Abstract
Complete sequences of myriad eukaryotic genomes, including several human genomes, are now available, and recent dramatic developments in DNA sequencing technology are opening the floodgates to vast volumes of sequence data. Yet, despite knowing for several decades that a significant proportion of cytosines in the genomes of plants and animals are present in the form of methylcytosine, until very recently the precise locations of these modified bases have never been accurately mapped throughout a eukaryotic genome. Advanced “next-generation” DNA sequencing technologies are now enabling the global mapping of this epigenetic modification at single-base resolution, providing new insights into the regulation and dynamics of DNA methylation in genomes.
Published in February 2001, the rough draft version of the human genome was widely heralded as the “book of life,” an ∼3-billion-letter code composed of just four letters within which is described our cellular and physiological complexity, and our genetic heritage (International Human Genome Sequencing Consortium 2001; Venter et al. 2001). Yet the book was missing proper formatting of some of the characters within its pages; omitted from this landmark volume was the elusive and dynamic fifth letter of the code: 5-methylcytosine. Accounting for ∼1%–6% of the nucleotides within mammalian and plant genomes (Montero et al. 1992), 5-methylcytosine, commonly referred to as DNA methylation, is a modified base that imparts an additional layer of heritable information upon the DNA code, which is important for regulating the underlying genetic information. For instance, DNA methylation is essential for viability and is involved in myriad biological processes, including embryogenesis and development, genomic imprinting, silencing of transposable elements, and regulation of gene transcription (Li et al. 1992; Bestor 2000; Lippman et al. 2004; Zhang et al. 2006; Reik 2007; Weber and Schübeler 2007; Zilberman et al. 2007; Lister et al. 2008). However, despite playing these critical roles in higher eukaryotes, identification of sites of DNA methylation throughout a genome has not been achieved until very recently. Consequently, the full extent to which DNA methylation regulates gene expression, chromatin structure, and yet-to-be-discovered processes has not been possible to ascertain.
As with analysis of conventional nucleotide polymorphisms, ultimately it will be important to understand the genome-wide distribution of 5-methylcytosine at the single-base resolution. In the past this has not been technically or economically feasible, but with the dramatic advances being made in high-throughput DNA sequencing it is now possible to map the sites of DNA methylation at single-base resolution throughout an entire genome. Previous investigations at a limited number of loci have reported significant correlation between the methylation state of CpGs within 1000 bases (Eckhardt et al. 2006). This has prompted the question of whether it was necessary to identify sites of DNA methylation at the single-base level (Down et al. 2008). However, numerous studies have demonstrated the critical importance of knowing the methylation status of individual CpG sites, which can vary even when in very close proximity to apparently invariable methylcytosines (Prendergast and Ziff 1991; Weaver et al. 2004, 2007). Inhibition of binding of the SP1 transcription factor and the insulator protein CTCF by cytosine DNA methylation within their binding elements has been extensively documented (Clark et al. 1997; Kitazawa et al. 1999; Mancini et al. 1999; Bell and Felsenfeld 2000; Hark et al. 2000; Inoue and Oishi 2005; Douet et al. 2007). It was recently demonstrated that RNA-directed DNA methylation of a single CpG located within a putative conserved intronic cis element of the Petunia floral homeotic gene pMADS3 caused ectopic expression of pMADS3 (Shibuya et al. 2009). Notably, both the epiallele and ectopic expression could be inherited in subsequent generation in the absence of the RNA trigger. Intriguingly, Weaver and colleagues discovered that high levels of maternal licking and grooming of rat pups are associated with lower cytosine methylation at a specific CpG in the promoter of the gene encoding glucocorticoid receptor (GR) in the hippocampus, whereas low levels of this maternal care are associated with high methylation at this same CpG (Weaver et al. 2004). Notably, the methylation status of another methylated cytosine only six bases downstream was not found to change. Moreover, increased methylation at the 5′ CpG, which is located within the binding site of the early growth response 1 (EGR1; also known as nerve growth factor-inducible protein A [NGFI-A]) transcription factor, results in a decrease in the binding of EGR1, inhibition of GR promoter activity, and lower EGR1-induced transcription of the GR gene (Weaver et al. 2007). Clearly, global identification of sites of DNA methylation at single-base resolution in combination with detailed maps of DNA–protein interactions throughout development and under diverse conditions will be critical to elucidating such complex processes.
In this review we discuss new approaches being used to identify sites of cytosine methylation throughout genomes that have been made possible by rapid advances in high-throughput DNA sequencing and discuss some of the challenges encountered and that lie ahead.
Initial studies of single-base detection of DNA methylation
A number of methods have been developed for genome-wide detection of sites of DNA methylation (for review, see Esteller 2007; Beck and Rakyan 2008). Widely used approaches include enzymatic digestion of methylated DNA followed by hybridization to high-density oligonucleotide arrays (Lippman et al. 2005; Martienssen et al. 2005; Vaughn et al. 2007) or two-dimensional gel electrophoresis (restriction landmark genomic scanning), and capture of methylated fragments of genomic DNA with methyl-binding domain proteins or antibodies specific to 5-methylcytosine (methylated DNA immunoprecipitation, MeDIP), followed by hybridization to arrays (Weber et al. 2005; Keshet et al. 2006; Zhang et al. 2006; Penterman et al. 2007; Zilberman et al. 2007). However, methods that rely on enzyme digestion are confined to recognition elements and consequently can only interrogate a very small subset of all sites of methylation or generate DNA probe fragments that are of such length that the precise location of the methylcytosine may not be identifiable. Furthermore, techniques such as MeDIP that rely on hybridization to oligonucleotide arrays are subject to several limitations including low resolution of detection, difficulty in discrimination of similar sequences, inability to determine the sequence context of DNA methylation sites, requirement for a dedicated array, and bias toward enrichment of sites containing relatively high levels of cytosine methylation.
