Abstract
The importance of gene regulation in the enzootic cycle of Borrelia burgdorferi, the spirochete that causes Lyme disease, is well established. B. burgdorferi regulates gene expression in response to changes in environmental stimuli associated with changing hosts. In this study, we monitored mRNA decay in B. burgdorferi following transcriptional arrest with actinomycin D. The time-dependent decay of transcripts encoding RNA polymerase subunits (rpoA and rpoS), ribosomal proteins (rpsD, rpsK, rpsM, rplQ, and rpsO), a nuclease (pnp), outer surface lipoproteins (ospA and ospC), and a flagellar protein (flaB) have different profiles and indicate half-lives ranging from approximately 1 min to more than 45 min in cells cultured at 35°C. Our results provide a first step in characterizing mRNA decay in B. burgdorferi and in investigating its role in gene expression and regulation.
INTRODUCTION
Lyme disease is caused by the bacterium Borrelia burgdorferi, a spirochete that cycles between Ixodes ticks and vertebrate hosts in nature. B. burgdorferi is acquired by a larval tick when the tick feeds on an infected vertebrate host. After acquiring the bacteria, the larval tick will remain infected as it molts to the nymphal and adult stages, and it can transmit B. burgdorferi during subsequent blood meals. Tick feeding initiates movement of B. burgdorferi from the tick midgut to the salivary glands and, ultimately, into the new vertebrate host (reviewed in reference 1). The incidence of Lyme disease in the United States has increased dramatically in the last decade, with 30,158 cases reported in 2010 (2).
Successful transmission from the tick vector to the vertebrate host depends on B. burgdorferi's ability to survive the transition between two very distinct environments, which requires a dramatic shift in gene expression pattern in this pathogen (reviewed in references 1 and 3). To better understand the life cycle of this spirochete and its virulence, much of the work on gene regulation in B. burgdorferi has been aimed at understanding how specific genes are affected by the environmental signals associated with switching hosts. mRNA levels are affected by temperature (215 genes) (4), tick feeding (24 genes) (5), and mammalian host factors (125 genes) (6). The alternative sigma factor RpoS is induced upon tick feeding (5) and is required for mammalian infection (7). Artificial induction of rpoS results in changes in transcript levels of 137 genes (8). Controlling gene expression is essential for B. burgdorferi to complete its enzootic life cycle. Regulation of mRNA levels in response to changes in the environmental stimuli associated with changing hosts depends on controlling transcription and is likely to depend upon the ability of the cell to degrade mRNA in a timely fashion. mRNA degradation in other bacteria is extensively regulated (reviewed in reference 9–13); however, mRNA degradation pathways have not yet been characterized in any Borrelia species.
B. burgdorferi is a slow-growing bacterium with generation times between 8 and 12 h at temperatures between 33 and 37°C in liquid media (14). mRNA decay rates in other bacterial species generally appear to be independent of doubling time (reviewed in reference 15). The average lifetime of mRNA in other bacteria is typically on the order of a few minutes (reviewed in reference 15); however, B. burgdorferi may have some extremely stable transcripts. In B. burgdorferi cultures killed by the antibiotic ceftriaxone, mRNA fragments are detectable for 14 days posttreatment, 11 days after the cells can be successfully revived and subcultured (16).
Phylogenetic analysis (Table 1) (17, 18) suggests that this spirochete has only a subset of the nucleases involved in mRNA degradation that are involved in mRNA decay pathways in either Bacillus subtilis or Escherichia coli (for recent reviews, see references 12, 13, 15, and 19–21). The limited subset of ribonucleases present in B. burgdorferi may influence degradation pathways and the persistence of mRNA transcripts.
Table 1.
Comparison of ribonucleases found in Escherichia coli, Bacillus subtilis, and Borrelia burgdorferia
| Nuclease | Presence in: |
Locus | ||
|---|---|---|---|---|
| E. coli | B. subtilis | B. burgdorferi | ||
| RNase E or G | + | − | − | − |
| RNase III | + | + | + | BB0705 |
| RNase II | + | − | − | − |
| RNase M5 | − | + | + | BB0626 |
| RNase Y | − | + | + | BB0504 |
| RNase J1 | − | + | +/− | BB0533 |
| RNase J2 | − | + | − | − |
| RNase Z | + | + | + | BB0755 |
The plus symbol indicates that a gene was previously annotated (18) and/or identified through performing a translated nucleotide database search using an E. coli K-12 (57) or B. subtilis 168 (58) query in tBLASTn (59), performed at the Comprehensive Microbial Resource website (60). The minus symbol indicates that no homolog was detected (RNase E, RNase II, and RNase J2). B. subtilis RNase J1 (555 amino acids) and B. burgdorferi B31 PhnP (762 amino acids) share homology of 124 amino acids (BLAST score, 73; identities, 31/124; positives, 54/124), indicated by +/−.
B. burgdorferi is naturally resistant to rifampin, the antibiotic that is used to arrest transcription in most studies of prokaryotic mRNA decay. Rifampin interacts with the beta subunit of RNA polymerase in the RNA exit channel, preventing transcript growth beyond 2 or 3 nucleotides (22). Schwartz and coworkers first determined that the sequence of the B. burgdorferi rpoB gene differs from E. coli's wild-type sequence at positions that are hot spots for rifampin-resistant mutations in E. coli and Mycobacterium tuberculosis (amino acid positions 500, 508, 518, 522, 531, 539, and 552 using E. coli numbering) (23).
In this study, we used actinomycin D as a transcription inhibitor to enable measurement of mRNA half-life (t1/2) in B. burgdorferi. The polypeptide antibiotic actinomycin D was used in 1962 to study mRNA turnover in B. subtilis (24) and is currently used extensively to arrest transcription in eukaryotes and archaea (25–28). We measured mRNA half-lives at 35°C for a variety of genes in B. burgdorferi, including genes that are differentially expressed during the enzootic cycle. These studies are the first to examine mRNA decay in B. burgdorferi and demonstrate diverse mRNA decay profiles. Our studies are a first step in characterizing mRNA decay mechanisms in this important pathogen.
MATERIALS AND METHODS
Borrelia burgdorferi strains and culture conditions.
