Abstract
CYP2A13, a human P450 enzyme preferentially expressed in the respiratory tract, is highly efficient in the metabolic activation of tobacco-specific nitrosamines. The aim of this study was to test the hypothesis that inflammation suppresses CYP2A13 expression in the lung, thus explaining the large interindividual differences in CYP2A13 levels previously found in human lung biopsy samples. We first demonstrated that the bacterial endotoxin lipopolysaccharide (LPS) and the proinflammatory cytokine IL-6 can suppress CYP2A13 messenger RNA (mRNA) expression in the NCI-H441 human lung cell line. We then report that an ip injection of LPS (1mg/kg), which induces systemic and lung inflammation, caused substantial reductions in CYP2A13 mRNA (~50%) and protein levels (~80%) in the lungs of a newly generated CYP2A13-humanized mouse model. We further identified two critical CYP2A13 promoter regions, one (major) between −484 and −1008bp and the other (minor) between −134 and −216bp, for the response to LPS, through reporter gene assays in H441 cells. The potential involvement of the nuclear factor NF-κB in LPS-induced CYP2A13 downregulation was suggested by identification of putative NF-κB binding sites within the LPS response regions and effects of an NF-κB inhibitor (pyrrolidine dithiocarbamate) on CYP2A13 expression in H441 cells. Results from gel shift assays further confirmed binding of NF-κB-like nuclear proteins of H441 cells to the major LPS response region of the CYP2A13 promoter. Thus, our findings strongly support the hypothesis that CYP2A13 levels in human lung can be suppressed by inflammation associated with disease status in tissue donors, causing underestimation of CYP2A13 levels in healthy lung.
Key Words: CYP2A, LPS, lung, inflammation, chemical carcinogenesis.
CYP2A13, a cytochrome P450 (P450) enzyme expressed preferentially in the respiratory tract (Koskela et al., 1999; Su et al., 2000; Zhu et al., 2006), is highly effective in the metabolic activation of many toxicants and carcinogens, including aflatoxin B1 (He et al., 2006), 4-aminobiphenyl (Nakajima et al., 2006), 3-methylindole (D’Agostino et al., 2009), and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a tobacco-specific lung procarcinogen (Su et al., 2000). Multiple lines of evidence suggest that CYP2A13 plays an important role in NNK-induced lung tumorigenesis, including its high efficiency toward NNK bioactivation (He et al., 2004; Jalas et al., 2005; Su et al., 2000), an epidemiological link between a reduced-activity CYP2A13 allele and decreased incidence of lung adenocarcinoma in light smokers (Wang et al., 2003), and a strong ability of transgenic CYP2A13 to mediate NNK-induced lung tumorigenesis in a CYP2A13-humanized mouse model (Megaraj et al., 2013).
CYP2A13 expression levels in human lung biopsy samples were found to vary drastically (Zhang et al., 2007). Differences in CYP2A13 expression levels in the lung may contribute to differing susceptibilities to tobacco smoke–induced lung cancer among individuals. Therefore, it is important to identify factors that dictate the large interindividual variations in CYP2A13 expression. In that regard, we have previously characterized the structure and function of the CYP2A13 gene promoter (Ling et al., 2007) and demonstrated the role of CCAAT/enhancer binding proteins (C/EBP) in the transcriptional regulation of CYP2A13. We have also identified two CYP2A13 variants (*2 and 7520C>G) that are associated with decreased allelic expression in human lung (D’Agostino et al., 2008; Wu et al., 2009; Zhang et al., 2004). Additionally, we obtained evidence supporting the involvement of epigenetic factors in CYP2A13 regulation and, based on the fact that C/EBP proteins can be regulated by inflammation, proposed that CYP2A13 expression might be modulated by lung inflammation (Ling et al., 2007).
The impact of infection and inflammation on hepatic P450 expression has been extensively studied (Aitken et al., 2006). However, only scant information is available on the modulation of pulmonary P450 expression by inflammation, although airway inflammation and cancer-related lung inflammation are common in people. In one study, the expression of several lung P450s, including CYP2E1, 2F2, and 2J6, was found to be suppressed in an allergic inflammation mouse model (Stoilov et al., 2006). However, there has been no previous report on the impact of inflammation on CYP2A13 expression.
In the present study, we sought to determine the effects of inflammation on CYP2A13 expression in vitro with use of a human lung cancer cell line (NCI-H441 or H441) known to express CYP2A13 under permissive conditions (Ling et al., 2007) and in vivo with use of a newly generated CYP2A13-humanized mouse model. We first determined whether the bacterial endotoxin lipopolysaccharide (LPS) and the proinflammatory cytokine IL-6 can suppress CYP2A13 expression in cultured H441 cells. We then determined whether an ip injection with LPS, which can induce systemic and lung inflammation, suppressed CYP2A13 expression in the lungs of the CYP2A13-humanized mice. We performed additional studies with H441 cells to identify critical CYP2A13 promoter regions for the response to LPS and to assess potential involvement of the nuclear factor NF-κB, which, along with C/EBP, had been previously reported to be involved in inflammation-induced suppression of hepatic P450 expression (Akira et al., 1993; Miyazawa et al., 1998). Our results, which revealed for the first time that CYP2A13 expression in the lung can be suppressed by inflammation, have important implications for assessing the risks of CYP2A13-mediated chemical toxicity in human lungs, as they suggest that CYP2A13 expression levels detected in lung biopsy samples may significantly underestimate the levels of CYP2A13 present in the lungs of a normal, healthy individual.
MATERIALS AND METHODS
Cell culture and treatments.
NCI-H441 cells (ATCC HTB-174) were incubated at 37°C with 5% CO2 in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum. Cells were seeded (2×105) into 12-well plates before treatment. To enable constitutive expression of CYP2A13, cells at 40–50% confluence were continuously exposed to 5-azacytidine (5-AzaC, a DNA demethylation agent) (2μM) (Sigma-Aldrich, St Louis, MO) for 72h. AzaC was added in dimethyl sulfoxide; the final concentration of dimethyl sulfoxide was 0.02% (wt/vol). Trichostatin A (TSA, a histone deacetylation inhibitor) (0.5μM) was added in ethanol at 48h after the addition of AzaC; the final concentration of ethanol was 0.05% (wt/vol). Cells were harvested 24h after TSA treatment. For determination of LPS or IL-6 effects, LPS (10 μg/ml) or IL-6 (100ng/ml) was added 48h after the addition of AzaC and 24h before cells were harvested.
Animals and treatments.
All procedures involving animals were approved by the Institutional Animal Care and Use Committee of the Wadsworth Center. LPS from Escherichia coli, serotype 0111:B4 (Sigma-Aldrich) was dissolved in 0.9% sterilized saline. CYP2A13-transgenic (Wei et al., 2012) and Cyp2a5-null (Zhou et al., 2010) mice, both on B6 background, obtained from breeding stocks maintained at Wadsworth Center were intercrossed to produce CYP2A13-humanized mice (CYP2A13+/−, CYP2A5−/−). Genotyping analysis for the Cyp2a5-null and CYP2A13-transgenic allele was performed as described (Wei et al., 2012; Zhou et al., 2010). Two-month-old male and female CYP2A13-humanized mice (25–30g) were either untreated or given a single ip injection of LPS at a dose of 0.5 or 1mg/kg and sacrificed at 12, 24, or 48h after dosing. Animal treatments were performed at fixed time, between 9:00 and 10:00 a.m. local time.
