Abstract

The ubiquitin signaling pathway consists of hundreds of enzymes that are tightly regulated for the maintenance of cell homeostasis. Parkin is an E3 ubiquitin ligase responsible for conjugating ubiquitin onto a substrate protein, which itself can be ubiquitinated. Ataxin-3 performs the opposing function as a deubiquitinating enzyme that can remove ubiquitin from parkin. In this work, we have identified the mechanism of interaction between the ubiquitin-like (Ubl) domain from parkin and three C-terminal ubiquitin-interacting motifs (UIMs) in ataxin-3. 1H–15N heteronuclear single-quantum coherence titration experiments revealed that there are weak direct interactions between all three individual UIM regions of ataxin-3 and the Ubl domain. Each UIM utilizes the exposed β-grasp surface of the Ubl domain centered around the I44 patch that did not vary in the residues involved or the surface size as a function of the number of ataxin-3 UIMs involved. Further, the apparent dissociation constant for ataxin-3 decreased as a function of the number of UIM regions used in experiments. A global multisite fit of the nuclear magnetic resonance titration data, based on three identical binding ligands, resulted in a KD of 669 ± 62 μM for each site. Our observations support a multivalent ligand binding mechanism employed by the parkin Ubl domain to recruit multiple UIM regions in ataxin-3 and provide insight into how these two proteins function together in ubiquitination–deubiquitination pathways.
Protein degradation is a tightly orchestrated process that is required to maintain cell homeostasis. When homeostasis is not maintained, disease-prone events such as protein misfolding and aggregation can frequently occur, the hallmarks of several disorders, including Alzheimer’s, Parkinson’s, and Huntington’s diseases. The ubiquitination proteolysis pathway is the primary mechanism for directing proteins to the proteasomal degradation machinery, eliminating unwanted proteins and preserving protein levels in the cell. A cascade of three proteins, ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligase (E3) enzymes, are utilized for covalently attaching the small protein ubiquitin to a substrate and building a polyubiquitin chain required for recognition by the proteasome. Of the three classes of proteins involved in ubiquitination, the E3 ubiquitin ligase proteins have been especially attractive targets for drug discovery because more than 1000 different homologues have been identified, suggesting that the E3 protein is a major contributor to the substrate specificity in the pathway.1
Parkin is a RING-inBetweenRING-RING (RBR) E3 ligase composed of 465 residues with multiple domains, including N-terminal Ubl (ubiquitin-like), UPD (unique-parkin domain), RING0, RING1, IBR (inBetweenRING), and RING22,3 domains. Specific mutations in the PARK2 gene, encoding parkin, have been linked to an autosomal recessive inheritance of Parkinson’s disease (PD). This form of PD afflicts approximately 5–10% of PD patients and is distinguished by an early onset age, typically earlier than 40 years old,4 but otherwise has many of the characteristics of the more common idiopathic form. One hypothesis for the function of parkin is that it regulates appropriate levels of its protein substrates, avoiding accumulations of aggregate-prone substrates such as α-synuclein, Aβ peptide, and Huntingtin protein.5 Other substrates have also been proposed for parkin such as synphillin-1,6 Pael-R,7 and cyclin E,8 although confirmation of these has not been resolved. One mechanism for recruitment of substrates by parkin is through its Ubl domain. Recent evidence shows that in addition to possible substrate recognition, the Ubl domain modulates the activity of parkin through interaction with the RBR region.9 Like ubiquitin, the Ubl domain has a β-grasp fold with an exposed domain hydrophobic face comprised of five β-sheets. The parkin Ubl domain has been previously shown to interact with proteins containing a short recognition motif termed a ubiquitin-interacting motif (UIM) found in proteasomal subunit S5a, the endocytosis protein Eps15, and ataxin-3, a deubiquitinating enzyme.1,10,11 The UIM region is composed of ∼20 residues and contains a conserved acidic stretch located prior to a five-residue (large–small–large–small–large side chain size) core motif, followed by a conserved serine residue.12,13
The multistep enzymatic reaction for the attachment of ubiquitin to a substrate protein can be reversed by ubiquitin specific proteases or deubiquitinating enzymes. This removal of ubiquitin from a substrate is thought to correct errors in the ubiquitin chain building process by ensuring the correct substrate, linkage, and chain length. This allows deubiquitinating enzymes to work with an E3 enzyme to edit substrate ubiquitination and adapt to constantly changing cellular environments.14,15 Ataxin-3 is a deubiquitinating enzyme that has a preference for editing K63-linked ubiquitin chains16−18 and plays a role in DNA repair and ERAD (endoplasmic reticulum-associated degradation).19,20 The deubiquitinating activity of ataxin-3 is controlled through its N-terminal Josephin domain that houses the catalytic site for activity. The C-terminus of ataxin-3 contains three UIM regions (UIM1–UIM3) thought to control the interaction with potential substrate proteins such as parkin,1 VCP (valosin-containing protein),21 and E6-AP (ubiquitin ligase E3A).22 The second and third UIM regions in ataxin-3 are separated by a variable glutamine tract5 that contributes to one of nine identified polyglutamine diseases causing Machado Joseph disease when the polyglutamine stretch exceeds a threshold of 45 residues.23
In this work, we identified the mechanism of interaction between the C-terminus of ataxin-3 and the Ubl domain of parkin. Our work presents evidence that all three UIM regions of ataxin-3 have direct weak interactions with the hydrophobic patch of the parkin Ubl domain consistent with a multivalent ligand binding mechanism. These observations provide a better understanding of the recognition of the UIM regions of ataxin-3 by parkin that contributes to an understanding of its role in deubiquitination activity.