B. burgdorferi strains A3-LS-flacp-ospC (29) and 297-LK-flacp-rpoS (8), in which either the ospC or rpoS gene, respectively, is under the control of an inducible lac promoter system, were a kind gift of Michael Gilbert. B. burgdorferi B31A, a high-passage strain, was the kind gift of Scott Samuels and Laura Hall. B. burgdorferi was grown at 35°C and 5% CO2 in Barbour-Stoenner-Kelley II (BSK-II) complete medium (in 6% rabbit serum) (Pel Freeze; Rogers, AR) (30) supplemented with the strain-appropriate antibiotics: A3-LS-flacp-ospC with 200 μg ml−1 kanamycin (Fisher Scientific, Fairlawn, NJ) and 50 μg ml−1 streptomycin (Fisher Scientific, Fairlawn, NJ), 297-LK-flacp-rpoS with 200 μg ml−1 kanamycin and 40 μg ml−1 gentamicin (Fisher Scientific, Fairlawn, NJ), and B31A with 25 μg ml−1 rifampin (Sigma-Aldrich, St. Louis, MO). Rifampin (Sigma-Aldrich, St. Louis, MO) was used with B31A cultures to prevent contamination by other bacteria. When used, the actinomycin D (TOCRIS Bioscience, Bristol, United Kingdom) concentration in cultures was 150 μg ml−1, and isopropyl β-d-1-thiogalactopyranoside (IPTG) (Fisher Scientific, Fairlawn, NJ) was added at 1 mM. Cultures were grown to mid-log phase (1 × 107 to 7 × 107 cells ml−1) before the start of each experiment (e.g., addition of actinomycin D or IPTG). Cell densities were determined using a Petroff-Hausser counting chamber under dark-field microscopy.
RNA isolation.
For RNA isolation, aliquots of 25 to 30 ml of B. burgdorferi culture, containing between 2.75 × 108 and 8.25 × 109 cells, were centrifuged at 4°C and 5,000 × g for 10 min, resuspended in 700 μl RLT buffer (Qiagen, Valencia, CA), placed in sterile 2-ml screw-cap tubes with approximately 0.25-ml sterile zirconia beads, and then subjected to shaking at 30 Hz for 5 min in a Qiagen tissue lyser II. RNA was purified using an RNeasy mini kit according to the manufacturer's protocol (Qiagen, Valencia, CA). RNA samples were treated with Turbo DNA-free DNase I (Applied Biosystems/Life Technologies, Grand Island, NY) by following the manufacturer's protocol. RNA concentration and quality were determined spectrophotometrically. Samples were diluted in nuclease-free water to a standard concentration of total RNA (usually 20 ng μl−1) and were stored at −20°C.
qRT-PCR.
RNA, purified form B. burgdorferi, was quantified with one-step reverse transcription and real-time quantitative PCR (qRT-PCR) using the EXPRESS one-step SYBR green ER kit (Invitrogen, Carlsbad, CA) on a Stratagene MX3000P thermocycler (Agilent Technologies, Inc., Wilmington, DE) using primers that amplify our genes of interest (see Table S1 in the supplemental material). qRT-PCRs were carried out according to the manufacturer's protocol using 40 ng total RNA per 20-μl reaction mixture unless otherwise specified. The absence of contaminating DNA was confirmed by performing PCR in the absence of reverse transcriptase. Cycle threshold (CT) values are the number of cycles required during amplification for the fluorescent signal to exceed background fluorescence levels and were obtained using the default settings in MxPro-Mx200P v4.10 software (Agilent Technologies, Inc., Wilmington, DE) in the Stratagene Mx3000P thermocycler. Efficiencies of the qRT-PCR for each primer set were determined using total RNA in a dilution series across two orders of magnitude. Efficiencies ranged from 87 to 120%.
Data analysis.
To determine the fraction of mRNA for a gene of interest that remains following antibiotic treatment, posttreatment CT values were compared to reference CT values (i.e., CTref) for the same transcript for RNA purified from the same cultures prior to antibiotic addition or from a control culture grown without antibiotic treatment. The fraction of remaining RNA (f) was measured as f = 2(CTref − CT), where CTref is the CT value determined for the mRNA from an antibiotic-free culture and CT is the value for the mRNA purified from culture at a given time after antibiotic addition. To determine the rate constant and half-life for each mRNA, the fraction of remaining RNA was plotted against time points from 0 to 45 min, and the data were globally fit using Sigma Plot 12.0 (Systat Software Inc.) to an equation for first-order decay, f = f0(e−kt), in which k is the first-order rate constant, f0 is the initial fraction of RNA, and t is time. Each rate constant was determined from a simultaneous global fit of data for all biological replicates and qRT-PCR replicates for each transcript. These fits assume that the rate constants are independent of the experimental variability present in the determination of CTref from individual samples.
RESULTS
To develop a method for studying mRNA decay in rifampin-resistant B. burgdorferi, we tested the effectiveness of actinomycin D in arresting transcription. We initially attempted to examine the effects of actinomycin D on total RNA synthesis by comparing levels of [3H]uridine incorporation in the presence and absence of actinomycin D (for example, see reference 31), but we observed very low levels of incorporation of [3H]uridine under all growth conditions (data not shown), preventing a direct measurement of the effect of actinomycin D on total RNA synthesis. We then tested whether actinomycin D could prevent induction of ospC from the IPTG-inducible flac promoter in A3-LS-flacp-ospC cells (29) that were cultured at 35°C. In these experiments, mid-log-phase cultures were treated with actinomycin D for 30 min prior to the addition of IPTG. B. burgdorferi cell density did not increase significantly following addition of actinomycin D, although cells retained motility for several hours after exposure at 35°C (data not shown). Three hours following actinomycin D treatment, RNA was purified from cultures, and ospC and flaB transcript levels were assayed by qRT-PCR using gene-specific primers (see Table S1 in the supplemental material). The amplification curves (Fig. 1) indicate that the fluorescence signal increases with the number of amplification cycles as expected. The amplification curve for ospC from cultures that lack actinomycin D and contain IPTG (Fig. 1A, open circles) are significantly shifted to a lower number of cycles relative to those obtained from control cultures lacking both actinomycin D and IPTG (closed circles), indicating that addition of IPTG increases ospC mRNA levels, as expected by transcriptional induction. For cultures treated with actinomycin D, there is little difference in the transcript levels resulting from the addition of IPTG (Fig. 1A, triangles), indicating that actinomycin D prevents induction of transcription of ospC from the flac promoter. In this strain, flaB is controlled by its natural promoter, which is not induced in the presence of IPTG. As expected, the flaB transcripts show no significant effect caused by the addition of IPTG for cultures where actinomycin D is present or absent (Fig. 1B).