Blood collection, lung homogenate preparation, and ELISA for IL-6 detection.
Postsacrifice, blood was collected from the left ventricle for preparation of serum. Sera and lung lobes were stored at −80°C until use. The frozen lungs were thawed, weighed, transferred to new tubes on ice containing 2ml of T-PER buffer containing protease inhibitor cocktail (Sigma-Aldrich), and homogenized on ice with a Polytron (Fisher Scientific, Pittsburgh, PA). Lung homogenates were centrifuged at 9000 × g for 10min at 4°C, and the supernatant was used for IL-6 determination. IL-6 levels were measured using a mouse IL-6 Duoset ELISA Development System (R&D Systems, Minneapolis, MN), using recombinant IL-6 as a standard.
RNA isolation and real-time PCR.
RNA isolation from H441 cells and mouse tissues and real-time PCR analysis were performed as described previously (Zhang et al., 2004, 2007). The primers for CYP2A13 and mouse serum amyloid P component (SAP) were CYP2A13 E5F (5′-acctggtgatgaccaccc-3′) and CYP2A13 E6R (5′-cgtggatgactgcctctg-3′) (Zhang et al., 2004), and mSAP-F (5′-tactgctttggatgtttgtcttcac-3′) and mSAP-R (5′-tcagcttcacatgattttcag-3′) (Charles et al., 2006). Mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and human β-actin were used as internal standards and amplified with primers mGAPDH-F (5′-tgtgaacggatttggccgta-3′) and mGAPDH-R (5′-tcgctcctggaagatggtga-3′) (Wei et al., 2012), and β-actin-F (5′-cctgactgactacctcatg-3′) and β-actin-R (5′-tccttctgcatcctgtcggca-3′) (Zhang et al., 2004). All reactions were performed in duplicate. The reverse transcription product from a CYP2A13-humanized mouse nasal tissue was used to construct standard curves. The relative levels of SAP or CYP2A13 messenger RNAs (mRNAs) in various total RNA preparations were normalized by the level of GAPDH or β-actin mRNA within the same sample.
Immunoblot analysis.
Mouse lung microsomes were prepared as described previously (Ding and Coon, 1990). Immunoblot analysis was performed using 15% NuPage gels (Invitrogen), and blots were probed with a rabbit anti-CYP2A5 polyclonal antibody (Gu et al., 1998). Heterologously expressed CYP2A13 (Su et al., 2000), contained in Sf9 microsomes, were used as positive controls. Immunoreactive bands were visualized using a chemiluminescence reagent kit (Amersham Biosciences, Piscataway, NJ). The intensities of detected bands were measured by a GS-710 Calibrated Imaging Densitometer (Bio-Rad, Hercules, CA). Calnexin, a marker protein for the endoplasmic reticulum, was detected as a loading control, using a rabbit antihuman calnexin antibody (GenScript).
Transient transfection and reporter gene assays.
NCI-H441 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. Cells were seeded (2×105) into 12-well plates 24h before transfection. The cells were washed twice with serum-free Modified Eagle Medium (MEM) and incubated with Opti-MEM (Invitrogen), prior to the addition of a luciferase reporter construct (p2A13_134, p2A13_216, p2A13_484, and p2A13_1008) (Ling et al., 2007) (0.6 µg), and an internal control reporter gene plasmid (pRL-TK; 0.1 µg), premixed with serum-free MEM containing Fugene 6 reagent (1.8 μl; Roche Applied Sciences). After 24h, the cells were cultured in RPMI 1640 with 10% fetal bovine serum and treated with LPS (10 μg/ml), pyrrolidine dithiocarbamate (PDTC; 100μM), or LPS (10 μg/ml)/PDTC (100μM) (Duan et al., 2012). Luciferase activities were determined 24h after LPS treatment using Luciferase Reporter Assay System (Promega, Madison, WI).
Nuclear protein isolation.
Nuclear extracts from H441 cells were prepared using a CelLytic NuCLEAR Extraction Kit (Sigma) according to the manufacturer’s protocol. Adherent cells from 70–90% confluent monolayer cultures were untreated or treated with LPS (10 μg/ml) and harvested 3h later. The nuclear protein preparations, in 20mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer, pH 7.9, containing 1.5mM MgCl2, 0.42M NaCl, 0.2mM EDTA, 25% glycerol, 1mM dithiothreitol (DTT), and a protease inhibitor cocktail [Sigma Catalog Number P8340, 1:100 dilution, containing 4-(2-aminoethyl)-benzenesulfonyl fluoride, pepstatin A, bestatin, leupeptin, aprotinin, and trans-epoxysuccinyl-l-leucyl-amido(4-guanidino)-butane] plus PMSF (0.1M, dissolved in isopropanol), were snap-frozen in aliquots in a dry ice/ethanol bath and stored at −80°C until use.
Electrophoretic mobility shift assays.
NF-κB consensus (5′-agttgaggggactttcccaggc-3′, sense), NF-κB mutant (5′-agttgaggcgactttcccaggc-3′, sense), C/EBP consensus (5′-tgcagattgcgcaatctgca-3′, sense), and C/EBP mutant (5′-tgcagagactagtctctgca-3′, sense) oligonucleotides were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The gel shift probe corresponding to −761 to −790bp of CYP2A13 promoter region was synthesized by Integrated DNA Technologies (Coralville, IA). Double-stranded oligonucleotide was end-labeled with γ-32P-ATP (Amersham Biosciences) using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). Gel shift assays were carried out by incubating nuclear extracts (5 μg protein) with the 32P-labeled DNA probe (~100,000 cpm) in a 25-μl binding reaction mixture containing 50mM Tris-HCl, 10mM MgCl2, 1mM DTT, 1mM EDTA, 12% glycerol, and 1.0 μg of poly(dI-dT) (Amersham Pharmacia Biotech). After incubation at room temperature for 30min, the samples were analyzed on 5% nondenaturing polyacrylamide gels that had been prerun at 150V for 1h in Tris-glycine buffer. After electrophoresis at 180V for 1h, the gels were dried and visualized by exposure to x-ray film (Kodak XAR) at −80°C for 24–36h. Each experiment was repeated three times.
Other methods and data analysis.
Protein concentrations were determined using a BCA protein assay kit (Pierce Biotechnology, Rockford, IL). Statistical significance was examined using Student’s t-test or one-way ANOVA followed by Dunnett’s or Tukey’s post hoc tests, as indicated in the figures.