Experimental Procedures
Molecular Biology
The DNA encoding the human parkin ubiquitin-like domain (residues 1–77) was inserted into the NdeI and BamHI sites of a pET44a vector (Novagen) as previously described.11 Parkin Ubl domain constructs were verified by DNA sequencing (Robarts Sequencing Facility). The cDNAs encoding full-length human ataxin-3 proteins with a 14- or 82-residue polyglutamine tract were provided by E. Fon (McGill University, Montreal, QC). A fragment containing all three UIM sequences from ataxin-3 (residues 194–361, UIM_123) was generated by inserting the corresponding sequence into the BamHI and XhoI sites of the pET21a vector (Novagen) that included an in-frame six-residue histidine tag. A similar approach was used for site-directed mutagenesis to incorporate a TEV cleavage site to allow for removal of the histidine tag from all ataxin-3 constructs. A C-terminal deletion construct, consisting of UIM1 and UIM2 (residues 194–261, UIM_12Δ3), was generated by encoding a stop site prior to the polyglutamine stretch. An N-terminal construct containing UIM1 (UIM_1Δ23) was generated in a similar fashion by placing a stop codon prior to the DNA sequence encoding UIM2.
Single functional UIM constructs (UIM_1–UIM_3) and a full-length ataxin-3 C-terminal region containing 45 consecutive glutamine residues were synthesized by DNA2.0 and cloned into the NdeI and XhoI sites of the pJexpress414 vectors. Conserved serine residues and hydrophobic residues within each UIM core region were substituted with alanine residues to generate UIM_1 (L249A, R250A, R251A, I253A, S2566A, L340A, Q341A, V344A, and S347A), UIM2 (L229A, Q230A, R231A, L233A, S236A, L340A, Q341A, V344A, and S347A), and UIM_3 (L229A, Q230A, R231A, L233A, S236A, L249A, R250A, R251A, I253A, and S256A). DNA sequencing was conducted to ensure the correct sequences of all ataxin-3 constructs (Robarts Research Institute).
Protein Expression and Purification
The human parkin Ubl domain was overexpressed in a BL21(DE3) Codon PlusRIL Escherichia coli strain. Bacterial cultures were diluted 1:100 (v/v) in LB medium and grown at 37 °C until an A600 of 0.6–0.7 had been reached. For 15N-labeled proteins, cells were grown in M9 medium using 15NH4Cl (1 g/L) as the sole nitrogen source. Cultures were grown with constant shaking overnight at 16 °C after the addition of 1 mM isopropyl 1-thio-β-d-galactopyranoside. Starter cultures and larger bacterial cultures contained the antibiotic ampicillin (50 μg/mL). Cells were lysed with either an EmulsiFlex C5 homogenizer (Avestin, Ottawa, ON) or a French press and centrifuged at 106255g for 1 h. Cell homogenates were filtered through a MILLEX HV 0.45 μm filter unit (Millipore, Billerica, MA) before being loaded onto a HiTrap Q XL column, using an AKTA FPLC system (GE Healthcare, Mississauga, ON). Binding buffers contained 25 mM Tris and 1 mM EDTA (pH 9.0), and elution buffers contained 25 mM Tris, 1 mM EDTA and 1 M NaCl (pH 9.0). The Ubl protein eluted in the flow-through fractions and was further purified on a size exclusion column (G75) at 4 °C in 50 mM Na2HPO4, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT (pH 8.0) with a flow rate of 6 mL/h.
All unlabeled and 15N-labeled human ataxin-3 UIM constructs were overexpressed in a manner similar to that used for the parkin Ubl domain. Clarified cell homogenates were applied to a Ni-NTA fast protein liquid chromatography affinity column (GE Healthcare). Loading and elution buffers contained 50 mM Na2HPO4, 500 mM NaCl, and 10 mM imidazole (pH 8). Elution buffers also contained 500 mM imidazole. Protein samples were dialyzed overnight at 4 °C in loading buffer with an addition of TEV protease (1.2 mg/L bacterial preparation) for cleavage and reloaded onto the Ni-NTA column. Cleaved ataxin-3 proteins eluted in the flow-through fractions. All ataxin-3 UIM constructs were purified using the same protocol.
Nuclear Magnetic Resonance (NMR) Spectroscopy
All NMR experiments were performed on a 600 MHz Varian Inova spectrometer (Biomolecular NMR Facility, University of Western Ontario) at 25 °C. Protein samples were extensively dialyzed into 10 mM KH2PO4, 150 mM NaCl, 1 mM EDTA, and 1 mM dithiothreitol (pH 7). Imidazole (100 μM) was added to serve as a pH indicator. Chemical shift perturbation experiments that monitored the interaction of parkin Ubl and ataxin-3 variants were globally fit using nonlinear 1:1 or 3:1 binding models. For the 1:1 model (eq 1) representing the protein (P; Ubl or ataxin-3) and ligand (L; ataxin-3 or Ubl)
| 1 |
the dissociation constants (KD) were determined using eq 2
| 2 |
where Pt is the total protein, Lt is the total ligand, and PL is the amount of complex formed. For a 3:1 multivalent ligand model, three possible binding sites were considered (eqs 3–5)
| 3 |
| 4 |
| 5 |
where PL1–PL3 represent binding of the Ubl domain (P) to three different UIM sites (L1–L3, respectively) on ataxin-3. For the sake of simplicity, we considered that binding to each site occurs with equal affinity (i.e., K1 = K2 = K3 = KD) and where weak binding does not result in multiple protein species occupying multiple ligand sites (i.e., P2L and P3L). The resulting equation (eq 6) (procedures of the Supporting Information) was used to globally fit this binding model where n is the number of identical sites (three).