Fig 1.
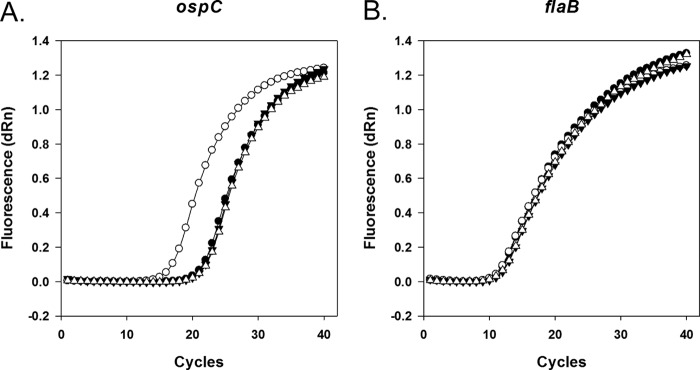
Actinomycin D prevents induction of transcription of ospC controlled by an IPTG-inducible promoter. Quantitative RT-PCR amplification curves are shown for ospC (A) and flaB (B) transcripts. A3-LS-flacp-ospC cultures at mid-log growth phase were split, and half of the initial culture was treated with actinomycin D (final concentration, 150 μg ml−1). Thirty minutes later, cultures were split again and treated with IPTG or used as controls lacking an inducer. Cells were cultured for an additional 3 h before RNA isolation and qRT-PCR amplification. ●, Lacking both actinomycin D and IPTG (CT ospC = 22.22; CT flaB = 12.42); ○, lacking actinomycin D but containing IPTG (CT ospC = 17.51; CT flaB = 12.30); ▼, containing actinomycin D but lacking IPTG (CT ospC = 22.57; CT flaB = 12.96); Δ, containing both actinomycin D and IPTG (CT ospC = 22.70; CT flaB = 12.87). Ten ng of total RNA was used in each reaction mixture. dRn, magnitude of the fluorescence signal over background fluorescence generated at each time point.
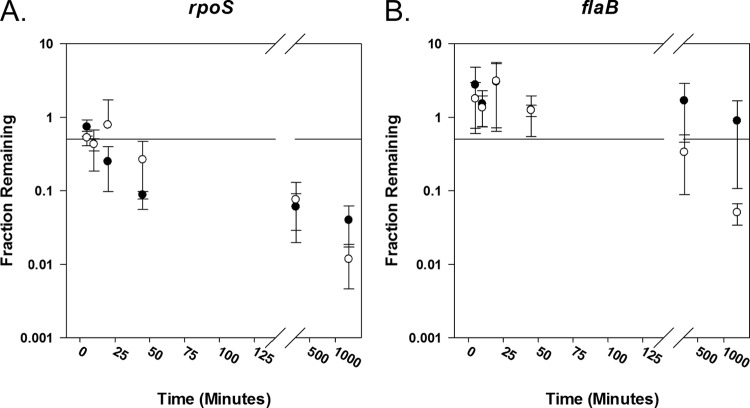
To assess whether we could measure mRNA degradation by arresting transcription with actinomycin D, we used qRT-PCR to compare rpoS mRNA degradation from an inducible strain (297-LK-flacp-rpoS) (8) when transcription was arrested by removing inducer (analogous to experiments performed in E. coli [32]) to that measured after actinomycin D addition. Decreasing mRNA levels will result in a higher number of amplification cycles to reach the threshold cycle (CT) where the fluorescent signal resulting from the amplification of the RNA is higher than that observed from the background. A complete summary of the CT values for the transcripts are provided in Table S2 in the supplemental material. We calculated the fraction of mRNA remaining by comparing the CT values of our experimental sample (removal of inducer or treatment with actinomycin D) to CT values of actinomycin D-free control cultures (33) that were treated analogously to the experimental conditions (pelleted, washed, and resuspended in media that contained inducer but not actinomycin D). The fraction of rpoS mRNA transcripts (Fig. 2A) and flaB mRNA transcripts (Fig. 2B) remaining following the addition of actinomycin D (open circles) or removal of inducer (filled circles) was plotted against time. Removal of inducer and the addition of actinomycin D both resulted in increases in the CT values of rpoS mRNA with time (see Table S2), indicating that the mRNA levels decreased with time (Fig. 2A). Within minutes, half of the RNA is degraded. In this rpoS-inducible strain, flaB expression is expected to be independent of the presence of inducer, and we observed that it is (Fig. 2B, filled circles). Following addition of actinomycin D, we observed that the CT values for flaB remained fairly constant for at least 45 min (see Table S2), suggesting that flaB mRNA is stable over this period of time (Fig. 2C, open circles). This was consistent with our observation that the amplification curve for flaB shifts only slightly to the right and CT values only modestly increase in strain A3-LS-flacp-ospC under actinomycin D treatment (Fig. 1B).
Fig 2.
Transcriptional arrest initiated by addition of actinomycin D or by the removal of inducer results in similar patterns of rpoS mRNA degradation in strain 297-LK-flacp-rpoS. Transcription of rpoS was induced by adding 1 mM (final concentration) IPTG to growing 297-LK-flacp-rpoS cultures. Twenty-four h later, cells were washed, split, and resuspended in media without inducer or with inducer plus 150 μg ml−1 actinomycin D. Aliquots from each culture were collected at time points 5, 10, 20, 45, 240, and 1,260 min after resuspension. The fraction of rpoS (A) and flaB (B) mRNA remaining at time points after transcriptional arrest brought about by removal of inducer (filled circles) or by addition of actinomycin D (empty circles) was determined. Means ± standard deviations are plotted against time (n = 2; with duplicate qRT-PCRs). A horizontal line representing 50% of the original mRNA level has been plotted as a reference in each graph. Raw data (CT values) are included in Table S1 in the supplemental material.