RESULTS
Effects of LPS and IL-6 on CYP2A13 expression in H441 human lung cells
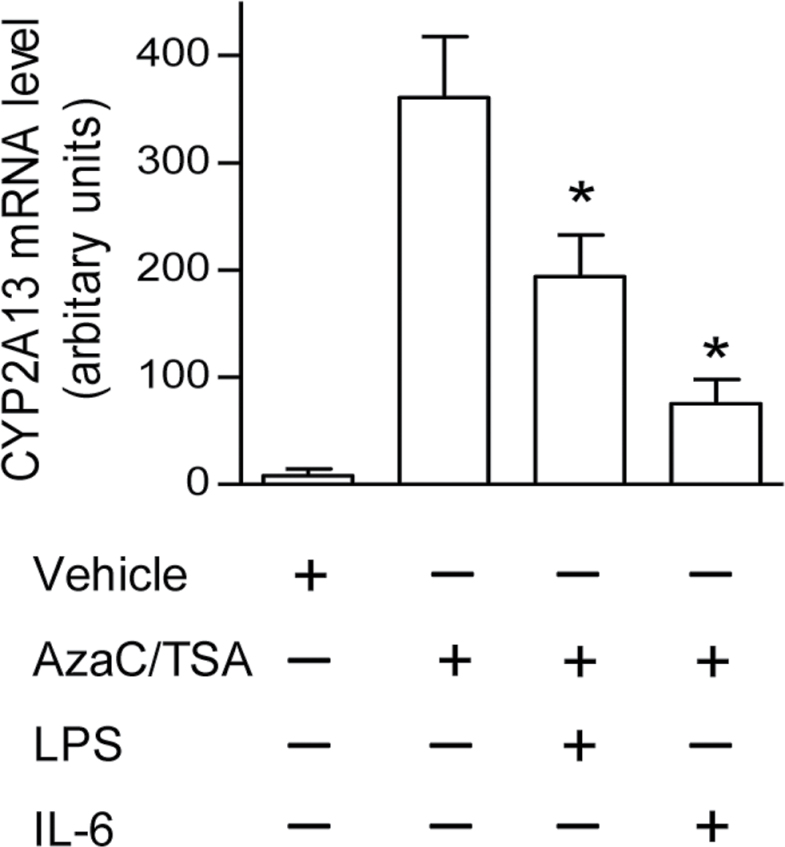
To test the hypothesis that inflammation suppresses CYP2A13 gene expression in human lung, we first examined effects of LPS on CYP2A13 expression in a human lung cell line (NCI-H441), under conditions (combined treatment with AzaC and TSA) that support CYP2A13 expression and transcriptional regulation (Ling et al., 2007). As shown in Figure 1, vehicle-treated H441 cells barely expressed CYP2A13. After combined treatment with AzaC and TSA, CYP2A13 expression was induced more than 10-fold. Further addition of the bacterial endotoxin, LPS, at 10 μg/ml, a dose effective for suppressing CYP1A1 expression in Hepa1c1c7 cells (Ke et al., 2001), caused CYP2A13 expression to decrease by ~50%, compared with cells treated with AzaC and TSA only, whereas addition of IL-6 at 100ng/ml, a concentration known to suppress CYP3A expression in hepatocytes (Sunman et al., 2004), suppressed CYP2A13 expression by ~75%. These data indicated that CYP2A13 expression can be suppressed by proinflammatory factors in the H441 human lung cell line model.
Fig. 1.
Effects of LPS and IL-6 on CYP2A13 expression in AzaC- and TSA-treated NCI-H441 cells. Where indicated, NCI-H441 cells were treated with dimethyl sulfoxide (0.02%) and ethanol (0.05%) as vehicle control, AzaC (2μM, 72h), and TSA (0.5μM, 24h) only (added to make the cells permissive of constitutive CYP2A13 expression), or AzaC/TSA plus LPS (10 μg/ml, 24h) or IL-6 (100ng/ml, 24h), as described under Materials and Methods section. RNA was isolated at 24h after LPS or IL-6 addition, for determination of CYP2A13 mRNA level using real-time PCR. Data represent means ± SD (n = 3) and were normalized by levels of β-actin. *p < 0.05, compared with cells treated with AzaC/TSA only; one-way ANOVA followed by Dunnett’s test.
Effects of LPS treatment on lung CYP2A13 expression in vivo in a CYP2A13-humanized mouse model
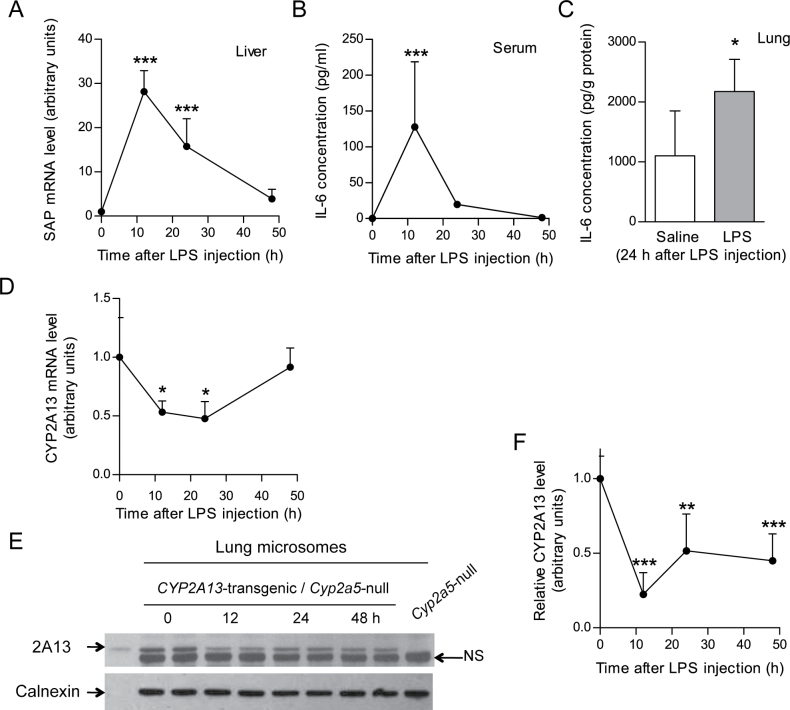
Establishment of a CYP2A13-transgenic mouse model with human CYP2A13 expression in the lung and nasal mucosa (Wei et al., 2012) made it possible to directly determine the in vivo effects of lung inflammation on CYP2A13 expression. CYP2A13 expression was suppressed by LPS injection, but the presence of the mouse CYP2A5 protein made it difficult to accurately determine the extent of the decreased expression of the CYP2A13 protein (data not shown). Therefore, a CYP2A13-humanized mouse model was produced, by intercrosses between CYP2A13-transgenic mouse and Cyp2a5-null mouse, allowing characterization of the effects of LPS treatment on human CYP2A13 expression.
The ability of a single dose of LPS (0.5–1mg/kg body weight, ip) to induce systemic and lung inflammation was confirmed by measuring serum and lung tissue levels of IL-6 as well as hepatic mRNA levels for SAP, an acute phase reactant induced by proinflammatory cytokines (Sunman et al., 2004). As shown in Figures 2A and 2B, hepatic SAP mRNA levels of LPS-treated CYP2A13-humanized mice were significantly elevated at 12 and 24h after treatment, whereas serum IL-6 levels were significantly increased at 12h after treatment. IL-6 levels in the lung were also higher in LPS-treated than in saline-treated groups, at 24h after LPS (Fig. 2C).
Fig. 2.