Recently, the MeDIP approach has been coupled with new sequencing technologies, providing sequence information on the immunoprecipitated DNA fragments, dubbed MeDIP-seq (Down et al. 2008). This technique has advantages over the use of arrays, such as yielding sequence-level information that aids in distinguishing highly similar sequences. However, the method largely remains susceptible to the same weaknesses mentioned above that are inherent in the utilization of an antibody or protein to capture large DNA molecules that contain methylation, for example, the bias in MeDIP toward CpG-rich sequences and low sensitivity for low CpG density regions, such as outside CpG islands (Irizarry et al. 2008; Lister et al. 2008; Tomazou et al. 2008).
A breakthrough in the high-resolution detection of DNA methylation was development of “bisulfite (BS) conversion” (Frommer et al. 1992), an experimental procedure in which treatment of DNA with sodium bisulfite under denaturing conditions converts cytosines, but not methylcytosines, into uracil via a sulfonation, deamination, desulfonation reaction. Subsequent synthesis of the complementary strand and sequencing allows determination of the methylation status of cytosines on each strand of the genomic DNA simply by observing whether the sequenced base at a cytosine position is a thymine (unmethylated) or a cytosine (methylated). Thus, BS conversion translates an epigenetic difference into a genetic one, offering an unparalleled assay for studying an epigenetic modification, and it is regarded as the gold-standard method of detecting cytosine methylation. Furthermore, repeatedly sampling a locus by sequencing independent template molecules can provide a digital measurement of the frequency that a cytosine is methylated.
BS sequencing has been extensively used for analysis of loci of interest by PCR amplification, cloning, and Sanger sequencing. For example, in a brute-force application of this technique, Eckhardt and colleagues analyzed up to 20 clones each of 2524 distinct regions of human chromosomes 6, 20, and 22 in 12 different tissues, assessing the methylation state of the CpG sites within these regions and identifying tissue-specific patterns of DNA methylation associated with differential transcript abundance (Eckhardt et al. 2006). Yet, this large undertaking covered only a tiny fraction of the human genome sequence. Clearly, reliance on locus-specific BS sequencing approaches to determine the presence or frequency of methylation rapidly becomes technically and financially impractical as the number of genomic loci being studied increases or presence of methylation at lower frequency is sought. Indeed, any method that relies on locus-specific amplification following BS treatment is impractical for scaling up to analysis of the entire genome, requiring synthesis of vast numbers of oligonucleotide primers. Moreover, a priori knowledge or assumption of the methylation state of the primer hybridization sites is required for successful base-pairing to the BS-converted sequences, or alternatively the use of degenerate primer sequences with concomitant reduction in amplification specificity. Clearly, dramatically higher sequencing throughput ultimately coupled to unbiased selection of genomic regions is necessary to avoid the limitations imposed by locus-specific cloning and Sanger sequencing in the detection of DNA methylation.
Single-base methylomes by genome-wide shotgun sequencing
Dramatic developments in high-throughput sequencing technologies are driving a paradigm shift in global single-base resolution DNA methylation analysis. Several “next-generation” DNA sequencing platforms are now available that, at the time of this writing, can yield several gigabases (Gb) of high-quality aligned sequence per 3- to 5-d run (for review, see Mardis 2008; Shendure and Ji 2008). For example, a single Illumina Genome Analyzer run currently produces over one hundred million distinct sequence reads (up to 76 nucleotides), which has recently been utilized for large-scale BS-sequencing studies to produce single-base resolution maps of the sites of DNA methylation (the “methylome”) for the entire Arabidopsis thaliana genome (Cokus et al. 2008; Lister et al. 2008) and for a select subset of sites in the mouse genome (Meissner et al. 2008). These experiments have provided the first high-resolution characterizations of the DNA methylome within the analyzed tissues and cell types, illuminating the genomic distribution and patterns of DNA methylation and its relationships with subsets of the transcriptome. Although each study utilized BS conversion and the same sequencing platform, each offers unique approaches to sequencing library production and data analysis and will be discussed below.
The new sequencing technologies have been designed with the general intention of sequencing a genome composed of four bases present in roughly similar proportions. However, after BS conversion, the DNA being sequenced is effectively composed of just three bases. The Illumina Genome Analyzer, which was used for all of the single-base resolution methylation studies mentioned above, encounters a high error rate when base-calling is performed on only BS-converted DNA. For this reason, it was necessary to utilize a single lane of each sequencing flowcell to sequence a control library, composed of all four bases. The Illumina analysis pipeline uses the control lane for autocalibration of the base-calling parameters to enable accurate base calling on the BS-converted libraries (Cokus et al. 2008; Lister et al. 2008). Additionally, Cokus et al. (2008) developed a multidimensional Gaussian mixture model to optimize the base calling performance.
Both Cokus et al. (2008) and Lister et al. (2008) analyzed not only wild-type A. thaliana but also a number of mutants deficient in enzymes required for the establishment, maintenance, and removal of DNA methylation. Cytosine methylation patterns are initiated and perpetuated through cell division by DNA methyltransferases, which catalyze the transfer of a methyl group to cytosine to form 5-methylcytosine. DNA methylation in mammals is thought to be predominantly in the CG sequence context; however, in plants it is commonly found in all sequence contexts (CG, CHG, CHH; where H = A, C, or T) (Bernstein et al. 2007; Henderson and Jacobsen 2007). DNA methylation is established by the de novo DNA methyltransferases DNMT3A and DNMT3B in mammals (Okano et al. 1999) and the orthologous DOMAINS REARRANGED METHYLTRANSFERASE 1/2 (DRM1/2) in plants that are targeted to methylate-specific genomic loci by small RNA molecules in the process of RNA-directed DNA methylation (Cao and Jacobsen 2002; Cao et al. 2003). Post-replicative maintenance of DNA methylation at CG sites is catalyzed by the DNA methyltransferase DNMT1 in mammals and its ortholog METHYLTRANSFERASE 1 (MET1) in plants, while the plant-specific DNA methyltransferase CHROMOMETHYLASE 3 is required for maintenance of DNA methylation at CHG sites in plants (Finnegan and Dennis 1993; Bartee et al. 2001; Bird 2002; Jackson et al. 2002; Kankel et al. 2003; Saze et al. 2003; Goll and Bestor 2005). Furthermore, plants possess the demethylase enzymes ROS1, DME, DML2, and DML3 that remove methylcytosines by a base-excision-mediated repair process (Gong et al. 2002; Penterman et al. 2007), and a recent study indicates that small RNA molecules can target the ROS1 demethylase to specific genomic target regions via the RNA-binding protein AT5G58130 (also known as ROS3) (Zheng et al. 2008).