We determined the mRNA decay profiles for transcripts encoding RNA polymerase subunits (rpoA and rpoS), ribosomal proteins (rpsD, rpsK, rpsM, rplQ, and rpsO), a nuclease (pnp), outer surface lipoproteins (ospA and ospC), and a flagellar protein (flaB) in B. burgdorferi. We grew B. burgdorferi B31A cells to low cell density (∼5 × 107 cells ml−1), arrested transcription by treating them with actinomycin D, and purified total RNA at subsequent time points. CT values for the genes of interest were determined by qRT-PCR using gene-specific primers (see Table S1 in the supplemental material). A complete summary of CT values is provided in Table S3.
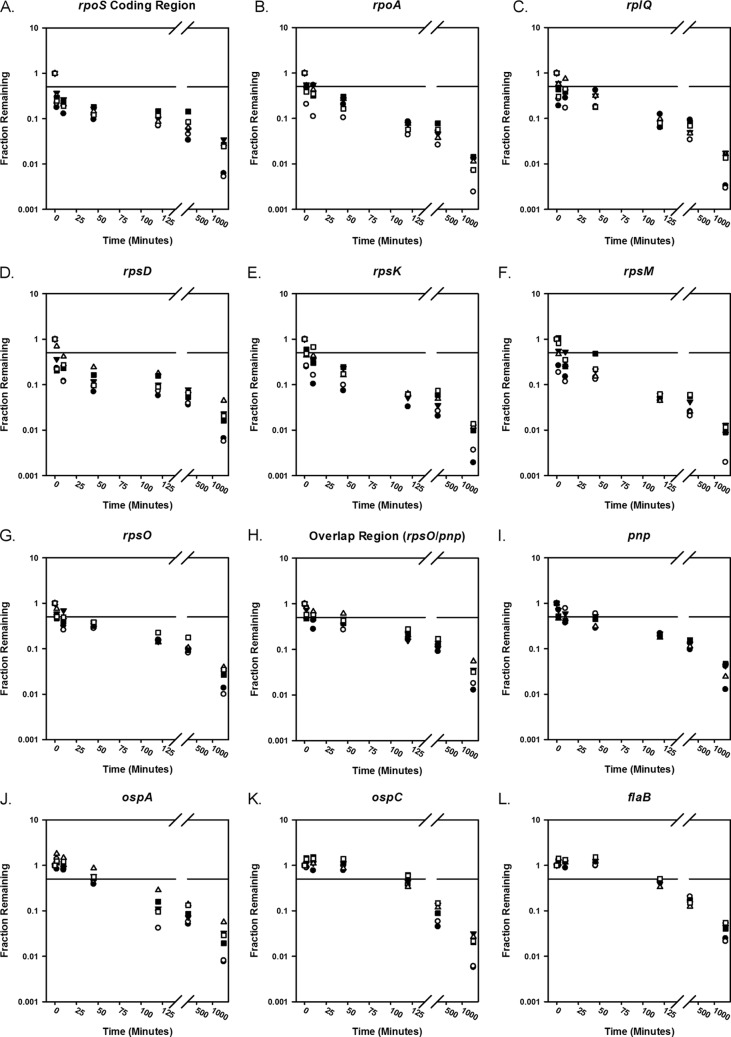
There are qualitative differences in the profiles of mRNA decay for the different genes. To monitor mRNA decay, the fraction of RNA remaining following transcription arrest was calculated by comparing CT values at given time points to the initial pretreatment value (33) and plotted against time (Fig. 3) in a semilogarithm plot. We observed at least two phases of mRNA decay for each transcript, as indicated by a changing slope in our plots (Fig. 3). Data from the first 45 min were fit as described in the experimental procedures to estimate the rate constant and half-life for each mRNA (Table 2). We inspected the decay curves visually to verify good agreement with the half-lives calculated from the best-fit first-order rate constants.
Fig 3.
mRNA decay curves for diverse genes in B. burgdorferi. The fraction of individual mRNA transcripts remaining at time points following the addition of actinomycin D. Three biological replicates (n = 3) were performed (represented by squares, circles, and triangles) with duplicate qRT-PCRs (open versus filled shapes). A horizontal line representing 50% of the original mRNA level has been plotted as a reference. (A) rpoS encoding alternative sigma factor RpoS (σs). (B) rpoA encoding the alpha subunit of RNA polymerase. (C) rplQ encoding ribosomal protein L17. (D) rpsD encoding ribosomal protein S4. (E) rpsK encoding ribosomal protein S11. (F) rpsM encoding ribosomal protein S13. (G) rpsO encoding ribosomal protein S15. (H) Region overlapping rpsO and pnp. (I) pnp encoding polynucleotide phosphorylase. (J) ospA encoding outer surface lipoprotein A. (K) ospC encoding outer surface lipoprotein C. (L) flaB encoding flagellar protein B.
Table 2.
Calculated half-lives of mRNA transcripts during the first 45 min of decay
| Gene type and name | Predicted first-order rate constant (min−1) | Half-life (min) |
|---|---|---|
| RNA polymerase subunits | ||
| rpoA | 0.087 ± 0.030 | 7.9 |
| rpoS (−171 UTR) | 0.75 ± 0.14 | 0.92 |
| rpoS coding sequence | 0.64 ± 0.12 | 1.1 |
| Ribosomal proteins | ||
| rplQ | 0.034 ± 0.014 | 20 |
| rpsD | 0.52 ± 0.12 | 1.3 |
| rpsK | 0.14 ± 0.043 | 5.1 |
| rpsM | 0.14 ± 0.041 | 5.0 |
| rpsO | 0.025 ± 0.008 | 27 |
| Overlap | ||
| rpsO and pnp | 0.019 ± 0.005 | 38 |
| RNase | ||
| pnp | 0.016 ± 0.006 | 44 |
| Outer surface lipoproteins | ||
| ospA | 0.014 ± 0.003 | 48 |
| ospC | —a | — |
| Flagellar protein | ||
| flaB | — | — |
—, not determined.
