Abstract
The brain must dynamically integrate, coordinate, and respond to internal and external stimuli across multiple time scales. Non-invasive measurements of brain activity with fMRI have greatly advanced our understanding of the large-scale functional organization supporting these fundamental features of brain function. Conclusions from previous resting-state fMRI investigations were based upon static descriptions of functional connectivity (FC), and only recently studies have begun to capitalize on the wealth of information contained within the temporal features of spontaneous BOLD FC. Emerging evidence suggests that dynamic FC metrics may index changes in macroscopic neural activity patterns underlying critical aspects of cognition and behavior, though limitations with regard to analysis and interpretation remain. Here, we review recent findings, methodological considerations, neural and behavioral correlates, and future directions in the emerging field of dynamic FC investigations.
Keywords: Functional connectivity, Resting state, Dynamics, Spontaneous activity, Functional MRI (fMRI), Fluctuations
Introduction
Until recently, most fMRI studies have implicitly assumed that the statistical interdependence of signals between distinct brain regions (functional connectivity, FC, Friston, 2011; for all abbreviations, see Table 1) is constant throughout recording periods of task-free experiments, as reflected in the analysis tools and metrics that are commonly applied to the data. While studies operating under this assumption have afforded exceptional developments in understanding large-scale properties of brain function, the resulting characterization ultimately represents an average across complex spatio-temporal phenomena. Accordingly, it has been proposed that quantifying changes in functional connectivity metrics over time may provide greater insight into fundamental properties of brain networks. Here, we discuss recent studies examining dynamic properties of resting-state FC. We consider the existing techniques for their evaluation, challenges and limitations with regard to methodology and interpretation, the electrophysiological basis of such dynamics, and information that these investigations could potentially reveal about brain organization and cognition that may fundamentally change the way we examine neuroimaging data.
Table 1.
Abbreviations used in the text.
| AI | Anterior insula |
| BLP | Band-limited power |
| BOLD | Blood–oxygen-level-dependent |
| CAP | Co-activation patterns |
| CBV | Cerebral blood volume |
| CNR | Contrast-to-noise ratio |
| dACC | Dorsal anterior cingulate cortex |
| DAN | Dorsal attention network |
| DMN | Default-mode network |
| EEG | Electroencephalography |
| FC | Functional connectivity |
| FEF | Frontal eye fields |
| fMRI | Functional MRI |
| GSR | Galvanic skin response |
| HRV | Heart-rate variability |
| ICA | Independent component analysis |
| ICN | Intrinsic connectivity network |
| InI | Inverse imaging |
| LAN | Language network |
| LFPs | Local field potentials |
| LGN | Lateral geniculate nucleus |
| LIP | Lateral intraparietal cortex |
| MCW | Maximal correlation windows |
| MDD | Major depressive disorder |
| MEG | Magnetoencephalography |
| MIP | Medial intraparietal cortex |
| MOT | Somatomotor network |
| mPFC | Medial PFC |
| MREG | Magnetic resonance encephalography |
| MRI | Magnetic resonance imaging |
| PCA | Principal component analysis |
| PET | Positron emission tomography |
| PFC | Prefrontal cortex |
| PPI | Psycho-physiological interactions |
| ROI | Region of interest |
| RS-fMRI | Resting-state fMRI |
| SC | Structural connectivity |
| sICA | Spatial ICA |
| SNR | Signal-to-noise ratio |
| TFM | Temporal functional modes |
| tICA | Temporal ICA |
| TPN | Task-positive network |
| TR | Repetition time |
| vACC | Ventral anterior cingulate cortex |
| VAN | Ventral attention network |
| VIS | Visual network |
| vlPFC | Ventral lateral PFC |
| VTA | Ventral tegmental area |
| WTC | Wavelet transform coherence |
Resting-state connectivity and static characterizations
The so-called “resting state” has received considerable attention in recent years and has been investigated with multiple modalities, including positron emission tomography (PET), magnetoencephalography (MEG), and electroencephalography (EEG), though the dominant approach is presently functional magnetic resonance imaging (fMRI). Resting-state fMRI (RS-fMRI) is a non-invasive method in which the FC and other properties of blood–oxygen-level-dependent (BOLD) signals are examined from scans acquired with no explicit task (Biswal et al., 1995; reviewed in Fox and Raichle, 2007). FC is quantified with metrics such as correlation, covariance, and mutual information between the time series of different regions, wherein the temporal and spatial scales examined are determined by the question of interest (Bressler and Menon, 2010; Bullmore and Sporns, 2009; Friston, 2011). It therefore represents an empirical characterization of the temporal relationship between regions, without indicating how the temporal covariation is mediated (Friston, 2011; Friston and Buchel, 2007). Various techniques for FC analysis have revealed sets of spatially distributed, temporally correlated brain regions (“intrinsic connectivity networks”, ICNs; also referred to as “resting-state networks”; Beckmann et al., 2005; Damoiseaux et al., 2006; Power et al., 2011; Yeo et al., 2011). While the neural underpinnings and functional role of spontaneous fluctuations and correlations remain unresolved (reviewed in Leopold and Maier, 2012), evidence suggests that ICNs relate to underlying neural activity (Britz et al., 2010; Brookes et al., 2011a,b; de Pasquale et al., 2010, 2012; Fox and Raichle, 2007; He et al., 2008; Laufs, 2008, 2010; Liu et al., 2011; Mantini et al., 2007; Musso et al., 2010; Nir et al., 2007, 2008; Shmuel and Leopold, 2008) and are likely shaped, but not fully determined, by structural connectivity (SC; for review, see Damoiseaux and Greicius, 2009). Patterns of FC observed at rest have also been shown to resemble those elicited by more traditional task-based paradigms or derived directly from task-data (Biswal et al., 1995; Calhoun et al., 2008; Fox et al., 2006; Laird et al., 2011; Smith et al., 2009; Vincent et al., 2007).
The duration and number of scans used for computing ICNs of a given subject vary considerably between studies. Presently, a typical acquisition in humans includes a single scan of approximately 5–10 min using a repetition time (TR) in the range of 2–3 s that allows for whole-brain coverage with standard imaging sequences. It has been suggested that correlation values within and between ICNs stabilize within 4–5 min of data (van Dijk et al., 2010), implying that most studies are adequately sampling the network activity despite relatively few data points. Indeed, most studies do converge on similar network patterns even across a variety of behavioral states (e.g. eyes closed, open, or open and fixating; Bianciardi et al., 2009; but see McAvoy et al., 2012) though there are also subtle, but important, differences in the patterns across both normal and diseased states (for reviews, see Greicius, 2008; Heine et al., 2012; Menon, 2011). The univariate and multivariate approaches typically applied to resting-state data (for review, see Cole et al., 2010) assume that the strength of interactions between regions is constant over time. For example, seed-based correlation approaches represent the relationship between two regions of interest as a single correlation coefficient that is calculated from the time series of the entire scan; temporal variations in this value will not be captured (see Fig. 1 for illustration). Another common technique, spatial independent component analysis, decomposes the fMRI data into a pre-specified number of components with maximal spatial independence. While this strategy removes the need for explicitly defining seed regions, it does not (without additional processing) account for changes in the strength of inter-regional interactions over time.
Fig. 1.
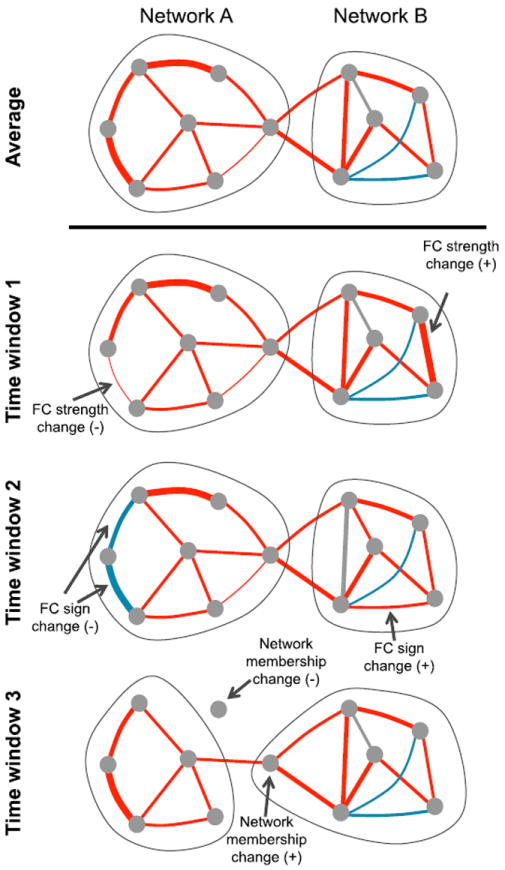
Time-varying changes in functional connectivity (FC). The schematic graph representation illustrates possible changes in connectivity properties (row 1). The FC strength between two nodes can change in magnitude (row 2), sign (row 3), or be lost/gained as the strength changes above or below a threshold, such that the node membership changes (row 3). Red edges, positive connections; blue edges, negative connections.
Examining the dynamics of functional connectivity
The assumption of stationarity provides a convenient framework in which to examine and interpret results. Approaches built upon these assumptions have produced a wealth of literature expanding our knowledge of large-scale brain networks. Yet, given the known dynamic, condition-dependent nature of brain activity (Rabinovich et al., 2012; von der Malsburg et al., 2010), it is natural to expect that FC metrics computed on fMRI data will exhibit variation over time. Indeed, FC has been demonstrated to exhibit changes due to task demands (Esposito et al., 2006; Fornito et al., 2012; Fransson, 2006; Sun et al., 2007), learning (Albert et al., 2009; Bassett et al., 2011; Lewis et al., 2009; Tambini et al., 2010), and large state transitions such as sleep (Horovitz et al., 2008, 2009), sedation (Greicius et al., 2008), and anesthesia (Boveroux et al., 2010; Peltier et al., 2005). Further, while between-subject variation is to be expected given its reported correlation with a variety of individual measures (IQ, personality, etc.; Adelstein et al., 2011; Song et al., 2008; van den Heuvel et al., 2009; Wei et al., 2011), within-subject FC has also been shown to vary considerably, even between different scans within the same imaging session (Honey et al., 2009; Liu et al., 2009; Meindl et al., 2010; Shehzad et al., 2009; Van Dijk et al., 2010). In fact, changes in both the strength and directionality of functional connections appear to vary not only between runs, but also at much faster time-scales (seconds–minutes) (Allen et al., in press; Chang and Glover, 2010; Handwerker et al., 2012; Jones et al., 2012; Kiviniemi et al., 2011; Sakoglu et al., 2010), a property that is not exclusive to humans (Hutchison et al., in press; Keilholz et al., 2013; Majeed et al., 2011).
Interpreting temporal variations in FC metrics (such as correlation) that are computed from fMRI time series is not necessarily straightforward. Low signal-to-noise ratio (SNR), changing levels of non-neural noise (e.g. from cardiac and respiratory processes and hardware instability), as well as variations in the BOLD signal mean and variance over time, can induce variations in FC metrics (see Issues and limitations section below). In addition, since functional networks can be spatially overlapping (i.e., the time series of a single node may have partial correlations with that of multiple networks), the FC between two regions that is attributed to their involvement in one particular network can appear to change if the time series of overlapping networks are not appropriately separated (Smith et al., 2012). It is also unclear the extent to which dynamic FC is best conceptualized as a multistable state space wherein multiple discrete patterns recur, akin to fixed points of a dynamic system, or whether it simply varies along a continuous state space. At present, studies have begun to identify discrete, reproducible patterns of FC and of the multivariate time series (refer to Reproducible patterns of sliding-window correlations, Single-volume co-activation patterns, and Repeating sequences of BOLD activity sections below), indicating some degree of multistability.
To gain insight into whether FC fluctuations can be attributed to neural activity or simply noise, it is necessary to compare changes in FC metrics to simultaneous measurement of neural or physiological processes and further, to examine whether the degree or pattern of variability can significantly differentiate between individuals or populations (refer to Interpreting fluctuations in BOLD functional connectivity section below). For example, studies are beginning to identify potential correlates of variations in resting-state FC in simultaneously recorded electrophysiological data (Allen et al., 2013; Chang et al., 2013b; Tagliazucchi et al., 2012b) as well as behavior (Thompson et al., in press), suggesting that variations in FC are to some degree of neuronal origin and perhaps linked with changes in cognitive or vigilance state. Disease-related alterations in the dynamic properties of FC have also been reported (Jones et al., 2012; Sakoglu et al., 2010), further suggesting a neural origin and raising the intriguing possibility that temporal features of FC could serve as a disease biomarker. Thus, while limitations of current analysis strategies and uncertainty surrounding the origins of dynamic FC advise caution when interpreting past and current findings, the existing results raise a series of important and exciting questions concerning network dynamics that may significantly expand our understanding of brain function.
Analysis strategies and findings
Below, we review analysis strategies that have been applied to characterize temporal variations in the spatiotemporal structure of BOLD signal fluctuations. Among these approaches, some are designed to capture pairwise variations in inter-regional synchrony (Sliding-window analysis and Time–frequency coherence analysis sections), while others focus on identifying changing patterns of synchrony at a multivariate level (Single-volume co-activation patterns, Repeating sequences of BOLD activity, and Independent component analysis sections). Pairwise approaches have been combined with clustering methods to identify, for instance, repeating configurations of correlations across multiple ROIs (Reproducible patterns of sliding-window correlations section). It should be noted that these analysis strategies are of an exploratory nature, and are not solidly grounded in neurobiological principles or models. Presently, it is not clear which classes of techniques will prove to be the most fruitful in characterizing functionally relevant dynamics. It should also be emphasized that temporal variation in FC metrics cannot be interpreted directly as non-stationarity2 of the underlying interactions between regions. In much of the literature exploring dynamic FC, the term ‘non-stationarity’ has been invoked in a technically incorrect sense, referring merely to the observed variability over time in the value of a given FC metric. Methods such as correlation and coherence, in fact, lack a proper model for resolving the underlying structure of network interactions (Smith et al., 2011), and moreover cannot distinguish between true variability in network interactions or variability due to stochastic noise (Handwerker et al., 2012; see also Issues concerning sliding-window analysis section below). Such issues must be considered when interpreting the results reviewed below, and the development of appropriate modeling techniques for dynamic FC will be an important future direction.
Sliding window analysis
To date, the most commonly used strategy for examining dynamics in resting-state FC has been a sliding window approach (Allen et al., in press; Chang and Glover, 2010; Handwerker et al., 2012; Hutchison et al., in press; Jones et al., 2012; Kiviniemi et al., 2011; Sakoglu et al., 2010). In this approach, a time window of fixed length (possibly with tapered/weighted edges) is selected, and data points within that window are used to calculate the FC metric of interest. The window is then shifted in time by a fixed number of data points (ranging from a single data point to the length of a window) that defines the amount of overlap between successive windows. This process results in quantification of the time-varying behavior of the chosen metric over the duration of the scan. Given a sufficient number of data points for robust calculation, any metric that could be applied to the entire scan can in principle be used in sliding window analysis; so far, the correlation coefficient has been the most commonly used metric. While there remain important concerns pertaining to the appropriate parameters and the validity of the approach (see Issues concerning sliding-window analysis section below), results indicate that sliding-window FC may capture phenomena of potential functional relevance.
Using a sliding-window approach (30 s, 60 s, 120 s, and 240 s windows; TR = 2 s; 1 data point shifts), Hutchison et al. (in press) reported (1) transient negative correlations between two nodes of a fronto-parietal network; (2) periods of high correlation between all nodes of the network alternating with periods of low correlation; and (3) the transient inclusion of new network nodes that were unobserved at longer window lengths. Such was the case for both anesthetized macaques and awake humans, implying that fluctuations in sliding-window correlation may not be driven solely by conscious processes such as attentional shifts, sensory processing, recollection, and planning.
Since it is difficult to interpret the existence of variability alone (and in fact, similar fluctuations can arise when applying a sliding-window analysis to randomly generated signals such as white noise), it is necessary to pose and formally test specific hypotheses. For example, one can ask whether group differences exist, or whether properties of dynamic FC differentiate across brain regions. In one of the earliest studies applying sliding-window FC, Sakoglu et al. (2010) found group differences between healthy controls and schizophrenic patients on ICA-derived time series during an auditory oddball task, suggesting that dynamic measures of FC may have clinical relevance (see Clinical applications section). In initial explorations of the relative magnitude of sliding-window correlation variability across different regions, Shen et al. (2013) showed that across a range of window sizes, regions with the most stable FC across time were those with bidirectional anatomical connections, followed in ranking by regions with unidirectional SC and then by regions that had no direct connections. Similarly, it has been reported that FC between bilateral homologues shows the least variability in connection strength over time, followed by the FC of nodes within sensory and motor networks, then between higher-order network nodes, and finally those regions not contained within the primary network derived using static approaches (Gonzalez-Castillo et al., 2012). The trend matches the node stability that is observed following clustering (Salvador et al., 2005), ICA (Abou-Elseoud et al., 2010), or hierarchical modular (Meunier et al., 2009) decompositions of RS-fMRI data. Such approaches have suggested a hierarchical organization of the brain (in which modules contain sub-modules that themselves contain further modules, spanning several topological scales; Meunier et al., 2010), and the above studies of FC variability suggest that pairs of regions at the smallest parcellation level are the most stable and robust. The regions possessing these stable pairwise connections, such as bilateral homologues, typically possess strong structural connections (e.g. via callosal fibers), participate in similar functional roles, and are phylogenetically preserved across species (Hutchison and Everling, 2012). In contrast, higher-order regions showing greater FC variability tend to be involved in a greater range of functions and have a high degree of flexibility (network and module membership changes). The heterogeneity of nodes and their pairwise dynamics within networks highlight the importance of considering the hierarchy and scale in which they are embedded.