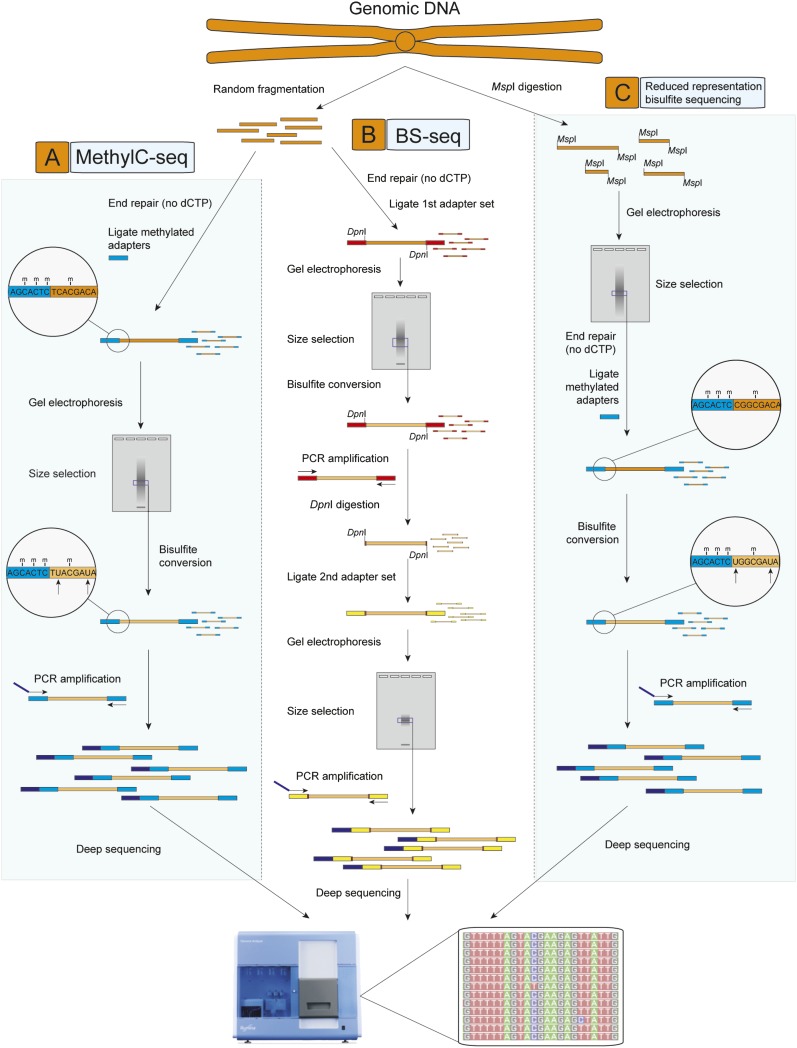
Cokus et al. (2008) generated a map of cytosine methylation at single-base resolution of the aerial tissues of A. thaliana, implementing a unique approach dubbed “BS-seq” to generate complex libraries of short BS-converted fragments of genomic DNA amenable to sequencing on the Illumina Genome Analyzer. In this method, purified genomic DNA was first fragmented and ligated to the first set of double-stranded adaptors that contained methylated adenine bases within DpnI restriction sites close to the site of ligation. After BS conversion, PCR was performed using primers complementary to the converted adapter sequences, yielding double-stranded DNA that was digested with DpnI to remove only the first adapter set. Sequencing adapters were subsequently ligated to the double-stranded BS-converted genomic DNA fragments, and PCR with primers complementary to the adapters was performed to yield a sequencing library (Fig. 1).
Figure 1.
Techniques for genome-wide sequencing of cytosine methylation sites. Three techniques used recently to generate bisulfite (BS) sequencing libraries compatible with next-generation sequencing are depicted. (A) MethylC-seq (Lister et al. 2008). Double-stranded universal adapter sequences in which all cytosines are methylated are ligated to fragmented genomic DNA. Sodium bisulfite treatment converts unmethylated cytosines to thymine, after which library yield enrichment by PCR with primers complementary to the universal adapter sequences produces the final library that can be sequenced. (B) BS-seq (Cokus et al. 2008). Ligation of a first set of double-stranded adaptors that contained methylated adenine bases within DpnI restriction sites close to the site of ligation with genomic DNA. After BS conversion, PCR is performed using primers complementary to the converted adapter sequences, yielding double-stranded DNA that is digested with DpnI to remove only the first adapter set. Sequencing adapters are subsequently ligated to the double-stranded BS-converted genomic DNA fragments, and PCR with primers complementary to the adapters performed to yield a sequencing library. (C) Reduced representation BS sequencing (RRBS) (Meissner et al. 2008). Genomic DNA is first digested by the methylation-insensitive MspI restriction enzyme, which cleaves the phosphodiester bond upstream of the CpG dinuclotide in its CCGG recognition element. Digested DNA is then separated by gel electrophoresis, and one or more specific size fractions are selected. The size-selected DNA is then end repaired, ligated to double-stranded methylated sequencing adapters (as described above for MethylC-seq), BS converted, and amplified by PCR with primers complementary to the adapter sequences.
Several computational filters were applied to the reads after sequencing, including removing sequences that likely mapped to multiple positions and potentially unconverted reads that contained at least three consecutive cytosines in the CHH context. Although sequence complexity is reduced after BS conversion, computational simulations indicated that sequence reads of just 31 bases can be uniquely mapped to ∼92% of the cytosines in the A. thaliana genome, and experimental results achieved very close to this theoretical maximum. Any genomic sequence that is unique at the sequence read-length after BS conversion can be interrogated for methylation status, overcoming cross-hybridization issues that can affect microarray signal specificity. Using reads of 31 bases, 2.6 Gb of sequence were retained post-filtering, covering ∼85% of the 43 million cytosines in the 119-Mb genome with an average coverage of 20× (Cokus et al. 2008).