The rpoS and rpsD transcripts have an initial rapid decay with a half-life of approximately 1 min (Fig. 3A and D and Table 2). rpoA, rplQ, rpsK, rpsM, and rpsO mRNA transcripts, the region spanning rpsO, and pnp, pnp, and ospA mRNA transcripts show slower degradation over the entire time course (Fig. 3B, C, E to J, and Table 2). These mRNA transcripts decay with half-lives of between 5 and 49 min, which is significantly longer than those of rpoS and rpsD. Interestingly, ospC and flaB levels are nearly unchanged for at least 45 min, and no half-lives could be estimated from these curves (Fig. 3K and L). After ∼45 min, the transcript levels begin decreasing.
The apparent rapid decay (t1/2 < 10 min) of the rpoA, rpsD, rpsK, rpsM, and rpoS transcripts, moderate decay (10 min < t1/2 < 50 min) of rplQ, ospA, pnp, and rpsO transcripts and the portion of the transcript overlapping rpsO and pnp, and relative stability of the ospC and flaB transcripts following actinomycin D addition to cell cultures grown at 35°C suggest that there are gene-specific determinants influencing mRNA decay, as expected. In all cases, the CT values at 22 h are substantially lower than the values obtained from no-template controls and minus reverse transcriptase controls (see Table S3 in the supplemental material). This suggests that RNA degradation is incomplete for all samples. For cultures grown at room temperature (24°C), even a smaller fraction of the mRNA is degraded following addition of actinomycin D (see Fig. S1).
DISCUSSION
RNA degradation is an important component of gene regulation. Although the importance of gene regulation in the enzootic cycle and in virulence of this bacterium is well recognized, this study is the first to investigate the kinetics of RNA degradation in the spirochete B. burgdorferi.
Measurement of mRNA decay requires a method for inhibiting new mRNA synthesis. Previous studies of mRNA decay in bacteria have arrested transcription by employing rifamycin antibiotics (34). Other mRNA decay studies of transcripts controlled by inducible promoters have arrested transcription by removing inducer from cultures to restore transcriptional repression (32, 35). B. burgdorferi is naturally rifampin resistant (23), and decay studies employing inducible promoters are limited to genes regulated by an inducible, often artificial, promoter. To circumvent these limitations, we demonstrated that actinomycin D is a suitable transcriptional inhibitor for mRNA decay studies in B. burgdorferi. We observed that treatment of B. burgdorferi with actinomycin D prior to addition of inducer prevents increases in mRNA levels controlled by an inducible promoter, indicating that it arrests transcription. We also observed similar patterns of decay when we compared the effects of removal of inducer to treatment with actinomycin D on the decay of an inducible transcript.
We measured the mRNA decay patterns for genes encoding RNA polymerase subunits, ribosomal proteins, a nuclease, outer surface lipoproteins, and a flagellar protein (Table 2). One of our most striking results was that transcripts are still detectable 22 h after actinomycin treatment of liquid cultures. Schwartz and coworkers also detected long-lived RNA fragments, suggesting that incomplete digestion of B. burgdorferi mRNA is common (16). Our results, together with those previously reported by the Schwartz laboratory, support using caution when using RT-PCR to identify viable B. burgdorferi, at least from liquid culture. It is also important to note that in considering mRNA degradation, a single endonucleolytic cleavage within the coding region is sufficient to inactivate an mRNA as a template for translation. This “functional” mRNA half-life is likely to be shorter than the “chemical” half-lives of the fragments we are detecting in our assays. It will be important, in future studies, to compare chemical and functional half-lives of important transcripts.
mRNA decay generally depends upon transcript-specific details (sequences, polyadenylation, secondary structures, ribosome occupancy, RNA binding proteins, etc.) as well as the concentrations, activities, and populations of nucleases in the cell (reviewed in reference 15). Although only a limited set of data has been collected, there is wide variation in the mRNA degradation profiles for the genes we tested. rpoS, rpoA, rpsD, rpsK, and rpsM mRNAs degrade on the time scale observed for most mRNAs in diverse bacteria (i.e., less than 10 min) (Fig. 3A, B, D, E, and F and Table 2); however, we observed that the degradation of ospC, ospA, rplQ, rpsO, pnp, and flaB fragments is relatively slow (20 min or greater) (Fig. 3C, G, H, I, J, K, and L and Table 2). Slow-growing prokaryotes generally have not shown a corresponding low rate of global mRNA decay (36–39); however, studies of mRNA decay in M. tuberculosis show correlations with growth rate and mRNA stability (40). Sherman and coworkers suggest that slower decay rates in mycobacteria conserve energy or provide a transcript memory associated with passage of the bacterium from one host to the next (40). Similar strategies may be useful for B. burgdorferi during its enzootic cycle.
We observe that the transcript decay is generally not first order; the rates of decay (determined by the slope of the decay curves shown in Fig. 3) all change after the first few hours (decreasing for rpoA, rpsD, rpsK, rpsM, rplQ, rpoS, pnp, and rpsO, the region overlapping rpsO, and pnp and ospA; increasing for flaB and ospC). For flaB and ospC, the increases in the degradation rates with time may simply result from differences in the affinities of nucleases for different transcripts. At early times, transcripts that are good nuclease substrates will be bound and processed. As these good substrates are used up, the effective nuclease concentration will increase to facilitate degradation of poorer substrates. The observed decreases in the rates of degradation of some of the transcripts at long times following transcriptional arrest may result from cellular global changes (e.g., the concentrations or activity of nucleases) or transcript-specific effects (e.g., ribosome occupancy). The significant differences in the decay profiles suggest that these transcripts will be useful for determining the transcript-specific details (e.g., nuclease recognition sequences and roles of secondary structure) that influence rates of mRNA decay in more detailed mechanistic studies.
mRNA lifetimes appear to be correlated with the function of the protein that they encode in E. coli (36), Lactococcus lactis (41), Prochlorococcus (39), and Bacillus cereus (42), although this correlation was not observed in B. subtilis (37). Generally, we observed rapid decay of transcripts encoding ribosomal proteins and RNA polymerase subunits in B. burgdorferi; however, rplQ and rpsO mRNA degradation was much slower than that observed for mRNAs encoding other ribosomal proteins (Table 2).
