Summary
The inherent cytotoxicity of aberrantly folded protein aggregates contributes substantially to the pathogenesis of amyloid diseases. It was recently shown that a class of evolutionary conserved proteins, called MOAG-4/SERF, profoundly alter amyloid toxicity via an autonomous but yet unexplained mode. We show that the biological function of human SERF1a originates from its atypical ability to specifically distinguish between amyloid and nonamyloid aggregation. This inherently unstructured protein directly affected the aggregation kinetics of a broad range of amyloidogenic proteins in vitro, while being inactive against nonamyloid aggregation. A representative biophysical analysis of the SERF1a:α-synuclein (aSyn) complex revealed that the amyloid-promoting activity resulted from an early and transient interaction, which was sufficient to provoke a massive increase of soluble aSyn amyloid nucleation templates. Therefore, the autonomous amyloid-modifying activity of SERF1a observed in living organisms relies on a direct and dedicated manipulation of the early stages in the amyloid aggregation pathway.
Graphical Abstract
Highlights
► SERF1a drives the assembly of amyloidogenic proteins ► SERF1a discriminates between amyloid and nonamyloid aggregation ► SERF1a acts through an early interaction with α-synuclein amyloid precursors ► SERF1a catalyzes the formation of transient α-synuclein “on-pathway” aggregates
The protein class called MOAG-4/SERF profoundly alters amyloid protein toxicity via an autonomous and previously unidentified pathway. Falsone and colleagues demonstrate that the amyloid-modifying ability of human SERF1a is driven by a transient interaction with early precursors in the amyloid pathway. As a consequence, SERF1a promotes amyloid aggregation of several amyloidogenic proteins, while being insensitive to unspecific protein aggregation. This identifies SERF1as a specialized amyloid factor, in support to its autonomous mode of action observed in cells.
Introduction
Amyloidogenic proteins are a class of polypeptides capable of assembling into insoluble fibers with a distinct cross beta-sheet structure (Eichner and Radford, 2011). Amyloids are under intensive scientific investigation because of their association with a series of highly prevalent and incurable neurodegenerative disorders, including Parkinson's disease, Alzheimer's disease, Huntington's disease, and prion-related encephalopathies (Chiti and Dobson, 2006). In degenerating neurons, amyloid fibers can occur as either intracellular or extracellular deposits, which are positive to the dyes Congo Red and Thioflavin T.
The multifactorial nature of these neurodegenerative disorders complicates efforts to clearly define a link between oligomers, fibers, and disease. It has been proposed that structurally unrelated amyloid proteins undergo similar structural rearrangements on their way to becoming mature amyloids and that the toxic species consist of intermediate protein aggregates (Glabe, 2006).
To identify cofactors that modulate intracellular amyloid formation, a genetic screen was performed and led to the discovery of MOAG-4/SERF (Modifier of aggregation-4/Small EDRK rich factor) as an evolutionary conserved class of amyloid-regulating proteins (van Ham et al., 2010). Knockdown of MOAG-4/SERF expression in eukaryotic cells suppressed aggregation of huntingtin (htt), α-synuclein (aSyn), and beta-amyloid (Aβ). This effect appears unrelated to other traditional aggregation-modifying pathways, such as the chaperone-folding machinery, proteasomal degradation, or autophagy, because their manipulation did not alter the activity of MOAG-4/SERF. However, the exact mechanism by which MOAG/SERF promotes amyloid formation remained unsolved.
To resolve this important issue, we explored the possibility that MOAG-4/SERF drives aggregation through a direct interaction with aggregation-prone disease proteins. To this end, we tested the effect of human SERF1a (short isoform) in an isolated in vitro system on a set of structurally unrelated amyloidogenic proteins.
Results
SERF1a Accelerates α-Synuclein Amyloidogenesis
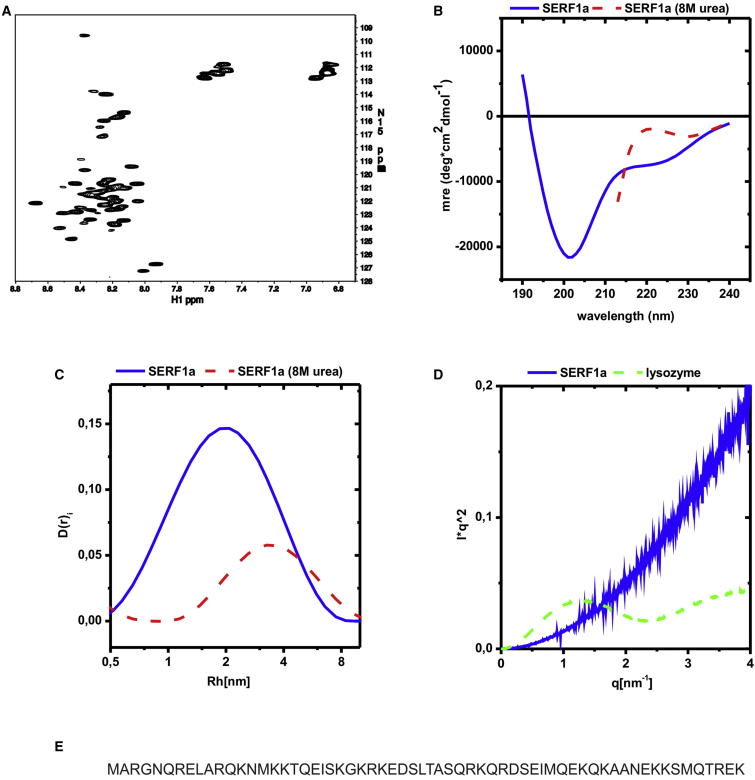
SERF1a is a basic (pI = 10.44), 7.4 kDa protein. A structural analysis identified the recombinant molecule as predominantly disordered (Figure 1).
Figure 1.
SERF1a Is Predominantly Disordered
(A) An 1H,15N-HSQC NMR spectrum of 100 μM SERF1a shows little signal dispersion of the proton peaks, with each signal located within a narrow proton range between 7.8 and 8.7 ppm. Such a distribution is typical for flexible proteins with low secondary structural contents (Tompa, 2010).
(B) The far-UV CD-spectrum of SERF1a (solid blue line) recorded between 190 and 240 nm, lacked any predominant alpha helix (minima at 222 and 208 nm) or beta sheet (minimum at 216 nm) signal. Instead, the curve was dominated by a strong negative signal around 200 nm and by a slow positive signal recovery below 200 nm. This is indicative for the predominance of conformational disorder (Tompa, 2010). The presence of a residual secondary structure was deducible by its disruption upon the addition of the chaotropic agent urea (dashed red line), which resulted in a positive signal shift (spectrum collected up to 210 nm because of the interfering strong background signal of the denaturant below this value). This was also reflected by secondary structural deconvolution analysis of the CD spectrum (Whitmore and Wallace, 2008; Sreerama et al., 1999), which yielded a predominant 76.8% random coil contents, and a residual 17.8% alpha helix and 6.2% beta sheet structure.
(C) These results were supported by dynamic light scattering (C), which provides the hydrodynamic radii of particles in solution. The measured mean radius for SERF1a was 2.08 nm (solid blue line), whereas globular proteins of similar molecular size possess much smaller Rh values (around 1.5 nm) (Uversky, 1993). In the presence of urea, Rh shifted to 3.16 nm (dashed red line), implying that, despite the lack of globularity, some residual structure was still present and that it was completely disrupted by this chaotrope.
(D) Small-angle X-ray scattering, a technique that measures the size and shape of a polymer, underscored the predominance of structural disorder (D). The Kratky plot of SERF1a increased monotonally without any detectable maximum (solid blue line), as expected for a macromolecule devoid of any well-defined structure (Tompa, 2010). In contrast, a well-defined maximum could be detected for the representative globular protein lysozyme (dashed green line).
(E) Primary structure of SERF1a (UniProt accession number O75920-2).
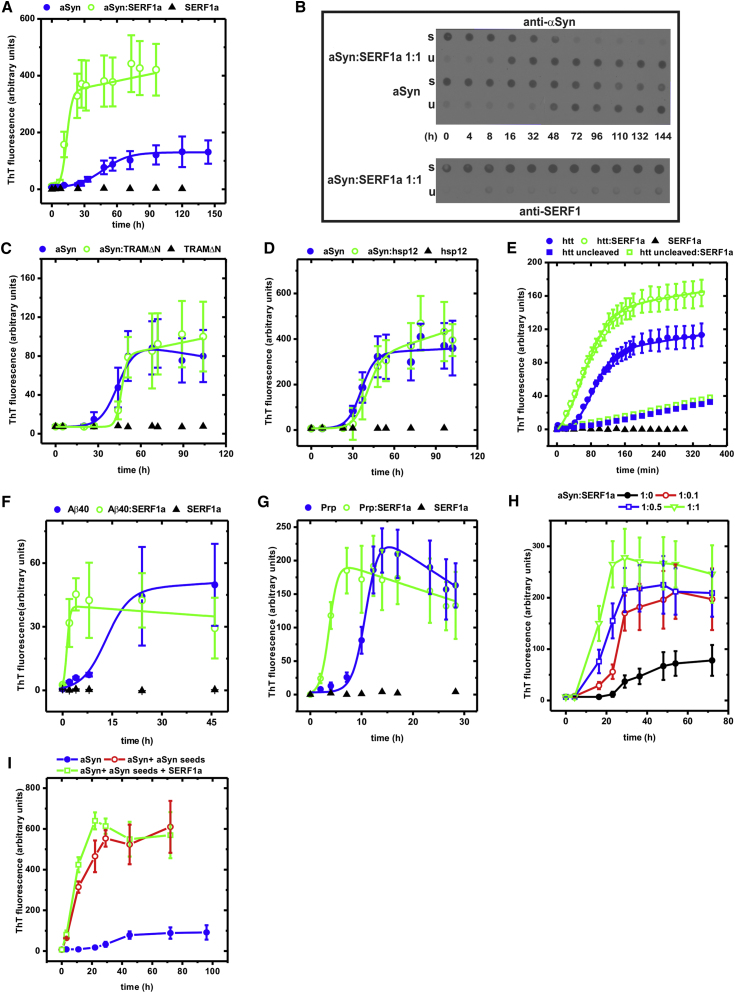
To look for a direct influence of SERF1a on the amyloid assembly, we measured the time-dependent amyloid conversion of the Parkinson's disease-associated protein aSyn, which aggregates into Lewy bodies (Spillantini et al., 1998). Monomeric aSyn was incubated in the absence or presence of SERF1a and amyloid formation was monitored by Thioflavin T (ThT) fluorescence. ThT exhibits a fluorescence emission maximum at 488 nm exclusively when bound to amyloid (LeVine, 1999). In the absence of SERF1a, aSyn converted into amyloid fibers with a half time of conversion tm = 50 hr, and an initial lag phase tl = 21 hr (Figure 2A and Table 1). Equimolar amounts of SERF1a accelerated half time of conversion to tm = 13 hr, and the lag phase was reduced by approximately 50% (tl = 10 hr). This caused a significant accumulation of ThT-reactive species, as indicated by the higher (approximately 4-fold) ThT fluorescence intensity as compared to aSyn alone.
Figure 2.
Amyloid-Promoting Properties of SERF1a
(A) ThT-monitored amyloid kinetics of aSyn in the absence (closed blue circle) and in the presence of SERF1a (open green circle).
(B) Dot-blot partition analysis/immunodetection of aSyn and SERF1a in sample aliquots drawn during the aSyn amyloid growth reaction. Upper (detection of aSyn): In the presence of equimolar amounts of SERF1a, the formation of unsoluble fibers was accelerated and after 48 hr aSyn relocated almost quantitatively from the soluble (s) into the unsoluble (u) fraction. In the absence of SERF1a, the formation of unsoluble aSyn species was slower. An amount of soluble aSyn was still detectable even at t ≥ 48 hr, indicating that the conversion was not quantitative. Lower (same samples; detection of SERF1a): SERF1a remained soluble during aSyn amyloid fiber growth, indicating that itself does not aggregate or coprecipitate with amyloid fibers (see also Figure S1A).