| 6 |
In both cases, chemical shift changes occurred in the fast exchange regime such that the observed chemical shift (δobs) was a weighted combination of the chemical shifts and populations for the free (δP and fP, respectively) and saturated protein (δPL and fPL, respectively) states (eq 7 and procedures in the Supporting Information). This equation can be recast such that the ratio of the change in chemical shift (Δδ = δobs – δP) and total chemical shift change (δTOT = δPL – δP) are related to the ratio of the complex (PL) and total protein (Pt) (eq 8).
| 7 |
| 8 |
Combining eqs 6 and 8, we plotted the chemical shift change (Δδ) as a function of total added ligand (Lt), at a constant total protein concentration (Pt), and fitted for KD and δTOT. A similar approach was used for the multivalent ligand model (procedures of the Supporting Information). All data were fit using the equations given above incorporated into Prism version 5.0b (GraphPad Software Inc., La Jolla, CA) or the titration analysis software built into NMRViewJ (version 8.0.rc4). Protein concentrations were determined by amino acid analysis (Hospital for Sick Children, Toronto, ON).
Results and Discussion
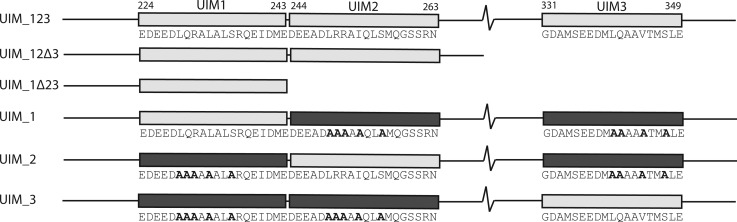
The C-terminus of ataxin-3 contains three UIMs that span residues 224–243 (UIM1), 244–263 (UIM2), and 331–349 (UIM3) (Figure 1). The region between UIM2 and UIM3 contains a polyglutamine region that is expanded in Machado Joseph disease. To identify the mechanisms used by ataxin-3 to recruit the parkin Ubl domain, we constructed a series of proteins comprised of different combinations of the UIM regions (Figure 1), including proteins that contained all three UIM regions (UIM_123), the first region only (UIM_1Δ23), and the first two UIM regions (UIM_12Δ3). Three proteins that tested the importance of an individual UIM within the context of the intact C-terminus were also designed. In these sequences, two of the UIM regions were rendered inactive by substituting key core UIM-defining residues and a conserved serine with alanine residues (Figure 1), shown to eliminate binding with a Ubl domain or ubiquitin.11,18,24 For example, UIM_1 contains only a functional first UIM sequence but retains the remainder of the C-terminus having appropriate alanine substitutions in UIM2 and UIM3.
Figure 1.
Schematic diagram of the UIM regions in ataxin-3. The boundaries of each UIM region are indicated above the full-length C-terminal construct (UIM_123) for UIM1 (E224–E243), UIM2 (D244–N263), and UIM3 (G331–L349). Other protein constructs that include deletion of UIM3 (UIM_12Δ3), deletion of UIM2 and UIM3 (UIM_1Δ23), or substitutions in the UIM regions (UIM_1–UIM_3) are shown below. Light gray rectangles indicate that the native UIM sequence is present, and dark gray rectangles represent UIM sequences with alanine substitutions. Specific residues substituted with alanine residues are represented in bold text.
The C-Terminus of Ataxin-3 Is Recruited by the Parkin N-Terminal Ubl Domain
As an initial step to identify the interactions between the C-terminus of ataxin-3 and the parkin Ubl domain, we monitored 1H–15N heteronuclear single-quantum coherence (HSQC) spectra of the 15N-labeled Ubl domain in the absence and presence of unlabeled UIM_123 from ataxin-3. In the absence of ataxin-3, the spectrum of the parkin Ubl domain is well-dispersed (Figure 2A), a pattern similar to that of the data previously reported.14 Upon addition of UIM_123, the 1H–15N HSQC spectrum of the 15N-labeled Ubl domain shows many resonances that shift compared to the original spectrum, indicative of an interaction between the two proteins. For example, Figure 2 shows residues F13 and V15 (β1), V43, K48, and E49 (β4), L61, Q64, I66, and V67 (β5), and V70 and R75 (C-terminus) in the parkin Ubl domain exhibit obvious chemical shift changes upon addition of the C-terminus of ataxin-3. The observed chemical shift changes were measured and plotted as a function of residue position (Figure S1 of the Supporting Information) to identify the binding surface for ataxin-3 on the Ubl domain. This analysis showed that most residues affected by ataxin-3 binding are found on the solvent-exposed hydrophobic patch of the Ubl domain (Figure 2B) that surrounds I44 and includes A46, I66, V67, and V70. Nearly all the affected residues fall on the β-grasp face of the Ubl domain (Figure 2B).
Figure 2.
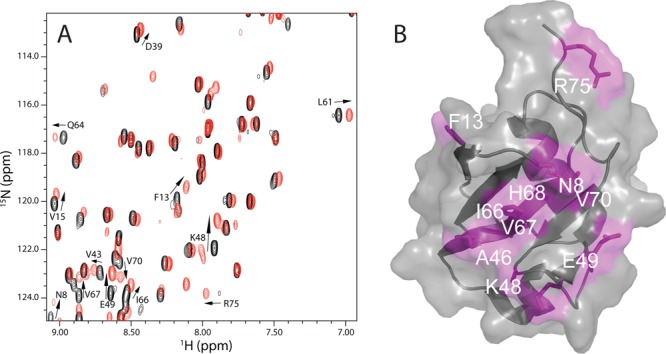
Ataxin-3 UIM regions interact with the parkin Ubl domain. (A) Region of 1H–15N HSQC spectra of the 15N-labeled Ubl domain (105 μM) in the absence (black) and presence (red) of excess ataxin-3 UIM_123 protein. Resonances that experienced the largest chemical shift changes upon UIM_123 addition are labeled, and the direction of the chemical shift change is denoted with an arrow. (B) Surface representation of the parkin Ubl domain (Protein Data Bank entry 1IYF) showing the binding surface for UIM_123. Residues that displayed chemical shift changes >0.5 standard deviation above the mean (0.078 ppm) are colored pink and labeled. The surface representation was created in PyMOL (PyMOL Molecular Graphics System, version 1.5.0.4, Schrödinger, LLC).