The enzootic cycle involves well-characterized changes in expression of rpoS, ospC, and ospA, and we investigated the decay of these transcripts to determine whether transcripts associated with gene expression in specific hosts have similar mRNA decay profiles. As ticks take on a blood meal, RpoS in B. burgdorferi cells increases (5) and influences the mRNA levels of virulence genes (43). ospC mRNA levels increase following feeding (5) and remain high during early mammalian infection. The precise function of OspC is not known, but it is required for mammalian infection (44). OspA is essential for both acquisition of B. burgdorferi by the tick and the spirochete's survival in the tick; OspA interacts with a tick midgut protein (45–47). OspA is synthesized by B. burgdorferi within unfed ticks and in culture at lower temperatures (e.g., 23°C), and OspA levels decrease upon temperature upshift in culture only if RpoS is expressed (7). Our data indicate that the mRNA decay profiles at 35°C of the genes essential for mammalian infection are quite different from each other (rpoS t1/2, ∼1 min; ospC t1/2, >50 min). Decay of the tick-specific ospA at 35°C (t1/2, 48 min) is more similar to that of ospC than that of rpoS, indicating that mRNA decay rates are independent of the gene's function in the enzootic cycle at one set of growth conditions. These longer-lived transcripts encoding outer surface lipoproteins may be important in determining the time scale for B. burgdorferi to transition between tick and vertebrate hosts. For example, the longer-lived transcripts may delay the reduction in OspA levels associated with initiation of B. burgdorferi migration from the tick midgut to salivary glands.
Correlations between RNA decay and gene function may result from operon structure rather than specific gene function (as proposed for B. subtilis [37]). Because the 5′ and 3′ ends of most genes in B. burgdorferi have not been mapped, the operon configurations were predicted (Fig. 4) using the Prokaryotic Operon Database (48) and Microbes Online Operon Predictions (49) and modified by 5′ end mapping data for rpoS (50) and rpsM (data not shown). We observed variability in the time scale of decay of genes that are expected to be found on a polycistronic transcript. Specifically, decay at the 5′ ends of transcripts is faster than at the 3′ ends (Fig. 4); e.g., compare 5′ rpsM (t1/2 of 5 min) to 3′ rplQ (t1/2 of 20 min). Similar observations have been reported previously for E. coli (36) and Prochlorococcus (39). Our limited data set suggests that there is a relationship between decay rates and operon configuration, but we are less confident that there is a correlation with decay rates and gene function. A wider data set evaluating decay of the entire transcriptome, especially under diverse conditions, will be required to determine whether there are more generally observed correlations with gene function or operon configuration.
Fig 4.
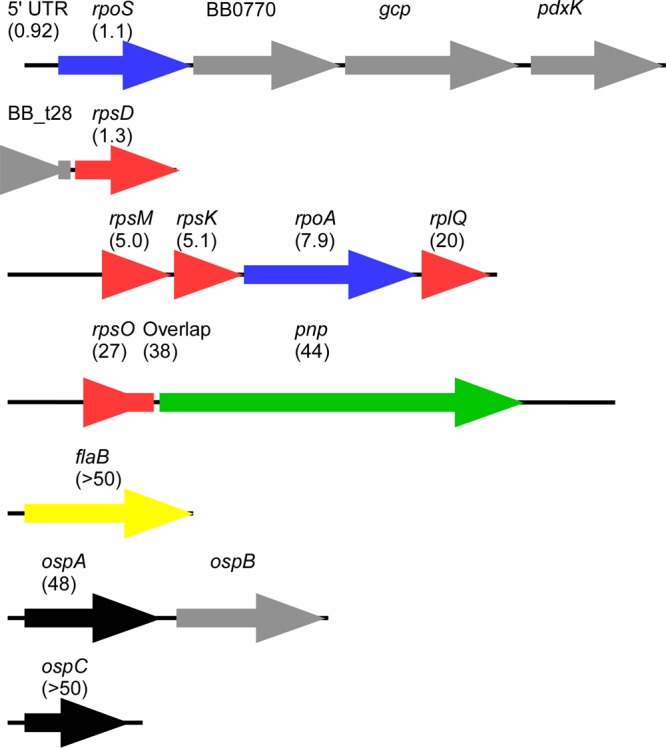
Comparison of mRNA half-lives in polycistronic transcripts. The expected operon configuration for genes used in this study are shown with the calculated half-life of the amplified product labeled in parentheses. Genes encoding RNA polymerase subunits are blue, ribosomal proteins are red, nucleases are green, outer surface lipoproteins are black, and flagellar proteins are yellow. Gene loci that were not tested are colored gray.
B. burgdorferi contains the orthologs to endoribonucleases RNase III and RNase Y but is missing an ortholog to RNase E (Table 1) (18), suggesting that its mechanisms of mRNA degradation are more similar to those observed in B. subtilis than those in E. coli. However, B. burgdorferi is missing close orthologs to the B. subtilis ribonucleases J1 and J2. Analysis of the roles of most ribonucleases (e.g., J1 and J2 in B. subtilis and RNase III and RNase Z in E. coli) indicates that they contribute to mRNA degradation pathways, at least for some genes (51–53). We propose that deletion or conditional expression of B. burgdorferi nucleases (Table 1) will help us determine which nucleases are most important in degrading these transcripts.
Although global changes in mRNA lifetime may occur as a result of environmental shifts, individual mRNA lifetimes in other bacteria have been identified from microarrays to be particularly influenced by specific environmental conditions, indicating that adaptation is mediated by mRNA decay (41, 54–56). We measured mRNA decay from cultures grown at room temperature. At room temperature, actinomycin D prevented induction of ospC in strain A3-LS-flacp-ospC, indicating that it serves to arrest transcription at this temperature (data not shown). B. burgdorferi cells grown at room temperature retained motility for more than 20 h following treatment with actinomycin D and showed some evidence of continued cell division (data not shown). We observed that the rate and extent of mRNA degradation of rpoS, ospC, ospA, flaB, pnp, and rpsO was much lower for cultures grown at room temperature (see Fig. S1 in the supplemental material), suggesting that the effects of temperature on mRNA degradation in B. burgdorferi are significant in the enzootic cycle.