Reproducible patterns of sliding-window correlations
The sliding-window approach can be used to search for the presence of reproducible, transient patterns of region-to-region correlation (“connectivity states”). For example, one may apply clustering methods to correlation (or covariance) matrices computed over windowed segments of the BOLD time series derived from voxels, regions-of-interest, or via ICA (Allen et al., in press). Such clustering approaches have resolved different connectivity patterns corresponding to the execution of distinct mental tasks (Gonzalez-Castillo et al., 2012), and have also been applied to data collected during rest, where subjects are expected to undergo spontaneous fluctuations in cognitive as well as vigilance states (Allen et al., in press; see Fig. 2). In Allen et al. (2012), distinct and repeatable patterns of FC determined from RS-fMRI data were observed, highlighting strong departures from average connectivity characterized over long time scales, and in particular calling into question descriptions of a single canonical DMN and its anti-correlation to a single “task-positive” network (TPN). More specifically, the examination of connectivity on finer temporal scales showed that the DMN regularly breaks into a number of constituents that can act in synchrony with both sensorimotor and attentional networks. Such observations suggest that dynamic FC can, to some extent, be conceived as a multistable process wherein the correlation patterns (and perhaps the underlying time courses) pass through multiple discrete states, rather than varying in a more continuous sense. The results also suggested that the averaged spatial pattern of FC might not actually resemble a state that occurs transiently within the scanning period (Allen et al., 2012; Hutchison et al., in press; Kiviniemi et al., 2011). The study of dynamic FC, and this finding in particular, raises the issue that the concept of a “network” is rather elusive, hinging (among other factors) upon the time-scale over which it is defined (Horwitz, 2003).
Fig. 2.
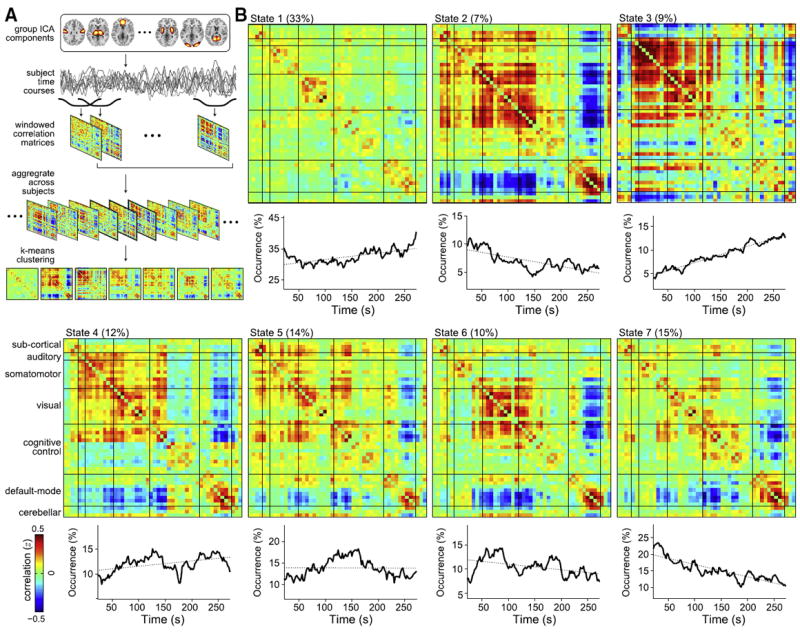
Detection of functional connectivity (FC) states with a sliding window/clustering approach. (A) An overview of the analysis. Group ICA is used to decompose resting-state data into intrinsic networks. Correlation matrices are computed from windowed portions of each subject’s component time series and the matrices are aggregated across subjects. K-means clustering is applied to the correlation matrices to find repeating patterns of connectivity, referred to as FC states. (B) Cluster centroids for FC States 1–7 show patterns in connectivity that are not apparent from stationary models. Below each centroid is the number of occurrences (in percentage units) of the state as a function of time. Linear fits (dotted lines) suggest a prominent increase in the appearance of State 3 over time, and decreases in the appearance of States 2 and 7. Adapted with permission from Allen et al. (in press).
Though clustering approaches provide a potentially powerful method for determining spontaneous changes in a subject’s internal state, there are a number of challenges and opportunities for development. Some difficulties are inherent to all studies of dynamics (see Issues and limitations section), such as obtaining enough data points in each windowed segment to robustly estimate covariance structure, as well as recording for long enough periods in each subject in order to study state transitions and variability at the level of the individual. Other challenges are more specific to clustering, chiefly the selection of algorithms (e.g., hierarchical or mean/medoid based) and associated free parameters (e.g., distance metric and the number of clusters into which to partition the data). Although initial work suggests that the results of the clustering procedure are not particularly sensitive to algorithmic parameters (Allen et al., in press), these results require replication in additional datasets. Alternative methods for identifying connectivity states are also being explored, and include using topological network descriptions as features in a clustering analysis (e.g., modularity or community membership (Bassett et al., 2011; Jones et al., 2012; Kinnison et al., 2012)), or formal models to detect change points in connectivity, as introduced by Cribben et al. (2012).
Single-volume co-activation patterns
It has been shown that canonical ICNs derived with methods such as seed-based correlation and ICA resemble the spatial pattern of BOLD activity in selected individual time frames. In other words, a given ICN resembles individual time frames in which the signal amplitude within its nodes is high, and in fact, spatial patterns resembling ICNs can be extracted from a small fraction of the total time frames of a resting state scan (Tagliazucchi et al., 2012c; Liu and Duyn, 2013). Motivated by this principle, and by the observation that volumes in which the signal intensity in one seed region are high can exhibit a variety of coactivation patterns among the remaining voxels, Liu et al. proposed clustering selected individual BOLD volumes of a resting-state scan based on spatial similarity (Liu and Duyn, 2013). Cluster centroids were defined as “co-activation patterns” (CAPs) intended to characterize a set of representative instantaneous configurations of BOLD activity. A large collection of resting-state data was decomposed into 30 CAPs, reflecting a repertoire of patterns across the population, but with notable differences from those derived using methods such as ICA (Liu and Duyn, 2013). Importantly, the seed-based correlation patterns computed over a given time interval in the scan reflect a summation of the CAPs occurring within the interval, such that changes in sliding-window correlations over time reflect the relative occurrences of distinct CAPs falling within the analysis window. These studies offer a novel conceptualization of time-varying correlations, and show that changes in sliding-window correlations or ICA identify changes in the systems that tend to have higher levels of spontaneous activity within a given temporal window.
Repeating sequences of BOLD activity
Extending from the observation that canonical network “states” can be captured at short time scales is the question of whether contiguous sequences of BOLD volumes (“spatiotemporal patterns”) reliably recur over different points in time. While the above section Single-volume coactivation patterns discusses repeatable occurrences of single-volume snapshots of BOLD activity, here we refer to repeatable sequences (consecutive volumes) of BOLD activity. Reproducible spatiotemporal patterns of BOLD fluctuations were first observed in the anesthetized rat using very fast sampling of a single slice (100 ms TR) and consisted primarily of bilateral ‘waves’ of high signal intensity that appeared to propagate from lateral to medial cortical areas (Majeed et al., 2009). Subsequent research has shown that similar patterns of lateral-to-medial propagation along the cortex of the rat can also be observed when cerebral blood volume (CBV), rather than BOLD contrast, is used (Magnuson et al., 2010). The authors have since developed an algorithm that identifies repeated occurrences of similar patterns across a scan (Majeed et al., 2011). Using this algorithm, patterns similar to those observed in the rat were also detected in humans (Majeed et al., 2011), alleviating concerns that the original patterns could be induced by the use of anesthesia. In human data, the patterns involved well-known areas of the DMN (posterior cingulate and anterior medial prefrontal cortex) and the TPN (superior parietal and premotor cortices), and were highly reproducible across subjects. Preliminary analysis of the contribution of the patterns to traditional measurements of FC suggests that they account for 25–50% of variance in the low frequency BOLD time courses, although this is likely to vary by species, network, and condition.
It may be the case that the observed patterns are part of the mechanism that coordinates the activity of large-scale functional networks. As signal-conduction delays bias long-range communication of distributed areas towards lower frequencies (see Schölvinck et al., 2013), Pan et al. used simultaneous imaging and recording to examine the relationship between the spontaneous BOLD fluctuations and infraslow electrical activity that is comparable in frequency (<1 Hz). The results showed significant correlation between BOLD and infraslow LFPs that was localized to the area near the recording electrode in the somatosensory cortex (Pan et al., 2013). When correlation between BOLD and infraslow LFPs was examined as a function of time lag, patterns of propagation along the cortex appeared, similar to those first described by Majeed et al., suggesting that the patterns may have an origin in infraslow oscillations. However, it remains unclear what the relationship is between the slowly propagating waves over the cortex (which would be regarded as a first-order description of the spatiotemporal activity) and the dynamic changes in FC (a second-order description). If, as preliminary results suggest, the waves of activity account for less than half of the variance in the BOLD signal, other processes arising from more localized variations in activity may also contribute. One very interesting possibility is that the patterns represent a sort of large-scale organization, and that variations in local activity are modulated and/or superimposed upon these patterns. If so, it may be possible to separate the large-scale and local components to maximize sensitivity to the process of interest. It will be critical to disentangle these and other (e.g. global signal changes, overlapping node membership) interacting elements to determine the most appropriate course of analysis and characterization of transient network properties.
Time–frequency analysis
A key limitation of sliding-window analysis is the use of a fixed sliding window size (see Issues concerning sliding-window analysis section). The window size governs the time-scale on which the analysis is performed; ideally, it is long enough to accommodate the relatively slow frequencies of the BOLD signal and estimate FC metrics with sufficient SNR, and yet short enough to be sensitive to transient changes in network connectivity. Yet, both the neurally relevant frequencies and the appropriate time scale for studying connectivity changes are presently open questions (see above and Issues and limitations sections). A time–frequency analysis can be applied to estimate the coherence and phase lag (time shift) between two time series as a function of both time and frequency. Implementing a time–frequency coherence analysis with the wavelet transform (wavelet transform coherence; WTC) provides a multi-resolution approach to time–frequency analysis (Torrence and Compo, 1998), circumventing the need to select a fixed sliding-window size. With the wavelet transform, the size of the effective analysis window (the scale of the wavelet) is varied in accordance with the natural time-scale of the frequencies in the signal: high frequencies (faster changes) are analyzed with shorter time windows, and progressively lower frequencies are analyzed with progressively longer time windows.
By providing a rich picture of the coherence across multiple time scales, the WTC lends itself well to exploratory analysis. One may characterize, for instance, the dominant frequencies at which regions or networks display coherence, as well as the extent to which magnitude and phase relationships between nodes fluctuate over time within a given band. The WTC has been applied to study the relationship between DMN and TPN with results indicating that anti-correlations appeared to be a transient, rather than stable, phenomenon (Chang and Glover, 2010). However, the vast amount of information produced by a WTC analysis – i.e., a time–frequency map for each pair of ROIs – presents challenges when scaling the analysis to multiple subjects and brain regions. One approach to handling this growth of information is to summarize the output along several potentially relevant dimensions, e.g. by forming a time-averaged coherence profile (Chang and Glover, 2010) or by quantifying the overall variability of coherence at selected frequency bands such as with standard deviation or mean-squared successive differences (Chang et al., 2011). Given sufficiently long time series, one can consider even higher-order aspects of variability, such as the rate at which coherence and phase are modulated. These metrics can be examined within and between groups of subjects, yielding features that can complement those of static analyses. For example, one can use a full four-dimensional (voxels × time) frequency decomposition to identify changes in spatiotemporal patterns (Miller and Calhoun, 2013). Future work will be necessary to determine the most informative metrics for a given hypothesis and subject population.
Independent component analysis
Since the late 1990s, spatial ICA (sICA) has been applied to fMRI as a data-driven approach that estimates networks from the entire spatio-temporal dataset at once (Beckmann and Smith, 2004; Calhoun and Adali, 2012; Calhoun et al., 2001; McKeown et al., 1998). One straightforward way to accommodate a degree of variability into ICA FC estimation is to perform sICA on a sliding-window basis. Kiviniemi et al. (2011) applied sICA successively to data using 108 s windows (60 data points; 1-s data point shifts). The FC profile of the DMN was found to vary over the sliding windows, never directly matching the whole-scan derived template. The highest inclusion of any DMN voxel across time was 82%, implying that no voxel is consistently connected to the primary network (at least in the DMN) over time, though statistical testing would be necessary to validate this claim.
Smith et al. (2012) applied temporal ICA (tICA) to regions of interest that were first defined by applying sICA, yielding a set of temporally independent modes (termed ‘temporal functional modes’ (TFMs)). These TFMs differed from networks commonly identified with seed-based correlation and sICA. In both the sICA and tICA models, the weights in a spatial map are constant over time, so neither method directly addresses the possibility of time-varying connection strengths (weights) between nodes. Relating to sliding-window FC analysis, Smith et al. report that when reconstructing node time series assuming fixed TFM connections and applying a sliding window analysis, a significant portion (~25%) of the variability in node correlations could be attributed to the spatially overlapping nature of functional networks. The impact of overlap (shared node membership due to time-series partial correlations) on estimates of dynamic FC highlights the need to consider the temporal relationships of nodes within the context of multiple interacting systems; i.e., brain regions playing unique roles within different functional networks.
Issues and limitations
Physiological noise and pre-processing
Since estimates of time-varying connectivity are based on relatively few time points, dynamic analysis is particularly sensitive to noise. Variations in the magnitude of noise levels across the scan, as well as non-neuronal events that generate strong spatially correlated signal fluctuations, can masquerade as “dynamics” of FC. It is therefore critical to reduce all known non-neural contributions to the fMRI time series during pre-processing.
Sources of noise in fMRI include scanner drift, head motion, and “physiological noise.” Physiological noise can arise from cardiac pulsation, shifts in the main magnetic field caused by motion of the body during respiration, and variations in the respiratory volume/rate and cardiac rate that evoke changes in BOLD contrast (Birn et al., 2008; Chang et al., 2009; Dagli et al., 1999; Shmueli et al., 2007). Variations in respiratory volume/rate and cardiac rate are of particular concern for resting-state analysis, as they reside predominantly in the low frequencies (<0.1 Hz) and tend to cause synchronous global modulations of the fMRI time series owing to their influence on arterial CO2 levels and cerebral blood flow (Chang and Glover, 2009; Peng et al., 2013; Wise et al., 2004). Head motion is also known to produce spurious, spatially structured artifacts in FC, and has been a topic of much controversy (Power et al., 2012; Satterthwaite et al., 2012; Van Dijk et al., 2012; Yan et al., 2013).
Since head motion and certain physiological events (such as a deep breath) are transient in nature, their adverse effects are lessened when resting-state data are analyzed using long time windows (as in conventional static analysis) to calculate FC. However, the impact on a dynamic analysis can be considerable: a slight head movement or a short deep breath will introduce strong signal fluctuations that can manifest as temporary changes in connectivity patterns. Successful denoising is extremely important for properly interpreting dynamic results, and recording respiration and cardiac events with a pneumatic belt and a plethysmograph is highly recommended. However, while a number of techniques have been developed for reducing noise (e.g. RETROICOR, RVHRCOR, PESTICA, CompCor, ME-ICA, censoring/“scrubbing” (Beall and Lowe, 2007; Behzadi et al., 2007; Chang et al., 2009; Glover et al., 2000; Kundu et al., 2012; Power et al., 2012)), residual noise inevitably remains, and dynamic studies of resting state would greatly benefit from a deeper understanding of how to properly remove noise from fMRI time series.
While many pre-processing steps commonly applied to resting-state fMRI data are equally applicable when performing dynamic FC analysis (e.g. spatial filtering, nuisance regression), certain steps require special consideration. For example, censoring or down-weighting time points with excessive motion or other known artifacts would affect a dynamic analysis due to its interruption of the temporal structure of the data; in a sliding-window analysis, it would result in different effective numbers of time points available within different windows. Regarding temporal filtering, one may apply additional high-pass filtering or similar detrending operation prior to sliding-window analysis if it is desired that changes in FC on the scale of the sliding window reflect only frequencies with periods smaller than the window size.
Issues concerning sliding-window analysis
A sliding-window analysis is a simple approach for exploring changes in FC (see Sliding window analysis section), but several critical issues must be considered in applying the method and interpreting results. One limitation is that most sources of noise in fMRI time series are non-stationary and can induce changes in FC over time (see Physiological noise and pre-processing section), and this noise may not be completely eliminated even with the most thorough pre-processing techniques. Secondly, white noise, as well as synthetic time series with statistical characteristics matching those of fMRI time series, can exhibit fluctuations in common FC metrics that are as large as those observed in actual fMRI data. As such, sliding-window analysis should be accompanied by hypotheses that are supported with appropriate statistical testing. For example, instead of asking simply whether (and by how much) FC varies over a scan, one might ask whether the range of sliding-window variability between particular regions is significantly different between two patient populations (where a positive finding would be most striking if no significant differences between populations were obtained by static FC analysis).