With single-base identification of methylcytosines, it was possible to categorize the amount and distribution of methylation in each sequence context, over- and underrepresented local sequence motifs associated with DNA methylation, and characterize the different methylation composition of diverse genomic environments including euchromatin and pericentromeric heterochromatin, gene bodies, telomeres, transposons, and various classes of repeat. Furthermore, by analyzing the sequences flanking sites of DNA methylation, it was evident that methylation in CG, CHG, and CHH sequence contexts each displayed different surrounding motifs that were enriched for methylation. Cokus and colleagues also conducted detailed analysis of the spatial patterning of sites of DNA methylation, identifying distinct correlations between proximal methylation in different contexts that suggest complex relationships between the various forms of methylation (Cokus et al. 2008). Furthermore, a periodicity of 167 nucleotides (nt) was discovered between sites of methylation, a spacing that is close to the internucleosome linker length in plants, possibly indicating that the linker sequences are more exposed to DNA methyltransferases or methylation is functionally related to nucleosome positioning. Adjacent sites of CHH methylation, which are initiated and maintained by the de novo plant DNA methyltransferase DRM2, frequently displayed a 10-base periodicity, equating to one turn of the DNA double helix. Interestingly, recent crystallization of DNMT3A, the mammalian ortholog of DRM2, with its regulatory factor DNA (cytosine-5-)-methyltransferase 3-like protein (DNMT3L), resolved a tetrameric complex with two active sites that could methylate CG sites separated by one helical turn of DNA (Jia et al. 2007). Thus, single-base resolution DNA methylation maps are able to reveal such fine-scale patterns indicative of conservation of the activity of these DNA methyltransferases between plants and mammals. Cokus et al. (2008) also performed limited BS-sequencing (90 megabases [Mb]) on a range of plants containing different combinations of genetic lesions in the DNA methyltransferases MET1, DRM1, DRM2, and CMT3, revealing the effect upon global DNA methylation patterns, methylation of different genomic features, and relationships between methylation in different sequence contexts. Finally, the authors also performed limited BS-sequencing (60 Mb) on genomic DNA from mouse germ cell tissues, uniquely mapping ∼66% of reads to the mouse genome in a demonstration of the technique's applicability to larger mammalian genomes.
In our study, sequencing of the BS-converted genome of A. thaliana isolated from flower buds was performed (Lister et al. 2008). We developed a method, dubbed “MethylC-seq,” in which fragmented genomic DNA is ligated to sequencing adapters where all cytosines are methylated. Subsequent BS conversion of the ligated genomic DNA does not convert the sequence of the methylated adapters, and amplification with primers complementary to the adapters yields a library amenable to sequencing (Fig. 1). We mapped reads of 49–56 bases to the A. thaliana genome, removing potential clonal reads that shared the same start site and reads that aligned to multiple positions in the genome, to retain over 39 million reads that yielded ∼2.0 Gb of unique MethylC-seq sequence. Approximately 79% of all cytosines in the genome were covered with at least two reads, with an average coverage of 16×, or 8× per strand of the genome. We identified over 2.2 million methylcytosines in the A. thaliana flower bud nuclear genomes, and, as also observed by Cokus et al. (2008), while the majority was identified in the CG context (55%), significant proportions were identified in the CHG and CHH contexts (23% and 22%, respectively). A parallel analysis using the methylcytosine immunoprecipitation methodology (Zhang et al. 2006) and hybridization to tiling microarrays with the same sample showed MethylC-seq to be significantly more sensitive, identifying 48.3% of the methylcytosines in regions not predicted as methylated by microarray-based detection, including genic, promoter, telomeric, and repetitive regions (Lister et al. 2008).
BS-seq/MethylC-seq generally yields many reads covering each cytosine, providing a digital read-out of the frequency at which that cytosine was methylated in the sample. Indeed, the frequency of methylation was found to have a distinct profile for each different context in A. thaliana, with CG methylation most commonly found at 80%–100%, while CHG was methylated at a wide range of frequencies and CHH methylated infrequently (∼30%) (Cokus et al. 2008; Lister et al. 2008). Similar principles apply to quantitation of DNA methylation levels at any particular cytosine by shotgun sequencing as they do in classical BS sequencing of cloned PCR products. Each nonclonal read can be counted as a localized assessment of the methylation state in one copy of the genome, and the granularity of the measurement is thus determined by sequence coverage. This measurement of methylation level from the shotgun BS sequencing agrees closely with conventional BS sequencing (Cokus et al. 2008). Of course, the cost to achieve a given coverage, and thus resolution of methylation level, depends on the size of a genome. It should be noted that the methylation state of a given stretch of genomic DNA, and thus the base composition after BS conversion, may have an impact upon the efficiency of PCR amplification during the sequencing library preparative and cluster amplification prior to sequencing. This may affect the relative representation of sequences that originate from the same genomic region but that possess different methylation states, which may be problematic for unbiased quantification of the level of DNA methylation at any given locus. However, quantitation of the methylation level in a tissue provides only the overall sum methylation state of the pooled genomes, yet in the context of a single cell the methylation state of a particular cytosine is binary. Advances in cell sorting, tissue microdissection, and sequencing from very low quantities of biological material will hopefully enable the focus to be shifted away from assessing average levels of methylation within a tissue to interrogating the changes that take place within few, or even single, cells.