The binding surface for ataxin-3 exhibited some similarities with other UIM-containing proteins for interaction with the parkin Ubl domain. For example, Eps15 and S5a, which each have two UIM motifs, utilize K48, A46, and I66 residues,11 centered around I44 in the Ubl domain, for binding (Figure S1 of the Supporting Information). On the other hand, residues V15, V56, V70, Q71, and R75 from the parkin Ubl domain display large chemical shift changes when ataxin-3 is added, not obvious in similar examinations of the interaction between the parkin Ubl domain and either Eps15 or S5a. This shows that although each UIM-containing protein uses the hydrophobic “I44 patch” as a main recruitment site, other residues appear to be more specialized for different UIM sequences.11 In particular, chemical shift perturbation plots showing the interaction of the parkin Ubl domain with all three UIM-containing proteins (Figure S1 of the Supporting Information) show the C-terminus of the Ubl domain appears to be more sensitive to ataxin-3 interactions than Eps15 or S5a. The utilization of the C-terminus of the parkin Ubl domain to recruit ataxin-3 is more similar to that of hPLIC2 and HHR23A13,25 and also bears some resemblance to that of ubiquitin upon interaction with the UIM1–UIM2 region of ataxin-3.26
In Machado Joseph disease, one of the contributing factors is an extension of the polyglutamine stretch between UIM2 and UIM3 to more than 45 continuous glutamine residues. In light of this, we attempted to examine the interaction of two versions of the C-terminus of ataxin-3 carrying a 45- or 82-polyglutamine residue insertion to determine how this might affect binding or the recognition surface on the parkin Ubl domain. Repeated attempts to produce these proteins lead to either inefficient expression or insoluble material not suitable for biophysical characterization.
The Parkin Ubl Domain Is Recognized by All Three UIMs from Ataxin-3
1H–15N HSQC titration experiments were utilized to investigate the specificity of the UIMs from ataxin-3 with the parkin Ubl domain. The backbone 1H, 15N, and 13C assignment for most of the ataxin-3 UIM_123 construct was completed using standard triple-resonance methods, although we were unable to assign resonances from the 15-residue polyglutamine region (Q291–Q305) and much of the region preceding UIM1 because of the redundancy of signals. The spectrum (Figure S2 of the Supporting Information) showed a tight arrangement of resonances having a 1H chemical shift range between 7.9 and 8.7 ppm. Because UIM regions frequently adopt α-helical conformations,13,27−29 and UIM1 and UIM2 have been shown to be α-helical,26 the NMR spectrum of UIM_123 is suggestive of α-helical content with a large amount of disordered structure from the regions adjoining the UIM regions. This is in agreement with chemical shift index analysis of the NMR assignment data that showed α-helical stretches for UIM1 and -2 but was inconclusive for UIM3 (Figure S3 of the Supporting Information). Incremental additions of the unlabeled parkin Ubl domain to 15N-labeled ataxin-3 UIM_123 revealed that resonances shifted in the fast exchange regime for residues found in all three UIM sequences of ataxin-3. For example, Figure 3 shows that A232 in UIM1, E246 and D248 in UIM2, and S335, V344, and T345 in UIM3 all undergo obvious chemical shift changes upon addition of the parkin Ubl domain. Using these data, the chemical shift changes for clearly resolved signals from UIM1 (A232, A234, and D241), UIM2 (D248), and UIM3 (S335, V344, and T345) were used to determine the dissociation constants for each UIM with the parkin Ubl domain. This approach showed that individually all three UIMs exhibited very weak binding to parkin with individual dissociation constants of >1000 μM. One of the disadvantages of this approach was the significant overlap of resonances from each UIM in the 1H–15N HSQC spectra of ataxin-3 UIM_123 in the absence and presence of Ubl domain protein that did not allow an extensive number of residues to be characterized. Further, this type of analysis did not take into account the competitive nature of binding of one UIM motif in the presence of others. Nevertheless, the data showed that each UIM binds weakly with the parkin Ubl domain.
Figure 3.

Three UIM regions in ataxin-3 interact with the parkin Ubl domain. Regions of the 1H–15N HSQC spectra of UIM_123 (73 μM) in the absence (black) and presence (cyan) of excess parkin Ubl domain. Resonances that experienced the largest chemical shift changes in UIM_123 upon addition of the parkin Ubl domain are labeled, and the direction of the chemical shift change is denoted with an arrow.
Our results differ from those of some other studies that show a particular UIM within a multi-UIM protein tends to be more selective for Ubl domain interaction. For example, the Ubl domains from parkin and hPLIC-2 have been shown to preferentially interact with UIM1 from the S5a subunit of the 19S proteasome,11,30 while the Ubl domain from hHR23a primarily interacts with UIM2.13 On the other hand, both UIMs from Eps15 appear to interact equally with the parkin Ubl domain.11 In all these cases, the Ubl domain binds to the individual UIM motifs with dissociation constants ranging from 100 to 400 μM. Our initial experiments suggest the interaction of an ataxin-3 UIM motif is much weaker.