Another issue concerns the choice of window size. Ideally, the window should be large enough to permit robust estimation of FC and to resolve the lowest frequencies of interest in the signal, and yet small enough to detect potentially interesting transients (Sakoglu et al., 2010). Empirically, window sizes around 30–60 s have been noted to produce robust results in conventional acquisitions; Shirer et al. reported that cognitive states may be correctly identified from covariance matrices estimated on as little as 30–60 s of data (Shirer et al., 2012), and topological descriptions of brain networks were found to stabilize at window lengths of roughly 30 s (Jones et al., 2012). As one shrinks the window size, the SNR of the estimated FC decreases since (1) there are fewer time points available for computing FC, and (2) the measurement is dominated by increasingly higher frequencies in the fMRI time series, where the SNR of the BOLD signal is substantially diminished due to the low-pass characteristics of the hemodynamic response. Hence, the overall variability in sliding-window FC tends to increase as window size shrinks, a phenomenon that is not unique to brain signals. Noise reduction strategies and fast acquisitions may help reduce noise (and, consequently, sliding-window variability; Marx et al., 2013). As an alternative to a fixed window size, one may estimate change points in FC to demarcate windows (Cribben et al., 2012) or use multi-scale approaches (such as described in Time–frequency analysis section).
A further consideration, which is relevant for both static and dynamic analyses, is how best to model fMRI data so as to reveal the relationships among a network of regions (Friston, 2011; Smith et al., 2011; Varoquaux and Craddock, 2013). While most of the studies reviewed in this article have used Pearson’s correlation as the FC metric, it has been shown that methods based on the precision matrix (inverse covariance, capturing partial correlations) may in certain cases better recover the underlying biological network structure (Smith et al., 2011; Varoquaux and Craddock, 2013) and are more compact by virtue of partialling out the effects of other nodes in the model. Future work may consider applying sliding-window analyses with other estimators of network structure. In sum, sliding-window analysis is a valuable tool for the investigation of dynamic FC, though appropriate processing, modeling, and statistical testing are crucial and caution is advised in interpreting the results.
Non-stationarity of BOLD signal time series
One challenge of interpreting apparent temporal variation in FC metrics is that the BOLD signal time series itself may be non-stationary. For instance, in an early investigation of time-resolved FC during a long (27 min) resting-state scan, the BOLD signal amplitude was found to increase when the subject became excessively drowsy (Fukunaga et al., 2006). In another study, the Hurst exponent of resting-state time series, related to frequency and autocorrelation properties, was found to be modulated by the difficulty of a preceding cognitive task, taking as long as 16 min to recover to baseline levels (Barnes et al., 2009).
Properties of the BOLD signal time series are intertwined with estimates of FC. For example, if the neuronal component of the BOLD signal transiently decreases in amplitude relative to background noise levels, estimates of its synchrony with other regions may decrease as a consequence of the reduced signal-to-noise ratio. For sliding window correlation analysis, changes in the power spectrum and temporal autocorrelation of a signal in a given time window can alter the statistical degrees of freedom, which (if not corrected or adjusted for) can also manifest as a change in correlation over time. Non-stationarity of one of both signals will impact the stationarity of the estimated relationship between them. Hence, characterizing the non-stationary behavior of the time series themselves will therefore contribute to a more complete understanding of temporal variability in FC metrics.
The approximate 1/f spectral distribution of fMRI time series also poses theoretical difficulties when choosing a window size, since at any chosen window size, (1) the FC metric will be dominated by the lowest resolvable frequency, which has the highest amplitude, and (2) relationships between signals in the higher frequencies, occurring on faster time-scales, likely exhibit non-stationarity within a window. Given these two effects, it may not be possible to adequately collapse the relationship between two 1/f-like signals into a single scalar measurement per sliding window. As an alternative, a multi-resolution analysis such as time–frequency analysis may provide a more complete characterization of the FC dynamics across the time scales. However, as discussed above, such analyses yield a large amount of information, with many possible ways of summarizing the output across time and no theory governing which features will be most relevant. It will be important to acknowledge these issues as the field proceeds to develop and interpret measurements of dynamic FC.
Significance testing
The generation of an appropriate null distribution is important for statistical testing of hypotheses involving dynamic FC (e.g., see Issues concerning sliding-window analysis section), and can often be accomplished straightforwardly using bootstrapping and permutation techniques (Efron and Tibshirani, 1986; Robinson et al., 2008). In these techniques, significance is determined by comparing an observed test statistic to a distribution of “bootstrap” test statistics that is generated under a model consistent with the null hypothesis. Specifying an appropriate null model is highly dependent on the precise question at hand, and the approaches used in existing studies differ. For example, to determine if FC (as quantified with a wavelet-based measure of coherence) exhibited more variability than what would be expected from a stationary relationship, Chang and Glover (2010) used vector autoregression to model the linear and stationary dependencies between the time series. The authors estimated model coefficients from observed time-series pairs and then synthesized thousands of surrogate time-series to obtain a distribution of coherence variability under the null hypothesis of a stationary relationship. In complementary work, Handwerker et al. (2012) tested the hypothesis that the frequency characteristics of sliding-window time series were dependent on the precise timing relationships between regional time courses by keeping the amplitude spectra of individual time series constant while randomizing the phase. Periodic changes in sliding window correlations remained even when phase randomization removed the timing relationships, implying that this phenomenon is not unique to BOLD signal data. Allen et al. (2012) also used phase randomization, but applied it to the sliding-window correlation time series (rather than the original regional BOLD time series) to test the hypothesis that FC states could be discriminated when the phase relationships of correlations across different brain regions were disrupted. As an alternative to phase randomization, Keilholz et al. (2013) permuted time courses across different sessions and subjects to create a null distribution that preserved temporal characteristics of the original data. Sakoglu et al. (2010) used bootstrapping across subjects to determine significance of group differences in the dynamic patients with schizophrenia versus controls in static versus dynamic functional network connectivity and found significant, but slightly less robust differences in the dynamic FC results. Developing appropriate null models that retain statistical properties (e.g., temporal autocorrelation and spatial structure) of original signals remains a challenge and an area for improvement in this emerging field. Reporting more quantitative measurements of effect size, instead of or in addition to results of test statistics/p-values, would also be desirable in trying to understand the potential importance of dynamic FC.
Interpreting fluctuations in BOLD functional connectivity
Given that fMRI time series are noisy and that each temporal observation in a dynamic analysis incorporates a relatively small number of independent data points, it is unsurprising that variation across time occurs and that the magnitude of variation will often not differ significantly from that obtained with simulated stochastic time series. Yet, even if the range of connectivity variation does not exceed that which can occur by chance, it may still be the case that the time-course of connectivity fluctuation tracks meaningful neural phenomena, as would be predicted from electrophysiological data (described below in the Electrophysiological precedents section). Since this is difficult to determine from the resting-state fMRI data alone, validation must come from concurrent independent measurements such as electrophysiology, systemic physiology, and behavior (described below in the Interpretation using concurrent independent measurements section, and Fig. 3).
Fig. 3.
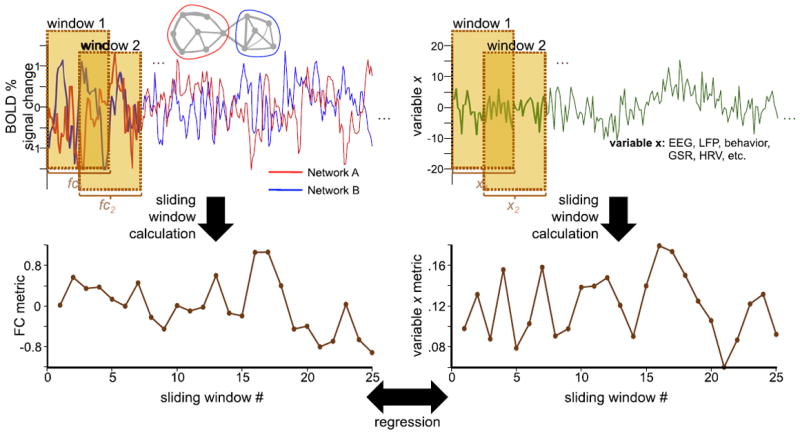
Evaluating the relationship between sliding-window functional connectivity (FC) and a concurrently measured variable. A sliding-window FC analysis is performed on pairs of BOLD signal time series, here derived from two networks “A” and “B” (upper left), producing a sequence of sliding-window FC values (lower left). Similarly, measurements of a neural, physiological, or behavioral variable “x” (upper right) can be computed in identical sliding windows (lower right). The two sliding window time series can then be compared using methods such as linear regression, as one way of determining whether the observed BOLD FC dynamics may be associated with variable “x”. EEG, electroencephalography; GSR, galvanic skin response; HRV, heart rate variability, LFP, local field potentials.
Electrophysiological precedents
Recent evidence suggests that modulation of neural activity may underlie observations of dynamic BOLD FC (Allen et al., 2013; Chang et al., 2013b; Tagliazucchi et al., 2012b). It is therefore possible that ICNs represent the hemodynamic manifestation of endogenous, self-organized neural dynamics that have been extensively characterized using electrophysiological recordings of single cells, LFPs, and surface EEG (e.g., Arieli et al., 1996; Boly et al., 2007; Eichele et al., 2008; Fukushima et al., 2012; for review see Deco et al., 2012; Rabinovich et al., 2012; Raichle, 2010; Ringach, 2009; Sadaghiani et al., 2010; Vogels et al., 2005; von der Malsburg et al., 2010). In the electrophysiological literature, it has long been appreciated that spontaneous activity demonstrates remarkable spatio-temporal structure (e.g. Kenet et al., 2003). Neural signals have been shown to continuously combine, dissolve, reconfigure, and recombine to form adaptive patterns of activity over various time scales (Rabinovich et al., 2012; von der Malsburg et al., 2010). Such processes are believed to underlie the flexibility and power of perception, cognition, and behavior, changing across time-scales to deal effectively with unpredictable aspects of current (and future) situations that cannot be reliably encapsulated within a fixed functional architecture. Given that electrophysiological recordings allow for a more direct examination of neural activity, their relevant features and findings may help to interpret the phenomenon of dynamic BOLD FC.
Oscillations of electrical activity constitute a mechanism through which populations of neurons can interact via synchronization (Akam and Kullmann, 2010) while providing a temporal frame of reference that may be exploited for sensory, cognitive, and motor processing (Arieli et al., 1996; Buzsaki, 2006; Engel et al., 2001; Llinás, 1988; Tsodyks et al., 1999; Varela et al., 2001; Womelsdorf et al., 2007). Their spatiotemporal patterns can self-organize and change within a fixed anatomic architecture (Womelsdorf et al., 2007). Recent multi-area recordings in the macaque and rodent brain have documented the dynamic formation of functional networks at multiple time scales (Siegel et al., 2012), manifesting as coherent fluctuations of LFPs. The time scales at which these networks emerge span all canonical frequency bands (Fig. 4). For example, long-range coherence in the gamma band has been found to index, among other functions, the coding for a prospective reward (Fujisawa and Buzsaki, 2011) or an attended, behaviorally relevant visual stimulus (Bosman et al., 2012; Gregoriou et al., 2009; Grothe et al., 2012). The emergence of transient synchrony in electrophysiological signals at multiple time scales could therefore underlie the temporal variations in BOLD signal connectivity in awake, conscious humans.
Fig. 4.
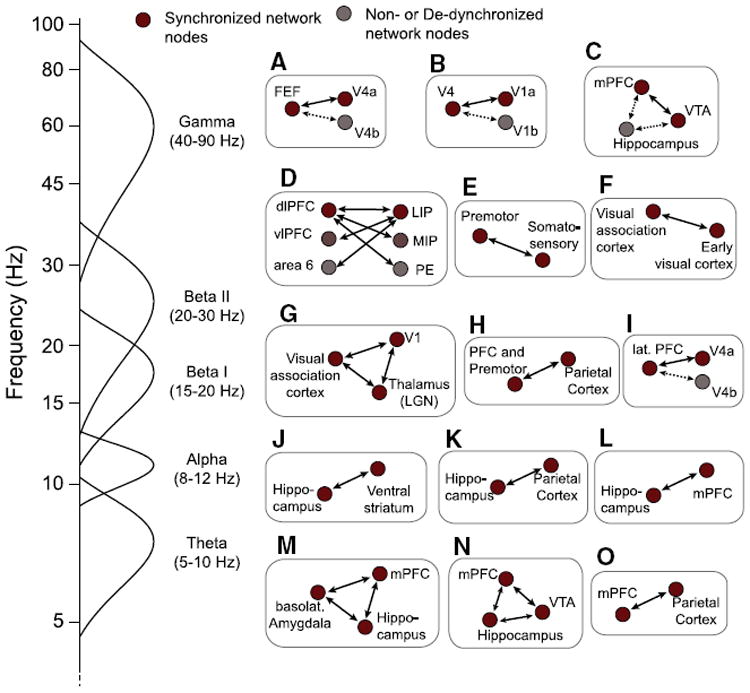
Overview of recently described functional networks emerging at fast time scales during specific cognitive states in large-scale synchronized local field potential (LFP) activity in animal studies. Each panel (A–H) sketches the brain areas that have been shown to engage in spatially selective coherent long-range networks during states that index visual attention, working memory, reward expectancies, memory retrieval, or sensorimotor integration. For the majority of examples coherent LFP states translated into synchronized spiking activity of individual cells. The selective overview of recently published example networks is described in detail in: A: Gregoriou et al. (2009), B: Bosman et al. (2012), Grothe et al. (2012), C: Fujisawa and Buzsaki (2011), D: Salazar et al. (2012), E: Brovelli et al. (2004), von Stein et al. (2000), Palva et al. (2010), F: Womelsdorf et al. (2007), G: Hughes et al. (2011), H: Pesaran et al. (2008), I: Liebe et al. (2012), J: Lansink et al. (2009), DeCoteau et al. (2007), K: Sirota et al. (2008), L: Benchenane et al. (2010), M: Popa et al. (2010), Lesting et al. (2011), N: Fujisawa and Buzsaki (2011), and O: Phillips et al. (in press). The sketched frequency axis (left) indicates the frequency range of the observed networks. For broadly distributed, selective neocortical and cortico–thalamic networks emerging at infra-slow (<0.3 Hz), slow (0.3–1 Hz), and delta (1–4 Hz) frequencies see, e.g., Timofeev et al. (2012).
Although the sluggish hemodynamic response likely prohibits the direct measurement of electrophysiological phase coherence in most frequency bands with fMRI, low-frequency BOLD fluctuations could instead reflect the amplitude (power) modulation of band-limited cortical activity (Britz et al., 2010; Leopold et al., 2003; Mantini et al., 2007; Musso et al., 2010; Nir et al., 2007; Shmuel and Leopold, 2008; for review see Schölvinck et al., 2013). In the macaque, it was shown that slow amplitude fluctuations, particularly of the gamma band, matched the frequencies of resting-state fluctuations (<0.1 Hz), shared a 1/f spectrum, and exhibited correlations with the BOLD signal time series (Leopold et al., 2003; Shmuel and Leopold, 2008). Recent studies with simultaneous LFP–fMRI recordings in rats, as well as in electrocorticography with humans, find that the spatial patterns of correlation between electrical recordings resemble those of BOLD FC (He et al., 2008; Keller et al., 2013; Nir et al., 2008; Pan et al., 2013). We can speculate that cross-frequency coupling (for reviews see Buzsaki and Watson, 2012; Jensen and Colgin, 2007; Lisman and Jensen, 2013; Siegel et al., 2012) may play a role in dynamic FC. The cycling of slow, widespread rhythms (e.g. 0.02–0.2 Hz; Vanhatalo et al., 2004) may alter the power/amplitude of the faster (nested) oscillations (delta through gamma) that are responsible for the temporal integration (and segregation) of distributed neural populations and brain areas, thereby changing the synchronization patterns which will then manifest as changes in BOLD FC.
Interpretation using concurrent independent measurements
Simultaneous EEG–fMRI
Scalp EEG offers a non-invasive, electrophysiological window into endogenous shifts in “brain states.” Resting-state EEG rhythms are themselves non-stationary, having ongoing fluctuations in amplitude and phase that track shifts in vigilance and cognitive states, and hence may be used as an independent variable with which to interrogate RS-fMRI data (for review see: Duyn, 2012; Laufs, 2008). Evidence has shown that fluctuations in EEG power correlate with the time series of ICNs, though the literature has not converged on consistent relationships between the two. For example, while some studies report a positive correlation between alpha power fluctuations and BOLD signals in the DMN (Mantini et al., 2007), others reported either weak or no correlation (Gonçalves et al., 2006; Knyazev et al., 2011; Laufs et al., 2003; Wu et al., 2010). Second, there is also evidence that fluctuations in the power of different frequencies of the EEG jointly contribute to the BOLD signal of RSNs (Mantini et al., 2007). While the majority of studies examine the EEG correlates of BOLD signal activity, recent studies have begun to relate features in the EEG to inter- and intra-subject variations in FC (Allen et al., 2013; Chang et al., 2013b; Hlinka et al., 2010; Lu et al., 2007; Scheeringa et al., 2012; Tagliazucchi et al., 2012b).