We also used MethylC-seq, at high read coverage (average of ∼6× for each BS-converted strand of the genome), to investigate and quantify the changes in the DNA methylome in a range of DNA methyltransferase mutants, identifying the subset of DNA methylation that required the activity of the different enzymes. The met1 mutant lost nearly all methylation in the CG context, but intriguingly new methylation in the CHG context was observed in the euchromatic regions of the chromosome. Furthermore, while gene-body CG methylation was effectively abolished, profiling of the distribution of DNA methylation within gene bodies in met1 revealed that CHG methylation was now distributed in a pattern very similar to wild-type CG gene body methylation, indicating compensation for the loss of CG methylation by the plant-specific CMT3 methyltransferase (Cokus et al. 2008; Lister et al. 2008). Interestingly, residual CHH methylation was observed in the drm1 drm2 cmt3 mutant, indicating that another methyltransferase may be present that can act in this context (Cokus et al. 2008; Lister et al. 2008). Additionally, we sequenced the methylome of a triple mutant defective in the DNA demethylase enzymes ROS1, DML2, and DML3, finding hundreds of discrete regions of hypermethylation throughout the genome of the ros1 dml2 dml3 mutant relative to wild type. Indeed, these hypermethylated regions were often located in gene promoters and 3′ UTRs, indicating that the demethylases actively protect these regions from DNA methylation, potentially to prevent interference with DNA binding proteins or the processes of transcriptional initiation and termination (Zhang et al. 2006; Penterman et al. 2007; Lister et al. 2008).
Single-base resolution maps of DNA methylation can be integrated with other cellular data sets to provide a multifaceted analysis of the cellular signals influencing DNA methylation patterns and the downstream effects of this modification upon transcription. As mentioned above, the process of RNA-directed DNA methylation in plant cells utilizes diverse pools of small RNAs (smRNAs) to target regions of the genome for methylation. To explore this relationship on a genome-wide level, we sequenced the cellular smRNA population from the same flower bud tissue and analyzed the overlap between these short effector molecules and DNA methylation (Lister et al. 2008). A high correlation between the presence of a smRNA and DNA methylation at the genomic locus was observed, and moreover it was found that the precise site of homology between a smRNA and the genomic DNA was specifically enriched for the presence of DNA methylation in a strand-specific manner. Furthermore, alteration of DNA methylation levels in the mutant lines had a dramatic effect upon proximal smRNA populations, with sites of hypomethylation displaying less smRNAs while hypermethylated regions suddenly showed a dramatic increase in small RNA density, likely indicating the presence of positive feedback systems affecting the abundance of smRNAs and DNA methylation.
Finally, to explore the relationship between changes in DNA methylation and gene expression, we developed a method for strand-specific RNA sequencing called mRNA-seq (Lister et al. 2008). The mRNA-seq method revealed that upon alteration of DNA methylation patterns, the abundance of hundreds of genes, transposons, and unannotated intergenic transcripts was altered (Lister et al. 2008). Notably, only by generating this sequence-based transcript information could the expression of distinct subfamily members of highly repeated transposon families be resolved, whereas cross-hybridization issues encountered with microarrays typically preclude such analysis.
Clearly, with the current state of new sequencing technologies, characterization of the sites of DNA methylation throughout a genome on the order of hundreds of megabases is practical. Furthermore, the demonstration by Cokus et al. (2008) that 66% of 31-base BS-seq reads mapped uniquely to the mouse genome indicates that the approach can be scaled to larger mammalian genomes. The projected significant increases in read length in the near future and the availability of paired-end sequencing will further increase the ability to uniquely map BS-converted sequence reads to large genomes. However, to achieve similar levels of coverage for a much larger genome, for example, the ∼3.2-Gb human genome, ∼25-fold more sequencing is required. Therefore, to overcome the large amounts of sequence required to sequence an entire mouse genome, Meissner et al. (2008) utilized an approach termed reduced representation BS sequencing (RRBS) (Meissner et al. 2005, 2008). In this method genomic DNA is first digested by the methylation-insensitive MspI restriction enzyme, which cleaves the phosphodiester bond upstream of the CpG dinuclotide in its CCGG recognition element. Digested DNA is then separated by gel electrophoresis, and one or more specific size fractions are selected. The size-selected DNA is then end repaired, ligated to methylated sequencing adapters (as described above for MethylC-seq), BS converted, and amplified by PCR with primers complementary to the adapter sequences (Fig. 1). The MspI digestion and size-selection yields a sequencing library that is enriched for CpG sites (Meissner et al. 2005, 2008). Indeed, computational analysis demonstrated that a selection of 40- to 220-bp MspI digestion products of mouse genomic DNA would maximally cover ∼1 million CpG sites at 36-base sequence read length, ∼5% of all CpG sites in the mouse genome, or only 1% of the entire genome (Jeddeloh et al. 2008; Meissner et al. 2008). Applying RRBS and mapping of the location of various histone modifications to mouse embryonic stem (ES) cells, ES cell-derived and primary neural progenitor cells (NPCs), and several other primary cell lines, Meissner et al. found that patterns of DNA methylation were more clearly reflected by the complement of histone modifications than CpG density. Furthermore, extensive changes in DNA methylation state in many regulatory regions located in CpG-poor sequences were identified during in vitro differentiation of the ES cells. Finally, the authors reported that cultured NPCs progressively became hypermethylated at a distinct set of high CpG density promoters as passage number increases, suggesting that progressive culturing may induce changes in epigenetic marks (Meissner et al. 2008).
Another approach recently reported combines digestion of genomic DNA with DNA methylation sensitive or insensitive restriction enzymes followed by high-throughput sequencing of the digestion fragments (Brunner et al. 2009). In a method dubbed Methyl-seq, these investigators digested genomic DNA from human ES cells, ES cell-derived cells, and fetal and adult liver cells with MspI, which digests at any 5′-CCGG-3′ site, and HpaII, which digests only at unmethylated 5′-CCGG-3′ sites. DNA fragments from the MspI and HpaII digests were then sequenced with an Illumina Genome Analyzer. Sequences in the MspI but not HpaII samples were classified as methylated, while sequences in the HpaII site only were from at least partially unmethylated regions of the genome. With between 3 and 10 million mapped reads for each digested sample, this approach assayed over 90,000 regions in the human genome, accounting for 65% of annotated CpG islands in high-, intermediate- and low-CpG promoters. The investigators identified changes in DNA methylation during cellular differentiation that were localized to low-density CpG promoters, H3K27me3-modified regions, and bivalent domains (Brunner et al. 2009).