Effects of LPS treatment on lung CYP2A13 expression in vivo in a CYP2A13-humanized mouse model. (A, B, and C) LPS effect on hepatic SAP mRNA level (A) and serum (B) and lung IL-6 protein level (C) in CYP2A13-humanized mice. Mice (2-month-old female) were injected ip with 1 (A and B) or 0.5 (C) mg/kg LPS, and serum and tissues were obtained at various time points as indicated for analysis. SAP mRNA levels were normalized by levels of GAPDH. IL-6 was detected by ELISA. Data represent means ± SD (n = 4–6). *p < 0.05 and ***p < 0.001, respectively, compared with 0h; one-way ANOVA followed by Dunnett's test (A and B); or with saline group (C); Student’s t-test. (D, E, and F) LPS effect on lung CYP2A13 mRNA (D) and protein (E and F) levels in CYP2A13-humanized mice. Mice (2-month-old female) were treated with 1mg/kg LPS. Lungs were obtained at various time points for analysis. Relative CYP2A13 mRNA levels (D) (normalized by levels of GAPDH) represent means ± SD (n = 4–6); *p < 0.05, compared with 0h group; one-way ANOVA followed by Dunnett's test. For immunoblot analysis, lung microsomal proteins (10 μg/lane) were prepared from tissues pooled from four mice per group and were analyzed in duplicate using a rabbit anti-CYP2A5 polyclonal antibody. In the representative blots shown (E), recombinant CYP2A13 protein was used as a positive control and lung microsome from a Cyp2a5-knockout mouse was used as a negative control, for identification of the CYP2A13 band. The position of a nonspecific band (NS) is indicated. Calnexin was detected as a loading control. For relative CYP2A13 protein levels (F), data represent means ± SD for results from three separate analyses, normalized by the amounts of calnexin detected in the same samples. **p < 0.01 and ***p < 0.001, respectively, compared with 0h; one-way ANOVA followed by Dunnett's test.
The effects of LPS-induced inflammation on CYP2A13 gene expression were examined at both mRNA and protein levels. LPS treatment resulted in ~50% reductions in CYP2A13 mRNA levels in lungs of the humanized mice, at 12 and 24h (Fig. 2D). The expression level had recovered by 48h after LPS, when serum levels of IL-6 were no longer detectable. The LPS treatment also lowered the levels of lung microsomal CYP2A13 protein (Figs. 2E and 2F); the decrease was greatest at 12h (~80%), and the level was still suppressed at 48h after treatment.
Notably, for immunoblot detection of CYP2A13, a rabbit anti-mouse CYP2A5 polyclonal antibody was employed, which also recognized an unknown mouse protein in lung microsomes (Fig. 2E). The identification of the CYP2A13 band was aided by the use of heterologously expressed CYP2A13 (positive control) and lung microsomal proteins from a Cyp2a5-null mouse (negative control). The intensity of the nonspecific band did not vary among the various samples, and it thus served as an additional loading control. The LPS treatment did not affect calnexin levels.
Only female mice were used in the experiments described in Figure 2. In preliminary studies with male mice, LPS also mediated suppression of lung CYP2A13 protein expression (data not shown).
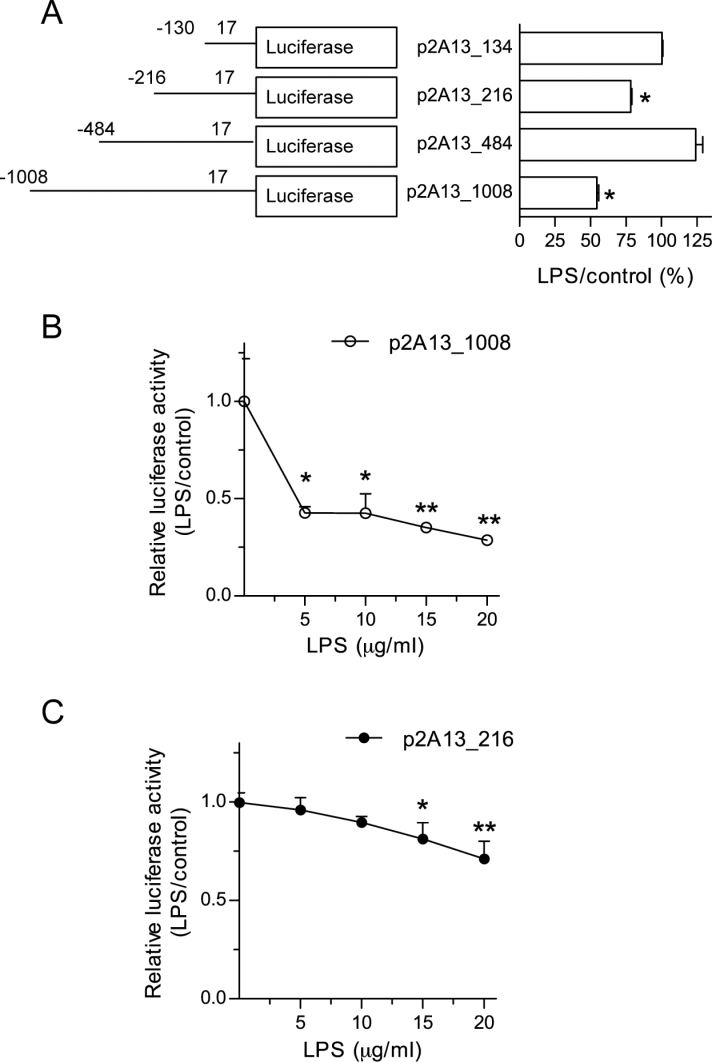
Identification of LPS response regions within CYP2A13 promoter
To provide mechanistic basis for LPS-mediated suppression of CYP2A13 expression, we searched for potential LPS response regions in the CYP2A13 proximal promoter region in human lung cells. H441 cells were transfected with four different CYP2A13 promoter activity reporter constructs (p2A13_1008, p2A13_484, p2A13_216, and p2A13_134, consisting of the indicated lengths in base pair of 2A13 promoter sequence as well as the first 17bp of coding sequence in the pGL3_Basic vector) (Ling et al., 2007). Luciferase reporter activity was then measured and compared between LPS-treated and untreated groups. As shown in Figure 3A, LPS treatment (10 μg/ml) decreased the activity of p2A13_1008 (by 46%) and, to a lesser extent, p2A13_216 (by 23%) (compared with untreated group), but not the activity of p2A13_484 or p2A13_134. These results suggest that the CYP2A13 proximal promoter region contain two separate LPS response elements, one (major) site located between −484 and −1008bp and the other (minor) site located between –134 and −216bp. Notably, the fact that deletion of −216 to −484 restores LPS’s effect on the promoter (Fig. 3A) suggested the presence of a repressor of LPS action in this region. The differential sensitivity of p2A13_1008 and p2A13_216 to LPS’s inhibitory effects was confirmed by a dose-response experiment, which showed 57% inhibition of p2A13_1008 (Fig. 3B), but no inhibition of p2A13_216 with 5 and 10 microgram/ml LPS (Fig. 3C), and 71 and 28% inhibition of p2A13_1008 and p2A13_216, respectively, with 20 μg/ml LPS.
Fig. 3.
Identification of LPS response regions within CYP2A13 promoter. (A) Effects of LPS on CYP2A13 promoter activity in NCI-H441 cells. NCI-H441 cells were transiently transfected with one of four CYP2A13 promoter constructs, at 24h prior to LPS treatment of the cells (at 10 μg/ml), and luciferase activities were determined 24h after the LPS treatment. Activities of firefly luciferase in cell lysates were normalized to those of cotransfected Renilla reniformis luciferase, and the results are shown as relative activities (LPS treated/untreated × 100) for each promoter construct. The amounts of CYP2A13 promoter sequence included in each construct are indicated. Data (means ± SD) represent three independent experiments. *p < 0.05, indicating significant decreases in promoter activity in LPS-treated relative to untreated cells (Student’s t-test). (B and C) Dose-response of the effects of LPS on CYP2A13 promoter activity. NCI-H441 cells were transfected with p2A13_1008 (B) or p2A13_216 (C) at 24h prior to LPS treatment at various doses, and luciferase activities were determined 24h after the LPS treatment. Data represent means ± SD (n = 3). *p < 0.05; **p < 0.01, one-way ANOVA followed by Dunnett’s multiple comparison test, compared with corresponding untreated cells.