(C and D) Amyloid kinetics of 100 μM aSyn in the absence (closed blue circle) and in the presence (open green circle) of equimolar amyloid-unrelated control proteins TRAMΔN (C) and hsp12 (D) (Ex/em slit widths = 10/20). Amyloidogenesis was insignificantly affected.
(E) Amyloid kinetics of GST-httQ53Ex1 in the absence (closed blue circle) and in the presence of SERF1a (open green circle). Fiber formation was initiated by the proteolytic cleavage of the GST-tag (Wacker et al., 2004). Without cleavage of the GST tag, amyloidogenesis did not occur (closed blue square in the absence of SERF1a; open green square in the presence of SERF1a). The weak signal increase is possibly due to autoproteolysis and the resulting formation of little amounts of amyloids.
(F) Amyloid kinetics of Aβ40 in the absence (closed blue circle) and in the presence of SERF1a (open green square).
(G) Amyloid kinetics of PrP in the absence (closed blue circle) and in the presence of SERF1a (open green square).
(H) Amyloid kinetics of 100 μM aSyn in the absence (closed black circle) and in the presence of different SERF1a ratios demonstrates that substoichiometric amounts of SERF1a are sufficient to enhance amyloidogenesis.
(I) A concentration of 1.5 μg/ml aSyn nucleation seeds, generated by ultrasonication of mature aSyn fibers, promotes the amyloid conversion of 100 μM aSyn (1.5 mg/ml) (open red circle). The addition of 100 μM SERF1a marginally influences seeded amyloidogenesis (open green square). (Closed blue circle) 100 μM aSyn in the absence of seeds.
See also Figure S1. The error bars are mean averages, including SEM of three independent experiments.
Table 1.
Amyloid Growth Parameters
| Protein | SERF1a | tm | tl | kapp |
|---|---|---|---|---|
| aSyn | − | 50.51 ± 3.19 hr | 21.94 ± 7.57 hr | 0.07 ± 0.014 hr−1 |
| + | 13.5 ± 1.37 hr | 10.94 ± 3.50 hr | 0.39 ± 0.076 hr−1 | |
| httQ53Ex1 | − | 81.00 ± 0.04 min | 20.35 ± 0.06 min | 0.036 ± 3.09 × 10−5 min−1 |
| + | 56.27 ± 0.35 min | 9.76 ± 0.42 min | 0.043 ± 2.96 × 10−4 min−1 | |
| Aβ40 | − | 12.99 ± 2.73 hr | 11.35 ± 5.97 hr | 0.28 ± 0.08 hr−1 |
| + | 1.58 ± 0.133 hr | 0.47 ± 0.169 hr | 1.79 ± 0.14 hr−1 | |
| PrP | − | 10.89 ± 0.13 hr | 8.33 ± 0.88 hr | 0.78 ± 0.07 hr−1 |
| + | 3.70 ± 0.13 hr | 1.99 ± 0.47 hr | 1.17 ± 0.2 hr−1 |
Amyloid growth kinetic parameters for proteins used in this study in the absence and in the presence of SERF1a: midpoint of amyloid growth, tm; initial lag phase, tl; apparent rate of amyloid formation, kapp. The values are mean averages, including the SEM of three independent measurements.
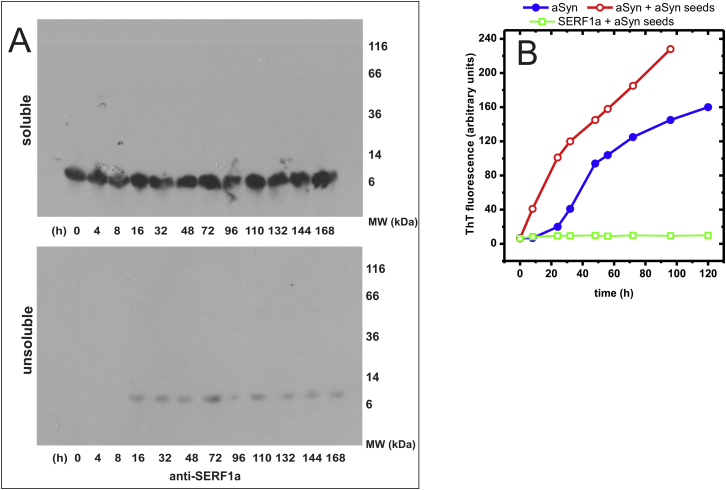
We could exclude that SERF1a might itself assemble into amyloid fibrils, as it was not reactive to ThT (Figure 2), even after several months of incubation at high concentrations (10 mM; data not shown). In addition, dot-blot partition analysis coupled to immunodetection showed that SERF1a remained in solution (Figure 2B, lower), whereas the aSyn amyloid fibers, as expected, partitioned into the insoluble fraction (Figure 2B, upper). SDS-PAGE analysis of the same samples showed that SERF1a did not incorporate into stable SDS-resistant oligomers (Figure S1A), which are typical for amyloid fiber intermediates (Bagriantsev et al., 2006). Finally, we excluded the possibility of heterogeneous cross-seeding (Yagi et al., 2005), as preformed aSyn nucleation seeds did not cause the formation of SERF1a fibers (Figure S1B).
Figure S1.
SERF1a Does Not Form SDS-Resistant and/or Unsoluble Aggregates, Related to Figure 2
(A) SDS-PAGE resolved partition analysis and immunodetection of SERF1a during the aSyn:SERF1a amyloid growth reaction (same samples as in Figure 2B). The two panels show that SERF1a localizes to the soluble fraction (upper panel), and minimally to the unsoluble fraction (lower panel), indicating that it does not precipitate, co-aggregate with aSyn or form SDS-resistant oligomers during amyloid growth (see also Figure 2B).
(B) 1.5 μg/ml aSyn nucleation seeds, generated by ultrasonication of mature aSyn fibers, promote the amyloid conversion of 100 μM aSyn (1.5 mg/ml) (open red circle), but not of 100 μM SERF1a (open green square). This excludes a hypothetical amyloidogenic cross-seeding of SERF1a, as observable between diverse amyloidogenic proteins (Yagi et al., 2005), in support of intrinsically nonamyloidogenic SERF1a. (Closed blue circle) 100 μM aSyn in the absence of seeds.
These results indicated that SERF1a was able to directly influence aSyn amyloid fiber assembly without becoming incorporated into the amyloid fibers. To determine the specificity of this effect we tested two control proteins TraMΔN from Escherichia coli and hsp12 from Saccharomyces cerevisiae. TraMΔN represents the globular, 8.1 kDa C-terminal domain (aa 57–127) of a bacterial conjugation component (Lu et al., 2008), whereas hsp12 is a 11.7 kDa intrinsically disordered heat shock protein without any aggregation-modifying activity (Welker et al., 2010). TraMΔN or hsp12 did not significantly alter aSyn amyloid growth (Figures 2C and 2D).
SERF1a Discriminates between Amyloid and Nonamyloid Aggregation
To address whether SERF1a is active on other amyloidogenic proteins, we tested its effect on human huntingtin (htt Ex1Q53; htt gene exon 1 with a 53 glutamine repeat sequence), human Aβ40, and full-length mouse prion protein (PrP), which are related to Huntington's disease, Alzheimer's disease, and transmissible spongiform encephalopathies (prion diseases), respectively (Chiti and Dobson, 2006). In all cases, the addition of SERF1a decreased the initial lag phase of amyloid growth and accelerated the half time of conversion, thus favoring the generation of ThT-reactive species (Figures 2E–2G and Table 1).
Therefore, SERF1a is active on a broad range of structurally diverse proteins/peptides, suggesting that this protein is a specialized amyloid-promoting factor. This is supported by SERF1a being inactive to different nonamyloid aggregation processes. First, this protein was unable to rescue “off-amyloid pathway” aggregates. These are stable, alternatively folded oligomers, which are nonamyloidogenic and do not grow into mature fibers (Cappai et al., 2005; Ehrnhoefer et al., 2008; Pham et al., 2009). SERF1a failed to process representative dopamine-induced “off-pathway” aSyn oligomers (Cappai et al., 2005; Pham et al., 2009) (isolated as described in Figure S2) into amyloid fibers or to disassemble them into monomers (Figures 3A–3C).
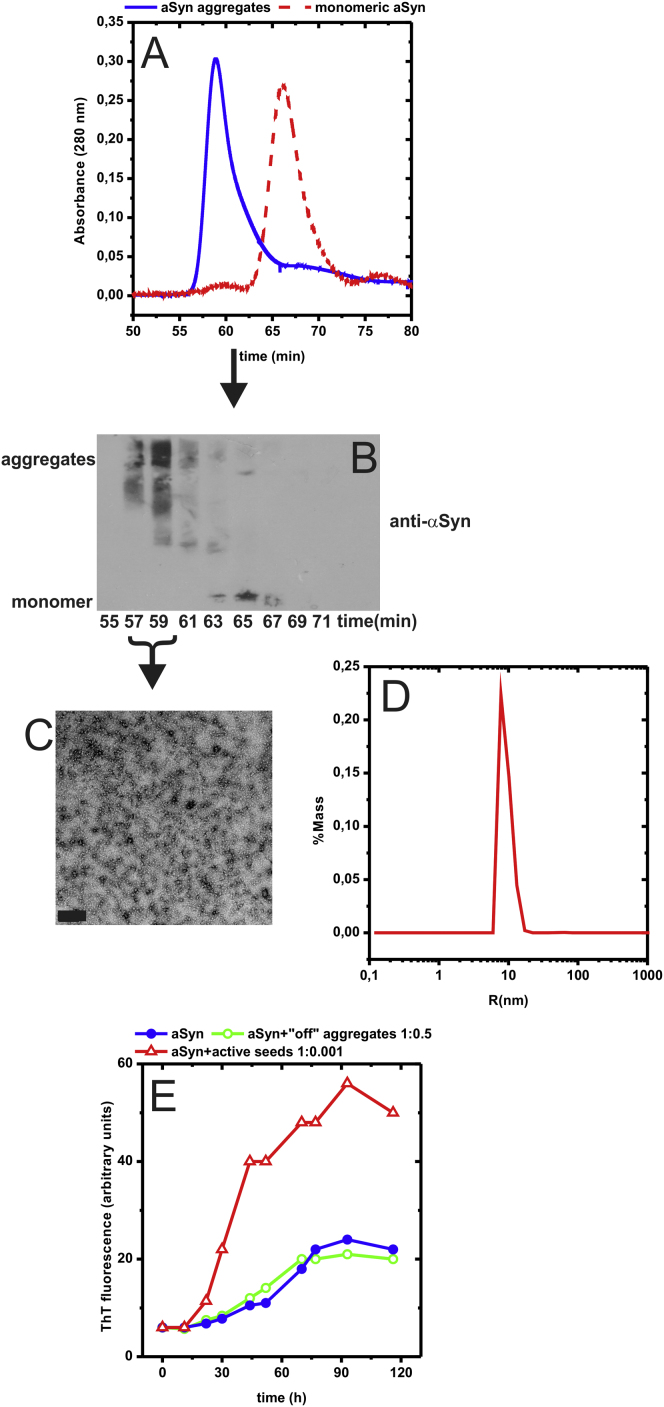
Figure S2.
Purification Scheme of Representative aSyn “Off-Pathway” Aggregates, Related to Figure 3
(A and B) The aggregates were produced by a well described reaction between aSyn and the neurotransmitter dopamine (Cappai et al., 2005; Pham et al., 2009). This small molecule induces the generation of stable, “dead-end” aggregates, which are apart from the amyloid pathway. These were purified by size exclusion chromatography, and eluted faster than monomeric aSyn (A). In agreement to literature (Cappai et al., 2005), the oligomers were SDS-resistant, as shown by SDS-PAGE/immunoblotting of the corresponding chromatographic fractions (B).
(C and D) TEM analysis (C) and dynamic light scattering (D) identified them as spheres with a diameter of approximately 20 nm.