Sequence complexity-reduction approaches such as RRBS or Methyl-seq clearly have the advantage that they enable many samples to be analyzed with less sequencing required for each, by interrogating a select subset of the genome. However, the enzymatic cleavage in the RRBS and Methyl-seq methods results in a bias toward regions that have a high CpG density such as CpG islands, at the expense of covering low CpG density regions. Combined with the restricted coverage inherent in RRBS, this bias potentially leads to selection against genomic regions of biological importance that may be affected by DNA methylation, such as enhancers.
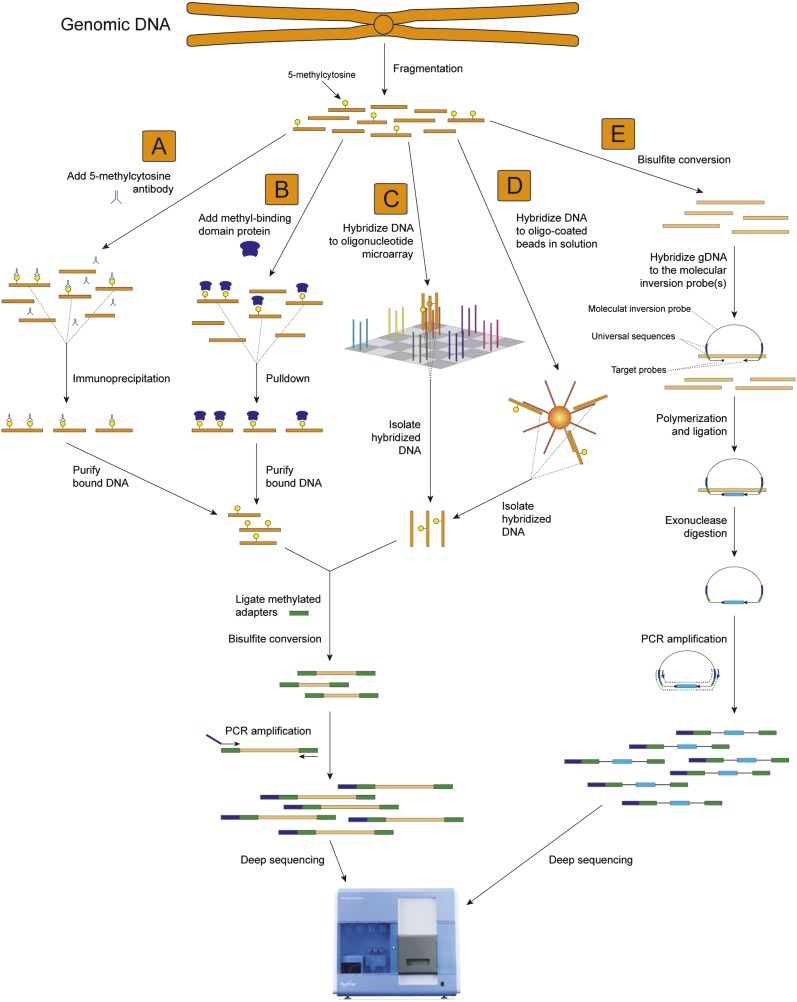
Several other sequence selection techniques that may be used prior to BS sequencing (Fig. 2; Garber 2008) include capture of specific sequences by hybridization to DNA molecules on arrays (Albert et al. 2007; Hodges et al. 2007; Okou et al. 2007) or bound to beads in solution (Bashiardes et al. 2005), with padlock or molecular inversion probes (Nilsson et al. 1994; Absalan and Ronaghi 2007; Ball et al. 2009; Deng et al. 2009), with proteins that bind to methylated DNA (Zhang et al. 2006), or with an antibody that binds to methylcytosines (MeDIP/mCIP) (Weber et al. 2005; Keshet et al. 2006; Zhang et al. 2006; Penterman et al. 2007; Zilberman et al. 2007). Recently, Down et al. (2008) performed MeDIP with mammalian male gametes followed by sequencing of the immunoprecipitated DNA using the Illumina Genome Analyzer, in a procedure called MeDIP-seq. While this procedure was able to generate a map of the likely methylated regions of the genome for this cell type, the lack of BS conversion meant that the investigators were not able to identify the sites of DNA methylation within the immunoprecipitated regions.
Figure 2.
Techniques for enrichment of methylated or target regions prior to BS sequencing. Five approaches that may be used to reduce the complexity of a sample before BS conversion and next-generation sequencing are depicted, targeting methylated regions or select target sequences. (A) MeDIP. Methylated fragments of genomic DNA are immunoprecipitated with an anti-5-methylcytosine antibody. Purified, immunoprecipitated DNA is ligated to double-stranded universal adapter sequences in which all cytosines are methylated. Sodium bisulfite treatment converts unmethylated cytosines to thymine, after which library yield enrichment by PCR with primers complementary to the universal adapter sequences produces the final library that can be sequenced. (B) MBD. Methylated fragments of genomic DNA are isolated from a complex mix of fragmented genomic DNA with a methyl binding domain protein, after which adapter ligation, BS conversion, and PCR enrichment are performed as in A. (C) Microarray capture. Target sequences within a complex mix of fragmented genomic DNA are captured by hybridization to specific oligonucleotides on the surface of a microarray. Following isolation of the hybridized genomic DNA, adapter ligation, BS conversion, and PCR enrichment are performed as in A. (D) Capture in solution. Specific target regions within a mix of fragmented genomic DNA are captured by hybridization to specific oligonucleotides attached to beads in solution. Following isolation of the hybridized genomic DNA, adapter ligation, BS conversion, and PCR enrichment are performed as in A. (E) Molecular inversion probe capture. Fragmented genomic DNA is BS converted, after which molecular inversion probes are added that are designed to hybridize to specific target sequences after conversion. Polymerization primed by the 3′ end of the molecular inversion probe followed by ligation generates a circular molecule that contains the target sequence and is not digested by subsequent exonuclease treatment. PCR using primers that hybridize to the ends of the molecular inversion probes allows amplification of the target region, to which double-stranded universal adapter sequences are ligated to produce a library that is sufficient for next-generation sequencing.