Bacteria rapidly respond to changes in environmental conditions using a combination of rapid degradation of mRNA and transcriptional control to modulate gene expression. This is observed even in slow-growing bacteria and appears to be independent of doubling time. (reviewed in reference 15). Gene regulation studies in B. burgdorferi have identified transcriptional regulation mediated by the alternative sigma factors RpoS and, to a lesser extent, RpoN, as well as a DNA binding protein (BosR) (reviewed in reference 3). Although B. burgdorferi has evolved with a limited subset of ribonucleases, DNA binding domains, or orthologs to transcriptional regulators found in other bacteria (3, 19), the mRNA levels of hundreds of genes are altered by changing environmental conditions. We expect that further understanding of the roles of mRNA degradation on gene expression and regulation during the enzootic cycle will be important in understanding the basic biology of this important pathogen.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to D. Scott Samuels and Laura Hall for providing strain B31A, Melissa Hargreaves for discussions about B. burgdorferi nucleases, and Mike Gilbert for providing inducible strains.
Research reported in this project was supported by Bates College, an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health, under grant number P20GM103423, and the National Center for Research Resources (P20RR016463).
The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 23 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00659-13.
REFERENCES
- 1.Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 10:87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) 2012. Summary of notifiable diseases–United States, 2010. MMWR Morb. Mortal. Wkly. Rep. 59:1–111 [PubMed] [Google Scholar]
- 3.Samuels DS. 2011. Gene regulation in Borrelia burgdorferi. Annu. Rev. Microbiol. 65:479–499 [DOI] [PubMed] [Google Scholar]
- 4.Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, Jasinskas A, Benach J, Katona L, Radolf J, Caimano M, Skare J, Swingle K, Akins D, Schwartz I. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narasimhan S, Santiago F, Koski RA, Brei B, Anderson JF, Fish D, Fikrig E. 2002. Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J. Bacteriol. 184:3122–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks CS, Hefty PS, Jolliff SE, Akins DR. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caimano MJ, Eggers CH, Gonzalez CA, Radolf JD. 2005. Alternate sigma factor RpoS is required for the in vivo-specific repression of Borrelia burgdorferi plasmid lp54-borne ospA and lp6.6 genes. J. Bacteriol. 187:7845–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 65:1193–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takayama K, Kjelleberg S. 2000. The role of RNA stability during bacterial stress responses and starvation. Environ. Microbiol. 2:355–365 [DOI] [PubMed] [Google Scholar]
- 10.Grunberg-Manago M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193–227 [DOI] [PubMed] [Google Scholar]
- 11.Condon C, Bechhofer DH. 2011. Regulated RNA stability in the Gram positives. Curr. Opin. Microbiol. 14:148–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva IJ, Saramago M, Dressaire C, Domingues S, Viegas SC, Arraiano CM. 2011. Importance and key events of prokaryotic RNA decay: the ultimate fate of an RNA molecule. Wiley Interdiscip. Rev. RNA 2:818–836. 10.1002/wrna.94 [DOI] [PubMed] [Google Scholar]
- 13.Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, Silva IJ, Viegas SC. 2010. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol. Rev. 34:883–923 [DOI] [PubMed] [Google Scholar]
- 14.Heroldová M, Nemec M, Hubálek Z. 1998. Growth parameters of Borrelia burgdorferi sensu stricto at various temperatures. Zentralbl. Bakteriol. 288:451–455 [DOI] [PubMed] [Google Scholar]
- 15.Evguenieva-Hackenberg E, Klug G. 2011. New aspects of RNA processing in prokaryotes. Curr. Opin. Microbiol. 14:587–592 [DOI] [PubMed] [Google Scholar]
- 16.Iyer R, Mukherjee P, Wang K, Simons J, Wormser GP, Schwartz I. 2013. Detection of Borrelia burgdorferi nucleic acids after antibiotic treatment does not confirm viability. J. Clin. Microbiol. 51:857–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaberdin VR, Singh D, Lin-Chao S. 2011. Composition and conservation of the mRNA-degrading machinery in bacteria. J. Biomed. Sci. 18:23. 10.1186/1423-0127-18-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb J, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, Van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 19.Lehnik-Habrink M, Lewis RJ, Mäder U, Stülke J. 2012. RNA degradation in Bacillus subtilis: an interplay of essential endo- and exoribonucleases. Mol. Microbiol. 84:1005–1017 [DOI] [PubMed] [Google Scholar]
- 20.Bechhofer DH. 2011. Bacillus subtilis mRNA decay: new parts in the toolkit. Wiley Interdiscip. Rev. RNA 2:387–394. 10.1002/wrna.66 [DOI] [PubMed] [Google Scholar]
- 21.Evguenieva-Hackenberg E, Klug G. 2009. Chapter 7. RNA degradation in archaea and gram-negative bacteria different from Escherichia coli. Prog. Mol. Biol. Transl. Sci. 85:275–317 [DOI] [PubMed] [Google Scholar]
- 22.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912 [DOI] [PubMed] [Google Scholar]
- 23.Alekshun M, Kashlev M, Schwartz I. 1997. Molecular cloning and characterization of Borrelia burgdorferi rpoB. Gene 186:227–235 [DOI] [PubMed] [Google Scholar]
- 24.Levinthal C, Keynan A, Higa A. 1962. Messenger RNA turnover and protein synthesis in B. subtilis inhibited by actinomycin D. Proc. Natl. Acad. Sci. U. S. A. 48:1631–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rattenbacher B, Bohjanen PR. 2012. Evaluating posttranscriptional regulation of cytokine genes. Methods Mol. Biol. 820:71–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham JR, Hendershott MC, Terragni J, Cooper GM. 2010. mRNA degradation plays a significant role in the program of gene expression regulated by phosphatidylinositol 3-kinase signaling. Mol. Cell. Biol. 30:5295–5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evguenieva-Hackenberg E, Wagner S, Klug G. 2008. In vivo and in vitro studies of RNA degrading activities in archaea. Methods Enzymol. 447:381–416 [DOI] [PubMed] [Google Scholar]
- 28.Bini E, Dikshit V, Dirksen K, Drozda M, Blum P. 2002. Stability of mRNA in the hyperthermophilic archaeon Sulfolobus solfataricus. RNA 8:1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert MA, Morton EA, Bundle SF, Samuels DS. 2007. Artificial regulation of ospC expression in Borrelia burgdorferi. Mol. Microbiol. 63:1259–1273 [DOI] [PubMed] [Google Scholar]