Examining the EEG correlates of within-scan changes in FC, Scheeringa et al. employed a psycho-physiological interaction analysis (PPI; Friston et al., 1997), which tests whether the regression slope of the relationship between brain regions differs between conditions of (in this case) low versus high alpha power. They reported an association between increases in alpha power and both decreases in BOLD connectivity within the visual cortex, and decreases in negative coupling between visual and default-mode regions (Scheeringa et al., 2012). Using a sliding-window analysis, both Chang et al. and Tagliazzucchi et al. demonstrated that alpha power tended to have an inverse relationship with BOLD FC, the former reporting this relationship with FC between the default-mode and dorsal attention networks (Chang et al., 2013b), and the latter with widespread AAL-atlas-defined region pairs (Tagliazucchi et al., 2012b). Both of these studies, as well as Allen et al. (2013), also observed that increases in the power of slower EEG oscillations showed the opposite behavior, correlating with increases in FC. Tagliazzucchi et al. additionally reported a correlation between increased gamma-band power and increased FC, Wu et al. (2010) showed that resting-state fMRI with eyes open versus eyes closed showed significant differences in network connectivity that was also correlated with EEG alpha power, and Allen et al. (2013) demonstrated that distinct FC states (see Reproducible patterns of sliding-window correlations section) were associated with reliable differences in EEG power spectra. Collectively, these studies imply that variability in BOLD FC to some degree reflects changes in neuronal synchrony, which may be driven largely by shifts in vigilance states. Recording EEG concurrently with resting-state scans may therefore be a desirable practice whenever feasible, as it may help to account for substantial within- (and between-) subject variance in FC.
Simultaneous LFP–fMRI
Animal models have played a critical role in the interpretation of both fMRI and FC mapping by allowing invasive, multimodal studies that can compare electrical measures of neural activity to the indirect response of the BOLD signal (Brinker et al., 1999; Logothetis et al., 2001; Lu et al., 2007; Magri et al., 2012; Pan et al., 2010, 2011; Schölvinck et al., 2010; Shmuel and Leopold, 2008). In healthy human subjects, simultaneous imaging and recording experiments are limited to the surface EEG, which has poor depth sensitivity and spatial resolution. In animal models, however, implanted electrodes can provide a localized measure of neural activity. The experiments are technically difficult and recordings are often limited to a single site. However, at least two groups have used multisite recording to investigate the neural basis of BOLD signal correlations (Lu et al., 2007; Pan et al., 2010, 2011). Lu et al. found that coherent delta oscillations in left and right somatosensory cortices behaved similarly to BOLD correlation when the level of anesthetic was varied. Pan et al. showed that while high frequency LFPs (particularly gamma) were most correlated with the local BOLD signal, the correlation between the band-limited power (BLP) of delta and theta bands from left and right somatosensory cortices best predicted BOLD correlation. In a subsequent study, The authors found that the sliding-window BOLD signal correlation between bilateral somatosensory cortices was significantly linked with the sliding-window BLP in the gamma, beta, and theta bands (Merritt et al., 2013).
Using simultaneous LFP–fMRI measurements in awake monkeys at rest, Schölvinck et al. reported that fluctuations in gamma LFP power measured from a single cortical site displayed spatially widespread cross-correlations with the fMRI (MION) signal (Schölvinck et al., 2010). This finding suggests that a component of the global fMRI signal is tightly linked with neural activity. Notably, a sliding-window analysis revealed that the strength of LFP–fMRI coupling was not constant, but instead varied considerably over time. The variations in LFP–fMRI correlation appeared to depend on the behavioral state of the animal, with stronger correlations during periods when the eyes were closed compared to open. Therefore, neurovascular coupling may itself be dynamic and subject to behavioral state, introducing yet another level of complexity when interpreting dynamics in FC.
Simultaneous measurement of physiological and autonomic states
Measurements of systemic physiological processes, such as galvanic skin response (GSR) and the respiratory and cardiac data that are often monitored during fMRI, can – in addition to their utility for noise reduction – potentially illuminate changes in autonomic processes or arousal that may underlie certain fluctuations in BOLD FC. For example, variability in the beat-to-beat interval of the cardiac cycle (heart rate variability; HRV) is a robust, non-invasive index of autonomic state (Task Force, 1996), and fluctuations in HRV across an fMRI scan can form a covariate with which to identify brain regions implicated in autonomic control (Critchley et al., 2003). States of different HRV levels can be readily connected to distinct states of autonomic nervous system activity; e.g., HRV is modulated by emotionally salient contexts (Jönsson and Sonnby-Borgström, 2003; Raz et al., 2012; Wallentin et al., 2011), and it is possible that fluctuations in BOLD FC are associated with resting-state fluctuations in autonomic nervous system tone (Chang et al., 2013a; Fan et al., 2012). In a study that explored this possibility, a sliding-window correlation analysis using seed regions implicated in salience and autonomic processing, the dorsal anterior cingulate cortex (dACC) and amygdala (Critchley et al., 2003; Dalton et al., 2005; Seeley et al., 2007), was performed to identify areas whose temporal variation in FC with these nodes significantly correlated with variations in HRV computed over the same sliding windows (Chang et al., 2013a). Thus, fluctuations in cardiac features (HRV) were used to identify potential autonomic correlates of fluctuations in FC (Fig. 3). A set of regions, including the brainstem, thalamus, putamen, and dorsolateral prefrontal cortex, was found to become more strongly coupled with the dACC and amygdala seeds during states of elevated HRV. Furthermore, dynamics of FC could be separated from those primarily related to BOLD signal fluctuations. The correspondence between changes in HRV and changes in FC suggests that fluctuations in autonomic tone, and their potential psychological correlates, contribute to variability in resting-state connectivity.
However, determining the relationship between autonomic processes and BOLD FC, especially the dynamic properties thereof, is complicated by the fact that fluctuations in autonomic processes are accompanied by changes in physiology that are known to alter BOLD signal dynamics. One may apply pre-processing strategies based on a priori models of the relationship between cardiac/respiratory fluctuations and non-neural modulation of the fMRI signal (i.e. that due to systemic changes in arterial CO2 and blood flow; see above section Physiological noise and pre-processing) such that after correction, the BOLD signal and its time-varying FC more closely reflect fluctuations in local neuronal metabolism. Residual effects may remain, presenting a caveat that must be considered when interpreting the results of such studies.
Relationship with behavioral response
Another approach to understanding and validating time-varying connectivity in human subjects is to link changes in FC to behavioral outputs. For example, if particular network configurations can be tied to better performance on a task, it provides evidence that the changes in connectivity are linked to brain function. The pre-stimulus amplitude of spontaneous BOLD activity has been found to be predictive of a subject’s ensuing behavior or perception (Fox et al., 2007; Hesselmann et al., 2008; see Sadaghiani et al., 2010 for review), and such findings have recently been extended to pre-stimulus measures of FC. Thompson et al. (2013) reported that greater anticorrelation between nodes of the DMN and TPN in a short window (12 s) centered a few seconds prior to task performance was predictive of a faster reaction time, both within and across subjects. The relative correlation between the two networks was a stronger predictor than the relative magnitude of the signal within the networks. This study contributed further evidence that the network relationships previously implicated in task performance using static analysis also predicted performance on much shorter time scales (Kelly et al., 2008). In addition, an ICA-based study showed that changes in DMN and a frontoparietal network were predictive of subsequent errors up to 30 s prior to the error (Eichele et al., 2008). Thus, behavioral outputs can be a powerful tool for determining the significance of BOLD dynamics.
Variability in MEG signals
MEG can provide complementary information with regard to FC dynamics (Dimitriadis et al., 2012). MEG directly captures electrophysiological activity through recordings of biomagnetic fields (Hämäläinen et al., 1993; Pizzella et al., 2001). Importantly, biomagnetic fields are not perturbed by changes in tissue conductivity, permittivity, and membrane boundaries to the extent that electric fields are, thereby allowing for a more straightforward localization of brain activity. Additionally, and in contrast with fMRI, the high temporal resolution of MEG enables the study of within- and across-network interactions and their modulation across frequency and over time on behaviorally relevant time-scales (Varela et al., 2001). Nevertheless, MEG suffers from low spatial resolution due to the inherent indetermination of the inverse problem. This leads to the emergence of spurious correlation, mainly affecting neighboring nodes (de Pasquale et al., 2012; Hauk et al., 2011).
Recent methodological advances have demonstrated that the slow BLP fluctuations of resting-state MEG signals have notable similarities to resting-state BOLD fMRI signals. For example, different spectral measures of interactions among nodes of ICN exhibited a peak at ~0.1 Hz (Brookes et al, 2011a,b; de Pasquale et al., 2010, 2012; Hipp et al., 2012; Liu et al., 2010) supporting the theory that the low frequency (~0.1 Hz) of spontaneous BOLD fluctuations is of neural origin. In addition, distinct ICNs estimated in MEG/EEG data (see Fig. 5A) appear to have distinctive spectral characteristics; it has been reported that stronger interactions were driven by alpha and beta band oscillations in the dorsal attention network (de Pasquale et al., 2010) and motor network (Brookes et al., 2011a,b), whereas in the default-mode network, interactions involved theta, alpha and beta oscillations (de Pasquale et al., 2010; Hipp et al., 2012).
Fig. 5.
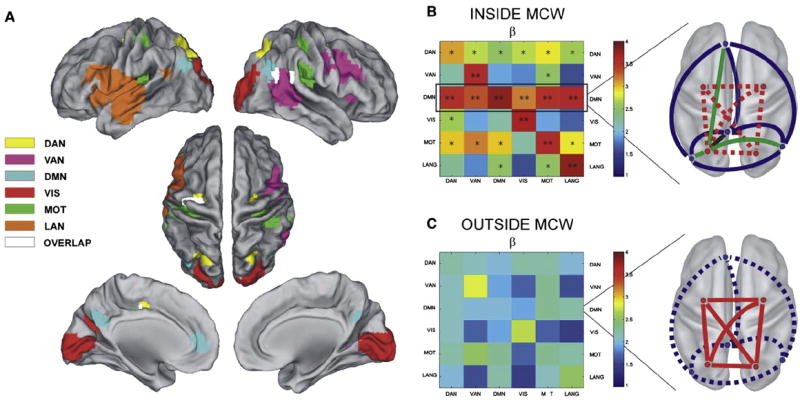
A) Spatial topography of MEG RSNs obtained in network-specific epochs of high internal connectivity (maximal correlation windows — MCWs): yellow: DAN; cyan: DMN; pink: ventral attention network (VAN); red: visual (VIS); green: somatomotor (MOT); orange: language (LAN); white: voxels shared across different networks. B) Left: within-MCW cross-network interaction estimated from the BLP in the β band. The matrix is not symmetric because the interaction is estimated for each network (row) during its respective MCWs. When internal connectivity is high, the DMN is the most strongly interacting network. Right: the DMN is strongly internally correlated (thick blue lines). Internal correlation within the DAN (red) is reduced and partially de-coupled (thin dotted red lines). Some nodes in the DAN (e.g., left PIPS) couple with nodes of the DMN (e.g., PCC) (thick green lines). C) Left: during periods outside MCWs, the overall interaction of the DMN with other networks (in the β band) is reduced and its centrality is no longer evident. Right: when the DMN’s internal connectivity is low, the DAN can have strong within-network connectivity, but little integration with other nodes or network occurs.
Adapted with permission from de Pasquale et al. (2012).
MEG studies of network dynamics have indicated that ICNs exhibit varying epochs of high and low internal coupling (de Pasquale et al., 2010). Subsequent studies have begun to uncover ‘rules’ governing the variability of FC within/across networks. For example, the DMN (especially its posterior cingulate node) was identified to be the network most strongly interacting with the other examined networks, specifically when its internal coherence is high (de Pasquale et al., 2012; Fig. 5B). This cross-network interaction requires a partial de-coupling of some nodes in other networks that become functionally coupled with the DMN. However, this functional relationship breaks apart when the DMN’s internal correlation is relatively lower (Fig. 5C). Such data suggest that the DMN assumes a role of transiently integrating systems, likely via beta-band synchronization, which might be linked to the transient periods of strong within network synchronization reported using fMRI (Hutchison et al., in press).
Modulation with conscious states
If dynamic FC is the cause or ongoing consequence of distributed mental activity, it follows that changes should be present across various conscious states in which the level of cognitive processing is significantly impacted. Specific state-dependent changes in static FC have been identified across a range of physiological (light and deep sleep, hypnosis, meditation), pharmacological (sedation, anesthesia), and pathological (coma-related states) alterations of consciousness (for review, see Heine et al., 2012; Tang et al., 2012). Studies of resting-state dynamics have revealed within-scan fluctuations in FC across species and under anesthesia (Hutchison et al., in press; Keilholz et al., 2013; Majeed et al., 2011; see also Sliding window analysis and Repeating sequences of BOLD activity sections above), which therefore cannot be fully attributed to conscious cognitive processing. This finding is supported by extensive electrophysiological studies in anesthetized animals showing that spontaneous dynamics are entrained rather than determined by sensory information (Arieli et al., 1996; Fiser et al., 2004; Kenet et al., 2003; Tsodyks et al., 1999, for reviews see Deco et al., 2012; Sporns, 2011), though as discussed above, it must be acknowledged that the presence of BOLD FC fluctuations does not, in itself, indicate functionally relevant dynamics or even true non-stationarity of neural interactions.
Further insight may be obtained by comparing changes in FC dynamics between states (particularly within the same species). Awake animal (Liang et al., 2011; Mantini et al., 2011) and anesthetized human (Boveroux et al., 2010; Kiviniemi et al., 2005; Greicius et al., 2008; Martuzzi et al., 2010; Peltier et al., 2005; Schrouff et al., 2011) investigations are both technically feasible. More mild alterations in state may also be examined; for instance, Rack-Gomer et al. report that caffeine ingestion induces differences in the variability of sliding-window correlations (Rack-Gomer and Liu, 2012). Approaches that can help control for drug-related confounds should be invoked whenever possible; for example, Långsjö et al. (2012) exploited the unique properties of the anesthetic dexmedetomidine that allows for a rapid return to consciousness from the unconscious state (with tactile or verbal stimulation) during constant dosing, ensuring a consistent dose between both states in a PET investigation of consciousness. Sleep may also offer unique opportunities for studying the functional relevance of FC dynamics (e.g. Horovitz et al., 2009).
Insights from large-scale network modeling
Recent empirical studies on the temporal dynamics of FC in the mammalian/human brain have unfolded in parallel with the development of several large-scale computational models of spontaneous neural dynamics. These models are explicitly rendered as neuronal networks whose elements (nodes) implement local or regional interactions among excitatory and inhibitory cell populations and whose connections (edges) represent inter-regional axonal pathways, for example derived from comprehensive tract tracing or diffusion imaging/tractography. A series of models of resting-state dynamics in macaque and human cortices (Deco et al., 2009; Ghosh et al., 2008; Honey et al., 2007), using different implementations for node dynamics and interaction terms, has yielded a largely consistent set of results (reviewed in Deco et al., 2011). First, resting-state brain activity is found to be constrained by the topology of the brain’s SC, most clearly expressed in the relation between SC and long-time averages of neuronal signal fluctuations. Second, over shorter time scales, functional interactions between nodes exhibit significant variability, fluctuating on multiple time scales. Third, these variable couplings give rise to a rich set of functional networks, a functional repertoire that is continually re-visited or rehearsed across time. Fourth, delays and noise jointly contributed to create a dynamic regime where the system was continually driven away from dynamic equilibrium, essentially resulting in dynamics that consisted of a series of transients during which the system explored a region of its state space around the point of dynamic equilibrium. And finally, that the models do not require the implementation of intrinsic or extrinsic drivers in order to trigger network fluctuations — the fluctuations are dynamic transients that result from the continuous interplay of stable (deterministic) and unstable (noisy) episodes. The emergence of additional time scales from faster dynamics has been observed in other high-dimensional systems, where they are described as “itinerant” or “metastable” dynamics (Kaneko and Tsuda, 2003). Taken together, computational modeling further supports that the existence of fluctuations in FC, and hence the time-dependent nature of FC, is neither a mystery nor an experimental artifact — but instead, a fundamental emergent feature of large-scale dynamics that may be exploited for neural computation (Rabinovich and Varona, 2011) and the maintenance of robust and flexible cognition (Deco and Corbetta, 2011).
A consequence of the dynamics that emerged within the modeled functional architecture was a notable time-dependence of network measures, such as node centrality, which is often used to characterize highly influential network elements. The model predicted that a given region’s status as a network hub varies across time (Honey et al., 2007). With time-dependent changes in hub centrality, it is also to be expected that there will be substantial alteration in other large-scale and node-specific graph measures such as degree, motif distribution, modularity, and efficiency. As these measures are closely interrelated, it will be important to disentangle (using both empirical and simulated data) whether these changes are a consequence of separate processes or reflect a more gross time-varying reorganization of the underlying architecture that exists on several topological scales (Meunier et al., 2010).
Possible mechanisms of action
It may be speculated that fluctuations in BOLD FC arise from changes in the organization and connectivity of neocortical micro-circuits themselves. Specifically, a change in large-scale FC and a change in the activation (depolarization) state of a local cortical column may interact through two principal, complementary routes: a local state change can be the source for long-range changes in FC, or the local state change is a reflection of distant influences and hence reflects the reconfiguration of a larger network.