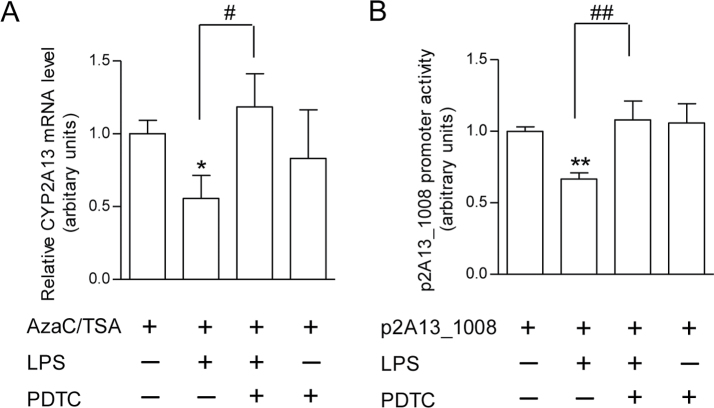
Potential role of NF-κB in LPS-induced suppression of CYP2A13 expression and CYP2A13 promoter activity
As NF-κB is known to be a major mediator of inflammation pathways, we further determined whether NF-κB plays a role in LPS-induced CYP2A13 suppression. As shown (Fig. 4), addition of PDTC, a chemical inhibitor of NF-κB, prevented LPS-mediated suppression of both the expression of endogenous CYP2A13 (Fig. 4A) and the promoter activity of the p2A13_1008 reporter gene in H441 cells (Fig. 4B). These results suggest that NF-κB is involved in inflammation-associated CYP2A13 repression.
Fig. 4.
Effects of PDTC, an NF-κB inhibitor, on LPS-induced suppression of CYP2A13 expression and CYP2A13 promoter activity. (A) Effects on CYP2A13 expression. NCI-H441 cells were treated with AzaC (72h)/TSA (24h) and LPS (10 μg/ml, 24h) as described in Figure 1, with or without cotreatment with PDTC (100μM, 24h). Cells were harvested 24h after LPS or LPS/PDTC treatment. Data represent means ± SD (n = 3) and were normalized by levels of β-actin. *p < 0.05; compared with cells treated with AzaC/TSA only; # p < 0.05; compared with cells treated with AzaC/TSA/LPS; one-way ANOVA, followed by Tukey’s test. (B) Effects on CYP2A13 promoter activity. NCI-H441 cells were transiently transfected with the p2A13_1008 vector at 24h prior to LPS (10 μg/ml), PDTC (100μM), or LPS plus PDTC treatment, and luciferase activities were determined 24h after the LPS or PDTC treatment. Data represent means ± SD (n = 3). **p < 0.01, compared with p2A13_1008 alone; ## p < 0.01, compared with p2A13_1008 plus LPS; one-way ANOVA, followed by Tukey’s test.
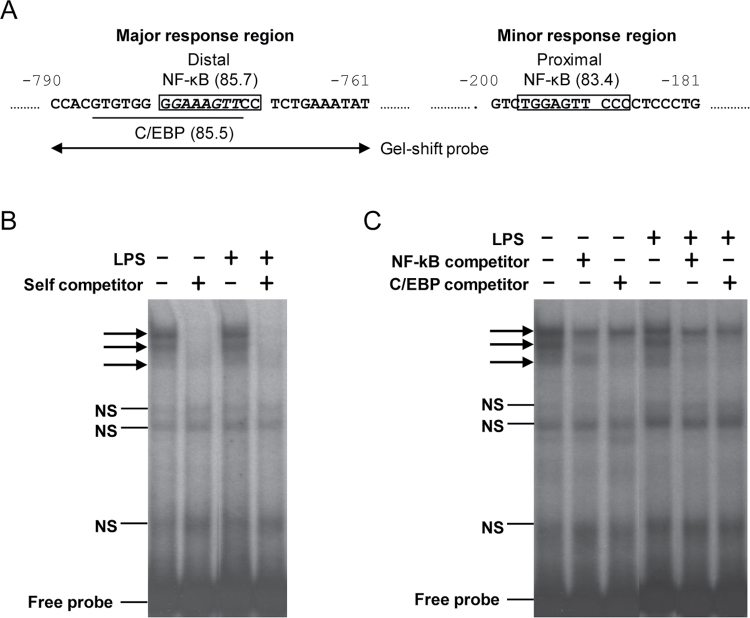
Analysis of the CYP2A13 proximal promoter region using the TFsearch program (http://www.cbrc.jp/research/db/TFSEARCH.html) revealed a putative NF-κB binding site (with a similarity score of 85.7) within the major LPS response region (at −771 to −780bp upstream of the ATG start codon) (Fig. 5A). Another putative NF-κB binding site (with a similarity score of 83.4) was found in the minor (more proximal) LPS response region, at ~200bp upstream of the ATG codon. The presence of these two putative NF-κB binding sites within the identified LPS response regions lends further support to potential functional importance of NF-κB in inflammation-associated CYP2A13 suppression. Interestingly, the distal NF-κB binding site has an eight-nucleotide overlap with a putative C/EBP binding site (with a similarity score of 85.5) (Fig. 5A). Because C/EBP, which is also important in LPS-mediated inflammation, has been implicated in transcriptional regulation of CYP2A13 in our previous study (Ling et al., 2007), we further explored the possibility that NF-κB may interact with C/EBP at this site.
Fig. 5.
Potential role of NF-κB in LPS-induced suppression of CYP2A13 expression and CYP2A13 promoter activity. (A) Predicted NF-κB binding sites in the proximal promoter region of CYP2A13. The two predicted binding sites are boxed, with the similarity scores shown in parentheses. The distal NF-κB binding site overlaps with a putative C/EBP binding site (underlined); the common sequence between the two is italicized. The sequence of the oligonucleotide probe used for gel shift assays is indicated by double-headed arrows. (B and C) Gel shift assays. H441 cells were either untreated or treated with LPS at 10 μg/ml and harvested 3h later for preparation of nuclear fractions. Nuclear proteins (5 μg) were incubated with 32P-labeled oligonucleotide probe at room temperature for 30min. The incubation mixtures were resolved by electrophoresis, and bands were visualized by autoradiography. Various unlabeled oligonucleotide probes were added in excess amounts in order to test their ability to compete with the radiolabeled probe, including the gel shift probe (i.e., self-competitor; 100-fold molar excess) (B), NF-κB consensus probe, and C/EBP consensus probe (100-fold molar excess) (C). The position of specific shift bands is indicated by arrows; nonspecific (NS) bands and free probe are also indicated.
We performed gel shift assays using a 30-bp probe (−761 to −790) that contained both the major NF-κB site and the C/EBP site (Fig. 5A) and nuclear extracts from control or LPS-treated H441 cells. As shown in Figure 5B, several shift bands were formed with nuclear proteins from either LPS-treated or untreated cells. However, only the three bands near the top of the gel, which were competed by excess amounts of unlabeled self-competitor probe, represented specific binding of nuclear proteins to the probe sequence. Of the three, the bottom two bands were also competed by excess amounts of unlabeled NF-κB and C/EBP consensus oligonucleotide probes in both LPS-treated and untreated cells (Fig. 5C), suggesting that the binding complexes contained both NF-κB-like and C/EBP-like transcription factors. On the other hand, the binding complex represented by the top band was not competed by excess amounts of either NF-κB or C/EBP consensus probe (Fig. 5C), and thus it unlikely contained any NF-κB or C/EBP family of transcription factors. In experiments not shown, addition of excess amounts of NF-κB or C/EBP probes that contained mutations at critical sites of the respective consensus sequences (Jiang and Zarnegar, 1997) failed to compete with the labeled 30-bp probe in the formation of the two lower bands, thus further confirming specificity of binding of the NF-κB-like and C/EBP-like transcription factors to the probe. Overall, the gel shift data support the idea that NF-κB interact with C/EBP to mediate LPS-induced suppression of CYP2A13 expression.