The Parkin Ubl Domain Recruits Individual Ataxin-3 UIM Regions
To identify the mechanism and relative importance of each UIM within ataxin-3 for parkin Ubl recruitment, experiments that examined changes in the NMR spectra of the 15N-labeled Ubl domain upon titration with either ataxin-3 proteins carrying UIM variants known to compromise Ubl binding or constructs lacking particular UIM regions were designed. In principle, these experiments should also show whether a particular UIM region in ataxin-3 interacts with a specific region of the parkin Ubl domain.
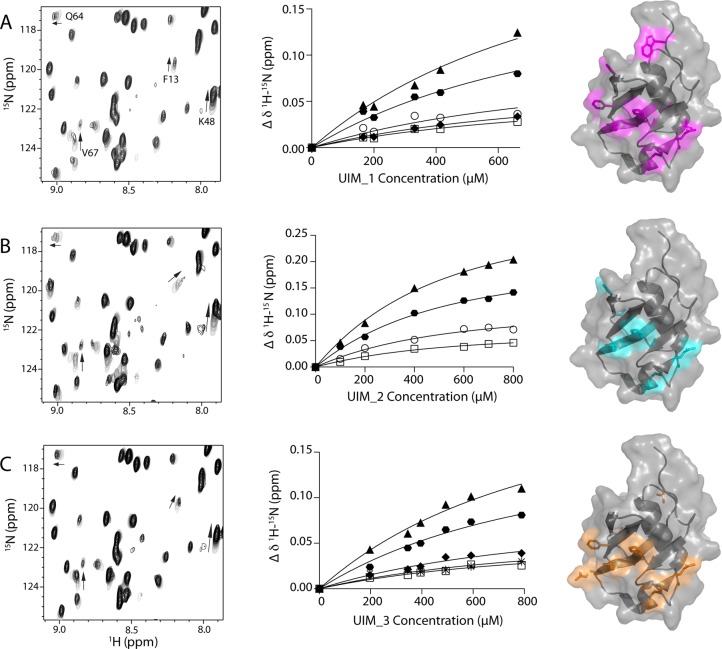
In the first of these experiments, conserved serine and hydrophobic core residues within each UIM region were substituted for alanine residues (Figure 1). This approach has been shown to eliminate binding of UIM to Ubl domains and ubiquitin in several other systems.11,18,24 The alanine substitutions favor α-helix formation in the UIM regions observed in other UIM structures.13,26 Further, the conserved serine in each UIM (S236, S256, or S347) is a hallmark of all UIM sequences27 and is absolutely essential for the interaction with Ubl domains and ubiquitin.1,27 The resulting protein constructs, UIM_1–UIM_3, each have only a single functioning UIM region allowing the interaction of each UIM to be determined in the context of the entire C-terminus of ataxin-3. In separate experiments, each UIM protein was titrated into the 15N-labeled parkin Ubl domain and the interaction monitored by analysis of the 1H–15N HSQC spectra (Figure 4). As with our previous experiments (Figure 3), these data showed that addition of each protein caused changes in the NMR spectrum of the parkin Ubl domain, indicating that all three of the UIM regions are involved in the interaction. Further, the data showed that each UIM region affected similar residues within the parkin Ubl structure (Figure 4) in a similar manner. For example, F13, K48, E49, and V67 consistently displayed the largest chemical shift changes upon titration with the ataxin-3 variants (Figure S4 of the Supporting Information). In addition, for each UIM construct, the affected resonances shifted in similar directions indicative of similar changes in the environment. This result is expected for the interaction of the common UIM motif found in each protein construct. The common chemical shift perturbations from each experiment were plotted as a function of UIM protein concentration (Figure 4). The results showed that each UIM region bound weakly to the parkin Ubl domain with statistically similar apparent dissociation constants (KD,UIM_1 = 840 ± 189 μM, KD,UIM_2 = 502 ± 41 μM, and KD,UIM_3 = 921 ± 165 μM). This indicates that each UIM within ataxin-3 appears to contribute equally to the interaction with the parkin Ubl domain.
Figure 4.
Individual UIM regions in ataxin-3 bind similarly to the parkin Ubl domain. Regions of 1H–15N HSQC spectra of the 15N-labeled parkin Ubl domain (100 μM) showing selected residues that exhibit changes in chemical shifts upon addition of (A) UIM_1, (B) UIM_2, and (C) UIM_3. The change in chemical shift as a function of increasing ataxin-3 protein concentration is shown for residues F13, K48, V67, and Q64. The middle panels show the binding curves for (A) UIM_1, (B) UIM_2, and (C) UIM_3 with the parkin Ubl domain derived from NMR data. The data were globally fit for 1:1 binding using eq 2 to determine the apparent dissociation constants; KD,UIM_I = 840 ± 189 μM, KD,UIM_2 = 502 ± 41 μM, and KD,UIM_3 = 921 ± 165 μM. In each case, fits were completed using residues K48 (▲), E49 (⬣), H68 (○), Q64 (◆), L61 (□), and V43 (∗). The right panel depicts the corresponding surface representations of the parkin Ubl domain (Protein Data Bank entry 1IYF) showing residues affected by binding of UIM_1 (purple), UIM_2 (cyan), and UIM_3 (orange). The surface representations were created in PyMOL (PyMOL Molecular Graphics System, version 1.5.0.4, Schrödinger, LLC).