It has been shown that the activation of a cortical microcircuit is reflected in the activity of the output cells that carry the change in excitation to distant, connected brain areas (Amzica and Steriade, 1995; Timofeev et al., 2012). The principal output cells in deep cortical layers of a cortical circuit form highly segregated subnetworks that are defined by their projection targets (reviewed in Krook-Magnuson et al., 2012), and various mechanisms may alter a local circuit’s activation in ways that influence the firing of deep-layer projection cells (Supplementary Table 1). A recent study suggests that modulation of the strength of local beta oscillations could be sufficient to trigger a dynamic reconfiguration of deep-layer cortical sub-networks (Canolty et al., 2012; Fig. 6A). For some deep-layer cells, increases in beta amplitude are accompanied by increased firing, while for others the relationship is reversed. One consequence of such a cell-specific beta-to-rate mapping is its potential to index switches in functional networks. As shown in Fig. 6B, such a change in the composition of co-activated cells in the cortical output layers predicts a change in the large-scale FC before and after the modulation of beta oscillations.
Fig. 6.
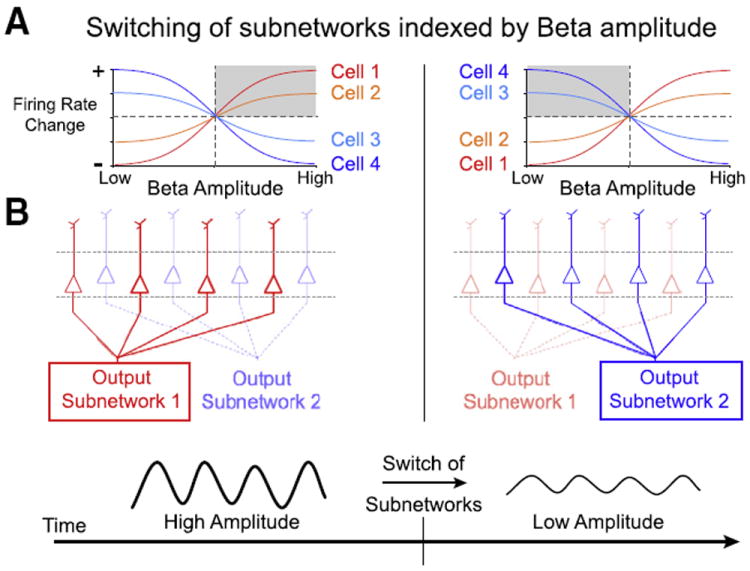
Proposed scheme of subnetwork modulation of deep layer cells by amplitude variations of beta oscillations. A) Canolty et al. (2012) have shown that in deep cortical layers, there is a highly robust sigmoidal relation between the firing rate of single cells and the amplitude of beta local field potential oscillations. Some cells (cells 1 and 2) have high firing rates during high beta amplitudes (gray shading, left panel), while other cells (cells 3 and 4) have a higher firing rate during suppressed beta amplitudes (gray shading, right panel). The amplitude-to-rate mapping is consistent across recording sessions. B) The cell-specific amplitude-to-rate mapping suggests that a state of high beta amplitude (left panels) coincides with the activation of a selected subnetwork of deep layer cells. When the beta amplitude changes, the subnetwork of cells with high firing rate switches (right panels). This hypothesis predicts that modulation of local oscillatory power indexes a change in the active projection sites, and a corresponding change in long-range functional network connectivity.
Beyond local cortical mechanisms, neuromodulatory nuclei – known to affect the activation level across distributed brain areas – may substantially contribute or regulate the dynamic reconfiguration of spatially circumscribed ICNs (Leopold et al., 2003). They include a variety of subthalamic nuclei and brainstem regions with an extensive range of neurotransmitters (Supplementary Table 1). Most of these nuclei connect bi-directionally with cortical sites and can promote transitions from DOWN- to UP-states during slow frequency oscillations, or induce transient functional connectivity through cortical and thalamo-cortical gating mechanisms (Timofeev et al., 2012). Presently, there only exists correlational evidence for both routes of network modulation and it is unknown which type of modulation occurs first (e.g. local beta amplitude increase or long distance induced beta phase alignment). Without more empirical evidence, the relationship between dynamic FC as well as the periods of metastable FC (i.e., distinct, recurring “states”) with that of the direct neural coordination observed with electrophysiology remains unclear. These investigations will be of great importance for the validation and interpretation of present and forthcoming results.
Clinical applications
Most, if not all, physiological and psychiatric diseases have disrupted large-scale functional and/or structural properties (for review see Greicius, 2008; Menon, 2011). Whether they are the cause or consequence of the disease is unclear, but it is possible that clinical populations exhibit significant changes in dynamic properties, and that the latter may in fact underlie many of the observed dysfunctions. Quantification of disrupted dynamics in clinical populations may lead to a better understanding of the disorder, more targeted drug treatment, and eventually, diagnostic or prognostic indicators. Moreover, an improved understanding of the link between disease and dynamics can further enhance our understanding of how dynamic network properties support normal brain function. Here we survey early work examining altered dynamics in schizophrenia, depression, and Alzheimer’s disease. We expect that as the field continues to expand, so too will the number diseases studied (e.g. Eckman et al., 2012; Leonardi et al., 2013).
Schizophrenia
There have been numerous reports of changes in static connectivity in schizophrenia (e.g. Calhoun et al., 2009). More recently, both Sakoglu et al. and Damaraju et al. have evaluated changes in the dynamic FC of patients with schizophrenia (Damaraju et al., 2012; Sakoglu et al., 2010). Using a sliding-window analysis, Sakoglu et al. reported that sensory, motor, and frontal networks had less engagement with other networks (including the DMN) when an auditory oddball task stimulus was presented, and that there were also significant differences in the time–frequency patterns of connectivity between schizophrenic patients compared to healthy controls. In Damaraju et al., the dynamic FC approached introduced by Allen et al. (in press) was applied to a large group (N > 300) of patients with schizophrenia and healthy controls. K-means clustering of dynamic FC states revealed similar FC states for both healthy controls and schizophrenic subjects over a range of cluster sizes (K = 2–9). However, the FC window states of healthy controls were found to switch more often, suggesting that patients with schizophrenia tend to linger in a state of “weak” and relatively “rigid” connectivity, while healthy controls dynamically switch between different FC states and are therefore probably faster in recruiting necessary resources in the face of changing task demands. Such a finding was not detectable using a static FC approach, and underscores the importance of evaluating dynamic changes in connectivity. More recently, similar results have been found in bipolar patients as well (Rashid et al., 2013).
Major depression
Transient FC as a marker of dynamic brain state changes would also be well in line with current concepts and findings of brain activity in depression. A recently proposed model conceptualizes major depressive disorder (MDD) as an imbalanced state shift that is biased towards being “stuck in a rut” (Holtzheimer and Mayberg, 2011), wherein negative mood states appear to be preferentially reached and withdrawn from with greater difficulty, mainly due to attentional and processing biases. Such a model suggests that alterations in static FC could be due to a bias towards more frequent down states, and would predict a decrease in mood-related brain dynamics due to the diminished capacity for self-induced mood changes. Initial findings have supported the notion that the resting-state dynamics of key brain structures in MDD may be critically altered and thus provide reasonable correlates of subjects’ decreased ability to flexibly react to external or internal cognitive demands (Hamilton et al., 2011; Horn et al., 2010). In such a context, a reduction of differentiated activation, corresponding to a reduction in the cognitive repertoire, would also possess counterparts in other time scales, such as the consistent reduction of alpha synchrony in resting state EEG (Allen and Cohen, 2010). More work is needed to understand and conceptualize the connections between fluctuating affective/autonomic states and brain network dynamics.
Alzheimer’s disease
Previous reports have found altered static FC measures in Alzheimer’s patients compared to healthy controls (Buckner et al., 2009; Greicius et al., 2004; Sorg et al., 2007; Supekar et al., 2008; reviewed in Greicius, 2008; Menon, 2011). Jones et al. (2012) studied impairments in the dynamics of spontaneous activity by examining time varying changes of a modularity metric (Q, see Rubinov and Sporns, 2011) applied to graphical representations of functional connectivity using a sliding-window approach. The authors reported differences in the “dwell time” within different sub-network configurations of the DMN between Alzheimer’s patients and age-matched healthy controls. More specifically, there was less time spent in brain states with strong posterior DMN region contributions and more time in states characterized by dorsal medial PFC component contributions. This is one of the first reports demonstrating RS-fMRI changes in Alzheimer’s patients beyond average FC metrics. Considering temporal features of FC may therefore provide a more accurate description of Alzheimer’s disease, potentially leading not only to a better understanding of the large-scale characterizations of the disease, but also to better diagnostic and prognostic indicators.
Future directions
Investigations of spontaneous FC changes within and between ICNs are now being undertaken at an accelerated pace, and results are being interpreted with cautious optimism. However, there are a number of concerns about – and direct challenges to – the very nature of the investigations, including the underlying causes, functional relevance, analysis, and interpretation, as outlined above. Early work has offered great promise in revealing aspects of dynamic FC at macroscopic scales, and there are multiple research directions that can further improve our understanding of this phenomenon (see Box 1).
Box 1. Open questions.
What is the neural origin, mechanism, and function of dynamic FC?
To what degree does dynamic FC represent conscious versus unconscious or autonomic processes?
What are the contributions to FC fluctuations from motion, physiological noise, and scanner noise and how are they most effectively removed?
What is the appropriate definition of a network and its nodes for quantifying dynamic FC?
Does FC undergo deterministic temporal sequences during rest or task?
Is there a finite set of network configurations (a “functional repertoire”) that is continually re-visited? Are these reproducible across subjects? How are they related to the underlying genotype? Do new configurations emerge during development or adulthood?
Is dynamic FC modulated by a central executive such as the prefrontal cortex, or is it self-organizing?
What roles do brainstem and subcortical structures play in regulating dynamic FC?
What are the implications of dynamic changes in FC for the creation and thresholding of network graphs, and for the interpretation of studies utilizing measures of temporal precedence such as Granger causality analysis?
Typical resting state acquisition parameters may not be optimized for exploring all aspects of dynamic FC changes. Scans of 5–10 min are likely not sufficient for considering the repertoire of states and their rate of change. While the optimal duration will depend upon the question of interest, it is advisable that the average length of scans be extended. Concurrent monitoring of physiological processes (respiration, cardiac, and even GSR) will be vital for evaluating FC dynamics, given their reported influence and the fact that fewer data points are averaged in a dynamic analysis. Concurrent recording of EEG as an index of mental state may also be desirable given the influence of vigilance state and sleep on FC (Tagliazucchi et al., 2012a), and for its utility as a complementary measure of neuro-electrical activity that can aid in modeling and interpreting dynamic BOLD FC.
There have been recent advances in MR pulse sequences and image reconstruction, such as multiband imaging (Larkman et al., 2001; Moeller et al., 2010; Feinberg et al., 2010), MR encephalography (MREG; Hennig et al., 2007), and inverse imaging (InI; Lin et al., 2006) that allow for higher temporal sampling rates (short TR). A high sampling rate may be beneficial for reducing structured physiological noise. However, the BOLD signal response to metabolic changes is filtered through a slow hemodynamic response, such that the benefit of decreased TR for depicting rapid neural events is unclear. Nevertheless, emerging evidence suggests that FC may be present in components of the fMRI signal above 0.1 Hz (Boubela et al., 2013; Lee et al., 2013; Niazy et al., 2011; van Oort et al., 2012), rendering this an intriguing avenue of future work.
While a number of strategies for analyzing dynamic FC were discussed above, there are many possible extensions and unexplored avenues. Techniques can be adopted from other fields such as electrophysiology (LFP analysis) or computer science (pattern recognition) that offer a wide variety of tools for dynamic data analysis. Visualization is also an important area for development, due to the multi-dimensional nature of the output from a dynamic analysis. Many authors are now submitting movies as supplementary material, displaying how FC varies over time. There is inherent value in having readers examine dynamics to identify patterns that may exist in the data and not detected by most algorithms. However, there must be a balance between data transparency and overwhelming the reader with information, hindering interpretation (Allen et al., 2012), and it will thus be fruitful to find ways of reducing the dimensionality while maintaining key features. Innovative methods for visualizing findings are under development (reviewed in Margulies et al., 2013); presenting the complexity of connectivity space through bundling similar edges and using surface-based glyphs (Böttger et al., in press), as well as dynamic network visualization tools developed in other disciplines (e.g. SONIA: http://www.stanford.edu/group/sonia/) offer a glimpse into possible avenues. Finally, it will be of critical importance to move from examining variation in FC with simple descriptive measures (such as correlation) to more complex, biologically informed generative models that can allow rigorous inference of non-stationarity functional network activity from fMRI data. Continued analysis of the temporal characteristics of spontaneous brain activity with direct electrophysiological measurements, as well as further comparative studies across states, species, disease models, pharmacological manipulations, and lesions, can help to inform such models and permit a deeper understanding spontaneous activity and the dynamics thereof.
Conclusion
Resting-state fMRI has brought significant attention to spontaneous brain activity, a subject that has long been appreciated in electrophysiological fields. Static analysis approaches will likely persist and continue to provide valuable information concerning normal and abnormal brain organization. However, if we desire a more comprehensive understanding of large-scale network activity, dynamic connectivity patterns must be considered and evaluated. Recent evidence suggests that these phenomena may be an intrinsic property of brain function with a neural origin. It is important to bear in mind, however, that the enterprise of dynamic FC is presently exploratory; a great number of questions remain in terms of methodological concerns and interpretation, and more effort towards developing methods and metrics for dynamic FC will be necessary before researchers can widely apply such tools in a more standardized sense. However, based upon the availability of proven analysis tools, data, and expertise, we expect dramatic progress to be made in this area in coming years. Indeed, it may represent an extraordinary new frontier in the study of brain connectivity.
Supplementary Material
Acknowledgments
The theme for this review was inspired from discussions and proceedings at the Neuro Bureau’s BrainHack (Leipzig, Germany) and the Biennial Resting-State Conference (Magdeburg, Germany). R.M.H. was supported by a Canadian Institute of Health Research postdoctoral fellowship, and C.C. was supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke, National Institutes of Health. We thank Steve Smith and four anonymous reviewers for helpful suggestions.
Footnotes
Formally, a non-stationary time series is one whose mean and covariance (or, in the strictest sense, all higher-order moments) are not constant over time.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neuroimage.2013.05.079.
Conflict of interest
The authors declare no conflicts of interest.