While these approaches are currently pragmatic and offer clear cost benefits for analysis of many samples and large genomes, they obviously suffer from the potential to miss important changes in DNA methylation that occur outside of the captured genomic regions. Furthermore, they require significant upfront costs and effort in development of the dedicated sequence capture effectors (e.g., molecular inversion probes or microarrays), which once synthesized are applicable to only a limited range of biological sources. Finally, techniques such as MeDIP/mCIP display a bias toward highly methylated regions and may miss a significant proportion of the genomic regions that contain DNA methylation (Cokus et al. 2008; Lister et al. 2008).
Several companies are developing new instruments that are claimed to deliver extraordinary reductions in the cost and time per base of sequence, with greatly increased read length and overall sequence output (e.g., Pacific Biosciences, http://www.pacificbiosciences.com; Complete Genomics, http://www.completegenomics.com; Visigen Biotechnologies, http://visigenbio.com; Intelligent Bio-systems, http://www.intelligentbiosystems.com) (Coombs 2008; Shendure and Ji 2008). With such advances, BS sequencing of many multi-gigabase-size genomes will very likely be economically feasible. Future development of sequence mapping algorithms to enable faster and more accurate mapping of BS-converted DNA to large and repetitive genomes will undoubtedly benefit whole-genome BS sequencing studies, for example, incorporating tolerance of C-T mismatches in the alignment scoring matrices. Increases in sequence read length will not only aid unambiguous alignment of sequences but will dramatically improve the ability to study allelic variation in DNA methylation through colocalization of genetic and epigenetic polymorphisms within a single read. Moreover, while cells within an organism possess the same genome sequence, numerous studies have reported differential cytosine methylation in distinct cell types, indicating that an organism's cells may display high variability, akin to its diverse transcriptomes (Futscher et al. 2002; Ching et al. 2005; Bibikova et al. 2006; Feldman et al. 2006; Oda et al. 2006; Weber and Schübeler 2007). Thus, with advances in sequencing technology it will be possible to explore this concept of a dynamic cytosine methylome via recording of this mark in each different cell type within an organism throughout development, under normal and disease states and in response to a variety of environmental influences.
Concluding remarks
The dramatic increase in sequencing throughput recently has ushered in a new era in the global detection of DNA methylation sites, opening the door to a plethora of detailed experiments investigating DNA methylation marks in plant and animal genomes. The frontier of high-throughput methylome sequencing will inevitably progress from only a small handful of studies to examination of the patterning and dynamics cytosine methylation in diverse samples and states, including nutrition/diet, various abiotic and biotic stresses, distinct cell types, mutants or diseases, and in large numbers of individuals from natural populations. Given the numerous observations of the variation in cytosine methylation patterns, it is possible that our cells possess multifarious temporal and spatial methylomes. Thus, it may eventuate that, with regard to mapping this “fifth base” of the genomic code, we are now not at the beginning of the end, but perhaps only at the end of the beginning.
Acknowledgments
We thank Dr. Brian Gregory for valuable input in the manuscript preparation. R.L. is supported by a Human Frontier Science Program Long-term Fellowship. This work was supported by grants from the National Institutes of Health, the National Science Foundation, the Department of Energy, and the Mary K. Chapman Foundation to J.R.E.
Footnotes
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083451.108.
References
- Absalan F., Ronaghi M. Molecular inversion probe assay. Methods Mol. Biol. 2007;396:315–330. doi: 10.1007/978-1-59745-515-2_20. [DOI] [PubMed] [Google Scholar]
- Albert T.J., Molla M.N., Muzny D.M., Nazareth L., Wheeler D., Song X., Richmond T.A., Middle C.M., Rodesch M.J., Packard C.J., et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- Ball M., Li J., Gao Y., Lee J., LeProust E.M., Park I., Xie B., Daley G., Church G.M. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartee L., Malagnac F., Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes & Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashiardes S., Veile R., Helms C., Mardis E.R., Bowcock A.M., Lovett M. Direct genomic selection. Nat. Methods. 2005;2:63–69. doi: 10.1038/nmeth0105-63. [DOI] [PubMed] [Google Scholar]
- Beck S., Rakyan V. The methylome: Approaches for global DNA methylation profiling. Trends Genet. 2008;24:231–237. doi: 10.1016/j.tig.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Meissner A., Lander E.S. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bestor T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- Bibikova M., Chudin E., Wu B., Zhou L., Garcia E.W., Liu Y., Shin S., Plaia T.W., Auerbach J.M., Arking D.E., et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–1083. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Brunner A.L., Johnson D.S., Kim S.W., Valouev A., Reddy T.E., Neff N.F., Anton E., Medina C., Nguyen L., Chiao E., et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009 doi: 10.1101/gr.088773.108. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S.E. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Cao X., Aufsatz W., Zilberman D., Mette M.F., Huang M.S., Matzke M., Jacobsen S.E. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 2003;13:2212–2217. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- Ching T.T., Maunakea A.K., Jun P., Hong C., Zardo G., Pinkel D., Albertson D.G., Fridlyand J., Mao J.H., Shchors K., et al. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat. Genet. 2005;37:645–651. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]
- Clark S.J., Harrison J., Molloy P.L. Sp1 binding is inhibited by mCpmCpG methylation. Gene. 1997;195:67–71. doi: 10.1016/s0378-1119(97)00164-9. [DOI] [PubMed] [Google Scholar]
- Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombs A. The sequencing shakeup. Nat. Biotechnol. 2008;26:1109–1112. doi: 10.1038/nbt1008-1109. [DOI] [PubMed] [Google Scholar]