- 30.Samuels DS. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hundt S, Zaigler A, Lange C, Soppa J, Klug G. 2007. Global analysis of mRNA decay in Halobacterium salinarum NRC-1 at single-gene resolution using DNA microarrays. J. Bacteriol. 189:6936–6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez PJ, Marchand I, Yarchuk O, Dreyfus M. 1998. Translation inhibitors stabilize Escherichia coli mRNAs independently of ribosome protection. Proc. Natl. Acad. Sci. U. S. A. 95:6067–6072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 34.Mohanty BK, Giladi H, Maples VF, Kushner SR. 2008. Analysis of RNA decay, processing, and polyadenylation in Escherichia coli and other prokaryotes. Methods Enzymol. 447:3–29 [DOI] [PubMed] [Google Scholar]
- 35.Kucharova V, Strand TA, Almaas E, Naas AE, Brautaset T, Valla S. 2013. Non-invasive analysis of recombinant mRNA stability in Escherichia coli by a combination of transcriptional inducer wash-out and qRT-PCR. PLoS One 8:e66429. 10.1371/journal.pone.0066429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein JA, Khodursky AB, Lin P, Lin-Chao S, Cohen SN. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. U. S. A. 99:9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hambraeus G, Von Wachenfeldt C, Hederstedt L. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269:706–714 [DOI] [PubMed] [Google Scholar]
- 38.Andersson AF, Lundgren M, Eriksson S, Rosenlund M, Bernander R, Nilsson P. 2006. Global analysis of mRNA stability in the archaeon Sulfolobus. Genome Biol. 7:R99. 10.1186/gb-2006-7-10-r99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steglich C, Lindell D, Futschik M, Rector T, Steen R, Chisholm SW. 2010. Short RNA half-lives in the slow-growing marine cyanobacterium Prochlorococcus. Genome Biol. 11:R54. 10.1186/gb-2010-11-5-r54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rustad TR, Minch KJ, Brabant W, Winkler JK, Reiss DJ, Baliga NS, Sherman DR. 2013. Global analysis of mRNA stability in Mycobacterium tuberculosis. Nucleic Acids Res. 41:509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Redon E, Loubière P, Cocaign-Bousquet M. 2005. Role of mRNA stability during genome-wide adaptation of Lactococcus lactis to carbon starvation. J. Biol. Chem. 280:36380–36385 [DOI] [PubMed] [Google Scholar]
- 42.Kristoffersen SM, Haase C, Weil MR, Passalacqua KD, Niazi F, Hutchison SK, Desany B, Kolsto A, Tourasse NJ, Read TD, Okstad OA. 2012. Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biol. 13:R30. 10.1186/gb-2012-13-4-r30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caimano MJ, Eggers CH, Hazlett KRO, Radolf JD. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 72:6433–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radolf JD, Caimano MJ. 2008. The long strange trip of Borrelia burgdorferi outer-surface protein C. Mol. Microbiol. 69:1–4 [DOI] [PubMed] [Google Scholar]
- 45.Yang XF, Pal U, Alani SM, Fikrig E, Norgard MV. 2004. Essential role for OspA/B in the life cycle of the Lyme disease spirochete. J. Exp. Med. 199:641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pal U, Li X, Wang T, Montgomery RR, Ramamoorthi N, DeSilva AM, Bao F, Yang X, Pypaert M, Pradhan D, Kantor FS, Telford S, Anderson JF, Fikrig E. 2004. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 119:457–468 [DOI] [PubMed] [Google Scholar]
- 47.Xu Q, Seemanapalli SV, McShan K, Liang FT. 2006. Constitutive expression of outer surface protein C diminishes the ability of Borrelia burgdorferi to evade specific humoral immunity. Infect. Immun. 74:5177–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taboada B, Ciria R, Martinez-Guerrero CE, Merino E. 2012. ProOpDB: prokaryotic operon database. Nucleic Acids Res. 40:D627–D631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Huang KH, Alm EJ, Arkin AP. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lybecker MC, Samuels DS. 2007. Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 64:1075–1089 [DOI] [PubMed] [Google Scholar]
- 51.Mäder U, Zig L, Kretschmer J, Homuth G, Putzer H. 2008. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol. Microbiol. 70:183–196 [DOI] [PubMed] [Google Scholar]
- 52.Stead MB, Marshburn S, Mohanty BK, Mitra J, Castillo LP, Ray D, Van Bakel H, Hughes TR, Kushner SR. 2011. Analysis of Escherichia coli RNase E and RNase III activity in vivo using tiling microarrays. Nucleic Acids Res. 39:3188–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perwez T, Kushner SR. 2006. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol. Microbiol. 60:723–737 [DOI] [PubMed] [Google Scholar]
- 54.Nakamura MM, Liew S, Cummings CA, Brinig MM, Dieterich C, Relman DA. 2006. Growth phase- and nutrient limitation-associated transcript abundance regulation in Bordetella pertussis. Infect. Immun. 74:5537–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barnett TC, Bugrysheva JV, Scott JR. 2007. Role of mRNA stability in growth phase regulation of gene expression in the group A Streptococcus. J. Bacteriol. 189:1866–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dressaire C, Picard F, Redon E, Loubière P, Queinnec I, Girbal L, Cocaign-Bousquet M. 2013. Role of mRNA stability during bacterial adaptation. PLoS One 8:e59059. 10.1371/journal.pone.0059059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 58.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi S, Codani J, Connerton IF, Cummings NJ, Daniel RA, Denizot F, Devine KM, Düsterhöft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, Haiech J, Harwood CR, Hénaut A, Hilbert H, Holsappel S, Hosono S, Hullo M, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S, Levine A, Liu H, Masuda S, Mauël C, Médigue C, Medina N, Mellado RP, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O'Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S, Parro V, Pohl TM, Portetelle D, Porwollik S, Prescott AM, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror SJ, Serror P, Shin B, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognoni A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa H, Zumstein E, Yoshikawa H, Danchin A. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256 [DOI] [PubMed] [Google Scholar]
- 59.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29:123–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.