References
- Abou-Elseoud A, Starck T, Remes J, Nikkinen J, Tervonen O, Kiviniemi V. The effect of model order selection in group PICA. Hum Brain Mapp. 2010;31:1207–1216. doi: 10.1002/hbm.20929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelstein JS, Shehzad Z, Mennes M, Deyoung CG, Zuo XN, Kelly C, Margulies DS, Bloomfield A, Gray JR, Castellanos FX, Milham MP. Personality is reflected in the brain’s intrinsic functional architecture. PLoS One. 2011;6:e27633. doi: 10.1371/journal.pone.0027633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akam T, Kullmann DM. Oscillations and filtering networks support flexible routing of information. Neuron. 2010;67:308–320. doi: 10.1016/j.neuron.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert NB, Robertson EM, Miall RC. The resting human brain and motor learning. Curr Biol. 2009;19:1023–1027. doi: 10.1016/j.cub.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JJ, Cohen MX. Deconstructing the “resting” state: exploring the temporal dynamics of frontal alpha asymmetry as an endophenotype for depression. Front Hum Neurosci. 2010;4:232. doi: 10.3389/fnhum.2010.00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Erhardt E, Calhoun VD. Data visualization in the neurosciences: overcoming the curse of dimensionality. Neuron. 2012;74:603–608. doi: 10.1016/j.neuron.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Damaraju E, Plis SM, Erhardt E, Eichele T, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. 2013 doi: 10.1093/cercor/bhs352. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Eichele T, Wu L, Calhoun VD. EEG signatures of functional connectivity states. Human Brain Mapping; Seattle, WA: 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amzica F, Steriade M. Disconnection of intracortical synaptic linkages disrupts synchronization of a slow oscillation. J Neurosci. 1995;15:4658–4677. doi: 10.1523/JNEUROSCI.15-06-04658.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arieli A, Sterkin A, Grinvald A, Aertsen A. Dynamics of ongoing activity: explanation of the large variability in evoked cortical responses. Science. 1996;273:1868–1871. doi: 10.1126/science.273.5283.1868. [DOI] [PubMed] [Google Scholar]
- Barnes A, Bullmore ET, Suckling J. Endogenous human brain dynamics recover slowly following cognitive effort. PLoS One. 2009;4:e6626. doi: 10.1371/journal.pone.0006626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. Dynamic reconfiguration of human brain networks during learning. Proc Natl Acad Sci U S A. 2011;108:7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EB, Lowe MJ. Isolating physiologic noise sources with independently determined spatial measures. Neuroimage. 2007;37:1286–1300. doi: 10.1016/j.neuroimage.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchenane K, Peyrache A, Khamassi M, Tierney P, Gioanni Y, Battaglia FP, Wiener SI. Coherent theta oscillations and reorganization of spike timing in the hippocampal–prefrontal network upon learning. Neuron. 2010;66:921–936. doi: 10.1016/j.neuron.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Bianciardi M, Fukunaga M, van Gelderen P, Horovitz SG, de Zwart JA, Duyn JH. Modulation of spontaneous fMRI activity in human visual cortex by behavioral state. Neuroimage. 2009;45:160–168. doi: 10.1016/j.neuroimage.2008.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birn RM, Smith MA, Jones TB, Bandettini PA. The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage. 2008;40:644–654. doi: 10.1016/j.neuroimage.2007.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Imaging. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Boly M, Balteau E, Schnakers C, Degueldre C, Moonen G, Luxen A, Phillips C, Peigneux P, Maquet P, Laureys S. Baseline brain activity fluctuations predict somatosensory perception in humans. Proc Natl Acad Sci U S A. 2007;104:12187–12192. doi: 10.1073/pnas.0611404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosman CA, Schoffelen JM, Brunet N, Oostenveld R, Bastos AM, Womelsdorf T, Rubehn B, Stieglitz T, De Weerd P, Fries P. Attentional stimulus selection through selective synchronization between monkey visual areas. Neuron. 2012;75:875–888. doi: 10.1016/j.neuron.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttger J, Schäfer A, Lohmann G, Villringer A, Margulies DS. Three-dimensional mean-shift edge bundling for the visualization of functional connectivity in the brain. IEEE Trans Vis Comput Graph. 2013 doi: 10.1109/tvcg.2013.114. in press. [DOI] [PubMed] [Google Scholar]
- Boubela RN, Kalcher K, Huf W, Kronnerwetter C, Filzmoser P, Moser E. Beyond noise: using temporal ICA to extract meaningful information from high-frequency fMRI signal fluctuations during rest. Front Hum Neurosci. 2013;7:168. doi: 10.3389/fnhum.2013.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveroux P, Vanhaudenhuyse A, Bruno MA, Noirhomme Q, Lauwick S, Luxen A, Degueldre C, Plenevaux A, Schnakers C, Phillips C, Brichant JF, Bonhomme V, Maquet P, Greicius MD, Laureys S, Boly M. Breakdown of within- and between-network resting state functional magnetic resonance imaging connectivity during propofol-induced loss of consciousness. Anesthesiology. 2010;113:1038–1053. doi: 10.1097/ALN.0b013e3181f697f5. [DOI] [PubMed] [Google Scholar]
- Bressler SL, Menon V. Large-scale brain networks in cognition: emerging methods and principles. Trends Cogn Sci. 2010;14:277–290. doi: 10.1016/j.tics.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Brinker G, Bock C, Busch E, Krep H, Hossmann KA, Hoehn-Berlage M. Simultaneous recording of evoked potentials and T2*-weighted MR images during somatosensory stimulation of rat. Magn Reson Med. 1999;41:469–473. doi: 10.1002/(sici)1522-2594(199903)41:3<469::aid-mrm7>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Britz J, Van De Ville D, Michel CM. BOLD correlates of EEG topography reveal rapid resting-state network dynamics. Neuroimage. 2010;52:1162–1170. doi: 10.1016/j.neuroimage.2010.02.052. [DOI] [PubMed] [Google Scholar]
- Brookes MJ, Woolrich M, Luckhoo H, Price D, Hale JR, Stephenson MC, Barnes GR, Smith SM, Morris PG. Investigating the electrophysiological basis of resting state networks using magnetoencephalography. Proc Natl Acad Sci U S A. 2011a;108:16783–16788. doi: 10.1073/pnas.1112685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes MJ, Hale JR, Zumer JM, Stevenson CM, Francis ST, Barnes GR, Owen JP, Morris PG, Nagarajan SS. Measuring functional connectivity using MEG. Methodology and comparison with fcMRI. Neuroimage. 2011b;56:1082–1104. doi: 10.1016/j.neuroimage.2011.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brovelli A, Ding M, Ledberg A, Chen Y, Nakamura R, Bressler SL. Beta oscillations in a large-scale sensorimotor cortical network: directional influences revealed by Granger causality. Proc Natl Acad Sci U S A. 2004;101:9849–9854. doi: 10.1073/pnas.0308538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews-Hanna JR, Sperling RA, Johnson KA. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J Neurosci. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Buzsaki G. Rhythms of the Brain. 1. Oxford University Press; USA: 2006. [Google Scholar]
- Buzsaki G, Watson BO. Brain rhythms and neural syntax: implications for efficient coding of cognitive content and neuropsychiatric disease. Dialogues Clin Neurosci. 2012;14:345–367. doi: 10.31887/DCNS.2012.14.4/gbuzsaki. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T. Multi-subject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev Biomed Eng. 2012;5:60–73. doi: 10.1109/RBME.2012.2211076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14:140–151. doi: 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Kiehl KA, Pearlson GD. Modulation of temporally coherent brain networks estimated using ICA at rest and during cognitive tasks. Hum Brain Mapp. 2008;29:828–838. doi: 10.1002/hbm.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Eichele T, Pearlson GD. Functional brain networks in schizophrenia: a review. Front Hum Neurosci. 2009;3:1–12. doi: 10.3389/neuro.09.017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canolty RT, Ganguly K, Carmena JM. Task-dependent changes in cross-level coupling between single neurons and oscillatory activity in multiscale networks. PLoS Comput Biol. 2012;8:e1002809. doi: 10.1371/journal.pcbi.1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Relationship between respiration, end-tidal CO2, and BOLD signals in resting-state fMRI. Neuroimage. 2009;47:1381–1393. doi: 10.1016/j.neuroimage.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Glover GH. Time–frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 2010;50:81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: the cardiac response function. Neuroimage. 2009;44:857–869. doi: 10.1016/j.neuroimage.2008.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Shen X, Glover GH. Proc. HBM; Quebec City, Canada: 2011. Behavioral correlates of temporal variations in brain network connectivity. [Google Scholar]
- Chang C, Metzger CD, Glover GH, Duyn JH, Heinze HJ, Walter M. Association between heart rate variability and fluctuations in resting-state functional connectivity. Neuroimage. 2013a;68:93–104. doi: 10.1016/j.neuroimage.2012.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Liu Z, Chen MC, Liu X, Duyn JH. EEG correlates of time-varying BOLD functional connectivity. Neuroimage. 2013b;72:227–236. doi: 10.1016/j.neuroimage.2013.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DM, Smith SM, Beckmann CF. Advances and pitfalls in the analysis and interpretation of resting-state fMRI data. Front Syst Neurosci. 2010;4:8. doi: 10.3389/fnsys.2010.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribben I, Haraldsdottir R, Atlas LY, Wager TD, Lindquist MA. Dynamic connectivity regression: determining state-related changes in brain connectivity. Neuroimage. 2012;61:907–920. doi: 10.1016/j.neuroimage.2012.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley HD, Mathias CJ, Josephs O, O’Doherty J, Zanini S, Dewar BK, Cipolotti L, Shallice T, Dolan RJ. Human cingulate cortex and autonomic control: converging neuroimaging and clinical evidence. Brain. 2003;126:2139–2152. doi: 10.1093/brain/awg216. [DOI] [PubMed] [Google Scholar]
- Dagli MS, Ingeholm JE, Haxby JV. Localization of cardiac-induced signal change in fMRI. Neuroimage. 1999;9:407–415. doi: 10.1006/nimg.1998.0424. [DOI] [PubMed] [Google Scholar]
- Dalton KM, Kalin NH, Grist TM, Davidson RJ. Neural–cardiac coupling in threat-evoked anxiety. J Cogn Neurosci. 2005;17:969–980. doi: 10.1162/0898929054021094. [DOI] [PubMed] [Google Scholar]
- Damaraju E, Turner J, Preda A, Van Erp T, Mathalon D, Ford JM, Potkin S, Calhoun VD. Static and Dynamic Functional Network Connectivity During Resting State in Schizophrenia. American College of Neuropsychopharmacology; Hollywood, CA: 2012. [Google Scholar]
- Damoiseaux JS, Greicius MD. Greater than the sum of its parts: a review of studies combining structural connectivity and resting-state functional connectivity. Brain Struct Funct. 2009;213:525–533. doi: 10.1007/s00429-009-0208-6. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pasquale F, Della Penna S, Snyder AZ, Lewis C, Mantini D, Marzetti L, Belardinelli P, Ciancetta L, Pizzella V, Romani GL, Corbetta M. Temporal dynamics of spontaneous MEG activity in brain networks. Proc Natl Acad Sci U S A. 2010;107:6040–6045. doi: 10.1073/pnas.0913863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pasquale F, Della Penna S, Snyder AZ, Marzetti L, Pizzella V, Romani GL, Corbetta M. A cortical core for dynamic integration of functional networks in the resting human brain. Neuron. 2012;74:753–764. doi: 10.1016/j.neuron.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Corbetta M. The dynamical balance of the brain at rest. Neuroscientist. 2011;17:107–123. doi: 10.1177/1073858409354384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa V, McIntosh AR, Sporns O, Kötter R. Key role of coupling, delay, and noise in resting brain fluctuations. Proc Natl Acad Sci U S A. 2009;106:10302–10307. doi: 10.1073/pnas.0901831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, McIntosh AR. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nat Rev Neurosci. 2011;12:43–56. doi: 10.1038/nrn2961. [DOI] [PubMed] [Google Scholar]
- Deco G, Jirsa V, Friston KJ. The dynamical and structural basis of brain activity. In: Rabinovich MI, Friston KJ, Varona P, editors. Principles of Brain Dynamics: Global State Interactions. 1. The MIT Press; Cambridge MA: 2012. [Google Scholar]
- DeCoteau WE, Thorn C, Gibson DJ, Courtemanche R, Mitra P, Kubota Y, Graybiel AM. Learning-related coordination of striatal and hippocampal theta rhythms during acquisition of a procedural maze task. Proc Natl Acad Sci U S A. 2007;104:5644–5649. doi: 10.1073/pnas.0700818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis SI, Laskaris NA, Tsirka V, Vourkas M, Micheloyannis S. An EEG study of brain connectivity dynamics at the resting state. Nonlinear Dyn Life Sci. 2012;16:5–22. [PubMed] [Google Scholar]
- Duyn JH. EEG–fMRI methods for the study of brain networks during sleep. Front Neurol. 2012;3:100. doi: 10.3389/fneur.2012.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckman M, Derrfuss J, Fiebach C. Finding spatio-temporal patterns in the dynamics of complex brain networks. Proc HBM, Beijing, China 2012 [Google Scholar]
- Efron B, Tibshirani R. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat Sci. 1986;1:54–74. [Google Scholar]
- Eichele T, Debener S, Calhoun VD, Specht K, Engel AK, Hugdahl K, Von Cramon DY, Ullsperger M. Prediction of human errors by maladaptive changes in event-related brain networks. Proc Natl Acad Sci. 2008;105:6173–6178. doi: 10.1073/pnas.0708965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AK, Fries P, Singer W. Dynamic predictions: oscillations and synchrony in top–down processing. Nat Rev Neurosci. 2001;2:704–716. doi: 10.1038/35094565. [DOI] [PubMed] [Google Scholar]
- Esposito F, Bertolino A, Scarabino T, Latorre V, Blasi G, Popolizio T, Tedeschi G, Cirillo S, Goebel R, Di Salle F. Independent component model of the default-mode brain function: assessing the impact of active thinking. Brain Res Bull. 2006;70:263–269. doi: 10.1016/j.brainresbull.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Fan J, Xu P, Van Dam NT, Eilam-Stock T, Gu X, Luo YJ, Hof PR. Spontaneous brain activity relates to autonomic arousal. J Neurosci. 2012;32:11176–11186. doi: 10.1523/JNEUROSCI.1172-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Gunther M, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One. 2010;20(5):e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser J, Chiu C, Weliky M. Small modulation of ongoing cortical dynamics by sensory input during natural vision. Nature. 2004;431:573–578. doi: 10.1038/nature02907. [DOI] [PubMed] [Google Scholar]
- Fornito A, Harrison BJ, Zalesky A, Simons JS. Competitive and cooperative dynamics of large-scale brain functional networks supporting recollection. Proc Natl Acad Sci U S A. 2012;109:12788–12793. doi: 10.1073/pnas.1204185109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8:700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proc Natl Acad Sci U S A. 2006;103:10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Raichle ME. Intrinsic fluctuations within cortical systems account for intertrial variability in human behavior. Neuron. 2007;56:171–184. doi: 10.1016/j.neuron.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Fransson P. How default is the default mode of brain function? Further evidence from intrinsic BOLD signal fluctuations. Neuropsychologia. 2006;44:2836–2845. doi: 10.1016/j.neuropsychologia.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Friston KJ. Functional and effective connectivity: a review. Brain Connect. 2011;1:13–36. doi: 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- Friston K, Buchel C. Functional connectivity: eigenimages and multivariate analyses. In: Friston KJ, Ashburner JT, Kiebel SJ, Nichols TE, Penny WD, editors. Statistical Parametric Mapping: the Analysis of Functional Brain Images. Elsevier: Amsterdam; 2007. pp. 492–507. [Google Scholar]
- Friston KJ, Buchel C, Fink CR, Morris J, Rolls E, Dolan R. Psychophysiological and modulatory interactions in neuroimaging. Neuroimage. 1997;6:218–229. doi: 10.1006/nimg.1997.0291. [DOI] [PubMed] [Google Scholar]
- Fujisawa S, Buzsaki G. A 4 Hz oscillation adaptively synchronizes prefrontal, VTA, and hippocampal activities. Neuron. 2011;72:153–165. doi: 10.1016/j.neuron.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga M, Horovitz SG, van Gelderen P, de Zwart JA, Jansma JM, Ikonomidou VN, Chu R, Deckers RH, Leopold DA, Duyn JH. Large-amplitude, spatially correlated fluctuations in BOLD fMRI signals during extended rest and early sleep stages. Magn Reson Imaging. 2006;24:979–992. doi: 10.1016/j.mri.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Fukushima M, Saunders RC, Leopold DA, Mishkin M, Averbeck BB. Spontaneous high-gamma band activity reflects functional organization of auditory cortex in the awake macaque. Neuron. 2012;74:899–910. doi: 10.1016/j.neuron.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Rho Y, McIntosh AR, Kötter R, Jirsa VK. Noise during rest enables the exploration of the brain’s dynamic repertoire. PLoS Comput Biol. 2008;4:e1000196. doi: 10.1371/journal.pcbi.1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med. 2000;44:162–167. doi: 10.1002/1522-2594(200007)44:1<162::aid-mrm23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Gonçalves SI, de Munck JC, Pouwels PJ, Schoonhoven R, Kuijer JP, Maurits NM, Hoogduin JM, Van Someren EJ, Heethaar RM, Lopes da Silva FH. Correlating the alpha rhythm to BOLD using simultaneous EEG/fMRI: inter-subject variability. Neuroimage. 2006;30:203–213. doi: 10.1016/j.neuroimage.2005.09.062. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Castillo J, Wu P, Robinson M, Handwerker D, Inati S, Bandettini P. Detection of task transitions on 45 mins long continuous multi-task runs using whole brain connectivity. Biennial Resting-State Conference; Magdeburg, Germany. 2012. [Google Scholar]