The Parkin Ubl Domain Uses a Multivalent Ligand Binding Mechanism to Recruit Ataxin-3
At least two possible mechanisms are possible for parkin Ubl domain recruitment of the UIM regions in ataxin-3. One method employs simultaneous or cooperative binding such that different portions of the Ubl domain recognize distinct UIM regions. This mechanism has recently been suggested for the binding of the first two UIM regions from ataxin-3 to ubiquitin where it is suggested that the UIMs adopt a more compact structure upon binding.26 A second mechanism employs a multi- or polyvalent ligand binding mode in which multiple UIM regions recognize a single binding surface on the parkin Ubl but only a single UIM occupies this site at any given time. Both mechanisms can give rise to a decrease in the apparent dissociation constant as the number of ligand sites increases.31−33 A recent example of this phenomenon is the recruitment of multiple phosphorylation sites on Sic1 using a single Cdc4 binding site.33
To identify whether different portions of the Ubl domain recognize distinct UIM regions, we constructed two additional proteins that lacked either the C-terminal UIM region (UIM_12Δ3) or both the second and third UIM regions (UIM_1Δ23) and compared these to the parent protein (UIM_123). Changes in the 1H–15N HSQC spectra of the 15N-labeled Ubl domain were determined upon titration of each protein (Figure 5). These experiments show that the addition of each UIM construct resulted in a very similar response in the parkin Ubl spectrum. For example, all UIM constructs elicited changes in the signals for similar residues (i.e., F13, K48, and R75). Further, the directions of the chemical shift changes were the same regardless of whether UIM_123, UIM_12Δ3, or UIM_1Δ23 was added (Figure 5). In fact, the residues affected were nearly identical to those observed in earlier experiments that utilized only a single intact UIM motif (Figure 4). The data also showed no evidence that additional residues in the Ubl domain underwent chemical shift changes upon addition of UIM_123 compared to the proteins with fewer UIM motifs (UIM_1Δ23 and UIM_12Δ3). Together, these data indicate that the parkin Ubl domain binding surface area for ataxin-3 does not increase as a function of the number of UIM regions, as expected if simultaneous or cooperative binding of multiple UIMs were occurring.
Figure 5.
Multiple UIM regions in ataxin-3 affect the same binding surface in the parkin Ubl domain. Regions of the 1H–15N HSQC spectra for the 15N-labeled Ubl domain titrated with (A) UIM_123, (B) UIM_12Δ3, and (C) UIM_1Δ23. Identical residues that experienced chemical shift changes are labeled, and arrows indicate the direction of the shift with increasing concentrations of the different UIM constructs. The dotted arrow in spectrum C indicates that R75 experiences a chemical shift visible at a lower contour level. The concentrations of the 15N-labeled parkin Ubl domain were 105, 100, and 40 μM for UIM_123, UIM_12Δ3, and UIM_1Δ23 experiments, respectively.
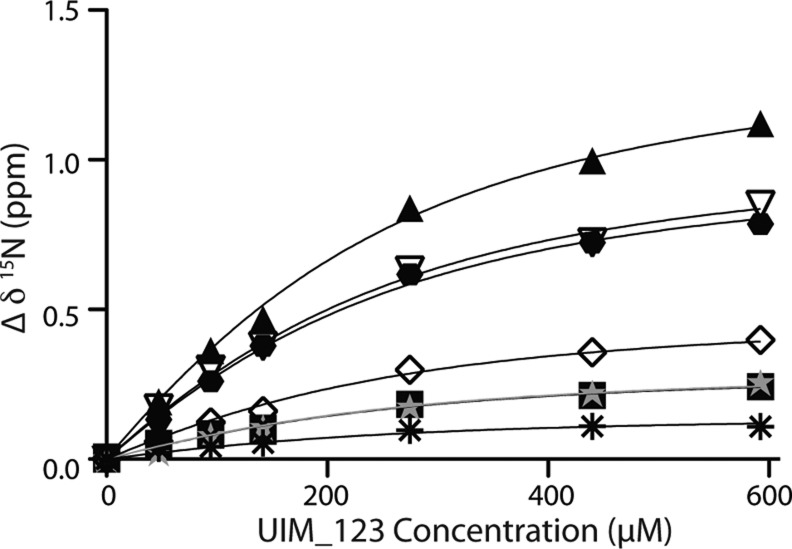
The observation that the three UIM sites in ataxin-3 bind equally to a single site of the parkin Ubl domain suggests a multi- or polyvalent ligand binding mode. This binding mechanism is characterized by multiple weak binding sites that have fast off rates, such that when the number of binding moieties increases, the apparent binding affinity may also increase.32 To provide further evidence for this mechanism, we fit the data for binding of UIM_123 to the parkin Ubl domain (Figure 5A) to a model for three identical binding sites in ataxin-3. This treatment of data resulted in a dissociation constant of 669 ± 62 μM for each UIM in ataxin-3 with the parkin Ubl domain (Figure 6). This value agrees well with results of experiments for the binding of the individual UIM regions (UIM_1–UIM_3) with the Ubl domain that provided an average dissociation constant of ∼750 μM (Figure 4). Using the same data, the apparent dissociation constant for binding between the two proteins was significantly lower (235.7 ± 14.0 μM), an indication that all three sites in ataxin-3 are individually recruited by the parkin Ubl domain.
Figure 6.
Multivalent ligand binding curves for the ataxin-3 UIM region binding to the parkin Ubl domain. Binding curves were generated by measuring the change in the 15N chemical shift from the data collected in Figure 5A. A KD of 669 ± 62 μM was determined using eq 6 and a global fit from changes in the chemical shift of K48 (▲), A46 (▽), E49 (⬣), V15 (◇), I44 (■), L50 (gray stars), and V43 (∗).