In other experiments not shown, gel shift assays were also performed for a probe that contained the proximal NF-κB binding site. However, the shift bands detected were not competed by excess amount of unlabeled NF-κB consensus probe (data not shown), suggesting that the putative proximal NF-κB binding site does not play a major role in the suppression of CYP2A13 expression by LPS.
DISCUSSION
We have shown for the first time that inflammation can suppress CYP2A13 expression in lung cells both in vitro and in vivo. The effect of inflammation is exerted at the level of transcription, given the similar effects of LPS on the expression of both the endogenous CYP2A13 gene and the transfected CYP2A13 reporter gene in H441 cells. The transcriptional suppression is mechanistically plausible, as indicated by the occurrence of binding sites for NF-κB and C/EBP in the CYP2A13 proximal promoter region. These binding sites, which interacted with NF-κB-like and C/EBP-like nuclear proteins in H441 cells in gel shift assays, were essential for the suppressive effects of LPS, while the potential role of NF-κB was further supported by the reversal of the LPS effects by an NF-κB chemical inhibitor, PDTC. These data strongly support the notion that CYP2A13 levels in human lung can be suppressed by inflammation associated with disease status in tissue donors. In that connection, NF-κB and C/EBP are both expressed in human lung, and their expression or binding activities appear to be modulated in airway epithelial cells by smoking (Didon et al., 2005; Isajevs et al., 2011).
LPS treatment is a widely used model of inflammation, which has been utilized previously to study the effects of inflammation on basal and inducible hepatic P450 expression (Richardson and Morgan, 2005). The effectiveness of LPS injection in inducing systemic and lung tissue inflammation in the CYP2A13-humanized mice was confirmed by LPS-mediated induction of IL-6, a proinflammatory cytokine, in serum and lung tissues. The humanized mice were on a C57BL/6 genetic background, which explains the relatively low magnitude of IL-6 increase following LPS injection, as C57BL/6 mice are poor responders to LPS (Corteling et al., 2002). Systemic inflammation was further supported by increases in the expression of SAP, a marker for acute phase response, in the liver, a major target organ of that response (Akiyama and Gonzalez, 2003).
The doses of LPS used in the present study (0.5–1.0mg/kg) were relatively low, as LPS at high doses (5mg/kg) can induce cellular injury and possibly nonspecific effects on gene expression (Gilmore et al., 2003). LPS has also been found to cause mucous cell metaplasia in mouse lung (Yanagihara et al., 2001). However, a single injection of LPS at 1mg/kg in the CYP2A13-humanized mouse did not result in any noticeable decrease in the expression of CYP2F2 or Clara cell secretory protein, which are molecular markers for Clara cells (Zemke et al., 2009), or any pathological changes in the lung (data not shown). Thus, the LPS treatment did not cause significant, if any, cellular injury or metaplasia in the CYP2A13-humanized mouse.
A downregulation of pulmonary CYP2A13 expression would lead to decreased bioactivation of many toxicants and procarcinogens that are CYP2A13 substrate, potentially leading to decreased risks of mutagenesis and carcinogenesis, particularly for those with high carcinogen exposure and high constitutive CYP2A13 expression. Thus, knowledge of the molecular mechanisms underlying the suppression of CYP2A13 expression by inflammation is valuable for designing strategies to reduce CYP2A13 expression and prevent CYP2A13-mediated toxicity in high-risk populations.
The availability of the CYP2A13-humanized mouse provided a unique opportunity to study the regulation of CYP2A13 expression in vivo. In the CYP2A13-humanized mouse, the CYP2A13 transgene was preferentially expressed in the respiratory tract (Wei et al., 2012), as was found for CYP2A13 expression in humans (Su et al., 2000). The absence of mouse CYP2A5 facilitated the analysis of CYP2A13 expression by immunoblotting. Notably, in addition to Cyp2a5, four other Cyp2a genes exist in mice; these and members of other Cyp2 gene subfamilies might have contributed to the nonspecific band detected by the anti-CYP2A5 polyclonal antibody on immunoblots.
The mechanisms underlying P450 regulation by inflammation are not well understood and may differ for individual P450s and tissues (Aitken et al., 2006). IL-6 is an important mediator of the acute phase responses associated with LPS administration; however, LPS can suppress hepatic P450 expression independent of IL-6, and LPS response elements have been identified in the promoters of P450 genes, such as CYP2C11 (Iber et al., 2000). In our study, although IL-6 expression was induced by LPS treatment in the CYP2A13-humanized mice, CYP2A13 expression in H441 cells was suppressed by either LPS or IL-6, a result suggesting that IL-6 signaling is involved in the repression of lung CYP2A13 expression by LPS. However, other cytokines might also be involved, including IL-1 and TNF-α, which can also be induced by LPS as well as by inflammasome activation (Eggesbo et al., 1994). Furthermore, it had been previously reported that aging might suppress CYP2A13 expression in human nasal mucosa (Getchell et al., 1993). The IKK/NF-κB signaling pathway, which is activated by oxidative stress in addition to inflammation, is one of the key mediators of aging (Tilstra et al., 2011). Thus, although the effects of reactive oxygen species on CYP2A13 expression were not investigated in the current study, it is conceivable that oxidative stress and aging might also suppress CYP2A13 expression in the lung.
Transcription factors that are activated by the LPS-induced acute phase response include NF-κB, activator protein-1, C/EBP, and cAMP response element binding protein, which are responsible for the gene expression changes during inflammation (Akiyama and Gonzalez, 2003; Halsey et al., 2007). Using reporter gene assays, we localized the LPS response elements in CYP2A13 promoter to two regions (from −130 to −216bp and from −484 to −1008bp). Notably, we cannot exclude the possibility that additional LPS response elements exist in more distal regions (upstream of −1.1kb) of the CYP2A13 promoter, which were not analyzed. We further confirmed, using gel shift assays, that the −761 to −790bp region, which contained a putative NF-κB binding site and a putative C/EBP binding site, interacted with NF-κB-like and C/EBP-like transcription factors in H441 cells. The fact that the sequences of the two binding sites had significant overlap, and that apparently the same set of proteins were competed by consensus oligonucleotide probes for either NF-κB or C/EBP, suggests that NF-κB and C/EBP may form heterodimeric or multimeric complexes at the composite binding site. In that regard, NF-κB and C/EBP family members are known to interact through direct contact (Stein et al., 1993).