- Gregoriou GG, Gotts SJ, Zhou H, Desimone R. High-frequency, long-range coupling between prefrontal and visual cortex during attention. Science. 2009;324:1207–1210. doi: 10.1126/science.1171402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius M. Resting-state functional connectivity in neuropsychiatric disorders. Curr Opin Neurol. 2008;21:424–430. doi: 10.1097/WCO.0b013e328306f2c5. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Kiviniemi V, Tervonen O, Vainionpaa V, Alahuhta S, Reiss AL, Menon V. Persistent default-mode network connectivity during light sedation. Hum Brain Mapp. 2008;29:839–847. doi: 10.1002/hbm.20537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe I, Neitzel SD, Mandon S, Kreiter AK. Switching neuronal inputs by differential modulations of gamma-band phase-coherence. J Neurosci. 2012;32:16172–16180. doi: 10.1523/JNEUROSCI.0890-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hämäläinen M, Hari R, Ilmoniemi R, Knuutila J, Lounasmaa O. Magnetoencephalography—theory, instrumentation, and applications to non-invasive studies of the working human brain. Rev Mod Phys. 1993;65:1–93. [Google Scholar]
- Hamilton JP, Chen G, Thomason ME, Schwartz ME, Gotlib IH. Investigating neural primacy in major depressive disorder: multivariate Granger causality analysis of resting-state fMRI time-series data. Mol Psychiatry. 2011;16:763–772. doi: 10.1038/mp.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DA, Roopchansingh V, Gonzalez-Castillo J, Bandettini PA. Periodic changes in fMRI connectivity. Neuroimage. 2012;63:1712–1719. doi: 10.1016/j.neuroimage.2012.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauk O, Wakeman DG, Henson R. Comparison of noisenormalized minimum norm estimates for MEG analysis using multiple resolution metrics. Neuroimage. 2011;54:1966–1974. doi: 10.1016/j.neuroimage.2010.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He BJ, Snyder AZ, Zempel JM, Smyth MD, Raichle ME. Electrophysiological correlates of the brain’s intrinsic large-scale functional architecture. Proc Natl Acad Sci U S A. 2008;105:16039–16044. doi: 10.1073/pnas.0807010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine L, Soddu A, Gómez F, Vanhaudenhuyse A, Tshibanda L, Thonnard M, Charland-Verville V, Kirsch M, Laureys S, Demertzi A. Resting state networks and consciousness: alterations of multiple resting state network connectivity in physiological, pharmacological, and pathological consciousness states. Front Psychol. 2012;3:295. doi: 10.3389/fpsyg.2012.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig J, Zhong K, Speck O. MR–encephalography: fast multi-channel monitoring of brain physiology with magnetic resonance. Neuroimage. 2007;34:212–219. doi: 10.1016/j.neuroimage.2006.08.036. [DOI] [PubMed] [Google Scholar]
- Hesselmann G, Kell CA, Eger E, Kleinschmidt A. Spontaneous local variations in ongoing neural activity bias perceptual decisions. Proc Natl Acad Sci U S A. 2008;105:10984–10989. doi: 10.1073/pnas.0712043105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipp JF, Hawellek DJ, Corbetta M, Siegel M, Engel AK. Large-scale cortical correlation structure of spontaneous oscillatory activity. Nat Neurosci. 2012;15:884–890. doi: 10.1038/nn.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlinka J, Alexakis C, Diukova A, Liddle PF, Auer DP. Slow EEG pattern predicts reduced intrinsic functional connectivity in the default mode network: an inter-subject analysis. Neuroimage. 2010;53:239–246. doi: 10.1016/j.neuroimage.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Holtzheimer PE, Mayberg HS. Stuck in a rut: rethinking depression and its treatment. Trends Neurosci. 2011;34:1–9. doi: 10.1016/j.tins.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Kötter R, Breakspear M, Sporns O. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc Natl Acad Sci U S A. 2007;104:10240–10245. doi: 10.1073/pnas.0701519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P. Predicting human resting-state functional connectivity from structural connectivity. Proc Natl Acad Sci U S A. 2009;106:2035–2040. doi: 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn DI, Yu C, Steiner J, Buchmann J, Kaufmann J, Osoba A, Eckert U, Zierhut KC, Schiltz K, He H, Biswal B, Bogerts B, Walter M. Glutamatergic and resting-state functional connectivity correlates of severity in major depression — the role of pregenual anterior cingulate cortex and anterior insula. Front Syst Neurosci. 2010;4:33. doi: 10.3389/fnsys.2010.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz SG, Fukunaga M, de Zwart JA, van Gelderen P, Fulton SC, Balkin TJ, Duyn JH. Low frequency BOLD fluctuations during resting wakefulness and light sleep: a simultaneous EEG–fMRI study. Hum Brain Mapp. 2008;29:671–682. doi: 10.1002/hbm.20428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz SG, Braun AR, Carr WS, Picchioni D, Balkin TJ, Fukunaga M, Duyn JH. Decoupling of the brain’s default mode network during deep sleep. Proc Natl Acad Sci U S A. 2009;106:11376–11381. doi: 10.1073/pnas.0901435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz B. The elusive concept of brain connectivity. Neuroimage. 2003;19:466–470. doi: 10.1016/s1053-8119(03)00112-5. [DOI] [PubMed] [Google Scholar]
- Hughes SW, Lorincz ML, Blethyn K, Kekesi KA, Juhasz G, Turmaine M, Parnavelas JG, Crunelli V. Thalamic gap junctions control local neuronal synchrony and influence macroscopic oscillation amplitude during EEG alpha rhythms. Front Psychol. 2011;2:193. doi: 10.3389/fpsyg.2011.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Everling S. Monkey in the middle: why non-human primates are needed to bridge the gap in resting-state investigations. Front Neuroanat. 2012;6:29. doi: 10.3389/fnana.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Gati JS, Everling S, Menon RS. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp. 2013 doi: 10.1002/hbm.22058. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen O, Colgin LL. Cross-frequency coupling between neuronal oscillations. Trends Cogn Sci. 2007;11:267–269. doi: 10.1016/j.tics.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Jones DT, Vemuri P, Murphy MC, Gunter JL, Senjem ML, Machulda MM, Przybelski SA, Gregg BE, Kantarci K, Knopman DS, Boeve BF, Petersen RC, Jack CR., Jr Non-stationarity in the “resting brain’s” modular architecture. PLoS One. 2012;7:e39731. doi: 10.1371/journal.pone.0039731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson P, Sonnby-Borgström M. The effects of pictures of emotional faces on tonic and phasic autonomic cardiac control in women and men. Biol Psychol. 2003;62:157–173. doi: 10.1016/s0301-0511(02)00114-x. [DOI] [PubMed] [Google Scholar]
- Kaneko K, Tsuda I. Chaotic itinerancy. Chaos. 2003;13:926. doi: 10.1063/1.1607783. [DOI] [PubMed] [Google Scholar]
- Keilholz SD, Magnuson ME, Pan WJ, Willis M, Thompson GJ. Dynamic properties of functional connectivity in the rodent. Brain Connect. 2013;3:31–40. doi: 10.1089/brain.2012.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CJ, Bickel S, Honey CJ, Groppe DM, Entz L, Craddock RC, Lado FA, Kelly C, Milham M, Mehta AD. Neurophysiological investigation of spontaneous correlated and anticorrelated fluctuations of the BOLD signal. J Neurosci. 2013;33:6333–6342. doi: 10.1523/JNEUROSCI.4837-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AM, Uddin LQ, Biswal BB, Castellanos FX, Milham MP. Competition between functional brain networks mediates behavioral variability. Neuroimage. 2008;39:527–537. doi: 10.1016/j.neuroimage.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Kenet T, Bibitchkov D, Tsodyks M, Grinvald A, Arieli A. Spontaneously emerging cortical representations of visual attributes. Nature. 2003;425:954–956. doi: 10.1038/nature02078. [DOI] [PubMed] [Google Scholar]
- Kinnison J, Padmala S, Choi JM, Pessoa L. Network analysis reveals increased integration during emotional and motivational processing. J Neurosci. 2012;32:8361–8372. doi: 10.1523/JNEUROSCI.0821-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiviniemi VJ, Haanpää H, Kantola JH, Jauhiainen J, Vainionpää V, Alahuhta S, Tervonen O. Midazolam sedation increases fluctuation and synchrony of the resting brain BOLD signal. Magn Reson Imaging. 2005;23:531–537. doi: 10.1016/j.mri.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Kiviniemi V, Vire T, Remes J, Elseoud AA, Starck T, Tervonen O, Nikkinen J. A sliding time-window ICA reveals spatial variability of the default mode network in time. Brain Connect. 2011;1:339–347. doi: 10.1089/brain.2011.0036. [DOI] [PubMed] [Google Scholar]
- Knyazev GG, Slobodskoj-Plusnin JY, Bocharov AV, Pylkova LV. The default mode network and EEG α oscillations: an independent component analysis. Brain Res. 2011;1402:67–79. doi: 10.1016/j.brainres.2011.05.052. [DOI] [PubMed] [Google Scholar]
- Krook-Magnuson E, Varga C, Lee SH, Soltesz I. New dimensions of interneuronal specialization unmasked by principal cell heterogeneity. Trends Neurosci. 2012;35:175–184. doi: 10.1016/j.tins.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Inati SJ, Evans JW, Luh WM, Bandettini PA. Differentiating BOLD and non-BOLD signals in fMRI time series using multi-echo EPI. Neuroimage. 2012;60:1759–1770. doi: 10.1016/j.neuroimage.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Eickhoff SB, Turner JA, Ray KL, McKay DR, Glahn DC, Beckmann CF, Smith SM, Fox PT. Behavioral interpretations of intrinsic connectivity networks. J Cogn Neurosci. 2011;23:4022–4037. doi: 10.1162/jocn_a_00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Långsjö JW, Alkire MT, Kaskinoro K, Hayama H, Maksimow A, Kaisti KK, Aalto S, Aantaa R, Jääskeläinen SK, Revonsuo A, Scheinin H. Returning from oblivion: imaging the neural core of consciousness. J Neurosci. 2012;32:4935–4943. doi: 10.1523/JNEUROSCI.4962-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansink CS, Goltstein PM, Lankelma JV, McNaughton BL, Pennartz CM. Hippocampus leads ventral striatum in replay of place-reward information. PLoS Biol. 2009;7:e1000173. doi: 10.1371/journal.pbio.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkman DJ, Hajnal JV, Herlihy AH, Coutts GA, Young IR, Ehnholm G. Use of multicoil arrays for separation of signal from multiple slices simultaneously excited. J Magn Reson Imaging. 2001;13:313–317. doi: 10.1002/1522-2586(200102)13:2<313::aid-jmri1045>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Laufs H. Endogenous brain oscillations and related networks detected by surface EEG-combined fMRI. Hum Brain Mapp. 2008;29:762–769. doi: 10.1002/hbm.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs H. Multimodal analysis of resting state cortical activity: what does EEG add to our knowledge of resting state BOLD networks? Neuroimage. 2010;52:1171–1172. doi: 10.1016/j.neuroimage.2010.05.034. [DOI] [PubMed] [Google Scholar]
- Laufs H, Kleinschmidt A, Beyerle A, Eger E, Salek-Haddadi A, Preibisch C, Krakow K. EEG-correlated fMRI of human alpha activity. Neuroimage. 2003;19:1463–1476. doi: 10.1016/s1053-8119(03)00286-6. [DOI] [PubMed] [Google Scholar]
- Lee HL, Zahneisen B, Hugger T, Levan P, Hennig J. Tracking dynamic resting-state networks at higher frequencies using MR–encephalography. Neuroimage. 2013;65:216–222. doi: 10.1016/j.neuroimage.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Leonardi N, Richiardi J, Van De Ville D. Functional connectivity eigennetworks reveal different brain dynamics in multiple sclerosis patients. IEEE 10th International Symposium on Biomedical Imaging: From Nano to Macro; San Francisco, CA, USA. 2013. [Google Scholar]
- Leopold DA, Maier A. Ongoing physiological processes in the cerebral cortex. Neuroimage. 2012;62:2190–2200. doi: 10.1016/j.neuroimage.2011.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold DA, Murayama Y, Logothetis NK. Very slow activity fluctuations in monkey visual cortex: implications for functional brain imaging. Cereb Cortex. 2003;13:422–433. doi: 10.1093/cercor/13.4.422. [DOI] [PubMed] [Google Scholar]
- Lesting J, Narayanan RT, Kluge C, Sangha S, Seidenbecher T, Pape HC. Patterns of coupled theta activity in amygdala–hippocampal–prefrontal cortical circuits during fear extinction. PLoS One. 2011;6:e21714. doi: 10.1371/journal.pone.0021714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Baldassarre A, Committeri G, Romani GL, Corbetta M. Learning sculpts the spontaneous activity of the resting human brain. Proc Natl Acad Sci U S A. 2009;106:17558–17563. doi: 10.1073/pnas.0902455106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N. Uncovering intrinsic connectional architecture of functional networks in awake rat brain. J Neurosci. 2011;31:3776–3783. doi: 10.1523/JNEUROSCI.4557-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebe S, Hoerzer GM, Logothetis NK, Rainer G. Theta coupling between V4 and prefrontal cortex predicts visual short-term memory performance. Nat Neurosci. 2012;15:456–462. doi: 10.1038/nn.3038. [DOI] [PubMed] [Google Scholar]
- Lin FH, Wald LL, Ahlfors SP, Hämäläinen MS, Kwong KK, Belliveau JW. Dynamic magnetic resonance inverse imaging of human brain function. Magn Reson Med. 2006;56:787–802. doi: 10.1002/mrm.20997. [DOI] [PubMed] [Google Scholar]
- Lisman JE, Jensen O. The theta–gamma neural code. Neuron. 2013;77:1002–1016. doi: 10.1016/j.neuron.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Duyn JH. Time-varying functional network information extracted from brief instances of spontaneous brain activity. Proc Natl Acad Sci U S A. 2013;110:4392–4397. doi: 10.1073/pnas.1216856110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Stufflebeam SM, Sepulcre J, Hedden T, Buckner RL. Evidence that functional asymmetry of the human brain is controlled by multiple factors. Proc Natl Acad Sci. 2009;111:746–754. doi: 10.1073/pnas.0908073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Fukunaga M, de Zwart JA, Duyn JH. Large-scale spontaneous fluctuations and correlations in brain electrical activity observed with magnetoencephalography. Neuroimage. 2010;51:102–111. doi: 10.1016/j.neuroimage.2010.01.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhu X-H, Zhang Y, Chen W. Neural origin of spontaneous hemodynamic fluctuations in rats under burst-suppression anesthesia condition. Cereb Cortex. 2011;21:374–384. doi: 10.1093/cercor/bhq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinás RR. The intrinsic electrophysiological properties of mammalian neurons: insights into central nervous system function. Science. 1988;242:1654–1664. doi: 10.1126/science.3059497. [DOI] [PubMed] [Google Scholar]
- Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412:150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- Lu H, Zuo Y, Gu H, Waltz JA, Zhan W, Scholl CA, Rea W, Yang Y, Stein EA. Synchronized delta oscillations correlate with the resting-state functional MRI signal. Proc Natl Acad Sci U S A. 2007;104:18265–18269. doi: 10.1073/pnas.0705791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnuson M, Majeed W, Keilholz SD. Functional connectivity in BOLD and CBV weighted resting state fMRI in the rat brain. J Magn Reson Imaging. 2010;32:584–592. doi: 10.1002/jmri.22295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magri C, Schridde U, Murayama Y, Panzeri S, Logothetis NK. The amplitude and timing of the BOLD signal reflects the relationship between local field potential power at different frequencies. J Neurosci. 2012;32:1395–1407. doi: 10.1523/JNEUROSCI.3985-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed W, Magnuson M, Keilholz SD. Spatiotemporal dynamics of low frequency fluctuations in BOLD fMRI of the rat. J Magn Reson Imaging. 2009;30:384–393. doi: 10.1002/jmri.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeed W, Magnuson M, Hasenkamp W, Schwarb H, Schumacher EH, Barsalou L, Keilholz SD. Spatiotemporal dynamics of low frequency BOLD fluctuations in rats and humans. Neuroimage. 2011;54:1140–1150. doi: 10.1016/j.neuroimage.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantini D, Perrucci MG, Del Gratta C, Romani GL, Corbetta M. Electrophysiological signatures of resting state networks in the human brain. Proc Natl Acad Sci U S A. 2007;104:13170–13175. doi: 10.1073/pnas.0700668104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantini D, Gerits A, Nelissen K, Durand JB, Joly O, Simone L, Sawamura H, Wardak C, Orban GA, Buckner RL, Vanduffel W. Default mode of brain function in monkeys. J Neurosci. 2011;31:12954–12962. doi: 10.1523/JNEUROSCI.2318-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies DS, Böttger J, Watanabe A, Gorgolewski KJ. Visualizing the human connectome. Neuroimage. 2013;80:445–461. doi: 10.1016/j.neuroimage.2013.04.111. [DOI] [PubMed] [Google Scholar]
- Martuzzi R, Ramani R, Qiu M, Rajeevan N, Constable RT. Functional connectivity and alterations in baseline brain state in humans. Neuroimage. 2010;49:823–834. doi: 10.1016/j.neuroimage.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx M, Pauly KB, Chang C. A novel approach for global noise reduction in resting-state fMRI: APPLECOR. Neuroimage. 2013;64:19–31. doi: 10.1016/j.neuroimage.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAvoy M, Larson-Prior L, Ludwikow M, Zhang D, Snyder AZ, Gusnard DL, Raichle ME, d’Avossa G. Dissociated mean and functional connectivity BOLD signals in visual cortex during eyes closed and fixation. J Neurophysiol. 2012;108:2363–2372. doi: 10.1152/jn.00900.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ. Analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl T, Teipel S, Elmouden R, Mueller S, Koch W, Dietrich O, Coates U, Reiser M, Glaser C. Test–retest reproducibility of the default-mode network in healthy individuals. Hum Brain Mapp. 2010;31:237–246. doi: 10.1002/hbm.20860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V. Large-scale brain networks and psychopathology: a unifying triple network model. Trends Cogn Sci. 2011;15:483–506. doi: 10.1016/j.tics.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Merritt M, Thompson G, Pan WJ, Magnuson ME, Keilholz S. The electrical basis of dynamic functional connectivity measured with sliding window correlation. Proc ISMRM. 2013:#2243. [Google Scholar]