(E) The isolated aggregates did not act as nucleation templates (E), consistent with their “off-pathway” nature. Even at a nearly stoichiometric ratio (100 μM monomeric aSyn:50 μM aggregate) they did not have any effect on aSyn amyloid growth (open green circle). As a control, little amounts of active nucleation seeds generated by ultrasonication of mature aSyn fibers, were sufficient to improve amyloidogenesis (100 μM monomeric aSyn:0.1 μM seed) (open upward red triangle).
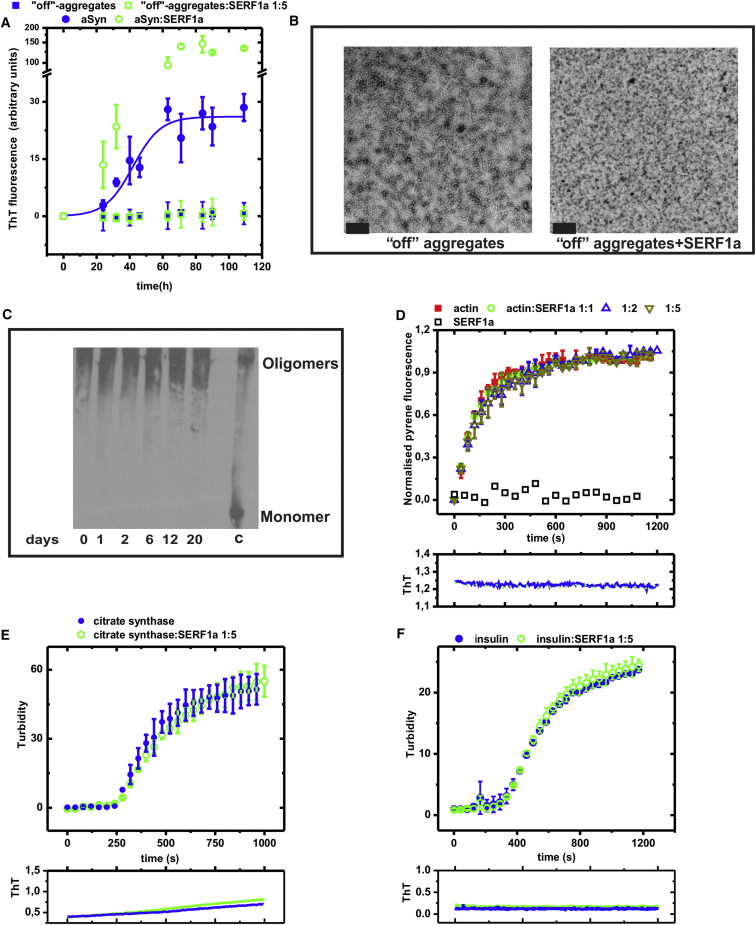
Figure 3.
SERF1a Does Not Affect Nonamyloid Protein Aggregation
(A) An excess of SERF1a (open green square) was unable to convert stable dopamine-induced “off-pathway” aSyn aggregates (closed blue square) (723 μg/ml, which is the mass equivalent of 50 μM monomeric aSyn) into amyloid fibers. As a control, the amyloid growth of 50 μM monomeric aSyn in the absence (closed blue circle) and in the presence (open green circle) of 50 μM SERF1a was measured.
(B) TEM images show no morphological difference for “off-pathways” aggregates before (left) and after agitation in the presence of SERF1a (right). Scale bars = 100 nm.
(C) SERF1a was not able to disassemble “off-pathway” aggregates into monomers: equimolar amounts of aggregates and SERF1a were incubated at 37°C; 10 μl aliquots were drawn and analyzed by SDS-PAGE and immunoblotting (anti-aSyn). SDS-resistant, high-molecular size aSyn oligomers were visible and stable over the whole experimental time range (20 days) despite the presence of SERF1a, which did not cause their disruption into monomeric aSyn. Lane c = control mixture of monomeric and oligomeric aSyn.
(D) SERF1a did not affect actin polymerization: the organized assembly of 10 μM actin, covalently attached to the fluorophore pyrene, was monitored by a polymerization-induced fluorescence change in the absence (closed red square) and in the presence of increasing concentrations of SERF1a (open blue circle, open upward green triangle, and open downward brown triangle).
(E and F) SERF1a did not influence unspecific protein aggregation: an excess of SERF1a (open green square) did not affect (E) heat-induced aggregation of citrate synthase (43°C) (closed blue circle) or (F) reductive aggregation of insulin (closed blue circle), as measured by changes of the solution's turbidity.
The lower graphs display changes in ThT fluorescence intensities for (D), (E), and (F), showing that ThT-reactive species did not accumulate during these reactions.
See also Figure S2. The error bars are mean averages, including SEM of three independent experiments.
Second, SERF1a did not influence the organized assembly of nonamyloid filaments, as it failed to affect the conversion of F-actin into G-actin (Figure 3D), despite both amyloid and actin polymerization imply nucleation, elongation, and maturation (Morris et al., 2009).
Third, SERF1a did not promote nonspecific aggregation, as shown for two conventional aggregation models. Turbidity measurements indicated that the addition of SERF1a did not alter the aggregation profile of citrate synthase (Buchner et al., 1998) (Figure 3E) and insulin (Scheibel et al., 1998) (Figure 3F). This also implied that SERF1a is not acting as a molecular chaperone, which is a protein class able to suppress nonspecific protein aggregation (Hartl et al., 2011).
These results all suggest that SERF1a has the atypical ability to specifically distinguish between amyloid and nonamyloid aggregation.
SERF1a Promotes the Generation of “On-Pathway” Aggregates
We analyzed the SERF1a-driven amyloid aggregation in more detail. We restricted our investigation to aSyn as a representative interaction partner, as this protein has a well-characterized mechanism of amyloidogenesis (Bertoncini et al., 2005; Dedmon et al., 2005; Fernández et al., 2004; Ullman et al., 2011). The interaction between SERF1a and aSyn caused a massive increase in soluble, high molecular weight aggregates. A size-exclusion chromatographic analysis showed that this type of aggregates was not detectable in the absence of SERF1a (Figure 4A). The aggregates were kinetically unstable and disappeared after a prolonged reaction time (3–4 days). Yet, after approximately 30 hr, they were sufficiently abundant and stable for their isolation (Figure S3). They consisted exclusively of ThT-reactive aSyn species (Figure S3B), and SERF1a did not coelute with them (Figures S3C and S3D), indicating that it did not stably associate with aggregating aSyn. Moreover, SERF1a did not bind to mature fibrils, as it barely colocalized into the insoluble fraction containing the aSyn aggregates (Figure 2B, lower, and Figure S1A). This is consistent with cellular studies attesting that the C. elegans ortholog MOAG-4 is excluded from the amyloid aggregates (van Ham et al., 2010).
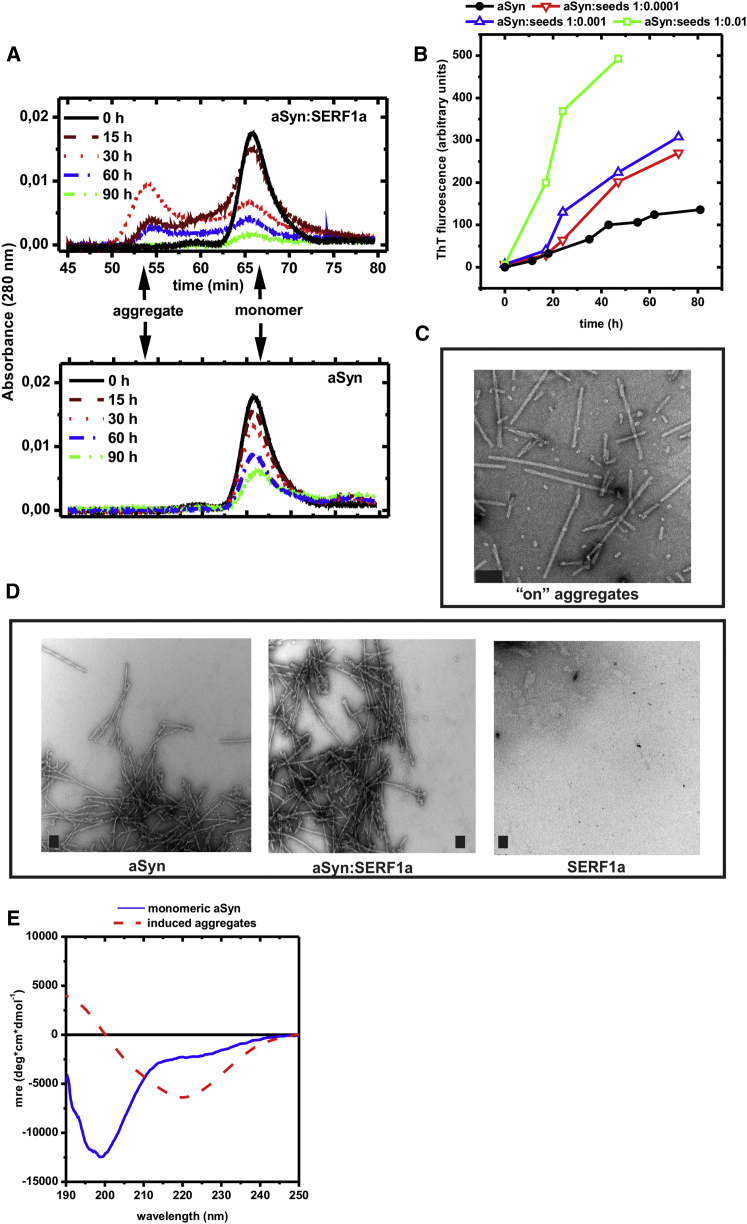
Figure 4.
SERF1a Induces Transiently Stable High Molecular Size aSyn Aggregates
(A) Size-exclusion chromatograms of soluble fractions of 150 μM aSyn, collected at different time points after agitation at 37°C, 1,400 rpm in the presence (upper) and in the absence (lower) of 100 μM SERF1a. Centrifuged samples were run on a Superdex 75 column, equilibrated with 50 mM Tris and 150 mM NaCl (pH 7.4) at 1 ml/min flow rate. SERF1a did not contribute to the absorbance signal, due to the lack of aromatic residues, and it was not present in the aggregate fraction (Figure S3C). Upper: SERF1a-induced aSyn aggregates accumulated over time, in concomitance to a decrease in monomeric aSyn, with a maximum accumulation around 30–40 hr (red dotted curve). After this time point, aggregates decreased in parallel to monomeric aSyn, indicating their consumption into unsoluble fibers (see also Figure 2B). Lower: In the absence of SERF1a, high molecular size aggregates were virtually undetectable, and the consumption of soluble monomeric aSyn was slower and less quantitative.
(B) The isolated aggregates acted as nucleation templates for aSyn fibrils: seeding a 100 μM (1.46 mg/ml) aSyn solution with different ratios of these aggregates caused an acceleration of fiber formation (closed black circle, open green square, open upward blue triangle, and open downward red triangle).
(C) Transmission electron micrograph of the isolated aSyn intermediates display a morphology consisting of small dots of 20–30 nm diameter and short rods of varying length (50–400 nm). Scale bars = 100 nm.
(D) Transmission electron micrographs of mature aSyn amyloid fibers obtained in the absence (left) and in the presence (middle) of SERF1a showed no morphological difference. SERF1a alone did not aggregate (right). Scale bars = 200 nm.
(E) Far-UV CD-spectrum of monomeric aSyn (solid blue line) and of soluble aSyn aggregates (solid red line). Although the spectrum of monomeric aSyn is typically unstructured (lack of predominant alpha helix or beta sheet signal, strong negative signal around 200 nm, and a slow positive signal recovery below 200 nm), the intermediate aggregates are structured, as shown by a prominent signal decrease around 215–220 nm and by a positive signal increase below 200 nm.
Signal deconvolution analysis (Whitmore and Wallace, 2008; Sreerama et al., 1999) yielded 9.9% alpha helix, 34.5% beta-sheet, and 56% random coil for the aSyn aggregates versus 6.4% alpha helix, 9.8% beta sheet, 82.9% random coil for monomeric aSyn, attesting a predominant structural rearrangement into beta-sheets. See also Figure S3.