Further studies are warranted to determine the precise mechanism for interactions between NF-κB and C/EBP at the CYP2A13 promoter, and whether or how they modulate CYP2A13 transcription in response to inflammation. NF-κB, which plays an important role in many biological processes, including inflammation, is involved in the LPS-mediated downregulation of several rat hepatic P450s, including CYP2C11, 2D5, 2B1, and 1A1 (Abdulla et al., 2005; Morgan, 2001). C/EBP, an essential transcription factor involved in inflammatory responses, is known to bind to CYP2A13 promoter and positively regulate CYP2A13 promoter activity (Ling et al., 2007). It is tempting to speculate that LPS-induced activation and nuclear translocation of NF-κB may change the structure and/or composition of the protein complex at the composite NF-κB/C/EBP binding site in the CYP2A13 promoter, leading to downregulation of CYP2A13 expression. Notably, LPS treatment did not lead to an appreciable increase in the overall abundance of the protein complex bound to the −761 to −790bp probe in gel shift assays, suggesting that the effect of LPS is more likely to modify the composition than to increase the overall abundance of proteins bound at this site.
In conclusion, we provide strong evidence that CYP2A13 is downregulated by inflammation. We also present initial mechanistic data suggesting the involvement of NF-κB and C/EBP in the underlying regulatory process. An important implication of our present finding is that CYP2A13 expression levels detected in lung biopsy samples (Zhang et al., 2007), which are often obtained from patients with lung diseases or other diseases with associated systemic inflammation, may significantly underestimate the levels of CYP2A13 present in the lungs of a normal, healthy individual. This scenario should be considered when assessing the role of CYP2A13 in chemical-induced lung toxicity. For activity assessment, greater emphasis should be placed on enzyme kinetics analysis, rather than on rate determination alone, when assessing lung microsomal P450 metabolic activity toward carcinogens, because the apparent K m values (compared with V max values) are less likely to be influenced by the decreases in P450 expression levels associated with inflammation and other disease status.
FUNDING
National Institutes of Health (CA092596 to X.D.).
ACKNOWLEDGMENTS
We thank Drs Xiuling Zhang, Jaime D’Agostino, Wei Yuan, and Donghong Gao of the Wadsworth Center for assistance with methods development during pilot studies. We gratefully acknowledge the use of the Biochemistry, Molecular Immunology, and Molecular Genetics core facilities of the Wadsworth Center.
REFERENCES
- Abdulla D., Goralski K. B., Del Busto Cano E. G., Renton K. W. (2005). The signal transduction pathways involved in hepatic cytochrome P450 regulation in the rat during a lipopolysaccharide-induced model of central nervous system inflammation. Drug Metab. Dispos. 33, 1521–1531 [DOI] [PubMed] [Google Scholar]
- Aitken A. E., Richardson T. A., Morgan E. T. (2006). Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu. Rev. Pharmacol. Toxicol. 46, 123–149 [DOI] [PubMed] [Google Scholar]
- Akira S., Taga T., Kishimoto T. (1993). Interleukin-6 in biology and medicine. Adv. Immunol. 54, 1–78 [DOI] [PubMed] [Google Scholar]
- Akiyama T. E., Gonzalez F. J. (2003). Regulation of P450 genes by liver-enriched transcription factors and nuclear receptors. Biochim. Biophys. Acta. 1619, 223–234 [DOI] [PubMed] [Google Scholar]
- Charles K. A., Rivory L. P., Brown S. L., Liddle C., Clarke S. J., Robertson G. R. (2006). Transcriptional repression of hepatic cytochrome P450 3A4 gene in the presence of cancer. Clin. Cancer Res. 12, 7492–7497 [DOI] [PubMed] [Google Scholar]
- Corteling R., Wyss D., Trifilieff A. (2002). In vivo models of lung neutrophil activation. Comparison of mice and hamsters. BMC Pharmacol. 2, 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino J., Zhang X., Wu H., Ling G., Wang S., Zhang Q. Y., Liu F., Ding X. (2008). Characterization of CYP2A13*2, a variant cytochrome P450 allele previously found to be associated with decreased incidences of lung adenocarcinoma in smokers. Drug Metab. Dispos. 36, 2316–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Agostino J., Zhuo X., Shadid M., Morgan D. G., Zhang X., Humphreys W. G., Shu Y. Z., Yost G. S., Ding X. (2009). The pneumotoxin 3-methylindole is a substrate and a mechanism-based inactivator of CYP2A13, a human cytochrome P450 enzyme preferentially expressed in the respiratory tract. Drug Metab. Dispos. 37, 2018–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didon L., Qvarfordt I., Andersson O., Nord M., Riise G. C. (2005). Decreased CCAAT/enhancer binding protein transcription factor activity in chronic bronchitis and COPD. Chest. 127, 1341–1346 [DOI] [PubMed] [Google Scholar]
- Ding X., Coon M. J. (1990). Immunochemical characterization of multiple forms of cytochrome P-450 in rabbit nasal microsomes and evidence for tissue-specific expression of P-450s NMa and NMb. Mol. Pharmacol. 37, 489–496 [PubMed] [Google Scholar]
- Duan Q., Wang X., Gong W., Ni L., Chen C., He X., Chen F., Yang L., Wang P., Wang D. W. (2012). ER stress negatively modulates the expression of the miR-199a/214 cluster to regulates tumor survival and progression in human hepatocellular cancer. PLoS One. 7, e31518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggesbo J. B., Hjermann I., Lund P. K., Joø G. B., Ovstebø R., Kierulf P. (1994). LPS-induced release of IL-1 beta, IL-6, IL-8, TNF-alpha and sCD14 in whole blood and PBMC from persons with high or low levels of HDL-lipoprotein. Cytokine. 6, 521–529 [DOI] [PubMed] [Google Scholar]
- Getchell M. L., Chen Y., Ding X., Sparks D. L., Getchell T. V. (1993). Immunohistochemical localization of a cytochrome P-450 isozyme in human nasal mucosa: Age-related trends. Ann. Otol. Rhinol. Laryngol. 102, 368–374 [DOI] [PubMed] [Google Scholar]
- Gilmore W. J., Hartmann G., Piquette-Miller M., Marriott J., Kirby G. M. (2003). Effects of lipopolysaccharide-stimulated inflammation and pyrazole-mediated hepatocellular injury on mouse hepatic Cyp2a5 expression. Toxicology. 184, 211–226 [DOI] [PubMed] [Google Scholar]
- Gu J., Zhang Q. Y., Genter M. B., Lipinskas T. W., Negishi M., Nebert D. W., Ding X. (1998). Purification and characterization of heterologously expressed mouse CYP2A5 and CYP2G1: Role in metabolic activation of acetaminophen and 2,6-dichlorobenzonitrile in mouse olfactory mucosal microsomes. J. Pharmacol. Exp. Ther. 285, 1287–1295 [PubMed] [Google Scholar]
- Halsey T. A., Yang L., Walker J. R., Hogenesch J. B., Thomas R. S. (2007). A functional map of NFkappaB signaling identifies novel modulators and multiple system controls. Genome Biol. 8, R104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. Y., Shen J., Ding X., Lu A. Y., Hong J. Y. (2004). Identification of critical amino acid residues of human CYP2A13 for the metabolic activation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco-specific carcinogen. Drug Metab. Dispos. 32, 1516–1521 [DOI] [PubMed] [Google Scholar]