Several studies have shown that some E3 ligases, including parkin, have a relationship with a deubiquitinating enzyme that prevents the ligase from rapid turnover via the proteasome.34−36 For example, the E3 ligase parkin is able to autoubiquitinate itself, thereby modulating its activity.9,36 However, in the presence of ataxin-3, autoubiquitination is inhibited likely through interception of the E3–ubiquitin conjugate in parkin by the deubiquitinating enzyme. This could be mediated by the Josephin domain of ataxin-3 shown to interact with the C-terminal RING1-IBR-RING2 (RBR) region of parkin or with the E2 enzyme.1,36 It has recently been shown that the Ubl domain acts as an autoinhibitory module by interacting with the RBR region of parkin,9 a finding supported by recent low-resolution X-ray data.37 For ubiquitination to proceed, structural studies37,38 have shown that a significant conformational change must occur, in part to relieve the interaction of the Ubl domain with the RBR region of parkin. The observed dissociation constant for the UIM regions of ataxin-3 is significantly weaker than that determined for the interaction of the Ubl domain with the RBR (3 μM),9 suggesting ataxin-3 would be an ineffective competitor of this interaction. This indicates that the interaction of the parkin Ubl domain with the ataxin UIM regions must occur following a conformational change in parkin, likely upon interaction with the E2 enzyme, an idea supported by models proposed by Durcan and co-workers.36,39 Alternatively, it has also been shown that PINK1 activation leads to self-association of parkin.40 It is intriguing to propose that the multiple UIM sites in ataxin-3 might be specific for multiple Ubl sites in a parkin oligomer.
Acknowledgments
We thank Dr. Edward Fon (McGill University) for the cDNAs of the full-length ataxin-3 proteins and Anne Rintala-Dempsey and Liliana Santamaria-Kisiel for maintenance of the Biomolecular NMR Facility (University of Western Ontario).
Supporting Information Available
Procedures describing equations used for binding of three UIM regions to the parkin Ubl domain, chemical shift perturbation plots for the parkin Ubl domain upon binding of UIM_123 compared to binding of Eps15 and S5a, the assigned 1H–15N HSQC spectrum for ataxin UIM_123, and chemical shift perturbation plots for the parkin Ubl domain upon binding of UIM_123, UIM_1, UIM_2, and UIM_3 proteins. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This research was supported by a research grant from the Canadian Institutes of Health Research (MOP-14606) and an award from the Canada Research Chairs program (GSS).
Supplementary Material
References
- Durcan T. M.; Kontogiannea M.; Thorarinsdottir T.; Fallon L.; Williams A. J.; Djarmati A.; Fantaneanu T.; Paulson H. L.; Fon E. A. (2011) The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum. Mol. Genet. 20, 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H.; Hattori N.; Kubo S.; Mizuno Y.; Asakawa S.; Minoshima S.; Shimizu N.; Iwai K.; Chiba T.; Tanaka K.; Suzuki T. (2000) Familial parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25, 302–305. [DOI] [PubMed] [Google Scholar]
- Kitada T.; Asakawa S.; Hattori N.; Matsumine H.; Yamamura Y.; Minoshima S.; Yokochi M.; Mizuno Y.; Shimizu N. (1998) Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608. [DOI] [PubMed] [Google Scholar]
- Mizuno Y.; Hattori N.; Kitada T.; Matsumine H.; Mori H.; Shimura H.; Kubo S.; Kobayashi H.; Asakawa S.; Minoshima S.; Shimizu N. (2001) Familial Parkinson’s disease. α-Synuclein and parkin. Adv. Neurol. 86, 13–21. [PubMed] [Google Scholar]
- Harris G. M.; Dodelzon K.; Gong L.; Gonzalez-Alegre P.; Paulson H. L. (2010) Splice isoforms of the polyglutamine disease protein ataxin-3 exhibit similar enzymatic yet different aggregation properties. PLoS One 5, e13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K. K.; Zhang Y.; Lim K. L.; Tanaka Y.; Huang H.; Gao J.; Ross C. A.; Dawson V. L.; Dawson T. M. (2001) Parkin ubiquitinates the α-synuclein-interacting protein, synphilin-1: Implications for Lewy-body formation in Parkinson disease. Nat. Med. 7, 1144–1150. [DOI] [PubMed] [Google Scholar]
- Imai Y.; Soda M.; Hatakeyama S.; Akagi T.; Hashikawa T.; Nakayama K. I.; Takahashi R. (2002) CHIP is associated with Parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol. Cell 10, 55–67. [DOI] [PubMed] [Google Scholar]
- Staropoli J. F.; McDermott C.; Martinat C.; Schulman B.; Demireva E.; Abeliovich A. (2003) Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron 37, 735–749. [DOI] [PubMed] [Google Scholar]
- Chaugule V. K.; Burchell L.; Barber K. R.; Sidhu A.; Leslie S. J.; Shaw G. S.; Walden H. (2011) Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke S. J.; Chai Y.; Marrs G. L.; Wen H.; Paulson H. L. (2005) Defining the role of ubiquitin-interacting motifs in the polyglutamine disease protein, ataxin-3. J. Biol. Chem. 280, 32026–32034. [DOI] [PubMed] [Google Scholar]
- Safadi S. S.; Shaw G. S. (2010) Differential interaction of the E3 ligase parkin with the proteasomal subunit S5a and the endocytic protein Eps15. J. Biol. Chem. 285, 1424–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K.; Falquet L. (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem. Sci. 26, 347–350. [DOI] [PubMed] [Google Scholar]
- Mueller T. D.; Feigon J. (2003) Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 22, 4634–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safadi S. S.; Shaw G. S. (2007) A disease state mutation unfolds the parkin ubiquitin-like domain. Biochemistry 46, 14162–14169. [DOI] [PubMed] [Google Scholar]