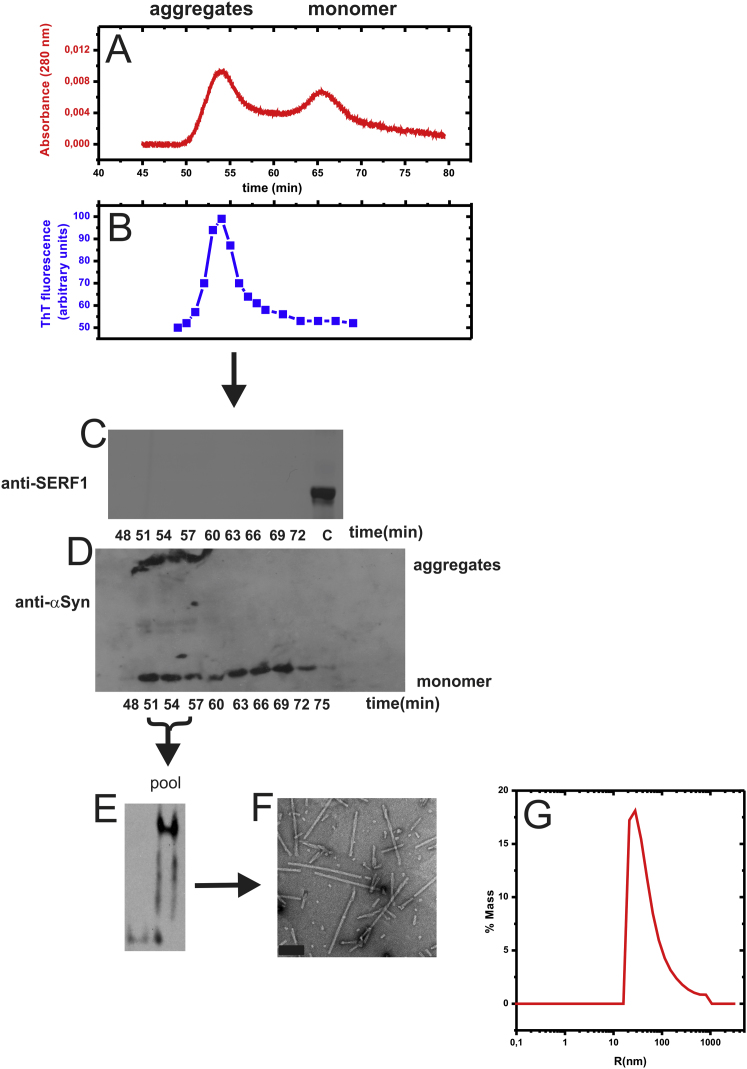
Figure S3.
Purification Scheme of SERF1a-Induced “On-Pathway” aSyn Aggregates, Related to Figure 4
(A) after approx. 30 hr agitation at 1400 rpm and 37°C, aggregates were isolated by size exclusion chromatography (see experimental procedures), where they eluted as a high molecular size peak.
(B) The fractions corresponding to this peak (50-60) were ThT-reactive (Ex/Em 442/482 nm, slit widths 20/20 nm).
(C–E) The aggregates did not contain SERF1a, as shown by SDS-PAGE/immunoblotting (c = SERF1a control), and they were composed exclusively of aSyn (D). They ran as a mixture of high molecular weight and monomeric species on SDS-PAGE, but as aggregates-only on a native gel (E), attesting their partial resistance to SDS.
(F) TEM image of the pooled aggregate fractions.
(G) Dynamic light scattering identified the aggregates as a polydisperse mixture of particles with a diameter between 20 and ≥ 400 nm.
The isolated aggregates acted as dose-dependent nucleation templates (“on-pathway” aggregates), because the conversion of monomeric aSyn into amyloid was significantly enhanced as the SERF1a:aSyn ratio was increased, and the initial lag phase was visibly reduced (Figure 4B). Transmission electron micrographs identified these aggregates as single dots and short-sized rods with a maximal length of 400 nm (Figure 4C). They were by far shorter than the tangles of mature amyloid fibrils (≥1 μM, Figure 4D), yet ThT-reactive (Figure S3B), and predominantly beta-sheet structured (Figure 4E), thus displaying peculiarities of elongating amyloid fibers. Their morphology was also completely different from that of the dopamine-stabilized “dead-end” aggregates (Figure 3B), which instead formed small spheres with a diameter of approximately 20 nm.
Such a measurable accumulation of transient “on-pathway” aggregates is significant, because these species are usually not abundant owing to their rapid conversion into mature fibers (Kim et al., 2009; Lashuel et al., 2002) and are therefore hard to capture.
This data led us to propose that the interaction between SERF1a and aSyn was transiently restricted to the initial stages of amyloidogenesis, serving to accelerate the generation of active, elongating amyloid intermediates (“on-pathway” aggregates), which drive the amyloid reaction as they are incorporated into the growing fibrils. Such an early effect was supported by SERF1a showing little influence on seeded (Figure 2I) compared to nonseeded amyloid growth (Figure 2A), suggesting a mode of action which preceded the preamyloid nucleation phase. This was reinforced by the ability of substoichiometric levels of SERF1a to still promote amyloidogenesis (Figure 2H), in support of a transient rather than a stable stoichiometric association. This finding is consistent with the effect of MOAG-4 on polyglutamine aggregation in cells (van Ham et al., 2010).
SERF1a Interacts with the C Terminus of α-Synuclein
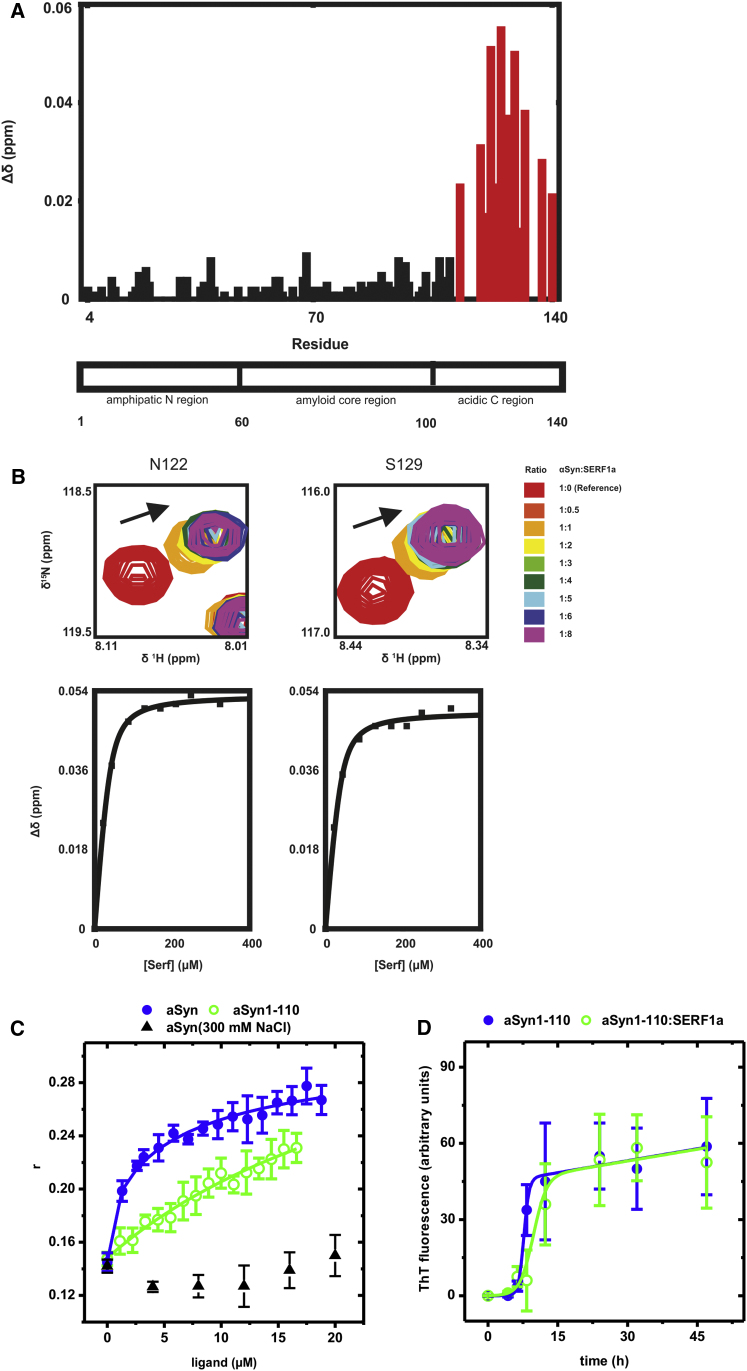
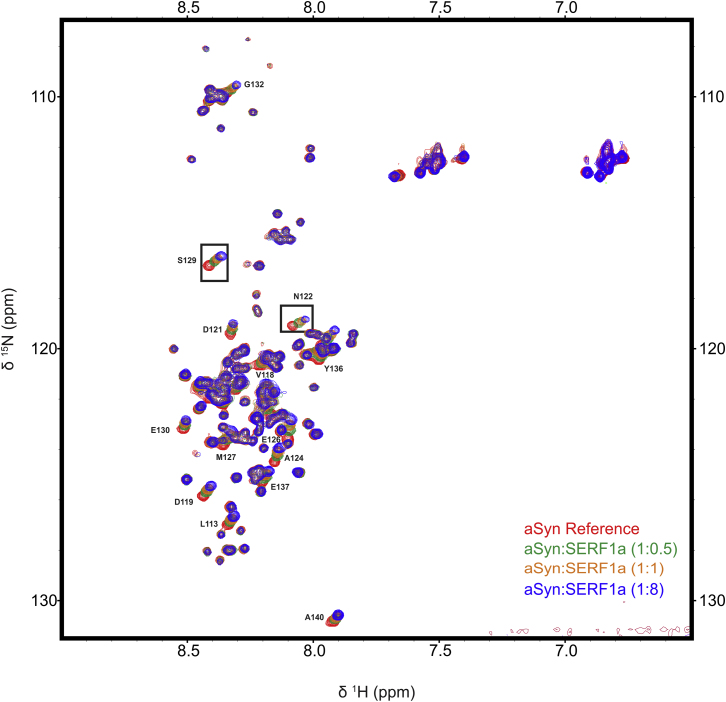
To define more accurately the details of this early and transient relationship, we investigated the mode of interaction between SERF1a and monomeric aSyn. We used 1H, 15N-HSQC NMR (heteronuclear single quantum coherence nuclear magnetic resonance) spectroscopy, because aSyn has been extensively studied by this technique (Chandra et al., 2003; Fernández et al., 2004; Rasia et al., 2005; Wang et al., 2011). The stepwise addition of SERF1a to a uniformly 15N-labeled monomeric aSyn sample caused substantial and saturable shift perturbations of the amino acid backbone signals located between C-terminal aSyn residues Gly111 and Ala140 (Figure 5A and Figure S4). Binding curves obtained from chemical shift titrations of the two well-resolved amino acids Asp122 and Ser129 yielded a dissociation constant for the interaction KD ∼7–10 μM (Figure 5B). This indicated that the interaction with SERF1a was localized to the acidic C-terminal region of aSyn.
Figure 5.
SERF1a Binds to the C-Terminal Region of aSyn
(A) aSyn 1H,15N-HSQC-NMR perturbation diagram showing chemical shift changes Δδ after the addition of SERF1a. The largest changes were localized to the C-terminal region, between the amino acids Gly111-Ala140 (red columns).
(B) Upper: Detail view of well-resolved and large chemical shift changes that occur for amino acid signals Asp122 and Ser129, upon titration with SERF1a (arrows). Lower panels: The corresponding binding curves yielded dissociation constants KD = 7.59 ± 1.87 μM (Asp122) and 9.81 ± 1.06 μM (Ser129).
