI] [PubMed] [Google Scholar]
- 32.Sezonov G, Joseleau-Petit D, D'Ari R. 2007. Escherichia coli physiology in Luria-Bertani broth. J. Bacteriol. 189:8746–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards GR, Patel MV, Lloyd CR, Vanderpool CK. 2013. Depletion of glycolytic intermediates plays a key role in glucose-phosphate stress in Escherichia coli. J. Bacteriol. 195:4816–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vogel J, Luisi BF. 2011. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9:578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43:880–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sittka AS, Lucchini K, Papenfort K, Sharma CM, Rolle K, Binnewise TT, Hinton JC, Vogel J. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 4:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson PJ, Giddens RA, Jones-Mortimer MC. 1977. Transport of galactose, glucose and their molecular analogues by Escherichia coli K12. Biochem. J. 162:309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papenfort K, Sun Y, Miyakoshi M, Vanderpool CK, Vogel J. 2013. Small RNA-mediated activation of sugar phosphate mRNA regulates glucose homeostasis. Cell 153:426–437 [DOI] [PMC free article] [PubMed] [Google Scholar]