- Deng J., Shoemaker R., Xie B., Gore A., Leproust E., Antosiewicz-Bourget J., Egli D., Maherali N., Park I., Yu J., et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douet V., Heller M.B., Le Saux O. DNA methylation and Sp1 binding determine the tissue-specific transcriptional activity of the mouse Abcc6 promoter. Biochem. Biophys. Res. Commun. 2007;354:66–71. doi: 10.1016/j.bbrc.2006.12.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down T., Rakyan V., Turner D., Flicek P., Li H., Kulesha E., Gräf S., Johnson N., Herrero J., Tomazou E., et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat. Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20, and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J., Dennis E.S. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993;21:2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., Molloy P.L., Paul C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futscher B.W., Oshiro M.M., Wozniak R.J., Holtan N., Hanigan C.L., Duan H., Domann F.E. Role for DNA methylation in the control of cell type specific maspin expression. Nat. Genet. 2002;31:175–179. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- Garber K. Fixing the front end. Nat. Biotechnol. 2008;26:1101–1104. doi: 10.1038/nbt1008-1101. [DOI] [PubMed] [Google Scholar]
- Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., David L., Zhu J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell. 2002;111:803–814. doi: 10.1016/s0092-8674(02)01133-9. [DOI] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Henderson I.R., Jacobsen S.E. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Hodges E., Xuan Z., Balija V., Kramer M., Molla M.N., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- Inoue S., Oishi M. Effects of methylation of non-CpG sequence in the promoter region on the expression of human synaptotagmin XI (syt11) Gene. 2005;348:123–134. doi: 10.1016/j.gene.2004.12.044. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Ladd-Acosta C., Carvalho B., Wu H., Brandenburg S.A., Jeddeloh J.A., Wen B., Feinberg A.P. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.P., Lindroth A.M., Cao X., Jacobsen S.E. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jeddeloh J.A., Greally J., Rando O.J. Reduced-representation methylation mapping. Genome Biol. 2008;9:231. doi: 10.1186/gb-2008-9-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankel M.W., Ramsey D.E., Stokes T.L., Flowers S.K., Haag J.R., Jeddeloh J.A., Riddle N.C., Verbsky M.L., Richards E.J. Arabidopsis MET1 cytosine methyltransferase mutants. Genetics. 2003;163:1109–1122. doi: 10.1093/genetics/163.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet I., Schlesinger Y., Farkash S., Rand E., Hecht M., Segal E., Pikarski E., Young R., Niveleau A., Cedar H., et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Kitazawa S., Kitazawa R., Maeda S. Transcriptional regulation of rat cyclin D1 gene by CpG methylation status in promoter region. J. Biol. Chem. 1999;274:28787–28793. doi: 10.1074/jbc.274.40.28787. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T.H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A.V., Black M., Vaughn M.W., Dedhia N., McCombie W.R., Lavine K., Mittal V., May B., Kasschau K.D., et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Lippman Z., Gendrel A.V., Colot V., Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat. Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- Lister R., O'Malley R.C., Tonti-Filippini J., Gregory B.D., Berry C.C., Millar A.H., Ecker J.R. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini D.N., Singh S.M., Archer T.K., Rodenhiser D.I. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene. 1999;18:4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- Mardis E.R. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- Martienssen R.A., Doerge R.W., Colot V. Epigenomic mapping in Arabidopsis using tiling microarrays. Chromosome Res. 2005;13:299–308. doi: 10.1007/s10577-005-1507-2. [DOI] [PubMed] [Google Scholar]
- Meissner A., Gnirke A., Bell G.W., Ramsahoye B., Lander E.S., Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero L.M., Filipski J., Gil P., Capel J., Martínez-Zapater J.M., Salinas J. The distribution of 5-methylcytosine in the nuclear genome of plants. Nucleic Acids Res. 1992;20:3207–3210. doi: 10.1093/nar/20.12.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M., Malmgren H., Samiotaki M., Kwiatkowski M., Chowdhary B.P., Landegren U. Padlock probes: Circularizing oligonucleotides for localized DNA detection. Science. 1994;265:2085–2088. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- Oda M., Yamagiwa A., Yamamoto S., Nakayama T., Tsumura A., Sasaki H., Nakao K., Li E., Okano M. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes & Dev. 2006;20:3382–3394. doi: 10.1101/gad.1470906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okou D.T., Steinberg K.M., Middle C., Cutler D.J., Albert T.J., Zwick M.E. Microarray-based genomic selection for high-throughput resequencing. Nat. Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- Penterman J., Zilberman D., Huh J.H., Ballinger T., Henikoff S., Fischer R.L. DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. 2007;104:6752–6757. doi: 10.1073/pnas.0701861104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast G.C., Ziff E.B. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Saze H., Mittelsten Scheid O., Paszkowski J. Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 2003;34:65–69. doi: 10.1038/ng1138. [DOI] [PubMed] [Google Scholar]
- Shendure J., Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Shibuya K., Fukushima S., Takatsuji H. RNA-directed DNA methylation induces transcriptional activation in plants. Proc. Natl. Acad. Sci. 2009;106:1660–1665. doi: 10.1073/pnas.0809294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomazou E., Rakyan V.K., Lefebvre G., Andrews R., Ellis P., Jackson D.K., Langford C., Francis M., Bäckdahl L., Miretti M., et al. Generation of a genomic tiling array of the human major histocompatibility complex (MHC) and its application for DNA methylation analysis. BMC Med. Genomics. 2008;1:19. doi: 10.1186/1755-8794-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn M.W., Tanurd Ić M., Lippman Z., Jiang H., Carrasquillo R., Rabinowicz P.D., Dedhia N., McCombie W.R., Agier N., Bulski A., et al. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5:e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Weaver I., Cervoni N., Champagne F., D'Alessio A., Sharma S., Seckl J., Dymov S., Szyf M., Meaney M. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Weaver I.C., D'Alessio A.C., Brown S.E., Hellstrom I.C., Dymov S., Sharma S., Szyf M., Meaney M. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: Altering epigenetic marks by immediate-early genes. J. Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Schübeler D. Genomic patterns of DNA methylation: Targets and function of an epigenetic mark. Curr. Opin. Cell Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Weber M., Davies J., Wittig D., Oakeley E., Haase M., Lam W., Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S.W., Chen H., Henderson I.R., Shinn P., Pellegrini M., Jacobsen S.E., et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zheng X., Pontes O., Zhu J., Miki D., Zhang F., Li W.X., Iida K., Kapoor A., Pikaard C.S., Zhu J.K. ROS3 is an RNA-binding protein required for DNA demethylation in Arabidopsis. Nature. 2008;455:1259–1262. doi: 10.1038/nature07305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D., Gehring M., Tran R.K., Ballinger T., Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]