- He X. Y., Tang L., Wang S. L., Cai Q. S., Wang J. S., Hong J. Y. (2006). Efficient activation of aflatoxin B1 by cytochrome P450 2A13, an enzyme predominantly expressed in human respiratory tract. Int. J. Cancer. 118, 2665–2671 [DOI] [PubMed] [Google Scholar]
- Iber H., Chen Q., Cheng P. Y., Morgan E. T. (2000). Suppression of CYP2C11 gene transcription by interleukin-1 mediated by NF-kappaB binding at the transcription start site. Arch. Biochem. Biophys. 377, 187–194 [DOI] [PubMed] [Google Scholar]
- Isajevs S., Taivans I., Svirina D., Strazda G., Kopeika U. (2011). Patterns of inflammatory responses in large and small airways in smokers with and without chronic obstructive pulmonary disease. Respiration. 81, 362–371 [DOI] [PubMed] [Google Scholar]
- Jalas J. R., Hecht S. S., Murphy S. E. (2005). Cytochrome P450 enzymes as catalysts of metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone, a tobacco specific carcinogen. Chem. Res. Toxicol. 18, 95–110 [DOI] [PubMed] [Google Scholar]
- Jiang J. G., Zarnegar R. (1997). A novel transcriptional regulatory region within the core promoter of the hepatocyte growth factor gene is responsible for its inducibility by cytokines via the C/EBP family of transcription factors. Mol. Cell. Biol. 17, 5758–5770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., Rabson A. B., Germino J. F., Gallo M. A., Tian Y. (2001). Mechanism of suppression of cytochrome P-450 1A1 expression by tumor necrosis factor-alpha and lipopolysaccharide. J. Biol. Chem. 276, 39638–39644 [DOI] [PubMed] [Google Scholar]
- Koskela S., Hakkola J., Hukkanen J., Pelkonen O., Sorri M., Saranen A., Anttila S., Fernandez-Salguero P., Gonzalez F., Raunio H. (1999). Expression of CYP2A genes in human liver and extrahepatic tissues. Biochem. Pharmacol. 57, 1407–1413 [DOI] [PubMed] [Google Scholar]
- Ling G., Wei Y., Ding X. (2007). Transcriptional regulation of human CYP2A13 expression in the respiratory tract by CCAAT/enhancer binding protein and epigenetic modulation. Mol. Pharmacol. 71, 807–816 [DOI] [PubMed] [Google Scholar]
- Megaraj V, Zhou X, Xie F, Liu Z, Yang W, Ding X. (2013) Role of CYP2A13 in the bioactivation and lung tumorigenicity of the tobacco-specific lung procarcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK): in vivo studies using a CYP2A13-humanized mouse model. Carcinogenesis. 10.1093/carcin/bgt269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa K., Mori A., Yamamoto K., Okudaira H. (1998). Transcriptional roles of CCAAT/enhancer binding protein-beta, nuclear factor-kappaB, and C-promoter binding factor 1 in interleukin (IL)-1beta-induced IL-6 synthesis by human rheumatoid fibroblast-like synoviocytes. J. Biol. Chem. 273, 7620–7627 [DOI] [PubMed] [Google Scholar]
- Morgan E. T. (2001). Regulation of cytochrome p450 by inflammatory mediators: Why and how? Drug Metab. Dispos. 29, 207–212 [PubMed] [Google Scholar]
- Nakajima M., Itoh M., Sakai H., Fukami T., Katoh M., Yamazaki H., Kadlubar F. F., Imaoka S., Funae Y., Yokoi T. (2006). CYP2A13 expressed in human bladder metabolically activates 4-aminobiphenyl. Int. J. Cancer. 119, 2520–2526 [DOI] [PubMed] [Google Scholar]
- Richardson T. A., Morgan E. T. (2005). Hepatic cytochrome P450 gene regulation during endotoxin-induced inflammation in nuclear receptor knockout mice. J. Pharmacol. Exp. Ther. 314, 703–709 [DOI] [PubMed] [Google Scholar]
- Stein B., Cogswell P. C., Baldwin A. S., Jr (1993). Functional and physical associations between NF-kappa B and C/EBP family members: A Rel domain-bZIP interaction. Mol. Cell. Biol. 13, 3964–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov I., Krueger W., Mankowski D., Guernsey L., Kaur A., Glynn J., Thrall R. S. (2006). The cytochromes P450 (CYP) response to allergic inflammation of the lung. Arch. Biochem. Biophys. 456, 30–38 [DOI] [PubMed] [Google Scholar]
- Su T., Bao Z., Zhang Q. Y., Smith T. J., Hong J. Y., Ding X. (2000). Human cytochrome P450 CYP2A13: Predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 60, 5074–5079 [PubMed] [Google Scholar]
- Sunman J. A., Hawke R. L., LeCluyse E. L., Kashuba A. D. (2004). Kupffer cell-mediated IL-2 suppression of CYP3A activity in human hepatocytes. Drug Metab. Dispos. 32, 359–363 [DOI] [PubMed] [Google Scholar]
- Tilstra J. S., Clauson C. L., Niedernhofer L. J., Robbins P. D. (2011). NF-κB in aging and disease. Aging Dis. 2, 449–465 [PMC free article] [PubMed] [Google Scholar]
- Wang H., Tan W., Hao B., Miao X., Zhou G., He F., Lin D. (2003). Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 63, 8057–8061 [PubMed] [Google Scholar]
- Wei Y., Wu H., Li L., Liu Z., Zhou X., Zhang Q. Y., Weng Y., D’Agostino J., Ling G., Zhang X, et al. (2012). Generation and characterization of a CYP2A13/2B6/2F1-transgenic mouse model. Drug Metab. Dispos. 40, 1144–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zhang X., Ling G., D’Agostino J., Ding X. (2009). Mechanisms of differential expression of the CYP2A13 7520C and 7520G alleles in human lung: Allelic expression analysis for CYP2A13 heterogeneous nuclear RNA, and evidence for the involvement of multiple cis-regulatory single nucleotide polymorphisms. Pharmacogenet. Genomics. 19, 852–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara K., Seki M., Cheng P. W. (2001). Lipopolysaccharide induces mucus cell metaplasia in mouse lung. Am. J. Respir. Cell Mol. Biol. 24, 66–73 [DOI] [PubMed] [Google Scholar]
- Zemke A. C., Snyder J. C., Brockway B. L., Drake J. A., Reynolds S. D., Kaminski N., Stripp B. R. (2009). Molecular staging of epithelial maturation using secretory cell-specific genes as markers. Am. J. Respir. Cell Mol. Biol. 40, 340–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Caggana M., Cutler T. L., Ding X. (2004). Development of a real-time polymerase chain reaction-based method for the measurement of relative allelic expression and identification of CYP2A13 alleles with decreased expression in human lung. J. Pharmacol. Exp. Ther. 311, 373–381 [DOI] [PubMed] [Google Scholar]
- Zhang X., D’Agostino J., Wu H., Zhang Q. Y., von Weymarn L., Murphy S. E., Ding X. (2007). CYP2A13: Variable expression and role in human lung microsomal metabolic activation of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. J. Pharmacol. Exp. Ther. 323, 570–578 [DOI] [PubMed] [Google Scholar]
- Zhou X., Zhuo X., Xie F., Kluetzman K., Shu Y. Z., Humphreys W. G., Ding X. (2010). Role of CYP2A5 in the clearance of nicotine and cotinine: Insights from studies on a Cyp2a5-null mouse model. J. Pharmacol. Exp. Ther. 332, 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. R., Thomas P. E., Lu G., Reuhl K. R., Yang G. Y., Wang L. D., Wang S. L., Yang C. S., He X. Y., Hong J. Y. (2006). CYP2A13 in human respiratory tissues and lung cancers: An immunohistochemical study with a new peptide-specific antibody. Drug Metab. Dispos. 34, 1672–1676 [DOI] [PubMed] [Google Scholar]