(C) Fluorescence anisotropy binding curves of Atto550-SERF1a upon titration with aSyn (closed blue circle; KD = 8.04 ± 2.09 μM) and aSyn1-110 (open green circle; nonsaturable binding, extrapolated KD > 1 mM). The interaction between Atto550-SERF1a with aSyn was abolished by increasing the ionic strength of the solution to 300 mM NaCl (closed upward black triangle), pointing to a predominantly hydrophilic binding mode.
(D) Amyloid fiber growth of the truncation mutant aSyn1-110 was insensitive to SERF1a: amyloid kinetics of aSyn1-110 in the absence (closed blue circle) and in the presence (open green circle) of SERF1a. Note that this truncation intrinsically improves the amyloid properties of aSyn (tm = 7.70 ± 0.18 hr; tl = 6.56 ± 0.83 hr; kapp = 1.75 ± 0.17 hr−; see also Hoyer et al., 2004 and Levitan et al., 2011).
See also Figure S4. The error bars are mean averages, including SEM of three independent experiments.
Figure S4.
1H,15N-HSQC NMR Titration Spectra of Alpha-Synuclein with SERF1a, Related to Figure 5
Overlay of four representative aSyn 1H,15N-HSQC NMR spectra recorded in the presence of increasing ratios of SERF1a. Amino acid residues with well resolved chemical shift perturbations are labeled. Chemical shift titrations of the two residues N122 and S129 are shown in detail in Figure 5B.
This binding mode was supported by fluorescence anisotropy, a technique that can measure changes in the rotational mobility of a fluorophore covalently attached to a protein upon complex formation (Lakowicz, 1999). The titration of site-specific fluorescence-labeled SERF1a with unlabeled full-length aSyn, led to a saturable increase of the anisotropy value r (Figure 5C). The resulting binding curve yielded KD = 8.04 ± 2.09 μM, in good agreement to the value previously obtained by NMR spectroscopy (Figure 5B). In contrast, the interaction was substantially altered for aSyn1-110, a truncation mutant lacking Gly111-Ala140, and binding to this mutant was no longer saturable (extrapolated KD value >1 mM) (Figure 5C).
According to current mechanistic models of aSyn amyloidogenesis, generic perturbations of the C-terminal region can expose the otherwise more protected amyloid core region, thereby promoting fibril formation (Bertoncini et al., 2005; Dedmon et al., 2005; Hoyer et al., 2004; Levitan et al., 2011; McClendon et al., 2009; Murray et al., 2003). This effect has been previously observed for charged ligands, such as metal ions or polyamines (Fernández et al., 2004; Nielsen et al., 2001; Rasia et al., 2005). They all bound to the C-terminal region (yet with a much weaker binding affinity than SERF1a: KD ∼7–10 μM for SERF1a versus several hundred micromolar for the other ligands) and thereby affected aSyn aggregation. We determined if this also applied to SERF1a and found that this protein was no longer able to influence fiber growth of the C-terminal truncation mutant aSyn1−110 (Figure 5D), which is in clear-cut contrast to full-length aSyn (Figure 2A). Thus, there existed a clear relationship between the amyloid-modifying activity of SERF1a and its binding to the C-terminal region of aSyn.
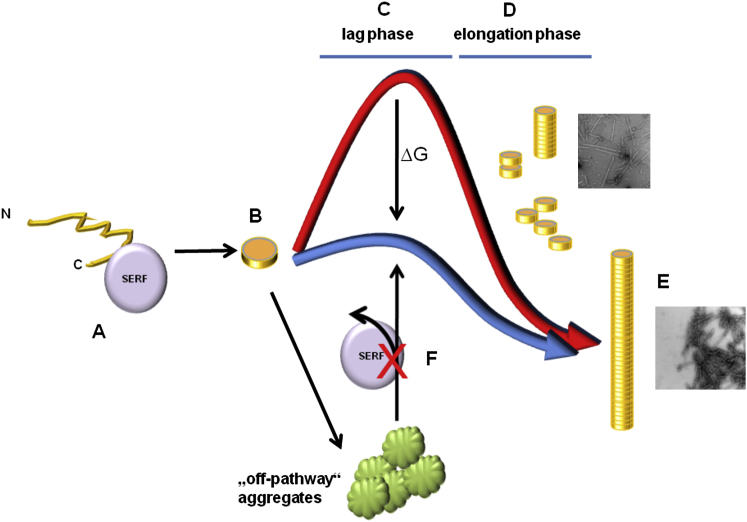
Based on these results, we have set up a model (Figure 6) wherein SERF1a promotes aSyn amyloid growth by directly interacting with the C-terminal region of the monomeric protein (Figure 6A). Such an early mode of action precedes the preamyloid nucleation phase (Figure 6B). Consequently, SERF1a promotes the “on-pathway” transition from a lag phase (Figure 6C) to amyloid fiber elongation (Figure 6D) and finally to mature amyloids (Figure 6E), while disfavoring “off-pathway” aggregation (Figure 6F).
Figure 6.
A Model for the Direct Amyloid-Modifying Ability of SERF1a
As illustrated for the amyloidogenic-representative aSyn, SERF1a binds directly to this protein (A), influencing early stages of amyloidogenesis (B). By facilitating the transition between lag phase (C) and elongation phase (D), the conversion of aSyn monomers into amyloid fibers (E) becomes improved, whereas nonamyloid aggregation remains unaffected (F).
Discussion
Our results provide clear evidence that the amyloid-promoting activity of SERF1a is driven by a transient and selective interaction with early precursors in the amyloid pathway. The selectivity of SERF1a for amyloid aggregation distinguishes this protein from other general modifiers of protein aggregation and explains the unique chaperone-, proteasome-, and autophagy-independent properties of the SERF proteins observed in cellular systems (van Ham et al., 2010).
According to our model, SERF1a binds transiently to the C-terminal region of aSyn. Perturbations within this region are reported to cause a stable exposure of the amyloid core, which is located in the middle region of the protein and is otherwise protected by intramolecular long-range interactions (Bertoncini et al., 2005; Dedmon et al., 2005; Hoyer et al., 2004; Levitan et al., 2011; McClendon et al., 2009; Murray et al., 2003). As a consequence, aSyn amyloid precursors can generate.
The C-terminal region is composed predominantly of polar and negatively charged amino acids and is therefore a reasonable interaction target for a positively charged protein, such as SERF1a (pI = 10.44). In accordance, a higher ionic strength (300 mM) abolishes this interaction. Other positively charged ligands, such as polyamines and divalent cations, bind to this region as well, but by orders of magnitude weaker than SERF1a (Fernández et al., 2004; Nielsen et al., 2001), in support of a more specific role of the latter.
The direct interaction of SERF1a with the C-terminal region of aSyn causes a massive increase in nucleation-active amyloid intermediates. Given that aggregating aSyn no longer interacts with SERF1a, we suppose that this initial short-lived association is sufficient to lower the thermodynamic barriers of amyloid transition by reducing the lag phase and thus leading to an earlier elongation phase. This is shown by the massive increase of “on-pathway” amyloid intermediates, which are able to autocatalytically assemble into mature fibers. We therefore propose that SERF1a directly affects aSyn amyloidogenesis by catalyzing the transition into amyloid-nucleating species, away from concurring “dead-end” folding events.
The general validity of this particular charge-driven interaction model remains to be demonstrated for other structurally diverse amyloidogenic proteins. Likewise, the representatives used in this study (aSyn, Aβ, htt, and PrP) do not share any charged sequence similarity. Yet, the ability by which SERF1a manages to collectively influence the early amyloid growth of these proteins (i.e., by generally decreasing the lag time of amyloid growth and by accelerating the half time of conversion, see Figure 2) leads to hypothesize the existence of mechanistically related processes that remain to be addressed in detail.
The inactivity of SERF1a on nonamyloid aggregation is in clear-cut contrast to general modifiers of protein aggregation, such as molecular chaperones, the autophagy pathway or the proteasome. This identifies SERF1as a specialized amyloid factor and supports its autonomous mode of action observed in cells (van Ham et al., 2010).
Experimental Procedures
Unless otherwise specified, all reagents were from Sigma-Aldrich, Vienna, Austria.
Protein Purification
Proteins were purified as described in the Extended Experimental Procedures.
Extended Experimental Procedures.
Protein Purification
Human SERF1a: Short Isoform of SERF1
the pFL-B02cl expression plasmid carrying the human SERF1A gene (GenBank accession number BC021174) was purchased from SourceBiosciences Lifesciences (Nottingham, UK) (clone no. EX-S0127-B2). The vector was transformed into E. coli BL21(De3)Star cells (Invitrogen), and streaked on a 100 μg/ml Ampicillin plate. One colony was used to inoculate an overnight LB/Amp culture (37°C, 150 rpm). After 16 hr, the overnight culture was diluted into pre-conditioned LB/Amp medium, and grown to an O.D. of 0.7-0.9 (37°C, 160 rpm). The gene expression was initiated by the addition of 1mM IPTG (Roth), and cells were harvested after 4 hr, and frozen at −80°C.
For protein purification, cell pellets from one liter culture were resuspended in 30 ml buffer containing 50 mM Tris, 800 mM NaCl, 3 mM NaN3, pH 7.4 supplemented with protease inhibitor cocktail tablets (Roche, Penzberg, Germany). The suspension was homogenized using an ultraturrax, cells were disrupted by sonification, centrifugation at 20,000 g for 40 min and then diluted to 300 ml with 50 mM Tris, 50 mM NaCl, 3 mM NaN3, pH 7.4 (buffer A). The supernatant was subjected to heat (80°C, 10 min.) under continuous stirring. During this step, most proteins precipitated, while SERF1a remained in the soluble fraction. Unsoluble material was separated by centrifugation (20,000 g, 40 min). The clear solution was loaded on a SP-Sepharose column (GE Life Sciences), which had been pre-equilibrated with buffer A. After the flow-through had passed the column, SERF1a was eluted by using a 50 mM to 1 M NaCl gradient, and collected over 90 min at a 1 ml/min flow rate. The protein eluted around 600 mM NaCl. As SERF1a does not contain any aromatic amino acid residues, the fractions containing the protein were identified by SDS-PAGE followed by Coomassie staining.
The collected fractions were loaded on a Superdex75 XK26/100 column (GE Life Sciences), and eluted with H2O at a flow rate of 2 ml/min. Fractions containing SERF1a were pooled, and the protein concentration was measured by using the BCA assay (Thermo Scientific, Rockford, IL). The protein was insensitive to Bradford Reagent. The identity of the purified protein was verified by immunoblotting and by mass spectrometry. The protein solution was finally aliquoted, and dry-frozen.
A sample of the reconstituted protein displayed the same NMR spectroscopic properties as non dry-frozen protein, purified without heat-precipitation, indicating that none of these steps had affected the structure of SERF1a (data not shown).
15N-labeled SERF1a was produced as above, except for growing transformed E. coli BL21(De3)Star cells in minimal medium (6.8g/l Na2HPO4, 3 g/l KH2PO4, 0.5 g/l NaCl, 1.5 g/l (15NH4)2SO4, 2 g/l glucose, 1 μg/l biotin, 1 μg/l thiamin, 100 μg/ml ampicillin and 1ml 1000x microsalts) instead of LB medium.
Atto550-Labeled SERF1a
a N- and C-terminal double cysteine SERF1a mutant (Cys-SERF1a-Cys) was engineered by standard PCR using the primers CACCATGTGTGCCCGTGGAAAT (fwd), and TCAACACTTTTCTCTTGTCTGCATAG (rev), and the pFL-B02cl plasmid carrying the human SERF1A gene as a template, and subsequently cloned into a pET101D expression vector by TOPO directional cloning (Invitrogen) according to the manufacturer's instructions. Bacterial growth, protein expression, and purification was same as for wild-type SERF1a, except that 1 mM DTT was added to the cation exchange buffer.