- Meunier D, Lambiotte R, Fornito A, Ersche KD, Bullmore ET. Hierarchical modularity in human brain functional networks. Front Neuroinform. 2009;3:37. doi: 10.3389/neuro.11.037.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Bullmore ET. Modular and hierarchically modular organization of brain networks. Front Neurosci. 2010;4:200. doi: 10.3389/fnins.2010.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R, Calhoun VD. Proc. HBM; Seattle, WA: 2013. Frequency space analysis reveals marked differences in whole brain resting state spatiotemporal activation patterns between schizophrenia patients and healthy controls. [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Uğurbil K. Multiband multislice GE–EPI at 7 Tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso F, Brinkmeyer J, Mobascher A, Warbrick T, Winterer G. Spontaneous brain activity and EEG microstates. A novel EEG/fMRI analysis approach to explore resting-state networks. Neuroimage. 2010;52:1149–1161. doi: 10.1016/j.neuroimage.2010.01.093. [DOI] [PubMed] [Google Scholar]
- Niazy RK, Xie J, Miller K, Beckmann CF, Smith SM. Spectral characteristics of resting state networks. Prog Brain Res. 2011;193:259–276. doi: 10.1016/B978-0-444-53839-0.00017-X. [DOI] [PubMed] [Google Scholar]
- Nir Y, Fisch L, Mukamel R, Gelbard-Sagiv H, Arieli A, Fried I, Malach R. Coupling between neuronal firing rate, gamma LFP, and BOLD fMRI is related to interneuronal correlations. Curr Biol. 2007;17:1275–1285. doi: 10.1016/j.cub.2007.06.066. [DOI] [PubMed] [Google Scholar]
- Nir Y, Mukamel R, Dinstein I, Privman E, Harel M, Fisch L, Gelbard-Sagiv H, Kipervasser S, Andelman F, Neufeld MY, Kramer U, Arieli A, Fried I, Malach R. Interhemispheric correlations of slow spontaneous neuronal fluctuations revealed in human sensory cortex. Nat Neurosci. 2008;11:1100–1108. doi: 10.1038/nn.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva S, Monto S, Palva JM. Graph properties of synchronized cortical networks during visual working memory maintenance. Neuroimage. 2010;49:3257–3268. doi: 10.1016/j.neuroimage.2009.11.031. [DOI] [PubMed] [Google Scholar]
- Pan W, Thompson G, Magnuson M, Majeed W, Jaeger D, Keilholz S. Simultaneous fMRI and electrophysiology in the rodent brain. J Vis Exp. 2010;42:1901. doi: 10.3791/1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Thompson G, Magnuson M, Majeed W, Jaeger D, Keilholz S. Broadband LFPs correlate with spontaneous fluctuations in fMRI signals in the rat somatosensory cortex under isoflurane anesthesia. Brain Connect. 2011;1:119–131. doi: 10.1089/brain.2011.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan WJ, Thompson GJ, Magnuson ME, Jaeger D, Keilholz S. Infraslow LFP correlates to resting-state fMRI BOLD signals. Neuroimage. 2013;26:288–297. doi: 10.1016/j.neuroimage.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier SJ, Kerssens C, Hamann SB, Sebel PS, Byas-Smith M, Hu X. Functional connectivity changes with concentration of sevoflurane anesthesia. NeuroReport. 2005;16:285–288. doi: 10.1097/00001756-200502280-00017. [DOI] [PubMed] [Google Scholar]
- Peng T, Niazy R, Payne SJ, Wise RG. The effects of respiratory CO(2) fluctuations in the resting-state BOLD signal differ between eyes open and eyes closed. Magn Reson Imaging. 2013;31:336–345. doi: 10.1016/j.mri.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Pesaran B, Nelson MJ, Andersen RA. Free choice activates a decision circuit between frontal and parietal cortex. Nature. 2008;453:406–409. doi: 10.1038/nature06849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JM, Vinck M, Everling S, Womelsdorf T. A long-range fronto-parietal 5- to 10-Hz network predicts “top-down” controlled guidance in a task-switch paradigm. Cereb Cortex. 2013 doi: 10.1093/cercor/bht050. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzella V, Della Penna S, Del Gratta C, Romani GL. SQUID systems for biomagnetic imaging. Supercond Sci Technol. 2001;14:R79–R114. [Google Scholar]
- Popa D, Duvarci S, Popescu AT, Lena C, Pare D. Coherent amygdalocortical theta promotes fear memory consolidation during paradoxical sleep. Proc Natl Acad Sci U S A. 2010;107:6516–6519. doi: 10.1073/pnas.0913016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM, Schlaggar BL, Petersen SE. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich MI, Varona P. Robust transient dynamics and brain functions. Front Comput Neurosci. 2011;5:24. doi: 10.3389/fncom.2011.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich MI, Friston KJ, Varona P, editors. Principles of Brain Dynamics: Global State Interactions. 1. The MIT Press; 2012. [Google Scholar]
- Rack-Gomer AL, Liu TT. Caffeine increases the temporal variability of resting-state BOLD connectivity in the motor cortex. Neuroimage. 2012;59:2994–3002. doi: 10.1016/j.neuroimage.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME. Two views of brain function. Trends Cogn Sci. 2010;14:180–190. doi: 10.1016/j.tics.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Rashid B, Damaraju E, Calhoun VD. Proc. HBM; Seattle, WA: 2013. Comparison of resting state dynamics in healthy, schizophrenia and bipolar disease. [Google Scholar]
- Raz G, Winetraub Y, Jacob Y, Kinreich S, Maron-Katz A, Shaham G, Podlipsky I, Gilam G, Soreq E, Hendler T. Portraying emotions at their unfolding: a multilayered approach for probing dynamics of neural networks. Neuroimage. 2012;60:1448–1461. doi: 10.1016/j.neuroimage.2011.12.084. [DOI] [PubMed] [Google Scholar]
- Ringach DL. Spontaneous and driven cortical activity: implications for computation. Curr Opin Neurobiol. 2009;19:439–444. doi: 10.1016/j.conb.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LF, de la Peña VH, Kushnir Y. Detecting shifts in correlation and variability with application to ENSO-monsoon rainfall relationships. Theor Appl Climatol. 2008;94:215–224. [Google Scholar]
- Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. Neuroimage. 2011;56:2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- Sadaghiani S, Hesselmann G, Friston KJ, Kleinschmidt A. The relation of ongoing brain activity, evoked neural responses, and cognition. Front Syst Neurosci. 2010;4:20. doi: 10.3389/fnsys.2010.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoglu U, Pearlson GD, Kiehl KA, Wang Y, Michael A, Calhoun VD. A method for evaluating dynamic functional network connectivity and task-modulation: application to schizophrenia. MAGMA. 2010;23(5-6):351–366. doi: 10.1007/s10334-010-0197-8. PMC pending #180300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar RF, Dotson NM, Bressler SL, Gray CM. Content-specific fronto-parietal synchronization during visual working memory. Science. 2012;338:1097–1100. doi: 10.1126/science.1224000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador R, Suckling J, Coleman MR, Pickard JD, Menon D, Bullmore E. Neurophysiological architecture of functional magnetic resonance images of human brain. Cereb Cortex. 2005 Sep;15(9):1332–1342. doi: 10.1093/cercor/bhi016. [DOI] [PubMed] [Google Scholar]
- Satterthwaite TD, Wolf DH, Loughead J, Ruparel K, Elliott MA, Hakonarson H, Gur RC, Gur RE. Impact of in-scanner head motion on multiple measures of functional connectivity: relevance for studies of neurodevelopment in youth. Neuroimage. 2012;60:623–632. doi: 10.1016/j.neuroimage.2011.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeringa R, Petersson KM, Kleinschmidt A, Jensen O, Bastiaansen MC. EEG alpha power modulation of FMRI resting-state connectivity. Brain Connect. 2012;2:254–264. doi: 10.1089/brain.2012.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölvinck ML, Maier A, Ye FQ, Duyn JH, Leopold DA. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci U S A. 2010;107:10238–10243. doi: 10.1073/pnas.0913110107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölvinck ML, Leopold DA, Brookes MJ, Khader PH. The contribution of electrophysiology to functional connectivity mapping. Neuroimage. 2013;80:297–306. doi: 10.1016/j.neuroimage.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrouff J, Perlbarg V, Boly M, Marrelec G, Boveroux P, Vanhaudenhuyse A, Bruno MA, Laureys S, Phillips C, Pélégrini-Issac M, Maquet P, Benali H. Brain functional integration decreases during propofol-induced loss of consciousness. Neuroimage. 2011;57:198–205. doi: 10.1016/j.neuroimage.2011.04.020. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD. Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad Z, Kelly AM, Reiss PT, Gee DG, Gotimer K, Uddin LQ, Lee SH, Margulies DS, Roy AK, Biswal BB, Petkova E, Castellanos FX, Milham MP. The resting brain: unconstrained yet reliable. Cereb Cortex. 2009;19:2209–2229. doi: 10.1093/cercor/bhn256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Hutchison RM, Bezgin G, Gati JS, Menon RS, Everling S, McIntosh AR. Resting-state functional connectivity dynamics are influenced by network structure; Toronto, ON. Canadian Association for Neuroscience Annual Meeting.2013. [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex. 2012;22:158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmuel A, Leopold DA. Neuronal correlates of spontaneous fluctuations in fMRI signals in monkey visual cortex: implications for functional connectivity at rest. Hum Brain Mapp. 2008;29:751–761. doi: 10.1002/hbm.20580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmueli K, van Gelderen P, de Zwart JA, Horovitz SG, Fukunaga M, Jansma JM, Duyn JH. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state fMRI BOLD signal. Neuroimage. 2007;38:306–320. doi: 10.1016/j.neuroimage.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel M, Donner TH, Engel AK. Spectral fingerprints of large-scale neuronal interactions. Nat Rev Neurosci. 2012;13:121–134. doi: 10.1038/nrn3137. [DOI] [PubMed] [Google Scholar]
- Sirota A, Montgomery S, Fujisawa S, Isomura Y, Zugaro M, Buzsaki G. Entrainment of neocortical neurons and gamma oscillations by the hippocampal theta rhythm. Neuron. 2008;60:683–697. doi: 10.1016/j.neuron.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Ramsey JD, Woolrich MW. Network modelling methods for fMRI. Neuroimage. 2011;54:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Van Essen DC, Feinberg DA, Yacoub ES, Ugurbil K. Temporally-independent functional modes of spontaneous brain activity. Proc Natl Acad Sci U S A. 2012;109:3131–3136. doi: 10.1073/pnas.1121329109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Zhou Y, Li J, Liu Y, Tian L, Yu C, Jiang T. Brain spontaneous functional connectivity and intelligence. Neuroimage. 2008;41:1168–1176. doi: 10.1016/j.neuroimage.2008.02.036. [DOI] [PubMed] [Google Scholar]
- Sorg C, Riedl V, Mühlau M, Calhoun VD, Eichele T, Läer L, Drzezga A, Förstl H, Kurz A, Zimmer C, Wohlschläger AM. Selective changes of resting-state networks in individuals at risk for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2007;104:18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. The non-random brain: efficiency, economy, and complex dynamics. Front Comput Neurosci. 2011;5:5. doi: 10.3389/fncom.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun FT, Miller LM, Rao AA, D’Esposito M. Functional connectivity of cortical networks involved in bimanual motor sequence learning. Cereb Cortex. 2007;17:1227–1234. doi: 10.1093/cercor/bhl033. [DOI] [PubMed] [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Comput Biol. 2008;4:e1000100. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, von Wegner F, Morzelewski A, Borisov S, Jahnke K, Laufs H. Automatic sleep staging using fMRI functional connectivity data. Neuroimage. 2012a;63:63–72. doi: 10.1016/j.neuroimage.2012.06.036. [DOI] [PubMed] [Google Scholar]
- Tagliazucchi E, von Wegner F, Morzelewski A, Brodbeck V, Laufs H. Dynamic BOLD functional connectivity in humans and its electrophysiological correlates. Front Hum Neurosci. 2012b;6:339. doi: 10.3389/fnhum.2012.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, Balenzuela P, Fraiman D, Chialvo DR. Criticality in large-scale brain fMRI dynamics unveiled by a novel point process analysis. Front Physiol. 2012c;3:15. doi: 10.3389/fphys.2012.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambini A, Ketz N, Davachi L. Enhanced brain correlations during rest are related to memory for recent experiences. Neuron. 2010;65:280–290. doi: 10.1016/j.neuron.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YY, Rothbart MK, Posner MI. Neural correlates of establishing, maintaining, and switching brain states. Trends Cogn Sci. 2012;16:330–337. doi: 10.1016/j.tics.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Task Force. Heart rate variability: standards of measurement, physiological interpretation and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation. 1996;93:1043–1065. [PubMed] [Google Scholar]
- Thompson G, Magnuson M, Merritt M, Schwarb H, Pan W, McKinley A, Tripp L, Schumacher E, Keilholz S. Short time windows of correlation between large scale functional brain networks predict vigilance intra-individually and inter-individually. Hum Brain Mapp. 2013 doi: 10.1002/hbm.22140. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeev I, Bazhenov M, Seigneur J, Sejnowski T. Neuronal synchronization and thalamocortical rhythms in sleep, wake and epilepsy. In: Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Jasper’s Basic Mechanisms of the Epilepsies. Bethesda, MD: 2012. [PubMed] [Google Scholar]
- Torrence C, Compo G. A practical guide to wavelet analysis. Bull Am Meteorol Soc. 1998;79:61–78. [Google Scholar]
- Tsodyks M, Kenet T, Grinvald A, Arieli A. Linking spontaneous activity of single cortical neurons and the underlying functional architecture. Science. 1999;286:1943–1946. doi: 10.1126/science.286.5446.1943. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Stam CJ, Kahn RS, Hulshoff Pol HE. Efficiency of functional brain networks and intellectual performance. J Neurosci. 2009;29:7619–7624. doi: 10.1523/JNEUROSCI.1443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KR, Hedden T, Venkataraman A, Evans KC, Lazar SW, Buckner RL. Intrinsic functional connectivity as a tool for human connectomics: theory, properties, and optimization. J Neurophysiol. 2010;103:297–321. doi: 10.1152/jn.00783.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk KRA, Sabuncu MR, Buckner RL. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage. 2012;59:431–438. doi: 10.1016/j.neuroimage.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oort E, Norris D, Smith SM, Beckmann C. Resting State Networks are Characterized by High Frequency BOLD Fluctuations. OHBM; Beijing: 2012. [Google Scholar]
- Vanhatalo S, Palva JM, Holmes MD, Miller JW, Voipio J, Kaila K. Infraslow oscillations modulate excitability and interictal epileptic activity in the human cortex during sleep. Proc Natl Acad Sci U S A. 2004;101:5053–5057. doi: 10.1073/pnas.0305375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela F, Lachaux JP, Rodriguez E, Martinerie J. The brainweb: phase synchronization and large-scale integration. Nat Rev Neurosci. 2001;2:229–239. doi: 10.1038/35067550. [DOI] [PubMed] [Google Scholar]
- Varoquaux G, Craddock RC. Learning and comparing functional connectomes across subjects. Neuroimage. 2013;80:405–415. doi: 10.1016/j.neuroimage.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Vincent JL, Patel GH, Fox MD, Snyder AZ, Baker JT, Van Essen DC, Zempel JM, Snyder LH, Corbetta M, Raichle ME. Intrinsic functional architecture in the anaesthetized monkey brain. Nature. 2007;447:83–86. doi: 10.1038/nature05758. [DOI] [PubMed] [Google Scholar]
- Vogels TP, Rajan K, Abbott LF. Neural network dynamics. Annu Rev Neurosci. 2005;28:357–376. doi: 10.1146/annurev.neuro.28.061604.135637. [DOI] [PubMed] [Google Scholar]
- von der Malsburg C, Phillips WA, Singer W, editors. Dynamic Coordination in the Brain: From Neurons to Mind. The MIT Press; 2010. [Google Scholar]
- von Stein A, Chiang C, König P. Top-down processing mediated by interareal synchronization. Proc Natl Acad Sci U S A. 2000;97:14748–14753. doi: 10.1073/pnas.97.26.14748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallentin M, Nielsen AH, Vuust P, Dohn A, Roepstorff A, Lund TE. Amygdala and heart rate variability responses from listening to emotionally intense parts of a story. Neuroimage. 2011;58:963–973. doi: 10.1016/j.neuroimage.2011.06.077. [DOI] [PubMed] [Google Scholar]
- Wei L, Duan X, Yang Y, Liao W, Gao Q, Ding JR, Zhang Z, Zeng W, Li Y, Lu G, Chen H. The synchronization of spontaneous BOLD activity predicts extra-version and neuroticism. Brain Res. 2011;1419:68–75. doi: 10.1016/j.brainres.2011.08.060. [DOI] [PubMed] [Google Scholar]
- Wise RG, Ide K, Poulin MJ, Tracey I. Resting fluctuations in arterial carbon dioxide induce significant low frequency variations in BOLD signal. Neuroimage. 2004;21:1652–1664. doi: 10.1016/j.neuroimage.2003.11.025. [DOI] [PubMed] [Google Scholar]
- Womelsdorf T, Schoffelen JM, Oostenveld R, Singer W, Desimone R, Engel AK, Fries P. Modulation of neuronal interactions through neuronal synchronization. Science. 2007;316:1609–1612. doi: 10.1126/science.1139597. [DOI] [PubMed] [Google Scholar]
- Wu L, Eichele T, Calhoun VD. Reactivity of hemodynamic responses and functional connectivity to different states of alpha synchrony: a concurrent EEG–fMRI study. Neuroimage. 2010;52:1252–1260. doi: 10.1016/j.neuroimage.2010.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan CG, Cheung B, Kelly C, Colcombe S, Craddock RC, Di Martino A, Li Q, Zuo XN, Castellanos FX, Milham MP. A comprehensive assessment of regional variation in the impact of head micromovements on functional connectomics. Neuroimage. 2013;76:183–201. doi: 10.1016/j.neuroimage.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zöllei L, Polimeni JR, Fischl B, Liu H, Buckner RL. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