- Fischer J. A. (2003) Deubiquitinating enzymes: Their roles in development, differentiation, and disease. Int. Rev. Cytol. 229, 43–72. [DOI] [PubMed] [Google Scholar]
- Burnett B.; Li F.; Pittman R. N. (2003) The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 12, 3195–3205. [DOI] [PubMed] [Google Scholar]
- Chai Y.; Berke S. S.; Cohen R. E.; Paulson H. L. (2004) Poly-ubiquitin binding by the polyglutamine disease protein ataxin-3 links its normal function to protein surveillance pathways. J. Biol. Chem. 279, 3605–3611. [DOI] [PubMed] [Google Scholar]
- Winborn B. J.; Travis S. M.; Todi S. V.; Scaglione K. M.; Xu P.; Williams A. J.; Cohen R. E.; Peng J.; Paulson H. L. (2008) The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 283, 26436–26443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Sawai N.; Kotliarova S.; Kanazawa I.; Nukina N. (2000) Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum. Mol. Genet. 9, 1795–1803. [DOI] [PubMed] [Google Scholar]
- Zhong X.; Pittman R. N. (2006) Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum. Mol. Genet. 15, 2409–2420. [DOI] [PubMed] [Google Scholar]
- Kobayashi T.; Tanaka K.; Inoue K.; Kakizuka A. (2002) Functional ATPase activity of p97/valosin-containing protein (VCP) is required for the quality control of endoplasmic reticulum in neuronally differentiated mammalian PC12 cells. J. Biol. Chem. 277, 47358–47365. [DOI] [PubMed] [Google Scholar]
- Mishra A.; Dikshit P.; Purkayastha S.; Sharma J.; Nukina N.; Jana N. R. (2008) E6-AP promotes misfolded polyglutamine proteins for proteasomal degradation and suppresses polyglutamine protein aggregation and toxicity. J. Biol. Chem. 283, 7648–765624. [DOI] [PubMed] [Google Scholar]
- Maciel P.; Costa M. C.; Ferro A.; Rousseau M.; Santos C. S.; Gaspar C.; Barros J.; Rouleau G. A.; Coutinho P.; Sequeiros J. (2001) Improvement in the molecular diagnosis of Machado-Joseph disease. Arch. Neurol. 58, 1821–1827. [DOI] [PubMed] [Google Scholar]
- Burnett B.; Li F.; Pittman R. N. (2003) The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 12, 3195–3205. [DOI] [PubMed] [Google Scholar]
- Walters K. J.; Kleijnen M. F.; Goh A. M.; Wagner G.; Howley P. M. (2002) Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Song A. X.; Zhou C. J.; Peng Y.; Gao X. C.; Zhou Z. R.; Fu Q. S.; Hong J.; Lin D. H.; Hu H. Y. (2010) Structural transformation of the tandem ubiquitin-interacting motifs in ataxin-3 and their cooperative interactions with ubiquitin chains. PLoS One 5, e13202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. D.; Wang B.; Alam S. L.; Higginson D. S.; Robinson H.; Sundquist W. I.; Hill C. P. (2003) Structure and ubiquitin binding of the ubiquitin-interacting motif. J. Biol. Chem. 278, 28976–28984. [DOI] [PubMed] [Google Scholar]
- Swanson K. A.; Kang R. S.; Stamenova S. D.; Hicke L.; Radhakrishnan I. (2003) Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J. 22, 4597–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K.; Tenno T.; Sugasawa K.; Jee J. G.; Ohki I.; Kojima C.; Tochio H.; Hiroaki H.; Hanaoka F.; Shirakawa M. (2004) Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. J. Biol. Chem. 279, 4760–4767. [DOI] [PubMed] [Google Scholar]
- Ko H. S.; Uehara T.; Tsuruma K.; Nomura Y. (2004) Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 566, 110–114. [DOI] [PubMed] [Google Scholar]
- Markin C. J.; Xiao W.; Spyracopoulos L. (2010) Mechanism for recognition of polyubiquitin chains: Balancing affinity through interplay between multivalent binding and dynamics. J. Am. Chem. Soc. 132, 11247–11258. [DOI] [PubMed] [Google Scholar]
- Kiessling L. L.; Gestwicki J. E.; Strong L. E. (2000) Synthetic multivalent ligands in the exploration of cell-surface interactions. Curr. Opin. Chem. Biol. 4, 696–703. [DOI] [PubMed] [Google Scholar]
- Mittag T.; Orlicky S.; Choy W.-Y.; Tang X.; Lin H.; Sicheri F.; Kay L. E.; Tyers M.; Forman-Kay J. D. (2008) Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 17772–17777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan T. M.; Fon E. A. (2011) Mutant ataxin-3 promotes the autophagic degradation of parkin. Autophagy 7, 233–234. [DOI] [PubMed] [Google Scholar]
- Kon N.; Kobayashi Y.; Li M.; Brooks C. L.; Ludwig T.; Gu W. (2010) Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29, 1270–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan T. M.; Kontogiannea M.; Bedard N.; Wing S. S.; Fon E. A. (2012) Ataxin-3 deubiquitination is coupled to Parkin ubiquitination via E2 ubiquitin-conjugating enzyme. J. Biol. Chem. 287, 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe J.-F.; Sauvé V.; Grenier K.; Seirafi M.; Yang M. Y.; Ménade M.; Al-Abdul-Wahid S.; Krett J.; Wong K.; Kozlov G.; Nagar B.; Fon E. A.; Gehring K. (2013) Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455. [DOI] [PubMed] [Google Scholar]
- Riley B. E.; Lougheed J. C.; Callaway K.; Velasquez M.; Brecht E.; Nguyen L.; Shaler T.; Walker D.; Yang Y.; Regnstrom K.; Diep L.; Zhang Z.; Chiou S.; Bova M.; Artis D. R.; Yao N.; Baker J.; Yednock T.; Johnston J. A. (2013) Structure and function of parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan T. M.; Fon E. A. (2013) Ataxin-3 and its E3 partners: Implications for Machado-Joseph disease. Front. Neurol. 4, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M.; Narendra D. P.; Jin S. M.; Tekle E.; Banerjee S.; Youle R. J. (2012) Pink1 drives parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J. Cell Biol. 200, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.