Site-specific N- and C-terminal fluorescence end-coupling of the reduced cysteine sites with the dye Atto550 maleimide (Atto-Tech, Siegen, Germany) was performed as specified by the manufacturer. In brief, lyophilized Cys-SERF1a-Cys was reconstituted in PBS, 12 mM TCEP, to a concentration of 10mg/ml. A DMF solution of Atto550 maleimide was added to a final concentration of 3 mM, the mixture was incubated 2 hr in the dark, and the reaction was terminated by the addition of freshly prepared GSH. The conjugated protein was purified on a Sephadex G-25 spin column (GE life sciences), dialyzed against water, lyophilised and stored at −80°C. The degree of labeling was 1.9, indicating that SERF1a was doubly labeled (N and C-terminally; SERF1a does not contain sequence specific cysteines).
Human α-synuclein was purified as described in Falsone et al.,2011. In brief, the expression plasmid carrying the SNCA gene was transformed into E. coli BL21(DE3)Star, and expressed by the addition of 1mM IPTG in LB/Amp culture medium (minimal medium for 15N-aSyn). After cell disruption, the supernatant was subjected to acid precipitation, centrifuged again, and aSyn was separated on a SourceQ column (GE Life Sciences), followed by a gel filtration step in H2O. The purified samples were aliquoted, and dry-frozen.
α-synuclein1-110
the truncation mutant lacking the C-terminal amino acids 111-140 was engineered by standard PCR using the primers CACCATGGATGTATTCATGAAA (fwd), and CTATTCCTGTGGGGCTCCTT (rev), and the SNCA gene as a template, and subsequently cloned into a pET101D expression vector by TOPO directional cloning (Invitrogen) according to the manufacturer's instructions. The vector was transformed into E. coli BL21(De3)Star cells (Invitrogen). Bacterial growth and protein expression were same as for wild-type aSyn. The purification was performed as described in Hoyer et al. (2004).
Human Huntingtin Q53Ex1, is the translational product of the HTT gene exon 1, containing a repetitive sequence of 53 glutamines (plasmid provided by Paul Muchowski, The Gladstone Institute, San Francisco, CA). The GST-fusion protein was purified according to Wacker et al. (2004). Briefly, The pellet from 1 l bacterial culture was resuspended in 50 mM KPi, 150 mM KCl, 1mM EDTA, pH 8.0, supplemented with protease inhibitor tablets, sonicated, and purified on GST-sepharose column (GE Lifesciences), followed by a gel filtration step on a Superdex 75 column equilibrated with 50mM Tris-HCl, 150mM NaCl, 1mM DTT, pH 7.0. The protein was concentrated to no more that 120 μM, and the concentration was measured by the Bradford assay (Bio-Rad, Hercules, CA).
The purified protein was stable only for a few days without any particular additives. It was therefore freshly prepared before each measurement, and used immediately after the purification.
Human Aβ40 was synthesized and purified by James I. Elliott (Yale University, New Haven, CT). Working samples were prepared as described in the experimental procedures under the subsection “kinetics of fiber growth.”
Mouse PrP (amino acids 23-230) was expressed in BL21 (DE3) according to previously reported methods (Coleman et al., 2009). In short, bacterial cells from 1.5 l of culture were lysed in 50 ml of washing buffer (10 mM Tris, pH 8.0) containing 20 U Benzonase, 500μl Protease Inhibitor cocktail and lysozyme (0.5 mg/ml) for 1 hr, then supplemented with 1% Nonidet P-40 substitute for further 20 min. incubation. Inclusion bodies were collected by centrifugation at 6,000 g for 15 min and resuspended and incubated in washing buffer containing 0.5 mg/ml lysozyme for 30 min. Inclusion bodies were pelleted by centrifugation at 6,000 g for 15 min and washed twice by resuspension in washing buffer. Inclusion bodies were solubilised by vigorous shaking, for 2 hr in 10ml of 8 M urea, 10 mM Tris, 10 mM Na2HPO4 10 mM reduced glutathione, pH 8.0) and clarified by centrifugation at 30,000 g for 15 min. The supernatant was passed through a Ni(II) bound 5ml HisTrap column (GE Healthcare), and eluted at low pH (pH 4.5). The PrP containing solution was desalted into 6 M urea, 10 mM Tris, pH 8.0 by Vivaspin-20 column, MWCO 10,000 Da (Sartorius, Göttingen, Germany) and was oxidised overnight, by the addition of 0.2 mM oxidised glutathione and 5 mM EGTA, at a protein concentration ≤ 0.4 mg/ml. Oxidised PrP protein was purified by reverse phase high pressure liquid chromatography using a Jupiter C4 (Phenomenex, Torrance, CA) column. The oxidised PrP fraction was lyophilised, and verified for purity and correct folding using SDS-PAGE and circular dichroism spectroscopy.
TraMΔN (amino acids 57-127) was purified as described by Lu et al., 2008.
Hsp12 was provided by Johannes Buchner (TU München, Munich, Germany).
NMR Spectroscopy
1H,15N-HSQC NMR titrations were carried out at 25°C on a Bruker Avance III (Bruker BioSpin, Karlsruhe, Germany) 700 MHz spectrometer equipped with a CTI cryoprobe head. A uniformly labeled 15N-aSyn solution and an unlabeled SERF1a solution were dialyzed overnight against 50 mM bis-Tris, 20 mM NaCl, and 3 mM NaN3 (pH 6.8). A 40 μM 15N-aSyn solution was titrated with increasing amounts of unlabeled SERF1a. Spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed with NMRview (Johnson and Blevins, 1994) or Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). Previously assigned aSyn backbone resonances were used (Falsone et al., 2009). Binding curves obtained from chemical shift titrations were fitted to the equation
| (Equation 1) |
where Δδobs is the observed chemical shift perturbation, Δδmax is the maximal chemical shift perturbation, and KD is the dissociation constant. 1H,15N-HSQC spectra of 15N-SERF1a were acquired at 25°C on a Varian Unity INOVA 600 MHz NMR spectrometer (Varian, Palo Alto, CA, USA) using an HCN triple-resonance probe with single axis z-gradients. Data were processed using NMRPipe (Delaglio et al., 1995) and analyzed in NMRView (Johnson and Blevins, 1994). Data were collected using samples consisting of 100 μM 15N-labeled protein in 50 mM NaPi, 50 mM NaCl, and 3 mM NaN3 (pH 6.5). 10% D2O were added for field-frequency locking.
Fluorescence Anisotropy
The fluorescence anisotropy of a 100 nM Atto550-SERF1a preparation in 50 mM bis-Tris, 20 mM NaCl, and 3 mM NaN3 (pH 6.8) was measured at 25°C on a LB50 spectrofluorimeter equipped with excitation and emission polarisers (PerkinElmer, Waltham, MA, USA), at an emission wavelength of 576 nm upon excitation at 554 nm. Slit widths were 15 and 20 nm for excitation and emission, respectively. The fluorescence anisotropy is defined as in Lakowicz (1999):
| (Equation 2) |
where IVV is the fluorescence intensity recorded with excitation and emission polarizers in vertical positions, and IVH is the fluorescence intensity recorded with the emission polarizer aligned in a horizontal position. The G factor is the ratio of the sensitivities of the detection system for vertically and horizontally polarized light G = IHV/IHH.
The Atto550-SERF1a solution was titrated against increasing amounts of aSyn or aSyn1-110 diluted in the same buffer. For each point, the anisotropy was recorded over 30 s and the mean r values for each measurement were used. Anisotropy changes were fitted by using the Levenberg-Marquardt algorithm to the equation (Falsone et al., 2009)
| (Equation 3) |
where r is the observed anisotropy, Δrmax is the maximal anisotropy change, and KD is the dissociation constant.
Circular Dichroism
SERF1a
Far-UV-circular dichroism spectra of 10 μM SERF1a in 20 mM NaPi and 50 mM NaF (pH 7.0) were recorded at 25°C on a J-715 spectropolarimeter (Jasco, Tokyo, Japan) with a response time of 4 s and with a data point resolution of 0.2 nm using a 1 mm quartz cuvette. Three scans were averaged to obtain smooth spectra.
aSyn
Far-UV-circular dichroism spectra of 10 μM aSyn or soluble aSyn aggregates in 10 mM NaPi and 20 mM NaF (pH 7.4) were recorded at 25°C on a J-715 spectropolarimeter (Jasco) with a response time of 4 s and with a data point resolution of 0.2 nm using a 1 mm quartz cuvette. Three scans were averaged to obtain smooth spectra.
Mean residue ellipticity calculations and secondary structure deconvolution analysis were performed on the DICHROWEB platform (Whitmore and Wallace, 2008) by applying the SELCON3 algorithm (Sreerama et al., 1999).
Dynamic Light Scattering
The Stokes radii of 5 mg/ml SERF1a or hen egg white lysozyme in 20 mM Tris and 150 mM NaCl (pH 7.4) were determined using a DLS equipment composed of a goniometer and a diode laser (Coherent Verdi V5, λ = 532 nm, Pmax = 5 W) with single-mode fiber detection optics (OZ from GMP, Zürich, Switzerland), an ALV/SO-SIPD/DUAL photomultiplier with pseudo-cross-correlation and an ALV 5000/E correlator with fast expansion (ALV, Langen, Germany). Measurements were carried out at a scattering angle of 90°. Correlation functions were collected for 60 s ten times and then averaged. From these functions, the average diffusion coefficient D was obtained by cumulant analysis (Koppel, 1972).
Samples of oligomeric aSyn in 20 mM Tris, 150 mM NaCl, and 3 mM NaN3 (pH 7.4) were measured on a DynaPro instrument (Protein Solutions, Lakewood, NJ, USA) in a 1.5 mm pathlength 45 μl quartz cuvette at 25°C. Samples were centrifuged before measuring.
Small Angle Light Scattering
SAXS equipment comprised a SAXSess camera (Anton-Paar, Graz, Austria) with high flux and low background, connected to an X-ray generator (Philips, PW1730/10) operating at 40 kV and 50 mA with a sealed-tube Cu anode. A Göbel mirror was used to convert the divergent polychromatic X-ray beam into a focused line-shaped beam of Cu Kα radiation (wavelength λ = 0.154 nm). The two-dimensional scattering pattern was recorded by a PI-SCX-fused fiber optic taper CCD camera from Princeton Instruments (Trenton, NJ, USA) and integrated into the one-dimensional scattering function I(q), where q is the length of the scattering vector, defined by q = (4π/λ)sin(θ/2), λ being the wavelength and θ being the scattering angle. The CCD detector used features a 2,084 × 2,084 array with 24 × 24 μm pixel size (chip size: 50 × 50 mm). The CCD is operated at −30°C with 10°C water-assisted cooling to reduce the thermally generated charge. Cosmic ray correction and background subtraction were performed on the two-dimensional image before further data processing. The temperature of the capillary and the metallic sample holder was controlled by a Peltier element.
Transmission Electron Microscopy
A protein suspension was placed onto carbon-coated grids, blot dried, and rinsed in distilled water for 1 min. Negative staining was performed by applying1% uranylaceate for 1 min and blotting the liquid with filter paper. Afterward the samples were air-dried and viewed with a Fei Tecnai 20 transmission electron microscope (Fei, Hillsbro, OR, USA) operated at an acceleration voltage of 120 kV. Images were recorded on a Gatan US1000 digital camera (Gatan, Pleasanton, CA, USA).
Kinetics of Amyloid Fiber Growth
General Settings
All ThT fluorescence measurements were done in triplicate on a Cary Eclipse fluorescence spectrometer (Agilent, Santa Clara, CA, USA) at 25°C and 482 nm upon excitation at 442 nm. Unless otherwise described, the photomultiplier voltage was 600 V. The slit widths were set as specified separately. The pH was controlled at the beginning and at the end of each reaction to exclude artificial fiber growth effects. Except for htt, a manual discontinuous setup was used for all measurements. As a standard procedure, all buffers were filtered through a 0.45 μm filter membrane, and all samples were centrifuged (13,000 g; 15 min; 4°C) before use.
The amyloid growth curves were fitted by using the Levenberg-Marquardt algorithm to the equation (Uversky et al., 2001)
| (Equation 4) |
where Fi is the initial fluorescence value, Ff is the final fluorescence value, tm is the midpoint of amyloid conversion, and mi and mf are correction values for the lag phase and the postgrowth fluorescence. The initial lag time tl corresponds to tm - 2/kapp, where kapp is the apparent rate constant for amyloid growth. The values are mean averages, including SEM of three independent measurements.
aSyn; aSyn1-110
Lyophilized protein was reconstituted in 50 mM TrisHCl, 150 mM NaCl, and 3 mM NaN3 (pH 7.4) (aSyn-working buffer) to a concentration of approximately 1 mM. A preparative gel filtration chromatography step (self-packed Superdex 75 column [GE Healthcare, Uppsala, Sweden]; 500 ml bed volume; flow rate 2 ml/min; elution buffer = aSyn-working buffer) was run to remove possible aggregates and degradation products that might artificially influence amyloid growth kinetics. The fractions corresponding to monomeric protein were pooled and concentrated in an Amicon Ultra-15 centrifugal filter unit with a 3 kDa cutoff (Merck Millipore, Billerica, MA, USA). Unless otherwise described, this solution was adjusted to 200 μM with aSyn-working buffer. An additional gel filtration chromatography analysis (Superdex 75 10/300; GE-Healthcare; flow rate 0.7 ml/min) was performed to confirm that aSyn was still homogenous and monomeric after the concentration procedure. A 100 μl volume of this same solution was mixed in a clean 1.5 ml Eppendorf plastic vial with 100 μl of a 200 μM SERF1a solution previously dialyzed against aSyn-working buffer, or with 100 μl working buffer alone, to a final volume of 200 μl. The vials were sealed with Parafilm, taped to the rack of an Eppendorf thermomixer 5436 (Eppendorf, Hamburg, Germany) to prevent their rotation during agitation, and shaken at 1,400 rpm, 37°C. Aliquots of 5 μl were taken at the indicated time points and diluted into 1 ml of a 5 μM ThT solution freshly prepared in aSyn-working buffer, and the ThT fluorescence of the samples was measured in a 1 ml quartz cuvette at room temperature. Ex/em slit widths = 10/10 nm.
GST-httQ53Ex1
Monomeric htt protein was freshly purified as described in Extended Experimental Procedures and used immediately. In a clean 1 ml quartz cuvette, 100 μl of a freshly prepared 120 μM htt solution in 50 mM Tris-HCl, 150 mM NaCl, and 1 mM DTT (pH 7.0) (htt-working buffer) was mixed with 100 μl of a 120 μM SERF1a solution that had been previously dialyzed against htt-working buffer, or with 100 μl htt-working buffer, to a volume of 200 μl. To this solution, 800 μl of a 5 μM ThT solution freshly prepared in htt-working buffer was added to a final volume of 1 ml. The cuvette was placed into a preheated (37°C) sample cell holder of a fluorescence spectrometer, and amyloid growth was initiated in situ by cleaving the GST tag with 10 μl PreScission protease (500 u; Invitrogen, Carlsbad, CA, USA). The cuvette was capped, and ThT fluorescence was measured with a continuous setup at 37°C without agitation, with a time step resolution of 2 min, and compared to uncleaved controls. Ex/em slit widths = 20/20 nm.
Aβ40
Lyophilized Aβ40 peptide was dissolved in hexafluoroisopropanol (HFIP; Fluka) to a concentration of 2 mg/ml, divided into 250 μg aliquots in 1.5 ml Eppendorf plastic vials, and HFIP was evaporated on a vacuum centrifuge. One aliquot was reconstituted in 250 μl milliQ H2O, dissolved by sonication for 10 min in an ice-cooled ultrasound water bath, centrifuged (13,000 rpm, 10 min), and the concentration of the supernatant was measured on a spectrophotometer at 214 nm absorbance wavelength (ε = 55,771 M−1 cm−1).
In a clean 1.5 ml Eppendorf plastic vial, 100 μl of a 200 μM Aμ40 solution was prepared in H2O and adjusted to 1× PBS / 3 mM NaN3 (Aβ-working buffer) by the addition of 10× PBS. To this solution, 100 μl of a 200 μM SERF1a solution prepared in Aβ-working buffer, or 100 μl Aβ-working buffer alone was added to a final volume of 200 μl. The plastic vials were sealed with Parafilm, taped to the rack of an Eppendorf thermomixer 5436 (Eppendorf), and agitated at 37°C and 300 rpm. Aliquots of 10 μl were taken at the indicated time points, diluted into 100 μl of a 20 μM ThT solution freshly prepared in Aβ-working buffer, and incubated for 5 min, and the fluorescence of the sample was measured in a 150 μl quartz cuvette at room temperature, with a photomultiplier setting of 650 V. Ex/em slit widths = 5/5 nm.
PrP
Amyloid fiber growth of mouse full-length Prp was measured by an adaptation of Breydo et al. (2008), under mild denaturing conditions and slightly acidic pH. These conditions did not compromise the amyloid-promoting effect of SERF1a (Figure 2G). A 136 μM PrP stock solution in 6 M GdnHCl (pH 6.0) was freshly prepared and used immediately. To prepare 200 μl of a 20 μM PrP reaction solution, 29.41 μl of the PrP stock solution was diluted in a clean 1.5 ml Eppendorf plastic vial with 109.32 μl H2O, 33.36 μl 6 M GdnHCl (pH 6.0), 20 μl 0.5 M MES (pH 6.0), and 3 mM NaN3. To this solution, 10 μl of 800 μM SERF1a prepared in 0.5 M MES (pH 6.0) and 3 mM NaN3 was added to a final concentration of 40 μM. No nucleation seeds were added. The plastic vials were sealed with Parafilm, taped to the rack of an Eppendorf thermomixer 5436 (Eppendorf), and agitated at 37°C and 1,000 rpm. Aliquots of 10 μl were taken at the indicated time points, diluted into 1 ml of a 10 μM ThT solution freshly prepared in 10 mM Na-acetate (pH 5.0), and measured in a 1 ml quartz cuvette at room temperature. Ex/em slit widths = 10/20 nm.
Partition Analysis
A 500 μl volume of a 200 μM aSyn solution in 50 mM TrisHCl, 150 mM NaCl, and 3 mM NaN3 (pH 7.4) were mixed with 500 μl of a 200 μM SERF1a solution in the same buffer, or with 500 μl buffer alone, sealed with Parafilm, taped to the rack of an Eppendorf thermomixer 5436 (Eppendorf), and agitated at 37°C and 1,400 rpm. After given time points, 50 μl samples were taken and frozen immediately at −20°C. Each sample was then centrifuged at 20,000 g, the unsoluble fraction was washed twice, and resuspended in 50 μl of 50 mM TrisHCl, 150 mM NaCl, and 3 mM NaN3 (pH 7.4). For soluble and unsoluble samples, 10 μl were used for dot-blotting, and 5 μl for PAGE-electrophoresis (see “Immunoblotting” section).
Immunoblotting
For dot-blotting, samples were directly applied onto a Hybond ECL nitrocellulose membrane (GE Lifesciencs) by using a Bio-Dot microfiltration apparatus (BioRad, Hercules, CA, USA) as specified by the manufacturer. In brief, samples were applied into slots presoaked with TBS, allowed to adsorb onto the membrane, and washed twice with TBST.
For electroblotting, samples were run on a SDS-PAGE gel (12% NuPAGE, Invitrogen) or an 8% native-PAGE gel (Tris [pH 7.6]) and transferred on a nitrocellulose membrane.
Membranes from dot-blots or gel electrotransfer were incubated 1 hr (room temperature) to overnight (4°C) in 5% skim milk/TBST, 3 mM NaN3, and shaken for 2 hr at room temperature with the respective primary antibody diluted in 1% skim milk/TBST, 3 mM NaN3 (1:2,000 anti-aSyn [produced in-house] and 1:200 anti-SERF1, K-13 clone [Santa Cruz Biotech, Santa Cruz, CA, USA]), and 1.30 hr with secondary antibody in 1% skim milk/TBST, 3 mM NaN3 (1:2,000 goat anti-rabbit (Santa Cruz Biotech) for aSyn and 1:1,000 mouse anti-goat [Santa Cruz Biotech] for SERF1a). The blots were then incubated with ECL reagent (Santa Cruz Biotech) and developed on a Kodak X-OMATIC film.
Actin Polymerization Assay
Actin polymerization was measured with a commercial assay (Cytoskeleton, Denver, CO, USA), by an adaptation of the manufacturer's instructions. The assay detects the fluorescence enhancement of pyrene-labeled G-actin during the conversion into polymeric F-actin. A 300 μl volume of a 10 μM actin solution in 5 mM Tris, 0.2 mM CaCl, 0.2 mM ATP, and 1 mM DTT (pH 8.0) were incubated for 1 hr and centrifuged. After baseline equilibration, the polymerization was induced by the addition of 30 μl polymerization buffer (500 mM KCl, 20 mM MgCl2, and 10 mM ATP) alone, or by the same volume of a SERF1a solution in polymerization buffer. The fluorescence was recorded on a Cary Eclipse fluorescence spectrometer at 25°C and 405 nm upon excitation at 360 nm. Ex/em slit = 2.5/5 nm.
The same measurements were repeated in the presence of 5 μM ThT, at ex/em wavelength 442/482 nm.
Citrate Synthase Aggregation Assay
A stock solution of citrate synthase from porcine heart was prepared as in Buchner et al. (1998). Citrate synthase was added to 40 mM HEPES-KOH (pH 7.5) preincubated at 43°C to a final concentration of 150 nM, in the absence of in the presence of SERF1a dissolved in the same reaction buffer. The aggregation was measured on a Cary Eclipse fluorescence spectrometer as an increase in turbidity, with excitation and emission wavelength both at 360 nm. Ex/em slit widths = 5/5 nm.
The same measurements were repeated in the presence of 5 μM ThT, at ex/em 442/482 nm wavelength.
Insulin Aggregation Assay
A 10 mg/ml human insulin stock solution was prepared in H20 by the dropwise addition of concentrated HCl until the dissolution was complete. A 30 μM insulin solution in 100 mM NaPi and 100 mM NaCl (pH 7.4) were incubated at 37°C, and aggregation was initiated by the addition of DTT to a final concentration of 20 mM (Scheibel et al., 1998) in the absence and in the presence of SERF1a. The turbidity was measured on a Cary Eclipse fluorescence spectrometer. Ex/em wavelengths were both 650 nm, low photomultiplier sensitivity. Ex/em slit widths = 2.5/5 nm.
The same measurements were repeated in the presence of 5 μM ThT, at ex/em 442/482 nm wavelength.
Isolation of Dopamine-Induced aSyn “Off-Pathway” Aggregates
Stable, SDS-resistant oligomers were produced by an adaptation of Cappai et al. (2005). A 300 μM aSyn solution in 50 mM Tris HCl, 150 mM NaCl, and 3 mM NaN3 (pH 7.4) were agitated with 5 mM dopamine in a 0.5 ml reaction volume for 72 hr at 37°C, 1,400 rpm. Unsoluble material was removed by centrifugation, and the oligomers were purified on a Superdex75 (GE Life Sciences, Uppsala, Sweden) column (self-packed XK16/100, 180 ml bed volume), at a flow rate of 1 ml/min, and a 1 ml sample collection volume. Oligomer-containing fractions were pooled and concentrated. The concentration of the oligomers was measured by the BCA assay. Monomeric, preweighed aSyn was used to generate the calibration curve. The final concentrations were kept around 3.5–4 mg/ml oligomeric aSyn.
Isolation of SERF-Induced Amyloid Intermediates
A 1 ml volume of a solution consisting of 150 μM aSyn and 100 μM SERF1a in 50 mM Tris, 150 mM NaCl, and 3 mM NaN3 (pH 7.4) was agitated at 1,400 rpm for 30–40 hr at 37°C. Longer agitation was avoided because of the decrease in the amount of soluble aggregates. Unsoluble material was separated by centrifugation, and the clear solution was immediately loaded on a Superdex75 (GE Life Sciences) gel filtration column (self-packed XK16/100, 180 ml bed volume), with a flow rate of 1 ml/min and a 1 ml sample collection volume. Chromatograms were recorded at an absorbance wavelength of 280 nm. At this wavelength, SERF1a was not visible because of the lack of aromatic residues and thus did not contribute to the chromatographic signal. Fractions containing aggregates eluted in the void volume. They were pooled and concentrated to 600–700 μg/ml. The concentration of the oligomers was measured by the BCA assay (Thermo Scientific, Rockford, IL, USA), using a calibration curve generated with monomeric, preweighed aSyn.
The freshly prepared oligomers were stored at 4°C and were used immediately for further studies. The preparations were stable for approximately 2 days.
Acknowledgments
This work was supported by the Austrian Science Fund (Project nr. P22400 to S.F.F. and P22630 to K.Z.). The transmission electron microscope was funded by the European Regional Development Fund (ERDF) with contributions of the Styrian Government and the Medical University of Graz. Thanks to Gertrude Havlicek for the TEM sample preparation. DICHROWEB is supported by grants to the BBSRC Centre for Protein and Membrane Structure and Dynamics (CPMSD).
Published online: July 26, 2012
Footnotes
Supplemental Information includes Extended Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.06.012.
This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
Supplemental Information
References
- Bagriantsev S.N., Kushnirov V.V., Liebman S.W. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- Bertoncini C.W., Jung Y.S., Fernandez C.O., Hoyer W., Griesinger C., Jovin T.M., Zweckstetter M. Release of long-range tertiary interactions potentiates aggregation of natively unstructured alpha-synuclein. Proc. Natl. Acad. Sci. USA. 2005;102:1430–1435. doi: 10.1073/pnas.0407146102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breydo L., Makarava N., Baskakov I.V. Methods for conversion of prion protein into amyloid fibrils. Methods Mol. Biol. 2008;459:105–115. doi: 10.1007/978-1-59745-234-2_8. [DOI] [PubMed] [Google Scholar]
- Buchner J., Grallert H., Jakob U. Analysis of chaperone function using citrate synthase as nonnative substrate protein. Methods Enzymol. 1998;290:323–338. doi: 10.1016/s0076-6879(98)90029-5. [DOI] [PubMed] [Google Scholar]
- Cappai R., Leck S.L., Tew D.J., Williamson N.A., Smith D.P., Galatis D., Sharples R.A., Curtain C.C., Ali F.E., Cherny R.A. Dopamine promotes alpha-synuclein aggregation into SDS-resistant soluble oligomers via a distinct folding pathway. FASEB J. 2005;19:1377–1379. doi: 10.1096/fj.04-3437fje. [DOI] [PubMed] [Google Scholar]
- Chandra S., Chen X., Rizo J., Jahn R., Südhof T.C. A broken alpha -helix in folded alpha -Synuclein. J. Biol. Chem. 2003;278:15313–15318. doi: 10.1074/jbc.M213128200. [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Dedmon M.M., Lindorff-Larsen K., Christodoulou J., Vendruscolo M., Dobson C.M. Mapping long-range interactions in alpha-synuclein using spin-label NMR and ensemble molecular dynamics simulations. J. Am. Chem. Soc. 2005;127:476–477. doi: 10.1021/ja044834j. [DOI] [PubMed] [Google Scholar]
- Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Ehrnhoefer D.E., Bieschke J., Boeddrich A., Herbst M., Masino L., Lurz R., Engemann S., Pastore A., Wanker E.E. EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 2008;15:558–566. doi: 10.1038/nsmb.1437. [DOI] [PubMed] [Google Scholar]
- Eichner T., Radford S.E. A diversity of assembly mechanisms of a generic amyloid fold. Mol. Cell. 2011;43:8–18. doi: 10.1016/j.molcel.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Falsone S.F., Kungl A.J., Rek A., Cappai R., Zangger K. The molecular chaperone Hsp90 modulates intermediate steps of amyloid assembly of the Parkinson-related protein alpha-synuclein. J. Biol. Chem. 2009;284:31190–31199. doi: 10.1074/jbc.M109.057240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández C.O., Hoyer W., Zweckstetter M., Jares-Erijman E.A., Subramaniam V., Griesinger C., Jovin T.M. NMR of alpha-synuclein-polyamine complexes elucidates the mechanism and kinetics of induced aggregation. EMBO J. 2004;23:2039–2046. doi: 10.1038/sj.emboj.7600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glabe C.G. Common mechanisms of amyloid oligomer pathogenesis in degenerative disease. Neurobiol. Aging. 2006;27:570–575. doi: 10.1016/j.neurobiolaging.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Hartl F.U., Bracher A., Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hoyer W., Cherny D., Subramaniam V., Jovin T.M. Impact of the acidic C-terminal region comprising amino acids 109-140 on alpha-synuclein aggregation in vitro. Biochemistry. 2004;43:16233–16242. doi: 10.1021/bi048453u. [DOI] [PubMed] [Google Scholar]
- Johnson B.A., Blevins R.A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kim H.Y., Cho M.K., Kumar A., Maier E., Siebenhaar C., Becker S., Fernandez C.O., Lashuel H.A., Benz R., Lange A., Zweckstetter M.J. Structural properties of pore-forming oligomers of alpha-synuclein. J. Am. Chem. Soc. 2009;131:17482–17489. doi: 10.1021/ja9077599. [DOI] [PubMed] [Google Scholar]
- Koppel D.E. Analysis of macromolecular polydispersity in intensity correlation spectroscopy: the method of cumulants. J. Chem. Phys. 1972;57:4814–4820. [Google Scholar]
- Lakowicz J.R. Principles of Fluorescence Spectroscopy. Springer; New York: 1999. Fluorescence anisotropy; pp. 308–310. [Google Scholar]
- Lashuel H.A., Petre B.M., Wall J., Simon M., Nowak R.J., Walz T., Lansbury P.T., Jr. Alpha-synuclein, especially the Parkinson's disease-associated mutants, forms pore-like annular and tubular protofibrils. J. Mol. Biol. 2002;322:1089–1102. doi: 10.1016/s0022-2836(02)00735-0. [DOI] [PubMed] [Google Scholar]
- LeVine H., 3rd Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- Levitan K., Chereau D., Cohen S.I., Knowles T.P., Dobson C.M., Fink A.L., Anderson J.P., Goldstein J.M., Millhauser G.L. Conserved C-terminal charge exerts a profound influence on the aggregation rate of α-synuclein. J. Mol. Biol. 2011;411:329–333. doi: 10.1016/j.jmb.2011.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Wong J.J., Edwards R.A., Manchak J., Frost L.S., Glover J.N. Structural basis of specific TraD-TraM recognition during F plasmid-mediated bacterial conjugation. Mol. Microbiol. 2008;70:89–99. doi: 10.1111/j.1365-2958.2008.06391.x. [DOI] [PubMed] [Google Scholar]
- McClendon S., Rospigliosi C.C., Eliezer D. Charge neutralization and collapse of the C-terminal tail of alpha-synuclein at low pH. Protein Sci. 2009;18:1531–1540. doi: 10.1002/pro.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris A.M., Watzky M.A., Finke R.G. Protein aggregation kinetics, mechanism, and curve-fitting: a review of the literature. Biochim. Biophys. Acta. 2009;1794:375–397. doi: 10.1016/j.bbapap.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Murray I.V., Giasson B.I., Quinn S.M., Koppaka V., Axelsen P.H., Ischiropoulos H., Trojanowski J.Q., Lee V.M. Role of alpha-synuclein carboxy-terminus on fibril formation in vitro. Biochemistry. 2003;42:8530–8540. doi: 10.1021/bi027363r. [DOI] [PubMed] [Google Scholar]
- Nielsen M.S., Vorum H., Lindersson E., Jensen P.H. Ca2+ binding to alpha-synuclein regulates ligand binding and oligomerization. J. Biol. Chem. 2001;276:22680–22684. doi: 10.1074/jbc.M101181200. [DOI] [PubMed] [Google Scholar]
- Pham C.L., Leong S.L., Ali F.E., Kenche V.B., Hill A.F., Gras S.L., Barnham K.J., Cappai R. Dopamine and the dopamine oxidation product 5,6-dihydroxylindole promote distinct on-pathway and off-pathway aggregation of alpha-synuclein in a pH-dependent manner. J. Mol. Biol. 2009;387:771–785. doi: 10.1016/j.jmb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Rasia R.M., Bertoncini C.W., Marsh D., Hoyer W., Cherny D., Zweckstetter M., Griesinger C., Jovin T.M., Fernández C.O. Structural characterization of copper(II) binding to alpha-synuclein: Insights into the bioinorganic chemistry of Parkinson's disease. Proc. Natl. Acad. Sci. USA. 2005;102:4294–4299. doi: 10.1073/pnas.0407881102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibel T., Weikl T., Buchner J. Two chaperone sites in Hsp90 differing in substrate specificity and ATP dependence. Proc. Natl. Acad. Sci. USA. 1998;95:1495–1499. doi: 10.1073/pnas.95.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillantini M.G., Crowther R.A., Jakes R., Hasegawa M., Goedert M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with lewy bodies. Proc. Natl. Acad. Sci. USA. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreerama N., Venyaminov S.Y., Woody R.W. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. Structure and Function of Intrinsically Disordered Proteins. CRC Press; Boca Raton, FL: 2010. Hydrodynamic techniques; pp. 43–84. [Google Scholar]
- Ullman O., Fisher C.K., Stultz C.M. Explaining the structural plasticity of α-synuclein. J. Am. Chem. Soc. 2011;133:19536–19546. doi: 10.1021/ja208657z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky V.N. Use of fast protein size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry. 1993;32:13288–13298. doi: 10.1021/bi00211a042. [DOI] [PubMed] [Google Scholar]
- Uversky V.N., Li J., Fink A.L. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson's disease and heavy metal exposure. J. Biol. Chem. 2001;276:44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- van Ham T.J., Holmberg M.A., van der Goot A.T., Teuling E., Garcia-Arencibia M., Kim H.E., Du D., Thijssen K.L., Wiersma M., Burggraaff R. Identification of MOAG-4/SERF as a regulator of age-related proteotoxicity. Cell. 2010;142:601–612. doi: 10.1016/j.cell.2010.07.020. [DOI] [PubMed] [Google Scholar]
- Wacker J.L., Zareie M.H., Fong H., Sarikaya M., Muchowski P.J. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat. Struct. Mol. Biol. 2004;11:1215–1222. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- Wang W., Perovic I., Chittuluru J., Kaganovich A., Nguyen L.T., Liao J., Auclair J.R., Johnson D., Landeru A., Simorellis A.K. A soluble α-synuclein construct forms a dynamic tetramer. Proc. Natl. Acad. Sci. USA. 2011;108:17797–17802. doi: 10.1073/pnas.1113260108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker S., Rudolph B., Frenzel E., Hagn F., Liebisch G., Schmitz G., Scheuring J., Kerth A., Blume A., Weinkauf S. Hsp12 is an intrinsically unstructured stress protein that folds upon membrane association and modulates membrane function. Mol. Cell. 2010;39:507–520. doi: 10.1016/j.molcel.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Whitmore L., Wallace B.A. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- Yagi H., Kusaka E., Hongo K., Mizobata T., Kawata Y. Amyloid fibril formation of alpha-synuclein is accelerated by preformed amyloid seeds of other proteins: implications for the mechanism of transmissible conformational diseases. J. Biol. Chem. 2005;280:38609–38616. doi: 10.1074/jbc.M508623200. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Coleman, B.M., Nisbet, R.M., Han, S., Cappai, R., Hatters, D.M., and Hill, A.F. (2009). Conformational detection of prion protein with biarsenical labeling and FlAsH fluorescence. Biochem. Biophys. Res. Commun. 380, 564–568. [DOI] [PubMed]
- Falsone, S.F., Leitinger, G., Karner, A., Kungl, A.J., Kosol, S., Cappai, R., and Zangger, K. (2011). The neurotransmitter serotonin interrupts α-synuclein amyloid maturation. Biochim. Biophys. Acta 1814, 553–561. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.